Abstract
Phasins are proteins that are proposed to play important roles in polyhydroxyalkanoate synthesis and granule formation. Here the phasin PhaP of Ralstonia eutropha has been analyzed with regard to its role in the synthesis of polyhydroxybutyrate (PHB). Purified recombinant PhaP, antibodies against PhaP, and an R. eutropha phaP deletion strain have been generated for this analysis. Studies with the phaP deletion strain show that PhaP must accumulate to high levels in order to play its normal role in PHB synthesis and that the accumulation of PhaP to low levels is functionally equivalent to the absence of PhaP. PhaP positively affects PHB synthesis under growth conditions which promote production of PHB to low, intermediate, or high levels. The levels of PhaP generally parallel levels of PHB in cells. The results are consistent with models whereby PhaP promotes PHB synthesis by regulating the surface/volume ratio of PHB granules or by interacting with polyhydroxyalkanoate synthase and indicate that PhaP plays an important role in PHB synthesis from the early stages in PHB production and across a range of growth conditions.
Polyhydroxyalkanoates (PHAs) are polyoxoesters that are synthesized intracellularly in diverse bacteria under conditions of limitation for a nutrient other than carbon (5). The polymers are water insoluble and accumulate as intracellular granules (5). Phasins are low-molecular-weight proteins that are proposed to promote PHA synthesis in cells (9, 10, 15, 18). Three different mechanisms for the function of phasins have been proposed. First, phasins may enhance PHA production by binding to granules and increasing the surface/volume ratio of the granules (18). Second, phasins may activate the rate of PHA synthesis by interacting directly with PHA synthase (18). Third, phasins may promote PHA synthesis indirectly by preventing growth defects associated with the binding of other cellular proteins to PHA granules (6, 18). Phasins have also been proposed to function as storage proteins (7), a role that could also conceivably affect PHA synthesis. Studies on the function of phasins in recombinant host strains (6, 9) and in vitro (3) have yielded anomalous results which hint that the timing and levels of expression of phasins may be crucial for their function. In order to understand the function of phasins, we have generated new tools and carried out a systematic study to examine the kinetics and amounts of the Ralstonia eutropha phasin PhaP in R. eutropha under a variety of growth conditions.
R. eutropha is well suited for studies on phasins, given that the strain produces a PHA, poly-[(R)-3-hydroxybutyrate] (PHB), under many standard cultivation conditions (5, 17) and is amenable to genetic manipulation (8, 12, 14). The first genetic analysis of the role of PhaP in PHB synthesis in R. eutropha was conducted by Wieczorek et al. (18). They reported the isolation of five phaP::Tn5 mutants that were claimed to be defective in the production of PhaP and to produce half as much PHB as the wild-type (wt) strain (18). None of these mutants, however, were actually shown to contain a Tn5 insertion in the phaP open reading frame (ORF), and none of the mutants were actually compared to a strictly isogenic wt control strain (mutants were generated in strain HF39 but were compared to strain Ae H16) (18). Furthermore, these phaP::Tn5 mutants were characterized for PHB production under only one set of cultivation conditions and without monitoring of the time course of PHB production (18). Thus, a number of fundamental questions about PhaP remain to be answered. What is the PHB production phenotype of mutants that are completely blocked for expression of PhaP? Does PhaP promote PHB synthesis only above a fixed threshold level of PHB or during synthesis of any amount of PHB? Does PhaP play a role in PHB synthesis only during a discrete period or throughout the entire process of PHB synthesis?
Here we report a study designed to address these questions. This study is based on the construction of a phaP deletion strain of R. eutropha and the use of this strain to analyze the role of PhaP in PHB synthesis over a range of cultivation conditions and time points. Immunoblot analyses demonstrate that the previously characterized phaP::Tn5 mutants actually produce small but detectable amounts of PhaP, whereas the phaP deletion strain is completely blocked for production of PhaP. PHB quantitation analyses indicate that PhaP plays an important role in PHB synthesis, whether cells are producing low, intermediate, or high levels of PHB, and that PhaP affects PHB synthesis beginning early in the process of PHB production. The implications of these results are discussed, and a refined model for the function of PhaP in PHB synthesis is presented.
