Abstract
Transmembrane protein 16F (TMEM16F) is a ubiquitously expressed Ca2+-activated phospholipid scramblase that also functions as a largely non-selective ion channel. Though recent structural studies have revealed the closed and intermediate conformations of mammalian TMEM16F (mTMEM16F), the open and conductive state remains elusive. Instead, it has been proposed that an open hydrophilic pathway may not be required for lipid scrambling. We previously identified an inner activation gate, consisting of F518, Y563, and I612, and showed that charged mutations of the inner gate residues led to constitutively active mTMEM16F scrambling. Herein, atomistic simulations show that lysine substitution of F518 and Y563 can indeed lead to spontaneous opening of the permeation pore in the Ca2+-bound state of mTMEM16F. Dilation of the pore exposes hydrophilic patches in the upper pore region, greatly increases the pore hydration level, and enables lipid scrambling. The putative open state of mTMEM16F resembles the active state of fungal scramblases and is a meta-stable state for the wild-type protein in the Ca2+-bound state. Therefore, mTMEM16F may be capable of supporting the canonical in-groove scrambling mechanism in addition to the out-of-groove one. Further analysis reveals that the in-groove phospholipid and ion transduction pathways of mTMEM16F overlap from the intracellular side up to the inner gate but diverge from each other with different exits to the extracellular side of membrane.
Significance
Transmembrane protein 16F (TMEM16F) plays important roles in regulation of membrane asymmetry and associated physiological processes. Extensive structure determination efforts have failed to capture a conductive state of the protein, hindering our understanding of TMEM16F activation and its dual functionality as both lipid scramblase and ion channel. A previous study discovered that mutations of inner activation gate residues to charged ones led to constitutively active scramblases. Leveraging this discovery, we performed atomistic simulations and identified a possible open and conductive state of TMEM16F in the Ca2+-bound state. Further analysis provides important new insights into the activation mechanism of TMEM16F and possible molecular basis of its lipid scramblase and ion channel function.
Introduction
The distribution of various lipids in the plasma membrane in eukaryotes is tightly regulated by ATP-driven flippases and flopasses, which catalyze lipid movements between two leaflets to generate and maintain asymmetric lipid distributions (1). Breakdown of lipid asymmetry is a key signaling mechanism involved in many physiological processes, including phagocytosis, blood coagulation, bone development, and viral infection (2, 3, 4). For example, externalization of signaling lipid phosphatidylserine disrupts cellular homeostasis and is essential for blood coagulation (5,6). Phospholipid scramblases are lipid channel proteins that promote passive phospholipid scrambling between two leaflets for regulated breakdown of lipid asymmetry (7,8). Transmembrane protein 16F (TMEM16) and Xkr families have been identified as the two major classes of scramblases that respond to intracellular Ca2+ concentration increase and caspase-3 cleavage, respectively (7,9, 10, 11). The TMEM16 family, also known as anoctamins or ANOs, consists of both Ca2+-activated Cl− channels (CaCCs), such as TMEM16A and -B, and Ca2+-activated phospholipid scramblases, such as TMEM16C–F, -G, -J, and -K (7,12, 13, 14, 15, 16, 17, 18, 19, 20). Among them, TMEM16F is one of the most well-characterized lipid scramblases, which, intriguingly, also functions as a largely non-selective ion channel (11,15,21,22). TMEM16F plays important roles in various physiological processes such as programmed cell death (23,24), blood coagulation (6,25), skeleton development (26), and cell fusion during entry of enveloped viruses such as HIV and Ebola (27,28). Loss-of-function mutations in TMEM16F cause Scott syndrome, a bleeding disorder associated with defective lipid scrambling in platelets (6,29,30).
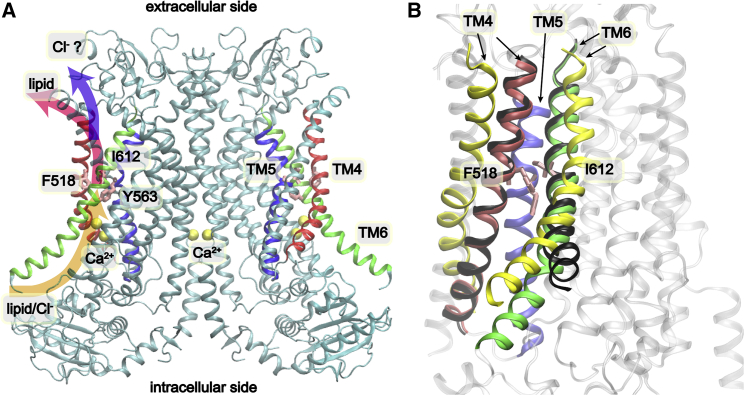
Extensive studies have been conducted to investigate the structure and function of TMEM16F since its identification in 2010 (11). Functional studies have revealed structure elements that are important for TMEM16F functions, including the Ca2+-binding site, pore-lining residues, and the dimerization interface (7,19). A key breakthrough was the recent determination of high-resolution structures of TMEM16 proteins, which provide a crucial structural basis for understanding their functions (31, 32, 33, 34, 35, 36, 37). Interestingly, these structures reveal that TMEM16 ion channels and lipid scramblases share a highly conserved structural organization. All TMEM16 proteins are double-barreled homodimers; each monomer contains 10 TM helices and harbors an independent membrane-facing conducting pore (Figs. 1 A and S1). These TM helices can be divided into two functional domains, including the pore-forming domain, TM3–8, and the dimerization and supporting domain, TM1, -2, -9, and -10. The pore-forming domain can be further divided into the regulatory (TM3–5) and Ca2+-binding activation modules (TM6–8) (38, 39, 40). The molecular mechanisms of Ca2+ activation appear conserved in the TMEM16 family proteins (32, 33, 34, 35, 36, 37). In the Ca2+-free state, TM3–7, especially TM4 and -6, pack tightly and create a narrow vestibule that is impermeable to ions and lipids. Ca2+ binding mainly induces large conformational transitions in TM6, such as straightening and vertical displacement in Aspergillus fumigatus TMEM16 (afTMEM16) (36) and helix unbending and extension in mammalian TMEM16A (mTMEM16A) (31,35). Specifically for murine TMEM16F, the TM6 helix is extended by two turns on the intracellular side (Fig. 1 B, green versus black cartoons), and the lower pore region can accommodate multiple lipid headgroups (Fig. 1 A, yellow arrow) (40). However, the narrowest point near the middle of the membrane-facing pore remains too constricted for either ions or lipid transduction (Fig. 1 A, red/blue arrows), indicative of a non-conductive conformational state. Instead, all available structures of Ca2+-bound mTMEM16 proteins resemble an intermediate non-conductive state of Ca2+-bound Nectria haematococca TMEM16 (nhTMEM16) (32).
Figure 1.
Overall structure of mTMEM16F. (A) Structure of Ca2+-bound mTMEM16F (PDB: 6QP6 (32)), where the pore-forming TM4, -5, and -6 are colored in red, blue, and green, respectively. The proposed lipid and ion conduction pathways are highlighted with arrows. Bounded Ca2+ ions are presented as yellow spheres. (B) Superposition of TM4–6 of Ca2+-bound mTMEM16F (colored in red, blue, and green, respectively), Ca2+-free mTMEM16F (black), and open nhTMEM16 (yellow). The PDB IDs used PDB: 6QP6, 6QPB (32), and 4WIS (34). All structures were aligned using the backbones of TM1–3 and TM7–10. To see this figure in color, go online.
At present, the open and conductive structures are only available for fungal scramblases nhTMEM16 and afTMEM16, two evolutionary remote homologs of mTMEM16F (34,36,41). In Ca2+-bound structures of both scramblases, TM4 and -6 move away from each other, which leads to a wider and largely hydrophilic groove at the protein-membrane interface (e.g., see Fig. 1 B). This TM movement during activation has been referred to as a “clam-shell” model and proposed to be conserved in TMEM16 family proteins (38, 39, 40). Based on structural and mutagenesis studies, a “credit card” model has been proposed as the lipid transport mechanism in TMEM16 family scramblases (20,42). Here, the membrane-facing groove is wide and hydrophilic enough that it provides a favorable pathway for the lipid headgroups to diffuse across the membrane, while the lipid tails remain anchored in the hydrophobic region of the bilayer. The credit card model has been observed directly in molecular dynamics (MD) simulations of nhTMEM16 (43,44). In addition to interactions between hydrophilic pore-lining residues and lipid headgroups, TMEM16F also induces substantial deformation and thinning of the membrane to significantly reduce the free-energy barrier for lipid diffusion, decreasing the phospholipid flip flop time constant from ∼1 h (spontaneous flip flop) to ∼2 × 10−5 s per lipid (45,46). It has been further suggested that the membrane-facing hydrophilic groove could function as both lipid and ion transduction pathways in nhTMEM16 based on simulation as well as experiment (44). Interestingly, Malvezzi et al. found that afTMEM16 was able to translocate phospholipids with polyethylene glycol headgroups (47), which are likely too large to fit in the groove. These observations suggest the existence of an “out-of-the-groove” transportation mechanism, where afTMEM16 may deform and thin the membrane around the groove such that lipids can diffuse across the membrane without strictly via the hydrophilic groove (47). It has been further shown that membrane thickness and the ability of the protein to thin the membrane can determine the scrambling activities, with and without an open hydrophilic pathway (48). The implication is that an open membrane-facing groove may not be required for scramblase activities. Curiously, recent cryoelectron microscopy (cryo-EM) structures of caspase-activated scramblases Xkr8 (49) and Xkr9 (50) do not reveal an open hydrophilic pathway, and our atomistic simulations of Xkr8 do not show significant membrane thinning (unpublished data).
