Abstract
The newly emerged BA.2.75 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant contains 9 additional mutations in its spike (S) protein compared to the ancestral BA.2 variant. Here, we examine the neutralizing antibody escape of BA.2.75 in mRNA-vaccinated and BA.1-infected individuals, as well as the molecular basis underlying functional changes in S. Notably, BA.2.75 exhibits enhanced neutralization resistance over BA.2 but less than the BA.4/5 variant. The G446S and N460K mutations of BA.2.75 are primarily responsible for its enhanced resistance to neutralizing antibodies. The R493Q mutation, a reversion to the prototype sequence, reduces BA.2.75 neutralization resistance. The impact of these mutations is consistent with their locations in common neutralizing antibody epitopes. Further, BA.2.75 shows enhanced cell-cell fusion over BA.2, driven largely by the N460K mutation, which enhances S processing. Structural modeling reveals enhanced receptor contacts introduced by N460K, suggesting a mechanism of potentiated receptor utilization and syncytia formation.
Keywords: SARS-CoV-2, Omicron, BA.2.75, neutralizing antibodies, cell-cell fusion, mRNAvaccine, mRNA booster
Newly emerged Omicron subvariants reignite concerns over escape from existing immunity. Qu and colleagues compare the immunity resistance and fusogenicity of BA.2.75 with prior variants. BA.2.75 exhibits stronger neutralization resistance than BA.2 but weaker than BA.4/5, as well as enhanced fusogenicity, which are largely driven by G446S and N460K, respectively.
Introduction
Emergence of the Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in late 2021 sparked an unprecedented wave of coronavirus disease 2019 (COVID-19) cases and exhibited robust evasion of vaccine- and infection-induced immunity (Gruell et al., 2022; Hoffmann et al., 2022). More recently, several subvariants of Omicron have been identified, which have driven subsequent waves of infection. The BA.1 subvariant, responsible for the initial Omicron wave, was replaced by BA.2, which displayed slightly enhanced transmissibility and resistance to BA.1-induced sera (Evans et al., 2022; Yamasoba et al., 2022b). BA.2 then evolved into several progeny subvariants, including the BA.2.12.1 variant, which subsequently became predominant (Centers for Disease Control and Prevention, 2022). Remarkably, the BA.4 and BA.5 variants, which bear identical spike (S) proteins and evolved from BA.2, are currently dominant worldwide, including in the US (Centers for Disease Control and Prevention, 2022). BA.4 and BA.5 bear an L452R mutation that is primarily responsible for further enhanced neutralizing antibody (nAb) resistance (Qu et al., 2022a; Qu et al., 2022b; Tuekprakhon et al., 2022). Recently, another distinct BA.2-derived subvariant, BA.2.75, has been identified. BA.2.75 is increasing in prevalence in southeast Asia and has been detected globally (Callaway, 2022). Notably, BA.2.75 bears 9 key S mutations including K147E, W152R, F157L, I210V, G257S, D339H, G446S, and N460K, as well as an R493Q reversion mutation (World Health Organization, 2022) (Figure 1 A). These mutations, particularly those in the receptor binding domain (RBD), have generated concern over further immune escape.
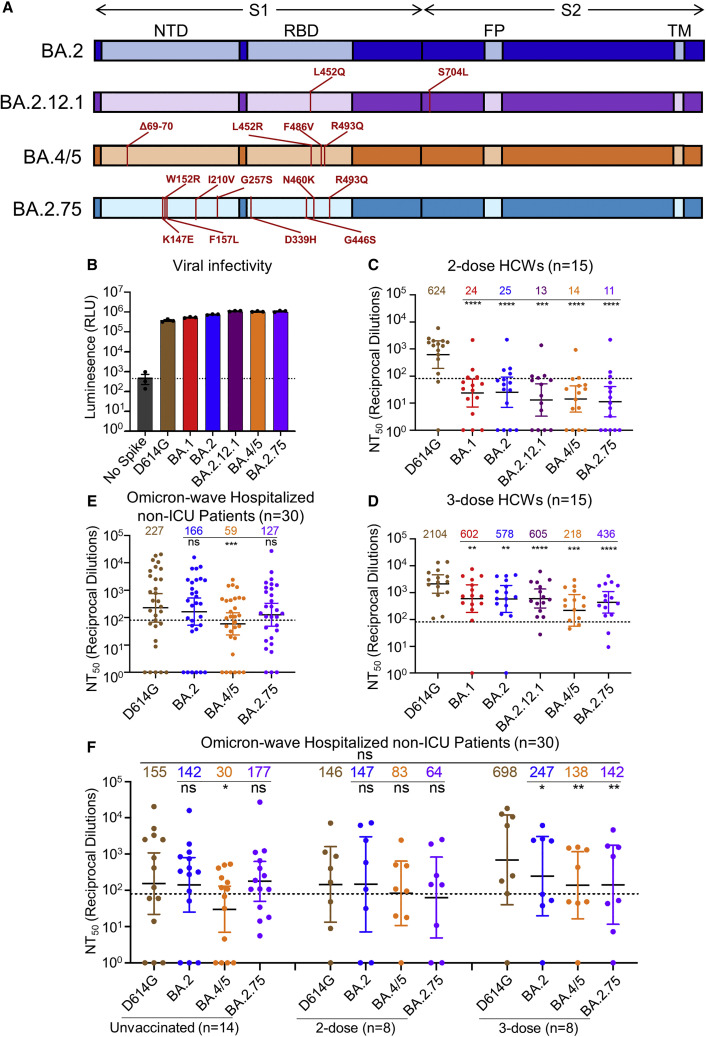
Figure 1.
BA.2.75 exhibits strong neutralization resistance to 2-dose and 3-dose mRNA vaccinee sera and Omicron-wave patient sera
(A) Schematic of BA.2-derived SARS-CoV-2 variants with mutations relative to the BA.2 background indicated. Highlighted are the S1 and S2 subunits, N-terminal domain (NTD), receptor binding domain (RBD), fusion peptide (FP), and transmembrane (TM) domain.
(B) Infectivity of pseudotyped lentivirus bearing S protein from SARS-CoV-2 variants of study; bars represent means ± standard error.
(C and D) Neutralizing antibody titers against lentivirus pseudotyped with S from individual SARS-CoV-2 variants for 15 health care workers for sera collected 3–4 weeks after second mRNA vaccination (C) or 1–12 weeks after homologous mRNA booster vaccination (D).
(E) Neutralizing antibody titers for sera collected from 30 COVID-19 patients hospitalized during the BA.1 pandemic wave.
(F) Neutralizing antibody titers against hospitalized BA.1 wave patients are divided by vaccination status.
(C–F) Dots indicate individual patient samples; bars represent geometric means with 95% confidence intervals; significance relative to D614G was determined by one-way repeated measures ANOVA with Bonferroni multiplicity correction. p values are displayed as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, and ns for not significant.
