Abstract
Phenotypic differences between planktonic bacteria and those attached to abiotic surfaces exist, but the mechanisms involved in the adhesion response of bacteria are not well understood. By the use of two-dimensional (2D) polyacrylamide gel electrophoresis, we have demonstrated that attachment of Escherichia coli to abiotic surfaces leads to alteration in the composition of outer membrane proteins. A major decrease in the abundance of resolved proteins was observed during adhesion of type 1-fimbriated E. coli strains, which was at least partly caused by proteolysis. Moreover, a study of fimbriated and nonfimbriated mutants revealed that these changes were due mainly to type 1 fimbria-mediated surface contact and that only a few changes occurred in the outer membranes of nonfimbriated mutant strains. Protein synthesis and proteolytic degradation were involved to different extents in adhesion of fimbriated and nonfimbriated cells. While protein synthesis appeared to affect adhesion of only the nonfimbriated strain, proteolytic activity mostly seemed to contribute to adhesion of the fimbriated strain. Using matrix-assisted laser desorption ionization–time of flight mass spectrometry, six of the proteins resolved by 2D analysis were identified as BtuB, EF-Tu, OmpA, OmpX, Slp, and TolC. While the first two proteins were unaffected by adhesion, the levels of the last four were moderately to strongly reduced. Based on the present results, it may be suggested that physical interactions between type 1 fimbriae and the surface are part of a surface-sensing mechanism in which protein turnover may contribute to the observed change in composition of outer membrane proteins. This change alters the surface characteristics of the cell envelope and may thus influence adhesion.
During starvation, bacteria undergo morphological and physiological changes which commonly increase their tendency to adhere to solid surfaces (23). Following microbial adhesion to solid surfaces, biofilms can develop on practically every material that comes in contact with aqueous liquids. The first stage of adhesion is governed by surface charges and energies and can to a large extent be understood by adhesion theories, such as the DLVO theory, the thermodynamic approach, and the extended DLVO theory (1, 14, 51, 52). However, in these models the spatial and temporal variability of the bacterial cell envelope is not considered, even though different cell surface structures that contact the surface give rise to different adhesion mechanisms (4, 8, 34, 35, 49). When approaching a surface, bacteria encounter totally different conditions than in the bulk water environment. A change in the environment generally induces structural and functional adaptations in the cell to enhance survival. As a result, biofilm bacteria often exhibit different phenotypes compared to those of their planktonic counterparts (4, 9–12). There is evidence that adhesion of bacteria to solid surfaces may trigger certain types of gene expression, such as flagella synthesis in Vibrio parahaemolyticus (8) and polysaccharide production in Pseudomonas aeruginosa (13, 50). In Escherichia coli, OmpR has been shown to regulate the production of curli, which is involved in colonization of inert surfaces (53). Apart from these examples, the differentiation process from planktonic to attached cells is not well understood.
As an interface between the environment and the interior of the cell, the bacterial cell envelope plays an important role in facilitating responses to change. In gram-negative bacteria, outer membranes are in close contact with the environment and contain a number of major proteins present in high copy number. Some of these proteins are highly regulated in response to growth, nutrient, and environmental conditions (19, 36, 39). An altered expression of outer membrane proteins (OMPs) implies a change in the outermost cell surface, which may have direct effects on adhesion. In fact, several OMPs have been suggested to be involved in adhesion of bacteria to various surfaces (2, 40, 45, 46, 55). Moreover, fimbriae have been discussed to play a role in nonspecific adhesion (34, 35, 38). Although type 1 fimbriae in E. coli do not facilitate initial adhesion, they stabilize the contact of the cell with hydrophobic surfaces (35) and, following initial attachment, they interact with the surface more than nonfimbriated cells do (34). It may be speculated that, in addition to conferring different physicochemical properties to the cell, fimbriae may be part of a surface-sensing mechanism that triggers adaptive changes in the cell which, in turn, may contribute to irreversible adhesion and colonization of surfaces. Similarly, in P-piliated uropathogenic E. coli, fimbria-mediated adherence to the specific host cell receptor was shown to induce the expression of virulence factors (31, 54).
The aim of this study was to determine whether E. coli undergoes changes in its cell envelope upon adhesion to abiotic surfaces. The role of protein synthesis and turnover in adhesion to abiotic surfaces was determined by the inhibitory effect of tetracycline or protease inhibitors. We also studied the compositions of OMPs by two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) in planktonic cells and cells attached to abiotic surfaces. To further reveal whether the contact of type 1 fimbriae with the surface influences the extent of the response, we compared OMP patterns among fimbriated and nonfimbriated mutants and their parental wild-type strains. Some of the proteins affected at their levels of abundance during adhesion of fimbriated cells were identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains were selected on the basis of their cell surface properties (Table 1). They were cultivated at 37°C in Luria-Bertani medium (22). When required, the medium was supplemented with either 50 μg of ampicillin ml−1, 25 μg of kanamycin ml−1, or 25 μg of tetracycline ml−1. Cells were grown overnight without shaking, harvested by centrifugation (12,100 × g for 10 min), washed in 0.2 M Tris-buffered saline (0.05 M Tris-HCl supplemented with 0.15 M NaCl; pH 7.5), and incubated in the same buffer for 24 h. Prior to the experiment, these cell suspensions were centrifuged (12,100 × g for 10 min) and resuspended in Tris (0.2 M; pH 7.5) at the appropriate cell concentration (approximately 3 × 108 cells ml−1 for adhesion experiments and approximately 1 × 1010 cells ml−1 for the isolation of OMPs). The capacity to express type 1 fimbriae was determined by mannose-sensitive agglutination of Saccharomyces cerevisae cells as described previously (34).
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| F-18 | Wild type | 24 |
| F-18fimA− | F-18 fimA Tcr | 24 |
| F-18fim+ | F-18(pPKL91) Apr | 25 |
| PC31 | gal tonA phx argF rel | 47 |
| MS7 | PC31 Δfim Kmr | 20 |
| MS7fim+ | MS7(pPKL4) Apr | 44 |
| Plasmids | ||
| pPKL4 | pBR322 carrying the fim operon, Apr Camr | 44 |
| pPKL91 | parB-stabilized version of pPKL9, carrying fimB, Apr Camr | 25 |
To assess the activity of bacteria incubated for 24 h in Tris (0.2 M; pH 7.5) as well as the inhibitory effect of tetracycline at the concentration used, the level of protein synthesis of planktonic cells was monitored in the presence or absence of tetracycline (100 μg ml−1) as described previously (33) by incorporation of [14C]leucine (final concentration, 5 μCi ml−1; Amersham) into material precipitable in 5% (wt/vol) trichloroacetic acid.
