Abstract
Plants utilize localized cell-surface and intracellular receptors to sense microbes and activate the influx of calcium, which serves as an important second messenger in eukaryotes to regulate cellular responses. However, the mechanisms through which plants decipher calcium influx to activate immune responses remain largely unknown. Here, we show that pathogen-associated molecular patterns (PAMPs) trigger calcium-dependent phosphorylation of CAM-BINDING PROTEIN 60-LIKE G (CBP60g) in Arabidopsis (Arabidopsis thaliana). CALCIUM-DEPENDENT PROTEIN KINASE5 (CPK5) phosphorylates CBP60g directly, thereby enhancing its transcription factor activity. TOUCH 3 (TCH3) and its homologs CALMODULIN (CAM) 1/4/6 and CPK4/5/6/11 are required for PAMP-induced CBP60g phosphorylation. TCH3 interferes with the auto-inhibitory region of CPK5 and promotes CPK5-mediated CBP60g phosphorylation. Furthermore, CPKs-mediated CBP60g phosphorylation positively regulates plant resistance to soil-borne fungal pathogens. These lines of evidence uncover a novel calcium signal decoding mechanism during plant immunity through which TCH3 relieves auto-inhibition of CPK5 to phosphorylate and activate CBP60g. The findings reveal cooperative interconnections between different types of calcium sensors in eukaryotes.
Activation of CAM-BINDING PROTEIN 60-LIKE G by the cooperative actions of calcium sensors regulates plant immunity against soil-borne fungal pathogens.
Introduction
Calcium ions (Ca2+), which encode signals in response to environmental changes and stresses, serve as an important second messenger in eukaryotes. Numerous stimuli trigger the cytosolic increase of Ca2+ (Klaus and Ikura, 2002; Bouché et al., 2005; Mccormack et al., 2005; Luan and Wang, 2021). Eukaryotic cells utilize calcium sensors, most of which harbor the helix-loop-helix EF-hand motif that binds to Ca2+ with a high affinity to transduce Ca2+ signals to regulate cellular processes and responses. Calmodulin (CaM/CAM) and CaM-like (CML) proteins, calcium-dependent protein kinases (CDPKs/CPKs), calcineurin B-like proteins (CBL), and CBL-interacting protein kinases (CIPKs) represent the three major types of sensors that receive Ca2+ signals in plant cells (Klaus and Ikura, 2002; Bouché et al., 2005; Mccormack et al., 2005; Luan, 2009; Yuan et al., 2017; Zipfel and Oldroyd, 2017; Luan and Wang, 2021). These Ca2+ signals are consequently delivered to diverse downstream effectors that decode signals from sensors and mount distinct and appropriate cellular responses, such as gene transcription, enzyme activation, protein folding and transport, and cytoskeletal rearrangement (Klaus and Ikura, 2002; Bouché et al., 2005; Mccormack et al., 2005; Luan, 2009; Luan and Wang, 2021).
CDPK calcium sensors rapidly respond to pathogen-associated molecular pattern (PAMP)-induced elevation of cytosolic Ca2+ and execute the kinase function via phosphorylating diverse substrates. In Arabidopsis thaliana, CPK4/CPK5/CPK6/CPK11 have been identified as Ca2+ sensors that are critical to transcriptional reprogramming during immune signaling (Boudsocq et al., 2010; Dubiella et al., 2013; Gao et al., 2013a; Kadota et al., 2014; Monaghan et al., 2014). CPK5 targets the NADPH oxidase RbohD and transcription factor WRKY33 to regulate the production of reactive oxygen species (ROS) and camalexin biosynthesis, respectively (Dubiella et al., 2013; Zhou et al., 2020). CPK28 phosphorylates PLANT U-BOX 25 (PUB25) and PUB26 to ubiquitinate BOTRYTIS-INDUCED KINASE 1 (BIK1) and trigger its degradation, thereby negatively regulating ROS production and plant immunity (Kadota et al., 2014, 2015; Monaghan et al., 2014). CAM is composed of 148 amino acids without catalytic activity. Upon binding to Ca2+, CAMs transduce signals to regulate a variety of cellular processes by targeting calmodulin-binding proteins (Mccormack et al., 2005). The Arabidopsis CAM type calcium sensor CAM7 binds to CYCLIC NUCLEOTIDE-GATED CHANNEL 2 (CNGC2) and CNGC4 and negatively controls their ion channel activity (Yu et al., 1998; Clough et al., 2000; Yu et al., 2000; Balagué et al., 2003; Jurkowski et al., 2004; Moeder et al., 2011; Tian et al., 2019; Luan and Wang, 2021). Plants encode some unique calmodulin-binding proteins that are absent in animals and fungi. CBP60g, a member of the plant-specific calmodulin-binding protein 60 family, has been shown to possess the ability to bind CAM and regulate plant immunity (Wang et al., 2009; Zhang et al., 2010b; Qin et al., 2018). CBL-CIPK interaction pairs belong to another type of calcium sensor and form a network for regulating plant responses to environmental stresses, nutrient sensing, and adaptation (Luan, 2009). Unraveling the mechanisms and specificities that decode diverse Ca2+ signals, within the plant’s sensory system and their interconnections with downstream effectors, remains an important challenge (Bouché et al., 2005; Mccormack et al., 2005; Luan and Wang, 2021).
Plants have evolved a sophisticated immune system to defend against invading microbes. Microbial infections trigger a rapid elevation of cytosolic Ca2+ in plant cells and consecutively initiate a set of calcium-dependent immune responses (Knoblauch et al., 2014; Choi et al., 2016; Sun and Zhang, 2020). Plant cell surface-localized pattern recognition receptors (PRRs) are used to sense conserved molecular signatures derived from microbes, namely pathogen-associated or microbe-associated molecular patterns (PAMPs/MAMPs), and activate PAMP-triggered immunity (PTI; Boller and Felix, 2009; Zhang et al., 2010a; Kang et al., 2015; Zhou and Zhang, 2020). Bacterial harpins, flagellin, peptidoglycans, and elongation factor-Tu are well-characterized PAMPs that elicit plant immune responses (Wei et al., 1992; He et al., 1993; Felix et al., 1999; Gómez-Gómez et al., 2002; Kunze et al., 2004; Zipfel et al., 2006; Erbs et al., 2008; Sun et al., 2017). Fungal cell wall components, such as chitin and oligosaccharide fragments (Felix et al., 1993; Hahn, 1996; Ron and Avni, 2004; Nürnberger et al., 2010; Zhang et al., 2014), and necrosis- and ethylene-inducing peptide 1 (Nep1)-like proteins (NLPs; Nürnberger et al., 1994; Sejalon-Delmas et al., 1997; Gaulin et al., 2006; Qutob et al., 2006; Brunner et al., 2014; Ma et al., 2015), have also been characterized as PAMPs. Once PRRs are activated, they trigger the rapid phosphorylation of receptor-like cytoplasmic kinases (RLCKs), such as Botrytis induced kinase 1 (BIK1) and PBS1-like proteins (PBLs), which then activate cyclic nucleotide-gated channels to trigger calcium influx (Tian et al., 2019).
Successful pathogens have evolved diverse pathogenic strategies. Delivering pathogenic effectors into plants by either evading or suppressing PTI activation is a major pathogenic strategy. To counteract this, plants have evolved intracellular nucleotide-binding domain (NBD) and leucine-rich repeat (LRR) receptors (NLRs) to sense effector activities and initiate effector-triggered immunity (ETI; Jones and Dangl, 2006; Zhou and Chai, 2008; Boller and He, 2009; Cui et al., 2009; Tamborski and Krasileva, 2020). Recent studies have discovered that some intracellular NLRs and helper NLRs form calcium channels to mediate Ca2+ transport into the cytoplasm, which activates calcium-dependent immune responses (Bi et al., 2021; Jacob et al., 2021). The activation of both PRRs and NLRs triggers calcium influx; however, the mechanism that underlies the decoding of calcium signal during immunity remains largely uncharacterized.
Verticillium dahliae is a soil-borne fungal pathogen that infects the roots of many plants causing Verticillium wilt. Very limited plant immune components against this pathogen have been identified. Tomato (Solanum lycopersicum) Ve1 confers resistance to V. dahliae strains carrying Ave1 (Schaible et al., 1951; Fradin et al., 2009). Studies on cotton (Gossypium hirsutum) identified several components that positively regulate resistance to V. dahliae (Qin et al., 2004; Gao et al., 2011; 2013b; Li et al., 2014; Sun et al., 2014; Yang et al., 2015; Li et al., 2016). Plant calmodulin-binding proteins (CBPs) have been shown to regulate plant development and responses to biotic and abiotic stresses (Reddy et al., 2002; Bouché et al., 2005). CBP60g is induced by pathogen infection and regulates the expression of a great number of defense-related genes (Bouché et al., 2005; Zhang et al., 2010b; Wang et al., 2011). We previously showed that V. dahliae delivers a secretory protein to inhibit CBP60g activity and promote virulence (Qin et al., 2018), further proving the role of CBP60g in plant immunity against soil-borne fungal pathogens. Plant immune mechanisms against soil-borne fungal pathogens remain unclear. We aim to determine the mechanisms by which plants decipher the calcium influx to activate downstream immune responses against soil-borne fungal pathogens.
Results
PAMP treatment induces calcium-dependent phosphorylation of CBP60g
The examination of the behavior of protoplast-expressed CBP60g, using Phos-tag SDS-PAGE followed by immunoblot, revealed a slower migration of CBP60g following treatment of flg22, which is an N-terminal peptide carrying PAMP activity of bacterial flagellin (Figure 1A). The appearance of this slower migration was clearly removed by lambda protein phosphatase (PPase) treatment, indicating elevated CBP60g phosphorylation upon flg22 stimulation (Figure 1A). 35S-promoter-driven CBP60g-FLAG (Pro35S:CBP60g-FLAG) transgenic plants were then constructed to verify PAMP-induced CBP60g phosphorylation in plants. A slower migration of transgenically produced CBP60g was also observed following the flg22 treatment (Figure 1B). Accordingly, when the sample was treated with protein PPase, it restored its normal migration (Figure 1B). In addition to the role in plant resistance to several Pseudomonas syringae strains (Wang et al., 2009; Zhang et al., 2010b), CBP60g also contributes to the resistance against fungal pathogen V. dahliae (Qin et al., 2018). Afterward, we examined whether PAMPs derived from V. dahliae could induce CBP60g phosphorylation. CBP60g-FLAG transgenic plants were treated with chitin and nlp20, a core peptide carrying the PAMP activity of V. dahliae NLPs. A slower migration of CBP60g was also detected in response to chitin and nlp20 treatments (Figure 1, C and D). The above results indicate that both bacterial and fungal PAMP recognition triggers CBP60g phosphorylation.
