Abstract
The degree to which developmental systems bias the phenotypic effects of environmental and genetic variation, and how these biases affect evolution, is subject to much debate. Here, we assess whether developmental variability in beetle horn shape aligns with the phenotypic effects of plasticity and evolutionary divergence, yielding three salient results. First, we find that most pathways previously shown to regulate horn length also affect shape. Second, we find that the phenotypic effects of manipulating divergent developmental pathways are correlated with each other as well as multivariate fluctuating asymmetry—a measure of developmental variability. Third, these effects further aligned with thermal plasticity, population differences and macroevolutionary divergence between sister taxa and more distantly related species. Collectively, our results support the hypothesis that changes in horn shape—whether brought about by environmentally plastic responses, functional manipulations or evolutionary divergences—converge along ‘developmental lines of least resistance’, i.e. are biased by the developmental system underpinning horn shape.
Keywords: developmental bias, developmental plasticity, fluctuating asymmetry, geometric morphometrics, integration, Onthophagus
1. Background
Organismal form and function emerge through the integration of genetic and environmental factors through development, a process prone to produce some phenotypic outcomes more often than others [1–5] (a phenomenon referred to as developmental variability [6]). If so, development has the potential to bias direction and strength of phenotypic effects resulting from environmental or genetic influences, which in turn may shape evolutionary trajectories so as to align preferentially along primary axes of developmental variability [6]. Yet, the role of developmental variability and the resulting bias in making some evolutionary trajectories more likely than others remains disputed. Here, we investigate how developmental variability in complex morphologies relates to environmental plasticity and micro- as well as macroevolutionary variation.
It has long been recognized that evolution is more likely to occur in some directions than others. Most prominently, the genetic variance–covariance matrix (G) has been shown to bias evolutionary change along ‘genetic lines of least resistance’ (i.e. gmax, [7–9]). Genetic covariances arise through pleiotropy and linkage disequilibrium and ultimately reflect heritable (co)variation in developmental outputs. However, while G reflects genetic biases in evolution, it does not contain information about whether or how these biases relate to developmental variability and its role in evolution [6]. A better method for studying developmental variability is to investigate the phenotypic variance and covariance induced by mutation through the study of the mutational matrix M (e.g. [10–12]). Because mutational covariances (the off-diagonal entries of M) are driven by pleiotropy, M provides insights into how genetic effects are funnelled through developmental systems to shape phenotypic outcomes. M thus indicates the phenotypic directions in which (random) genetic perturbations are likely to have the strongest effects. However, developmental variability may be influenced by, but does not strictly require genetic (co)variation. By quantifying naturally occurring fluctuating asymmetry (FA), we here take an alternative approach to study how developmental variability relates to the effects of genetic and environmental perturbations and evolutionary divergence.
Despite generally strong stabilizing selection on developmental outputs, interactions occurring during ontogeny across levels of organization—from gene regulation and cellular transduction to tissue formation and endocrine signals—induce stochastic developmental variation, even in the absence of genetic or environmental perturbation [13,14]. Focusing on this intrinsic developmental stochasticity, we here take advantage of multivariate FA in trait shape to quantify developmental variability independent of environmental and genetic variation [15–17]. In bilaterally symmetric organisms, the left and the right side of the same individual share the exact same genotype and environment, yet, there are often slight differences between both sides that can be attributed to developmental interactions (after accounting for measurement error and directional asymmetry, and assuming somatic mutational effects are negligible, see [18]). Measurements of FA thus allow to disentangle genetic and environmental (co)variation from developmental (co)variation. FA therefore provides insights into alternative—but naturally occurring—morphological variants produced by the same developmental system. As such, FA reveals the degree and type of trait (co)variation (or integration, see [19]) that is produced by a developmental system itself and thus provides information about a trait's developmental architecture and variability. If this developmental variability biases the effects of genetic and environmental perturbations on phenotype formation, we expect FA to predict trait variation across several levels of biological organization. We test this hypothesis by quantifying developmental variability in the horns of dung beetles and by assessing the degree to which it predicts the phenotypic effects of (i) developmental genetic variation induced by functional genetic manipulations, (ii) environmental perturbations caused by thermal plasticity and (iii) evolutionary divergences among populations and species.
Males of the bull-headed dung beetle Onthophagus taurus develop a pair of exaggerated and strongly nutrition-sensitive head horns (see figure 1). Horn length has a sigmoidal scaling relationship with a steep threshold that separates small ‘minor’ males that only develop minute horns from large ‘major’ males that develop a pair of elongated head horns used in male–male combat ([20], see figure 1). The developmental processes and pathways involved in the nutritional plasticity and evolution of beetle horn morphology—including the doublesex, hedgehog, insulin and serotonin signalling pathways (reviewed in [21])—have been heavily investigated. However, so far, most studies have focused on measurements of horn length, which only capture one aspect of morphological variation. This has two major implications. First, other aspects of beetle horn morphology, such as components of shape, continue to be poorly understood despite strong intra- and interspecific variation (but see [22–24]). Second, the degree to which functional genetic manipulations recapitulate plastic and genetic variation in horn morphology remains entirely unclear. Using a multivariate approach, we here aim to quantify similarities between developmental and evolutionary covariation in horn shape while controlling for the effects of horn size and scaling.
Figure 1.