Assay used to determine the effect of protein synthesis and turnover on adhesion.
Adhesion was measured as described previously (34). AT-cut 5-MHz quartz crystals (Maxtek Inc., Torrance, Calif.), coated with an evaporated gold film, were used as surfaces. To render surfaces hydrophilic or hydrophobic, crystals were treated prior to the experiment as described previously (34). Bacterial suspensions were added to a final concentration of approximately 3 × 108 cells ml−1 in Tris (0.2 M; pH 7.5). After 60 min of incubation at 22°C, surfaces were rinsed with Tris (0.2 M; pH 7.5) to avoid further attachment of bacteria and remaining surface-associated bacteria were incubated for an additional hour.
To study the influence of protein synthesis or proteolytic activity on adhesion in this system, tetracycline (100 μg ml−1) or a mixture of protease inhibitors inhibiting a broad spectrum of serine and cysteine proteases (Complete, EDTA free, concentration recommended by the manufacturer; Boehringer Mannheim), respectively, were added to the bacteria immediately before they were allowed to adhere to the surface. As a control, adhesion of dead bacteria, killed by the addition of 0.18 mM H2O2, was measured. To determine whether tetracycline or the molecules contained in the protease inhibitor mix have a surface-active rather than a physiological effect on adhesion, their influence on adhesion of dead cells was also studied as described above.
Microscopy.
The cell concentration in the experiments was determined by acridine orange (AO) direct counts. Aliquots of bacterial suspensions were diluted in Tris (0.2 M; pH 7.5), and l-ml samples were stained with an equal volume of filter-sterilized AO solution (100 μg of AO ml−1, 2% [vol/vol] formaldehyde in phosphate-buffered saline) for 5 min and passed through a 0.2-μm-pore-size Nucleopore filter (prestained with Sudan Black). Filters were viewed at a magnification of ×1,250 in a Zeiss epifluorescence microscope.
Attached bacteria were counted directly on the quartz crystal surfaces. Crystals were stained for 5 min in filter-sterilized AO solution. The surfaces were briefly dipped into filter-sterilized deionized water to remove excess stain and then air dried. Surfaces were examined by epifluorescence microscopy at a magnification of ×500. For every sample, a minimum of 30 fields of view were counted. All cells were counted regardless of color.
Adhesion assay used for the isolation of outer membranes of attached cells.
Hydrophilic and hydrophobic acid-cleaned glass beads (diameter, 0.5 mm) contained in miniature scintillation vials were used as surfaces and were freshly prepared prior to each experiment as described previously (35). Scintillation vials were filled with 8 g of glass beads. To each vial, 2 ml of bacterial suspensions (final concentration, approximately 1010 cells ml−1) was added to cover the glass beads. In parallel experiments, bacteria were allowed to attach in the presence of divalent cations (5 mM MgCl2), tetracycline (100 μg ml−1), or the protease inhibitor mix (concentration recommended by the manufacturer). As a control, 2 ml of bacterial suspension was added to empty vials. These samples served as planktonic cells. The suspensions were incubated for 1 h at 22°C to allow for surface contact. Subsequently, the suspension was sucked out of the glass-bead-filled samples to remove planktonic cells and replaced by 2 ml of sterile Tris (0.2 M; pH 7.5). Remaining surface-associated cells were incubated for an additional hour. Planktonic samples were incubated for 2 h without exchange of the buffer solution, which resulted in protein patterns identical to those of planktonic cells exposed to a buffer exchange after 1 h or those of unattached cells that remained in the supernatants of samples containing glass beads (data not shown).
Preparation of outer membranes.
After the adhesion experiment, the buffer solution was removed from the vials, glass beads with attached bacteria were resuspended in 2 ml of distilled water (dH2O), and samples were immediately put on ice. In order to inhibit further protein synthesis and protease activity, tetracycline at a final concentration of 100 μg ml−1 and protease inhibitors (concentration recommended by the manufacturer, Complete, EDTA free; Boehringer Mannheim) were added to the samples. Outer membranes were isolated as described by Filip et al. (16). Briefly, cells were detached and/or disrupted by sonication on ice (20 1-min cycles separated by 30-s intervals to allow for cooling; probe amplitude, 14 μm; Soniprep 150; MSE Scientific Instruments). The cell extracts were transferred to new tubes and centrifuged (12,100 × g for 10 min) to remove whole cells, which was confirmed by examination of the supernatant in the microscope. To solubilize the cytoplasmic membrane, Sarkosyl (N-lauroyl sarcosine) was added to the supernatant at a final concentration of 2%. After incubation of the mixture at 22°C for 30 min, the sample was centrifuged at 38,000 × g for 40 min at 4°C. The pellet consisting of outer membranes was washed once in 1 ml of ice-cold dH2O (38,000 × g for 10 min) and resuspended in dH2O. The protein concentration in the outer membrane fractions was determined either spectrophotometrically at 280 nm or with a NanoOrange protein quantification kit (Molecular Probes). Bovine serum albumin was used as a standard in both assays. Outer membranes were stored at −20°C until use.
To exclude the possibility that protein patterns may be affected by a loss of proteins during the isolation procedure, a quantitative measure for the total cell debris in each cell extract was obtained by measuring the optical densities at 600 nm (OD600) for all samples, which were directly proportional to the OD600 of the initial cell suspensions. The protein amount was determined for the whole-cell protein extract, and the fraction of Sarkosyl-soluble proteins and the fraction of Sarkosyl-insoluble proteins were determined by measuring the OD280 of each fraction. Similarly as described in reference 40, the ratio of the OD280 to the OD600, i.e., the protein amount of each fraction related to the total amount of cell debris, was calculated for each sample, and the resulting ratios for attached cells were compared with those for planktonic cells.
High-resolution 2D-PAGE with IPGs in the first dimension.
Isoelectric focusing in the first dimension was performed mainly as described previously (17), with the modifications reported by Norbeck and Blomberg (32). Equivalent amounts of lyophilized outer membrane preparations (8 μg) were resolved directly in 500 μl of rehydration buffer (7 M urea, 2 M thiourea, 1% [vol/vol] NP-40, 13 mM dithiothreitol, 1% [vol/vol] Pharmalyte 3-10 [Amersham Pharmacia Biotech], and 0.01% [wt/vol] bromophenol blue). This protein solution was then used to reswell the nonlinear immobilized-pH-gradient (IPG) strips (pH 3 to 10/NL; Amersham Pharmacia Biotech) overnight at room temperature as described previously (43). Focusing was subsequently performed at 20°C, under mineral oil, for about 40,000 V · h.
