Abstract
Chimpanzees (Pan troglodytes) harbor rich assemblages of malaria parasites, including three species closely related to P. falciparum (sub-genus Laverania), the most malignant human malaria parasite. Here, we characterize the ecology and epidemiology of malaria infection in wild chimpanzee reservoirs. We used molecular assays to screen chimpanzee fecal samples, collected longitudinally and cross-sectionally from wild populations, for malaria parasite mitochondrial DNA. We found that chimpanzee malaria parasitism has an early age of onset and varies seasonally in prevalence. A subset of samples revealed Hepatocystis mitochondrial DNA, with phylogenetic analyses suggesting that Hepatocystis appears to cross species barriers more easily than Laverania. Longitudinal and cross-sectional sampling independently support the hypothesis that mean ambient temperature drives spatiotemporal variation in chimpanzee Laverania infection. Infection probability peaked at ~24.5 °C, consistent with the empirical transmission optimum of P. falciparum in humans. Forest cover was also positively correlated with spatial variation in Laverania prevalence, consistent with the observation that forest-dwelling Anophelines are the primary vectors. Extrapolating these relationships across equatorial Africa, we map spatiotemporal variation in the suitability of chimpanzee habitat for Laverania transmission, offering a hypothetical baseline indicator of human exposure risk.
Subject terms: Ecological epidemiology, Microbial ecology
Chimpanzees are known to suffer from malaria parasitism. Here, the prevalence and epidemiology of several malaria parasites is examined in a wild chimpanzee population.
Introduction
African great apes harbor a wide diversity of malaria parasites, including seven species closely related to Plasmodium falciparum (sub-genus Laverania), the most prevalent and malignant malaria parasite of humans1–6. Although the emergence of P. falciparum in the human population has been traced to a gorilla parasite6–8, Laverania species appear to exhibit considerable host-specificity in wild ape populations9–11. Chimpanzees (Pan troglodytes)—the most abundant and widely distributed great ape species12—host three Laverania parasites (P. reichenowi, P. gaboni, and P. billcollinsi)2,5,7,9,13–15, which are primarily transmitted by forest-dwelling Anopheles mosquitoes (An. marshallii, An. moucheti, An. vinckei) that feed promiscuously upon both humans and wild apes16,17. Although habitats of humans and chimpanzees often interface and humans are therefore likely to be exposed to these zoonotic parasites, three surveys of rural human populations in West Central Africa have failed to document cross-species transmission10,11,18, suggesting that strong molecular (e.g., binding affinity of parasite invasion ligand Rh5 to host receptor basigin) as well as other barriers govern the host-specificity of the Laverania19–22. Cross-species transmission of Laverania has, however, recently been documented among apes living in captive environments23, suggesting that a high magnitude of exposure can overcome molecular barriers to infection.
The capacity to identify potential exposure hotspots is currently limited because the ecological conditions that mediate Plasmodium transmission among chimpanzee hosts remain largely unexplored. Previous studies employing non-invasive sampling and molecular diagnostics have shown that the prevalence of chimpanzee Laverania infection varies regionally within equatorial Africa (generally ~30–50% among chimpanzee sub-species, though some sites exhibit an absence of infection)7,24. Infections are also temporally dynamic, manifesting seasonal and inter-annual trends in prevalence25. However, to our knowledge, no study has quantified the ecological drivers of this variation in infection probability. Doing so would offer a mechanistic explanation of these spatiotemporal dynamics and could pave the way to explicitly quantify the magnitude of human exposure in ecologically permissive settings.
Although the ecological drivers of chimpanzee Laverania infection remain largely unknown, both laboratory and field studies have revealed that ambient temperature is a strong determinant of Plasmodium transmission dynamics in human populations26–29. Ambient temperature influences both the population dynamics of the Anopheles mosquito vector and the rate of parasite development therein (i.e., the duration of sporogony)26,30,31. While higher temperatures accelerate the rate of parasite development within the vector, mosquito survival tends to decline above a particular temperature threshold26,32. Taken together, empirical studies indicate that transmission of P. falciparum peaks at a mean temperature of ~25 °C in human populations27,33. In essence, this optimal temperature represents the value at which the maximal number of mosquito vectors survive long enough to harbor the development of infectious parasites, and deviation from this value is associated with a reduction in the magnitude of transmission. Thus, spatial variation in ambient temperature defines the suitability of a particular locale for transmission, and temporal variation in this parameter underlies the seasonality of infection27,34–36.
Additional ecological variables can influence transmission dynamics. Intra-day fluctuations in ambient temperature can amplify or dampen transmission relative to that which would be expected at a particular mean temperature28,37. The effect of precipitation on malaria transmission is more complex. Rainfall is necessary to support breeding habitat for aquatic mosquito larvae (e.g., transitory oviposition sites)26,29, whereas high levels of precipitation may flush breeding sites, resulting in larval mortality38. Finally, given that chimpanzee malaria parasites are transmitted by forest-dwelling Anophelines16,17, the availability of forested habitat is also likely to be a necessary determinant of infection39, though the relationship between deforestation and human malaria parasitism has proven to be complex40.
In this study, we characterize the ecology and epidemiology of malaria infection in wild chimpanzee reservoirs and test the hypothesis that ambient temperature constitutes a critical driver of spatiotemporal variation in chimpanzee Laverania prevalence. We used molecular diagnostic assays to screen chimpanzee fecal specimens, collected via both longitudinal and cross-sectional sampling strategies (Fig. 1), for malaria parasite mitochondrial DNA. These analyses revealed that the epidemiology of chimpanzee malaria parasitism is characterized by early age of onset and seasonality of infection. In addition to Plasmodium parasites, we amplified Hepatocystis (a malaria parasite of monkeys and bats41) from a subset of samples. While Laverania parasites exhibit considerable host-specificity, phylogenetic analyses revealed that Hepatocystis appears to cross species barriers more easily. We also analyzed the relationship between environmental variables and fecal parasite rates to define the ecological niche of the chimpanzee Laverania. Longitudinal and cross-sectional sampling strategies each independently supported the hypothesis that mean ambient temperature drives spatiotemporal variation in Laverania infection probability among chimpanzees. Infection probability peaked at ~24.5 °C among chimpanzee populations, consistent with the empirical transmission optimum of P. falciparum. Forest cover was also positively correlated with spatial variation in Laverania prevalence, consistent with the observation that forest-dwelling Anophelines constitute their primary vectors. Finally, we extrapolated these relationships across equatorial Africa to define spatiotemporal variation in the suitability of chimpanzee habitat for Laverania transmission, offering a hypothetical baseline indicator of human zoonotic exposure risk.
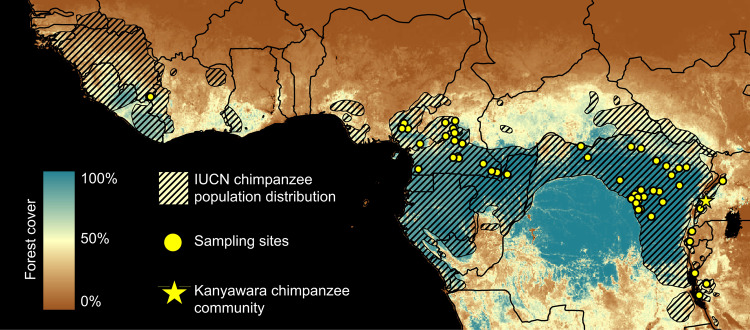
Fig. 1. Geographic location of wild chimpanzee sampling sites.
Study sites are shown in relation to the geographic range of chimpanzees (Pan troglodytes). A total of N = 3314 chimpanzee fecal samples were analyzed in this study. N = 878 fecal samples were collected longitudinally from 54 members of the Kanyawara chimpanzee community in Kibale National Park, Uganda (yellow star). N = 2436 additional fecal samples were collected from 55 sampling sites across equatorial Africa (yellow circles). The coloration of the map corresponds to spatial variation in the percentage of forest cover (derived from ref. 62).
Results
Characterization of malaria infection within the Kanyawara chimpanzee community
Malaria prevalence varies both spatially and temporally among wild chimpanzee populations7,25, but no analysis has elucidated the ecological underpinnings of these dynamics. To investigate the epidemiology of Laverania infection in wild chimpanzees, we first employed a high-density, longitudinal sampling strategy. Between June 2013 and August 2016, we collected 878 fecal samples from the Kanyawara chimpanzee community living in Kibale National Park, western Uganda (N = 1–29 samples per individual; median = 18 samples; Supplementary Table 1). This community of wild chimpanzees—which has been under continuous observation since 198742—is habituated to human observation, and all sampled inhabitants are identifiable by both morphology and by microsatellite genotype43. Samples were collected from 32 female (N = 496 samples) and 22 male (N = 382 samples) chimpanzees and stored in RNAlater at −20 °C until DNA extraction. At the time of sampling, subjects ranged in age between 0.3 and an estimated 55.8 years of age (median age at sampling: 16.6 years; Supplementary Table 1). Fecal samples were collected under direct observation and only when a positive identification was achievable. To evaluate identification reliability, DNA extracted from a subset of fecal samples (N = 44) was genotyped at 19 nuclear microsatellite loci, as described previously44. Microsatellite genotyping was successful in 97.7% of these samples (43/44), suggesting that fecal DNA was of sufficient quality for subsequent molecular analyses. Of the successfully genotyped samples, we observed a misidentification rate of 2.3% (1/43), indicating that identification fidelity was sufficient for analysis of demographic predictors of infection.
To quantify variation in Plasmodium infection within and among the Kanyawara chimpanzees, DNA was extracted from fecal samples and screened for malaria parasites using a single genome amplification (SGA) strategy via nested PCR, which targeted a 956-bp segment of the apicomplexan cytochrome B (cytB) mitochondrial gene as described previously3,7. To increase the sensitivity of this approach, we adopted an intensified PCR protocol, whereby all samples were screened in eight independent PCR reactions, as previously described4. All positive reactions were confirmed via direct sequencing without interim cloning. Of these 878 samples, 31.1% (N = 217) tested positive for one or more malaria parasite species in at least one replicate. Because the sensitivity of this assay to detect parasite DNA has been shown in humans to be greater in blood than in matched fecal samples, this value likely underestimates the frequency of blood-stage infection18. However, Loy et al.18 also revealed that human fecal samples that tested positive by this approach exhibited a 26-fold increase in parasite DNA copy number in the blood relative to fecal samples that tested negative. Thus, it is likely that the positive fecal samples identified in our study reflect chimpanzees that harbor elevated parasitemia, which is a correlate of both morbidity45–47 and transmission potential48,49, at least in humans.