We previously combined experiment and atomistic simulation and identified three hydrophobic residues in the middle of the putative lipid permeation pathway of mTMEM16F, namely F518 (TM4), Y563 (TM5), and I612 (TM6), that act as an inner activation gate to control the lipid scrambling activity (40). Bulky mutations of these residues reduce the scramblase activity, whereas replacing the inner gate residues by either hydrophilic or charged amino acids leads to gain-of-function scramblases. In particular, lysine substitution of either F518 or Y563 leads to constitutively active scramblases even in the absence of Ca2+ (40). Homology modeling suggested that charged mutations likely destabilized the hydrophobic inner gate to promote activation (40). In addition, the effect may also be partially attributed to charge repulsion between the introduced positive charges on TM4/5 and conserved K616 on TM6. Notably, a lysine substitution in mTMEM16A L543 (corresponding to the F518 in mTMEM16F) confers novel Ca2+-activated lipid scrambling activity to the CaCC (40), suggesting that the inner gate and activation mechanism may be conserved within mTMEM16 proteins. The discovery of these activation mutations provides an opportunity to leverage atomistic simulations to discover possible open state(s) and study the molecular basis of mTMEM16F activation and lipid and ion transport mechanisms.
In this work, we performed large-scale atomistic simulations of the inner gate charged mutants of mTMEM16F to study the activation mechanism and conducting properties. The results show that both constitutively active mutants (F518K and Y563K) lead to open and conductive states that resemble those of nhTMEM16. Free-energy analysis suggests that the open state of the groove is a meta-stable state even for the wild-type (WT) mTMEM16F in the Ca2+-bound state. We further analyzed the activation transition and structural properties of the activated state, as well as lipid and ion permeation pathways of mTMEM16F.
Materials and methods
Atomistic simulations
The cryo-EM structures of mouse TMEM16F in the Ca2+-bound (PDB: 6QP6) were used in all simulations reported in this work. The missing short loops in the cytosolic domain (I197-N227 and N428-Q437) were rebuilt using the ProMod3 tool with Swiss-PDB server (51). The missing N- and C-terminal segments as well as a long loop in the cytosolic domain (P150-P186) are presumably dynamic and thus not included in current simulations. Residues before and after the missing segments are capped with either an acetyl group (for N-terminus) or an N-methyl amide (for C-terminus). To minimize the effects of missing loop residues on the cytosolic domain, the backbones of the structured region of the cytosolic domain (D72-E78, T90-R116, L123-H129, W132-L148, F187-R191, P206-V220, K243-P247, L874-T875) were harmonically restrained with a force constant of 1.0 kcal/(mol×Å2) during all simulations.
As summarized in Table S1, 6 sets of atomistic simulations were performed. The first four sets involved the mTMEM16F WT, F518K, Y563K, and F518K/Y563K mutants. The initial structure of Ca2+-bound mTMEM16F was taken from PDB: 6QP6 (32). All initial mTMEM16F structures were first inserted in model lipid bilayers and then solvated with TIP3P water using the CHARMM-GUI web server (52). The cytoplasmic leaflet contains a ratio of 1:1 for palmitoyl-oleoyl phosphatidylserine (POPS):phosphatidylethanolamine (POPE) and the exoplasmic leaflet contains 100% phosphatidylcholine (POPC). All systems were neutralized, and 150 mM KCl was added. The final simulation boxes contain about 795 lipid molecules, with a total of ∼316,000 atoms and dimensions of ∼165 × 165 × 135 Å3. The CHARMM36m all-atom force field (53) and the CHARMM36 lipid force field (54) were used. All simulations were performed using CUDA-enabled versions of Gromacs 2019.4 (55,56). Electrostatic interactions were described by using the particle mesh Ewald algorithm (57) with a cutoff of 12 Å. The cutoff of van der Waals interactions was set at 12 Å with a smooth switching function starting at 10 Å. Covalent bonds to hydrogen atoms were constrained by the SHAKE algorithm (58), and the MD time step was set at 2 fs. The temperature was maintained at 298 K using the Nose-Hoover thermostat (59,60). The pressure was maintained semi-isotopically at 1 bar at membrane lateral directions using the Parrinello-Rahman barostat algorithm (61).
All systems were first minimized for 5,000 steps using the steepest descent algorithm, followed by a series of equilibration steps where the positions of heavy atoms of the protein/lipid were harmonically restrained with force constants gradually decreased from 10 to 0.1 kcal/(mol×Å2) (52). In the last equilibration step, only protein heavy atoms were harmonically restrained with a force constant of 0.1 kcal/(mol×Å2), and the system was equilibrated 10 ns under constant particle number, pressure, and temperature conditions. All production simulations were performed under constant particle number, pressure, and temperature conditions at 298 K and 1 bar.
Umbrella sampling free-energy calculations
Umbrella sampling simulations (62) were used to calculate the potentials of mean force (PMFs) of opening transitions. The distance between Cα atoms of inner gate residues F/K518 and I612 reaction coordinate was chosen as the order parameter. Umbrella sampling windows were placed at 1 Å intervals and covered 5.5 to 16.5 Å. For the F518K/Y563K mutant, the initial structures of each window were taken from sim 10. The same set of initial structures was used for umbrella sampling of the WT channel, except that reverse mutation and additional equilibration steps were performed before sampling. Specifically, systems were first minimized for 5,000 steps using the steepest descent algorithm, followed by six equilibration processes (with 2,500 steps for each equilibration process) where the positions of heavy atoms of the protein/lipid were harmonically restrained with force constants gradually decreased from 10 to 0.1 kcal/(mol×Å2). Two monomers within the dimer were restrained and sampled independently in each window, and the data were combined in subsequent analysis. The force constant of the harmonic umbrella potentials was 0.24 kcal/(mol×Å2), and the sampling time for each window was 40 ns. The final PMFs were calculated using the weighted histogram analysis method (63). Note that two PMFs were calculated for two pores within the dimer, respectively, from each set of umbrella sampling. Two sets of umbrella sampling were performed, with independently selected initial structures, to better evaluate convergence. The final free-energy profile was the average of the four individual ones (2 monomer × 2 umbrella sampling), and the error bars were estimated as the standard deviation among individual profiles.
Meta-dynamics simulation of ion permeation pathways
Well-tempered meta-dynamics simulations (64) were used to generate many ion permeation events through the pore to explore possible ion permeation pathways of the WT mTMEM16F protein. To generate an open structure of WT TMEM16F, reverse mutation was performed on a representative fully open snapshot from simulations of the mTMEM16F F518K/Y563K mutant (Table S1, sim 10). Subsequent simulations (Table S1, sims 13–15) demonstrated that the open state of WT mTMEM16F was stable (Fig. S2, right row). The initial structure for meta-dynamics was taken from sim 13. Meta-dynamics simulations were performed using plumed 2.5.3 plugin (65) (Table S1, meta 1 and 2). Systems were first minimized for 5,000 steps using the steepest descent algorithm, followed by six equilibration processes (with 2,500 steps for each equilibration process) where the positions of heavy atoms of the protein/lipid were harmonically restrained with force constants gradually decreased from 10 to 0.1 kcal/(mol×Å2). The backbones of the structured region of the cytosolic domain were harmonically restrained with a force constant of 1.0 kcal/(mol×Å2). A single Cl− ion inside the intracellular vestibule was randomly selected, and its z coordinate was used as the collective variable in meta-dynamics simulations. The height of the Gaussians was set to 0.01 kcal/mol with a deposition time τ of 2 ps and a bias factor of 15. Note that two Cl− ions were subjected to independent meta-dynamics sampling in each run, one in each of the two pores within the dimer. To prevent the selected Cl− ions from escaping the pore region, a flat-bottom harmonic potential was imposed between the intracellular and extracellular boundaries with a force constant of 10 kcal/(mol×Å2), such that multiple reversible permeation events could be generated for each ion. A permeation event is identified whenever the ion diffuses from one of the two boundaries and reaches the other. The length of each meta-dynamics simulations was 400 ns, which was enough to generate ∼10 permeation events.