In the second dimension, polyacrylamide gels were used with a ratio of 12.5% acrylamide monomer to 2.1% bisacrylamide (Duracryl, 0.65% bis; Genomic Solutions, Inc., Ann Arbor, Mich.) containing 0.1% (wt/vol) sodium dodecyl sulfate, 0.37 M Tris base, and 0.27 M Tris-HCl. First-dimensional IPG strips were equilibrated for 30 min in 10 ml of equilibration solution (3% [wt/vol] sodium dodecyl sulfate, 50 mM dithiothreitol, 0.3 M Tris base, 0.075 M Tris-HCl, and 0.01% [wt/vol] bromophenol blue). The strips were loaded on top of the second-dimension gels by submerging the strips in a warm gel overlay agarose solution (0.5% [wt/vol] agarose, SeaPlaque; FMC) dissolved in gel buffer. Electrophoresis in the second dimension was performed at a limiting power of 16,000 mW (maximum voltage, 500 V) per gel for about 6 h until the dye front reached the bottom of the gel.
Visualization of protein spots and image analysis.
Protein spots were visualized by silver staining (analytical gels) using a modified procedure of Morrisey (30) or by staining with Coomassie blue (preparative gels) (42). Subsequently, gels were scanned in an AGFA white-light scanner at a resolution of 200 by 200 μm and the raw images were processed using the 2D software PDQuest (Protein and DNA Imageware Inc., Huntington Station, N.Y.). Following background subtraction and spot detection, the gel patterns were matched to each other by visual comparison.
Tryptic digestion of proteins.
Protein spots were cut out from Coomassie blue-stained gels, placed in separate siliconized tubes, and gently macerated with a handheld mixer. The dye was removed by adding 85 μl of 25 mM NH4HCO3 in 50% CH3CN, and the tube was vortexed for 30 min before the supernatant was removed. This procedure was repeated twice. Destained gel pieces were dried for 40 min in a HetoSic speedvac system. Subsequently, 15 μl of trypsin (10 μg ml−1; Promega, Scandinavian Diagnostics Services, Falkenberg, Sweden) in 25 mM NH4HCO3, pH 8, was added. The dried samples were rehydrated on ice for 60 min and subsequently incubated for 16 h at 37°C. Peptides were extracted from the gel by adding approximately 15 μl of 5% CF3COOH in 75% CH3CN to the tube. The samples were then vortexed for 30 min and centrifuged for 2 min at 15,000 × g. Trypsin-digested proteins were stored at −20°C until use.
Identification of proteins by MALDI-TOF MS.
Mass spectra were obtained using a TofSpec E (Micromass, Manchester, England) time-lag focusing MALDI-TOF mass spectrometer equipped with a reflecton. Samples were prepared by mixing 0.5 μl of the tryptic digest with 0.5 μl of the matrix solution directly on the MS target, where they were left to dry in air. The matrix used was α-cyano-4-hydroxy-cinnamic acid (Aldrich Chemie, Steinheim, Germany) at 10 mg ml−1 in CH3CN-water (ratio, 1:1).
Monoisotopic peptide masses obtained from mass spectra were used to identify proteins through database searches using MS-Fit available at the website http://prospector.ucsf.edu.
RESULTS
Involvement of protein synthesis and turnover in interactions of fimbriated and nonfimbriated E. coli strains with abiotic surfaces.
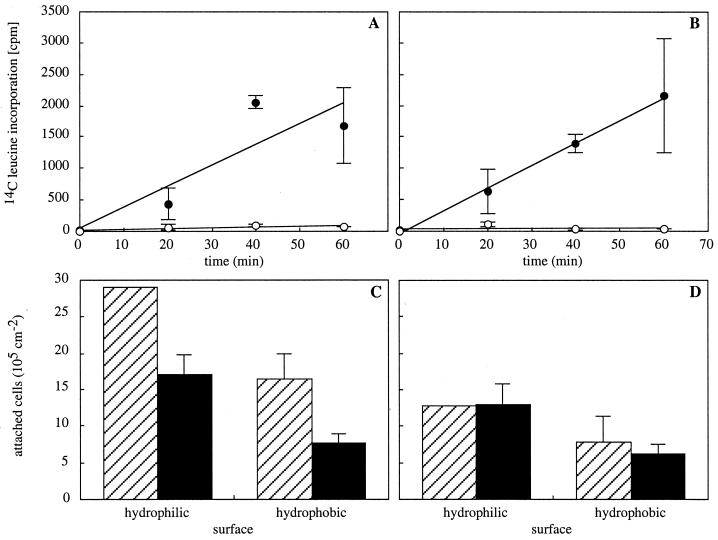
After 24 h of incubation in Tris (0.2 M; pH 7.5), strains MS7 and MS7fim+ displayed equivalent rates of protein synthesis as assessed by the incorportion of [14C]leucine into trichloroacetic acid-precipitable material (Fig. 1A and B). Tetracycline (100 μg ml−1) effectively inhibited protein synthesis of both strains (Fig. 1A and B). To test whether adhesion of fimbriated and nonfimbriated bacterial cells to abiotic surfaces requires protein synthesis, the numbers of attached cells to hydrophilic and hydrophobic surfaces were determined after 2 h of adhesion in the presence or in the absence of tetracycline. Similarly, the involvement of proteolytic activity was tested in the presence and in the absence of a mixture of protease inhibitors (Complete; Boehringer Mannheim). Exposure of cells to tetracycline or protease inhibitors did not affect their viability, as determined from the number of CFU.
FIG. 1.
Involvement of protein synthesis in adhesion. Protein synthesis occurs in cells starved in Tris for 24 h (filled circles) and is inhibited by tetracycline (100 μg ml−1; open circles) as shown for E. coli strains MS7 (A) and MS7fim+ (B). The numbers of attached cells per square centimeter after 2 h of adhesion to hydrophilic and hydrophobic surfaces of strain MS7 (C) and strain MS7fim+ (D) are shown in the absence (hatched bars) or presence (black bars) of tetracycline. Data shown are means (± standard deviations) of results from two to four experiments.