Figure 1.
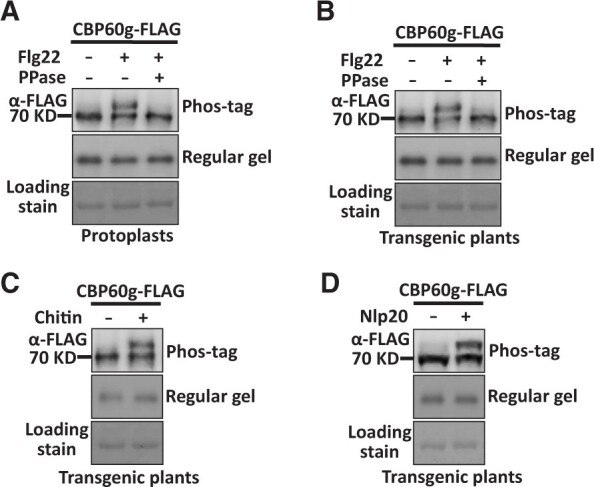
PAMP treatment induces phosphorylation of CBP60g. A, Flg22 induces phosphorylation of CBP60g in protoplasts. Col-0 protoplasts were transfected with Pro35S:CBP60g-FLAG and treated with or without 2 µM flg22. Total protein extracted from protoplasts was treated with or without 2 unit/µL PPase as indicated. B, Flg22 induces phosphorylation of CBP60g in transgenic plants. Pro35S:CBP60g-FLAG transgenic plants were treated with H2O or 2 µM flg22 for 5 min and total protein extracted from leaves was treated with or without 2 unit/µL PPase as indicated. C and D, Chitin or nlp20 induces phosphorylation of CBP60g in transgenic plants. Pro35S:CBP60g-FLAG transgenic plants were treated with H2O, 250 µg/mL chitin, or 10 µM nlp20 as indicated for 5 min. Total protein was extracted from treated leaves. Protein samples were separated by Phos-tag SDS-PAGE and subjected to anti-FLAG immunoblot. Coomassie brilliant blue staining of the filter indicates loading of the protein. The experiments were repeated 3 times with similar results.
Pathogen recognition triggers activation of calcium channels and cytosolic increase of Ca2+ (Luan, 2009; Luan and Wang, 2021). We used the aequorin-transformed Arabidopsis reporter line (Col-0AEQ) to monitor cytoplasmic calcium in response to V. dahliae recognition (Knight et al., 1991; Tian et al., 2019). The results showed that the V. dahliae treatment induced calcium influx in Arabidopsis (Supplemental Figure S1). To investigate whether calcium signals are involved in PAMP-induced CBP60g phosphorylation, CBP60g-FLAG transgenic plants were pretreated with calcium channel inhibitor LaCl3, treated with or without PAMP, and then examined for CBP60g phosphorylation. As shown in Figure 2A, the LaCl3 treatment completely blocked PAMP-induced CBP60g phosphorylation, suggesting that calcium influx is required in this process. CNGC2 and CNGC4 are phosphorylated by BIK1 and form an active calcium channel, allowing calcium ions to enter the cytoplasm (Tian et al., 2019). We found that PAMP-induced CBP60g phosphorylation was weakened in the bik1 pbl1 mutant (Figure 2B). Moreover, PAMP-induced CBP60g phosphorylation was significantly impaired in the cngc2 mutants (Supplemental Figure S2) compared to wild-type (WT) plants (Figure 2C). The above results indicate that a calcium signal is required for PAMP-induced CBP60g phosphorylation. Thus, perception of PAMPs induces calcium-dependent phosphorylation of CBP60g.
Figure 2.
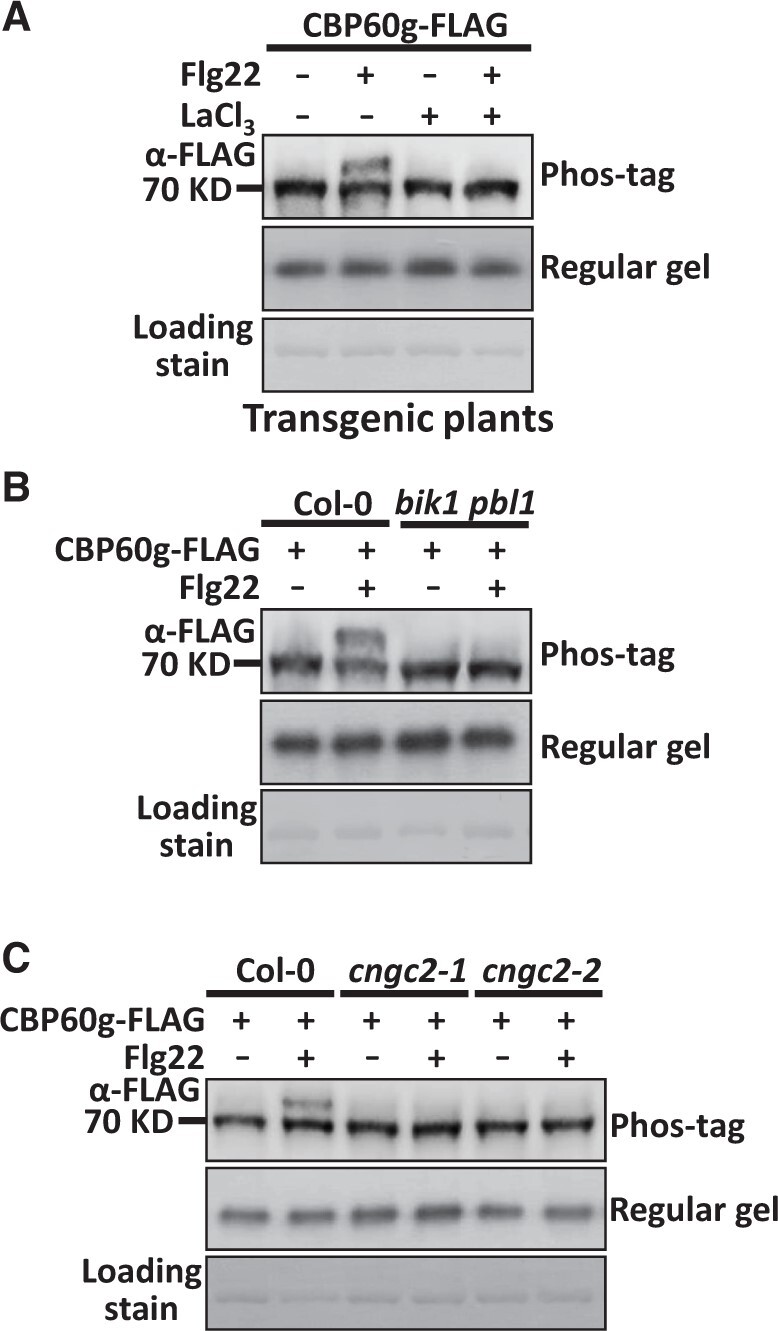
The interruption of CNGC activity inhibits PAMP-induced phosphorylation of CBP60g. A, Treatment of LaCl3 blocks flg22-induced phosphorylation of CBP60g. Pro35S:CBP60g-FLAG transgenic plants were pretreated with 10 µM LaCl3 or H2O before 2 µM flg22 treatment. B, Flg22-induced phosphorylation of CBP60g is impaired in the bik1 pbl1 mutant. C, Flg22-induced phosphorylation of CBP60g is impaired in the cngc2 mutant. Protoplasts isolated from WT or mutant plants were transfected with Pro35S:CBP60g-FLAG, and then treated with or without 2 µM flg22 for 5 min. Total protein was extracted and protein samples were separated by SDS-PAGE and subjected to anti-FLAG immunoblot. CBB staining of the filter indicates loading of the protein. The experiments were repeated 3 times with similar results.
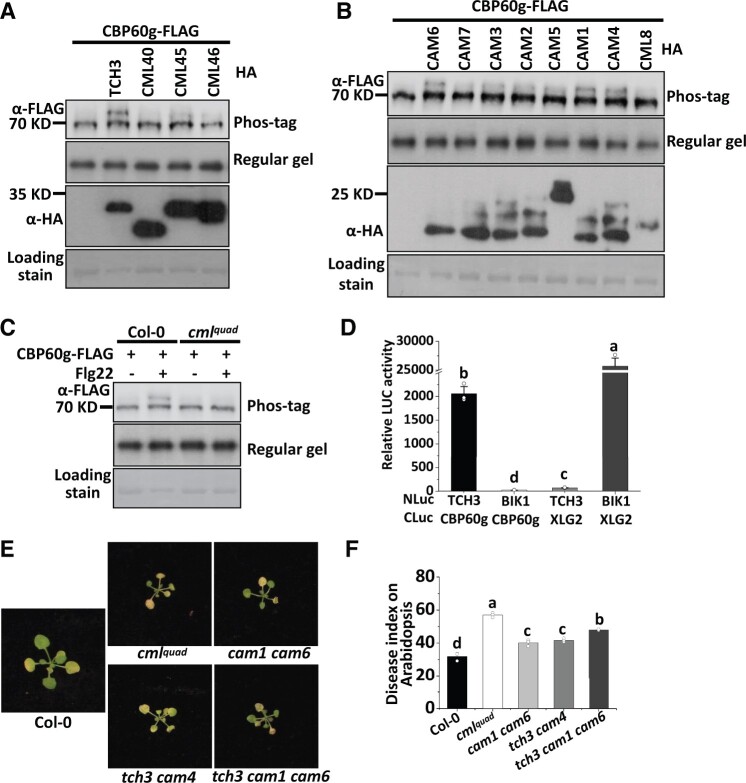
TCH3 and CAM1/4/6 contribute to PAMP-induced phosphorylation of CBP60g and V. dahliae resistance
To cope with calcium influx, plants employ different types of calcium sensors that decode calcium signals. To study the mechanism involved in calcium-dependent phosphorylation of CBP60g, we examined the role of calcium sensors in PAMP-induced phosphorylation of CBP60g. The Arabidopsis genome encodes 7 CAMs and more than 50 CMLs (Mccormack et al., 2005). Among them, TOUCH 3/TCH3 (also named CML12; Sistrunk et al., 1994; McCormack and Braam, 2003), CML40, CML45, and CML46 have been reported to be strongly upregulated in response to PAMP treatment (Supplemental Figure S3; Chen et al., 2009; Li et al., 2015). We next examined the role of these calcium sensors in PAMP-induced phosphorylation of CBP60g. These CMLs were then individually co-expressed with CBP60g-FLAG in protoplasts. Over-expression of TCH3, but not CML40, CML45, and CML46, induces CBP60g phosphorylation in the absence of PAMP treatment (Figure 3A), suggesting that TCH3 plays a role in CBP60g phosphorylation. A single mutation of TCH3 gene (Supplemental Figure S2; Gleeson et al., 2012) did not obviously impair PAMP-induced CBP60g phosphorylation (Supplemental Figure S4); however, a functional redundancy of TCH3 with its homologs was speculated. We subsequently examined whether the over-expression of homologs related to TCH3 could induce CBP60g phosphorylation. CAMs were individually coexpressed with CBP60g-FLAG, among which CAM1, CAM4, and CAM6 induced CBP60g phosphorylation in the absence of PAMP treatment (Figure 3B). CAM2 and CAM3 may be also involved in inducing CBP60g phosphorylation but have a minor contribution. Since CAM1 and CAM4 are isoforms and differ from CAM6 by only 5 amino acids, we generated a quadruple mutant (cmlquad) with mutations in TCH3, CAM1, CAM4, and CAM6 by CRISPR-Cas9 gene-editing approach. Protoplasts isolated from the cmlquad mutant were transfected with CBP60g-FLAG and induced with flg22. We observed a substantial impairment of CBP60g phosphorylation induced by PAMP in the cmlquad mutant (Figure 3C), as well as less pronounced reductions of PAMP-induced CBP60g phosphorylation in the cam1 and cam6 mutants and cam1 cam6 and tch3 cam4 double mutants (Supplemental Figure S4). These results indicate that TCH3 and closely associated CAMs are required for PAMP-induced CBP60g phosphorylation.
