Abstract
In the cyanobacterium Synechococcus elongatus, cell division is regulated by a circadian clock. Deletion of the circadian clock gene, kaiC, abolishes rhythms of gene expression and cell division timing. Overexpression of the ftsZ gene halted cell division but not growth, causing cells to grow as filaments without dividing. The nondividing filamentous cells still exhibited robust circadian rhythms of gene expression. This result indicates that the circadian timing system is independent of rhythmic cell division and, together with other results, suggests that the cyanobacterial circadian system is stable and well sustained under a wide range of intracellular conditions.
Two particularly important periodic biological events are circadian rhythms and the cell division cycle (CDC). The CDC operates in all growing organisms, and circadian rhythms are widely found in organisms from prokaryotic cyanobacteria to essentially all eukaryotes, including protista, fungi, plants, and animals up to human beings (7, 13, 20, 38, 41). Fundamental characteristics of circadian rhythms which define and distinguish them from other periodic phenomena in living organisms are (i) they are endogenous and genetically determined, (ii) the rhythms continue with a ∼24-h period under constant conditions that is entrainable by environmental cycles, and (iii) the period length is compensated for changes in ambient temperature over a wide range of physiologically relevant temperatures (3, 12, 37, 42). Despite the fact that circadian rhythms display features that are not shared by the CDC oscillator (such as temperature compensation), some researchers have suggested that there might be a bidirectional interdependent linkage between these two oscillating systems (13, 14, 23). In the case of other circadian rhythms, there is evidence of such bidirectional linkage in that some outputs can feed back onto the central oscillator (36). For circadian rhythm-CDC coupling, other researchers have favored an alternative hypothesis that the circadian clock mechanism oscillates independently of the status of the CDC, but the CDC is dependent on the phase of the circadian clock such that cell division is gated to occur only in specific circadian phases (13, 14, 16, 35).
Cyanobacteria are the simplest organisms in which circadian rhythms have been clearly documented (20). Over the past 15 years, many circadian rhythms including rhythms of photosynthetic activity, nitrogen fixation, global gene expression, and—most relevant for this study—cell division have been found in several cyanobacterial species. In fact, among photosynthetic organisms, our knowledge of clock components and interactions is most highly advanced in the unicellular cyanobacterium Synechococcus elongatus. The kai genes that are intimately involved in circadian timekeeping in Synechococcus have been cloned (18), and their homologs have been found in other cyanobacterial species as well as in archaea (6, 20, 21, 22, 29, 33). Deletion of any one of the kai genes does not affect viability (in single-strain cultures) but causes arhythmicity. As had been suggested for other model organisms such as mice, flies, and fungi (17, 28, 30, 43), Ishiura et al. (18) proposed for cyanobacteria that transcriptional and/or translational control of circadian clock genes by their own products (negative feedback regulation) is essential for circadian timekeeping. Biochemical and biophysical analyses of the processes by which the kai genes and their products are involved in circadian timekeeping are under way (19, 20).
We previously reported that cell division in S. elongatus is gated by a circadian oscillator (35). In light-dark (LD) cycles, division occurs only in the light phase. In constant light (LL), where growth is apparently continuous, the cells divide in the subjective day and late subjective night but are kept from dividing in the early subjective night by the circadian oscillator. In this study, we demonstrate that the same kai-dependent clock that regulates gene expression also controls cell division. The bacterial cell division gene ftsZ is expressed with a circadian pattern in Synechococcus. Most importantly, overexpression of FtsZ protein stops the cells from dividing while they continue to grow (resulting in filamentous cells) but does not affect circadian rhythms of gene expression. These results indicate that the circadian pacemaker that gates cell division and gene expression in Synechococcus oscillates stably and independently of feedback from the CDC.
MATERIALS AND METHODS
Strains and culture conditions.
S. elongatus PCC 7942 (wild type; also known as Anacystis nidulans R2 or Synechococcus sp. strain PCC 7942) and derivative strains were grown in modified BG-11 medium (15) at 30°C. Depending on the antibiotic resistance of each strain, spectinomycin (20 μg/ml), kanamycin (12.5 μg/ml), and/or chloramphenicol (10 μg/ml) was added to the medium. Continuous culturing and measurement of cell density of cultures were performed as previously described (35, 46).
Cloning and sequencing of ftsZ.