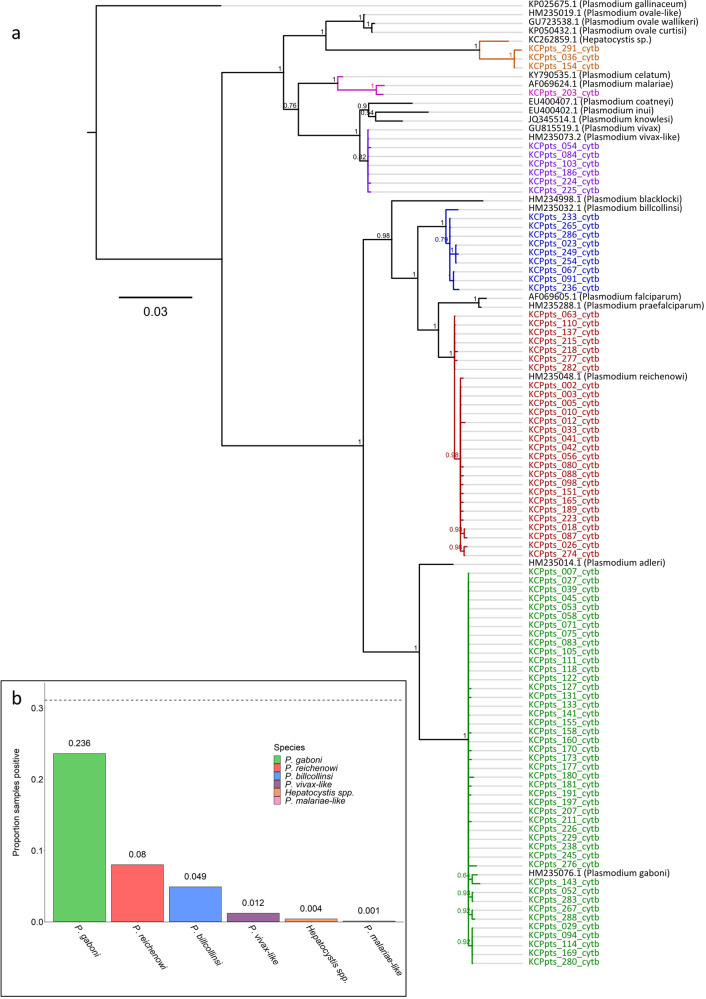
To diagnose the parasite species responsible for each infection, we aligned the newly generated sequences to a set of previously published cytB reference sequences (Supplementary Data 1) and constructed a Bayesian phylogenetic tree using MrBayes (version 3.2.6)50 (Fig. 2a). Parasite species identity was confirmed via NCBI nucleotide BLAST. As expected, the majority of positive samples were attributable to chimpanzee Laverania parasites (23.6% P. gaboni; 8.0% P. reichenowi; 4.9% P. billcollinsi; Fig. 2b). Two additional members of the primate malaria clade were amplified in a minority of samples (1.2% P. vivax-like; 0.1% P. malariae-like; Fig. 2b), and 6.0% of samples were found to harbor multiple parasite species. These results demonstrate that the Kanyawara chimpanzees carry an assemblage of malaria parasites that is largely representative of the parasite diversity in chimpanzees across equatorial Africa2,7,9,51.
Fig. 2. Richness of malaria parasites isolated from the Kanyawara chimpanzee community.
a Bayesian phylogeny generated from a representative subset of the SGA-derived cytB sequences generated in this study. Sequences were derived from N = 878 fecal samples, collected from the Kanyawara chimpanzee community in Kibale National Park, western Uganda, between 2013 and 2016. Color corresponds to malaria parasite species, inferred from the phylogenetic relationships of sequences to previously published reference sequences (green: P. gaboni, red: P. reichenowi, blue: P. billcollinsi, violet: P. vivax-like, orange: Hepatocystis spp., magenta: P. malariae). Newly generated sequences are listed in Supplementary Data 3, and previously published reference sequences used in this figure are listed in Supplementary Data 1. b Distribution of parasite species isolated from the Kanyawara chimpanzees. Malaria parasites were amplified in 31.1% of samples (dotted line). A majority of parasites amplified were members of the sub-genus Laverania (i.e., relatives of P. falciparum): P. gaboni (23.6%), P. reichenowi (8.0%), and P. billcollinsi (4.9%). Members of the primate malaria clade were amplified in a minority of samples: P. vivax-like (1.2%) and P. malariae-like (0.1%). In addition, Hepatocystis was amplified from 0.4% of samples.
Evolutionary relationships of chimpanzee Hepatocystis parasites
In addition to the Plasmodium species highlighted above, three samples (0.4%) harbored parasite sequences attributable to Hepatocystis spp., a clade of mammalian malaria parasites primarily isolated from Old World monkeys and bats1,41,52. Despite its divergent nomenclature, the Hepatocystis clade is more closely related to the primate malaria clade (e.g., P. vivax, P. malariae, P. ovale) than either is to Laverania41,53. Previous studies have similarly identified Hepatocystis in chimpanzee fecal samples14. However, to our knowledge, no study has evaluated the evolutionary relationships of these parasites relative to the large diversity of previously described Hepatocystis lineages to evaluate whether these species constitute (A) a novel Hepatocystis clade endemic to chimpanzee hosts or (B) the product of multiple independent cross-species transmission events originating in other mammals.
To discriminate between these possibilities, we analyzed the phylogenetic relationships of 20 great ape Hepatocystis sequences (i.e., three Kanyawara-derived sequences and 17 unpublished sequences amplified fortuitously during previous molecular epidemiological studies of ape malaria parasites1,2,7) relative to 56 previously published Hepatocystis cytB sequences (Supplementary Data 2) isolated from a wide range of hosts, including African and Asian monkeys, bats, and other mammals (Fig. 3). To determine the species origin of the ape fecal samples, we sequenced host mitochondrial D loop fragments and discarded sequences from samples from which this gene did not amplify. Of the 17 unpublished sequences, 15 were derived from chimpanzee fecal samples, while one was amplified from a dried blood spot (LIpts50094), collected from a captive chimpanzee living in the Limbe Wildlife Centre near Douala, Cameroon (Supplementary Data 3). Still another fecal sample contained human mtDNA sequences. The human-derived sample was collected at the Bafwabula field site in the Democratic Republic of the Congo in January 2007. While this would be the first report of a Hepatocystis infection in a human, we cannot exclude the presence of low-level primate DNA in the fecal sample, potentially resulting from the consumption of monkey bushmeat as the source of the Hepatocystis amplicon.
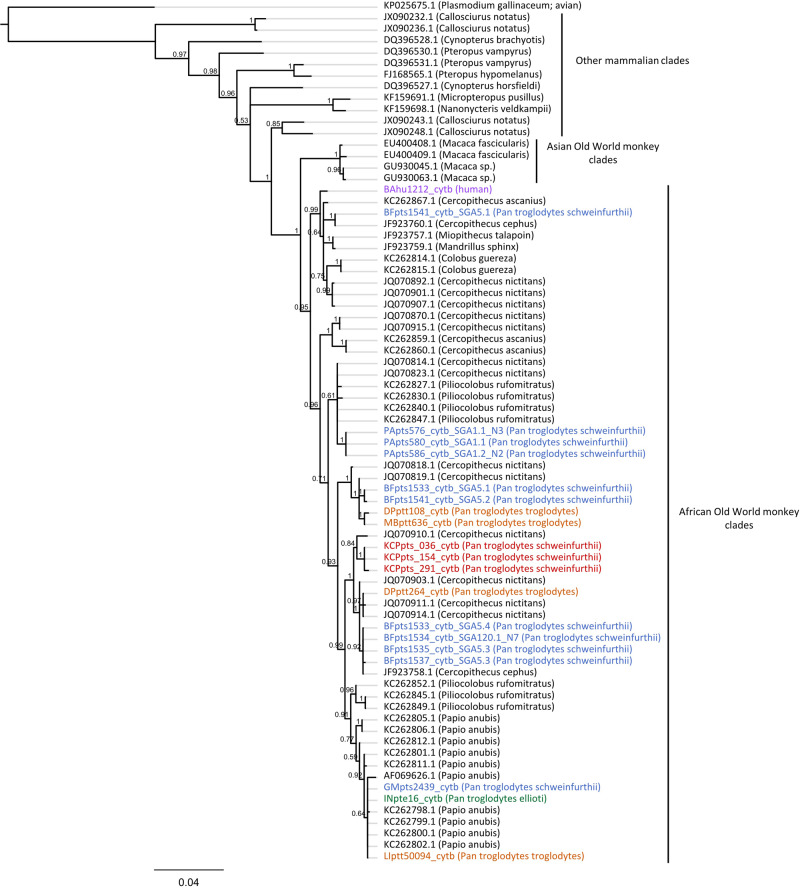
Fig. 3. Phylogeny of Hepatocystis parasites isolated from chimpanzees and one putative human host.
Bayesian phylogeny generated from an alignment of N = 19 SGA-derived Hepatocystis cytB sequences (965 bp) produced in this study and N = 56 previously published Hepatocystis sequences from African Old World monkeys, Asian Old World monkeys, and other mammals. Chimpanzee parasite lineages cluster within a wide range of African Old World monkey parasites, suggesting a capacity to cross species boundaries. Sequences are annotated with respect to mammalian host species. Tip labels corresponding to Hepatocystis lineages isolated from Kanyawara fecal samples are colored red, and one human sample is colored violet. The remaining tip labels are colored with respect to chimpanzee sub-species (blue: Pan troglodytes schweinfurthii; orange: Pan troglodytes troglodytes; green: Pan troglodytes ellioti). Newly generated sequences are listed in Supplementary Data 3, and previously published reference sequences used in this figure are listed in Supplementary Data 2.