Analysis
Unless stated otherwise, snapshots were extracted every 50 ps after 200 ns of all equilibrium MD trajectories for analysis. For lipid contact probability calculation, we used the stable state of trajectories from both WT and mutants, i.e., 0.5–1 μs in WT simulations and 2.5–4 μs in mutant simulations. To calculate the phospholipid head contact probability, a protein residue is considered in contact with the lipid headgroup if the minimal heavy atom distance is no larger than the standard cutoff of 4.2 Å. Molecular illustrations were prepared using VMD (66). Pore profiles were calculated using program HOLE (67). The helix tilt angle is defined as the angle between the helical principal axis and the z axis (membrane normal).
Results
Inner gate charged mutations can induce spontaneous mTMEM16F pore opening
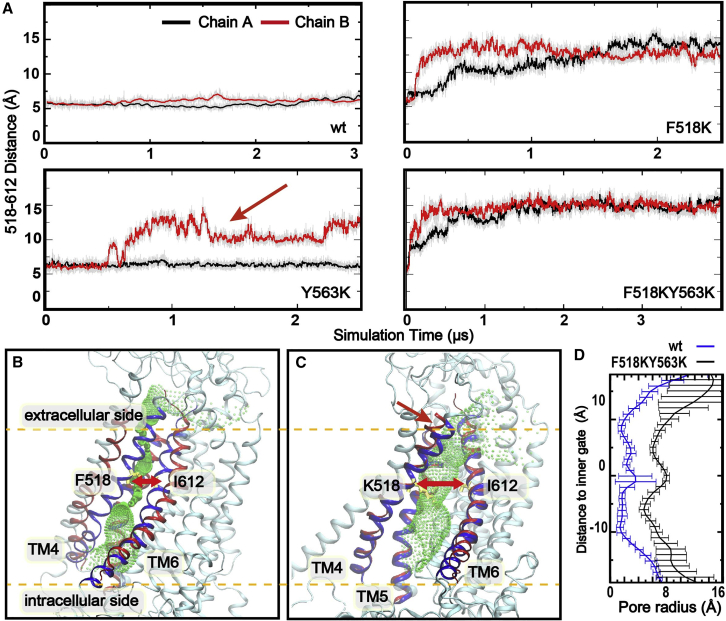
Sets of three multi-μs atomistic simulations were performed for mTMEM16F inner gate charged mutants, F518K and Y563K, together with the WT protein to evaluate if these charged mutations would promote pore-opening transitions (Table S1). We first examined the distance between Cα atoms of inner gate residues 518 (F/K, TM4) and I612 (TM6), which reflects the level of pore opening at the most constricted inner gate region during the simulations. Of note, the two membrane-facing pores within the same dimer act independently. As summarized in Figs. 2 A and S2, both F518K and Y563K single mutations consistently promoted the pore dilation, widening the TM4-TM6 separation from ∼5 to over 10 Å in 6 out of 12 pores within the two sets of 3 independent simulations (sims 4–6 and 7–9). In contrast, no spontaneous pore opening was observed in the three parallel WT simulations of the same simulation length (3 μs) (Figs. 2 A and S2, sims 1–3). This observation suggests that a single charged mutation, either F518K or Y563, is indeed able to induce spontaneous inner gate opening, consistent with experimental observations that F518K and Y563K mutants are constitutively active under physiological conditions. Nonetheless, there is substantial heterogeneity in the opening transitions sampled in theses simulations. The dilated states of the pore remain dynamic in all cases, as reflected by the substantial fluctuation in the inner gate residue 518-612 separation distance (Figs. 2 and S2). It is also clear that the more widely open states (with ∼14 Å separation, e.g., red arrow in Fig. 2 A) were only transiently sampled with a single charged mutation within the current simulation timescale.
Figure 2.
Spontaneous mTMEM16F pore opening induced by inner gate charged mutations. (A) Distance between the Cα atoms of inner gate residues 518 and 612 as a function of the simulation time (Table S1, sims 1, 4, 7, and 10). The red arrow highlights a region where the 518-612 distance shows substantial fluctuations. All traces plot 2.5 ns (50 frame) running averages for clarity, and the raw data (recorded every 50 ps) are represented as gray lines. (B and C) Representative structures of the closed (sim 1, chain A at 1 μs) and open (sim 4, chain A at 4 μs) pore conformations of mTMEM16F. TM4, -5, and -6 are colored in blue, while those of the open state of nhTMEM16 are shown as a red cartoon as a reference. The pore profiles calculated by the HOLE program (67) are illustrated as green dots. (D) Average pore radius profiles calculated by HOLE for the WT (blue trace) and F518K/Y563K mutant (black trace). The averages were calculated from the last 100 ns of WT and F518K/Y563K mutant simulations (sims 1 and 10) with one snapshot every 1 ns, and the error bars plot the standard deviations within the 100 snapshots. To see this figure in color, go online.
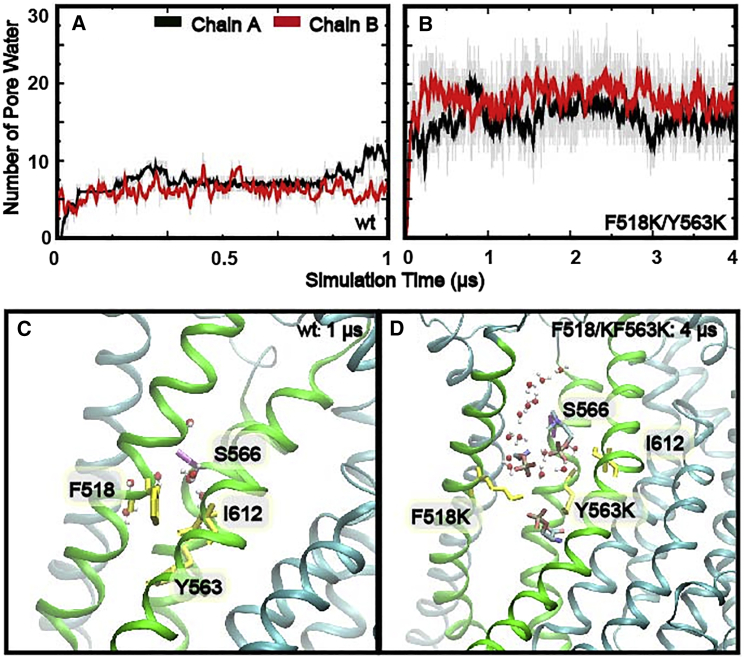
To better sample the fully open state and facilitate its characterization, we performed additional simulations with double charged mutations, F518K/Y563K (Table S1, sims 10–12), which should enhance the repulsive electrostatic force to promote more rapid opening transitions and further stabilize the fully open state. Indeed, during F518K/Y563K mutant simulations, spontaneous pore opening occurred more readily as the inner gate residue 518-612 distance increased rapidly from ∼5 to ∼15 Å within 0.5 μs and remained around 14 Å in all three independent simulations (Figs. 2 A and S2, sims 10–12; Video S1). It should also be noted that despite a slight increasing of pore dilation, the open state from the double charged mutant is similar to the most widely open states of the two single charged TMEM16F mutants (Fig. S3). Two representative conformations were extracted from the simulation trajectories to illustrate the structural differences between the closed and open states of the pore (Fig. 2 A and C). Clearly, inner gate charged mutations lead to an expansion of the entire membrane-facing pore (Fig. 2 D). In particular, expansion of the upper pore region exposes the hydrophilic groove formed by TM3–6, which connects to the lower pore region though the disengaged inner gate region to provide a continuous and largely hydrophilic membrane-facing groove (Fig. 2 C, green mesh). The dilation of the pore is accompanied by a significant increase in pore hydration (and lipid headgroup occupancy; see below), especially in the narrower upper pore region. As shown in Fig. 3, the number of water molecules within 6 Å radius around the pore-centering residue S566 increased from ∼5 to ∼20. Together with the physical expansion, the open pore seems ready to support the permeation of the large lipid headgroups across the membrane core. For example, lipid headgroups can readily penetrate into the inner poor region (Fig. 3 D).
Figure 3.
Hydration of the upper pore region. (A and B) Number of water molecules within 6 Å around S566 located at the center of the upper pore region (see C and D) as a function of simulation time. The water counts for chains A and B are shown using black and red lines, respectively. All traces plot 10 ns (10 frame) running averages for clarity, and the raw data (hydration property analyzed every 1 ns) are represented as gray lines. (C and D) Representative snapshots showing the hydration of the upper pore region in the closed and open states (WT: sim 1 chain A at 1 μs; F518K/Y563K: sim 10 chain A at 1 μs). Water molecules are represented as sticks and balls. Lipid headgroups are shown as sticks. TM4, -5, and -6 are colored in green, while the other TMs are colored in cyan. To see this figure in color, go online.