Plasmid isolation, restriction digestion, ligation, transformation, and Southern blotting were performed essentially as described by Sambrook et al. (40). To make the ftsZ gene probe for screening genomic libraries, genomic PCR was performed with the primers FZTM1 (5′-CCT GAA TTC AAY ACN GAY GNC ARG C-3′) and FZTM2 (5′-CCT GAA TTC GTN CCN GTN CCN CCN CCC CAT-3′). These primers were the same as those used by Zhang and coworkers for cloning ftsZ from Anabaena (11, 47). PCR mixtures of 100 μl contained 1 μmol of Tris-HCl (pH 9), 5 μmol of KCl, 150 nmol of MgCl2, 1 μg of Triton X-100, 20 nmol of each of the deoxynucleoside triphosphates, 50 pmol of each primer, 75 ng of genomic DNA, and 2.5 U of Taq DNA polymerase (Promega, Madison, Wis.). PCR was performed on a Perkin-Elmer DNA thermal cycler (Applied Biosystems, Foster City, Calif.) with an initial hot start at 95°C followed by 30 cycles at 95°C (1 min), 55°C (1 min), and 74°C (2 min) and a final polymerization step of 74°C for 7 min. One major 220-bp DNA fragment was amplified by PCR. This PCR product was cleaved with EcoRI, cloned into the EcoRI site of pBluescript II vector (Stratagene, La Jolla, Calif.), and sequenced. The resulting sequence showed strong similarity to known ftsZ sequences, and the PCR product was used as a probe for genomic Southern analyses and for screening a cosmid library. To screen the cosmid library (provided by Susan Golden), hybridization was performed to a digoxigenin-labeled PCR probe (∼20 ng/ml) in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% N-lauroylsarcosine–0.2% sodium dodecyl sulfate–1% blocking reagent (Roche, Indianapolis, Ind.) at 68°C for 16 h, and hybridization of the probe on the filters was detected by the immuno-chemiluminescence method as recommended by the manufacturer (Roche). DNA fragments containing the ftsZ gene were isolated from identified cosmid clones, subcloned into a plasmid vector (pMT111 or pMT115), and sequenced. Plasmid templates for DNA sequencing were sequenced from the insert ends with T3, T7, M13 primers or with custom oligonucleotide primers. All double-stranded DNA templates were prepared with a Wizard Plasmid Miniprep kit (Promega) or by PCR and sequenced on an ABI377 DNA sequencer (Applied Biosystems). Database searches for similarity with other proteins were performed with BLASTP and TBLASTN (1) at the National Center for Biotechnology Information, National Institutes of Health, via the Internet.
RNA analyses.
Cells were grown in 24-hour LD cycles (LD 12:12) and harvested. The culture was mixed with crushed ice to immediately chill the cells, and then the cell suspension was centrifuged at 15,000 × g at 4°C for 5 min. The cell pellets were frozen in liquid nitrogen and stored at −80°C. Total RNA was isolated by a modification of the method of Chomczynski and Sacchi (9). Five milliliters of TRI-Reagent (Molecular Research Center, Cincinnati, Ohis) was added to the frozen cell pellet in 13-ml centrifuge tubes, and the samples were vortexed with 2 g of glass beads for 5 min at room temperature. The homogenates were centrifuged at 10,000 × g at 4°C for 10 min; then the supernatants (∼3.6 ml of each) were transferred into new tubes to which 360 μl of 1-bromo-3-chloropropane was added and vortexed vigorously for 15 s. The mixture was incubated at room temperature for 10 min and then spun at 10,000 × g for 15 min (4°C). About 2.2 ml of the aqueous phase was transferred to a new tube, and 2.2 ml of isopropyl alcohol was added and kept on ice for 10 min. Total RNA was collected by centrifugation at 12,000 × g at 4°C for 8 min, washed with 75% ethanol, dried, dissolved in water, and stored at −80°C.
For Northern analyses, RNA was separated by electrophoresis in 1.0% agarose gels under denaturing conditions. After blotting onto a nylon membrane, an ftsZ-specific RNA probe was used to hybridize to ftsZ mRNA on the membrane.
For primer extension, the 5′ ends of primers FZPE1 (5′-ATC GAA CCC CGA CAG AGA GCC GTC AC-3′) and FZPE2 (5′-ATC GGC ATA GGG TCG GTC AT-3′) were labeled with [γ-32P]ATP (<5,000 Ci/mmol; NEN Life Science Products, Boston, Mass.) using T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.). Ninety-eight micrograms of total RNA and 2.4 pmol of 32P-labeled primer (∼106 cpm) in 30 μl of hybridization buffer (40 mM piperazine-N,N'-bis[2-ethanesulfonic acid] [PIPES], 1 mM EDTA, 0.4 M NaCl, 80% formamide [pH 6.4]) were denatured by heating at 85°C for 10 min and then incubated at 32°C overnight. RNA-primer hybrids were recovered by ethanol precipitation, air dried, and dissolved in 20 μl of reverse transcriptase reaction mixture containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 20 U of RNase inhibitor (Ambion, Austin, Tex.), and 100 U of SuperScript II RNase H− reverse transcriptase (Life Technologies, Gaithersburg, Md.). The reaction was performed at 42°C for 1 h and terminated by heating at 70°C for 10 min. RNA in the mixture was removed with RNase A. Ten micrograms of salmon sperm DNA was added as a carrier, and cDNAs were extracted with phenol-chloroform-isoamyl alcohol (25:24:1), precipitated with ethanol, and then analyzed by electrophoresis through 6% polyacrylamide–7 M urea gels. The sequencing ladders to determine the endpoints of primer extensions were prepared with the same 32P-labeled primer using pMT115 as the template by cycle sequencing (Seq Therm EXCELL II kit; Epicentre Technologies, Madison, Wis.). The gels were dried and then exposed to X-ray film (X-Omat AR; Eastman Kodak, Rochester, N.Y.) without intensifying screens at −80°C for 1 to 3 days.