Figure 3.
TCH3 and CAM1/4/6 contribute to PAMP-induced CBP60g phosphorylation and resistance to V. dahliae. A, Expression of TCH3 induces phosphorylation of CBP60g. Protoplasts isolated from WT plants were transfected with Pro35S:CBP60g-FLAG alone or together with HA fusions of TCH3, CML40, CML45, or CML46 as indicated. B, Expression of homologs of TCH3 induces phosphorylation of CBP60g. Protoplasts isolated from WT plants were transfected with Pro35S: CBP60g-FLAG alone or together with HA fusions of CAM1-7 or CML8 as indicated. C, CBP60g phosphorylation induced by flg22 is significantly compromised in the cmlquad mutant. Protoplasts isolated from WT or mutant plants were transfected with Pro35S:CBP60g-FLAG and treated with or without 2 µM flg22 for 5 min. Protein samples were separated by Phos-tag SDS-PAGE and subjected to anti-FLAG immunoblot. CBB staining of the filter indicates loading of the protein. D, TCH3 interacts with CBP60g. Nicotiana benthamiana leaves infiltrated with Agrobacterium strains carrying Pro35S:NLuc-TCH3, Pro35S:CLuc-CBP60g, Pro35S:CLuc-XLG2, or Pro35S:NLuc-BIK1 as indicated were subjected to luciferase complementation assay. Infiltrated N. benthamiana leaves were sliced into strips 2 d post-infiltration, and relative luminescence was determined by Microplate Luminometer. Error bars indicate SD of three biological repeats. Student’s t test was carried out to determine the significance of difference. Different letters indicate significant differences. NLuc stands for N-terminal fragment of firefly luciferase; CLuc stands for C-terminal fragment of firefly luciferase. E and F, The cmlquad mutant is compromised in resistance to V. dahliae. Disease symptoms (E) and index analyses (F) of Arabidopsis WT, cmlquad, cam1 cam6, tch3 cam4, or tch3 cam1 cam6 mutant plants infected with V. dahliae. The plants were photographed and subjected to disease index analyses 3–4 weeks post-inoculation. Disease indexes were evaluated with three replicates generated from 24 plants for each inoculum. Error bars indicate SD of three biological replicates. Student’s t test was carried out to determine the significance of difference. Different letters indicate significant differences.
CBP60g harbors a CAM binding motif and has been shown to bind CAM in vitro (Zhang et al., 2010b). A split-luciferase assay was then conducted to test putative interactions between CBP60g and TCH3 or its closely related CAMs. Coexpression of the N terminus of luciferase (NLuc)-tagged TCH3 and C terminus of luciferase (CLuc)-tagged CBP60g resulted in much higher luciferase activity compared to the co-expression of NLuc-TCH3 and CLuc-XLG2, or NLuc-BIK1 and CLuc-CBP60g (Figure 3D and Supplemental Figure S5E), indicating a specific interaction between TCH3 and CBP60g. Similarly, CAM1, CAM4, and CAM6 interact with CBP60g (Supplemental Figure S5, A–D). Levels of NLuc- and CLuc-fusion proteins were detected by immunoblot (Supplemental Figure S5F). Thus, CBP60g interacts with TCH3 and its closely related CAMs.
We previously showed that CBP60g contributes to plant resistance to V. dahliae (Qin et al., 2018). We subsequently examined whether the cmlquad mutant exhibits an altered resistance to V. dahliae. The cmlquad, tch3 cam1 cam6, cam1 cam6, and tch3 cam4 mutant and wild-type plants were infected with V. dahliae and subjected to disease symptom analyses. As shown in Figure 3, E and F, the tch3 cam1 cam6, cam1 cam6, and tch3 cam4 mutants exhibited attenuated resistance and the cmlquad mutant displayed a significantly compromised resistance to V. dahliae when compared to WT plants. Thus, TCH3 and its closely related CAMs contribute to PAMP-induced phosphorylation of CBP60g and resistance to V. dahliae.
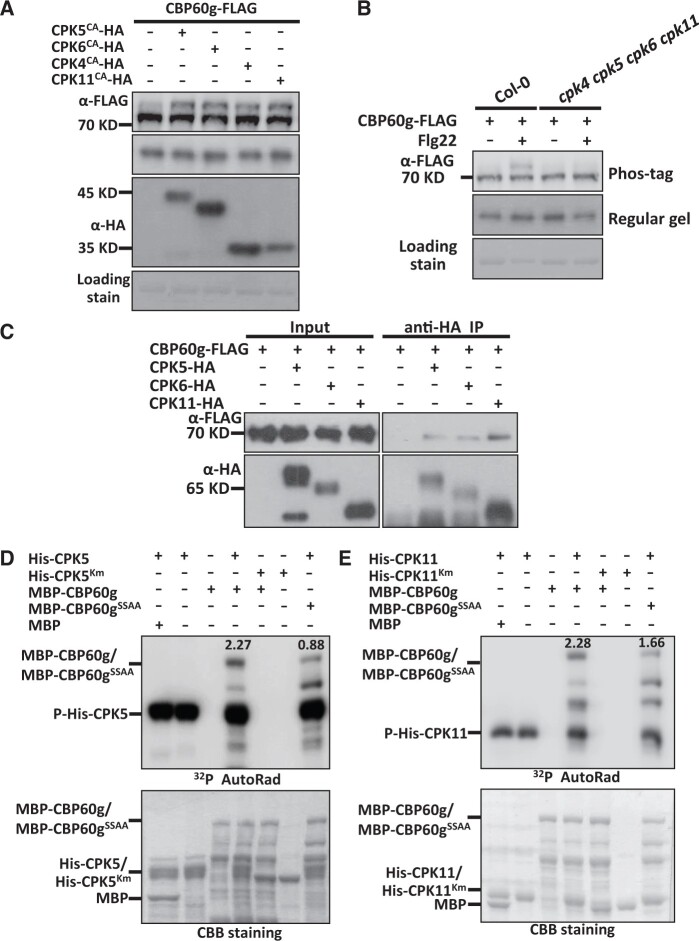
CPK5 directly phosphorylates CBP60g
Although the expression of TCH3 promotes CBP60g phosphorylation, TCH3 exhibits neither auto-phosphorylation nor kinase activity towards CBP60g (Supplemental Figure S6), suggesting that TCH3 is not capable of phosphorylating CBP60g directly. We then sought to explore the kinases responsible for calcium-dependent phosphorylation of CBP60g. CDPKs are another group of calcium sensors that regulate plant responses to stresses and development (Cheng et al., 2002; Harper and Harmon, 2005; Boudsocq et al., 2010). CPK4, CPK5, CPK6, and CPK11 are auto-inhibitory protein kinases and function redundantly in PTI signaling (Boudsocq et al., 2010). To investigate whether CDPKs contribute to PAMP-induced CBP60g phosphorylation, genes encoding constitutive active forms of CPK4 (CPK4CA), CPK5 (CPK5CA), CPK6 (CPK6CA), and CPK11 (CPK11CA) were constructed and co-expressed with CBP60g-FLAG in protoplasts. As shown in Figure 4A, CPK4CA, CPK5CA, CPK6CA, and CPK11CA induced CBP60g phosphorylation in the absence of PAMP treatment. Moreover, PAMP-induced CBP60g phosphorylation was significantly impaired in the cpk4 cpk5 cpk6 cpk11 mutant compared with that in WT plants (Figure 4B), which indicated that CPK4, CPK5, CPK6, and CPK11 are required for PAMP-induced phosphorylation of CBP60g and raised the possibility for CDPK-mediated CBP60g phosphorylation.
Figure 4.
CPK5 and CPK11 interact with and directly phosphorylate CBP60g. A, Expression of genes encoding constitutive active forms of CPK4, CPK5, CPK6, or CPK11 induces phosphorylation of CBP60g. Protoplasts isolated from WT plants were transfected with Pro35S:CBP60g-FLAG alone or together with constructs encoding HA-tagged CPK4CA, CPK5CA, CPK6CA, or CPK11CA as indicated. B, CBP60g phosphorylation induced by flg22 is significantly compromised in the cpk4 cpk5 cpk6 cpk11 mutant. Protoplasts isolated from WT or mutant plants were transfected with Pro35S:CBP60g-FLAG and treated with or without 2 µM flg22 for 5 min as indicated. Protein samples were separated by Phos-tag SDS-PAGE and subjected to anti-FLAG or anti-HA immunoblot. CBB staining of the filter indicates loading of the protein. C, CBP60g interacts with CPK5/6/11. Arabidopsis protoplasts were transfected with CBP60g-FLAG alone or together with CPK5/6/11-HA as indicated. Protein was extracted 16 h post-transfection and immunoprecipitated with anti-HA. Anti-FLAG or anti-HA immunoblot was used to detect the presence of CBP60g-FLAG or CPK5/6/11-HA in the purified complex, respectively. D and E, CBP60g is phosphorylated by CPK5 and CPK11 in vitro. MBP-CBP60g or MBP-CBP60gSSAA (CBP60gS450AS495A mutant form) was incubated with His-CPK5 or His-CPK5Km (D) and His-CPK11 or His-CPK11Km (E) as indicated and subjected to in vitro phosphorylation assay. CPK5Km and CPK11Km indicate the kinase-inactive mutant form of CPK5 and CPK11, respectively. CBB staining indicates loading of the protein. The ratio of phosphorylation signal to protein amount was shown at the top of phosphor band. The experiments were repeated 3 times with similar results.