Construction and characterization of R. eutropha phaP deletion strain.
To test the importance of PhaP in PHB production in R. eutropha, we began by constructing an R. eutropha strain in which the phaP ORF has been deleted (Table 1). A fragment of DNA that spans 0.8 kb immediately upstream and 0.3 kb immediately downstream of the phaP ORF but that lacks the phaP ORF was constructed by PCR. The construct was confirmed by sequencing, cloned into a gene replacement vector, and used to accomplish precise deletion of the phaP ORF in the wt strain Ae H16 through homologous recombination by a standard technique (11, 14). Successful construction of the resulting phaP deletion strain Re1052 was established based on PCR and Southern hybridization analyses of the strain.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| R. eutropha | ||
| Ae H16 | Wild type, gentamicin resistant, also termed DSM 428 and ATCC 17699 | ATCC 17699 |
| H2262 | phaP::Tn5 strain derived from HF39, partial Tn5 approx. 0.1 kb upstream of phaP ORF | 18 |
| H2271 | phaP::Tn5 strain derived from HF39, Tn5 insertion 26 bp upstream of phaP ORF | 18 |
| H2275 | phaP::Tn5 strain derived from HF39, Tn5 insertion 156 bp upstream of phaP ORF | 18 |
| Re1052 | phaP precise deletion gene replacement strain, derived from Ae H16/pGY63 | This study |
| E. coli | ||
| BL21(DE3) | Strain used for pET expression system | 16 |
| S17-1 | Strain for conjugative transfer of plasmids into R. eutropha for gene replacement | 13 |
| Plasmids | ||
| pET5a | T7 expression system plasmid, confers ampicillin resistance | Promega |
| pGY63 | phaP precise deletion fragment cloned into pJQ200mp18Km | This study |
| pGY101 | phaP expression plasmid, corresponding to phaP ORF cloned into pET5a | This study |
| pJQ200mp18 | Gene replacement vector, carries sacB, oriV, oriT, traJ, confers gentamicin resistance | 11 |
| pJQ200mp18Km | Derivative of pJQ200mp18, gentamicin resistance gene disrupted, confers kanamycin resistance | This study |
| pUT-miniTn5-Km | Source of kanamycin resistance gene for pJQ200mp18Km | 1 |
Development of cultivation conditions for production of PHB to low, intermediate, or high levels in the wt strain.
The role of PhaP in PHB synthesis under cultivation conditions that result in a wide range of PHB concentrations is not known. To address this issue, we developed a standard approach to accomplish the accumulation of PHB to low, intermediate, or high levels in the wt strain based on the use of the growth media tryptic soy broth-dextrose free (TSB), PHA(med), and PHA(high), respectively. TSB is a nutrient-rich medium (Becton-Dickinson Microbiology Systems, Cockeysville, Md.). PHA(med) and PHA(high) are based on a minimal medium (8) supplemented with fructose (0.5 or 1%, respectively) and ammonium chloride (0.1 or 0.01%, respectively). The approach involves the use of a single TSB starter culture for inoculation of the three types of growth media in 5-ml aliquots in test tubes at an initial optical density at 600 nm (OD600) of 1.0 and cultivation for 48 h. The amount of PHB in cells at 48 h was determined by the sulfuric acid–high-pressure liquid chromatography method (4). In a typical analysis PHB accumulates to 2.6% cell dry weight (cdw), 58% cdw, and 81% cdw for the wt strain cultivated in TSB, PHA(med), and PHA(high), respectively (limit of detection, PHB < 0.1% cdw).
Immunoblot analyses indicate that R. eutropha wt strain produces PhaP under conditions that promote production of PHB to low, intermediate, or high levels.