Construction of the luciferase reporter strains and in vivo luminescence measurement.
To construct ftsZ promoter (ftsZp)-bacterial luciferase transcriptional fusions, the vectors pAM1580 (2) and pAM1583I (2) (modified by M. Izumo [unpublished data]) were used for construction of the luxAB reporter strain. These vectors carry a promoterless luxAB gene set and transfer inserts to the neutral sites of the Synechococcus chromosome by homologous recombination. The upstream region of the ftsZ gene was isolated after restriction digestion of the ftsZ clone or amplified by PCR using Pfu DNA polymerase (Stratagene) and cloned into the unique StuI site upstream of the luxAB genes in the vectors. The constructs were linearized with NdeI to avoid homologous single recombination and then introduced into neutral site I (8) or neutral site II (2) of the wild-type Synechococcus chromosome by homologous double recombination (2, 15). Antibiotic-resistant colonies were selected, and correct transformations were confirmed by genomic PCR or Southern analysis. In vivo luminescence was measured as previously described by Kondo et al. (24, 26) and Mori et al. (35).
Overexpression of ftsZ in E. coli and cyanobacteria.
Using primers TRCFZ1 (5′-TCG AGC TCA AGG AGG AAT AAC ATA TGA CCG ACC CTA TGC CGA TC-3′) and TRCFZ2 (5′-TGG GAT CCC ATA TGC TAG GGT CGG TTT TGA ATT TTC CG-3′) and the cosmid 8B5 as a template, we amplified a 1,215-bp SacI/BamHI fragment which contains the ftsZ gene (underlines denote created SacI and BamHI sites). An alternative ribosome binding site was introduced just upstream of the ftsZ open reading frame (ORF). This 1,215-bp SacI/BamHI fragment was inserted into the SacI-BamHI site downstream of the trc promoter of p322-Ptrc-ΔNdeI, a derivative of p322-Ptrc (27), yielding plasmid pMT411. A 3.4-kb BglII segment of pMT411 that contains laclq, trcp::ftsZ, and an rrnB operon region (as a transcription terminator) was isolated and inserted into the BamHI site of either pAM1313 for targeting to neutral site I (2, 24) or pTS2K-ΔNdeI, a derivative of pTS2K (27), for targeting to neutral site II. The constructs were introduced into either neutral site I or II on the chromosome of cyanobacterial strains by double crossover. The FtsZ protein was overexpressed in cyanobacterial strains in liquid or on solid (1.5% agar) BG-11 medium containing 0.5 or 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were observed under a light microscope, and the microscopic images (bright field with 20× or 40× objectives) were captured by a charge-coupled device camera.
Nucleotide sequence accession number.
The nucleotide sequence of the S. elongatus ftsZ gene and flanking regions has been deposited in the DDBJ/EMBL/GenBank databases under accession number AF076530.
RESULTS AND DISCUSSION
Circadian rhythm of cell division regulated by kaiC-dependent clock.
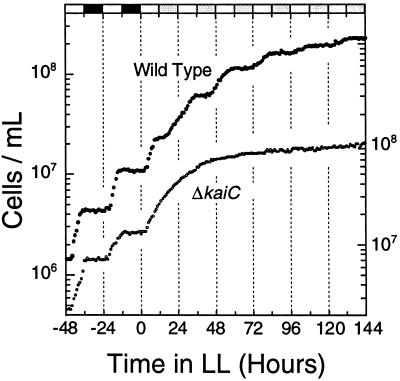
In a previous study (35), we showed that S. elongatus cells exhibit a rhythm of cell division cycling in continuous cultures of rapidly growing cells. The period length of the cell division rhythm is altered in the same manner by mutations (25) that change the period length of the promoter activity rhythm of the psbAI gene (24, 35). The mutations in strains C22a and C27a (previously named SP22 and LP27, respectively), which altered period lengths of circadian rhythms of both cell division (35) and psbAI promoter activity (25), have been mapped to the circadian clock gene kaiC in the cluster of kai genes (18). We therefore examined the cell division phenotype of a kaiC-deficient strain that is arhythmic for the psbAI promoter rhythm. The wild-type (AMC149) and kaiC deletion (ΔkaiC [46]) strains were grown in batch cultures. In both the wild-type and ΔkaiC strains, cell division occurred in the day phases of LD 12:12 cycles (Fig. 1). After transfer to LL, the wild-type strain exhibited a stepwise growth curve; the cells divided in the subjective day and stopped dividing in the early subjective night (Fig. 1), as we reported previously (35). In contrast, in the ΔkaiC strain, cell density increased without apparent rhythmicity until the culture reached stationary phase (Fig. 1). Lack of a cell division rhythm in the ΔkaiC strain indicates that the same kaiC-dependent clock that regulates global gene expression regulates cell division and supports our prior finding using point mutations of the kaiC gene (35).