This prompted us to test putative interactions between CDPKs and CBP60g. CBP60g-FLAG was transfected alone or along with CPK5-HA in protoplasts and immunoprecipitated with anti-HA antibody. Immunoprecipitates were further detected with anti-FLAG and anti-HA immunoblot. CPK5 was found to be coimmunoprecipitated with CBP60g, indicating that CPK5 interacts with CBP60g in plants (Figure 4C). Similarly, CPK6 and CPK11 also interact with CBP60g in plants (Figure 4C). An in vitro phosphorylation assay was then performed to determine whether CBP60g is a direct substrate of CDPKs. Recombinant CPK5 and CPK11 WT and kinase-deficient forms, CPK5Km and CPK11Km, were purified and incubated with recombinant MBP-CBP60g protein. By itself, purified MBP-CBP60g did not exhibit autophosphorylation activity, whereas co-incubation with purified CPK5 and CPK11 resulted in MBP-CBP60g phosphorylation. However, CPK5Km and CPK11Km failed to phosphorylate MBP-CBP60g (Figure 4, D–E). Thus, CBP60g is a direct substrate of CPK5 and CPK11 in vitro.
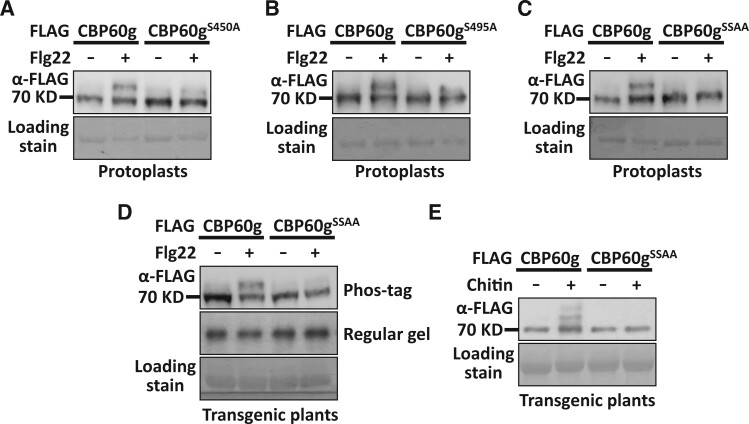
To characterize CBP60g residues involved in CPK5-induced phosphorylation, recombinant CPK5 was purified and incubated with MBP-CBP60g to induce phosphorylation. The products were subjected to mass spectrometry analyses. Meanwhile, CBP60g-FLAG was expressed in protoplasts and treated with PAMP. After immunoprecipitation, CBP60g-FLAG was subjected to mass spectrometry analyses to characterize in planta phosphorylation sites. Ser450 was identified as a residue that is phosphorylated by CPK5 and an in planta phosphorylation site (Supplemental Figure S7). Serine 450 was then mutated and subjected to mobility shift assay. A partially compromised PAMP-induced phosphorylation was observed when Serine 450 was mutated to Alanine (CBP60gS450A; Figure 5A). This indicates that Serine 450 contributes to PAMP-induced CBP60g phosphorylation and suggests the existence of additional phosphorylation sites. CBP60g was then subjected to the Arabidopsis Protein Phosphorylation Site Database (PhosPhAt 4.0) predictor (Heazlewood et al., 2008; Durek et al., 2010; Zulawski et al., 2013) and a few putative phosphorylation sites were predicted. Site-directed mutagenesis followed by Phos-tag mobility shift assay was then conducted to characterize critical sites that are required for PAMP-induced CBP60g phosphorylation. In addition to Serine 450, mutation of Serine 495 residue to Alanine (CBP60gS495A) compromised PAMP-induced CBP60g phosphorylation (Figure 5, A and B and Supplemental Figure S8). Corroborating this, double mutation of Ser450 and Ser495 to Alanine (CBP60gSSAA) significantly impaired PAMP-induced CBP60g phosphorylation (Figure 5, C–E). Accordingly, the double mutation of Serine 450 and Serine 495 compromised phosphorylation of CBP60g by CDPKs as shown in the in vitro phosphorylation assay (Figure 4, D–E). Thus, CPK5 and CPK11 interact with and directly phosphorylate CBP60g.
Figure 5.
Ser450 and Ser495 are required for PAMP-induced full phosphorylation of CBP60g. A, Ser450 is required for CBP60g full phosphorylation induced by flg22 in protoplasts. Protoplasts isolated from WT plants were transfected with Pro35S:CBP60g-FLAG or Pro35S:CBP60gS450A-FLAG, treated with or without flg22. B, Ser495 is required for CBP60g full phosphorylation induced by flg22 in protoplasts. Protoplasts isolated from WT plants were transfected with Pro35S:CBP60g-FLAG or Pro35S:CBP60gS495A-FLAG, treated with or without flg22. C, Double mutation of Ser450 and Ser495 largely impaired flg22-induced full phosphorylation of CBP60g in protoplasts. Protoplasts were transfected with Pro35S:CBP60g-FLAG or Pro35S:CBP60gSSAA-FLAG, treated with or without flg22. D-E, Double mutation of Ser450 and Ser495 largely impaired flg22-induced (D) and chitin-induced (E) full phosphorylation of CBP60g in transgenic plants. Pro35S:CBP60g-FLAG or Pro35S:CBP60gSSAA-FLAG transgenic plants were treated with or without 2 µM flg22, or 250 µg/mL chitin. Total protein was extracted from protoplasts or leaves of transgenic plants. Protein samples were subjected to Phos-tag SDS-PAGE followed by anti-FLAG immunoblot. CBB staining of the filter indicates loading of the protein. SSAA stands for double mutation of Ser450A and Ser495A residues. The experiments were repeated 3 times with similar results.
TCH3 promotes CBP60g phosphorylation in a CDPK-dependent manner
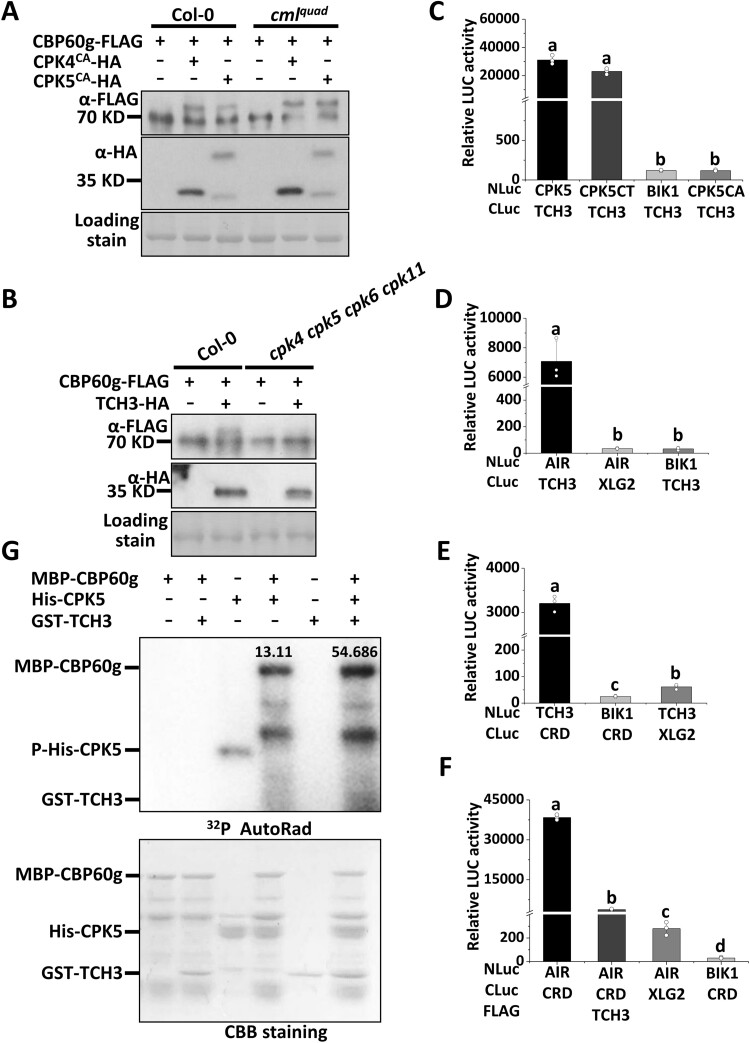
As PAMP-induced CBP60g phosphorylation is both TCH3/CAM- and CDPK-dependent, we further investigated putative interconnections between TCH3/CAM- and CDPK-dependent CBP60g phosphorylation. Co-expression of the full-length form of CPK5 with CBP60g stimulates CBP60g phosphorylation. This induced CBP60g phosphorylation was compromised in the cmlquad mutant (Supplemental Figure S9). However, CBP60g phosphorylation induced by the constitutive active form of CPK5/CPK4 (CPK5CA/CPK4CA) remains intact in the cmlquad mutant (Figure 6A), suggesting that TCH3/CAM1/CAM4/CAM6 do not act downstream of CPK5/CPK4 in inducing CBP60g phosphorylation. By contrast, TCH3-promoted CBP60g phosphorylation was significantly impaired in the cpk4 cpk5 cpk6 cpk11 mutant (Figure 6B), suggesting that CDPKs function downstream of TCH3 to induce CBP60g phosphorylation. The results indicate that TCH3 promotes CDPK-mediated CBP60g phosphorylation, thus confirming the genetic interconnections between TCH3- and CDPK-dependent phosphorylation of CBP60g.
Figure 6.