Here we characterize the BA.2.75 S protein by examining its sensitivity to neutralizing antibodies from mRNA-vaccinated and/or boosted health care workers (HCWs), as well as from Omicron-wave-hospitalized COVID-19 patients. In addition, we examine BA.2.75 infectivity, S processing, and fusogenicity. Mutational analysis revealed the N460K as a key driver of enhanced fusogenicity, while the G446S and N460K mutations were primarily responsible for reduced neutralization sensitivity of BA.2.75 compared to BA.2. Moreover, we find that the R493Q reversion mutation enhances the neutralization sensitivity of BA.2.75. These findings inform our understanding of SARS-CoV-2 evolution and will aid in addressing the ongoing threat of emerging SARS-CoV-2 variants.
Results
BA.2.75 exhibits enhanced neutralization resistance over BA.2
We first sought to characterize sensitivity to vaccine-induced immunity of the BA.2.75 variant. Utilizing our previously reported pseudotyped lentivirus assay (Zeng et al., 2020), we examined nAb titers for 15 Ohio State University Wexner Medical Center HCWs in serum samples collected 3–4 weeks after vaccination with a second dose of Moderna mRNA-1273 (n = 7) or Pfizer/BioNTech BNT162b2 (n = 8) vaccine, and 1–12 weeks after vaccination with a homologous booster dose (see STAR Methods). Patient sera were examined for nAb titers against lentivirus pseudotyped with S from ancestral SARS-CoV-2 S bearing only the D614G mutation, as well as S from BA.1, BA.2, BA.2.12.1, BA.4/5, and BA.2.75 (Figure 1A). All S constructs were functional and produced comparably infectious lentivirus pseudotypes (Figure 1B).
Notably, all Omicron sublineages, including BA.2.75, exhibited strong resistance to 2-dose-induced immunity compared to D614G (p < 0.0001), with only 1–2 HCW samples exhibiting 50% neutralization titers (NT50) above the limit of quantification (NT50 = 80) (Figure 1C). In contrast, administration of a booster dose recovered the nAb response against all Omicron subvariants (Figures 1D and S1A–S1H). In serum from the boosted individuals, BA.2.75 exhibited 4.8-fold (p < 0.0001) lower neutralization than D614G, with somewhat stronger neutralization resistance than BA.2 and BA.2.12.1, which were neutralized 3.6-fold (p < 0.01) and 3.5-fold (p < 0.001) less efficiently than D614G, respectively (Figure 1D). However, BA.2.75 showed higher neutralization sensitivity than BA.4/5, which had 9.7-fold (p < 0.001) lower neutralization than D614G (Figure 1D).
We also examined the nAb response in a cohort of non-intensive care unit (ICU) COVID-19 patients (n = 30) hospitalized at the Ohio State University Wexner Medical Center during the Omicron wave of the pandemic. These patient samples were collected between early February and early March of 2022, representing a BA.1 dominant period in Ohio. Overall, the nAb titers of the Omicron-wave patients were much lower than those of boosted HCWs, and BA.2.75 exhibited neutralization resistance modestly higher than BA.2 (by 44.0%, p > 0.05) but much lower than BA.4/5 (3.8-fold; p < 0.001) relative to D614G (Figures 1E and S1I). This cohort of Omicron-wave patients included 14 unvaccinated patients, 8 patients vaccinated with 2 doses of Moderna mRNA-1273 (n = 4) or Pfizer/BioNTech BNT162b2 (n = 4), and 8 patients vaccinated and boosted with Pfizer/BioNTech BNT162b2. We found that, while BA.2.75 was neutralized comparably to BA.2 and D614G for unvaccinated patients, BA.2.75 was neutralized 2.3-fold less efficiently than D614G in 2-dose vaccinated patients (p > 0.05) and 4.9-fold less efficiently for 3-dose vaccinated patients (p < 0.01), respectively (Figure 1F). The boosted HCWs with breakthrough infection exhibited higher nAb titers overall (Figure 1F), as would be expected.
BA.2.75 neutralization is modulated by G446S, N460K, and R493Q mutations
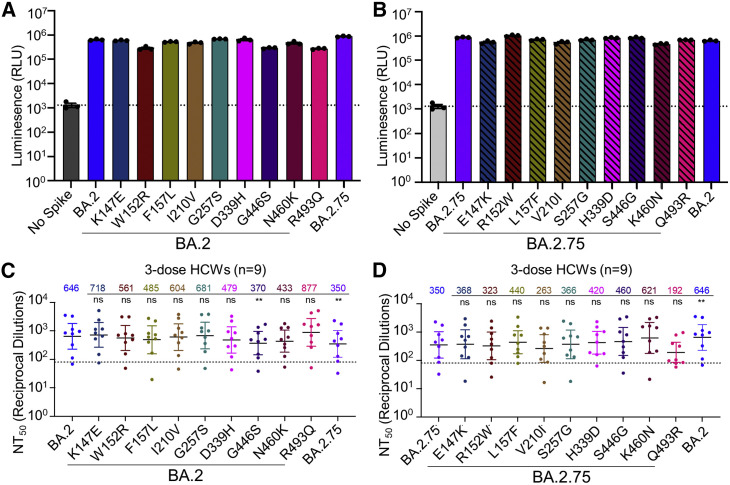
To understand the determinants of BA.2.75 neutralization resistance, we examined all nine point mutations in the BA.2 background, as well as nine corresponding reversion mutations in the background of BA.2.75. None of these single mutations substantially impacted lentiviral pseudotype infectivity (Figures 2 A and 2B). We then examined the neutralization sensitivity of these mutants to sera from 9 HCWs collected 1–12 weeks after homologous booster vaccination with Moderna mRNA-1273 (n = 2) or Pfizer/BioNTech BNT162b2 (n = 7). When the G446S mutation was introduced to BA.2, a slight but significant reduction in sensitivity to 3-dose mRNA vaccine-induced nAbs was observed (42.7%, p < 0.01), which was comparable to BA.2.75 (Figure 2C). Introduction of a S446G reversion mutation into BA.2.75 enhanced neutralization sensitivity by 31.4%, albeit the change was not statistically significant (p = 0.055) (Figure 2D). Interestingly, introduction of a R493Q mutation into BA.2 increased neutralization sensitivity by 35.8% (p > 0.05), while introduction of the Q493R reversion mutation into BA.2.75 reduced neutralization sensitivity by 45.1% (p > 0.05) (Figures 2C and 2D). Of note, the N460K mutation also substantially increased neutralization resistance of BA.2 by 33.0% (p > 0.05), whereas the K460N reversion mutation in BA.2.75 was 77.4% (p = 0.069) more neutralization sensitive (Figures 2C and 2D). Thus, the G446S and N460K mutations in BA.2.75 are largely responsible for its enhanced neutralization resistance, while the R493Q reversion mutation in BA.2.75 at least partially restores neutralizing epitopes found in the prototype SARS-CoV-2, which were otherwise abolished in BA.2.