These phylogenetic analyses revealed that all ape-derived Hepatocystis isolates clustered within a clade of Hepatocystis lineages derived from African monkey hosts, and no isolate clustered with sequences from Asian monkeys, bats, or other mammals (Fig. 3). These ape lineages were phylogenetically interspersed among African monkey isolates. This pattern contrasts starkly with the topology of the Laverania phylogeny, which consists of largely host-specific clades despite extensive sympatry of hosts. Based on these data, it is possible that the great ape-derived Hepatocystis lineages are the result of multiple independent cross-species transmission events, suggesting that these parasites may cross species barriers promiscuously54. However, it is also possible that some, or all, of the fecal-derived chimpanzee Hepatocystis sequences resulted from the consumption of Hepatocystis-infected monkeys. Nevertheless, the fact that one Hepatocystis sequence was derived from a chimpanzee blood sample indicates productive infection in at least some cases.
Ecological determinants of Laverania infection among the Kanyawara chimpanzees
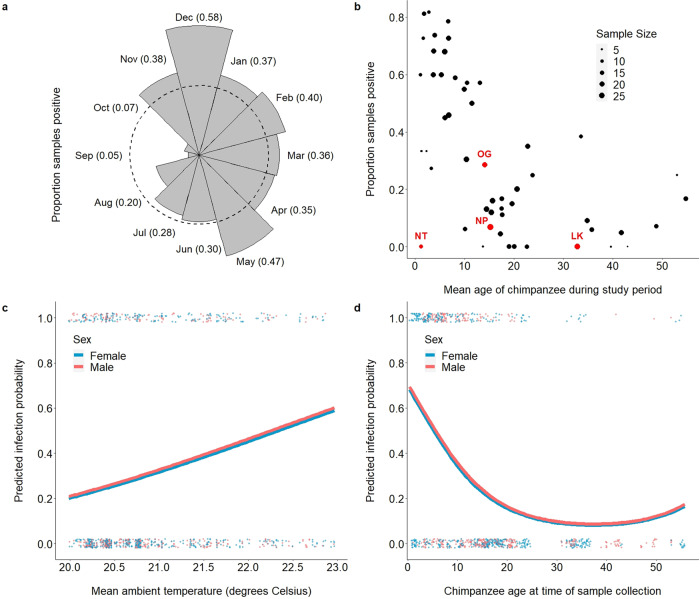
Given the high prevalence of Laverania infection harbored by the Kanyawara chimpanzees (Fig. 2b), we evaluated the extent to which the incidence of these parasites varied temporally among chimpanzee hosts. Between November and May, the proportion of positive samples was greater than the dataset mean (31.1%) in 9 out of 11 sampling months, and that samples were more likely to test positive for malaria parasites during this part of the year (N = 404 samples; positive samples expected = 125.6 ± 9.3; positive samples observed = 163; binomial test, p < 0.0001; Fig. 4a). By contrast, the proportion of samples that tested positive was greater than the dataset mean in only 3 out of 13 sampling months between June and October, and samples were generally less likely to test positive for malaria parasites during these months (N = 474 samples; positive samples expected = 147.4 ± 10.1; positive samples observed = 110; binomial test, p < 0.0001; Fig. 4a). Taken together, these results highlight a seasonal pattern of infection.
Fig. 4. Longitudinal analysis of malaria parasitism in the Kanyawara cohort of wild chimpanzees.
a The proportion of chimpanzee fecal samples (N = 878) that tested positive for malaria parasites varied by month of sampling (dashed line corresponds to dataset mean). Bar length corresponds to proportion of samples that tested positive during a given month of sampling (monthly mean is listed in parentheses). Samples collected between November and May tended to be more likely to test positive for malaria parasites, while samples collected between June and October tended to be less likely. b Demographic variables also influenced infection probability. Samples collected from younger chimpanzees were generally more likely to test positive for malaria parasites than were samples collected from older chimpanzees. This result highlights the early age of infection onset (including the youngest sample in the dataset, collected from a 3-month-old female), indicative of a high magnitude of ongoing transmission. Despite this observation, a subset of chimpanzees (e.g., NT) deviated from this trend, potentially indicative of resistance to infection. c Predicted infection probabilities, derived from the Kanyawara GLMM (Table 2), demonstrate that seasonality of infection is partially driven by variation in mean ambient temperature. Mean ambient temperature (measured directly via weather monitoring stations) was positively correlated with infection probability across the range of temperature values observed at this sampling site (20.0–23.0 °C; p < 0.0001). Raw data (binary) are plotted as dots and stratified vertically for visualization. d Infection probabilities predicted by the Kanyawara GLMM also demonstrated that the youngest study subjects tended to be the most likely to test positive for malaria parasites (p < 0.0001). Raw data (binary) are plotted as dots and stratified vertically for visualization.
To evaluate the ecological drivers of infection probability, daily measurements of ambient temperature (minimum, maximum) and rainfall were collected from Kanyawara during the study period. We defined mean ambient temperature as the average of minimum and maximum air temperature estimates recorded during the 30 days prior to sample collection and defined intra-day temperature variation as the average difference between minimum and maximum temperature estimates during the same period. Mean ambient temperature during sample collection ranged from 20.0 to 23.0 °C (median: 20.7 °C) and intra-day temperature variation ranged from 7.9 to 15.6 °C (median: 11.0 °C). Mean daily rainfall during the 30 days prior to sample collection ranged from 0 to 14.5 mm per day (median: 3.0 mm).
To discern the ecological drivers of this seasonal pattern of infection, we used a generalized linear mixed model (GLMM)55 with binomial error structure and logit link function to evaluate the relationship between climatic variation and the probability of Plasmodium infection among the Kanyawara chimpanzees. For each sample, we specified malaria infection (binary) as the outcome variable, mean ambient temperature (quadratic) and rainfall as fixed effects, chimpanzee individual as a random effect, and age (quadratic), sex, and intra-day temperature variation as covariates (Table 2). A full model that included all predictor variables and covariates provided a more parsimonious fit to the data than did a null model containing only random effects (likelihood ratio test: χ2 = 64.5, d.f. = 6, p < 0.0001; ΔAIC = 52.5). As predicted, ambient temperature was positively correlated with the probability of infection, though the quadratic term had no effect and was omitted from the best-fit model (Fig. 4c; Table 2). Unexpectedly, we found that rainfall was negatively correlated with infection probability (Table 2). These results highlight ambient temperature as an important climatic predictor of seasonal variation in chimpanzee Plasmodium infection probability for this community.
Table 2.
Longitudinal analysis of malaria parasitism among the Kanyawara chimpanzees.
| Parametera | Estimate | Std Err | z | P-value |
|---|---|---|---|---|
| Intercept | −13.2 | 3.17 | −4.17 | <0.0001 |
| Ecological | ||||
| Mean ambient temperature | 0.834 | 0.174 | 4.80 | <0.0001 |
| (Mean ambient temperature)2 | NSb | NSb | NSb | NSb |
| Intra-day temperature variation | −0.466 | 0.085 | −5.48 | <0.0001 |
| Precipitation | −0.096 | 0.035 | −2.75 | 0.006 |
| Demographic | ||||
| Age | −26.5 | 4.63 | −5.73 | <0.0001 |
| (Age)2 | 15.4 | 4.45 | 3.47 | 0.0005 |
| Sex (male) | 0.073 | 0.319 | 0.23 | 0.819 |
aOutput of longitudinal GLMM corresponding to the probability ape malaria parasite infection relative to ecological and demographic predictor variables of interest. Model based upon 878 fecal samples collected longitudinally from 54 individual wild chimpanzees of the Kanyawara cohort in western Uganda. Note that predictor variables were scaled to augment model convergence. All samples in the longitudinal analysis of the Kanyawara cohort were screened eight times for malaria parasites using an intensified SGA methodology, as described previously3,4,7. See “Methods” for model specification details.
bMean ambient temperature2 did not improve the fit of the longitudinal GLMM and was, therefore, omitted from the final, most parsimonious, model.
Demographic variables also influenced infection probability. As reported previously13, fecal samples of younger chimpanzees (including the youngest chimpanzee in the dataset, 3.7 months old at time of sampling) were more likely to test positive for malaria parasites than were older individuals (Fig. 4b, d; Table 2). This finding is consistent with the epidemiology of P. falciparum in humans, in which children under five years of age experience the bulk of morbidity and mortality before gradually developing immunity to the parasite upon repeated exposure56–58. Interestingly, however, while most chimpanzees adhered to this pattern, some notable outliers were evident. For example, the fourth youngest chimpanzee in the dataset (NT; mean age: 1.28 years; Fig. 4b) tested positive for Plasmodium infection in 0/12 samples (model prediction: 7.6 ± 2.8 positive samples; p < 0.0001). Although the father of this chimpanzee (OG) exhibited an unremarkable pattern of infection (positive in 6/15 samples), her mother (NP) tested positive for Plasmodium in 1/29 samples, and her maternal grandfather (LK) was positive in 0/25 samples (Fig. 4b). It remains unclear whether such striking outliers are completely refractory to infection or are capable of controlling parasitemia such that it is below the limit of detection in feces. Future studies will be necessary to evaluate whether this pattern is attributable to genetic (e.g., erythrocyte receptor polymorphism) or other factors.
Taken together, these results demonstrate that the epidemiology of chimpanzee Laverania infection is characterized by high prevalence, early age of onset, and seasonality of infection, consistent with an elevated magnitude of active transmission, as opposed to rare, exclusively chronic infection. Given the promiscuous biting preferences of ape malaria vectors17, these results provide complementary, albeit indirect, support for the assertion that humans living on the borders of chimpanzee habitat with active transmission may be exposed to their parasites and that the apparent host-specificity of the Laverania may be more attributable to cellular—rather than ecological—barriers to infection.
Ecological analysis of spatiotemporal variation in malaria among chimpanzees across equatorial Africa
To evaluate the generalizability of our longitudinal results of the Kanyawara chimpanzee community, we analyzed 2436 additional fecal samples collected between November 2000 and May 2016 from 55 sampling sites distributed across wild chimpanzee habitat in equatorial Africa (N = 1–258 samples per site; median = 29; Fig. 1), including both previously published (N = 1936) and unpublished (N = 500) samples (Table 1). As above, samples were screened for parasite mtDNA using a SGA strategy targeting a 956-bp cytB amplicon, as described previously3.