TCH3 targets the autoinhibitory region (AIR) of CPK5 and promotes CDPK-dependent CBP60g phosphorylation. A, CPK4CA/CPK5CA-induced phosphorylation of CBP60g remains intact in the cmlquad mutant. B, TCH3-induced phosphorylation of CBP60g is significantly compromised in the cpk4 cpk5 cpk6 cpk11 mutant. Protoplasts isolated from WT or mutant plants as indicated were transfected with Pro35S:CBP60g-FLAG alone or together with CPK4CA/CPK5CA-HA or TCH3-HA, treated with or without 2 µM flg22 for 5 min. Protein samples extracted from protoplasts were separated by Phos-tag SDS-PAGE and subjected to anti-FLAG or anti-HA immunoblot. C, TCH3 interacts with the C-terminal of CPK5. Nicotiana benthamiana leaves infiltrated with Pro35S:NLuc-TCH3, Pro35S:CLuc-CPK5, Pro35S:CLuc-CPK5CT, Pro35S:CLuc-CPK5CA, Pro35S:CLuc-XLG2, or Pro35S:NLuc-BIK1 as indicated were subjected to luciferase complementation assay. D, TCH3 interacts with CPK5-AIR (AIR) in N. benthamiana. Nicotiana benthamiana leaves infiltrated with Pro35S:NLuc-AIR, Pro35S:CLuc-TCH3, Pro35S:CLuc-XLG2, or Pro35S:NLuc-BIK1 as indicated were subjected to luciferase complementation assay. E, TCH3 interacts with CPK5-CRD (CRD) in N. benthamiana. Nicotiana benthamiana leaves infiltrated with Pro35S:NLuc-CRD, Pro35S:CLuc-TCH3, Pro35S:CLuc-XLG2, or Pro35S:NLuc-BIK1 as indicated were subjected to luciferase complementation assay. F, TCH3 impairs the interaction between CPK5-CRD (CRD) and AIR. N.b. leaves infiltrated with Pro35S:NLuc-AIR, Pro35S:CLuc-CRD, Pro35S:TCH3-FLAG, Pro35S:CLuc-XLG2, or Pro35S:NLuc-BIK1 as indicated were subjected to luciferase complementation assay. Infiltrated N. benthamiana leaves were sliced into strips 2 d post infiltration, and relative luminescence was determined by Microplate Luminometer. Error bars indicate SD of three biological repeats. Student’s t test was carried out to determine the significance of difference. Different letters indicate significant differences. G, TCH3 enhances CPK5 activity towards CBP60g in vitro. MBP-CBP60g was incubated with His-CPK5 or together with GST-TCH3 as indicated and subjected to in vitro phosphorylation assay. CBB staining indicates loading of the protein. The ratio of phosphorylation signal over protein amount is shown above the phosphor band. The experiments were repeated 3 times with similar results.
TCH3 interrupts the interaction between CPK5-CRD and CPK5-AIR
The above results indicated potential modulation of CPK5 by TCH3. CDPKs are autoinhibitory protein kinases harboring an N-terminal protein kinase domain and a C-terminus, which usually contains an autoinhibitory region (AIR) and a C-terminal regulatory domain (CRD; Harper et al., 2004; Hamel et al., 2014). Split luciferase assay indicated a specific interaction between TCH3 and CPK5 (Figure 6C and Supplemental Figure S10A). Moreover, this interaction occurred between the C terminus of CPK5 (CPK5CT) and TCH3 (Figure 6C and Supplemental Figure S10A). CPK5CT was further divided into a CRD and an AIR, and both interact with TCH3 (Figure 6, D–E and Supplemental Figure S10, B–C). It has been proposed that CRD is prebound to AIR to keep CDPK in its autoinhibitory form. Upon calcium influx, binding of calcium ions induces a conformational change in the CRD, leading to the relief of CDPK autoinhibition (Harper et al., 2004; Wernimont et al., 2010; Hamel et al., 2014). An interaction between CPK5-CRD and CPK5-AIR was observed, which is consistent with the proposed model (Harper et al., 2004; Hamel et al., 2014; Figure 6F and Supplemental Figure S10D). Furthermore, expression of TCH3 was found to significantly attenuate this interaction (Figure 6F and Supplemental Figure S10D). An in vitro phosphorylation assay was subsequently performed to determine whether TCH3 can enhance CPK5 activity toward CBP60g. As shown in Figure 6G and Supplemental Figure S11, the addition of TCH3 increased the CPK5-mediated phosphorylation of CBP60g in the presence of calcium ions. Moreover, an in vitro phosphorylation assay showed that TCH3 promotes CPK5 autophosphorylation in the presence of calcium ions (Supplemental Figure S11), indicating that TCH3 stimulates CPK5 activity to promote CBP60g phosphorylation. These observations thus indicate that TCH3 interferes with CPK5 CRD-AIR complex formation, thereby leading to an enhancement of CPK5-mediated CBP60g phosphorylation. Thus, the results indicate both genetic and physical interactions between TCH3 and CPK5 in CBP60g activation.
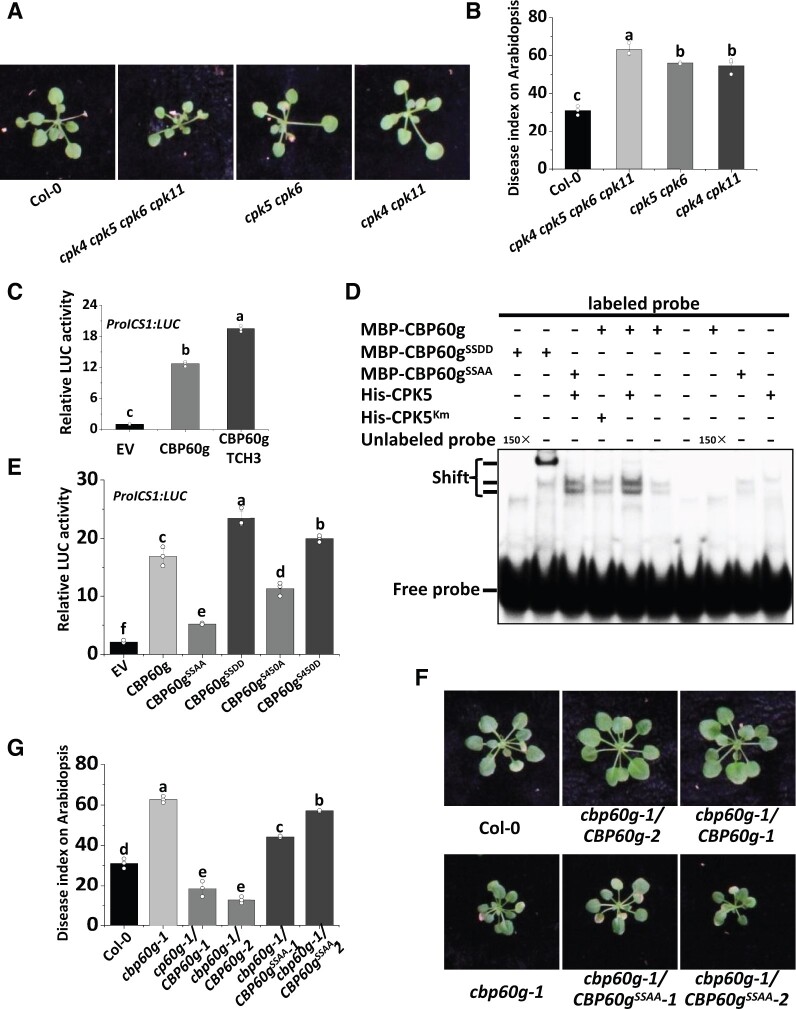
Phosphorylation of CBP60g regulates CBP60g activity and resistance to V. dahliae
To determine whether CDPK-mediated phosphorylation of CBP60g contributes to plant resistance to V. dahliae, cpk5 cpk6, cpk4 cpk11, and cpk4 cpk5 cpk6 cpk11 quadruple mutant plants were root-infected with V. dahliae. Both cpk5 cpk6 and cpk4 cpk11 mutant plants were found to exhibit attenuated resistance to V. dahliae (Figure 7, A and B). The markedly compromised resistance in the cpk4 cpk5 cpk6 cpk11 quadruple mutant plants (Figure 7, A and B) indicates that these CDPKs positively regulate plant resistance to V. dahliae. In support of the contribution to fungal resistance, the phosphorylation of CBP60g induced by fungal-derived PAMPs was observed to be notably compromised in the cmlquad and cpk4 cpk5 cpk6 cpk11 quadruple mutant plants (Supplemental Figure S12).
Figure 7.
Phosphorylation of CBP60g regulates CBP60g activity and resistance to V. dahliae. A and B, The cpk4 cpk11, cpk5 cpk6, and cpk4 cpk5 cpk6 cpk11 mutants are compromised in resistance to V. dahliae. Disease symptoms (A) and index analyses (B) of Arabidopsis WT, cpk4 cpk11, cpk5 cpk6, or cpk4 cpk5 cpk6 cpk11 mutant plants infected with V. dahliae. The plants were photographed and subjected to disease index analyses. Disease indexes were evaluated with three replicates generated from 24 plants for each inoculum. Error bars indicate SD of three biological replicates. Student’s t test was carried out to determine the significance of difference. Different letters indicate significant differences. C, TCH3 enhances CBP60g-activated ProICS1:LUC reporter activity. ProICS1:LUC reporter was transfected alone, or together with CBP60g and TCH3, as indicated. Pro35S:RLuc was co-transfected as internal control. Error bars indicate SD of three biological repeats. Student’s t test was carried out to determine the significance of difference. Different letters indicate significant differences. D, CPK5 enhances the DNA-binding activity of CBP60g. MBP-CBP60g, MBP-CBP60gSSAA, or MBP-CBP60gSSDD were incubated with [γ-32P]ATP-labeled 60-bp double-stranded DNA probe within the ICS1 promoter, and subjected to EMSA. Unlabeled probe was used as competitors (150×) for binding. MBP-CBP60g was preincubated with or without His-CPK5 or His-CPK5Km as indicated for 30 min at room temperature as indicated before EMSA. The experiments were repeated 3 times with similar results. E, Ser450 and Ser495 are required for transcription activator activity of CBP60g. ProICS1:LUC was transfected alone or together with CBP60g, CBP60gS450A, CBP60gS450D, CBP60gSSAA, or CBP60gSSDD as indicated. LUC reporter activity was determined 16 h post-transfection. Error bars indicate SD of three biological repeats. Student’s t test was carried out to determine the significance of difference. Different letters indicate significant differences between CBP60g, CBP60gS450A, CBP60gS450D, CBP60gSSAA, or CBP60gSSDD. F and G, Ser450 and Ser495 are required for resistance to V. dahliae. Disease symptoms (F) and index analyses (G) of Arabidopsis WT, cbp60g-1 mutant, cbp60g-1/CBP60g-FLAG, and cbp60g-1/CBP60gSSAA-FLAG transgenic plants infected with V. dahliae. The plants were photographed and subjected to disease index analyses. Disease indexes were evaluated with three replicates generated from 24 plants for each inoculum. Error bars indicate SD of three biological replicates. Student’s t test was carried out to determine the significance of difference. Different letters indicate significant differences.