Figure 2.
The G446S, N460K, and R493Q mutations modulate BA.2.75 neutralization sensitivity
(A) Relative infectivity of lentivirus pseudotyped with BA.2 S with single mutations from BA.2.75 lineage-defining mutations; bars represent mean ± standard error.
(B) Relative infectivity of lentivirus pseudotyped with BA.2.75 S with single reversion mutations to remove BA.2.75 lineage-defining mutations; bars represent mean ± standard error.
(C and D) Neutralizing antibody titers against lentivirus pseudotyped with S from BA.2 with single mutations from BA.2.75 lineage-defining mutations (C) or BA.2.75 with single reversion mutations to remove BA.2.75 lineage-defining mutations (D) for sera collected from 9 health care workers 1–12 weeks after homologous mRNA booster vaccination. Dots indicate individual patient samples; bars represent geometric means with 95% confidence intervals; significance relative to D614G was determined by one-way repeated measures ANOVA with Bonferroni multiplicity correction. p values are displayed as ∗∗p < 0.01, and ns for not significant.
BA.2.75 exhibits enhanced syncytia formation and S processing compared to BA.2
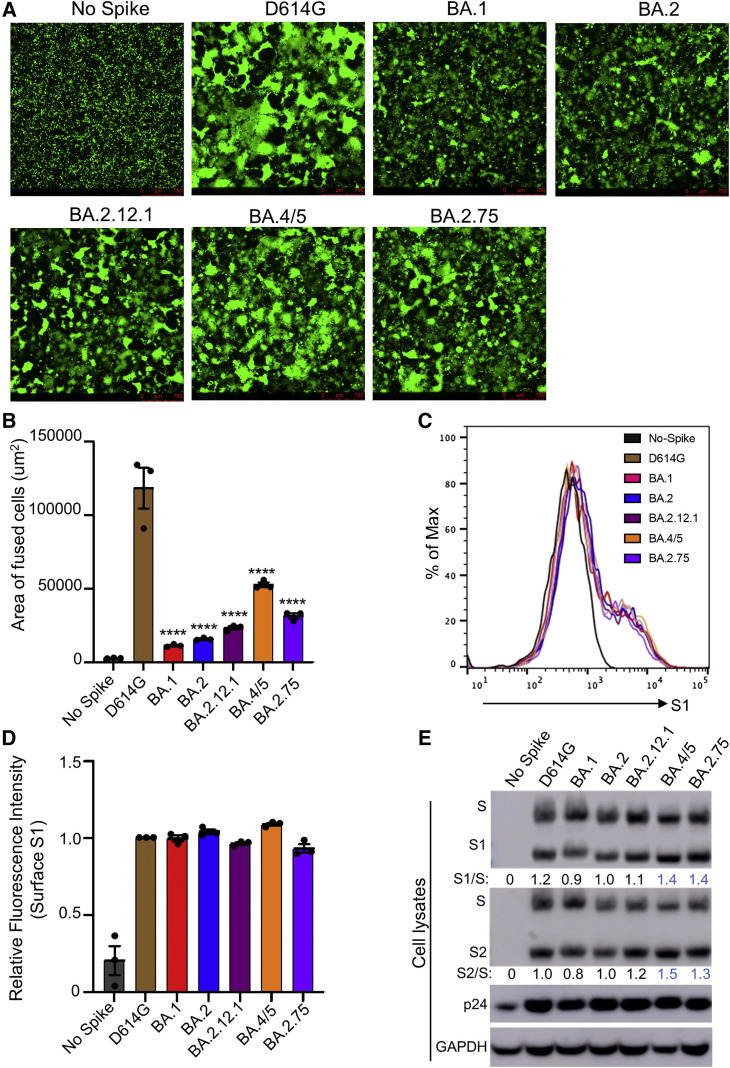
We next sought to characterize key features of the BA.2.75 S protein, including the ability to mediate cell-cell fusion. HEK293T cells stably expressing human angiotensin I converting enzyme 2 (ACE2) (HEK293T-ACE2 cells) were transfected to express the green fluorescent protein (GFP) and variant SARS-CoV-2 S proteins. As previously reported (Zeng et al., 2021), all Omicron sublineages exhibited reduced fusogenicity compared to the ancestral D614G S (Figures 3 A and 3B). However, BA.2.75 exhibited enhanced syncytia formation compared to BA.2, with mean syncytia size 2.0-fold higher than BA.2 (p < 0.0001) (Figures 3A, 3B, and S2A); this was despite similar surface expression, as examined by flow cytometry (Figures 3C and 3D).
Figure 3.
BA.2.75 exhibits enhanced cell-cell fusion and S processing
(A) Fluorescence images displaying syncytia formation are presented for HEK293T-ACE2 cells 48 h after co-transfection with a GFP expression construct and SARS-CoV-2 variant S proteins. Scale bars represent 150 μm.
(B) Quantification of syncytia formation in (A) displays the mean syncytia size; bars represent mean ± standard error, with significance relative to D614G determined by one-way ANOVA with Bonferroni multiplicity correction. p values are displayed as ∗∗∗∗p < 0.0001.
(C) Histogram displays of the surface staining of HEK293T cells expressing S proteins, which were detected by an anti-S1 antibody (T62).
(D) Quantification of relative surface expression as shown in (C); bars represent mean ± standard error.
(E) Pseudotyped lentivirus producer cell lysate was assessed for processing of S by probing with anti-S1 (T62), anti-S2, anti-HIV-1 Gag (anti-p24), and anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase). Band intensities were quantified in ImageJ and the ratio of S1/S or S2/S is displayed relative to the S1/S or S2/S ratio of BA.2.
To determine if the enhanced fusogencity phenotype might be related to alterations in processing of S protein, we examined cell lysates from the pseudotyped lentivirus producer cells. As shown in Figure 3E, BA.2.75 spike exhibited enhanced processing, as reflected in the ratio of S1 or S2 subunit to full-length S, which was ∼30%–40% higher than BA.2. Consistent with its enhanced fusion, BA.4/5 showed the highest S processing among Omicron variants (Figures 3A, 3B, 3E, and S2A).