Table 1.
Chimpanzee fecal samples analyzed in this study.
| Field sites tested | Field sites | Field sites positive | Samples | Samples positive | References |
|---|---|---|---|---|---|
| Longitudinal analysis | |||||
| Kanyawara (KCP) | 1 | 1 | 878 | 273 | This study |
| Pan-African analysis | |||||
| BO, GI, GM, KB, KY, MH, NB, NY, UG | 9 | 1 | 500 | 6 | This study |
| AM, AN, AZ, BA, BB, BD, BF, BG, BI, BL, BQ, CP, DG, DP, EB, EK, EN, EP, GO, GT, IS, KA, KO, KS, LB, LH, LU, MB, MD, MF, MK, MP, MT, MU, ON, OP, PA, PO, SL, UB, VM, WA, WB, WE, WL, YW | 46 | 31 | 1936 | 390 | 1,2,7 |
| Total | 55 | 32 | 2436 | 396 | |
| Combined dataset | 56 | 33 | 3314 | 669 |
Key: Amunyala (AM), Ango (AN), Azunu (AZ), Babingi (BI), Bafwaboli (BA), Bafwasende (BF), Belgique (BQ), Bondo-Bili (BD), Bongbola (BL), Bossou (BO), Boumba Bek (BB), Budongo (BG), Campo Ma’an (CP), Diang (DG), Doumo Pierre (DP), E’kom (EK), Ebo (EB), Engali (EN), Epulu (EP), Gishwati (GI), Goalougo Triangle (GT), Gombari (GO), Gombe (GM), Isiro (IS), Kabuka (KA), Kagwene (YW), Kibale Kanyawara (KCP), Kibale Ngogo (KB), Kisangani (KS), Kotakoli (KO), Kyambura Gorge (KY), Liabelem Highlands (LH), Lobéké (LB), Lubutu (LU), Mahale (MH), Makombe (MK), Mamfé (MF), Manbele (MB), Mbam et Djerem (MD), Metep (MP), Minta (MT), Munbgere (MU), Nimba (NB), Nyungwe (NY), Onga (ON), Opienge (OP), Parisi (PA), Poko (PO), Somalomo (SL), Ubangi (UB), Ugalla (UG), Vome (VM), Walengola (WL), Wamba (WB), Wanie-Rukula (WA), Wassa Emtse (WE).
Although direct measurements of climate were not available from all sites, satellite remote sensing technology can estimate climatic variables59. To estimate ambient temperature at each sampling site, we extracted daily Land Surface Temperature (LST) estimates from the MODerate Resolution Imaging Spectroradiometer (MODIS) thermal sensor on board the NASA-Terra satellite at 1 km2 (downsampled to 10 km2) spatial and 1-day temporal resolution (MOD11A1v005; http://modis.gsfc.nasa.gov/)60. We derived minimum and maximum air temperature estimates by applying the transformation outlined in Weiss et al.36. As above, mean ambient temperature was defined as the average of minimum and maximum air temperature estimates recorded during the 30 days prior to sample collection and intra-day temperature variation as the average difference between minimum and maximum temperature estimates during the same period. Rainfall estimates were derived from the Global Precipitation Climatology Project (GPCP V2.3; https://www.esrl.noaa.gov/psd/)61 at 1-day temporal resolution and 1-degree spatial resolution. Forest cover estimates were derived from high-resolution global maps of tree canopy cover (30 m, downsampled to 10 km), published by Hansen et al.62. The resulting dataset of 2436 samples included only those data points for which all predictor variables were available. Mean ambient temperature during the 30 days prior to sample collection ranged from 9.8 to 27.3 °C (median: 23.8 °C). Mean daily rainfall during the 30 days prior to sample collection ranged from 0 to 14.8 mm per day (median: 4.0 mm). Intra-day temperature variation during the 30 days prior to sample collection ranged from 6.0 to 13.4 °C (median: 8.8 °C). Forest cover at sampling sites ranged from 16 to 100% (median: 90%).
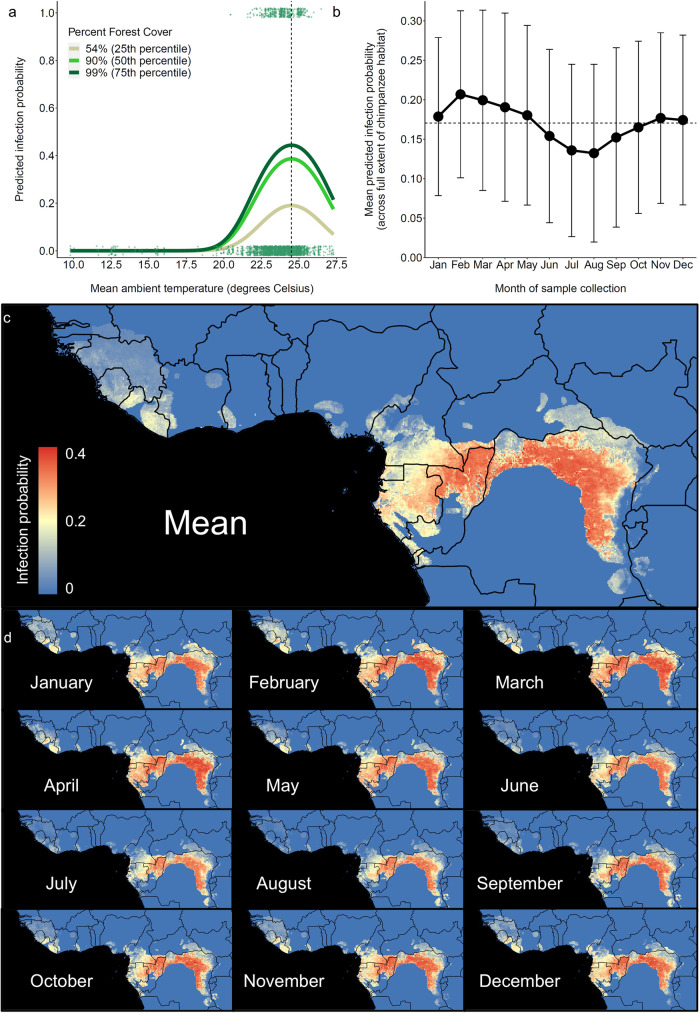
We used a GLMM with binomial error structure and logit link function to evaluate the relationship between ecological variation and the probability of Laverania infection among wild chimpanzees across equatorial African sampling sites. We coded Plasmodium infection (binary) as the outcome variable, mean ambient temperature (quadratic), rainfall, and percent forest cover as fixed effects, sampling site as a random effect, and daily temperature variation and number of replicates (i.e., 1 or 8–10, depending upon whether samples were screened using the conventional SGA method3 or the intensified method4, respectively) as covariates (Supplementary Table 1). A full model that included all predictor variables and covariates provided a more parsimonious fit to the data than did a null model containing only random effects (likelihood ratio test: χ2 = 37.9, d.f. = 5, p < 0.0001; ΔAIC = 27.9). As predicted, ambient temperature was positively correlated with infection probability at low-temperature values and negatively correlated with infection at high-temperature values (Fig. 5a; Table 3). Infection probability peaked at ~24.5 °C, consistent with the ~25 °C transmission optimum of P. falciparum27,33. In addition, forest cover was positively correlated with infection probability (Fig. 5a; Table 3), while rainfall did not influence infection and was omitted from the best-fit model (Table 3). As expected, the sensitivity of the assay was enhanced by usage of the intensive PCR protocol (i.e., by screening samples in 8–10 replicates versus one replicate; Table 3).
Fig. 5. Ecological niche modeling of malaria parasitism in wild chimpanzee reservoirs across equatorial Africa.
a Analysis of N = 2436 wild chimpanzee fecal samples collected from 55 sampling sites across equatorial Africa demonstrate that mean ambient temperature (inferred from MODIS remote sensing datasets; see Methods) and forest cover62 are critical determinants of chimpanzee Laverania epidemiology. The pan-African GLMM (Table 3) demonstrates that (1) infection probability peaks ~24.5 °C (dotted vertical line), consistent with the empirical transmission optimum of P. falciparum27, and (2) forest cover is positively correlated with infection probability (p = 0.001), consistent with the observation that forest-dwelling Anophelines constitute the primary vectors of these parasites16,17. Color corresponds to three categories of forest cover: 54% (25th percentile), 90% (50th percentile), and 99% (75th percentile). Raw data (binary) are plotted as dots and stratified vertically for visualization. b, c Given the ecological relationships identified in this study, we extrapolated infection probabilities across chimpanzee habitat in equatorial Africa. We derived composite rasters from mean ambient temperature and intra-day temperature variation measurements recorded between 2000 and 2017 across the African continent. Using these composite temperature rasters and the Hansen et al.62 forest cover dataset, we projected the predicted probabilities of chimpanzee Laverania infection (derived from the pan-African model) across the spatial extent of chimpanzee habitat. The monthly mean pixel value of each raster is plotted, and error bars correspond to standard deviation of pixel values on each raster. d Stratification by month highlights the seasonality of these infections. Across chimpanzee habitat, infection probability peaks between the months of January and May, and infection probability declines between June and September. Because mosquito vectors of ape malaria parasites readily bite humans17, these maps can serve as a baseline proxy for spatiotemporal variation in the risk of human exposure in areas where humans and apes overlap.
Table 3.
Ecological modeling of malaria parasitism among chimpanzee hosts.
| Parametera | Estimate | Std Err | z | P-value |
|---|---|---|---|---|
| Intercept | −2.54 | 0.338 | −7.50 | <0.0001 |
| Ecological | ||||
| Mean ambient temperature | 0.355 | 0.127 | 2.79 | 0.005 |
| (Mean ambient temperature)2 | −0.164 | 0.063 | −2.60 | 0.009 |
| Intra-day temperature variation | 0.208 | 0.099 | 2.09 | 0.036 |
| Precipitation | NSb | NSb | NSb | NSb |
| Percent forest cover | 0.032 | 0.010 | 3.20 | 0.001 |
| Technical | ||||
| Number of replicates (8)c | 1.164 | 0.500 | 2.33 | 0.020 |
aOutput of pan-African GLMM corresponding to the probability ape malaria parasite infection relative to ecological predictor variables of interest. Model was based upon 2436 fecal samples collected at 55 sampling sites in wild chimpanzee habitat across equatorial Africa. Note that predictor variables were scaled to augment model convergence. See “Methods” for model specification details.
bPrecipitation did not improve the fit of the longitudinal GLMM and was, therefore, omitted from the final, most parsimonious, model.
cSamples in the pan-African analysis were screened either in either one or eight replicates.