The movie was generated based on sim10. Chain A is shown in the front view by cartoons with TMs 3-6 colored in yellow, red, blue and green, respectively, and the other TMs in cyan. The phosphate atoms in lipid headgroups are represented as transparent orange spheres. The two bound Ca2+ ions are shown as two red spheres. The inner gate residues, K518, K563 and I612, are shown as yellow sticks. Note that, as the pore opens up, it becomes fully occupied with lipid headgroups.
Open conformation and activation mechanism of TMEM16 scramblases are likely conserved
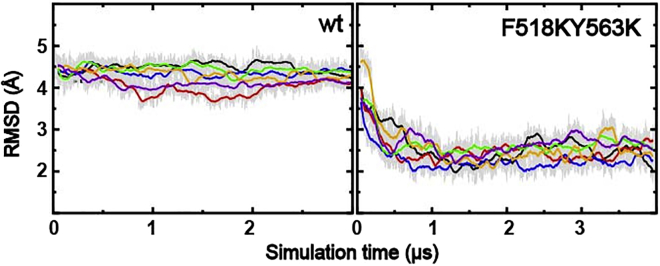
As shown in Fig. 2 C, the open state of the pore identified in our simulations of mTMEM16F double charged mutants (blue cartoon) is highly similar to the known activated state of nhTMEM16 (red cartoon) (34). During the simulations, the backbone root-mean-squared distance of TM4–6 regions between the mTMEM16F F518K/Y563K mutant and nhTMEM16 decreased from ∼4.5 to ∼2 Å as the pore undergoes opening transitions, while the root-mean-square distance remains stable above 4 Å in all three simulations of WT mTMEM16F (Fig. 4). The most notable difference between the open states of mTMEM16F and nhTMEM16 is at the upper pore region of TM4 (M507-F518) (Fig. 2 C, red arrow), likely due to two additional proline (P332 and P341) in the nhTMEM16 sequence. Nonetheless, the overwhelming similarity of the open conformations strongly suggest that the activation and ion and lipid permeation mechanisms are likely conserved among various TMEM16 scramblases.
Figure 4.
Backbone root-mean-squared distance of TM4–6 with respect to nhTMEM16 as a function of simulation time. The results were calculated for each monomer from simulations 1–3 and 10–12 (see Table S1) (6 independent traces). All traces plot 10 ns (10 frame) running averages for clarity, and the raw data (analyzed every 1 ns) are represented as gray lines. To see this figure in color, go online.
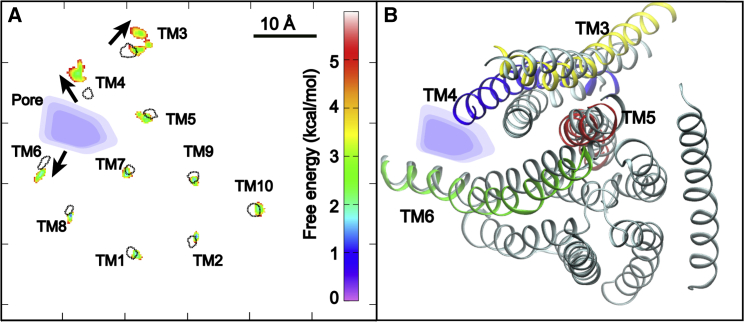
To further analyze the predicted activation transition, we calculated the distributions of centers of mass (COMs) of all 10 TMs in the closed and open states. As shown in Fig. 5, the activation mainly involves movements of TM3, -4, and -6, while the rest show minimal movements. Specifically, COMs of both TM3 and -6 shift ∼2 Å away from each other. TM4 shows the largest movement, shifting ∼4 Å outward and away from the rest of TMs (Fig. 5 A). Importantly, TM helices maintained their origin orientations during activation, and changes in the tilting angles before and after activation were within 2° for all the TMs (Table S2). These movements not only lead to larger separation between TM4 and -6 and thus dilation of the pore but also slightly increase the separation of TM3 and -4. Though no inner gate residues reside on TM3, its movement is apparently required for TM4 movement and thus mTMEM16F activation. We note that TM3 is part of a specific phosphatidylinositol-(4,5)-bisphosphate (PIP2) binding site near the cytosolic membrane interface that was previously shown to play a key role in allosteric regulation of TMEM16A CaCCs (39). It is known that PIP2 also regulates TMEM16F channel activities (68,69). Notably, TM3 is the only helix that displayed substantial heterogeneity in our simulations (Fig. 4 A) and appears to sample multiple conformations in the activated state of mTMEM16F. Such elevated dynamics could be consistent with a potential role of TM3 in allosteric regulation of mTMEM16F activation by PIP2 or other cofactors.
Figure 5.
TM movements of mTMEM16F activation induced by F518K/Y563K inner gate mutations. (A) Distributions of centers of mass of all 10 TMs along membrane lateral directions (x, y) in the closed and activated states. The distributions for the activated state (sims 10–12, 1–2.5 μs) are plotted as colored free-energy maps (in kcal/mol), while those for the closed state (sims 1–3, 2.5–3 μs) are shown as dashed contours (at level: 5 kcal/mol). The approximate pore location is also indicated by blue shadows. The arrows highlight three TMs with the largest movements. (B) Representative snapshots showing the orientation of pore-forming TMs in the closed (sim 1, 1 μs, gray cartons) and open states (sim 10, 4 μs). TM3–6 are colored in yellow, blue, red, and green, respectively, in the open state. To see this figure in color, go online.
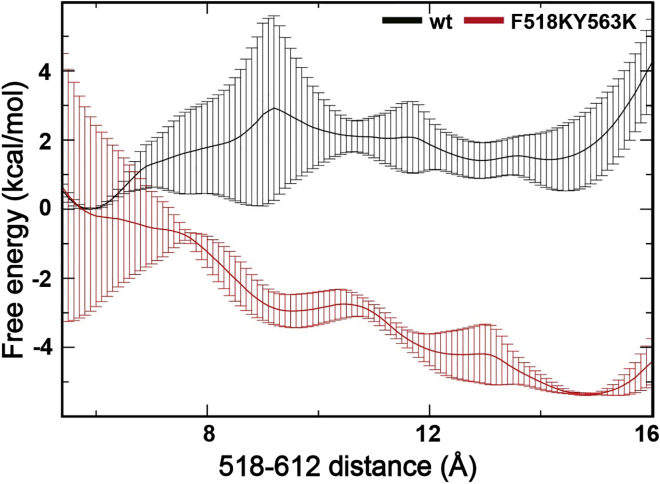
To evaluate if the predicted open state is accessible for the WT protein, we calculated the free-energy profiles of the open-close transition of the pore in the Ca2+-bound state, using the distance between Cα atoms of inner gate residue F518 and I612 as the order parameter. The results show that the open state (∼15 Å; Fig. 6, black trace) is ∼1.5 kcal less stable than the closed state (∼5.5 Å) in the WT protein, suggesting an open-state population of only ∼8% at room temperature. Introduction of charges at the inner gate in the F518K/Y563K mutant significantly stabilizes the open state, which is now ∼−5 kcal/mol more stable than the closed state, increasing the open-state population to over 99.9%. We caution that accurate calculation of free-energy profiles of large-scale conformational transitions of a large and complex protein like mTMEM16F is extremely challenging. Large uncertainties persist even with an aggregated 160 ns sampling per umbrella sampling window, reflected in substantial error bars in Fig. 6. Nonetheless, the free-energy profiles further confirm that the open states of the pore are likely similar between the WT and mutant channels, both having a characteristic separation between inner gate residues F/K518 and I612 at ∼15 Å. That is, the inner gate charged mutations likely increase the stability of the open state without significantly perturbing its conformational properties.
Figure 6.
Averaged free-energy profiles of pore opening of WT and F518K/Y563K mutant mTMEM16F. The distance between Cα atoms of inner gate residues F/K518 and I612 is chosen as the order parameter. The profiles were calculated using umbrella sampling (see materials and methods), and individual profiles from both monomers from two independent umbrella sampling calculations are shown in Fig. S4. To see this figure in color, go online.
It is curious that existing structures show that the pore of mTMEM16F remains closed even in the presence of Ca2+ and other activators such as PIP2 (33). This may be due to the open state of mTMEM16F being less stable than the closed state and the pore only opening transiently. A relatively low population of the open state (∼8%), coupled with the apparent dynamic nature of the open state (reflected as the broad meta-stable open state basin; Fig. 6), could explain why cryo-EM structural studies have failed to resolve the open state. We further tested if the WT channel would undergo spontaneous closure when initiated from reverse mutation of the fully open conformation of the double charge mutant (Table S1, sims 13–15). The results suggest that the open state persists after 1 μs simulations (Fig. S2), suggesting it is at least a meta-stable state. We note that spontaneous closure of the pore will likely be very slow and beyond the μs timescale due to the need for expelling the lipid headgroups from the dilated groove. For example, we only observed a single lipid diffusion event out of ∼30 μs of simulations (Table S1, sims 4–12), which lasted essentially the full 4 μs (see below).