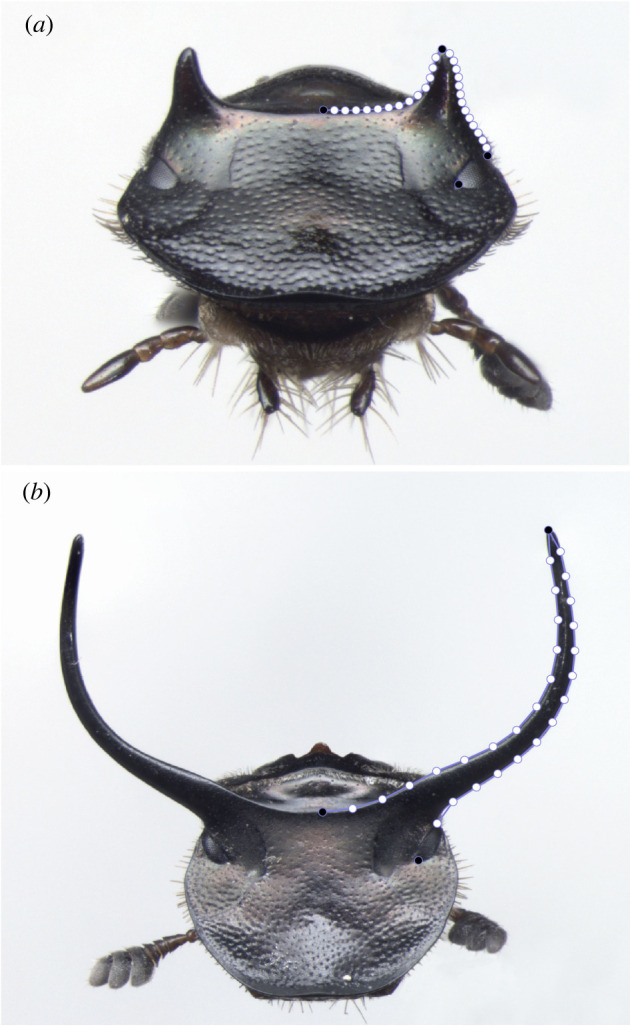
Males of the bull-headed dung beetle develop into minor (a) and major (b) horn morphologies. Shape variation was quantified using four landmarks (black circles) and 28 semi-landmarks (open circles). (Online version in colour.)
Specifically, we use geometric morphometrics to investigate the developmental genetic basis of horn shape variation and the role of developmental variability in its evolution. First, we assess patterns of naturally occurring FA in horn shape in wild-caught individuals as an estimate of developmental variability. Second, we reanalyse previous functional genetic manipulations of serotonin signalling [25], Notch signalling [23], limb patterning [26], sex determination [27], insulin signalling [28], Fat signalling and histone(de)acetylation [29] to test whether genes previously shown to affect horn length also impact horn shape, and assess the extent to which developmental variability predicts the phenotypic effects of functional genetic manipulations. Lastly, we ask whether developmental variability also predicts environmentally plastic responses and evolutionary differentiation on different evolutionary scales. Taken together, our data suggest that the architecture of developmental systems predicts the effects of functional genetic manipulations, environmental plasticity, as well as evolutionary divergence—a finding consistent with the hypothesis that developmental bias shapes genetic, environmental and evolutionary (co)variation over long timespans.
2. Material and methods
(a) . Data collection
To investigate the evolution and development of horn shape, we used a two-dimensional geometric morphometric approach. For each of the datasets described below (FA, functional genetic manipulations, environmental plasticity and evolutionary divergence), we took pictures of beetle head horns using a digital camera (Scion) mounted on a Leica MZ-16 stereomicroscope. The final dataset included measurements for several wild-caught and laboratory-reared populations of Onthophagus taurus, one wild-caught population of the closely related Onthophagus illyricus, as well as laboratory-reared individuals of the more distantly related Digitonthophagus gazella (see electronic supplementary material, table S1 for a description of treatment and morph-specific sample sizes).
TpsDig2 [30] was used to quantify horn shape using four landmarks and 26 semi-landmarks (following [23,24]; see figure 1). The landmarks of all 1044 specimens were subjected simultaneously to a Procrustes analysis in the R-package geomorph [31]. The position of semi-landmarks was optimized by minimizing bending energy. Centroid size was extracted as a shape-independent measure of overall structural size [32]. These Procrustes coordinates were then used for further analysis for each dataset separately (described below). Because horn shape shows strong morph-specific allometric variation [23], male morph (minor/intermediate versus major) and centroid size were included as fixed effects and as interactions with treatment. Because ‘intermediate’ males are very rare, we pooled intermediate males with minor males.
(b) . Fluctuating asymmetry as an estimate of developmental variability
To assess FA, we measured the shape of the left and right horn of 63 wild-caught major males collected in North Carolina and Virginia, USA. Photographs and measurements were taken twice to account for measurement error due to positioning and digitization (rendering a total number of 252 measurements for each of the 60 variables (30 two-dimensional landmarks)). The statistical significance of FA was assessed by testing for an individual-by-side interaction using a Procrustes ANOVA (using the individual-by-side-by-measurement as error term). The FA-component, i.e. the specimen-specific deviation from symmetry adjusted for directional asymmetry, was then extracted (using the function bilat.symmetry in the R-package geomorph [31]).
(c) . Functional genetic manipulations
To test whether developmental variability predicts the effects of developmental genetic perturbations, we tested how functional genetic manipulations affect horn shape. To this end, we measured individuals generated for several previous functional genetic experiments that investigated the effects of RNA interference (RNAi) or pharmacological manipulations of several major developmental pathways in the development of horn length. We chose these pathways because their previously documented function in the development of horn length raised the possibility that they may also be affecting horn shape, thereby putting us in a position to compare and contrast these effects to those of developmental variability as assessed by FA. Specifically, we chose functional genetic manipulations of the following pathways: (i) serotonin signalling: inhibition of serotonin biosynthesis through the application of alpha-methyl-p-tyrosine (AMPT) [25], (ii) Notch signalling: RNAi-mediated gene expression knockdown of serrate (ser) [23], (iii) limb patterning: Distal-less (Dll) RNAi [26], (iv) sex determination: doublesex (dsx) RNAi [27], (v) insulin signalling: Forkhead box, subgroup O (foxo) RNAi [28], (vi) Fat signalling: dachs (d) (Hu et al. in preparation) and (vii) histone (de)acetylation: histone deacetylase 3 (HDAC3) RNAi (Hu et al. in preparation). For each functional manipulation, we acquired Procrustes coordinates (as described above) for individuals subjected to RNAi or pharmacological treatments as well as the respective control injections. Procrustes ANOVAs (with randomized residual permutation procedure and type II sums of squares, as implemented in the R packages geomorph and RRPP [33]) were used to test for the effects of each developmental manipulation on horn shape. We fitted horn shape (Procrustes variables) as a function of log centroid size (CS), treatment (T), morph (M) and all interactions as follows:
where S represents the matrix of Procrustes shape variables (30 two-dimensional landmarks), is a vector of intercepts, is the vector of partial regression coefficients for centroid size, for treatment and for morph, respectively. is the error term. Non-significant interactions were removed. For each analysis, we only included those control individuals that were reared simultaneously with the treatment groups (e.g. we only compared Dll dsRNA-injected animals to control animals that were reared for the same experiment (i.e. [26]), and not all buffer-injected animals generated across studies).