Finally, we extrapolated these relationships between ecological variables and the probability of chimpanzee Laverania infection across equatorial Africa to identify hypothetical exposure hotspots. To characterize ecological variation across equatorial Africa, we derived composite rasters from mean ambient temperature and intra-day temperature variation measurements recorded between March 2000 and February 2017 across the African continent at 1-degree spatial and 1-month temporal resolution. Using these composite temperature rasters and the Hansen et al.62 forest cover dataset, we projected the predicted probabilities of chimpanzee Laverania infection (derived from the pan-African model) across the spatial extent of chimpanzee habitat (as defined by the International Union for the Conservation of Nature, IUCN; Fig. 5c). The resulting risk map indicates spatial variation in the suitability of chimpanzee habitat for Laverania transmission across all seasons during an average year. Stratification of these results by month of sampling highlights temporal variation in infection probability and predicts elevated transmission between January and May, as well as a low transmission season between June and September, for much of the region (Fig. 5b, d). This risk map also highlights spatial variation in the seasonality of transmission (e.g., high levels of sustained transmission in Central Africa, contrasting a more sharply seasonal pattern of infection in habitat toward the edges of the chimpanzee habitat range). Although inter-annual variation would be expected to cause deviations from these estimates, this risk map offers a general baseline prediction for follow-up studies of exposure and transmission.
Discussion
Although infection of wild chimpanzees with Laverania parasites is well-documented, the ecological drivers of spatiotemporal variation in prevalence remain largely unexplored. Our analyses define the ecological niche of chimpanzee Laverania parasites and support the hypothesis that variation in mean ambient temperature determines infection dynamics. Extrapolation of these relationships across equatorial Africa offers a framework for the estimation of transmission intensity in chimpanzees and thereby helps quantify the risk of human exposure to these parasites.
Ambient temperature influences numerous elements of the Plasmodium life cycle, including the population dynamics of the Anopheline vector and the rate of parasite development26–31. Both longitudinal and cross-sectional sampling strategies independently support the conclusion that mean ambient temperature is correlated with the probability of Laverania infection (Figs. 4c and 5a, Table 2), which peaks at ~24.5 °C in wild chimpanzee populations, close to the ~25.0 °C empirical transmission optimum of P. falciparum in humans27,33. The consistency of this result is underscored by the fact that these two approaches use non-overlapping datasets and alternative methods of temperature measurement (Kanyawara: direct measurement; pan-African: remote sensing). In addition to mean ambient temperature, intra-day temperature variation—which has also been shown to influence the epidemiological processes at the parasite-mosquito interface28,37—was correlated with infection probability in both datasets (Tables 2 and 3). However, the direction of this correlation differed in these analyses, which suggests a more complicated relationship with malaria infection (e.g., countervailing effects of temperature fluctuation when observed at mean ambient temperature extremes28 or vector-specific effects37). Taken together, these results indicate that the temperature-sensitive epidemiology of the chimpanzee Laverania closely resembles that of P. falciparum, suggesting that conserved mechanisms govern the epidemiology of these taxa.
Forest cover was also positively correlated with infection probability (Fig. 5a and Table 3) which is consistent with the finding that forest-dwelling Anopheline species (An. vinckei, An. moucheti) constitute the primary vectors of these parasites16,17 and supports a recent analysis of malaria parasitism across non-human primates39. Although these signatures of forest-dependent transmission suggest that human encroachment into undisturbed habitat may pose a greater exposure risk than does habitat fragmentation, these relationships are likely complex40 and warrant further investigation at fine-grained spatial scales.
Extrapolation of these relationships across equatorial Africa highlights spatial and temporal variation in the ecological suitability of chimpanzee habitat for Laverania transmission (Fig. 5b–d). Although we found two distinct transmission seasons: (1) a “high season” between January and May, and (2) a “low season” between June and September, the amplitude and character of this seasonal pattern likely varies regionally. For example, northern Democratic Republic of the Congo and northern Republic of the Congo are predicted to support high levels of transmission across seasons, while Cameroon and Gabon will have infection probability dropping between June and August. Still other countries, such as Rwanda and Tanzania, are predicted to harbor very low transmission intensity, a projection supported by the absence of infection in samples collected from these countries. Although these patterns are expected to vary between years as a function of local climatic anomalies, this map offers a baseline set of predictions for longitudinal follow-up studies of Laverania epidemiology.
In addition to highlighting variation in Laverania infection among wild chimpanzees, these maps offer a rough proxy for the estimation of human exposure risk. Our analyses suggest that the epidemiology of chimpanzee Laverania infection is defined by (1) high prevalence (Fig. 2), (2) early age of onset (Fig. 4b, d), and (3) seasonality of infection (Figs. 4a, c and 5a–d). These results are consistent with a high magnitude of ongoing transmission among chimpanzee hosts. In addition, human landing capture experiments conducted in Gabon have demonstrated that the Anopheline vectors of ape malaria parasites readily bite humans when given the opportunity17. Given that fecal parasite rates are indicative of blood stage parasitemia18, which correlates with transmission potential48,49, these observations suggest that the per capita probability of human exposure is likely to be greatest in areas where the magnitude of transmission among chimpanzees is similarly elevated. Accordingly, these habitat suitability maps highlight predicted human exposure hotspots, which can inform the design of zoonotic surveillance efforts that quantify exposure directly (e.g., via the measurement of entomological inoculation rates and serological exposure). Future studies should prioritize rural human populations living near forests containing Plasmodium-infected apes demonstrating active transmission in the Republic of Congo and the Democratic Republic of the Congo, as well as parts of southern Cameroon and eastern Gabon, between the months of January and May. Since gorillas are infected with the precursor of P. falciparum, this ape species in particular should be included in epidemiological studies. Future efforts that fine-tune and extend this risk map (e.g., via the inclusion of human population density, ape population density, suitable mosquito vectors, and range overlap data layers) are necessary to operationalize these predictions.
Although it is likely that human exposure to chimpanzee malaria parasites occurs with regularity in parts of equatorial Africa, chimpanzee-to-human Laverania transmission has never been documented. Strong molecular barriers to cross-species Laverania transmission have been identified19,20,22 and two surveys of rural human populations in Gabon and Cameroon10,11, as well as an extensive survey of indigenous hunter gatherers in Cameroon18, have failed to reveal evidence of zoonotic transmission. The strong host-specificity of the Laverania species contrasts with the lack of host-specificity of Hepatocystis parasites (Fig. 3). Though Laverania and Hepatocystis parasites each achieve high prevalence in hosts that occupy sympatric distributions in equatorial Africa, cross-species transmission of the former has not yet been convincingly documented23,51,52. While Hepatocystis parasites are known to cross species barriers rather promiscuously54, our study suggests that chimpanzees are susceptible to monkey Hepatocystis infection (Fig. 3). Although some, or all, Hepatocystis sequences derived from chimpanzee fecal samples may have resulted from the consumption of Hepatocystis-infected monkeys, we also amplified one Hepatocystis sequence from the blood of a captive chimpanzee, providing evidence for a productive infection. This is the first description of a chimpanzee Hepatocystis infection and suggests that cross-species transmission of Hepatocystis between monkeys and chimpanzees (and potentially humans) may have been underestimated. Future studies that screen human and chimpanzee populations for this parasite using Hepatocystis-specific primers will be necessary to examine the frequency of productive cross-species transmission.
The analyses presented here may also catalyze future studies of host-parasite coevolution in wild chimpanzee hosts. Although malaria pathogenesis ranks amongst one of the strongest selective pressures to confront human populations during the past 50,000 years63,64, little is known about the pathogenicity of the Laverania in chimpanzees. While some case studies have documented several characteristic symptoms—such as fever, anemia, elevated parasitemia, and possibly mortality—in Plasmodium unexposed captive chimpanzees upon initial exposure to these parasites45,65, other studies have produced equivocal results66. If Laverania parasites are routinely pathogenic in wild chimpanzees, natural selection could plausibly have favored the evolution of mechanisms of resistance. Three closely related individuals in our sample had exceptionally high frequencies of testing negative for malaria parasites, raising the possibility that they shared a genetic polymorphism conferring resistance. It is also conceivable that chimpanzees have evolved a series of behavioral responses (e.g., medicinal plant use67) to mitigate malaria pathogenesis. Future studies that synthesize molecular and behavior approaches at long-term study sites, such as Kanyawara, will facilitate investigation of these hypotheses.
Although efforts to control malaria have achieved considerable success during recent decades, the emergence of non-human primate malaria parasites in Southeast Asia (P. knowlesi68,69) and South America (P. simium70) have raised concerns that zoonoses could rollback these hard-fought gains. While the ape Laverania appear to exhibit considerable host-specificity on contemporary timescales, there is nevertheless a strong impetus to critically evaluate the zoonotic potential of these parasites. Chimpanzees and gorillas live in close proximity to humans in parts of equatorial Africa, two pandemic malaria parasites (P. falciparum, P. vivax) have emerged from African great ape reservoirs in the past1,7, and evidence of contemporary human-to-ape transmission of P. falciparum has been documented23,51,71,72. In addition, chimpanzee and gorilla Laverania parasites have transcended primate species barriers in captivity and vectors of ape malaria parasites exhibit promiscuous biting preferences17. Accordingly, it is likely that human exposure occurs with regularity in parts of equatorial Africa. Given finite resources, it is critical that any effort to evaluate the zoonotic potential of the chimpanzee Laverania adopt a targeted approach. Our results offer an ecological framework that will facilitate the design of surveillance efforts by highlighting where and when human exposure is most likely to occur.