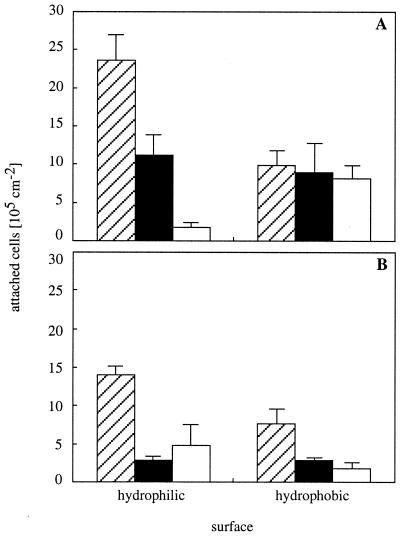
For the nonfimbriated strain MS7, inhibition of protein synthesis led to decreased numbers of attached cells on both hydrophilic and hydrophobic surfaces (Fig. 1C), with 59% ± 10% of cells being left on hydrophilic and 45% ± 3% of cells being left on hydrophobic surfaces. For the fimbriated strain MS7fim+, however, no significant change in the number of attached cells was observed in the presence of tetracycline (Fig. 1D). Inhibition of proteolytic activity during adhesion generally led to significantly lower numbers of attached cells of both strains, except for those of MS7 on hydrophobic surfaces (Fig. 2). The effect was about twice as strong for strain MS7fim+, with 20% ± 4% of cells being left on hydrophilic and 43% ± 5% of cells being left on hydrophobic surfaces, compared to strain MS7, with which 47% ± 12% of cells were left on hydrophilic and 89% ± 39% of cells were left on hydrophobic surfaces. In comparison, adhesion of dead fimbriated cells was 34% ± 19% on hydrophilic and 24% ± 11% on hydrophobic surfaces, while 8% ± 3% and 83% ± 17% of dead nonfimbriated cells remained on hydrophilic and hydrophobic surfaces, respectively. Adhesion of dead bacteria was not affected by the addition of tetracycline or protease inhibitors, which excludes a surface-active effect of these molecules.
FIG. 2.
Involvement of protein turnover in adhesion. The numbers of attached cells per square centimeter after 2 h of adhesion to hydrophilic and hydrophobic surfaces of strain MS7 (A) and strain MS7fim+ (B) are shown in the absence (hatched bars) or presence (black bars) of protease inhibitors. White bars represent the numbers of attached dead cells. Data shown are means (± standard deviations) of results from two to four experiments.
Effects of bacterial adhesion to abiotic surfaces on the compositions of OMPs.
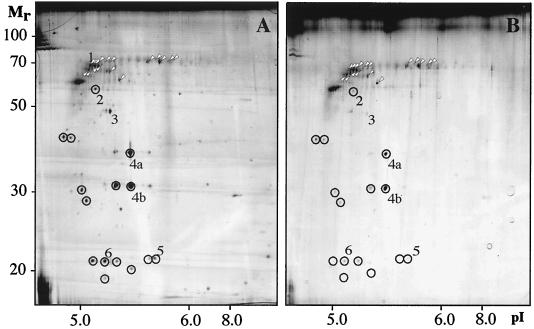
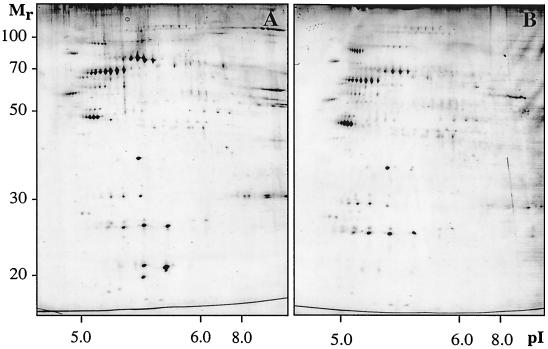
Since protein synthesis and turnover appeared to be involved in adhesion, we addressed whether a change in the composition of surface-exposed proteins occurred during adhesion. OMPs were extracted from planktonic and attached cells, and the protein concentration (OD280) relative to the OD600 unit was determined for the outer membranes of attached and planktonic cells. There was no reduction in the OD280/OD600 ratios for attached cells compared to those for planktonic cells (data not shown). Equivalent amounts (8 μg) of protein based on the OD280s of the cell extracts were analyzed by 2D-PAGE. The resulting protein patterns differed between planktonic cells and cells attached to abiotic surfaces as shown for the type 1-fimbriated strain PC31 (Fig. 3). Corresponding observations were made for F-18, a wild-type strain with a different genetic background (data not shown). Furthermore, there was no significant difference between the protein patterns of cells attached to hydrophilic (data not shown) and cells attached to hydrophobic surfaces (Fig. 3B). The amounts of proteins resolved by gel electrophoresis decreased to a large extent in cells attached to surfaces. The levels of the 38 most dominant proteins resolved on 2D gels were compared between cells in the planktonic and cells in the attached state. In comparison to the total levels of these proteins, 17 proteins showed at least a twofold relative increase (Fig. 3), 15 proteins showed at least a twofold relative decrease (Fig. 3), and the relative levels of abundance of 6 proteins remained unchanged. A relative increase in protein levels was observed only for proteins of high molecular weight. The observed changes in the OMP pattern were not counteracted by an increased amount of divalent cations (5 mM Mg2+) during adhesion of strain PC31 to hydrophobic surfaces (data not shown).
FIG. 3.
2D gel analysis of OMPs abundant after 2 h of adhesion to hydrophobic surfaces in planktonic cells (A) and attached cells (B) of E. coli strain PC31. Gel analyses have been repeated three times to confirm reproducibility, and representative results are shown. Proteins with relatively increased levels are indicated by white arrowheads, and proteins with relatively decreased levels are indicated by circles.
Identification of OMPs affected in their abundance during adhesion.
Some of the OMPs of strain PC31 resolved by 2D-PAGE were cut out from preparative gels and were digested with trypsin. The generated peptides were analyzed by MALDI-TOF MS, and proteins were identified by comparison to sequence databases (Table 2). Seven protein spots could be identified, two of which were mass isoforms of the same protein, OmpA. All suggested proteins had the highest molecular-weight search scores in the MS-Fit runs.
TABLE 2.
Identification of proteins by MALDI-TOF MS peptide mass fingerprinting
| Protein no.a | Mr | Proteinb | Mowsec score | No. of masses matched (total) | Accession no.d |
|---|---|---|---|---|---|
| 1 | 68,407.4 | BtuB | 5.96 × 108 | 13 (16) | 416728 |
| 2 | 54,000.3 | TolC | 1.87 × 1010 | 13 (21) | 882565 |
| 3 | 43,283.8 | EF-Tu | 4.50 × 107 | 10 (14) | 119201 |
| 4a | 37,200.9 | OmpA | 4.52 × 106 | 12 (14) | 129135 |
| 4b | 37,200.9 | OmpA | 1.41 × 107 | 12 (13) | 129135 |
| 5 | 20,964.0 | Slp | 4.53 × 104 | 6 (9) | 585997 |
| 6 | 18,602.7 | OmpX | 8.53 × 103 | 6 (11) | 730225 |
Numbers refer to designations of protein spots in Fig. 2A.