FIG. 1.
Circadian rhythm of cell division in batch cultures of S. elongatus. Cell number data for the wild-type strain (●) and a clock-null ΔkaiC strain (○). The wild-type and ΔkaiC (46) strains were grown in LD 12:12 and transferred into LL (45 μE/m2/s) at time zero. The last two LD cycles preceding LL are illustrated by the bars on the upper abscissa (white, light; black, darkness; gray, subjective night phases of LL). The left ordinate is for the wild-type strain, and the right ordinate is for the ΔkaiC strain.
Cloning the ftsZ gene from Synechococcus.
Although the circadian gating of cell division in cyanobacteria has been clearly demonstrated (35) (Fig. 1), the molecular bases for gating the CDC are still largely unknown. The protein FtsZ is ubiquitous in bacteria and is also found in chloroplasts. In bacteria, a cytoskeletal element called the Z ring is formed in the middle of the cell (5, 39), and septation occurs through the action of the Z ring. The FtsZ proteins assemble into the Z ring and are known to be essential for cell division (4, 10). Our previous study indicated that DNA replication occurs continuously and randomly throughout the circadian cycle within a population of rapidly growing cells but that cytokinesis is gated by the circadian clock such that division is forbidden in the early subjective night (35). Consequently, we hypothesized that genes related to cytokinesis (or septation) might play a role in the circadian gating of cell division; therefore, we cloned the ftsZ gene from Synechococcus and investigated its temporal expression patterns.
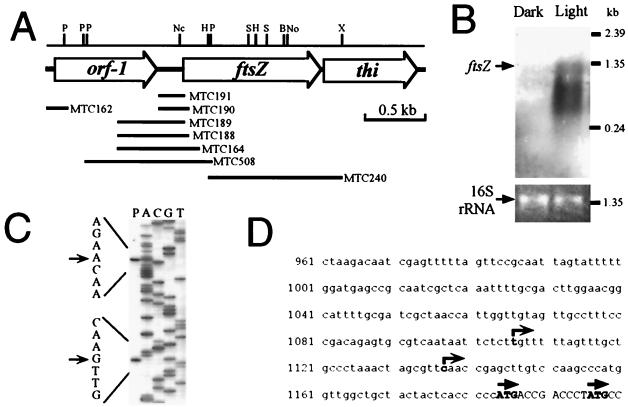
To make the ftsZ gene probe for screening genomic libraries, PCR was performed with the primer set that was used for cloning ftsZ from Anabaena (11, 47). One major 220-bp DNA fragment was amplified by genomic PCR. This PCR product was sequenced and found to be very similar to a region of known ftsZ genes. Therefore, the PCR product was used as a probe for genomic Southern analysis and for screening a cosmid library. Genomic Southern analyses confirmed that the ftsZ gene was a single-copy gene (data not shown). Thirteen positive clones were obtained from the 650 clones of the cosmid library, and cosmid 8B5 was used for further characterization. The ftsZ ORF was localized within cosmid 8B5 and sequenced. The predicted amino acid sequence of S. elongatus FtsZ shows strong similarity to FtsZ proteins of other bacteria (74% identity to Anabaena FtsZ, 72% identity to Synechocystis sp. strain PCC 6803 FtsZ, and 48% identity to E. coli FtsZ) and chloroplasts (61% identity to Arabidopsis plastid FtsZ). DNA sequence analysis upstream and downstream of Synechococcus ftsZ demonstrated that the ftsZ gene is flanked by an unknown gene (homologous to ORF sll1632 located just upstream of ftsZ in Synechocystis sp. strain PCC 6803 [21]) and a homolog of the thiD gene (37% identity to the putative amino acid sequence of the thiD gene from Salmonella enterica serovar Typhimurium [45] encoding phosphomethylpyrimidine kinase) (Fig. 2A). Unlike in E. coli (39), the ftsZ genes of S. elongatus and Synechocystis sp. strain PCC 6803 are not organized in a cluster with other cell division genes (e.g., ddlb-ftsQ-ftsA-ftsZ-envA). Northern blotting analysis shows the maximum size of detectable ftsZ transcript is 1.3 kb and the ftsZ gene is expressed more strongly in dividing cells during the day (Fig. 2B). Primer extension indicated at least two putative transcriptional start sites (Fig. 2C and D), and those sites were confirmed by S1 analysis (data not shown). Reporter analysis (see below) indicated that essential promoter elements are located within the 167-bp 5′ region (strain MTC191). These findings indicate the lack of conservation between cyanobacteria and other bacteria in the flanking regions of the ftsZ gene.