To test whether functional genetic manipulations of different pathways had similar or divergent morphological effects on each morph, we quantified the shape deformation caused by each manipulation using separate multiple multivariate models. For each manipulation and morph, we fitted horn shape (Procrustes variables) as a function of treatment (T; i.e. dsRNA/pharmacological injection versus buffer injection) and log centroid size (CS), as follows:
where S represents a matrix of Procrustes shape variables and and are the vectors of partial coefficients relating to the shape deformations due to size and treatment, respectively. The treatment vector then describes the effect on horn shape caused by the manipulation of a specific pathway (e.g. serotonin signalling) while accounting for the confounding effects of allometric variation. If significant, the partial effects of treatment for each developmental manipulation were extracted and compared to each other. The similarity between shape effects was quantified using pairwise vector correlations as follows:
where the numerator denotes the dot product of the two vectors of partial coefficients for the effect of treatments j (e.g. dsxRNAi) and k (e.g. foxoRNAi) on shape, while the denominator represents their norms [34,35]. This vector correlation provides a quantitative assessment for the alignment between the effects of the two developmental manipulations being compared. Values close to 1 indicate that manipulations induce very similar shape changes. If r is close to 0, the shape effects of different manipulations are unrelated. Bias-corrected and accelerated bootstrap (BCa) confidence intervals were calculated based on 9999 non-parametric bootstrap replicates.
(d) . Comparing the phenotypic effects of developmental perturbations with fluctuating asymmetry
To test whether the shape changes due to FA aligned with developmental manipulations we used two complementary approaches. First, we extracted the variance–covariance matrix of the FA shape component D, which summarizes developmental covariance. To test whether the treatment effects of functional genetic manipulations align with D, we computed pairwise vector correlations between the vectors of coefficients related to treatment effects (e.g. ) and the first two principal components of D that together accounted for 64 per cent of the total variation. We then tested whether the observed vector correlations were larger than expected under a uniform distribution (see [36]). Although our dataset contains 60 landmark variables, Procrustes superimposition and semi-landmark sliding results in the loss of four dimensions for position, scaling, and rotation as well as approximately half a dimension for each semi-landmark, leading to the loss of 30 out of all 60 dimensions. We thus used the expected distribution of correlation coefficients under a unifrom distribution in 30 dimensions to assess the significance of our observed vector correlations.
Second, we quantified the amount of FA variance in the direction of the shape change vectors associated with the functional genetic manipulation as follows:
where β is the shape deformation vector and D is the variance–covariance matrix of the FA component (see [37] for a similar approach). This gives the amount of FA variation in the direction of the shape change caused by the developmental manipulation (this is equivalent to estimates of evolvability that quantify the amount of variation in the genetic (co)variance matrix G in the direction of a selection vector, e.g.: [38]). If developmental manipulations cause shape deformations primarily in directions of D, we expect to be large (i.e. close to the first eigenvalue of D). To test whether is larger than expected under a null model, we used two complementary approaches. First, we permuted the FA shape component 10 000 times. For each iteration, we calculated D and , and tested whether our observed value is larger than expected under this null model. Second, we generated 10 000 random variance–covariance matrices with the same dimensionality and trace as our original D. The first 30 eigenvalues of the simulated matrices were forced to be identical to those of D while the remaining eigenvalues were set to zero (using the R-package clusterGeneration [39]). We then tested whether the observed relationship between and D is larger than between and the simulated matrices.
(e) . Environmental plasticity
To test whether developmental variability is aligned with the effect of environmental plasticity, we compared FA to the effect of rearing temperature. To this end, we measured horn shape in males from four populations throughout the North American range that were reared at 19°C or 27°C (see [40]). Because very few minor males developed at 27°C, we were unable to test for temperature effects on minor/intermediate horn shape. A Procrustes ANOVA was used to test for the effects of rearing temperature (Tem) on horn shape while accounting for population differences (Pop) and log centroid size (CS) as follows:
We then extracted the partial coefficients of the temperature effect and compared it to the effects of functional genetic manipulations and FA as described above. The effect of population was not significant, indicating that the recently established populations in North America do not differ in horn shape.
(f) . Evolutionary divergence
We investigated the extent to which developmental variability predicts evolutionary divergence at three different levels. First, we tested for intraspecific population differences among O. taurus collected in the Eastern US and Western Australia [41], the two most divergent populations in terms of static horn length allometry and life history [42]. These animals were reared simultaneously under laboratory conditions (note that the analysis of thermal plasticity (above) did not indicate significant levels of divergence among populations in the recently established North American populations). Second, we investigated macroevolutionary divergence in horn shape between O. taurus and its sister species O. illyricus (collected at Monte Cucco, Italy in 2021). Both taxa are morphologically very similar although they diverged ca 3–4 Ma [43]. Lastly, to investigate horn divergence on a larger evolutionary scale, we also compared O. taurus to D. gazella, a species in a different genus that also develops a paired set of head horns with a similar morphology [24].
To test for evolutionary divergence at the population, sister species and genus level, we used Procrustes ANOVAs fitting horn shape as a function of evolutionary lineage (L) (i.e. American versus Australian population of O. taurus, O. taurus versus O.illyricus or O. taurus versus D. gazella) taking log centroid size and male morph, as well as all interactions into account. We then extracted partial coefficients of population or species divergence for each morph from the following model:
These partial coefficients were compared to the effects of functional genetic manipulations, FA and environmental plasticity using vector correlations as described above.
3. Results and discussion
(a) . Developmental variability in dung beetle horn shape
To quantify developmental variability, we investigated patterns of FA. Horn shape of major males showed significant levels of FA (individual-by-side interaction tested against the individual-by-side-by-measurement interaction: F62,126 = 3.29, p < 0.001, n = 252), indicating environment and genotype-independent developmental variation. FA was related to the curvature of the horn, suggesting that random developmental perturbations are most likely to cause changes in horn curvature, rather than other components of horn shape. If such developmental bias shapes plasticity and evolution, we expect the latter to fall along major axes of FA variation.