Designations of proteins: BtuB, vitamin B12 receptor protein; EF-Tu, elongation factor Tu; Slp, starvation lipoprotein; TolC, OMP part of the Tol transport system.
Mowse, molecular-weight search.
Nonredundant protein database at the National Center for Biotechnology Information.
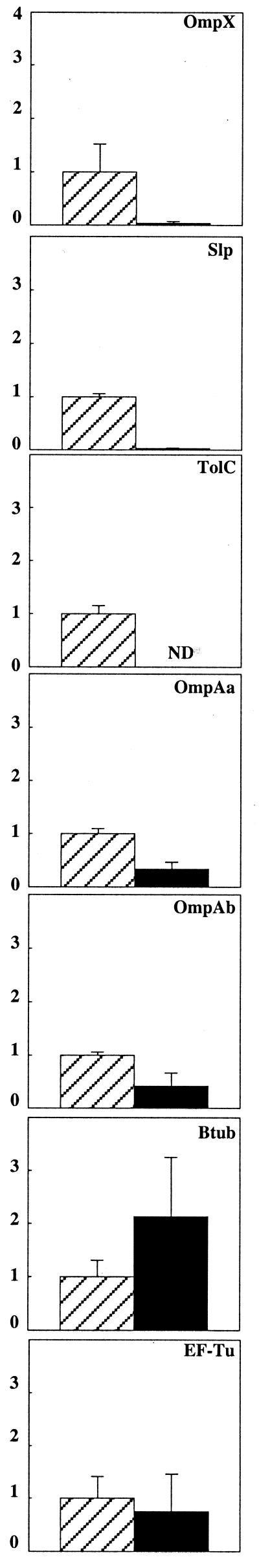
Most of the identified proteins belong to different classes of OMPs. There were two β-barrel proteins (OmpA and OmpX), one trimeric barrel protein (TolC), one monomeric channel protein (BtuB), and one lipoprotein (Slp). Moreover, one membrane-associated protein not belonging to the outer membrane was identified (EF-Tu). These proteins may also be classified according to the changes in their relative levels during adhesion. The levels of one group of proteins, consisting of OmpX, Slp, and TolC, were strongly reduced and that of OmpA was moderately reduced, whereas those of BtuB and EF-Tu remained unchanged (Fig. 4).
FIG. 4.
Relative levels of abundance of proteins resolved by 2D-PAGE and identified by MALDI-TOF MS. Data are based on normalized comparisons, where values in planktonic cells are set at 1. Data are means of results of three experiments ± standard deviations. ND, not detected.
Effect of type 1 fimbria-mediated surface contact on changes in the OMP composition.
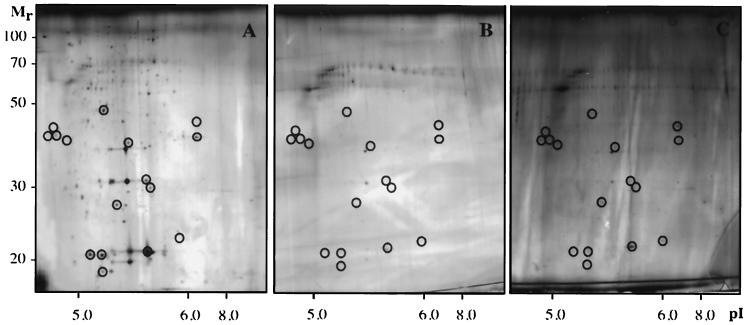
To study a possible role of type 1 fimbriae in the adhesion response leading to altered compositions of OMPs, we analyzed outer membrane fractions of nonfimbriated and fimbriated mutant strains. Planktonic cells of the nonfimbriated strain MS7 exhibited a different pattern of OMPs from those of the parental wild-type strain and the fimbriated mutant MS7fim+ (Fig. 5A). Moreover, in comparison with the total levels of the 42 most dominant proteins of strain MS7, the abundance of 1 protein was relatively increased (>2-fold) and the levels of 6 proteins were relatively decreased (>2-fold) in cells attached to surfaces (Fig. 5), while the levels of 35 proteins remained unchanged. In contrast, adhesion of the fimbriated strain MS7fim+ led to changes in the composition of OMPs that were similar to, but even more pronounced, than those of the wild type (Fig. 6).
FIG. 5.
2D gel analysis of OMPs abundant after 2 h of adhesion to hydrophobic surfaces in planktonic cells (A) and attached cells (B) of the nonfimbriated E. coli strain MS7. Gel analyses were repeated three times to confirm reproducibility, and representative results are shown.
FIG. 6.
2D gel analysis of OMPs abundant after 2 h of adhesion to hydrophobic surfaces in planktonic cells (A), attached cells (B), and cells attached in the presence of protease inhibitors (concentration recommended by the manufacturer, Complete; Boehringer Mannheim) (C) of the fimbriated E. coli strain MS7fim+. Proteins with relatively decreased levels during adhesion, which are restored in the presence of protease inhibitors, are indicated by circles. Gel analyses were repeated three times to confirm reproducibility, and representative results are shown.
Effect of protease activity on changes in the OMP composition upon type 1 fimbria-mediated surface contact.
To assess the involvement of protein synthesis and turnover in the changes of OMP patterns of attached fimbriated cells, outer membranes were isolated from the fimbriated mutant strain MS7fim+ attached to hydrophobic surfaces for 2 h in the presence of tetracycline (100 μg ml−1) or protease inhibitors (concentration recommended by the manufacturer, Complete; Boehringer Mannheim). The presence of tetracycline during the adhesion process did not inhibit changes in the OMP pattern as revealed by 2D analysis (data not shown). However, addition of protease inhibitors gave rise to an increased abundance of several OMPs (Fig. 6C) compared to levels in attached cells without protease inhibitors added (Fig. 6B).