FIG. 2.
Primary structure and expression of the S. elongatus ftsZ gene. (A) Physical map of the ftsZ gene and its flanking regions. Segments used for constructing luxAB reporter strains are indicated by horizontal bars with the names of the transformed strains. P, PstI; Nc, NcoI; H, HindIII; S, SalI; B, BssHI; No, NotI; X, XhoI. (B) Northern blot analysis of the ftsZ gene. To show equivalent loading of the two lanes in the gel, ethidium bromide staining of the 16S rRNA band is shown. Cells were grown in LD 12:12 cycles and harvested in the day and night. In LD cycles, cyanobacteria divide only in the day (Fig. 1). (C) Determination of putative transcriptional start sites of the ftsZ gene by primer extension. The transcription start site was confirmed by S1 mapping (not shown). (D) Sequence of the promoter region of ftsZ. Putative transcriptional (right-angle arrows) and translational (straight arrows) start sites are indicated.
Circadian rhythms of ftsZp activity.
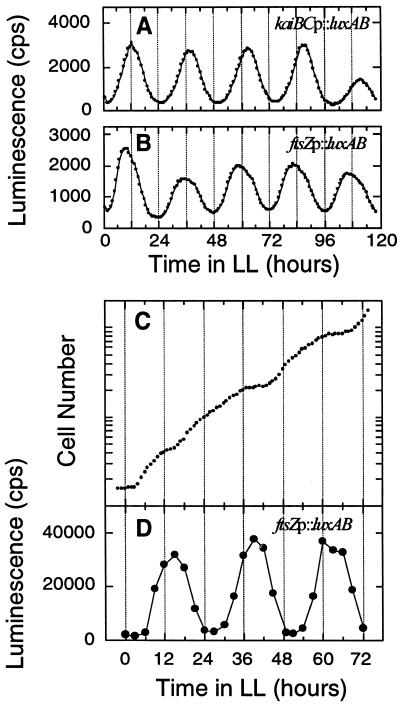
To monitor the expression patterns of ftsZ, we constructed transcriptional fusion strains with the ftsZ promoter linked to a bacterial luciferase gene set. The upstream regions of the ftsZ gene were isolated and transcriptionally fused to the luxAB gene set (the DNA segments fused to luxAB are indicated in Fig. 2A), and the reporter constructs were introduced into neutral sites of the wild-type Synechococcus chromosome. Rhythmic fluctuations of in vivo luminescence from the luxAB reporter strain MTC508 are shown in Fig. 3B and D. The rhythms of luminescence peaked at the end of the subjective day (or the beginning of subjective night), with troughs near the subjective dawn both in slowly growing batch cultures (doubling time [DT] > 24 h [Fig. 3B]) and in continuously diluted cultures of rapidly dividing cells (DT ∼ 12 h [Fig. 3C and D]). The phasing of this promoter rhythm was similar to that expressed by the kaiBC promoter, a class I gene (18, 31, 32). In batch cultures of the other reporter strains (MTC164, MTC188, MTC189, MTC190, and MTC191) in which shorter pieces of the 5′ region of ftsZ were fused to luxAB (Fig. 2A), luminescence rhythms were as observed from strain MTC508 (data not shown). Therefore, the activity of the ftsZ promoter appears to be under the control of the circadian clock in Synechococcus.
FIG. 3.
Rhythms of luminescence in ftsZp::luxAB reporter strain MTC508. (A and B) In vivo luminescence from 3-ml batch liquid cultures of kaiBCp::luxAB (A) and ftsZp::luxAB (B) strains was measured as described by Kondo et al. (24, 26). (C and D). The ftsZp::luxAB cells were grown in LD 12:12 (125 μE/m2/s) and then released into LL, and continuous dilution of the culture was started. Cell density of the culture was monitored every hour, and a growth curve calculated from the actual cell density and the dilution rate (dilution rate = 43.3 ml of medium exchanged every hour in a total volume of 780 ml) is plotted in panel C. The average doubling time of this culture was 12.1 h. From the culture in panel C, 1 ml of cell suspension was withdrawn every 3 h and for measurement of luminescence (D) as described by Mori et al. (35).
We observed that ftsZp activity was highest when cells in the population were not dividing, which is somewhat different from the case for E. coli (5, 34) and other bacteria (39). This result suggests that expression of ftsZ might not be a rate-limiting step of the cell division cycle in Synechococcus. Alternatively, assuming that the turnover of the FtsZ protein is rapid enough that its abundance patterns are similar to that of ftsZp activity, FtsZ may negatively regulate cell division in cyanobacteria just as overexpression of FtsZ inhibits cell division in E. coli (44).
Overexpression of FtsZ in cyanobacteria halts division.