Recombinant PhaP protein, expressed in Escherichia coli by the use of the pET expression system (16) and purified to near homogeneity by passage directly through an anion-exchange chromatography column, was used for generation and purification of rabbit anti-PhaP polyclonal antibodies. The wt strain was cultivated in TSB, PHA(med), and PHA(high) for 48 h and was analyzed for CFU, OD600, and PhaP accumulation. The phaP deletion strain was analyzed in parallel as a negative control. CFU measurements indicated that cells in all cultures remained viable over the course of the cultivation (data not shown). Results for immunoblot and OD600 measurements are shown in Fig. 1.
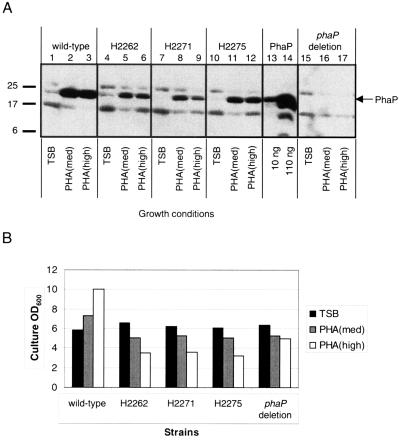
FIG. 1.
(A) Accumulation of PhaP in wt and phaP mutant R. eutropha strains. Proteins were separated by sodium dodecyl sulfate–15% polyacrylamide gel electrophoresis and were subjected to immunoblot analysis for detection of PhaP. Cells from R. eutropha cultures were harvested after cultivation for 48 h in TSB, PHA(med), and PHA(high). Bacterial samples correspond to cells from 10 μl of culture diluted to an OD600 of 1.0. Purified PhaP was included as a control. Note that although all samples are shown together here, the phaP deletion strain was actually analyzed on a separate gel (which also included purified PhaP). Molecular mass standards are indicated (kDa). (B) Measurements of OD600 for R. eutropha strains after cultivation for 48 h in TSB, PHA(med), and PHA(high). Data are extrapolated from measurements on 10-fold dilutions of cultures.
The results indicate that PhaP accumulates to detectable levels in the wt strain during cultivation under all three conditions (Fig. 1A, lanes 1, 2, and 3). The phaP deletion strain produces no PhaP, as expected (Fig. 1A, lanes 15, 16, and 17); no signal was detected, even upon prolonged exposure of blots to film. The results further indicate that PhaP accumulates in the wt strain to low levels during cultivation in TSB and to much higher levels during cultivation in PHA(med) and PHA(high). The wt strain exhibits relatively modest differences in growth (Fig. 1B) but dramatic differences in PHB accumulation from TSB to PHA(high), consistent with the report of Wieczorek et al. (18) that PhaP accumulation is regulated by PHB production.
Immunoblot analyses indicate that the three existing phaP::Tn5 strains H2262, H2271, and H2275 each produce small but detectable amounts of PhaP.
Wieczorek et al. (18) reported that the three phaP::Tn5 mutants H2262, H2271, and H2275 fail to produce PhaP despite the fact that none of the three strains actually contains Tn5 insertions in the phaP ORF. Here we analyzed PhaP accumulation in these strains as well. Immunoblot analyses indicate that all three strains produce low but detectable amounts of PhaP (Fig. 1A, lanes 4 through 12). The possibility that this result was due to contamination of cultures or stocks was ruled out by replication of the immunoblot results in an independent analysis and by confirmation of the genotypes of the strains by PCR analyses.
PHB quantitation and electron microscopy analyses indicate that the phaP deletion mutant exhibits defects in PHB production and granule formation that are similar to the defects exhibited by the phaP::Tn5 strains.