DISCUSSION
In this study, we have demonstrated that attachment of E. coli cells directly or indirectly triggers reactions in the cell which cause major changes in the compositions of OMPs. While in the nonfimbriated strain MS7 surface-induced changes were seen only in a small number of proteins, the OMP compositions in the fimbriated strains PC31 and MS7fim+ changed to a much larger extent. Although equivalent amounts of protein were analyzed on each 2D gel, it is striking that the fraction of OMPs resolved by 2D analysis decreased in attached fimbriated cells but not in their nonfimbriated counterparts. Planktonic cells of nonfimbriated and fimbriated cells differ in their OMP patterns. Since equivalent amounts of protein were applied on all gels, this may indicate that fimbriae are a dominant part of the outer membrane and that in nonfimbriated cells a higher number of otherwise less dominant proteins appear on the gel. Changes were seen mainly as a reduction in the level of proteins in attached cells, whereas no OMPs that were strongly (>5-fold) increased or unique for adhesion could be resolved by 2D analysis. However, the protein concentration of the outer membrane fraction per OD600 unit, i.e., per total amount of cell debris in each sample, was not reduced in attached cells compared to levels in planktonic cells. Therefore, a possible loss of proteins during the isolation procedure can be excluded. Furthermore, the addition of Mg2+ ions during adhesion of strain PC31 did not counteract the major decrease of OMPs, which may further exclude the possibility that the adhesion process might result in the extraction of divalent cations, thus leading to an increased solubilization of proteins.
It may be argued that the reduction in OMP levels may not occur after the cells have become attached but that this begins in unattached cells, which gradually become attached after achieving a certain threshold level of OMP reduction. However, considering the facts that bacteria were starved and adjusted to the buffer system for 24 h prior to adhesion experiments and that unattached cells, removed from the samples 1 h before outer membranes were isolated from attached cells, showed OMP patterns comparable to those of planktonic cells, the observed changes must specifically occur in response to adhesion.
It is important to note, however, that the protein patterns revealed present only a partial picture of the total OMP fraction. Since it is known that heating proteins in the presence of urea causes carbamoylation, leading to the appearance of multiple-charge isoforms, protein samples were not heated prior to electrophoresis. This, in turn, causes certain OMPs, e.g., OmpC and OmpF, to form high-molecular-weight aggregates that do not enter the gel. This may also explain the absence of FimA from the 2D gels.
A possible explanation for the changes observed may be that the relative composition of OMPs changes in attached cells. The major decrease in the level of resolved OMPs may indicate an increased relative abundance of OMPs that are not resolved by 2D analysis. However, since the rate of protein synthesis of bacteria under the experimental conditions applied was low and its inhibition of protein synthesis by tetracycline did not affect the changes seen in attached fimbriated cells, this possibility is unlikely. It may also be taken into account that either all cells have a low rate of protein synthesis or most cells are inactive and only a few cells have a high rate of protein synthesis. On the other hand, the addition of protease inhibitors during adhesion of the fimbriated mutant strain MS7fim+ partly blocked the surface-induced alterations in the levels of several OMPs, which suggests that proteolytic activity contributes to the changes seen.
These results are in agreement with the observation that protein synthesis and protein degradation seem to play different roles in the adhesion of fimbriated or nonfimbriated cells. For the latter, inhibition of both protein synthesis and protein degradation caused a decrease in the number of attached cells, except on hydrophobic surfaces. However, for fimbriated cells inhibition of protein synthesis had no significant effect on adhesion whereas proteolytic degradation appeared to be involved to a high degree. As a surface-active effect of tetracycline and protease inhibitors could be excluded, these results support the idea that protein turnover is required for adhesion of fimbriated cells. With respect to the changes seen in the compositions of OMPs, we find it likely that the different mechanisms of initial surface contact of fimbriated and nonfimbriated cells not only influence strength of binding due to physicochemical characteristics but further induce different responses leading to distinct changes in the outer membrane. Our results indicate that type 1 fimbria-mediated surface contact may activate one or more proteases involved in adhesion, directly or indirectly leading to decreased levels of abundance of several OMPs. Interestingly, activation of a proteolytic system has also been suggested to be involved in shortening fimbriae during adhesion of E. coli to establish a closer contact with host cells (31). Furthermore, it is known that there are two different but overlapping regulatory pathways which respond to misfolded proteins in the bacterial cell envelope, both of which induce several OMP folding and degrading factors. First, the Cpx pathway is activated by misfolded proteins associated with the periplasmic face of the inner membrane (41) or by overproduction of fimbrial subunits (18). This signaling system has recently been suggested to be involved in adhesion (15), and it is proposed that the main function of this pathway is to monitor the assembly of surface organelles such as fimbriae (41). Second, the ςE pathway plays an important role in responding to alterations in OMP expression. For instance, activity of ςE is partly regulated by the production of OmpX (26), the level of which has in this study been shown to be strongly decreased during adhesion.
Some proteins resolved by 2D-PAGE have been identified by MALDI-TOF MS, all of which, except EF-Tu, were OMPs. While the relative levels of abundance of EF-Tu and BtuB were unaffected by adhesion, the level of OmpA, one of the major proteins in the outer membrane of E. coli which forms small nonspecific diffusion channels with low permeability, was moderately decreased. This protein is required for structural integrity of the outer membrane and the generation of a normal cell shape. It is also involved in cell-cell interactions (5) and invasion of E. coli (37). We identified OmpA as two mass isoforms with sizes of approximately 30 and 40 kDa and an abnormal mobility on SDS-polyacrylamide gels, also described previously (28), which may be related to the heat modifiability of this protein.
The levels of the proteins OmpX, Slp, and TolC showed the greatest decreases during adhesion. The integral membrane protein OmpX belongs to a family of virulence-related OMPs, such as OmpX in Enterobacter cloacae or Ail in Yersinia enterocolitica, which are involved in serum resistance or adhesion to host cells (27). However, the function of OmpX in E. coli is still unknown. Slp is a lipoprotein which stabilizes the outer membrane during carbon starvation or stationary phase (3). TolC is a trimeric OMP that is involved in signal sequence-independent extracellular secretion of proteins and toxic compounds in gram-negative bacteria (48). Interestingly, it is also required for proper expression of other OMP genes (29). Thus, it is possible that the strongly reduced level of TolC during adhesion may affect the expression of OMPs in attached cells. In agreement with our results, both ompX and tolC have been identified as members of the mar regulon (6, 7), which has recently been shown to be repressed in attached cells compared to levels in planktonic cells (21).