Methods
Chimpanzee samples
In this study, we used a combination of longitudinal and cross-sectional sampling strategies to collect 3314 fecal samples from wild chimpanzees for molecular analyses of malaria parasitism. For longitudinal analyses, 878 fecal samples were collected from 54 chimpanzees inhabiting the Kanyawara chimpanzee community, located in Kibale National Park, western Uganda (Fig. 1; Supplementary Table 1). This cohort of wild chimpanzees was habituated to human observation by RWW and has been under continuous direct observation since 1987. All Kanyawara study subjects are identifiable by both morphological appearance and microsatellite genotype. At this field site, researchers and field assistants conduct focal follows of individual chimpanzees on a daily basis and record individual-level behavior, party-level behavior, social affiliation data, and biological samples, among other parameters. Fecal samples—collected only upon direct observation of defecation and only when a positive identification was certain—were preserved in RNAlater (1:1 vol/vol) and stored at −20 °C until exportation and DNA extraction. For cross-sectional analyses, 2436 chimpanzee fecal samples were collected from 55 chimpanzee field sites, distributed across equatorial Africa (Fig. 1; Supplementary Table 2). This dataset included 1936 wild chimpanzee samples, previously collected for molecular studies of simian retroviruses73–78 or malaria parasites1,7, and 500 samples that were newly collected for these analyses (Table 1). Samples were only included in this dataset if collection dates were recorded and corresponding ecological variables (see below) were available. This cross-sectional dataset comprised a combination of samples that were either collected from habituated chimpanzees occupying long-term research sites or opportunistically from non-habituated chimpanzees during ape and biodiversity surveys. Samples were collected in RNAlater (1:1 vol/vol), transported at ambient temperature, and stored at −80 °C. For both longitudinal and cross-sectional surveys, DNA was extracted using the QIAamp Stool DNA minikit (Qiagen, Valencia, CA). Confirmation of host species was obtained either by direct observation of defecation (longitudinal analyses) or via amplification and sequencing of host mitochondrial DNA73–78 (cross-sectional analyses).
Molecular epidemiological studies of wild-living chimpanzees at the Bafwaboli (BA) field site also yielded a fecal sample from an unknown human, which was identified by mtDNA analysis. This human fecal sample as well as a dried blood spot sample collected from a captive chimpanzee (50094) at the Limbe Wildlife Centre in Cameroon were both positive for Hepatocystis mtDNA sequences (965 bp) when screened for Laverania infections using cross-reactive PCR primers (Supplementary Data 3).
Microsatellite analyses
To evaluate the accuracy of Kanyawara chimpanzee identification, we used a semi-nested multiplex PCR protocol, as described previously43,44 (with slight modifications), to genotype 44 fecal samples at 19 polymorphic microsatellite loci. Briefly, all microsatellite loci were first amplified in tandem via an initial multiplexing step. For each sample, 5 μL of extracted DNA was added to a 20 μL multiplex PCR reaction containing all 19 primer pairs at 0.15 mM concentration, 1X Expand Long Template Buffer without MgCl2, 1.75 mM MgCl2, 110 μM of each dNTP, 16 μg bovine serum albumin (BSA), and 0.5 U of Expand Long Template Taq DNA polymerase. Thermocycling was performed with an initial denaturation step for 5 min at 95 °C, 30 cycles of 30 s at 95 °C, 90 s at 58 °C, and 30 s at 72 °C, followed by a final extension at 72 °C for 30 min. Next, a singleplex PCR was performed for each locus in 10 μL volume by mixing 5 μL of the multiplex reaction (diluted 1:100), 0.25 mM of the corresponding fluorescently labeled (FAM, HEX, or NED) forward primer, 0.25 mM nested reverse primer, 1X Expand Long Template Buffer without MgCl2, 0.875 mM MgCl2, 110 μM of each dNTP, 8 μg BSA, and 0.25 U of Expand Long Template Taq DNA polymerase. Thermocycling was performed with an initial denaturation step for 5 min at 95 °C, 30 cycles of 30 s at 95 °C, 90 s at primer-specific annealing temperatures, and 30 s at 72 °C, followed by a final extension at 72 °C for 30 min. Amplification products were electrophoresed on an ABI PRISM 3100 Genetic Analyzer and sized relative to an HD400 (ROX) size standard using GeneMapper 5.0 (Applied Biosystems). This procedure was repeated up to six times for loci that failed to amplify. The microsatellite genotype derived from each of 44 samples was then compared to those of the Kanyawara chimpanzees, and incorrectly attributed samples were recorded as such.
Conventional PCR and single genome amplification
To quantify variation in malaria infection, fecal DNA was screened for malaria parasites using a conventional nested PCR, targeting a 956-bp segment of the apicomplexan mitochondrial cytB gene, using external primers DW2 (5′-TAATGCCTAGACGTATTCCTGATTATCCAG-3′) and DW4 (5′-TGTTTGC TTGGGAGCTGTAATCATAATGTG-3′) in the first-round PCR, and internal primers Pfcytb1 (5′-CTCTATTAATTTAGTTAAAGCACA-3′) and PLAS2a (5′-GTGGTAATTGACATCCWATCC-3′) in the second round, as described previously3,7. To increase the sensitivity of this approach, a subset of fecal samples were subjected to an intensified PCR protocol, whereby samples were screened in 8–10 independent PCR reactions, as previously described4. All Kanyawara fecal samples and a subset of pan-African samples (Supplementary Data 3) were subjected to this intensified PCR protocol. To minimize PCR-induced errors, we applied a single genome amplification (SGA) approach to each fecal sample found to be positive by conventional or intensive PCR, as described previously1,3,7. Briefly, fecal DNA from positive samples was end point diluted in 96-well plates until <30% of wells tested positive. Given a Poisson distribution, this dilution will yield a single amplifiable template per positive reaction >80% of the time. All reactions that were identified to be positive using this SGA methodology were sequenced directly without interim cloning, yielding sequences that were derived from a single template.
Phylogenetic analyses
For analysis of malaria parasitism within the Kanyawara chimpanzee community, we trimmed sequences to 863 bps and used Geneious aligner (version 10) to align sequences to a set of phylogenetically informative reference sequences, corresponding to a representative diversity of simian and human malaria parasites. Sequences shorter than 863 bps were omitted from phylogenetic analyses. Alignments were then visually inspected and sequences containing ambiguities were removed from subsequent analyses. We used jModelTest (version 2.0)79 to identify the best-fit nucleotide substitution model and MrBayes (version 3.2.6)50 to generate Bayesian posterior probabilities, using a chain length of 5.5 million and 10% burn-in. Convergence was achieved when the average deviation of split frequencies was <0.01. Parasite species identity was inferred via NCBI nucleotide BLAST and confirmed using the phylogenetic relationships generated above. To analyze the evolutionary relationships of chimpanzee Hepatocystis isolates, we used the methodology outlined above, except sequences were trimmed to 815 bps to allow for comparison to a greater diversity of Hepatocystis reference sequences. In this case, chimpanzee and human Hepatocystis isolates were aligned to a representative diversity of previously published Hepatocystis isolates, spanning parasites of African Old World monkeys, Asian Old World Monkeys, and other mammals.
Estimation of ecological parameters
We estimated ecological variables using two alternative methodologies: direct measurement and remote sensing. For longitudinal analyses of the Kanyawara chimpanzee community, CAC recorded measurements of ambient air temperature (minimum, maximum) and rainfall twice daily during the study period via local weather monitoring stations installed at the field site. Mean ambient temperature was calculated as the average of minimum and maximum ambient air temperature measurements and intra-day temperature variation was calculated as the difference between these values. For each sample, we defined mean ambient temperature, intra-day temperature variation, and rainfall as the average of measurements of the corresponding variable during the 30 days prior to sample collection. For cross-sectional analyses, ecological variables were estimated using remote sensing datasets. For each sample, we downloaded daytime and nighttime Land Surface Temperature (LST) measurements from the MODerate Resolution Imaging Spectroradiometer (MODIS) thermal sensor on board the NASA-Terra satellite at 1 km2 spatial resolution and 1-day temporal resolution (MOD11A1v005; http://modis.gsfc.nasa.gov/)60 at the geospatial point of sample collection. We subsequently performed quality control to remove low quality or missing data (e.g., resulting from cloud cover) and downsampled spatial resolution to 10 km2 to increase applicability of variables across the sampling site. We then converted these daytime and nighttime LST estimates to minimum and maximum ambient temperature using the equations presented in Weiss et al.36 (with corrected typographical error; Daniel Weiss, pers. comm.):
| 1 |
| 2 |
where Tmin is minimum ambient temperature, Tmax is maximum ambient temperature, LSTnight is the nighttime LST estimate, LSTday is the daytime LST estimate, and Daylength is the number of daylight hours on the day of sampling. Given these values, we calculated mean ambient temperature as the average of minimum and maximum ambient air temperature measurements and intra-day temperature variation was calculated as the difference between these values, as above. Rainfall estimates were derived from the Global Precipitation Climatology Project (GPCP V2.3)61 at 1-day temporal resolution and 1-degree spatial resolution (https://www.esrl.noaa.gov/psd/). Again, for each sample, we defined mean ambient temperature, intra-day temperature variation, and rainfall as the average of measurements of the corresponding variable during the 30 days prior to sample collection. Forest cover estimates were derived from high-resolution global maps of tree canopy cover (30 m, downsampled to 10 km), published by Hansen et al.62. The resulting dataset included only those data points for which all predictor variables were available (N = 2436 samples).
Statistics and reproducibility
We developed two separate generalized linear mixed models to identify the ecological and demographic determinants of Laverania prevalence in wild chimpanzee hosts. For longitudinal analyses (i.e., the Kanyawara GLMM), we specified a generalized linear mixed model (GLMM)55 with binomial error structure and logit link function to evaluate the relationship between climatic variation and the probability of Plasmodium infection among the Kanyawara chimpanzees. For each sample, we specified malaria infection (binary) as the outcome variable, mean ambient temperature (quadratic) and rainfall as fixed effects, chimpanzee individual as a random effect, and age (quadratic), sex, and intra-day temperature variation as covariates (Table 2). Similarly, for cross-sectional analyses (i.e., the pan-African GLMM), we specified a GLMM with binomial error structure and logit link function to evaluate the relationship between climatic variation and the probability of Plasmodium infection among wild chimpanzees inhabiting sampling sites across equatorial Africa. In this case, we coded Plasmodium infection (binary) as the outcome variable, mean ambient temperature (quadratic), rainfall, and percent forest cover as fixed effects, sampling site as a random effect, and daily temperature variation and number of replicates (i.e., 1 or 8–10, depending upon whether samples were screened using the conventional SGA method3 or the intensified method4, respectively) as covariates (Table 3). Variables in the Pan-African analysis were mean-centered to improve model stability. In each case, we used likelihood ratio tests to compare the fit of the full GLMM (containing all predictor variables, covariates, random effect, and intercept) to that of the null model (containing only random effect and intercept). Predictor variables that did not improve the fit of each model were subsequently excluded from the final models. These statistical analyses were performed in R (version 3.4.3) using the lme4 package80.