The observation that the phaP::Tn5 strains produce small but detectable amounts of PhaP raised the possibility that the phaP deletion strain might exhibit more severe defects in terms of PHB production and granule formation in comparison to the three phaP::Tn5 strains. However, comparison of PHB production by the wt strain Ae H16, the phaP deletion strain, H2271, and H2275, as cultivated in PHA(high) for 48 h, indicates that the phaP deletion and phaP::Tn5 strains exhibit similar defects in PHB production (PHB accumulated to 81, 58, 50, and 45% cdw, respectively, and to 1.7, 0.64, 0.38, and 0.33 mg/ml of culture, respectively). Analysis of the phaP deletion strain by electron microscopy indicates that PHB accumulates as a single large granule per cell (data not shown), as has been reported for the phaP::Tn5 mutant strains (18), rather than as the many small granules typical of wt cells. These observations suggest that the production of small amounts of PhaP is functionally equivalent to the complete absence of PhaP and that PhaP must accumulate to high levels to function normally.
PHB quantitation analyses indicate that the phaP deletion strain exhibits defects in PHB production under a range of cultivation conditions.
We proceeded to test the role of PhaP in PHB synthesis under a range of cultivation conditions. The wt and phaP deletion strains were cultivated in parallel in TSB, PHA(med), and PHA(high) as described above, except that cultivations were scaled up to 200-ml cultures in 1-liter baffled flasks. At specific time points aliquots were removed from the cultures for measurements of OD600, CFU, cdw, and PHB. CFU measurements indicated that cells in all cultures remained viable over the course of the experiment (data not shown). Results for measurements of cdw and PHB are shown in Fig. 2. OD600 measurements matched cdw measurements and thus are not shown.
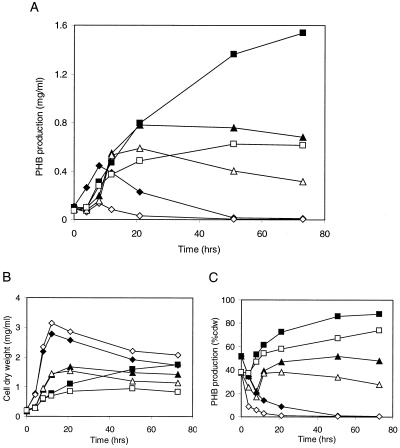
FIG. 2.
Comparisons of PHB production and cdws for wt and phaP deletion strains. Symbols: closed diamonds, wt strain TSB; open diamonds, phaP deletion strain TSB; closed triangles, wt strain PHA(med); open triangles, phaP deletion strain PHA(med); closed squares, wt strain PHA(high); open squares, phaP deletion strain PHA(high). (A) PHB accumulation (mg/ml of culture) in wt and phaP deletion strains as determined over a time course of 75 h. (B) The cdws for wt and phaP deletion strains. (C) PHB accumulation (% cdw) in wt and phaP deletion strains.
The phaP deletion strain exhibited defects in PHB production relative to the wt strain under all growth conditions (Fig. 2). The wt strain cultivated in TSB, PHA(med), and PHA(high) accumulated PHB to maximum levels of 0.44, 0.78, or 1.5 mg per ml of culture, respectively. For comparison, the phaP deletion strain accumulated PHB to maximum levels of 0.13, 0.59, or 0.62 mg per ml of culture, respectively. Both strains exhibited the same trends of PHB accumulation under each of the growth conditions, but the phaP deletion strain produced approximately half as much PHB as the wt strain under each condition. Also, the phaP deletion strain cultivated in PHA(high) exhibited a gradually decreasing rate of production of PHB relative to the wt strain rather than reaching a maximal level of PHB and abruptly halting production. These observations suggest that PhaP plays an important role in PHB production under each of the growth conditions and that this role begins early in the PHB production period.