(b) . Correlated effects of functional manipulations and fluctuating asymmetry indicate developmental bias
All functional manipulations that were previously shown to affect horn length also affected horn shape, although the variance explained by treatment effects was often small in the multivariate models (see electronic supplementary material, table S2). These effects were often morph-specific and in some cases also affected nutritional scaling itself (see electronic supplementary material, table S2). Especially strong effects were found when manipulating dsx expression in minor males and d, HDAC3, and Dll in major males (figures 2 and 3). While some manipulations increased the relative width of the horn in major males (e.g. HDAC3RNAi, dRNAi and DllRNAi), a vast majority affected its curvature (figures 2 and 3). This was also evident when quantitatively comparing shape change vectors (figure 4). Almost all manipulations found in minor males correlated with each other. Similarly strong alignments were found in major males. This alignment could be explained if all functional genetic manipulations ultimately target the same developmental genetic, cellular transduction or metabolic pathway. However, the RNAi and pharmacological targets selected in this study manipulate products that do not, at least on the surface, function in the same process. For instance, Dll encodes a transcription factor that patterns proximo-distal axis formation by determining distal identity and is expressed in very few select tissue regions (e.g. the tip of the horn, [26]), foxo encodes a transcription factor that is best known for regulating growth responses to nutrient depletion across the entire body (e.g. [44]), HDAC3 encodes an enzyme that alters heterochromatin accessibility by removing acyl groups, dsx encodes an alternatively spliced transcription factor that conveys somatic sex identity [45] and in males is expressed in the entire horn primordium, whereas serotonin is a biogenic amine that has primarily been studied in its function as a neurotransmitter [46]. When considering the diversity of biochemical products targeted by our manipulations, the correspondence in phenotypic changes seen across treatments can therefore not be explained through their participation in a simple, singular developmental pathway. However, developmental systems giving rise to complex traits such as beetle horns often integrate inputs from diverse sources. Hence, although gene products may perform independent functions on the level of organelles, cells or tissues, they may well act in combination and/or through each other in the formation of larger structures. For instance, DSX may help determine which cells in the developing horn primordium are responsive to FOXO and which are not by designating a larger or smaller population of cells as male depending on nutritional conditions (somatic sex determination in insects is typically mosaic, with most epidermal cells not possessing any identity; [47]). Alternatively, or in addition, DSX and/or DLL may determine in a region-specific manner how HDAC3 shapes chromatin configurations in horn primordial cells, thereby regulating the accessibility of genes such as ser and d to other transcription factors. If so, the similarity in phenotypic effects across diverse manipulations would reflect the action of a tightly integrated network of developmental interactions that is more likely to affect the curvature of the horn than any other morphological aspect. In other words, the phenotypic effects of functional genetic manipulations might be driven by developmental variability.
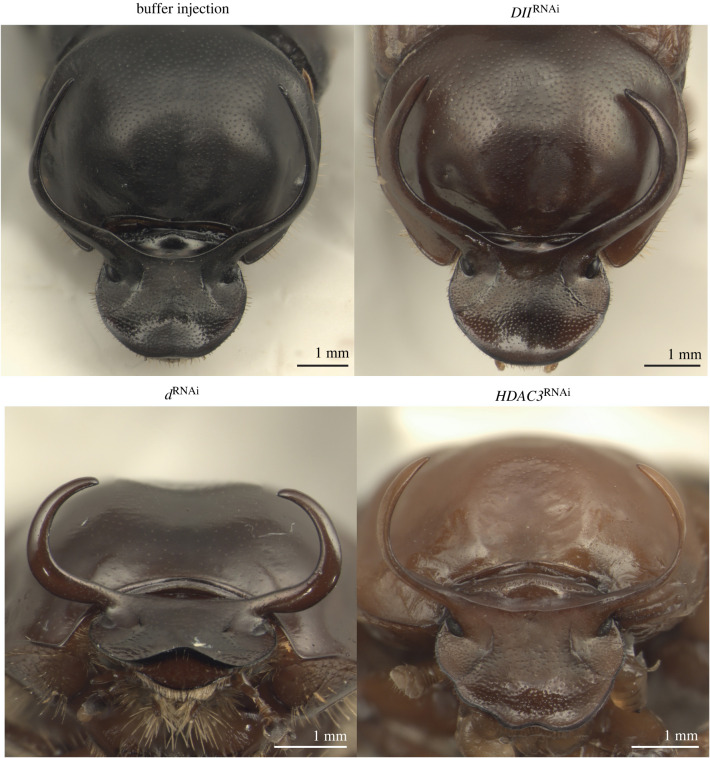
Figure 2.
Examples of the effect of functional genetic manipulations on major male horn shape. RNAi-mediated gene expression knockdown of Distal-less (DllRNAi), dachs (dRNAi) and Histone deacetylase 3 (HDAC3RNAi) had strong effects on horn shape and strongly increased the curvature of the horn compared to buffer-injected control animals. (Online version in colour.)
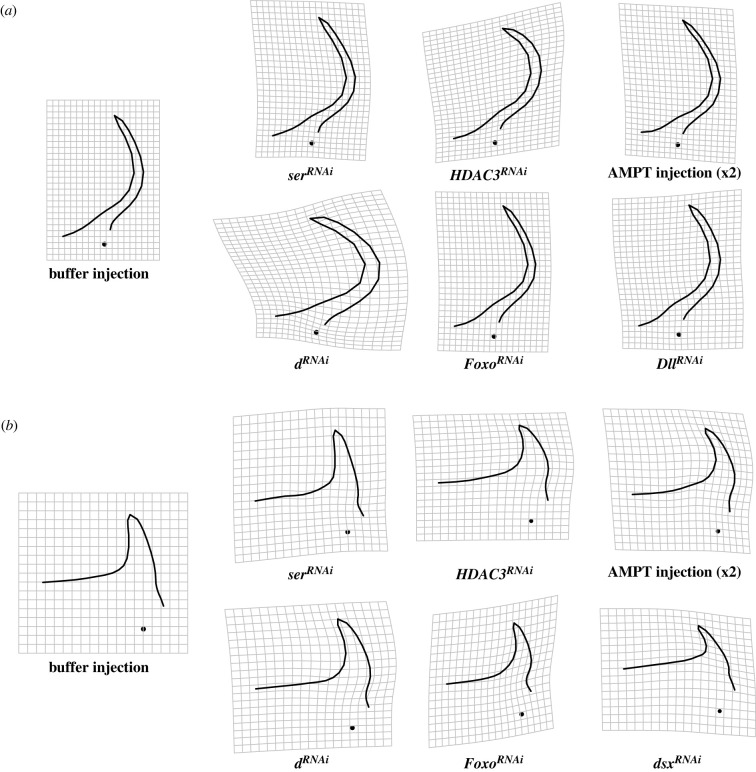
Figure 3.