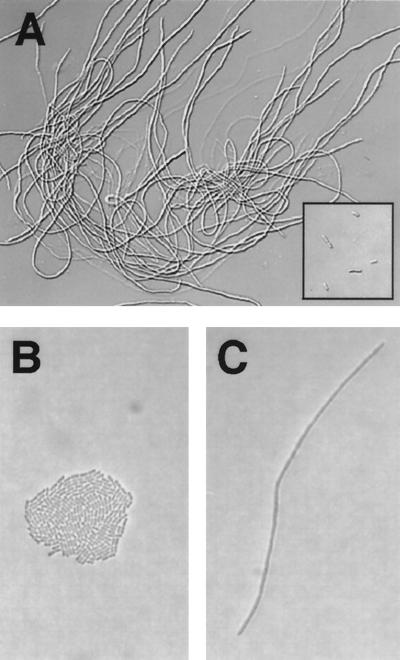
The FtsZ protein was overexpressed in trcp::ftsZ transformants in liquid BG-11 medium containing 0.5 mM IPTG. As was also found for E. coli (44), overexpression of FtsZ causes the cyanobacterial cells to become filamentous (Fig. 4A). When the FtsZ protein was overexpressed in trcp::ftsZ cells growing on solid (1.5% agar) BG-11 medium containing 1 mM IPTG, single cells formed a long filament (Fig. 4C), whereas colonies formed from single cells of the trcp::null strain on the same medium (Fig. 4B). Microscopic examination confirmed that the filamentous cells did not form septa. Figures 4A and C indicate that cell division, but not cell growth, was stopped when FtsZ was overexpressed from the trc promoter.
FIG. 4.
Cell division of growing cyanobacteria is stopped by overexpression of FtsZ, resulting in filamentous cells. (A) The trcp::ftsZ strain was grown for 101 h in liquid BG-11 medium supplemented with 0.5 mM IPTG. The insert in the bottom right corner of panel A shows trcp::null cells as a control under the same conditions. (B and C) The trcp::ftsZ and trcp::null cells were also grown on solid (1.5% agar) BG-11 medium supplemented with 1 mM IPTG for 48 h. A colony of trcp::null cells (B) and a filamentous trcp::ftsZ cell (C) are shown. Presumably both the colony in (panel B) and the filament in (panel C) were derived from a single initial cell.
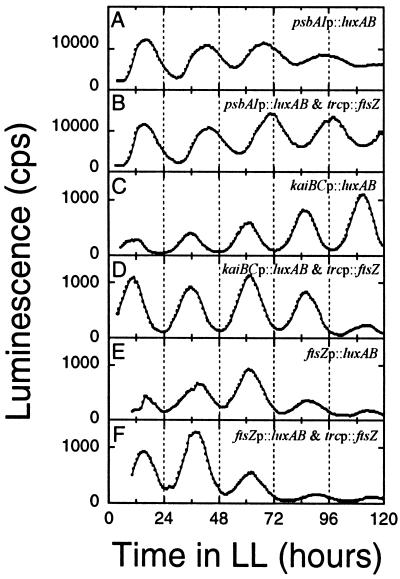
Circadian rhythms of gene expression in nondividing cyanobacteria.
To determine whether circadian rhythms of gene expression persist in nondividing cells, we transformed three different luxAB reporter strains with the trcp::ftsZ construct. Figure 5 shows luminescence patterns that report rhythms of promoter activities of the psbAI, kaiBC, and ftsZ genes in Synechococcus. Whether or not cell division was stopped by overexpression of FtsZ, the expression patterns of all three genes maintained robust circadian fluctuations for at least 4 to 5 days. Rather surprisingly, overexpression of FtsZ protein did not appear to affect the level of ftsZp activity, implying that there is little or no feedback of FtsZ abundance on the ftsZ promoter.
FIG. 5.
Luminescence rhythms in dividing and nondividing cyanobacteria in liquid cultures. In vivo luminescence was monitored as described by Kondo et al. (24) in the following reporter strains: (A) psbAIp::luxAB; (B) FtsZ overexpression in the psbAIp::luxAB strain; (C) kaiBCp::luxAB; (D) FtsZ overexpression in the kaiBCp::luxAB strain; (E) ftsZp::luxAB; (B) FtsZ overexpression in the ftsZp::luxAB strain. FtsZ protein was overexpressed continuously with 0.5 mM IPTG in panels B, D, and F, and filamentous morphology in those cultures was confirmed microscopically.
It might be hypothesized that there is a different clockwork that regulates the expression of these three promoters from the clockwork that gates cell division, and therefore the overexpression of FtsZ would not be expected to impinge on the gene expression patterns. However, the data in Fig. 1 show that cell division is regulated by the same kaiC-dependent clock that controls the gene expression patterns (18). Therefore, the data in Fig. 4 and 5 demonstrate that the circadian clock that regulates gene expression and gates cell division is not affected by halting cell division.