CBP60g is a master transcription regulator that activates ICS1, FMO1, and a number of defense-related genes. Dual reporter analyses revealed that transient expression of CBP60g in Arabidopsis protoplasts significantly enhanced the expression of ProICS1:LUC (firefly luciferase; Qin et al., 2018). We have shown that TCH3 promotes CDPK-dependent CBP60g phosphorylation (Figure 6B) and observed that co-expression of TCH3 enhances ProICS1:LUC induction by CBP60g (Figure 7C). Treatment of chitin, a well-characterized fungal-derived PAMP, enhanced the DNA-binding activity of CBP60g in vivo (Supplemental Figure S13). These findings prompted us to examine the contribution of phosphorylation to the transcription factor activity of CBP60g. The results of an electrophoretic mobility shift assay (EMSA) revealed that the pre-incubation of CPK5, but not its kinase-inactive form CPK5Km, with CBP60g increased the binding activity of CBP60g to a 60-bp DNA fragment (probe) within the ICS1 promoter (Figure 7D). CBP60gSSAA exhibited a compromised enhancement whereas CBP60gSSDD (double mutation of Ser450 and Ser495 to Aspartic acid) further enhanced this binding activity (Figure 7D). Further reporter analyses indicated that expression of CBP60gS450A or CBP60gSSAA was associated with defects in the activation of ProICS1:LUC, whereas that of CBP60gS450D (mutation of Ser450 to Aspartic acid) or CBP60gSSDD enhanced ProICS1:LUC activation (Figure 7E). These findings thus indicate that CDPK-mediated phosphorylation of CBP60g is critical for the transcription factor activity of CBP60g. We did not observe transactivation activity of CBP60g, nor did PAMP treatment stimulate the transactivation activity of CBP60g (Supplemental Figure S14A). In addition, PAMP treatment did not obviously alter the protein–protein interactions between TCH3 and CPK5 (Supplemental Figure S14B). We subsequently developed cbp60g-1/CBP60g-FLAG and cbp60g-1/CBP60gSSAA-FLAG plants and assessed the resistance of these transgenic lines to V. dahliae. We observed that CBP60g-FLAG, but not CBP60gSSAA-FLAG, could restore resistance of the cbp60g-1 mutant against V. dahliae (Figure 7, F and G), indicating that CBP60g Ser450 and Ser495 are critical residues required for full resistance against V. dahliae. Expression of CBP60g-FLAG and CBP60gSSAA-FLAG in transgenic lines was confirmed by immunoblot (Supplemental Figure S15). The results indicate that PAMP-induced phosphorylation of CBP60g positively regulates CBP60g activity and resistance to V. dahliae (Figure 7 and Supplemental Figure S16). The transcriptional induction of CBP60g during immunity has been well documented in previous studies (Chen et al., 2009; Zhang et al., 2010b; Li et al., 2015). Either V. dahliae infection or PAMP treatment up-regulates CBP60g transcription to a similar extent in the WT, cpk4 cpk5 cpk6 cpk11, and cmlquad mutant plants (Supplemental Figure S17), suggesting that transcriptional induction of CBP60g during immunity is independent of CPK4/5/6/11 and TCH3/CAM1/4/6. These lines of evidence support dual layers of CBP60g regulation during immunity, reinforcing its role as a master immune regulator (Figure 8).
Figure 8.
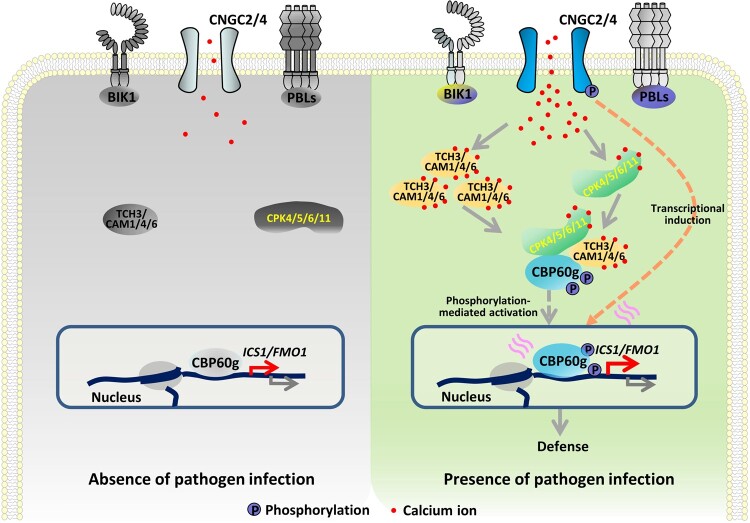
Model for TCH3- and CPK5-dependent activation of CBP60g. Plants deploy PRRs to sense the invasion of pathogens. Perception of PAMPs triggers activation of calcium channels and a cytosolic increase of Ca2+, leading to activation of calcium sensors, such as CDPKs. Perception of PAMPs also induces the expression of the gene encoding the calcium sensor TCH3. TCH3 directly binds to CBP60g and enhances its activity. TCH3 targets the autoinhibitory region of CPK5 and enhances CPK5-mediated phosphorylation of CBP60g. Alternatively, binding of TCH3 to CBP60g may also contribute to CBP60g phosphorylation by CPKs. Phosphorylation of CBP60g enhances its transcription factor activity and regulates the expression of defense-related genes and thereby triggers defense against V. dahliae. In addition to post-translational phosphorylation-mediated activation, CBP60g is transcriptionally induced by PAMP perception or pathogen infection.
Discussion
Sensing environmental changes and stresses triggers the cytosolic increase of Ca2+ in eukaryotic cells. During plant–microbe interactions, calcium influx is rapidly induced upon pathogen infection; however, the mechanisms involved in decoding calcium signals in plant immunity remain largely uncharacterized. Flowering plants have a greater variety of calcium sensors than those in animals and fungi, including many CMLs and CDPKs (Mccormack et al., 2005; Luan and Wang, 2021), suggesting the existence of more complicated decoding mechanisms within the sensor system and interconnections with binding targets. In this study, we revealed calcium-dependent phosphorylation and activation of CBP60g triggered by PAMP perception. Two types of calcium sensors are both required for PAMP-induced CBP60g phosphorylation. TCH3 interacts with CBP60g and promotes CBP60g phosphorylation; however, it does not phosphorylate CBP60g directly. Conversely, CPK5 and its homologs phosphorylate CBP60g directly to enhance its DNA-binding activity and are also required for TCH3-induced CBP60g phosphorylation. TCH3 interacts with both CPK5-AIR and CPK5-CRD and disrupts the association between CPK5-AIR and CPK5-CRD. Thus, the findings support a model in which TCH3 and Ca2+ cooperate to relieve autoinhibition of CPK5 to phosphorylate and activate CBP60g. Moreover, a novel role of TCH3 in plant immunity has been identified, and a cooperative action of different types of calcium sensors in decoding immune-related calcium signals has been revealed (Figure 8). Additionally, the findings revealed both genetic and biochemical interconnections within the calcium sensor system, thus representing a novel action mode of calcium sensors.
Verticillium dahliae is a soil-borne fungal pathogen that infects many plants and causes severe disease. Plants use LysM PRRs to recognize chitin oligomers, which are well-characterized fungal PAMPs shown to elicit PTI immune responses. The mutation of the gene encoding the Arabidopsis LysM receptor CERK1 compromised plant resistance against V. dahliae. Moreover, V. dahliae secretes a polysaccharide deacetylase to deacetylate chitin oligomers and suppress their recognition by CERK1 (Gao et al., 2019). The evidence proved that CERK1-mediated PTI plays an important role in V. dahliae resistance. As a typical response following PAMP perception, the activation of calcium influx is observed during V. dahliae infection (Supplemental Figure S1).
Several plant CMLs have been shown to respond to pathogen recognition. The soybean (Glycine max) CaM (SCaM)-4 and SCaM-5 are rapidly induced by Fusarium solani or Phytophthora parasitica elicitor preparations in an intracellular Ca2+-dependent manner (Heo et al., 1999). Tobacco mosaic virus (TMV) infection induced the expression of tobacco CaM (NtCaM)-13 (Yamakawa et al., 2001). Tobacco CML ACRE-31 and its tomato ortholog APR134 are induced by Cladosporium fulvum and P. syringae pv. tomato strains, respectively. Silencing of APR134 compromised tomato resistance to Pst pv. tomato strain (Durrant et al., 2000; Mysore et al., 2002). Tobacco regulator of gene silencing CaM (rgs-CaM) is induced upon viral infection (Anandalakshmi et al., 2000). In Arabidopsis, several CMLs are induced following flg22 treatment (Chen et al., 2009; Li et al., 2015). CAM7 gates CNGC2/4 to regulate calcium influx. As other types of calcium sensors, CPK5 and CPK28 have been shown to regulate immunity. For instance, CPK5 positively regulates ROS and PTI by phosphorylating RbohD whereas CPK28 negatively regulates PTI by phosphorylating PUB25/26 to ubiquitinate BIK1 and trigger its degradation (Kadota et al., 2014, 2015; Monaghan et al., 2014). Our genetic evidence and molecular analyses revealed genetic and physical interactions between TCH3 and CPK5 in phosphorylating CBP60g. The interconnection and cooperation between the different types of calcium sensors provide versatility and flexibility to eukaryotes to cope with various calcium signatures triggered by environmental changes and stresses.
In animals, CAM-dependent protein kinases (CaMKs) are activated by binding of CaM to the AIR. However, the Arabidopsis genome does not encode conventional CaMKs (Hrabak et al., 2003; Harper et al., 2004). Here, we provided evidence for binding of TCH3 to CPK5-AIR and CPK5-CRD. Furthermore, TCH3 interrupts CPK5-AIR and CPK5-CRD complex formation. It is possible that the interruption of interaction between CPK5-AIR and CPK5-CRD by TCH3 induces a conformational change, thus changing CPK5 from the auto-inhibitory form to its active state. This supports a model in which TCH3 promotes CPK5 activation to activate CBP60g during plant immunity (Figure 8).
SARD1 is the homolog related to CBP60g in the plant-specific CBP60 family and displays a functional redundancy with CBP60g regarding immunity. However, SARD1 has been shown to lack a CAM binding motif and thus is deficient in CAM binding (Zhang et al., 2010b), suggesting a functional divergence in calcium-mediated signaling between SARD1 and CBP60g. This study identified two residues harbored by CBP60g and required for its full phosphorylation but not harbored by SARD1. This may be one of the functional divergence mechanisms that CBP60g and SARD1 exhibit in immunity. Furthermore, the genetic evidence indicated that increased CPK5 signaling activates SARD1 transcription during systemic acquired resistance (SAR; Guerra et al., 2020). The mutation of SARD1 compromised CPK5-activated N-hydroxy-L-pipecolic acid accumulation and SAR, and a post-translational modification of SARD1 mediated by CPK5 was suspected (Guerra et al., 2020). Single mutation of CBP60g Ser450 residue exhibited detectable deficiency in transcriptional activation activity of CBP60g (Figure 7E), phosphorylation by CPK5, and resistance to V. dahliae (Supplemental Figure S17). Double mutation of Ser450 and Ser495 only partially impaired CPK5-mediated CBP60g phosphorylation (Figure 4D), suggesting the existence of additional residues for CPK5-mediated full phosphorylation of CBP60g. Whether SARD1 is a substrate of CDPKs or other kinases during plant immunity remains to be further investigated.