Effect of functional genetic manipulations on major (a) and minor (b) horn shape. Deformation grids show changes relative to the average shape of control-injected individuals (the effect of serotonin signalling manipulation (AMPT injection) on horn shape was magnified twofold). DllRNAi and dsxRNAi could only be assessed in majors or minor males, respectively.
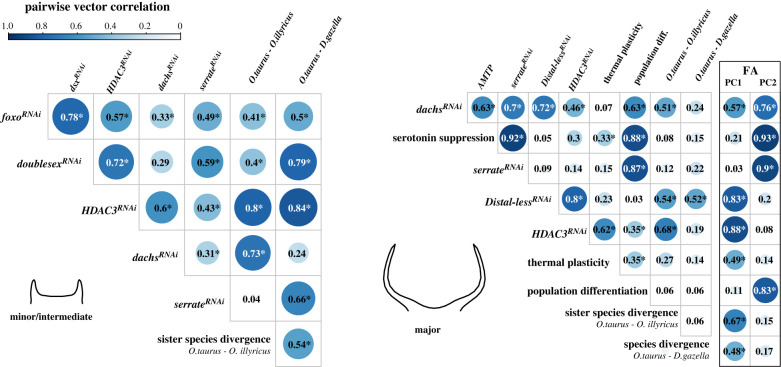
Figure 4.
Pairwise correlations between the effects of functional genetic manipulations, plasticity, evolutionary divergence and FA on horn shape (PC1 (explaining 39.4% of the total variation) and PC2 (24.8%) of the covariance matrix of the FA shape component). Only manipulations that had significant effects in the respective morph are shown. Correlations larger than expected under a uniform distribution in 30-dimensional space are indicated with an asterisk (see [36]). (Online version in colour.)
To assess whether functional manipulations align with the main axes of developmental variability, we next tested for an alignment with FA. Surprisingly, the effects of all developmental genetic manipulations were aligned with either the first or the second PC of the fluctuating asymmetric shape component (explaining 39.4 and 24.8% of the total variation, respectively; figure 4) and were more strongly aligned with the developmental (co)variance matrix D than expected by chance (all p < 0.001). This indicates that naturally occurring developmental covariance due to FA predicts the phenotypic effects of developmental manipulations induced by RNAi or pharmacological treatments. The similarities between the phenotypic effects of functional genetic manipulations of various, seemingly unrelated pathways and the association with FA therefore suggest that developmental variability shapes and channels the effects of developmental genetic perturbations.
(c) . Environmental plasticity and evolution along ‘developmental lines of least resistance’
To assess whether developmental variability also predicted environmental plasticity and heritable variation, we next investigated thermal plasticity and population and species divergence in horn shape. We found significant horn shape differences due to thermal plasticity (F1,97 = 3.00, p = 0.033, electronic supplementary material, table S3), population differentiation in major males between American and Australian populations (F1,66 = 13.86, p = 0.001; but not in minor males: F1,40 = 2.31, p = 0.100, electronic supplementary material, table S3), and species differences between O. taurus and its syntopic sister species O. illyricus (major males: F1,355 = 51.59, p < 0.001; minor males: F1,163 = 227.32, p < 0.001). These effects were again related primarily to the curvature of the horn (see figure 5) and were aligned with the effects of various developmental manipulations (figure 4). For instance, in major males, the effects of gene expression knockdowns of d, Dll and ser not only correlated with the effects of serotonin biosynthesis inhibition, but also the changes associated with population differentiation between North American and Australian populations that were first established in the late 1960s [48,49]. Similarly, the effects of DllRNAi and HDAC3RNAi correlated not just with each other but also with the interspecific differences between O. taurus and its sister species O. illyricus. While these alignments may indicate that the pathways are causally involved in shaping plastic or genetic divergence, it is also possible that the developmental bias discussed above might shape these overlapping patterns.
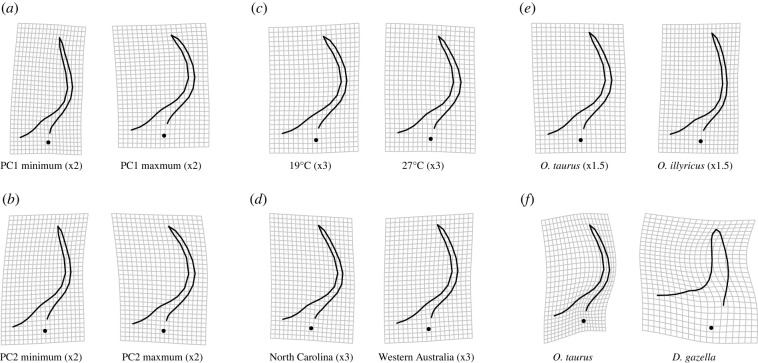
Figure 5.
Phenotypic changes associated with FA (PC1 (a) and PC2 (b)), thermal plasticity (c) and evolutionary divergence on different levels for major horn shape (d–f), (d) population divergence in O. taurus, (e) divergence between sister species and (f) divergence between distantly related speices. Pictures show deformations relative to the average shape (magnification is indicated).
To assess the potential role of developmental variability in shaping evolution on longer timescales, we next tested whether FA aligned with macroevolutionary divergences. We found that the vector of evolved differences between the sister species O. taurus and O. illyricus aligned with the direction of the first principal component of the developmental (co)variance matrix D (r = 0.67) and was more closely aligned with D than expected by chance (p < 0.001) even though the two species diverged ca 3–4 Ma [43]. This is consistent with the hypothesis that developmental architectures bias macroevolutionary divergence. Furthermore, D as well as functional genetic effects were related to evolved morphological difference between O. taurus and D. gazella, members of two lineages that shared their last common ancestor about 38 Ma [50]. These findings recapitulate patterns in other species where mutational covariation predicts evolutionary divergences across millions of years (e.g. [51]) and systems where developmental integration predicts phenotypic covariation among individuals [15–17,52]. Collectively, our results support the hypothesis that changes in horn shape—whether brought about by plastic responses to environmental change, functional genetic perturbations, or short and long-term evolutionary divergences—are biased by the developmental system underpinning horn shape.