Enhanced syncytia formation and processing of BA.2.75 is determined by the N460K mutation
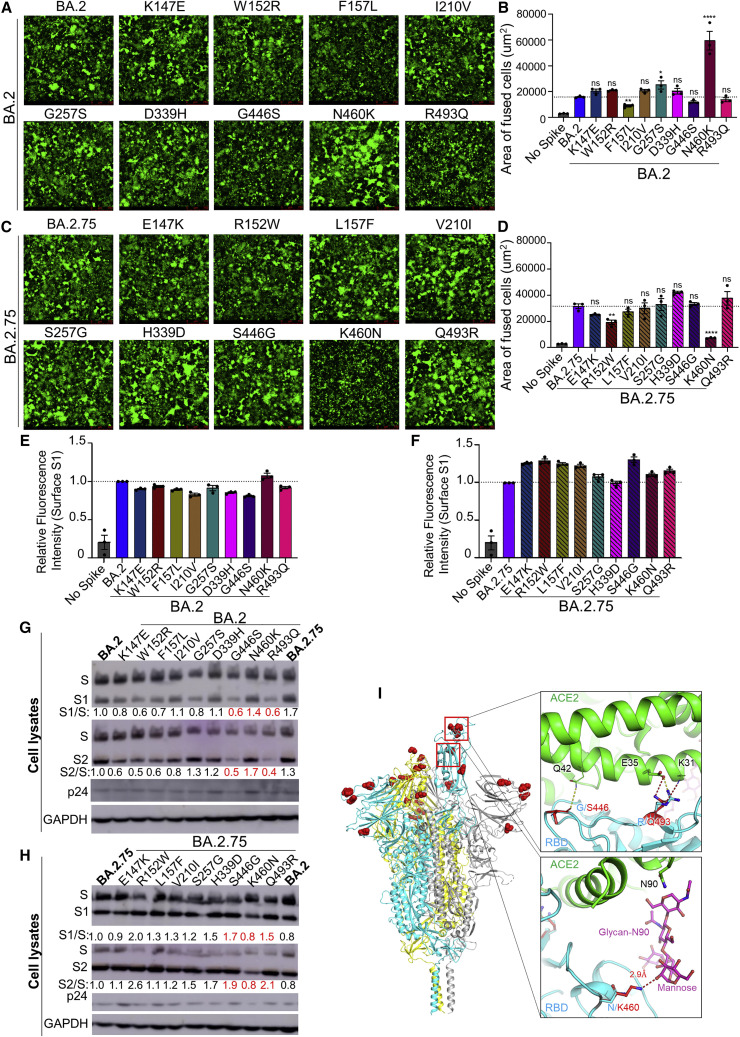
We further characterized the impact of BA.2.75-defining mutations on S fusogenicity and processing. Introduction of the N460K mutation into the BA.2 S drastically enhanced cell-cell fusion, with mean syncytia size 3.8-fold (p < 0.0001) higher than BA.2 (Figures 4A, 4B, and S2B). Conversely, introduction of the K460N reversion mutation into BA.2.75 significantly reduced cell-cell fusion, with mean syncytia size 4.3-fold (p < 0.0001) lower than BA.2.75 (Figures 4C, 4D, and S2C). We found that F157L and G257S in the BA.2 background, as well as the R152W reversion mutant in the BA.2.75 background, also exhibited modestly altered fusion activity (Figures 4A–4D). Importantly, the differences in membrane fusion between these mutants were not due to the surface expression level of S, as examined by flow cytometry (Figures 4E, 4F S2D, and S2E). Consistent with enhanced fusion activity, introduction of the N460K mutation into the BA.2 S protein enhanced processing of S into the S1 and S2 subunits, as reflected in a S1/S ratio 40% higher than in BA.2 (Figure 4G); a similar 70% increase in S2/S ratio was also observed (Figure 4G). Conversely, introduction of a K460N reversion mutation into BA.2.75 reduced S protein processing by 20% (Figure 4H). Thus, the N460K mutation in BA.2.75 enhances S processing, consistent with increased fusogenicity.
Figure 4.
The N460K mutation determines enhanced cell-cell fusion and S processing of BA.2.75
(A) Fluorescence images displaying syncytia formation are presented for HEK293T-ACE2 cells 48 h after co-transfection with a GFP expression construct and BA.2 single mutant S proteins. Scale bars represent 150 μm.
(B) Quantification of syncytia formation in (A) displays the mean syncytia size; bars represent mean ± standard error, with significance relative to D614G determined by one-way ANOVA with Bonferroni multiplicity correction with p values displayed as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, and ns for not significant.
(C) Fluorescence images displaying syncytia formation are presented for HEK293T-ACE2 cells 48 h after co-transfection with a GFP expression construct and BA.2.75 single reversion mutant S proteins. Note that the negative control for Figures 4A and 4C corresponds to that shown in Figure 3A, which were from the same experiment. Scale bars represent 150 μm.
(D) Quantification of syncytia formation in (C) displays the mean syncytia size; bars represent mean ± standard error, with significance relative to D614G determined by one-way ANOVA with Bonferroni multiplicity correction with p values displayed as ∗∗p < 0.01, ∗∗∗∗p < 0.0001, and ns for not significant.
(E and F) Quantification of relative S surface expression in transfected HEK293T cells for BA.2 single mutants (E) or BA.2.75 reversion mutants (F), as examined by flow cytometry; bars represent mean ±standard error.
(G) Pseudotyped lentivirus producer cell lysate was assessed for processing of S from BA.2 single mutants by probing with anti-S1 (T62), anti-S2, anti-HIV-1 p24, and anti-GAPDH. Band intensities were quantified in ImageJ and the ratios of S1/S and S2/S are displayed relative to the S1/S and S2/S ratios of BA.2.
(H) Pseudotyped lentivirus producer cell lysate was assessed for processing of S from BA.2.75 reversion mutants by probing with anti-S1, anti-S2, anti-HIV-1 p24, and anti-GAPDH. Band intensities were quantified in ImageJ and the ratios of S1/S and S2/S are displayed relative to the S1/S and S2/S ratios of BA.2.75.
(I) Structural modeling of Omicron BA.2.75 spike protein viewed as a ribbon. Mutations of BA.2.75 specific mutants are highlighted by red spheres. The RBD of the cyan spike protomer is in an “up” conformation. Upper inset: the mutation G446S reduces the backbone flexibility and possibly stabilizes the hydrogen bond between its carbonyl group and the residue Q42 on ACE2 receptor (green); the mutation R493Q abolishes the salt bridge interaction with the E35 on ACE2 receptor and potentially forms two hydrogen bonds with E35 and K31. Lower inset: the mutation N460K enables formation of a hydrogen bond with the glycan-N90 on ACE2 receptor (green).