Materials and methods
Plant materials and constructs
Arabidopsis thaliana plants used in this study include the Col-0 wild-type and the cbp60g-1 (Zhang et al., 2010b), tch3-1 (Gleeson et al., 2012), cpk4 cpk11, cpk5 cpk6, cpk4 cpk5 cpk6 cpk11 (Zhou et al., 2020), bik1 pbl1 (Zhang et al., 2010a) mutants. tch3-2, cngc2-1, cngc2-2 mutant, cam1, cam6, cam1 cam6, tch3 cam4, tch3 cam1 cam6, and cmlquad mutants were generated using CRISPR-Cas9 (Supplemental Figure S2). Arabidopsis was grown under a 12-h-fluorescent light (60 µEm−2s−1)/12-h-dark cycle. Constructs used in this study include Pro35S:CBP60g-FLAG, ProICS1:LUC (Qin et al., 2018), pCambia1300-BIK1-NLuc, and pCambia1300-XLG2-CLuc (Chen et al., 2008). pCambia1300-FLAG, pCambia1300-CLuc (Chen et al., 2008), and pCambia1300-HA-NLuc (Liang et al., 2016) plasmid vectors were used to generate transient expression or transgenic expression constructs. CBP60g or CBP60gSSAA was cloned into pCambia1300-35S-FLAG (Zhang et al., 2010a), transformed into WT, cbp60g-1, cmlquad or cpk4 cpk5 cpk6 cpk11 mutant plants to generate transgenic lines. The CAM1, CAM2, CAM3, CAM4, CAM5, CAM6, CAM7, CML8, TCH3, CML40, CML45, CML46, CPK5, CPK6, CPK11, and CBP60g coding sequences were amplified by RT-PCR from cDNA isolated from Col-0 plants and cloned into the pUC-FLAG and pUC-HA vectors to generate transient expression constructs. Site-directed mutagenesis was used to generate CBP60gSSAA, CPK5Km, and CPK11Km and these altered genes were introduced into pMAL-C2X, pET28a, or pCambia1300-FLAG. Constitutive active form of CPK4, CPK5, CPK6, CPK11 were generated by amplification of the truncated constitutive active version from the corresponding CDPKs. Primer sequences are provided in Supplemental Data Set S1.
Transient expression in protoplasts
Arabidopsis protoplasts isolated from the leaves of 5-week-old plants were transfected with the constructs as indicated (Zhang et al., 2010a). Twelve hours after transfection, protoplasts were harvested and used for experiments. For immunoblot analyses, protoplasts were treated with or without PAMPs as indicated for 5 min. Total protein was extracted with extraction buffer (50 mM HEPES [pH 7.5], 150 mM KCl, 1 mM EDTA, 1 mM DTT, 1% Triton X-100, 1×complete protease inhibitors) and subjected to anti-FLAG (Sigma F3165, 1:10,000 dilution) or anti-HA (Roche 11666606001, 1:5,000 dilution) immunoblot. For PPase treatment, total protein was treated with lambda PPase (New England Biolabs) according to manufacturer’s instructions. For dual reporter analyses, the protoplasts were incubated overnight under faint light and LUC activity was recorded by a microplate luminometer (Berthhold Centro XS3).
Phos-tag SDS PAGE and immunoblot
An SDS-PAGE gel was supplied with 20 mM Phos-tag and 40 mM MnCl2 and used for Phos-tag SDS PAGE electrophoresis. After electrophoresis, the gels were washed 3 times with 10 mM EDTA before protein transfer according to the manufacturer (Apexbio F4002)’s instructions. Membrane carrying the transferred protein was subjected to immunoblot.
Split-luciferase complementation assay
Agrobacterium tumefaciens GV3101 strain carrying NLuc- or CLuc-tagged constructs was infiltrated into leaves of 4-week-old Nicotiana benthamiana plants. LUC activity in leaves was examined 2 d post infiltration. Cut pieces of N. benthamiana leaves were treated with 1 mM luciferin and kept in dark for 5 min to quench the fluorescence. The quantitative LUC activity was then determined by microplate luminometer. The expression of CLuc-tagged proteins or HA-NLuc-tagged proteins was detected by anti-CLuc (Sigma L2164, 1:5,000 dilution) or anti-HA (Roche 11666606001) immunoblot, respectively.
Coimmunoprecipitation assay in protoplasts
Five-week-old Arabidopsis plants were used for protoplast isolation. CBP60g-FLAG was transfected alone or co-transfected with CDPKs-HA into Arabidopsis protoplasts. Total protein was extracted with extraction buffer. For anti-HA IP, total protein was incubated with 2 μg of anti-HA antibody (Roche 11666606001) together with protein A agarose at 4°C for 4 h. The bound agarose was washed, collected, and boiled for 5 min with 1×protein loading buffer. Immunoprecipitates were separated by 8% SDS-PAGE and the presence of CBP60g-FLAG, CPK5-HA, CPK6-HA, or CPK11-HA was detected by anti-FLAG (Sigma F3165) or anti-HA (Roche 11666606001) immunoblot, respectively.
In vitro phosphorylation assay
The His- and MBP-fused recombinant proteins were expressed in Escherichia coli strain and purified using Ni-NTA agarose (Qiagen) or amylose resin (New England Biolabs), respectively, according to the manufacturer’s instructions. In vitro phosphorylation reactions were performed in reaction buffer containing 20 mM Hepes (pH7.5), 10 mM MgCl2, 5 mM CaCl2, and 1 mM DTT. His-CPK5, His-CPK5Km, His-CPK11, or His-CPK11Km was incubated with MBP-CBP60g, respectively, in reaction buffer supplied with 0.1 mM ATP and 5 µCi of [γ-32P]ATP at 28°C for 30 min. The reactions were stopped by adding 4×SDS loading buffer, and the phosphorylation of recombinant proteins was analyzed by autoradiography.
Reporter assay in protoplasts
ProICS1:LUC construct was transfected alone or co-transfected with CBP60g-FLAG, CBP60gSSAA-FLAG, CBP60gSSDD-FLAG, CBP60gS450A-FLAG, CBP60gS450D-FLAG, or TCH3-FLAG, as indicated into Arabidopsis protoplasts. The protoplasts were incubated overnight under faint light and LUC activity was determined by using the Dual-Luciferase Reporter System (Promega) following the manufacturer’s instructions.
Electrophoretic mobility shift assays (EMSAs)
A 60-bp probe within the DNA fragment 7 (Zhang et al., 2010b) in the ICS1 promoter was labeled with [γ-32P]ATP using T4 polynucleotide kinase. Binding reactions were carried out in a 20 μL volume of reaction buffer (10 mM Tris-HCl [pH 7.5], 50 mM KCl, 1mM DTT, 1 μL 50 ng/μL poly[dI-dC]) for 30 min at room temperature. Labeled DNA probe (2 fmol) was incubated with MBP-CBP60g. 150× unlabeled DNA probe was used for competition. MBP-CBP60g or MBP-CBP60gSSAA was pre-incubated with or without His-tagged CPK5 or CPK5Km as indicated for 30 min at room temperature before DNA binding. The reaction was stopped by adding DNA loading buffer and the samples were separated by an 8% native PAGE gel. After electrophoresis, the gel was autoradiographed.
Infection assay
Arabidopsis plants were infected by V. dahliae Ls17 via the root-dip inoculation method (Gao et al., 2010). A conidial suspension of 107/mL V. dahliae Ls17 was used as the inoculums. The disease grade was classified as follows: Grade 0 (no symptoms), 1 (0–25% wilted leaves), 2 (25–50%), and 3 (50–75%). The disease index was calculated as (sum [number of plants × disease grade])/([total number of plants] × [maximal disease grade]) × 100 as previously reported (Qin et al., 2018).
Calcium influx assay
Seedlings of the aequorin-transformed Arabidopsis reporter line (ColAEQ) were grown on 1/2 MS medium for 10 d. Plant roots were sliced into 0.5 cm pieces and placed in 96-well plates containing 100 μL water, incubated with 10 μM coelenterazine in dark for 4 h. The samples were then treated with a conidial suspension of 107/mL V. dahliae. Luminescence was recorded at intervals of 2 min by microplate luminometer.
Mass spectrometric analysis
Recombinant MBP-CBP60g was incubated in kinase reaction buffer with CPK5-His at 28°C for 30 min and separated by SDS-PAGE and stained with Coomassie brilliant blue. CBP60g-FLAG was transfected into Arabidopsis protoplasts isolated from 5-week-old plants, treated with flg22 for 5 min. Total protein was extracted from protoplasts and immunoprecipitated with anti-FLAG (Sigma F3165). Immunoprecipitates were separated by SDS-PAGE and silver stained. Mass spectrometric data analysis was performed by Beijing Qinglian Biotech Co., Ltd. Database searches were performed on an in-house Mascot server (Matrix Science Ltd., London, UK) against CBP60g protein sequence. All identified phosphorylated peptides were manually checked to exclude false-positives.
RNA isolation and RT-qPCR
Total RNA was isolated with the TRIzol reagent (Invitrogen) and used for cDNA synthesis with SuperScript III First-Strand Synthesis System for RT-qPCR (Invitrogen) following the instructions provided by the manufacturer. The RT-qPCR was performed with the SYBR Premix Ex Taq kit (TaKaRa) following standard protocols.
Chromatin immunoprecipitation (ChIP)
Arabidopsis protoplasts isolated from 5-week-old plants were transfected with CBP60g-FLAG and used for ChIP analyses. Twelve hours after transfection, protoplasts were treated with or without 250 µg/mL chitin for 4 h. Cells were collected and cross-linked with 1% formaldehyde for 20 min and quenched by glycine for 5 min. Nuclei were then extracted and subjected to ChIP. Bioruptor was used for sonication and DNA was eluted with 1% SDS and 0.1 M NaHCO3 at 65°C overnight. Anti-FLAG antibody (Sigma M8823) was used for ChIP and eluted DNA was subjected to qPCR analyses. The relative enrichment fold changes were calculated by normalizing percent input of each primer pair against the control gene primer.
Transactivation activity assay
Arabidopsis protoplasts were transfected with the indicated reporter and constructs. Twelve hours after transfection, protoplasts were collected and the LUC activity was determined by using Dual-Luciferase® Reporter system (Promega) following manufacture’s instruction.
Statistical analysis
Student’s t test was used to determine whether the difference between the two groups of data is statistically significant (Supplemental Data Set S2). Different letters above the data columns are used to indicate differences that are statistically significant.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CBP60g (At5g26920), TCH3 (At2g41100), CPK5 (At4g35310), CPK4 (At4g09570), CPK6 (AT2G17290), CPK11 (At1g35670), CML40 (At3g01830), CML45 (At3g29000), CML46 (At5g39670), CAM1 (At5g37780), CAM2 (At2g41110), CAM3 (At3g56800), CAM4 (At1g66410), CAM5 (At2g27030), CAM6 (At5g21274), CAM7 (At3g43810), CML8 (At4g14640), CNGC2 (At5g15410), ICS1 (At1g74710).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Verticillium dahliae treatment induces calcium influx in Arabidopsis.