It is unclear what mechanisms are responsible for the alignment between developmental variability and evolutionary divergence. One possibility is that developmental variability shapes the genetic covariation within a population (i.e. the G matrix), which, in turn, shapes evolutionary trajectories. If so, evolution along genetic lines of least resistance [8] may be underpinned by ‘developmental lines of least resistance’. That is, the degree to which genetic variation at a locus is able to contribute to phenotypic changes is channelled by the developmental systems within which this genetic variation contributes to interactions among component parts. The observation that FA correlates with evolutionary divergences on the level of populations and species further suggests that evolution does not ‘override’ the effects of developmental covariation.
Lastly, while developmental bias may be an emerging property of the mechanics of development, it is of course also an evolved property. This raises the possibility that the alignments between developmental variability and genetic divergence reported here could result from developmental biases shaped by prior periods of selection [6,12,53]. Beetle horns function in direct male–male combat and, at least horn length, is related to the likelihood of winning [20]. Horn morphology matches ecology [54] and, in some species, horn shape matches fighting styles [55,56]. Horn shape is thus very likely to be under strong (nonlinear) selection. Future studies in closely related species and estimates of multivariate selection on horn shape will be necessary to assess whether developmental variability biases responses to selection, or whether, in addition, developmental variability evolves such as to align with axes of variation favoured by selection.
4. Conclusion
Our multivariate analysis of developmental, environmental and evolutionary variation in horn shape indicates that developmental variability of complex traits aligns with plastic and micro- as well as macroevolutionary variation. This is consistent with a major role of developmental variability (or bias) in shaping phenotypic variation, plasticity and evolutionary changes over considerable timespans. The finding that FA aligns with the effects of functional genetic manipulations suggests that FA, which has mostly been discussed in terms of developmental instability and integration [16,19], may be a useful tool to study the role of developmental bias in evolution. More broadly, our study adds to the growing literature suggesting that developmental architectures influence evolutionary dynamics. Future research will be necessary to investigate whether developmental variability is adaptive and shaped by selection.
Acknowledgements
We would like to thank two anonymous reviewers for helpful comments on previous versions of this manuscript. We also thank David Berger and David Polly for insightful discussions about the statistical analyses.
Data accessibility
All data and code underlying this study will be made publicly available on the Dryad Digital Repository: https://doi.org/10.5061/dryad.ksn02v77n [57].
Electronic supplementary material is available online [58].
Authors' contributions
P.T.R.: conceptualization, data curation, formal analysis, funding acquisition, methodology, visualization, writing—original draft and writing—review and editing; Y.H.: data curation, methodology and validation; A.P.M.: conceptualization, funding acquisition, project administration, resources and writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This work was supported by two postdoc fellowships by the Swiss National Science Foundation (grant nos. P2ZHP3_184003 and P400PB_199257 to P.T.R.). Additional support was provided by National Science Foundation (grant nos. IOS 1256689 and 1901680) as well as grant 61369 from the John Templeton Foundation. The opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the National Science Foundation or the John Templeton Foundation.
References
- 1.Maynard Smith J, Burian R, Kauffman S, Alberch P, Campbell J, Goodwin B, Lande R, Raup D, Wolpert L. 1985. Developmental constraints and evolution: a perspective from the Mountain Lake Conference on Development and Evolution. Q. Rev. Biol. 60, 265-287. ( 10.1086/414425) [DOI] [Google Scholar]
- 2.Kavanagh KD, Shoval O, Winslow BB, Alon U, Leary BP, Kan A, Tabin CJ. 2013. Developmental bias in the evolution of phalanges. Proc. Natl Acad. Sci. USA 110, 18 190-18 195. ( 10.1073/pnas.1315213110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fritz JA, Brancale J, Tokita M, Burns KJ, Hawkins MB, Abzhanov A, Brenner MP. 2014. Shared developmental programme strongly constrains beak shape diversity in songbirds. Nat. Commun. 5, 3700. ( 10.1038/ncomms4700) [DOI] [PubMed] [Google Scholar]
- 4.Hendrikse JL, Parsons TE, Hallgrimsson B. 2007. Evolvability as the proper focus of evolutionary developmental biology. Evol. Dev. 9, 393-401. ( 10.1111/j.1525-142X.2007.00176.x) [DOI] [PubMed] [Google Scholar]
- 5.Brattstrom O, Aduse-Poku K, van Bergen E, French V, Brakefield PM. 2020. A release from developmental bias accelerates morphological diversification in butterfly eyespots. Proc. Natl Acad. Sci. USA 117, 27 474-27 480. ( 10.1073/pnas.2008253117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uller T, Moczek AP, Watson RA, Brakefield PM, Laland KN. 2018. Developmental bias and evolution: a regulatory network perspective. Genetics 209, 949-966. ( 10.1534/genetics.118.300995) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lande R. 1979. Quantitative genetic analysis of multivariate evolution, applied to brain:body size allometry. Evolution 33, 402-416. ( 10.1111/j.1558-5646.1979.tb04678.x) [DOI] [PubMed] [Google Scholar]
- 8.Schluter D. 1996. Adaptive radiation along genetic lines of least resistance. Evolution 50, 1766-1774. ( 10.1111/j.1558-5646.1996.tb03563.x) [DOI] [PubMed] [Google Scholar]