The results argue against models whereby PhaP promotes PHB synthesis indirectly by preventing deleterious effects associated with PHB production in R. eutropha cells or by functioning as a storage protein. Comparisons of cdw measurements indicate that the wt and phaP deletion strains exhibit comparable growth during the first 8 h of cultivation in TSB (Fig. 2). During this same period, however, the phaP deletion strain exhibits a defect in PHB production relative to the wt strain (Fig. 2). These observations indicate that PhaP can promote PHB synthesis independent of any effects that PhaP may have on growth, at least during cultivation in TSB and perhaps under all cultivation conditions.
Quantitative immunoblot analyses indicate that PhaP accumulation parallels PHB production in wt strain under a range of cultivation conditions.
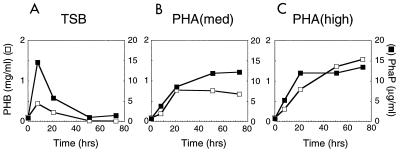
The model emerging from these studies suggests that PhaP will accumulate to high levels under each set of growth conditions during periods of PHB production. Quantitative immunoblot analyses of PhaP were therefore conducted as part of the analysis described above. The results indicate that PhaP accumulation does parallel PHB production in the wt strain under each of the cultivation conditions (Fig. 3). In general, as levels of PHB increase, levels of PhaP increase. Also, for cultivation in TSB in particular, as levels of PHB decrease, levels of PhaP decrease. Levels of PhaP and PHB are not strictly correlated across the three cultivation conditions, though. The ratio of PhaP to PHB is higher during cultivation in TSB versus PHA(high). Also, during cultivation in PHA(high), PhaP levels seem to reach a plateau within 21 h, while PHB levels continue to increase throughout the 75-h cultivation period. These observations may reflect some degree of physiological control over PhaP accumulation.
FIG. 3.
Comparison of levels of PhaP versus PHB for wt strain cultivated over a time course of 72 h. Symbols: closed squares, PhaP (μg/ml of culture); open squares, PHB (mg/ml of culture). (A) TSB growth medium; (B) PHA(med) growth medium; (C) PHA(high) growth medium.
Refined models for the role of phasins in PHA production.
Our results are consistent with two of the previously proposed models for the role of PhaP, that PhaP promotes PHB synthesis by regulating the ratio of surface area to volume of PHB granules (18) and that PhaP promotes PHB synthesis by interaction with PHA synthase (18). The key new observation is that PhaP promotes PHB synthesis throughout the period of PHB production and across a range of cultivation conditions. Within the context of the first model, PhaP may bind the surface of PHB granules throughout cultivation and prevent their coalescence. Within the context of the second model, PhaP may interact with PHA synthase, triggering a conformational change. Independent of which model applies, PhaP does not play a specialized role only for production of PHB to very high levels in cells but instead plays a more fundamental role in PHB synthesis.
Nucleotide sequence accession numbers.
During sequencing analyses of the phaP region (total of 1,663 bp, spanning from 852 bp upstream to 232 bp downstream of the phaP ORF) we discovered errors in the original published sequence (18). We have submitted the corrected sequence to GenBank (accession no. AF314206). A subset of these errors, specifically those which occur within the phaP ORF, have been reported previously (2).
Acknowledgments
We thank Ute Müh, Björn Junker, Jimmy Jia, Joon Ho Choi, and Wei Yuan for useful discussions and Alexander Steinbüchel for sending us the phaP::Tn5 strains H2262, H2271, and H2275.
We acknowledge the NIH award of BRS Shared Instrumentation Grant No. S10 RR05734-01 and the MIT Biomedical Microscopy Laboratory and assistance from Patricia Reilly. G.Y. is a DOE-Energy Biosciences Research Fellow of the Life Sciences Research Foundation. This work was supported by NIH Grant GM 49171 to A.J.S. and J.S.