Pathway and mechanism of lipid translocation
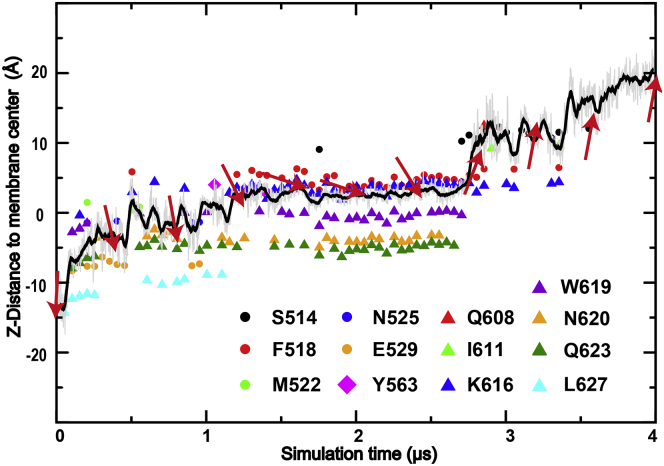
The predicted open state of mTMEM16F is apparently conductive to phospholipids. As shown in Fig. S5, the dilated pore is fully accessible to the headgroup, and mTMEM16F induces substantial membrane thinning near the pore region as also observed previously (43). Interestingly, a full translocation of a POPE molecule from the inner leaflet to the extracellular side was observed during the simulation of mTMEM16F F518K/Y563K mutant (sim 12) (Video S2). In Fig. 7, the z-position of the COM of the POPE headgroup is depicted to visualize how the phospholipid headgroup migrated across the membrane, together with the lipid orientation and headgroup-contacting residues. Consistent with the pore profile shown in Fig. 2 D, the main bottleneck for lipid permeation is around the inner gate, where the dilated pore is the narrowest. The permeating headgroup remained in this region for over 2 μs before it eventually managed to flip the orientation and rapidly diffused toward the extracellular side around 2.8 μs. Such an extended plateau was probably also required to allow the hydrophobic alkyl tails to diffuse from the lower to upper leaflet within the hydrophobic membrane interior. The observation here is fully consistent with the credit card model, as also observed in previous studies of the nhTMEM16 scramblase (43,44). It is worth noting that the open state in both mutated and WT channels (sims 10–12 and 13–15) led to significant disruption of lipid tails. The resulting mixing of lipid tails from upper and lower leaflets should facilitate lipid scrambling, similar to what was observed in a previous simulation of nhTMEM16 (70). However, in nhTMEM16, lipid tails have been observed to penetrate into the extracellular side of the translocation groove, whereas only headgroups were observed in this region in the current simulations of mTMEM16F, apparently due to the hydrophilic nature of the extracellular groove.
Figure 7.
A spontaneous phospholipid translocation event observed in the simulation of mTMEM16F F518K/Y563K mutant. The z-distance of the lipid headgroup center of mass to the membrane center is shown as the black line as a function of simulation time. The z-position was analyzed every 1 ns (gray trace), and 20 ns running average is plotted. The phospholipid orientations at various time points are illustrated as red arrows, with the headgroup being the arrow head and the tail as the arrow tail. The lipid headgroup contacting residues along the trajectory are shown by different colors and shapes drawn at corresponding z-positions. To see this figure in color, go online.
Chain A is shown in the front view. TMs, phosphate atoms and inner gate residues are shown in sticks and colored according to atom types. The phosphate atoms of the passing lipid are shown with larger orange spheres, while the tail is shown in stick.
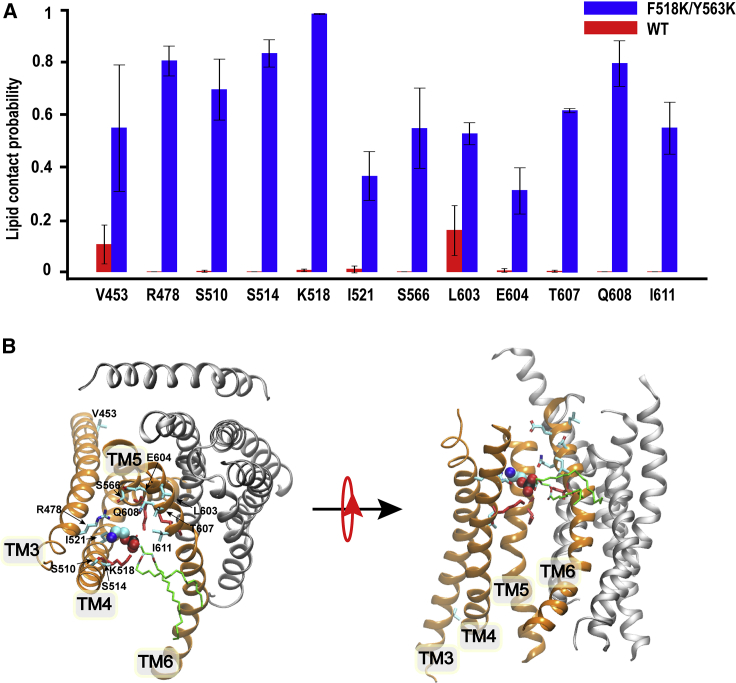
To further characterize the lipid transduction pathway and scrambling mechanism, we analyzed pore-lining residues that form close contacts with phospholipid headgroups in both the closed and open states of mTMEM16F by calculating contact probabilities from all trajectories (see materials and methods). An increased contact probability for a given phospholipid contacting site may indicate its role as a potential anchoring point or a waystation for phospholipid translocation. Fig. 8 A summarizes all 12 pore-lining residues, namely V453, R478, S510, S514, K518, I521, S566, L603, E604, T607, Q608, and I611, whose phospholipid head contact probabilities are >0.3 and at least three times larger in F518K/Y563K mutant simulations than in WT simulations. It is worth noting that 9 out of these 12 residues are charged and polar ones and that all but one (V453) locate in the upper pore region above the inner gate (Fig. 8 B). Therefore, a major consequence of the pore dilation (as induced by inner gate charged mutations here) is the exposure of the upper pore hydrophilic patches, which increases the pore hydration level (Fig. 3) and, importantly, provides favorable interactions with lipid headgroups to lower the free-energy barrier for lipid transduction. Note that even though the lower pore region also undergoes substantial expansion during activation (Fig. 2), it is well accessible to phospholipid headgroups even before full activation, which was also observed in our previous simulations (40).
Figure 8.
Lipid headgroup contact probabilities of pore-lining residues. (A) Lipid headgroup contact probabilities of key residues, selected as those with probability >0.3 and increased at least 3-fold in the activated state. Error bars show the standard deviations among independent simulations. (B) The top and side views of a monomer of mTMEM16F scramblase with a POPE interacting with upper pore region (sim 12). The lipid headgroups are presented as green spheres and the tails as green sticks. Key headgroup contacting residues (see A) and the lipid headgroup are shown with C, N, O, and P atoms colored in cyan, blue, red, and black, respectively. The lipid tail is shown by green sticks. The inner gate residues are shown in red sticks. To see this figure in color, go online.
Ion and lipid transduction pathways only partially overlap
As discussed above, an intriguing property of mTMEM16F is that it can moonlight as a non-selective CaCC (11,21,22,71). An important question is whether ions and lipids share the same pathway through mTMEM16F. As shown in Fig. 2 D, the minimal average pore radius of the WT mTMEM16F along the pore is nearly <2 Å, too narrow to accommodate hydrated ions (which require a pore radius >5 Å). The ion impermeability of the WT protein is also consistent with a recent simulation study showing that the pore region remained dehydrated and occluded to ions (72). The open state of the pore, however, has a minimum radius of ∼6 Å and is well hydrated (Fig. 3), clearly capable of supporting the passage of fully hydrated ions. The large opening of the activated mTMEM16F pore is also apparently consistent with the non-selective nature of its channel function.
To further probe ion permeation pathway of the WT channel and its relationship with phospholipid translocation, we performed meta-dynamics simulations of the WT protein in the activated state (Table S1, meta 1 and 2). The conformation of activated WT mTMEM16F was generated using reverse mutation from the open state derived from simulations of the mTMEM16F F518K/Y563K mutant (see materials and methods). During meta-dynamics simulations, bias potentials were only accumulated along the z axis. The selected Cl− ion was allowed to freely explore the membrane lateral directions and follow various possible pathways across the membrane (see materials and methods). A total 19 independent Cl− passing events were observed during the two meta-dynamics runs, including 9 permeation events from the extracellular side to the intracellular side and 10 events in the opposite direction (e.g., see Video S3). As summarized in Fig. 9, Cl− permeation pathways that are highly consistent with the pore profile were predicted by HOLE (Fig. 2). In addition, comparison of the distribution of Cl− with the lipid headgroup density map (Fig 9, orange mesh) suggests that the ion and lipid headgroup permeation pathways overlap on the intracellular side and both go through the dilated inner gate region but diverge in the upper pore region on the extracellular side (Fig. 9 A and B). Specifically, while the lipid headgroups apparently exit toward membrane above the inner gate, as expected in the credit card model, ions exit through the extracellular vestibule walled by TM4–6 (the region with blue spheres but no orange meshes in Fig. 9 A and B). A closer inspection revealed several polar and charged residues, including S593, E594, E595, D597, and K574, in the extracellular vestibule that provide a highly hydrophilic and well-hydrated environment, allowing it to effectively serves as a hub for Cl− before it enters the inner gate or exits to bulk solvent region (Fig. 9 C). We further note that this ion permeation pathway is highly similar to the one observed for mTMEM16A CaCCs when activated by both Ca2+ and PIP2 binding (39).