- 9.Careau V, Wolak ME, Carter PA, Garland T. 2015. Evolution of the additive genetic variance–covariance matrix under continuous directional selection on a complex behavioural phenotype. Proc. R. Soc. B 282, 20151119. ( 10.1098/rspb.2015.1119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camara MD, Pigliucci M. 1999. Mutational contributions to genetic variance–covariance matrices: an experimental approach using induced mutations in Arabidopsis thaliana. Evolution 53, 1692-1703. [DOI] [PubMed] [Google Scholar]
- 11.Houle D, Fierst J. 2012. Properties of spontaneous mutational variances and covariances for wing size and shape in Drosophila melanogaster. Evolution 67, 1116-1130. ( 10.1111/j.1558-5646.2012.01838.x) [DOI] [PubMed] [Google Scholar]
- 12.Jones AG, Arnold SJ, Burger R. 2007. The mutation matrix and the evolution of evolvability. Evolution 61, 727-745. ( 10.1111/j.1558-5646.2007.00071.x) [DOI] [PubMed] [Google Scholar]
- 13.Klingenberg CP. 2019. Phenotypic plasticity, developmental instability, and robustness: the concepts and how they are connected. Front. Ecol. Evol. 7, 1-15. ( 10.3389/fevo.2019.00056) [DOI] [Google Scholar]
- 14.Møller AP, Swaddle JP. 1997. Asymmetry, developmental stability and evolution. Oxford, UK: Oxford University Press. [Google Scholar]
- 15.Klingenberg CP, McIntyre GS. 1998. Geometric morphometrics of developmental instability: analyzing patterns of fluctuating asymmetry with Procrustes methods. Evolution 52, 1363-1375. ( 10.1111/j.1558-5646.1998.tb02018.x) [DOI] [PubMed] [Google Scholar]
- 16.Goswami A, Binder WJ, Meachen J, O'Keefe FR. 2015. The fossil record of phenotypic integration and modularity: a deep-time perspective on developmental and evolutionary dynamics. Proc. Natl Acad. Sci. USA 112, 4891-4896. ( 10.1073/pnas.1403667112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badyaev AV, Foresman KR. 2004. Evolution of morphological integration. I. Functional units channel stress-induced variation in shrew mandibles. Am. Nat. 163, 868-879. ( 10.1086/386551) [DOI] [PubMed] [Google Scholar]
- 18.Klingenberg CP. 2015. Analyzing fluctuating asymmetry with geometric morphometrics: concepts, methods, and applications. Symmetry 7, 843-934. ( 10.3390/sym7020843) [DOI] [Google Scholar]
- 19.Klingenberg CP. 2014. Studying morphological integration and modularity at multiple levels: concepts and analysis. Phil. Trans R. Soc. B 369, 20130249. ( 10.1098/rstb.2013.0249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moczek AP, Emlen DJ. 2000. Male horn dimorphism in the scarab beetle, Onthophagus taurus: do alternative reproductive tactics favour alternative phenotypes? Anim. Behav. 59, 1273-1273. ( 10.1006/anbe.1999.1342) [DOI] [PubMed] [Google Scholar]
- 21.Casasa S, Schwab DB, Moczek AP. 2017. Developmental regulation and evolution of scaling: novel insights through the study of Onthophagus beetles. Curr. Opin. Insect Sci. 19, 52-60. ( 10.1016/j.cois.2016.11.004) [DOI] [PubMed] [Google Scholar]
- 22.Emlen DJ, Marangelo J, Ball B, Cunningham CW. 2005. Diversity in the weapons of sexual selection: horn evolution in the beetle genus Onthophagus (Coleoptera: Scarabaeidae). Evolution 59, 1060-1084. ( 10.1111/j.0014-3820.2005.tb01044.x) [DOI] [PubMed] [Google Scholar]
- 23.Crabtree JR, Macagno ALM, Moczek AP, Rohner PT, Hu Y. 2020. Notch signaling patterns head horn shape in the bull-headed dung beetle Onthophagus taurus. Dev. Genes Evol. 230, 213-225. ( 10.1007/s00427-020-00645-w) [DOI] [PubMed] [Google Scholar]
- 24.Rohner PT, Linz DM, Moczek AP. 2021. Doublesex mediates species-, sex-, environment- and trait-specific exaggeration of size and shape. Proc. R. Soc. B 288, 20210241. ( 10.1098/rspb.2021.0241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwab DB, Newsom KD, Moczek AP. 2020. Serotonin signaling suppresses the nutrition-responsive induction of an alternate male morph in horn polyphenic beetles. J. Exp. Zool. A Ecol. Integr. Physiol. 333, 660-669. ( 10.1002/jez.2413) [DOI] [PubMed] [Google Scholar]
- 26.Moczek AP, Rose DJ. 2009. Differential recruitment of limb patterning genes during development and diversification of beetle horns. Proc. Natl Acad. Sci. USA 106, 8992-8997. ( 10.1073/pnas.0809668106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kijimoto T, Moczek AP, Andrews J. 2012. Diversification of doublesex function underlies morph-, sex-, and species-specific development of beetle horns. Proc. Natl Acad. Sci. USA 109, 20 526-20 531. ( 10.1073/pnas.1118589109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casasa S, Moczek AP. 2018. Insulin signalling's role in mediating tissue-specific nutritional plasticity and robustness in the horn-polyphenic beetle Onthophagus taurus. Proc. R. Soc. B 285, 20181631. ( 10.1098/rspb.2018.1631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y, Crabtree JR, Macagno ALM, Moczek AP. In preparation. Histone deacetylases regulate organ-specific growth in horned beetles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohlf FJ. 2009. Tpsdig. 2.14. Department of Ecology and Evolution, State University of New York. See http://life.bio.sunysb.edu/morph/index.html. [Google Scholar]
- 31.Adams DC, Collyer ML, Kaliontzopoulou A, Balken EK. 2021. Geomorph: software for geometric morphometric analyses. R package version 4.0. See https://cran.r-project.org/package=geomorph. [Google Scholar]
- 32.Klingenberg CP. 2016. Size, shape, and form: concepts of allometry in geometric morphometrics. Dev. Genes Evol. 226, 113-137. ( 10.1007/s00427-016-0539-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collyer ML, Adams DC. 2019. RRPP: linear model evaluation with randomized residuals in a permutation procedure. R package version 0.4.0. See https://CRAN.R-project.org/package=RRPP. [Google Scholar]
- 34.Claude J. 2008. Morphometrics with R. New York, NY: Springer. [Google Scholar]
- 35.Rohner PT, Roy J, Schafer MA, Blanckenhorn WU, Berger D. 2019. Does thermal plasticity align with local adaptation? An interspecific comparison of wing morphology in sepsid flies. J. Evol. Biol. 32, 463-475. ( 10.1111/jeb.13429) [DOI] [PubMed] [Google Scholar]