Geospatial mapping
Given the ecological relationships identified in the Pan-African GLMM, outlined above, we extrapolated infection probabilities across wild chimpanzee habitat in equatorial Africa. To characterize ecological variation across equatorial Africa, we derived composite rasters from mean ambient temperature and intra-day temperature variation measurements recorded between March 2000 and February 2017 across the African continent at 1-degree spatial and 1-month temporal resolution. Using these composite temperature rasters and the Hansen et al.62 forest cover dataset, we projected the predicted probabilities of chimpanzee Laverania infection (derived from the pan-African model) across the IUCN-designated spatial extent of chimpanzee habitat, according to the following equation:
| 3 |
where intercept is −2.538 for samples screened using conventional PCR (i.e., one replicate) and −1.374 for those screened using intensive PCR (i.e., 8–10 replicates), and AT, TV, and FC are each corrected by subtracting the means of the chimpanzee dataset (23.4 for AT, 9.1 for TV, and 77.2 for FC). All geospatial analyses were performed in ArcMap (v10.6 Environmental Systems Research Institute Inc.).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by grants from the National Institutes of Health R01AI091595, R01AI120810, R01AI050529, and P30AI045008 (B.H.H.); R01HL139337 (M.T.D.), the National Geographic Society (E.J.S.), the International Primatological Society (E.J.S.), and the American Society of Primatologists (E.J.S.), as well as fellowships from Harvard University (E.J.S.) and the National Science Foundation (E.J.S.). We thank Dr. Kristen Skillman for help in putting the manuscript together, as well as two referees for their review and thoughtful comments on this manuscript.
Author contributions
E.J.S. wrote the manuscript, generated all figures and tables, collected chimpanzee fecal samples from Kibale National Park, Budongo Forest, and Kyambura Gorge, extracted DNA from fecal samples, performed molecular diagnostic assays, performed chimpanzee microsatellite genotyping analyses, performed phylogenetic analyses, performed all statistical analyses, extracted all remote sensing climate data, extracted all forest cover data, and performed all geospatial mapping, in the laboratories of B.H.H. and M.T.D. W.L., Y.L., and B.H.H. performed molecular diagnostic assays for all other field sites. J.N.N., M.P., S.K., A.E.P., E.V.L., C.M.S., D.B.M., A.K.P., F.A.S., M.K.G., N.S., C.A., K.Z., K.K., C.A.C, R.C., A.R., M.A.H., and R.W.W. facilitated chimpanzee fecal sample collection from field sites across equatorial Africa. N.D.W. provided chimpanzee dried blood spot samples from the Limbe sanctuary in Cameroon. C.A.C. facilitated the collection of climate data within Kibale National Park. R.W.W., M.T.D., and B.H.H. offered revisions of the manuscript. All authors contributed to the intellectual content of this article.
Peer review
Peer review information
Communications Biology thanks Virginie Rougeron and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Caitlin Karniski and Luke R. Grinham. Peer reviewer reports are available.
Data availability
Plasmodium and Hepatocystis mtDNA sequence generated for this study have been deposited at NCBI GenBank under the accession numbers MW228501-MW228790 and OL691962-OL691978, respectively (Supplementary Data 4). All other datasets generated in this study are available from the corresponding authors upon request.
Code availability
All statistical analyses were performed using open-source packages in R (version 3.4.3) as referenced in the methods section. The scripts used to implement these analyses are available from the corresponding authors upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Manoj T. Duraisingh, Email: mduraisi@hsph.harvard.edu
Beatrice H. Hahn, Email: bhahn@pennmedicine.upenn.edu
Richard W. Wrangham, Email: wranghamrichard@gmail.com
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-03962-0.
References
- 1.Liu W, et al. African origin of the malaria parasite Plasmodium vivax. Nat. Commun. 2014;5:3346. doi: 10.1038/ncomms4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu W, et al. Multigenomic delineation of Plasmodium species of the Laverania subgenus infecting wild-living chimpanzees and gorillas. Genome Biol. Evolution. 2016;8:1929–1939. doi: 10.1093/gbe/evw128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu, W. et al. Single genome amplification and direct amplicon sequencing of Plasmodium spp. DNA from ape fecal specimens. Protocol Exchange 1–14 (2010).
- 4.Liu W, et al. Wild bonobos host geographically restricted malaria parasites including a putative new Laverania species. Nat. Commun. 2017;8:1635. doi: 10.1038/s41467-017-01798-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prugnolle F, et al. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc. Natl Acad. Sci. USA. 2010;107:1458–1463. doi: 10.1073/pnas.0914440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharp PM, Plenderleith LJ, Hahn BH. Ape origins of human malaria. Annu. Rev. Microbiol. 2020;74:39–63. doi: 10.1146/annurev-micro-020518-115628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu W, et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467:420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otto TD, et al. Genomes of all known members of a Plasmodium subgenus reveal paths to virulent human malaria. Nat. Microbiol. 2018;3:687–697. doi: 10.1038/s41564-018-0162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boundenga L, et al. Diversity of malaria parasites in great apes in Gabon. Malar. J. 2015;14:1–8. doi: 10.1186/s12936-015-0622-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Délicat-Loembet L, et al. No evidence for ape Plasmodium infections in humans in gabon. Plos One. 2015;10:e0126933. doi: 10.1371/journal.pone.0126933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sundararaman SA, et al. Plasmodium falciparum-like parasites infecting wild apes in southern Cameroon do not represent a recurrent source of human malaria. Proc. Natl Acad. Sci. USA. 2013;110:7020–7025. doi: 10.1073/pnas.1305201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Junker J, et al. Recent decline in suitable environmental conditions for African great apes. Diversity Distrib. 2012;18:1077–1091. doi: 10.1111/ddi.12005. [DOI] [Google Scholar]
- 13.de Nys HM, et al. Age-related effects on malaria parasite infection in wild chimpanzees. Biol. Lett. 2013;9:20121160. doi: 10.1098/rsbl.2012.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Nys HM, et al. Malaria parasite detection increases during pregnancy in wild chimpanzees. Malar. J. 2014;13:413. doi: 10.1186/1475-2875-13-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser M, et al. Wild chimpanzees infected with 5 Plasmodium species. Emerg. Infect. Dis. 2010;16:1956–1959. doi: 10.3201/eid1612.100424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paupy C, et al. Anopheles moucheti and Anopheles vinckei are candidate vectors of ape Plasmodium parasites, including Plasmodium praefalciparum in Gabon. PLoS ONE. 2013;8:e57294. doi: 10.1371/journal.pone.0057294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makanga, B. et al. Ape malaria transmission and potential for ape-to-human transfers in Africa. Proc. Natl Acad. Sci. USA113, 5329–5334 (2016). [DOI] [PMC free article] [PubMed]
- 18.Loy, D. E. et al. Investigating zoonotic infection barriers to ape Plasmodium parasites using faecal DNA analysis. Int. J. Parasitol.48, 531–542 (2018). [DOI] [PMC free article] [PubMed]
- 19.Martin M, Rayner J, Gagneux P, Barnwell J, Varki A. Evolution of human–chimpanzee differences in malaria susceptibility: Relationship to human genetic loss of N-glycolylneuraminic acid. Proc. Natl Acad. Sci. USA. 2005;102:12819–12824. doi: 10.1073/pnas.0503819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scully EJ, Kanjee U, Duraisingh MT. Molecular interactions governing host-specificity of blood stage malaria parasites. Curr. Opin. Microbiol. 2017;40:21–31. doi: 10.1016/j.mib.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sundararaman SA, et al. Genomes of cryptic chimpanzee Plasmodium species reveal key evolutionary events leading to human malaria. Nat. Commun. 2016;7:11078. doi: 10.1038/ncomms11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wanaguru M, Liu W, Hahn BH, Rayner JC, Wright GJ. RH5-Basigin interaction plays a major role in the host tropism of Plasmodium falciparum. Proc. Natl Acad. Sci. USA. 2013;110:20735–20740. doi: 10.1073/pnas.1320771110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ngoubangoye B, et al. The host specificity of ape malaria parasites can be broken in confined environments. Int. J. Parasitol. 2016;46:737–744. doi: 10.1016/j.ijpara.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Mapua MI, et al. A comparative molecular survey of malaria prevalence among Eastern chimpanzee populations in Issa Valley (Tanzania) and Kalinzu (Uganda) Malar. J. 2016;15:423. doi: 10.1186/s12936-016-1476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu DF, et al. Seasonal and inter-annual variation of malaria parasite detection in wild chimpanzees. Malar. J. 2018;17:1–5. doi: 10.1186/s12936-018-2187-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Craig M, le Sueur D, Snow B. A climate-based distribution model of malaria transmission in sub-Saharan Africa. Parasitol. Today. 1999;15:105–111. doi: 10.1016/S0169-4758(99)01396-4. [DOI] [PubMed] [Google Scholar]
- 27.Mordecai EA, et al. Optimal temperature for malaria transmission is dramatically lower than previously predicted. Ecol. Lett. 2013;16:22–30. doi: 10.1111/ele.12015. [DOI] [PubMed] [Google Scholar]
- 28.Paaijmans KP, et al. Influence of climate on malaria transmission depends on daily temperature variation. Proc. Natl Acad. Sci. USA. 2010;107:15135–15139. doi: 10.1073/pnas.1006422107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parham PE, Michael E. Modeling the effects of weather and climate change on malaria transmission. Environ. Health Perspect. 2010;118:620–626. doi: 10.1289/ehp.0901256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaPointe DA, Goff ML, Atkinson CT. Thermal constraints to the sporogonic development and altitudinal distribution of avian malaria Plasmodium relictum in Hawai’i. J. Parasitol. 2010;96:318–324. doi: 10.1645/GE-2290.1. [DOI] [PubMed] [Google Scholar]
- 31.Vanderberg, J. P. & Yoeli, M. Effects of temperature on sporogonic development of Plasmodium berghei. J. Parasitol.52, 559–564 (1966). [PubMed]
- 32.Macdonald, G. The Epidemiology and Control of Malaria (Oxford University Press, 1957).