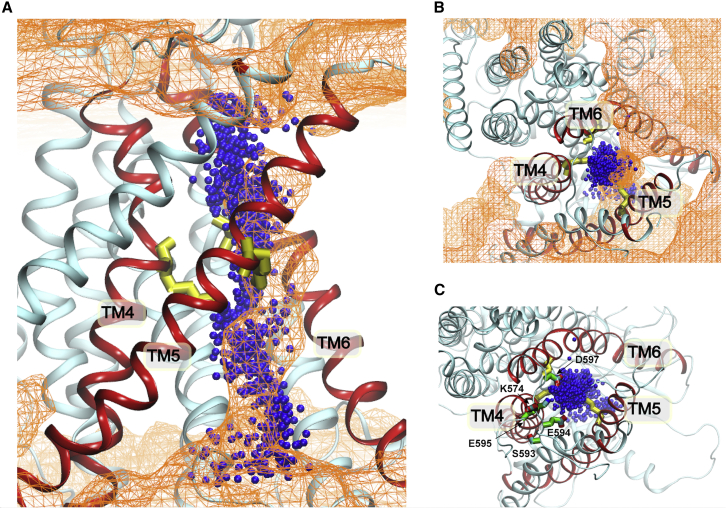
Figure 9.
Phospholipid and ion permeation pathways of mTMEM16F. (A and B) The side (A) and top (B) views of ion positions (blue beads) as they permeated through the open pore from all 19 transition events sampled by meta-dynamics simulations (Table S1, sim meta 1 and 2). The orange mesh plots the density of the phosphate group derived from simulations of F518K/Y563K mutant scramblase with an isovalue of 0.001 atom/Å3. Inner gate residues are shown as yellow sticks. (C) Several polar and charged residues (S593, E594, E595, D597, and K574) in the outlet of ion permeation pathway (shown as sticks with C, O, and N colored in green, red, and blue). To see this figure in color, go online.
Chain A is shown in the front view and elements, such as TMs, phosphate atoms and inner gate residues, are presented with the same scheme as in Movie S1. The passing chloride ion is shown as a green sphere along with its hydration shell. Note that the ion resides in the intracellular and extracellular vestibules for extended periods before and after crossing the hydrophobic inner gate region (the major free energy barrier).
Discussions
Recent structural studies of mTMEM16F have provided a crucial basis for understanding its dual functionality as both scramblases and ion channels and revealed key features of Ca2+-dependent activation (31, 32, 33, 34, 35, 36, 37). Yet, these structures do not include an open and conductive conformation, preventing one from a deeper understanding of the molecular mechanisms of mTMEM16F activation and ions and lipids transduction. Leveraging the previous discovery of constitutively active inner gate charged mutations, namely F518K and Y563K, we performed multi-μs atomistic simulations and were able to reproducibly observe spontaneous pore-opening transitions of mTMEM16F, providing key new insights into its possible activation. Free-energy analysis suggests that charge mutations mainly stabilize the open state, but do not appear to significantly affect the degree of pore opening, as measured by the distance between Cα atoms of inner gate residues F/K518 and I612 (Fig. 6). The predicted open state is only a meta-stable state for WT mTMEM16F, with an estimated occupancy of ∼8%. The low occupancy together with the dynamic nature of the open state could explain why existing structural studies failed to resolve an open state of mTMEM16F even in the Ca2+-bound state.
During the opening transition, TM4 and -6 moved away from one and another, increasing the minimal pore radius to ∼6 Å. Importantly, the fully dilated pore of mTMEM16F predicted by the simulations resembles the open state of the fungi homologs nhTMEM16, suggesting that the activation mechanism is likely conserved among TMEM16 family proteins. While mTMEM16F activation mainly involves TM4 and -6 movements, our simulations also reveal a significant shift of TM3 to accommodate the pore opening. Importantly, TM3 remains dynamic in the activate state. Our previous studies of TMEM16A have revealed that binding of PIP2 to the cytosolic side of TM3–5 allosterically regulates the pore open-close equilibrium in CaCC channel run down (38). Atomistic simulations further revealed that PIP2 binding induces movement of both TM3 and -4 and leads to a similar pore-opening process as observed here (39). The dynamics of TM3 in the activated state of mTMEM16F therefore suggest that TM3 may also be involved in allosteric regulation of TMEM16F.
The predicted open state of mTMEM16F pore is well hydrated and capable of supporting both ion and lipid transduction. Analysis of lipid headgroup contacting probabilities of pore-lining residues revealed that dilation of the pore mainly increased the accessibility of upper pore hydrophilic patches to facilitate lipid translocation, supporting the credit card model (20,42). This mechanism is confirmed by a spontaneous lipid translocation event observed during one of the simulations (sim 12). We note that only small headgroup lipids (POPC, POPS, and POPE) were used in the current study, and the current study does not exclude the out-of-groove model. Additional simulations will be necessary to evaluate if lipids with larger headgroups may follow the out-of-groove pathways (47). Nonetheless, given the low occupancy (∼8%) of the predicted open state for WT TMEM16F in the Ca2+-bound state, the physiological importance and relevance of the predicted in-groove pathway need to be further evaluated, especially considering structural and functional evidence supporting a critical role of membrane thinning in TMEM16 scramblases (48). It is possible that both in-groove and out-of-groove mechanisms co-exit and complement one and another. For example, Feng et al. suggested that the large distortion of membrane could allow lipid to bypass a closed pore and directly migrate from one leaflet to another near the protein surface (33). Interestingly, some residues found important in the out-of-groove model are also involved in the credit card model, e.g., E604 and R478 (33). Falzone et al. showed that increasing membrane thickness could inhibit scrambling despite an open membrane-facing hydrophilic pathway in afTMEM16 (48). On the other hand, discoveries of small-molecule blockers of lipid transport emphasize the involvement of a well-defined pore where the blockers could bind rather than a relatively flat protein surface that may not support strong and specific small molecular binding (73, 74, 75).
Further meta-dynamic simulations were performed to generate a large number of ion permeation events to probe possible ion pathways. The results show that ion and phospholipid permeation of mTMEM16F employ similar physical pathways on the intracellular side but follow diverged to exit to the extracellular side (Fig. 9). The extracellular vestibule walled by TM4–6 is lined with several polar and charged residues and is well hydrated, which seems to be the preferred exit route for ions once they cross over the inner gate region (Fig. 9 C). Interestingly, some blockers could inhibit both Cl− channel and scramblase activity of mTMEM16F, while other blockers show different inhibition effects toward ion and lipid translocation (73, 74, 75). These experimental observations support that ion and lipid pathways do not fully overlap. This also raises the question of whether ions might co-transport with lipid headgroups (15,44). In the meta-dynamics simulations, even though the groove was fully occupied by lipid headgroups, only Cl− permeation events were observed without any co-transportation of lipids. That is, the presence of lipid headgroups may facilitate the ion conduction by increasing hydrophilicity of the pore region, but the conduction of ions and lipids are not required to be coupled.
It has also been proposed that the ion conductive state in mTMEM16 scramblase is analogous to those of TMEM16 CaCCs such as the mTMEM16A chloride channel (7). However, mTMEM16F exhibits distinct ion conductance properties, such as the I-V curve of mTMEM16A being linear while that of mTMEM16F is outwardly rectifying (76,77). Also, unlike mTMEM16 CaCCs, mTMEM16F has been reported to transport cations such as Na+, K+, and Ca2+ (77,78). Despite many similarities in sequence, structure, and activation mechanism, there are differences in details of conformational changes involved in activation of mTMEM16F and mTMEM16A. Previous simulations suggested that PIP2-induced opening of the mTMEM16A pore mainly involves a lever-like movement of TM4, followed by dilation of the upper pore region and conformational changes in other TMs (39). The putative open pore of mTMEM16F found in our double charged mutant is wider than that of mTMEM16A, which is capable of accommodating larger substrates such as lipid headgroups and is less selective on substrate types. This is supported by the previous observation that a single inner gate mutation (L547K) activates novel lipid scramblase activity in mTMEM16A, which could be due to it bringing further dilation on the upper pore region (40). However, additional computational and experimental studies are needed to further resolve the molecular determinant of similarities and differences in activities among different TMEM16 family proteins. We also caution that the activation mechanism and open-state conformations revealed that using the current gain-of-function mutations may not fully reflect those of the WT protein.