In summary, we suggest that type 1 fimbriae may play an important role in sensing a surface and causing adaptive changes in the cell, indicated by the alteration in the composition of OMPs. Physical interactions between type 1 fimbriae and a solid surface may elicit a physiological response that leads to structural changes in the outer membranes of attached cells. While proteolysis is partly involved in the response, it is likely that other mechanisms exist which still have to be described. Since adhesion may be regarded as a complex stimulus including different independent but interconnected signals, it remains to be shown whether the adhesion response observed is due to a direct effect of surface contact or caused by surface-related indirect effects. Studies in progress will isolate mutations in some of the identified genes to determine more precisely the role of a change in OMP expression during adhesion of E. coli to abiotic surfaces.
ACKNOWLEDGMENTS
We thank Per Klemm and Karen Krogfeldt for supplying strains.
This work was financially supported by a grant to M.H. from the Foundation for Strategic Research through the Marine Science and Technology (MASTEC) Program. Financial support for the protein identification part was obtained by K.-A.K. from the Swedish Medical Research Council (grants 3967 and 10435), the Swedish Research Council for Engineering Sciences, the Knut and Alice Wallenberg Foundation, and the Ingabritt and Arne Lundberg Foundation.
REFERENCES
- 1.Absolom D R, Lamberti F V, Policova Z, Zingg W, van Oss C J, Neumann A W. Surface thermodynamics of bacterial adhesion. Appl Environ Microbiol. 1983;46:90–97. doi: 10.1128/aem.46.1.90-97.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achouak W, Pages J-M, de Mot R, Molle G, Heulin T. A major outer membrane protein of Rahnella aquatilis functions as a porin and root adhesin. J Bacteriol. 1998;180:909–913. doi: 10.1128/jb.180.4.909-913.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander D M, St. John A C. Characterization of the carbon starvation-inducible and stationary phase-inducible gene slp encoding an outer membrane lipoprotein in Escherichia coli. Mol Microbiol. 1994;11:1059–1071. doi: 10.1111/j.1365-2958.1994.tb00383.x. [DOI] [PubMed] [Google Scholar]
- 4.Allison D G, Sutherland I W. The role of exopolysaccharides in adhesion of freshwater bacteria. J Gen Microbiol. 1987;133:1319–1327. [Google Scholar]
- 5.Anthony K G, Sherburne C, Sherburne R, Frost L S. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol Microbiol. 1994;13:939–953. doi: 10.1111/j.1365-2958.1994.tb00486.x. [DOI] [PubMed] [Google Scholar]
- 6.Aono R, Tsukagoshi N, Yamamoto M. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J Bacteriol. 1998;180:938–944. doi: 10.1128/jb.180.4.938-944.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbosa T M, Levy S B. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J Bacteriol. 2000;182:3467–3474. doi: 10.1128/jb.182.12.3467-3474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belas R, Simon M, Silverman M. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J Bacteriol. 1986;167:210–218. doi: 10.1128/jb.167.1.210-218.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brözel V S, Strydom G M, Cloete T E. A method for the study of de novo protein synthesis in Pseudomonas aeruginosa after attachment. Biofouling. 1995;8:195–201. [Google Scholar]
- 10.Costerton J W, Cheng K J, Geesey G G, Ladd T I, Nickel J C, Dasgupta M, Marrie T J. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 11.Dagostino L, Goodman A E, Marshall K C. Physiological responses induced in bacteria adhering to surfaces. Biofouling. 1991;4:113–119. [Google Scholar]
- 12.Dalton H, Poulsen L K, Halasz P, Angles M L, Goodman A E, Marshall K C. Substratum-induced morphological changes in a marine bacterium and their relevance to biofilm structure. J Bacteriol. 1994;176:6900–6906. doi: 10.1128/jb.176.22.6900-6906.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies D G, Chakrabarty A M, Geesey G G. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Environ Microbiol. 1993;59:1181–1186. doi: 10.1128/aem.59.4.1181-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derjaguin B V, Landau L. Theory of the stability of strongly charged hydrophobic sols and of the adhesion of strongly charged particles in solutions of electrolytes. Acta Physicochim URSS. 1941;14:633–662. [Google Scholar]
- 15.Dorel C, Vidal O, Prigent-Combaret C, Vallet I, Lejeune P. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol Lett. 1999;178:169–175. doi: 10.1111/j.1574-6968.1999.tb13774.x. [DOI] [PubMed] [Google Scholar]
- 16.Filip C, Fletcher G, Wulff J L, Earhart C F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Görg A. High-resolution two-dimensional electrophoresis of proteins using immobilized pH gradients. In: Celis J E, editor. Cell biology: a laboratory handbook. New York, N.Y: Academic Press, Inc.; 1994. pp. 231–242. [Google Scholar]
- 18.Jones C H, Danese P N, Pinkner J S, Silhavy T J, Hultgren S J. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 1997;16:6394–6406. doi: 10.1093/emboj/16.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufmann A, Stierhof Y-D, Henning U. New outer membrane-associated protease of Escherichia coli K-12. J Bacteriol. 1994;176:359–367. doi: 10.1128/jb.176.2.359-367.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klemm P, Jørgensen B J, van Die I, de Ree H, Bergmans H. The fim genes responsible for synthesis of type 1 fimbriae in Escherichia coli, cloning and genetic organization. Mol Gen Genet. 1985;199:410–414. doi: 10.1007/BF00330751. [DOI] [PubMed] [Google Scholar]
- 21.Maira-Litran T, Allison D G, Gilbert P. Expression of the multiple antibiotic resistance operon (mar) during growth of Escherichia coli as a biofilm. J Appl Microbiol. 2000;88:243–247. doi: 10.1046/j.1365-2672.2000.00963.x. [DOI] [PubMed] [Google Scholar]
- 22.Maniatis T E, Fritsch F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 23.Marshall K C. Biofilms: an overview of bacterial adhesion, activity, and control at surfaces. ASM News. 1992;58:202–207. [Google Scholar]
- 24.McCormick B A, Franklin D P, Laux D C, Cohen P S. Type 1 pili are not necessary for colonization of the streptomycin-treated mouse large intestine by type 1-piliated Escherichia coli F-18 and E. coli K-12. Infect Immun. 1989;57:3022–3029. doi: 10.1128/iai.57.10.3022-3029.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCormick B A, Klemm P, Krogfelt K A, Burghoff R L, Pallesen L, Laux D C, Cohen P S. Escherichia coli F-18 phase locked ‘on’ for expression of type 1 fimbriae is a poor colonizer of the streptomycin-treated mouse large intestine. Microb Pathog. 1993;14:33–43. doi: 10.1006/mpat.1993.1004. [DOI] [PubMed] [Google Scholar]