- 36.Klingenberg CP, Marugan-Lobon J. 2013. Evolutionary covariation in geometric morphometric data: analyzing integration, modularity, and allometry in a phylogenetic context. Syst. Biol. 62, 591-610. ( 10.1093/sysbio/syt025) [DOI] [PubMed] [Google Scholar]
- 37.Johansson F, Watts PC, Sniegula S, Berger D. 2021. Natural selection mediated by seasonal time constraints increases the alignment between evolvability and developmental plasticity. Evolution 75, 464-475. ( 10.1111/evo.14147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen TF, Houle D. 2008. Measuring and comparing evolvability and constraint in multivariate characters. J. Evol. Biol. 21, 1201-1219. ( 10.1111/j.1420-9101.2008.01573.x) [DOI] [PubMed] [Google Scholar]
- 39.Qiu W, Joe H. 2020. Clustergeneration: random cluster generation (with specified degree of separation). R package version 1.3.7. See https://CRAN.R-project.org/package=clusterGeneration [Google Scholar]
- 40.Rohner PT, Moczek AP. 2020. Rapid differentiation of plasticity in life history and morphology during invasive range expansion and concurrent local adaptation in the horned beetle Onthophagus taurus. Evolution 74, 2059-2072. ( 10.1111/evo.14045) [DOI] [PubMed] [Google Scholar]
- 41.Macagno ALM, Edgerton TJ, Moczek AP. 2021. Incipient hybrid inferiority between recently introduced, diverging dung beetle populations. Biol. J. Linn. Soc. 132, 931-944. ( 10.1093/biolinnean/blaa228) [DOI] [Google Scholar]
- 42.Moczek AP. 2009. Phenotypic plasticity and the origins of diversity: a case study on horned beetles. In Phenotypic plasticity of insects: mechanisms and consequences (eds Whitman DW, Ananthakrishnan TN), pp. 81-134. Enfield, UK: Science Publishers, Inc. [Google Scholar]
- 43.Pizzo A, Roggero A, Palestrini C, Cervella P, Del Pero M, Rolando A. 2006. Genetic and morphological differentiation patterns between sister species: the case of Onthophagus taurus and Onthophagus illyricus (Coleoptera, Scarabaeidae). Biol. J. Linn. Soc. 89, 197-211. ( 10.1111/j.1095-8312.2006.00674.x) [DOI] [Google Scholar]
- 44.Zeng B, Huang Y, Xu J, Shiotsuki T, Bai H, Palli SR, Huang Y, Tan A. 2017. The FOXO transcription factor controls insect growth and development by regulating juvenile hormone degradation in the silkworm, Bombyx mori. J. Biol. Chem. 292, 11 659-11 669. ( 10.1074/jbc.M117.777797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kopp A. 2012. Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 28, 175-184. ( 10.1016/j.tig.2012.02.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berger M, Gray JA, Roth BL. 2009. The expanded biology of serotonin. Annu. Rev. Med. 60, 355-366. ( 10.1146/annurev.med.60.042307.110802) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinett CC, Vaughan AG, Knapp JM, Baker BS. 2010. Sex and the single cell. II. There is a time and place for sex. PLoS Biol. 8, e1000365. ( 10.1371/journal.pbio.1000365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fincher G, Woodruff R. 1975. A European dung beetle, Onthophagus taurus Schreber, new to the U.S. (Coleoptera: Scarabaeidae). Coleopterists Bullet. 29, 349-350. [Google Scholar]
- 49.Tyndale-Biscoe M. 1996. Australia's introduced dung beetles: original release and redistributions. Canberra ACT, Australia: CSIRO Entomology. [Google Scholar]
- 50.Breeschoten T, Doorenweerd C, Tarasov S, Vogler AP. 2016. Phylogenetics and biogeography of the dung beetle genus Onthophagus inferred from mitochondrial genomes. Mol. Phylogenet. Evol. 105, 86-95. ( 10.1016/j.ympev.2016.08.016) [DOI] [PubMed] [Google Scholar]
- 51.Houle D, Bolstad GH, van der Linde K, Hansen TF. 2017. Mutation predicts 40 million years of fly wing evolution. Nature 548, 447-450. ( 10.1038/nature23473) [DOI] [PubMed] [Google Scholar]
- 52.Willmore KE, Klingenberg CP, Hallgrimsson B. 2005. The relationship between fluctuating asymmetry and environmental variance in rhesus macaque skulls. Evolution 59, 898-909. ( 10.1111/j.0014-3820.2005.tb01763.x) [DOI] [PubMed] [Google Scholar]
- 53.Cheverud J. 1984. Quantitative genetics and developmental constraints on evolution by selection. J. Theor. Biol. 110, 155-171. ( 10.1016/S0022-5193(84)80050-8) [DOI] [PubMed] [Google Scholar]
- 54.Emlen DJ. 2001. Costs and the diversification of exaggerated animal structures. Science 291, 1534-1536. ( 10.1126/science.1056607) [DOI] [PubMed] [Google Scholar]
- 55.Madewell R, Moczek AP. 2006. Horn possession reduces maneuverability in the horn-polyphenic beetle, Onthophagus nigriventris. J. Insect. Sci. 6, 1-10. ( 10.1673/2006_06_21.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCullough EL, Tobalske BW, Emlen DJ. 2014. Structural adaptations to diverse fighting styles in sexually selected weapons. Proc. Natl Acad. Sci. USA 111, 14 484-14 488. ( 10.1073/pnas.1409585111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohner PT, Hu Y, Moczek AP. 2022. Data from: Developmental bias in the evolution and plasticity of beetle horn shape. Dryad Digital Repository. ( 10.5061/dryad.ksn02v77n) [DOI] [PMC free article] [PubMed]
- 58.Rohner PT, Hu Y, Moczek AP. 2022. Developmental bias in the evolution and plasticity of beetle horn shape. Figshare. ( 10.6084/m9.figshare.c.6210975) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Rohner PT, Hu Y, Moczek AP. 2022. Data from: Developmental bias in the evolution and plasticity of beetle horn shape. Dryad Digital Repository. ( 10.5061/dryad.ksn02v77n) [DOI] [PMC free article] [PubMed]
- Rohner PT, Hu Y, Moczek AP. 2022. Developmental bias in the evolution and plasticity of beetle horn shape. Figshare. ( 10.6084/m9.figshare.c.6210975) [DOI] [PMC free article] [PubMed]
Data Availability Statement
All data and code underlying this study will be made publicly available on the Dryad Digital Repository: https://doi.org/10.5061/dryad.ksn02v77n [57].
Electronic supplementary material is available online [58].