Structural modeling
To understand how BA.2.75 mutations contribute to functional changes, we created models of ectodomain of BA.2.75 by itself, as well as ectodomain or RBD of BA.2.75 spike protein in complexes with the ACE2 receptor using homology modeling using Omicron BA.1 spike and its ACE2 complex (PDB: 7TNW and 7TO4) and BA.2 RBD-ACE2 complex (PDB: 7XB0) as templates (Figures 4I and S3). The G446S mutation does not appear to alter main-chain interactions with the Q42 receptor residue; however, this mutation could reduce backbone flexibility, thus potentially stabilizing the specific interaction with ACE2, as well as spike integrity. The R493Q mutation would abolish a strong salt bridge interaction with the E35 residue on the ACE2 receptor, which could reduce receptor binding affinity; however, this effect may be offset by the formation of two new hydrogen bonds between the Q493 residue on spike and residues E35 and K31 on ACE2. Finally, N460K forms a new hydrogen bond with the glycan-N90 on ACE2 through an elongated side chain that reaches out to the alpha-1,3 mannose molecule on the N-linked glycan of the receptor residue N90, and this would likely enhance receptor binding affinity of BA.2.75. In addition, N460K could form a salt bridge with D240 (Cao et al., 2022) and hydrogen bond with T415 on RBD, respectively, which would enhance intra-RBD hydrophilic interactions, resulting in reduced flexibility of ACE2-contacting loops on the receptor binding motif (RBM) therefore facilitating receptor engagement.
Discussion
The BA.2.75 subvariant is the latest in a series of Omicron variants to be identified. BA.2.75 has an alarming nine additional S mutations compared with BA.2, and preliminary reports suggest a slight growth advantage (Callaway, 2022; World Health Organization, 2022). These features portend that BA.2.75 could potentially overtake the BA.4/5 subvariants as the dominant circulating strain. Given this concern, it is critical to examine key features and novel phenotypes of BA.2.75, especially in its S protein. In this study, we show that BA.2.75 exhibits an increased neutralization resistance compared to ancestral BA.2 but has significantly lower neutralization resistance than BA.4/5 for 3-dose mRNA-vaccinated HCWs as well as for hospitalized Omicron-wave patients. Critically, we demonstrate that the G446S and N460K mutations in the S protein of BA.2.75 underlie its enhanced neutralization resistance, while the R493Q mutation in BA.2.75, which is a reversion mutation, sensitizes it to neutralization. These findings could reflect the emergence of compensatory mutations to improve S function while maintaining neutralization resistance. Notably, the G446S mutation occurs in an epitope bound by class III neutralizing antibodies, rather than class II neutralizing antibodies that target the epitope of the R493Q mutation (Greaney et al., 2021). Structural analysis suggests that the side chain addition by G446S creates a steric clash with the CDR region of class III neutralizing antibodies, thus potentially hampering their recognition (Liu et al., 2022; Wang et al., 2022a). Hence, the exchange of these mutations may alter the susceptibility of BA.2.75 to class II and class III nAbs.
We further demonstrate that BA.2.75 exhibits enhanced S-mediated cell-cell fusion compared to BA.2, albeit to a lesser extent than BA.4/5. This enhanced triggering of BA.2.75 S-mediated fusion may reflect improved receptor utilization that is not present in earlier Omicron subvariants, consistent with several recent preprints (Cao et al., 2022; Saito et al., 2022; Wang et al., 2022a). Critically, we find that the N460K mutation present in BA.2.75 is essential for the enhanced fusion phenotype. This may relate to enhanced processing of N460K-containing S in virus producing cells, which would prime more cell surface-associated S for membrane fusion. While structural modeling did not provide an immediate explanation, the N460K mutation might enhance receptor utilization through a hydrogen bond with a receptor glycan N90, and/or through stabilization of the receptor binding motif (RBM) by enhancing intra-domain hydrophilic interactions with D240 and T415 residues. It is worth noting that this glycan interaction is mediated by a terminal mannose molecule, so it may not be easily observed in conditions of protein overexpression where glycosylation is often insufficient, and in cryo-EM where glycan moieties are often less visible. In addition, ACE2 N90-glycan has been shown to be critical for Omicron BA.2 receptor recognition by interacting with RBD residue T415 by crystal structure (Li et al., 2022). The proximity of N460K, D240, T415 and ACE2 N90-glycan enables a strong hydrophilic interaction network to stabilize local conformation and RBD-receptor interactions. G446S, on the other hand, may reduce the flexibility of loop 440–450, potentially enhancing overall spike thermostability, which likely decreases S processing efficiency. Furthermore, G446 is not well resolved in many apo spike structures, in line with its flexible local conformation. A more stable backbone loop conformation produced by the G446S mutation may reduce the energy cost for receptor engagement through hydrogen bond formation with Q42. Lastly, the loss of a strong salt bridge interaction by the R493Q mutation is offset by the addition of two potential hydrogen bonds to the adjacent receptor residues, which could explain its modestly decreased fusion efficiency and processing. The contributions of these key residues to BA.2.75 replication kinetics in physiologically relevant human lung and airway epithelial cells needs to be carefully investigated. Further characterization of emerging SARS-CoV-2 variants will continue to aid our understanding of key features of SARS-CoV-2 evolution, spike biology, and immune evasion. Continued analysis of emerging variants also will improve ongoing public health responses and any potential reformulation of SARS-CoV-2 mRNA vaccine boosters.