- 26.Mecsas J, Rouviere P E, Erickson J W, Donohue T J, Gross C A. The activity of ςE, an Escherichia coli heat-inducible ς-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993;7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 27.Mecsas J, Welch R, Erickson J W, Gross C A. Identification and characterization of an outer membrane protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including Ail of Yersinia enterocolitica. J Bacteriol. 1995;177:799–804. doi: 10.1128/jb.177.3.799-804.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molloy M P, Herbert B R, Walsh B J, Tyler M I, Traini M, Sanchez J-C, Hochstrasser D F, Williams K L, Gooley A A. Extraction of membrane proteins by differential solubilization for separation using two-dimensional gel electrophoresis. Electrophoresis. 1998;19:837–844. doi: 10.1002/elps.1150190539. [DOI] [PubMed] [Google Scholar]
- 29.Morona R, Reeves P. The tolC locus of Escherichia coli affects the expression of three major outer membrane proteins. J Bacteriol. 1982;150:1016–1023. doi: 10.1128/jb.150.3.1016-1023.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrisey J H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- 31.Mulvey M A, Lope-Boado Y S, Wilson C L, Roth R, Parks W C, Heuser J, Hultgren S J. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 32.Norbeck J, Blomberg A. Two-dimensional electrophoretic separation of yeast proteins using a non-linear wide range (pH 3–10) immobilized pH gradient in the first dimension: reproducibility and evidence for isoelectric focusing of alkaline (pI > 7) proteins. Yeast. 1997;13:1519–1534. doi: 10.1002/(SICI)1097-0061(199712)13:16<1519::AID-YEA211>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 33.Nyström T, Mården P, Kjelleberg S. Relative changes in incorporation rates of leucine and methionine during starvation survival of two bacteria isolated from marine waters. FEMS Microbiol Ecol. 1986;38:285–292. [Google Scholar]
- 34.Otto K, Elwing H, Hermansson M. Effect of ionic strength on the initial interactions of Escherichia coli with surfaces studied on-line by a novel quartz crystal microbalance. J Bacteriol. 1999;181:5210–5218. doi: 10.1128/jb.181.17.5210-5218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otto K, Elwing H, Hermansson M. The role of type-1 fimbriae in adhesion of Escherichia coli to hydrophilic and hydrophobic surfaces. Colloids Surf B. 1999;15:99–111. [Google Scholar]
- 36.Overbeeke N, Lugtenberg B. Expression of outer membrane protein e of Escherichia coli K12 by phosphate limitation. FEBS Lett. 1980;112:229–232. doi: 10.1016/0014-5793(80)80186-4. [DOI] [PubMed] [Google Scholar]
- 37.Prasadarao N V, Wass C A, Weiser J N, Stins M F, Huang S-H, Kim K S. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect Immun. 1996;64:146–153. doi: 10.1128/iai.64.1.146-153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pratt L A, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 39.Pratt L A, Silhavy T J. Porin regulon of Escherichia coli. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 105–127. [Google Scholar]
- 40.Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J Bacteriol. 1999;181:5993–6002. doi: 10.1128/jb.181.19.5993-6002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raivio T L, Silhavy T J. The ςE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr Opin Microbiol. 1999;2:159–165. doi: 10.1016/S1369-5274(99)80028-9. [DOI] [PubMed] [Google Scholar]
- 42.Rosenfeld J, Capdevielle J, Guillemot J C, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- and two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez J-C, Rouge V, Pisteur M, Ravier F, Tonella L, Moosmayer M, Wilkins M R, Hochstrasser D F. Improved and simplified in-gel sample application using reswelling of dry immobilized pH gradients. Electrophoresis. 1997;18:324–327. doi: 10.1002/elps.1150180305. [DOI] [PubMed] [Google Scholar]
- 44.Schembri M A, Pallesen L, Connell H, Hasty D L, Klemm P. Linker insertion analysis of the FimH adhesin of type 1 fimbriae in an Escherichia coli fimH-null background. FEMS Microbiol Lett. 1996;137:257–263. doi: 10.1111/j.1574-6968.1996.tb08115.x. [DOI] [PubMed] [Google Scholar]
- 45.Schröder W, Moser I. Primary structure analysis and adhesion studies on the major outer membrane protein of Campylobacter jejuni. FEMS Microbiol Lett. 1997;150:141–147. doi: 10.1111/j.1574-6968.1997.tb10362.x. [DOI] [PubMed] [Google Scholar]
- 46.Sperandio V, Girón J A, Silveira W D, Kaper J B. The OmpU outer membrane protein, a potential adherence factor of Vibrio cholerae. Infect Immun. 1995;63:4433–4438. doi: 10.1128/iai.63.11.4433-4438.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stentebjerg-Olesen B, Pallesen L, Bogø Jensen L, Christiansen G, Klemm P. Authentic display of a cholera toxin epitope by chimeric type 1 fimbriae: effects of insert position and host background. Microbiology. 1997;143:2027–2038. doi: 10.1099/00221287-143-6-2027. [DOI] [PubMed] [Google Scholar]
- 48.Thanabalu T, Koronakis E, Hughes C, Koronakis V. Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 1998;17:6487–6496. doi: 10.1093/emboj/17.22.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van der Mei H C, Weerkamp A H, Busscher H J. Physico-chemical surface characteristics and adhesive properties of Streptococcus salivarius strains with defined cell surface structures. FEMS Microbiol Lett. 1987;40:15–19. [Google Scholar]
- 50.Vandevivere P, Kirchman D L. Attachment stimulates exopolysaccharide synthesis by a bacterium. Appl Environ Microbiol. 1993;59:3280–3286. doi: 10.1128/aem.59.10.3280-3286.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Oss C J. Energetics of cell-cell and cell-biopolymer interactions. Cell Biophys. 1989;14:1–16. doi: 10.1007/BF02797387. [DOI] [PubMed] [Google Scholar]
- 52.Verwey E J W, Overbeek J T G. Theory of the stability of lyophobic colloids. Amsterdam, The Netherlands: Elsevier; 1948. [Google Scholar]
- 53.Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilm on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J P, Normark S. Induction of gene expression in Escherichia coli after pilus-mediated adherence. Science. 1996;273:1234–1236. doi: 10.1126/science.273.5279.1234. [DOI] [PubMed] [Google Scholar]
- 55.Zhao S, Meng J, Doyle M P, Meinersman R, Wang G, Zhao P. A low molecular weight outer-membrane protein of Escherichia coli O157:H7 associated with adherence to INT407 cells and chicken caeca. J Med Microbiol. 1996;45:90–96. doi: 10.1099/00222615-45-2-90. [DOI] [PubMed] [Google Scholar]