Author contributions
Z.J. and J.C., conception and design of the study; Z.J. and J.H., performing the simulation and analysis; Z.J., J.H., and J.C., analysis and interpretation of data, drafting and revising the manuscript.
Acknowledgments
The authors thank Dr. Huanghe Yang for initial discussions that led to the conception of this study. All simulations were performed on the pikes GPU cluster housed in the Massachusetts Green High-Performance Computing Cluster (MGHPCC). This work was supported by National Institutes of Health grant R35 GM144045 (J.C.).
Declaration of interests
The authors declare no competing interests.
Editor: Philip Biggin.
Footnotes
Zhiguang Jia and Jian Huang contributed equally to this work.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.08.011.
Supporting material
References
- 1.Balasubramanian K., Schroit A.J. Aminophospholipid asymmetry: a matter of life and death. Annu. Rev. Physiol. 2003;65:701–734. doi: 10.1146/annurev.physiol.65.092101.142459. [DOI] [PubMed] [Google Scholar]
- 2.Pittoni V., Valesini G. The clearance of apoptotic cells: implications for autoimmunity. Autoimmun. Rev. 2002;1:154–161. doi: 10.1016/s1568-9972(02)00032-0. [DOI] [PubMed] [Google Scholar]
- 3.Clarke R.J., Hossain K.R., Cao K. Physiological roles of transverse lipid asymmetry of animal membranes. Biochim. Biophys. Acta Biomembr. 2020;1862:183382. doi: 10.1016/j.bbamem.2020.183382. [DOI] [PubMed] [Google Scholar]
- 4.Bevers E.M., Williamson P.L. Getting to the outer leaflet: physiology of phosphatidylserine exposure at the plasma membrane. Physiol. Rev. 2016;96:605–645. doi: 10.1152/physrev.00020.2015. [DOI] [PubMed] [Google Scholar]
- 5.Sivagnanam U., Palanirajan S.K., Gummadi S.N. The role of human phospholipid scramblases in apoptosis: an overview. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:2261–2271. doi: 10.1016/j.bbamcr.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Yang H., Kim A., et al. Jan L.Y. TMEM16F forms a Ca2+-activated cation channel required for lipid scrambling in platelets during blood coagulation. Cell. 2012;151:111–122. doi: 10.1016/j.cell.2012.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falzone M.E., Malvezzi M., et al. Accardi A. Known structures and unknown mechanisms of TMEM16 scramblases and channels. J. Gen. Physiol. 2018;150:933–947. doi: 10.1085/jgp.201711957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bevers E.M., Williamson P.L. Phospholipid scramblase: an update. FEBS Lett. 2010;584:2724–2730. doi: 10.1016/j.febslet.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 9.Nagata S., Suzuki J., et al. Fujii T. Exposure of phosphatidylserine on the cell surface. Cell Death Differ. 2016;23:952–961. doi: 10.1038/cdd.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki J., Denning D.P., et al. Nagata S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science. 2013;341:403–406. doi: 10.1126/science.1236758. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki J., Umeda M., et al. Nagata S. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 2010;468:834–838. doi: 10.1038/nature09583. [DOI] [PubMed] [Google Scholar]
- 12.Tian Y., Schreiber R., Kunzelmann K. Anoctamins are a family of Ca2+-activated Cl− channels. J. Cell Sci. 2012;125:4991–4998. doi: 10.1242/jcs.109553. [DOI] [PubMed] [Google Scholar]
- 13.Whitlock J.M., Hartzell H.C. Anoctamins/TMEM16 proteins: chloride channels flirting with lipids and extracellular vesicles. Annu. Rev. Physiol. 2017;79:119–143. doi: 10.1146/annurev-physiol-022516-034031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williamson P. Phospholipid scramblases. Lipid Insights. 2015;8:41–44. doi: 10.4137/LPI.S31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitlock J.M., Hartzell H.C. A Pore Idea: the ion conduction pathway of TMEM16/ANO proteins is composed partly of lipid. Pflügers Archiv. 2016;468:455–473. doi: 10.1007/s00424-015-1777-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi S., Pang C., et al. An H. Recent progress in structural studies on TMEM16A channel. Comput. Struct. Biotechnol. J. 2020;18:714–722. doi: 10.1016/j.csbj.2020.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pedemonte N., Galietta L.J.V. Structure and function of TMEM16 proteins (Anoctamins) Physiol. Rev. 2014;94:419–459. doi: 10.1152/physrev.00039.2011. [DOI] [PubMed] [Google Scholar]
- 18.Galietta L.J.V. The TMEM16 protein family: a new class of chloride channels? Biophys. J. 2009;97:3047–3053. doi: 10.1016/j.bpj.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Picollo A., Malvezzi M., Accardi A. TMEM16 proteins: unknown structure and confusing functions. J. Mol. Biol. 2015;427:94–105. doi: 10.1016/j.jmb.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalienkova V., Clerico Mosina V., Paulino C. The groovy TMEM16 family: molecular mechanisms of lipid scrambling and ion conduction. J. Mol. Biol. 2021;433:166941. doi: 10.1016/j.jmb.2021.166941. [DOI] [PubMed] [Google Scholar]
- 21.Malvezzi M., Chalat M., et al. Accardi A. Ca2+-dependent phospholipid scrambling by a reconstituted TMEM16 ion channel. Nat. Commun. 2013;4:2367–2369. doi: 10.1038/ncomms3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scudieri P., Caci E., et al. Galietta L.J.V. Ion channel and lipid scramblase activity associated with expression of TMEM16F/ANO6 isoforms. J. Physiol. 2015;593:3829–3848. doi: 10.1113/JP270691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ousingsawat J., Wanitchakool P., et al. K. J. C. d. Kunzelmann, and disease Contribution of TMEM16F to pyroptotic cell death. Cell Death Dis. 2018;9:300–311. doi: 10.1038/s41419-018-0373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Kruchten R., Mattheij N.J.A., et al. Bevers E.M. Both TMEM16F-dependent and TMEM16F-independent pathways contribute to phosphatidylserine exposure in platelet apoptosis and platelet activation. Blood. 2013;121:1850–1857. doi: 10.1182/blood-2012-09-454314. [DOI] [PubMed] [Google Scholar]
- 25.Fujii T., Sakata A., et al. Nagata S. TMEM16F is required for phosphatidylserine exposure and microparticle release in activated mouse platelets. Proc. Natl. Acad. Sci. USA. 2015;112:12800–12805. doi: 10.1073/pnas.1516594112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehlen H.W.A., Chinenkova M., et al. Vortkamp A. Inactivation of anoctamin-6/Tmem16f, a regulator of phosphatidylserine scrambling in osteoblasts, leads to decreased mineral deposition in skeletal tissues. J. Bone Miner. Res. 2013;28:246–259. doi: 10.1002/jbmr.1751. [DOI] [PubMed] [Google Scholar]
- 27.Zaitseva E., Zaitsev E., et al. Chernomordik L.V. Fusion stage of HIV-1 entry depends on virus-induced cell surface exposure of phosphatidylserine. Cell Host Microbe. 2017;22:99–110.e7. doi: 10.1016/j.chom.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Younan P., Iampietro M., et al. Bukreyev A. Role of transmembrane protein 16F in the incorporation of phosphatidylserine into budding Ebola virus virions. J. Infect. Dis. 2018;218:S335–S345. doi: 10.1093/infdis/jiy485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zwaal R.F.A., Comfurius P., Bevers E.M. Scott syndrome, a bleeding disorder caused by defective scrambling of membrane phospholipids. Biochim. Biophys. Acta. 2004;1636:119–128. doi: 10.1016/j.bbalip.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Castoldi E., Collins P.W., et al. Bevers E.M. Compound heterozygosity for 2 novel TMEM16F mutations in a patient with Scott syndrome. Blood. 2011;117:4399–4400. doi: 10.1182/blood-2011-01-332502. [DOI] [PubMed] [Google Scholar]
- 31.Paulino C., Kalienkova V., et al. Dutzler R. Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM. Nature. 2017;552:421–425. doi: 10.1038/nature24652. [DOI] [PubMed] [Google Scholar]
- 32.Alvadia C., Lim N.K., et al. Paulino C. Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F. Elife. 2019;8:e44365. doi: 10.7554/eLife.44365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng S., Dang S., et al. Cheng Y. Cryo-EM studies of TMEM16F calcium-activated ion channel suggest features important for lipid scrambling. Cell Rep. 2019;28:1385–1579.e564. doi: 10.1016/j.celrep.2019.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunner J.D., Lim N.K., et al. Dutzler R. X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature. 2014;516:207–212. doi: 10.1038/nature13984. [DOI] [PubMed] [Google Scholar]