The results described herein have several important ramifications. First, there does not appear to be feedback from the cell division output rhythm back upon the central oscillator in Synechococcus. This is different than the case with some circadian rhythms (e.g., locomotor activity) in higher organisms, where induced activity of an output can change the phase or period of the central oscillator (36), Second, the periodicity of the circadian system is precise and stable whether the cells are dividing rapidly (DT = 12 h [see also references 26 and 35]), dividing slowly, or not dividing at all. These different division states must have an important impact on the intracellular milieu, but the circadian mechanism appears to be unaltered. This result is consistent with our previous observation that the circadian timing mechanism of S. elongatus is impervious to conditions of metabolic repression, either by extended darkness or by inhibition of protein synthesis (46). Apparently, the circadian clockwork is well buffered and stable against significant changes of the intracellular milieu. Finally, the persistence of the gene expression rhythms when cell division is stopped clearly indicates that the circadian clockwork gates cell division, but its timing circuit is not dependent on the CDC in cyanobacteria.
ACKNOWLEDGMENTS
We thank D. G. Adams and C. C. Zhang for providing the Anabaena ftsZ clone, S. S. Golden for providing the cosmid library of Synechococcus genomic DNA and the luxAB vectors, J. Lutkenhaus for providing E. coli anti-FtsZ antibodies, and S. Kutsuna, M. Ishiura, and T. Kondo for providing p322-Ptrc and pTS2K vectors. We are grateful for advice from T. Kondo and M. Ishiura in early stages of this project.
This work was supported by the National Science Foundation (grant MCB-9874371).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Andersson C R, Tsinoremas N F, Shelton J, Lebedeva N V, Yarrow J, Min H, Golden S S. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 2000;305:527–542. doi: 10.1016/s0076-6879(00)05511-7. [DOI] [PubMed] [Google Scholar]
- 3.Barkai N, Leibler S. Biological rhythms: circadian clocks limited by noise. Nature. 2000;403:267–268. doi: 10.1038/35002258. [DOI] [PubMed] [Google Scholar]
- 4.Bi E F, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 5.Bramhill D. Bacterial cell division. Annu Rev Cell Dev Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- 6.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 7.Bünning E. The physiological clock. 3rd ed. New York, N.Y: Springer-Verlag; 1973. [Google Scholar]
- 8.Bustos S A, Schaefer M R, Golden S S. Different and rapid responses of four cyanobacterial psbA transcripts to changes in light intensity. J Bacteriol. 1990;172:1998–2004. doi: 10.1128/jb.172.4.1998-2004.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Corton J C, Ward J E, Jr, Lutkenhaus J. Analysis of cell division gene ftsZ (sulB) from gram-negative and gram-positive bacteria. J Bacteriol. 1987;169:1–7. doi: 10.1128/jb.169.1.1-7.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doherty H M, Adams D G. Cloning and sequence of ftsZ and flanking regions from the cyanobacterium Anabaena PCC 7120. Gene. 1995;163:93–96. doi: 10.1016/0378-1119(95)00416-4. [DOI] [PubMed] [Google Scholar]
- 12.Edery I. Circadian rhythms in a nutshell. Physiol Genomics. 2000;3:59–74. doi: 10.1152/physiolgenomics.2000.3.2.59. [DOI] [PubMed] [Google Scholar]
- 13.Edmunds L N., Jr . Cellular and molecular bases of biological clocks. New York, N.Y: Springer-Verlag; 1988. [Google Scholar]
- 14.Ehret C F, Wille J J. The photobiology of circadian rhythms in protozoa and other eukaryotic microorganisms. In: Halldal P, editor. Photobiology of microorganisms. New York, N.Y: Wiley; 1970. pp. 369–416. [Google Scholar]
- 15.Golden S S, Brusslan J, Haselkorn R. Genetic engineering of the cyanobacterial chromosome. Methods Enzymol. 1987;153:215–231. doi: 10.1016/0076-6879(87)53055-5. [DOI] [PubMed] [Google Scholar]
- 16.Goto K, Johnson C H. Is the cell division cycle gated by a circadian clock? the case of Chlamydomonas reinhardtii. J Cell Biol. 1995;129:1061–1069. doi: 10.1083/jcb.129.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardin P E, Hall J C, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 18.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson C R, Tanabe A, Golden S S, Johnson C H, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 19.Iwasaki H, Kondo T. The current state and problems of circadian clock studies in cyanobacteria. Plant Cell Physiol. 2000;41:1013–1020. doi: 10.1093/pcp/pcd024. [DOI] [PubMed] [Google Scholar]
- 20.Johnson C H, Golden S S. Circadian programs in cyanobacteria: adaptiveness and mechanism. Annu Rev Microbiol. 1999;53:389–409. doi: 10.1146/annurev.micro.53.1.389. [DOI] [PubMed] [Google Scholar]