Limitations of this study include a relatively small sample size for the boosted HCWs and Omicron-wave patients and the utilization of pseudotyped lentivirus for the neutralization assay rather than an authentic virus assay. However, our results for neutralization resistance are in accordance with several recent preprints (Cao et al., 2022; Gruell et al., 2022; Saito et al., 2022; Sheward et al., 2022; Wang et al., 2022b; Xie et al., 2022; Yamasoba et al., 2022a) and the lentiviral pseudotyped neutralization assay has been previously confirmed using authentic SARS-CoV-2 (Zeng et al., 2020), as well as confirmed by numerous laboratories in the field. Further, the HCWs exhibited a wide range in time between booster administration and sample collection. Additionally, our study hasn’t examined the neutralization resistance of BA.2.75 to the nAbs induced by the new bivalent boosters, which have recently received an emergency use authorization (EUA) from the U.S. Food and Drug Administration. It is important to emphasize that while structural modeling is useful and may provide possible explanations to the observations from functional analyses, it is not as accurate as experimental approaches such as X-ray crystallography and cryo-EM. Further structure-based functional mutagenesis analysis will be necessary to delineate the impact of BA.2.75 mutations. Future studies will focus on the biology and replication characteristics of BA.2.75 using variants isolated from human COVID-19 patients.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-SARS-CoV-2 S1 | Sino Biological | Cat# 40150-T62, RRID:AB_2920715 |
| anti-SARS-CoV-2 S2 | Sino Biological | Cat# 40590-T62, RRID: AB_2857932 |
| anti-p24 | NIH | Cat# ARP-1513, RRID: AB_2832923 |
| anti-GAPDH | Santa Cruz Biotechnology | Cat# sc-47724, RRID: AB_627678 |
| anti-Rabbit IgG (whole molecule)–FITC antibody | Sigma-Aldrich | Cat# F9887, RRID: AB_259816 |
| anti-Mouse IgG (whole molecule)–Peroxidase | Sigma-Aldrich | Cat# A5278, RRID: AB_258232 |
| anti-Rabbit IgG (whole molecule)–Peroxidase | Sigma-Aldrich | Cat# A9169, RRID: AB_258434 |
| Bacterial and virus strains | ||
| E.coli DH5α | Invitrogen | Strain #: DH5α |
| E.coli Stable 2 | Invitrogen | Strain #: Stable 2 |
| Biological samples | ||
| 2-dose HCWs Sera | (Evans et al., 2022; Qu et al., 2022a) | N/A |
| 3-dose HCWs Sera | (Evans et al., 2022; Qu et al., 2022a) | N/A |
| Omicron-wave Infected Patient Sera | (Evans et al., 2022; Qu et al., 2022a) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Polyethylenimine (PEI) | Thermo Fisher Scientific | Cat# BMS1003 |
| Dulbecco’s Modified Eagles Medium (DMEM) | Sigma-Aldrich | Cat#: 11965-092 |
| Fetal Bovine Serum (FBS) | Thermo Fisher Scientific | Cat#: F1051 |
| 0.05% Trypsin + 0.53 mM EDTA | Corning | Cat# 25-052-CI |
| Penicillin-Streptomycin | HyClone | Cat#: SV30010 |
| Protease Inhibitor Cocktail | Sigma-Aldrich | Cat# P8340 |
| Immobilon Crescendo Western HRP substrate | Millipore | Cat# WBLUR0500 |
| QIAprep Spin Miniprep Kit | QIAGEN | Cat# 27106 |
| Coelenterazine | GoldBio | Cat#: CZ2.5, CAS: 55779-48-1 |
| Deposited data | ||
| NT50 Values and De-identified patient data | SeroNet Coordinating Center, NCI, NIH | N/A |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | Cat#: CRL-11268, RRID: CVCL_1926 |
| HEK293T-ACE2 | BEI Resources | Cat#: NR-52511, RRID: CVCL_A7UK |
| Recombinant DNA | ||
| pNL4-3-inGluc | David Derse, NCI, NIH (Mazurov et al., 2010) |
N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_D614G | GenScript Biotech (Evans et al., 2022) |
N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.1 | GenScript Biotech (Evans et al., 2022) |
N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.12.1 | GenScript Biotech (Qu et al., 2022) |
N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.4/5 | GenScript Biotech (Qu et al., 2022) |
N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75 | GenScript Biotech This paper |
N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2_K147E | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2_W152R | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2_F157L | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2_I210V | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2_G257S | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2_D339H | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2_G446S | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2_N460K | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2_R493Q | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75_E147K | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75_R152W | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75_L157F | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75_V210I | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75_S257G | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75_H339D | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75_S446G | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75_K460N | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75_Q493R | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2 | GenScript Biotech (Evans et al., 2022) |
N/A |
| Software and algorithms | ||
| GraphPad Prism | Version 9.0.0 | GraphPad |
| ImageJ | (Schneider et al., 2012) | https://imagej.nih.gov/ij/ |
| FlowJo | FlowJo, LLC | https://www.flowjo.com/ |
| SWISS-MODEL | (Guex et al., 2009) | https://swissmodel.expasy.org/ |
| programs Coot | (Emsley et al., 2010) | https://www2.mrc-lmb.cam.ac.uk/personal/ pemsley/coot/ |
| PyMOL | Warren DeLano and Sarin Bromberg | https://pymol.org/ |
| Leica Application Suite X | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| Other | ||
| Cytation 5 Imaging Reader | BioTek | N/A |
| L-70 Optima Ultracentrafuge | Beckman | N/A |
| Amersham Imager 600 | GE Healthcare Life Sciences | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Dr. Shan-Lu Liu (liu.6244@osu.edu).
Materials availability
Plasmids generated in this study are available upon request made to the lead contact.
Experimental model and subject details
Patient information
Sera were collected from the Ohio State University Wexner Medical Center health care workers (HCWs) under approved institutional review board (IRB) protocols (2020H0228 and 2020H0527). Demographic information was self-reported and all subjects provided informed consent. Sera from 15 HCWs were collected 3-4 weeks after vaccination with a second dose of Moderna mRNA-1273 (n = 7) or Pfizer/BioNTech BNT162b2 (n = 8) vaccine, and 1-12 weeks after vaccination with a homologous booster dose; the relative long duration of sample collection for the booster could contribute to bigger variability in NT50 titer. These HCWs ranged in age from 32 to 56 years (median 37 years) and included 6 female and 9 male HCWs. Analysis by age and gender could not be performed due to low sample number.
Sera were collected from patients 30 hospitalized for COVID-19 at the Ohio State University Wexner Medical Center under an approved IRB protocol (2020H0527). Sera were collected between early February and early March of 2022, during the Omicron wave (BA.1 was circulating) in Ohio. Patients included 14 unvaccinated patients, 8 patients vaccinated with 2 doses of Moderna mRNA-1273 (n = 4) or Pfizer/BioNTech BNT162b2 (n = 4), and 8 patients vaccinated and boosted with Pfizer/BioNTech BNT162b2. This cohort included 11 female and 19 male patients. Patients ranged in age from 28 to 78 years (median 62 years).
Cell lines and maintenance
HEK293T (ATCC CRL-11268, RRID: CVCL_1926) and HEK293T-ACE2 (BEI NR-52511, RRID: CVCL_A7UK) cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (Cibco, 11965-092) supplemented with 10% Fetal Bovine Serum (Signa, F1051) and 1% penicillin/streptomycin (HyCline, SV30010). Cells were maintained at 5% CO2 and 37°C.
Method details
Plasmids
Pseudotyped lentivirus was produced using a pNL4-3-inGluc lentivirus vector comprised of a ΔEnv HIV-1 backbone bearing a Gaussia luciferase reporter gene driven by a cytomegalovirus (CMV) promoter (Goerke et al., 2008; Zeng et al., 2020). SARS-CoV-2 S constructs bearing N- and C-terminal Flag tags were synthesized and cloned into a pcDNA3.1 vector by GenScript (Piscataway, NJ) by Kpn I and BamH I restriction enzyme cloning.
Pseudotyped lentivirus production and infectivity
Pseudotyped lentivirus was produced by transfecting HEK293T cells with pNL4-3-inGluc and S construct in a 2:1 ratio using polyethylenimine transfection. Pseudotyped lentivirus was collected at 48 hr and 72 hr after transfection. Collections were pooled and used to infect HEK293T-ACE2 cells to assess pseudotyped lentivirus infectivity. 48 hr and 72 hr after infection, infected cell culture media was assessed for Gaussia luciferase activity by combining 20 μL of media with 20 μL of Gaussia luciferase substrate (0.1 M Tris pH 7.4, 0.3 M sodium ascorbate, 10 μM coelenterazine). Luminescence was then immediately measured by a BioTek Cytation5 plate reader using BioTek Gen5 Microplate Reader and Imager Software (Winooski, VT).