- 35.Dang S., Feng S., et al. Jan L.Y. Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature. 2017;552:426–429. doi: 10.1038/nature25024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falzone M.E., Rheinberger J., et al. Accardi A. Structural basis of Ca2+-dependent activation and lipid transport by a TMEM16 scramblase. Elife. 2019;8:e43229. doi: 10.7554/eLife.43229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bushell S.R., Pike A.C.W., et al. Carpenter E.P. The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K. Nat. Commun. 2019;10:3956. doi: 10.1038/s41467-019-11753-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le S.C., Jia Z., et al. Yang H. Molecular basis of PIP2-dependent regulation of the Ca(2+)-activated chloride channel TMEM16A. Nat. Commun. 2019;10:3769. doi: 10.1038/s41467-019-11784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia Z., Chen J. Specific PIP2 binding promotes calcium activation of TMEM16A chloride channels. Commun. Biol. 2021;4:259. doi: 10.1038/s42003-021-01782-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le T., Jia Z., et al. Yang H. An inner activation gate controls TMEM16F phospholipid scrambling. Nat. Commun. 2019;10:1846. doi: 10.1038/s41467-019-09778-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalienkova V., Clerico Mosina V., et al. Paulino C. Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM. Elife. 2019;8:e44364. doi: 10.7554/eLife.44364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi T., Menon A.K. Transbilayer lipid asymmetry. Curr. Biol. 2018;28:R386–R391. doi: 10.1016/j.cub.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 43.Bethel N.P., Grabe M. Atomistic insight into lipid translocation by a TMEM16 scramblase. Proc. Natl. Acad. Sci. USA. 2016;113:14049–14054. doi: 10.1073/pnas.1607574113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang T., Yu K., et al. Tajkhorshid E. Lipids and ions traverse the membrane by the same physical pathway in the nhTMEM16 scramblase. Elife. 2017;6:e28671. doi: 10.7554/eLife.28671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe R., Sakuragi T., et al. Nagata S. Single-molecule analysis of phospholipid scrambling by TMEM16F. Proc. Natl. Acad. Sci. USA. 2018;115:3066–3071. doi: 10.1073/pnas.1717956115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allhusen J.S., Conboy J.C. The ins and outs of lipid flip-flop. Acc. Chem. Res. 2017;50:58–65. doi: 10.1021/acs.accounts.6b00435. [DOI] [PubMed] [Google Scholar]
- 47.Malvezzi M., Andra K.K., et al. Accardi A. Out-of-the-groove transport of lipids by TMEM16 and GPCR scramblases. Proc. Natl. Acad. Sci. USA. 2018;115:E7033–E7042. doi: 10.1073/pnas.1806721115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Falzone M.E., Feng Z., et al. Accardi A. TMEM16 scramblases thin the membrane to enable lipid scrambling. Nat. Commun. 2022;13:2604. doi: 10.1038/s41467-022-30300-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakuragi T., Kanai R., et al. Nagata S. The tertiary structure of the human Xkr8–Basigin complex that scrambles phospholipids at plasma membranes. Nat. Struct. Mol. Biol. 2021;28:825–834. doi: 10.1038/s41594-021-00665-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Straub M.S., Alvadia C., et al. Dutzler R. Cryo-EM structures of the caspase-activated protein XKR9 involved in apoptotic lipid scrambling. Elife. 2021;10:e69800. doi: 10.7554/eLife.69800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwede T., Kopp J., et al. Peitsch M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J., Cheng X., et al. Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theor. Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang J., Rauscher S., et al. MacKerell A.D., Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klauda J.B., Venable R.M., et al. Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hess B., Kutzner C., et al. Lindahl E. Gromacs 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theor. Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 56.Abraham M.J., Murtola T., et al. Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 57.Darden T., York D., Pedersen L. Particle mesh Ewald: an N-log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 58.Ryckaert J.-P., Ciccotti G., Berendsen H.J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 59.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 60.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 61.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 62.Torrie G.M., Valleau J.P. Non-physical sampling distributions in Monte-carlo free-energy estimation - umbrella sampling. J. Comput. Phys. 1977;23:187–199. [Google Scholar]
- 63.Kumar S., Rosenberg J.M., et al. Kollman P.A. THE weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 64.Barducci A., Bussi G., Parrinello M. Well-tempered metadynamics: a smoothly converging and tunable free-energy method. Phys. Rev. Lett. 2008;100:020603. doi: 10.1103/PhysRevLett.100.020603. [DOI] [PubMed] [Google Scholar]
- 65.Tribello G.A., Bonomi M., et al. Bussi G. Plumed 2: new feathers for an old bird. Comput. Phys. Commun. 2014;185:604–613. [Google Scholar]
- 66.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 67.Smart O.S., Neduvelil J.G., et al. Sansom M.S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 68.Ye W., Han T.W., et al. Jan L.Y. Phosphatidylinositol-(4, 5)-bisphosphate regulates calcium gating of small-conductance cation channel TMEM16F. Proc. Natl. Acad. Sci. USA. 2018;115:E1667–E1674. doi: 10.1073/pnas.1718728115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aoun J., Hayashi M., et al. Hoque K.M. Anoctamin 6 contributes to Cl- secretion in accessory cholera enterotoxin (Ace)-stimulated diarrhea: an essential role for phosphatidylinositol 4, 5-bisphosphate (PIP2) signaling in cholera. J. Biol. Chem. 2016;291:26816–26836. doi: 10.1074/jbc.M116.719823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee B.-C., Khelashvili G., et al. Accardi A. Gating mechanism of the extracellular entry to the lipid pathway in a TMEM16 scramblase. Nat. Commun. 2018;9:3251. doi: 10.1038/s41467-018-05724-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kunzelmann K., Nilius B., et al. Heemskerk J.W.M. Molecular functions of anoctamin 6 (TMEM16F): a chloride channel, cation channel, or phospholipid scramblase? Pflügers Archiv. 2014;466:407–414. doi: 10.1007/s00424-013-1305-1. [DOI] [PubMed] [Google Scholar]
- 72.Kostritskii A.Y., Machtens J.-P. Molecular mechanisms of ion conduction and ion selectivity in TMEM16 lipid scramblases. Nat. Commun. 2021;12:2826. doi: 10.1038/s41467-021-22724-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suzuki T., Suzuki J., Nagata S. Functional swapping between transmembrane proteins TMEM16A and TMEM16F. J. Biol. Chem. 2014;289:7438–7447. doi: 10.1074/jbc.M113.542324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han L., Miao Y., et al. Wang X. The binding of autotaxin to integrins mediates hyperhomocysteinemia-potentiated platelet activation and thrombosis. Blood Adv. 2021;6:46–61. doi: 10.1182/bloodadvances.2021004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Öhlinger T., Müllner E.W., et al. Salzer U. Lysophosphatidic acid-induced pro-thrombotic phosphatidylserine exposure and ionophore-induced microvesiculation is mediated by the scramblase TMEM16F in erythrocytes. Blood Cells Mol. Dis. 2020;83:102426. doi: 10.1016/j.bcmd.2020.102426. [DOI] [PubMed] [Google Scholar]
- 76.Nguyen D.M., Chen L.S., et al. Chen T.-Y. Comparison of ion transport determinants between a TMEM16 chloride channel and phospholipid scramblase. J. Gen. Physiol. 2019;151:518–531. doi: 10.1085/jgp.201812270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shimizu T., Iehara T., et al. Okada Y. TMEM16F is a component of a Ca2+-activated Cl− channel but not a volume-sensitive outwardly rectifying Cl− channel. Am. J. Physiol. Cell Physiol. 2013;304:C748–C759. doi: 10.1152/ajpcell.00228.2012. [DOI] [PubMed] [Google Scholar]
- 78.Ye W., Han T.W., et al. Jan L.Y. Dynamic change of electrostatic field in TMEM16F permeation pathway shifts its ion selectivity. Elife. 2019;8:e45187. doi: 10.7554/eLife.45187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The movie was generated based on sim10. Chain A is shown in the front view by cartoons with TMs 3-6 colored in yellow, red, blue and green, respectively, and the other TMs in cyan. The phosphate atoms in lipid headgroups are represented as transparent orange spheres. The two bound Ca2+ ions are shown as two red spheres. The inner gate residues, K518, K563 and I612, are shown as yellow sticks. Note that, as the pore opens up, it becomes fully occupied with lipid headgroups.
Chain A is shown in the front view. TMs, phosphate atoms and inner gate residues are shown in sticks and colored according to atom types. The phosphate atoms of the passing lipid are shown with larger orange spheres, while the tail is shown in stick.
Chain A is shown in the front view and elements, such as TMs, phosphate atoms and inner gate residues, are presented with the same scheme as in Movie S1. The passing chloride ion is shown as a green sphere along with its hydration shell. Note that the ion resides in the intracellular and extracellular vestibules for extended periods before and after crossing the hydrophobic inner gate region (the major free energy barrier).