- 21.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A Y, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 22.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Venter J C, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 23.Klevecz R R. Quantized generation time in mammalian cells as an expression of the cellular clock. Proc Natl Acad Sci USA. 1976;73:4012–4016. doi: 10.1073/pnas.73.11.4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo T, Strayer C A, Kulkarni R D, Taylor W, Ishiura M, Golden S S, Johnson C H. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90:5672–5676. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondo T, Tsinoremas N F, Golden S S, Johnson C H, Kutsuna S, Ishiura M. Circadian clock mutants of cyanobacteria. Science. 1994;266:1233–1236. doi: 10.1126/science.7973706. [DOI] [PubMed] [Google Scholar]
- 26.Kondo T, Mori T, Lebedeva N V, Aoki S, Ishiura M, Golden S S. Circadian rhythms in rapidly dividing cyanobacteria. Science. 1997;275:224–227. doi: 10.1126/science.275.5297.224. [DOI] [PubMed] [Google Scholar]
- 27.Kutsuna S, Kondo T, Aoki S, Ishiura M. A period-extender gene, pex, that extends the period of the circadian clock in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1998;180:2167–2174. doi: 10.1128/jb.180.8.2167-2174.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lakin-Thomas P L. Circadian rhythms: new functions for old clock genes. Trends Genet. 2000;16:135–142. doi: 10.1016/s0168-9525(99)01945-9. [DOI] [PubMed] [Google Scholar]
- 29.Leipe D D, Aravind L, Grishin N V, Koonin E V. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 2000;10:5–16. [PubMed] [Google Scholar]
- 30.Liu Y, Heintzen C, Loros J, Dunlap J C. Regulation of clock genes. Cell Mol Life Sci. 1999;55:1195–205. doi: 10.1007/s000180050366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Tsinoremas N F, Johnson C H, Lebedeva N V, Golden S S, Ishiura M, Kondo T. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Golden S S, Kondo T, Ishiura M, Johnson C H. Bacterial luciferase as a reporter of circadian gene expression in cyanobacteria. J Bacteriol. 1995;177:2080–2086. doi: 10.1128/jb.177.8.2080-2086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lorne J, Scheffer J, Lee A, Painter M, Miao V P. Genes controlling circadian rhythm are widely distributed in cyanobacteria. FEMS Microbiol Lett. 2000;189:129–133. doi: 10.1111/j.1574-6968.2000.tb09218.x. [DOI] [PubMed] [Google Scholar]
- 34.Margolin W. Themes and variations in prokaryotic cell division. FEMS Microbiol Rev. 2000;24:531–548. doi: 10.1111/j.1574-6976.2000.tb00554.x. [DOI] [PubMed] [Google Scholar]
- 35.Mori T, Binder B, Johnson C H. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci USA. 1996;93:10183–10188. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mrosovsky N. Locomotor activity and non-photic influences on circadian clocks. Biol Rev. 1996;71:343–372. doi: 10.1111/j.1469-185x.1996.tb01278.x. [DOI] [PubMed] [Google Scholar]
- 37.Pittendrigh C S. On temperature independence in the clock system controlling emergence time in Drosophila. Proc Natl Acad Sci USA. 1954;40:1018–1029. doi: 10.1073/pnas.40.10.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pittendrigh C S. Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol. 1993;55:16–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 39.Rothfield L, Justice S, Garcia-Lara J. Bacterial cell division. Annu Rev Genet. 1999;33:423–448. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Sweeney B M. Rhythmic phenomena in plants. 2nd ed. San Diego, Calif: Academic Press; 1987. [Google Scholar]
- 42.Sweeney B M, Hastings J W. Effects of temperature upon diurnal rhythms. Cold Spring Harbor Symp Quant Biol. 1960;25:87–104. doi: 10.1101/sqb.1960.025.01.009. [DOI] [PubMed] [Google Scholar]
- 43.Wager-Smith K, Kay S A. Circadian rhythm genetics: from flies to mice to humans. Nat Genet. 2000;26:23–27. doi: 10.1038/79134. [DOI] [PubMed] [Google Scholar]
- 44.Ward J E, Jr, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985;42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- 45.Webb E, Febres F, Downs D M. Thiamine pyrophosphate negatively regulates transcription of some thi genes of Salmonella typhimurium. J Bacteriol. 1996;178:2533–2538. doi: 10.1128/jb.178.9.2533-2538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y, Mori T, Johnson C H. Circadian clock-protein expression in cyanobacteria: rhythms and phase setting. EMBO J. 2000;19:3349–3357. doi: 10.1093/emboj/19.13.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang C C, Huguenin S, Friry A. Analysis of genes encoding the cell division protein FtsZ and a glutathione synthetase homologue in the cyanobacterium Anabaena sp. PCC 7120. Res Microbiol. 1995;146:445–455. doi: 10.1016/0923-2508(96)80290-7. [DOI] [PubMed] [Google Scholar]