Lentivirus neutralization assay
Pseudotyped lentivirus neutralization assays were performed as previously described (Zeng et al., 2020). Patient or HCW sera were 4-fold serially diluted in complete DMEM and pseudotyped lentivirus was added to neutralize for 1 hr (final dilutions: 1:80, 1:320, 1:1280, 1:5120, 1:20480, and no serum control). The pseudotyped lentivirus/sera mixtures were then transferred to HEK293T-ACE2 cells for infection. Then 48 hr and 72 hr after infection, infected cell media was assayed for Gaussia luciferase activity by combining 20 μL of cell culture media with 20 μL of Gaussia luciferase substrate. Luminescence was read immediately by a BioTek Cytation5 plate reader using BioTek Gen5 Microplate Reader and Imager Software (Winooski, VT). NT50 values were determined by least-squares-fit, non-linear regression in GraphPad Prism 9 (San Diego, CA).
Spike surface expression
HEK293T cells used to produce pseudotyped lentivirus were singularized by incubation in phosphate buffer saline (PBS) with 5 mM ethylenediaminetetraacetic acid (EDTA) at 37°C for 5 min and fixed 72 hr after transfection by incubation in 3.7% formaldehyde in PBS for 10 min. Cells were then stained with rabbit anti-S1 primary antibody (Sino Biological, 40150-T62) and anti-rabbit-IgG-fluorescein isothiocyanate (FITC) secondary antibody (Sigma, F9887). Samples were analyzed by a Life Technologies Attune NxT flow cytometer and data was processed using FlowJo v7.6.5 (Ashland, OR).
Syncytia formation
HEK293T-ACE2 cells were transfected with SARS-CoV-2 S constructs and a GFP expression construct. Cells were then imaged at 4x magnification 24 hr and 48 hr after transfection with a Leica DMi8 confocal microscope. Syncytia size was quantified using Leica Applications Suit X (Wetzlar, Germany) image analysis software. Three images were taken per sample with representative images being displayed.
Spike processing and incorporation
Pseudotyped lentivirus producing HEK293T cells were lysed by incubating in RIPA lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1mM EDTA, Nonidet P-40, 0.1% sodium dodecyl sulfate (SDS)) supplemented with protease inhibitor (Sigma, P8340) on ice for 30 min. Cell debris was pelleted and cell lysate was dissolved in 5x sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) Laemmli buffer (312.5 mM Tris-HCl pH 6.8, 10% SDS, 25% glycerol, 0.5% bromophenol blue, 10% β-mercaptoethanol). Pseudotyped lentivirus was purified by ultracentrifugation through a 20% sucrose cushion at 28,000 rpm and 4°C using a Beckman L-80 ultracentrifuge with TW-41 rotor. Pelleted pseudotyped lentivirus was resuspended in 1x SDS-PAGE Laemmli buffer. Cell lysate and purified virus were run on a 10% acrylamide SDS-PAGE gel and were transferred to a polyvinylidene difluorid (PVDF) membrane. Membranes were blotted with anti-S1 (Sino Biological, 40150-T62), anti-S2 (Sino Biological, 40590-T62), anti-p24 (NIH ARP-1513), and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Santa Cruz Biotech, sc-47724) with anti-mouse-IgG-horse radish peroxidase (HRP) (Sigma A5278) and anti-rabbit-IgG-HRP (Sigma, A9169) secondary antibodies. Blots were imaged with Immobilon Crescendo Western HRP substrate (Millipore, WBLUR0500) on a GE Amersham Imager 600. Band intensities were quantified using ImageJ (Bethesda, MD) image analysis software.
Homology modeling
Structural modeling of Omicron BA.2.75 spike ectodomain by itself, ectodomain, or RBD in complexes with ACE2 receptor was conducted on SWISS-MODEL server using cryo-EM structures of SARS-CoV2 Omicron BA.1 strain spike ectodomain and ACE2 complex, as well as Omicron BA.2 strain spike RBD in complex with ACE2 (PDB 7TNW, 7TO4 and 7XB0, respectively) as templates. Glycan model building, residue examination, and rotamer adjustment were carried out manually with programs Coot (Emsley et al., 2010) and PyMOL (pymol.org).
Quantification and statistical analysis
NT50 values were determined by least-squares-fit, non-linear-regression in GraphPad Prism 9 (San Diego, CA). NT50 values were log10 transformed for hypothesis testing to better approximate normality. Throughout, multiplicity was addressed by the use of Bonferroni corrections. Statistical analyses were performed using GraphPad Prism 9 (San Diego, CA) and are referenced in the figure legends and include one-way ANOVA (Figures 3B, 4B, and 4D), one-way repeated measures ANOVA (Figures 1C–1F, 2C, and 2D), and a paired, two-tailed Student’s t test with Welch’s correction was used (Figures S1A–S1F). Syncytia sizes were quantified by Leica Applications Suit X (Wetzlar, Germany). Band intensities (Figures 3E, 4G, and 4H) were quantified by ImageJ (Bethesda, MD) image analysis software.
Acknowledgments
We thank the NIH AIDS Reagent Program and BEI Resources for providing important reagents for this work and Xue Zou for assistance. We also thank the Clinical Research Center/Center for Clinical Research Management of The Ohio State University Wexner Medical Center and The Ohio State University College of Medicine in Columbus, Ohio, specifically Francesca Madiai, Dina McGowan, Breona Edwards, Evan Long, and Trina Wemlinger, for logistics, collection, and processing of samples. In addition, we thank Sarah Karow, Madison So, Preston So, Daniela Farkas, and Finny Johns in the clinical trials team of The Ohio State University for sample collection and other supports. This work was supported by a fund provided by an anonymous private donor to OSU. S.-L.L., R.J.G., L.J.S., and E.M.O. were supported by the National Cancer Institute of the NIH under award no. U54CA260582. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. J.P.E. was supported by Glenn Barber Fellowship from The Ohio State University College of Veterinary Medicine. R.J.G. was additionally supported by the Robert J. Anthony Fund for Cardiovascular Research and the JB Cardiovascular Research Fund, and L.J.S. was partially supported by NIH R01 HD095881. K.X. was supported by a Path to K award from The Ohio State University Office of Head Start and the Center for Clinical & Translational Science. The content is solely the responsibility of the authors and does not necessarily represent the official views of the university or the Center for Clinical & Translational Science.