- 33.Ryan SJ, et al. Mapping physiological suitability limits for malaria in Africa under climate change. Vector-Borne Zoonotic Dis. 2015;15:718–725. doi: 10.1089/vbz.2015.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gemperli A, et al. Mapping malaria transmission in West and Central Africa. Tropical Med. Int. Health. 2006;11:1032–1046. doi: 10.1111/j.1365-3156.2006.01640.x. [DOI] [PubMed] [Google Scholar]
- 35.Gething PW, et al. Modelling the global constraints of temperature on transmission of Plasmodium falciparum and P. vivax. Parasites Vectors. 2011;4:92. doi: 10.1186/1756-3305-4-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiss DJ, et al. Air temperature suitability for Plasmodium falciparum malaria transmission in Africa 2000–2012: a high-resolution spatiotemporal prediction. Malar. J. 2014;13:171. doi: 10.1186/1475-2875-13-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyons CL, Coetzee M, Chown SL. Stable and fluctuating temperature effects on the development rate and survival of two malaria vectors, Anopheles arabiensis and Anopheles funestus. Parasites Vectors. 2013;6:104. doi: 10.1186/1756-3305-6-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paaijmans KP, Wandago MO, Githeko AK, Takken W. Unexpected high losses of Anopheles gambiae larvae due to rainfall. PLoS One. 2007;2:e1146. doi: 10.1371/journal.pone.0001146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faust C, Dobson AP. Primate malarias: diversity, distribution and insights for zoonotic Plasmodium. One Health. 2015;1:66–75. doi: 10.1016/j.onehlt.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tucker Lima JM, Vittor A, Rifai S, Valle D. Does deforestation promote or inhibit malaria transmission in the Amazon? A systematic literature review and critical appraisal of current evidence. Philos. Trans. R. Soc. Lond. Ser. B, Biol. Sci. 2017;372:20160125. doi: 10.1098/rstb.2016.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borner J, et al. Phylogeny of haemosporidian blood parasites revealed by a multi-gene approach. Mol. Phylogenetics Evolution. 2016;94:221–231. doi: 10.1016/j.ympev.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 42.Emery Thompson M, Muller MN, Machanda ZP, Otali E, Wrangham RW. The Kibale Chimpanzee Project: over thirty years of research, conservation, and change. Biol. Conserv. 2020;252:108857. doi: 10.1016/j.biocon.2020.108857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langergraber KE, Mitani JC. & Vigilant, L. The limited impact of kinship on cooperation in wild chimpanzees. Proc. Natl Acad. Sci. USA. 2007;104:7786–7790. doi: 10.1073/pnas.0611449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arandjelovic M, et al. Two-step multiplex polymerase chain reaction improves the speed and accuracy of genotyping using DNA from noninvasive and museum samples. Mol. Ecol. Resour. 2009;9:28–36. doi: 10.1111/j.1755-0998.2008.02387.x. [DOI] [PubMed] [Google Scholar]
- 45.Herbert A, et al. Malaria-like symptoms associated with a natural Plasmodium reichenowi infection in a chimpanzee. Malar. J. 2015;14:220. doi: 10.1186/s12936-015-0743-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torres, J. R. Therapy of Infectious Diseases 597–613 (2003).
- 47.Trampuz A, Jereb M, Muzlovic I, Prabhu RM. Clinical review: severe malaria. Crit. Care. 2003;7:315. doi: 10.1186/cc2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akim NI, et al. Dynamics of P. falciparum gametocytemia in symptomatic patients in an area of intense perennial transmission in Tanzania. Am. J. Tropical Med. Hyg. 2000;63:199–203. doi: 10.4269/ajtmh.2000.63.199. [DOI] [PubMed] [Google Scholar]
- 49.Mackinnon MJ, Read AF. Genetic relationships between parasite virulence and transmission in the rodent malaria Plasmodium chabaudi. Evolution. 1999;53:689–703. doi: 10.1111/j.1558-5646.1999.tb05364.x. [DOI] [PubMed] [Google Scholar]
- 50.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 51.Prugnolle F, et al. African monkeys are infected by Plasmodium falciparum nonhuman primate-specific strains. Proc. Natl Acad. Sci. USA. 2011;108:11948–11953. doi: 10.1073/pnas.1109368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ayouba A, et al. Ubiquitous Hepatocystis infections, but no evidence of Plasmodium falciparum-like malaria parasites in wild greater spot-nosed monkeys (Cercopithecus nictitans) Int. J. Parasitol. 2012;42:709–713. doi: 10.1016/j.ijpara.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): Evolution of life-history traits and host switches. Mol. Phylogenetics Evolution. 2008;47:261–273. doi: 10.1016/j.ympev.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 54.Thurber MI, et al. Co-infection and cross-species transmission of divergent Hepatocystis lineages in a wild African primate community. Int. J. Parasitol. 2013;43:613–619. doi: 10.1016/j.ijpara.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baayen, R. H. Analyzing Linguistic Data: A Practical Introduction to Statistics (Cambridge University Press, 2008).
- 56.Stanisic DI, et al. Acquisition of antibodies against Plasmodium falciparum merozoites and malaria immunity in young children and the influence of age, force of infection, and magnitude of response. Infect. Immun. 2015;83:646–660. doi: 10.1128/IAI.02398-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor RR, Allen SJ, Greenwood BM, Riley EM. IgG3 antibodies to Plasmodium falciparum merozoite surface protein 2 (MSP2): increasing prevalence with age and association with clinical immunity to malaria. Am. J. Tropical Med. Hyg. 1998;58:406–413. doi: 10.4269/ajtmh.1998.58.406. [DOI] [PubMed] [Google Scholar]
- 58.World Malaria Report (World Health Organization, 2015).
- 59.Shaman J. Letter to the Editor: Caution needed when using gridded meteorological data products for analyses in Africa. Eur. Surveill. 2014;19:20930. doi: 10.2807/1560-7917.ES2014.19.41.20930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tatem AJ, Goetz SJ, Hay SI. Terra and Aqua: new data for epidemiology and public health. Int. J. Appl. Earth Observation Geoinf. 2004;6:33–46. doi: 10.1016/j.jag.2004.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adler RF, et al. The version-2 global precipitation climatology project (GPCP) monthly precipitation analysis (1979–present) J. Hydrometeorol. 2003;4:1147–1167. doi: 10.1175/1525-7541(2003)004<1147:TVGPCP>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hansen MC, et al. High-resolution global maps of 21st-century forest cover change. Science. 2013;342:850–853. doi: 10.1126/science.1244693. [DOI] [PubMed] [Google Scholar]
- 63.Carter R, Mendis KN. Evolutionary and historical aspects of the burden of malaria. Clin. Microbiol. Rev. 2002;15:564–594. doi: 10.1128/CMR.15.4.564-594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am. J. Hum. Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tarello W. A fatal Plasmodium reichenowi infection in a chimpanzee? Rev. de. Med. Veterinaire. 2005;156:503–505. [Google Scholar]
- 66.Taylor DW, et al. Parasitologic and immunologic studies of experimental Plasmodium falciparum infection in nonsplenectomized chimpanzees (Pan troglodytes) Am. J. Tropical Med. Hyg. 1985;34:36–44. doi: 10.4269/ajtmh.1985.34.36. [DOI] [PubMed] [Google Scholar]
- 67.Krief S, Martin M, Grellier P, Kasenene J, Sevenet T. Novel antimalarial compounds isolated in a survey of self-medicative behavior of wild chimpanzees in Uganda. Antimicrobial Agents Chemother. 2004;48:3196–3199. doi: 10.1128/AAC.48.8.3196-3199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cox-Singh J, et al. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin. Infect. Dis. 2008;46:165–171. doi: 10.1086/524888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh B, Daneshvar C. Human infections and detection of Plasmodium knowlesi. Clin. Microbiol. Rev. 2013;26:165–184. doi: 10.1128/CMR.00079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brasil, P. et al. Outbreak of human malaria caused by Plasmodium simium in the Atlantic Forest in Rio de Janeiro: a molecular epidemiological investigation. Lancet Global Health5, e1038–e1046 (2017). [DOI] [PubMed]
- 71.Krief, S. et al. On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from bonobos. PLoS Pathog.6, e1000765 (2010). [DOI] [PMC free article] [PubMed]
- 72.Pacheco MA, Cranfield M, Cameron K, Escalante AA. Malarial parasite diversity in chimpanzees: the value of comparative approaches to ascertain the evolution of Plasmodium falciparum antigens. Malar. J. 2013;12:328. doi: 10.1186/1475-2875-12-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Etienne L, et al. Noninvasive follow-up of simian immunodeficiency virus infection in wild-living nonhabituated western lowland gorillas in Cameroon. J. Virol. 2012;86:9760–9772. doi: 10.1128/JVI.01186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keele BF, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keele BF, et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Y, et al. Eastern chimpanzees, but not bonobos, represent a simian immunodeficiency virus reservoir. J. Virol. 2012;86:10776–10791. doi: 10.1128/JVI.01498-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Neel C, et al. Molecular epidemiology of simian immunodeficiency virus infection in wild-living gorillas. J. Virol. 2010;84:1464–1476. doi: 10.1128/JVI.02129-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rudicell RS, et al. Impact of simian immunodeficiency virus infection on chimpanzee population dynamics. PLoS Pathog. 2010;6:1–17. doi: 10.1371/journal.ppat.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bates, D. & Maechler, M. Lme4: linear mixed-effects models using s4 classes. Cran R Project Website (2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Plasmodium and Hepatocystis mtDNA sequence generated for this study have been deposited at NCBI GenBank under the accession numbers MW228501-MW228790 and OL691962-OL691978, respectively (Supplementary Data 4). All other datasets generated in this study are available from the corresponding authors upon request.
All statistical analyses were performed using open-source packages in R (version 3.4.3) as referenced in the methods section. The scripts used to implement these analyses are available from the corresponding authors upon request.