REFERENCES
- 1.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanley S Z, Pappin D J, Rahman D, White A J, Elborough K M, Slabas A R. Re-evaluation of the primary structure of Ralstonia eutropha phasin and implications for polyhydroxyalkanoic acid granule binding. FEBS Lett. 1999;447:99–105. doi: 10.1016/s0014-5793(99)00235-5. [DOI] [PubMed] [Google Scholar]
- 3.Jossek R, Reichelt R, Steinbüchel A. In vitro biosynthesis of poly(3-hydroxybutyric acid) by using purified poly(hydroxyalkanoic acid) synthase of Chromatium vinosum. Appl Microbiol Biotechnol. 1998;49:258–266. doi: 10.1007/s002530051166. [DOI] [PubMed] [Google Scholar]
- 4.Karr D B, Waters J K, Emerich D W. Analysis of poly-beta-hydroxybutyrate in Rhizobium japonicum bacteroids by ion-exclusion high-pressure liquid-chromatography UV detection. Appl Environ Microbiol. 1983;46:1339–1344. doi: 10.1128/aem.46.6.1339-1344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madison L L, Huisman G W. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev. 1999;63:21–53. doi: 10.1128/mmbr.63.1.21-53.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maehara A, Ueda S, Nakano H, Yamane T. Analyses of a polyhydroxyalkanoic acid granule-associated 16-kilodalton protein and its putative regulator in the pha locus of Paracoccus denitrificans. J Bacteriol. 1999;181:2914–2921. doi: 10.1128/jb.181.9.2914-2921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCool G J, Cannon M C. Polyhydroxyalkanoate inclusion body-associated proteins and coding region in Bacillus megaterium. J Bacteriol. 1999;181:585–592. doi: 10.1128/jb.181.2.585-592.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peoples O P, Sinskey A J. Poly-beta-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC) J Biol Chem. 1989;264:15298–15303. [PubMed] [Google Scholar]
- 9.Pieper-Fürst U, Madkour M H, Mayer F, Steinbüchel A. Identification of the region of a 14-kilodalton protein of Rhodococcus ruber that is responsible for the binding of this phasin to polyhydroxyalkanoic acid granules. J Bacteriol. 1995;177:2513–2523. doi: 10.1128/jb.177.9.2513-2523.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pieper-Fürst U, Madkour M H, Mayer F, Steinbüchel A. Purification and characterization of a 14-kilodalton protein that is bound to the surface of polyhydroxyalkanoic acid granules in Rhodococcus ruber. J Bacteriol. 1994;176:4328–4337. doi: 10.1128/jb.176.14.4328-4337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 12.Schubert P, Steinbüchel A, Schlegel H G. Cloning of the Alcaligenes eutrophus genes for synthesis of poly-beta- hydroxybutyric acid (PHB) and synthesis of PHB in Escherichia coli. J Bacteriol. 1988;170:5837–5847. doi: 10.1128/jb.170.12.5837-5847.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon S, Priefer T, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagensis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 14.Slater S, Houmiel K L, Tran M, Mitsky T A, Taylor N B, Padgette S R, Gruys K J. Multiple beta-ketothiolases mediate poly(beta-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J Bacteriol. 1998;180:1979–1987. doi: 10.1128/jb.180.8.1979-1987.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinbüchel A, Aerts K, Babel W, Föllner C, Liebergesell M, Madkour M H, Mayer F, Pieper-Fürst U, Pries A, Valentin H E, Wieczorek R. Considerations on the structure and biochemistry of bacterial polyhydroxyalkanoic acid inclusions. Can J Microbiol. 1995;41:94–105. doi: 10.1139/m95-175. [DOI] [PubMed] [Google Scholar]
- 16.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 17.Taidi B, Anderson A J, Dawes E A, Byrom D. Effect of carbon source and concentration on the molecular mass of poly(3-hydroxybutyrate) produced by Methylobacterium extorquens and Alcaligenes eutrophus. Appl Microbiol Biotechnol. 1994;40:786–790. [Google Scholar]
- 18.Wieczorek R, Pries A, Steinbüchel A, Mayer F. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J Bacteriol. 1995;177:2425–2435. doi: 10.1128/jb.177.9.2425-2435.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]