Author contributions
S.-L.L. conceived and directed the project. P.Q. performed most of the experiments. J.P.E. assisted in experiments and contributed data processing and analyses. C.C. and R.J.G. provided clinical samples. P.Q., J.P.E., and S.-L.L. wrote the paper. K.X. performed homology modeling. Y.-M.Z., L.J.S., E.M.O., and K.X. provided insightful discussion and revision of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: September 28, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2022.09.015.
Supplemental information
Data and code availability
-
•
NT50 values and de-identified patient information will be deposited to the National Cancer Institute SeroNet Coordinating Center. Additionally, NT50 values and de-identified patient information reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Callaway E. Will “Centaurus” be the next global coronavirus variant? Indian cases offers clues. Nature. 2022;608:462–463. doi: 10.1038/d41586-022-02154-4. [DOI] [PubMed] [Google Scholar]
- Cao Y., Song W., Wang L., Liu P., Yue C., Jian F., Yu Y., Yisimayi A., Wang P., Wang Y., et al. Characterizations enhanced infectivity antibody evasion Omicron BA.2.75. Preprint at bioRxiv. 2022 doi: 10.1101/2022.07.18.500332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention COVID data tracker. United States Department of Health and Human Services. 2022 https://covid.cdc.gov/covid-data-tracker [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J.P., Zeng C., Qu P., Faraone J., Zheng Y.M., Carlin C., Bednash J.S., Zhou T., Lozanski G., Mallampalli R., et al. Neutralization of SARS-CoV-2 Omicron sub-lineages BA.1, BA.1.1, and BA.2. Cell Host Microbe. 2022;30 doi: 10.1016/j.chom.2022.04.014. 1093.e3–1102.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerke A.R., Loening A.M., Gambhir S.S., Swartz J.R. Cell-free metabolic engineering promotes high-level production of bioactive Gaussia princeps luciferase. Metab. Eng. 2008;10:187–200. doi: 10.1016/j.ymben.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Barnes C.O., Weisblum Y., Schmidt F., Caskey M., Gaebler C., Cho A., Agudelo M., Finkin S., et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun. 2021;12:4196. doi: 10.1038/s41467-021-24435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Schommers P., Lehmann C., Kurth F., Sander L.E., Klein F. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat. Med. 2022;28:477–480. doi: 10.1038/s41591-021-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C., Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30:S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Krüger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.S., Winkler M.S., et al. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2022;185 doi: 10.1016/j.cell.2021.12.032. 447.e11–456.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Liao H., Meng Y., Li W., Han P., Liu K., Wang Q., Li D., Zhang Y., Wang L., et al. Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1. Cell. 2022;185 doi: 10.1016/j.cell.2022.06.023. 2952.e10–2960.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Mazurov D., Ilinskaya A., Heidecker G., Lloyd P., Derse D. Quantitative comparison of HTLV-1 and HIV-1 cell-to-cell infection with new replication dependent vectors. PLOS Pathog. 2010;6:e1000788. doi: 10.1371/journal.ppat.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu P., Faraone J., Evans J.P., Zou X., Zheng Y.M., Carlin C., Bednash J.S., Lozanski G., Mallampalli R.K., Saif L.J., et al. Neutralization of the SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 subvariants. N. Engl. J. Med. 2022;386:2526–2528. doi: 10.1056/NEJMc2206725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu P., Faraone J.N., Evans J.P., Zheng Y.M., Yu L., Ma Q., Carlin C., Lozanski G., Saif L.J., Oltz E.M., et al. Durability of booster mRNA vaccine against SARS-CoV-2 BA.2.12.1, BA.4, and BA.5 subvariants. N. Engl. J. Med. 2022 doi: 10.1056/NEJMc2210546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Tamura T., Zahradnik J., Deguchi S., Tabata K., Kimura I., Ito J., Nasser H., Toyoda M., Nagata K., et al. Preprint at bioRxiv. 2022 doi: 10.1101/2022.08.07.503115. [DOI] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheward D.J., Kim C., Fischbach J., Muschiol S., Ehling R.A., Björkström N.K., Karlsson Hedestam G.B., Reddy S.T., Albert J., Peacock T.P., Murrell B. Evasion of neutralizing antibodies Omicron sublineage BA.2.75. Preprint at bioRxiv. 2022 doi: 10.1101/2022.07.19.500716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuekprakhon A., Nutalai R., Dijokaite-Guraliuc A., Zhou D., Ginn H.M., Selvaraj M., Liu C., Mentzer A.J., Supasa P., Duyvesteyn H.M.E., et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185 doi: 10.1016/j.cell.2022.06.005. 2422.e13–2433.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Iketani S., Li Z., Guo Y., Yeh A.Y., Liu M., Yu J., Sheng Z., Huang Y., Liu L., Ho D.D. Antigenic characterization of the SARS-CoV-2 Omicron subvariant BA.2.75. Cell Host Microbe. 2022 doi: 10.1016/j.chom.2022.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ai J., Li X., Zhao X., Wu J., Zhang H., He X., Zhao C., Qiao R., Li M., et al. Preprint at bioRxiv. 2022 doi: 10.1101/2022.08.04.502716. [DOI] [Google Scholar]
- World Health Organization Tracking SARS-CoV-2 variants. 2022. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants [PubMed]
- Xie X., Zou J., Liu M., Ren P., Shi P.Y. Neutralization of SARS-CoV-2 Omicron sublineages by 4 doses of mRNA vaccine. Preprint at bioRxiv. 2022 doi: 10.1101/2022.07.29.502055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasoba D., Kimura I., Kosugi Y., Uriu K., Fujita S., Ito J., Sato K., The Genotype to Phenotype Japan (G2P-Japan) Consortium Preprint at bioRxiv. 2022 doi: 10.1101/2022.07.14.500041. [DOI] [Google Scholar]
- Yamasoba D., Kimura I., Nasser H., Morioka Y., Nao N., Ito J., Uriu K., Tsuda M., Zahradnik J., Shirakawa K., et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell. 2022;185 doi: 10.1016/j.cell.2022.04.035. 2103.e19–2115.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Evans J.P., Pearson R., Qu P., Zheng Y.M., Robinson R.T., Hall-Stoodley L., Yount J., Pannu S., Mallampalli R.K., et al. Neutralizing antibody against SARS-CoV-2 spike in COVID-19 patients, health care workers, and convalescent plasma donors. JCI Insight. 2020;5:e143213. doi: 10.1172/jci.insight.143213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Evans J.P., Qu P., Faraone J., Zheng Y.M., Carlin C., Bednash J.S., Zhou T., Lozanski G., Mallampalli R., et al. Neutralization and Stability of SARS-CoV-2 Omicron Variant. Preprint at bioRxiv. 2021 doi: 10.1101/2021.12.16.472934. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
NT50 values and de-identified patient information will be deposited to the National Cancer Institute SeroNet Coordinating Center. Additionally, NT50 values and de-identified patient information reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.