Supplemental Figure S2. gRNA targeted sequences and mutation or deletion sites of mutants generated by CRISPR-Cas9 gene-editing approach.
Supplemental Figure S3. PAMP treatment induces the expression of TCH3 in Arabidopsis.
Supplemental Figure S4. TCH3 and CAM1/4/6 contribute to PAMP-induced CBP60g phosphorylation.
Supplemental Figure S5. CAM1, CAM4, CAM6, and TCH3 interact with CBP60g.
Supplemental Figure S6. TCH3 does not phosphorylate CBP60g.
Supplemental Figure S7. PAMP induces CBP60g phosphorylation at Ser450 residue.
Supplemental Figure S8. A screen for putative residues contributing to PAMP-induced CBP60g phosphorylation.
Supplemental Figure S9. Stimulation of CBP60g phosphorylation by full-length form of CPK5 was compromised in the cmlquad mutant.
Supplemental Figure S10. Levels of NLuc- and CLuc-tagged proteins as indicated in N. benthamiana.
Supplemental Figure S11. TCH3 promotes CPK5 autophosphorylation and phosphorylation of CBP60g in the presence of calcium ions.
Supplemental Figure S12. TCH3/CAM1/4/6 and CPK4/5/6/11 are required for CBP60g phosphorylation induced by chitin.
Supplemental Figure S13. PAMP treatment enhanced the DNA-binding activity of CBP60g.
Supplemental Figure S14. PAMP or LaCl3 treatment did not obviously induce transactivation activity of CBP60g and alter TCH3-CPK5 interaction.
Supplemental Figure S15. Levels of CBP60g and CBP60gSSAA in transgenic plants.
Supplemental Figure S16. Single mutation of CBP60g Ser450 residue exhibited deficiency in phosphorylation by CPK5 and resistance to V. dahliae.
Supplemental Figure S17. Verticillium dahliae infection or PAMP treatment up-regulates the transcription of CBP60g.
Supplemental Data Set S1. Sequences of primers used in this study.
Supplemental Data Set S2. Results of t tests for the data presented in each figure.
Supplementary Material
Acknowledgments
The authors are grateful to Huishan Guo for providing Verticillium dahliae strains and vectors, Xiangzong Meng for providing cpk4 cpk5 cpk6 cpk11 mutant and reporter constructs; Xiaoming Zhang and Ningkun Liu for technical assistance; Bo Li for providing reporter constructs; Yang Zhao for providing Col-0AEQ reporter line; Peichen Zhao for Phos-tag SDS-PAGE analysis, Lei Su and Haiyun Wang for microscopy analysis.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31922075, 32172504), the National Key R&D Program of China (2021YFD1400800), and the Strategic Priority Research Program of Chinese Academy of Sciences (Grant No. XDPB16) and the Youth Innovation Promotion Association of the Chinese Academy of Sciences to J. Z.
Conflict of interest statement. The authors declare no conflict of interest.
Contributor Information
Lifan Sun, State Key Laboratory of Plant Genomics, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China; CAS Center for Excellence in Biotic Interactions, University of Chinese Academy of Sciences, Beijing 100049, China.
Jun Qin, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, Shaanxi 712100, China.
Xiaoyun Wu, State Key Laboratory of Plant Genomics, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China; CAS Center for Excellence in Biotic Interactions, University of Chinese Academy of Sciences, Beijing 100049, China.
Jinghan Zhang, State Key Laboratory of Plant Genomics, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China; School of Life Sciences, Hebei University, Baoding, Hebei 710023, China.
Jie Zhang, State Key Laboratory of Plant Genomics, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China; CAS Center for Excellence in Biotic Interactions, University of Chinese Academy of Sciences, Beijing 100049, China.
J.Z. and L.S. designed the experiments and wrote the article; L.S., J.Q., X.W., and J.H.Z. performed the experiments and analyzed the data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Jie Zhang (zhangjie@im.ac.cn).
References
- Anandalakshmi R, Marathe R, Ge X, Herr JM Jr, Mau C, Mallory A, Pruss G, Bowman L, Vance VB (2000) A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290: 142–144 [DOI] [PubMed] [Google Scholar]
- Balagué C, Lin B, Alcon C, Flottes G, MalmstroM S, KoHler C, Neuhaus G, Pelletier G, Roby GD (2003) HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15: 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi G, Su M, Li N, Liang Y, Song D, Xu J, Hu M, Wang J, Zou M, Deng Y, et al. (2021) The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184: 3528–3541 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Boller T, He SY (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Yellin A, Snedden WA, Fromm H (2005) Plant-specific calmodulin-binding proteins. Annu Rev Plant Biol 56: 435–466 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, Mccormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J (2010) Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner F, Rosahl S, Lee J, Rudd JJ, Geiler C, Kauppinen S, Rasmussen G, Scheel D, Nürnberger T (2014) Pep-13, a plant defense-inducing pathogen-associated pattern from phytophthora transglutaminases. EMBO J 21: 6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, Cai R, Zuo J, Tang X, Li X (2009) Ethylene INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 REPRESS SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SH, Willmann MR, Chen HC, Sheen J (2002) Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129: 469–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WG, Hilleary R, Swanson SJ, Kim SH, Gilroy S (2016) Rapid, long-distance electrical and calcium signaling in plants. Annu Rev Plant Biol 67: 287–307 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Fengler KA, Yu IC, Lippok B, Bent AF (2000) The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci USA 97: 9323–9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Xiang T, Zhou JM (2009) Plant immunity: a lesson from pathogenic bacterial effector proteins. Cell Microbiol 11: 1453–1461 [DOI] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte C (2013) Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci USA 110: 8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek P, Schmidt R, Heazlewood JL, Jones A, MacLean D, Nagel A, Kersten B, Schulze WX (2010) PhosPhAt: The Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res 38: 828–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE, Jones JD (2000) cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12: 963–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbs G, Silipo A, Aslam S, Castro CD, Liparoti V, Flagiello A, Pucci P, Lanzetta R, Parrilli M, Molinaro A (2008) Peptidoglycan and muropeptides from pathogens Agrobacterium and Xanthomonas elicit plant innate immunity: structure and activity. Chem Biol 15: 438–448 [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Felix G, Regenass M, Boller T (1993) Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: Induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J 18: 265–276 [Google Scholar]
- Fradin EF, Zhao Z, Ayala J, Castroverde C, Nazar RN, Robb J, Thomma L (2009) Genetic dissection of Verticillium Wilt resistance mediated by tomato Ve1. Plant Physiol 150: 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Zhang BS, Zhao JH, Huang JF, Jia PS, Wang S, Zhang J, Zhou JM, Guo HS (2019) Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat Plants 5: 1167–1176 [DOI] [PubMed] [Google Scholar]
- Gao F, Zhou BJ, Li GY, Jia PS, Li H, Zhao YL, Zhao P, Xia GX, Guo HS (2010) A glutamic acid-rich protein identified in Verticillium dahliae from an insertional mutagenesis affects microsclerotial formation and pathogenicity. PLoS One 5: e15319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, McCormack M, Sheen J, et al. (2013a). Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PloS Pathog 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Li F, Li M, Kianinejad AS, Dever JK, Wheeler TA, Li Z, He P, Shan L (2013b). Cotton GhBAK1 mediates Verticillium Wilt resistance and cell death. J Integr Plant Biol 55: 586–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wheeler T, Li Z, Kenerley CM, Shan L (2011) Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium Wilt. Plant J 66: 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulin E, Dramé N, Lafitte C, Torto-Alalibo T, Martinez Y, Ameline-Torregrosa C, Khatib M, Mazarguil H, Villalba-Mateos F, Kamoun S, et al. (2006) Cellulose binding domains of a phytophthora cell wall protein are novel pathogen-associated molecular patterns. Plant Cell 18: 1766–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson L, Squires S, Bisgrove S (2012) The microtubule associated protein END BINDING 1 represses root responses to mechanical cues. Plant Sci 187: 1–9 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T (2002) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18: 277–284 [DOI] [PubMed] [Google Scholar]
- Guerra T, Schilling S, Hake K, Gorzolka K, Sylvester FP, Conrads B, Westermann B, Romeis T (2020) Calcium-dependent protein kinase 5 links calcium signaling with N-hydroxy-l-pipecolic acid- and SARD1-dependent immune memory in systemic acquired resistance. New Phytol 225: 310–325 [DOI] [PubMed] [Google Scholar]
- Hahn MG (1996) Microbial elicitors and their receptors in plants. Annu Rev Phytopathol 34: 387–412 [DOI] [PubMed] [Google Scholar]
- Hamel LP, Sheen J, Séguin A (2014) Ancient signals: comparative genomics of green plant CDPKs. Trends Plant Sci 19: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Harmon A (2005) Plants, symbiosis and parasites: a calcium signalling connection. Nat Rev Mol Cell Biol 6: 555–566 [DOI] [PubMed] [Google Scholar]
- Harper JF, Breton G, Harmon A (2004) Decoding Ca(2+) signals through plant protein kinases. Annu Rev Plant Biol 55: 263–288 [DOI] [PubMed] [Google Scholar]
- He SY, Huang HC, Collmer A (1993) Pseudomonas syringae pv. Syringae harpinpss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell 73: 1255–1266 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, Schulze WX (2008) PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res 36: 1015–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo WD, Lee SH, Kim JC (1999) Involvement of specific calmodulin isoforms in salicylic acid-independent activation of plant disease resistance responses. Proc Natl Acad Sci USA 96: 766–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, et al. (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob P, Kim NH, Wu F, El-Kasmi F, Chi Y, Walton WG, Furzer OJ, Lietzan AD, Sunil S, Kempthorn K, et al. (2021) Plant “helper” immune receptors are Ca2+-permeable nonselective cation channels. Science 373: 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jurkowski GI, Smith RK, Yu IC, Ham JH, Bent AF (2004) Arabidopsis dnd2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol Plant Microbe Interact 17: 511–520 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones J, Shirasu K, Menke F, Jones A (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell 54: 43–55 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Shirasu K, Zipfel C (2015) Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol 56: 1472–1480 [DOI] [PubMed] [Google Scholar]
- Kang S, Yang F, Li L, Chen H, Chen S, Zhang J (2015) The Arabidopsis transcription factor BRASSINOSTEROID INSENSITIVE1-ETHYl methanesulfonate-suppressor1 is a direct substrate of MITOGEN-ACTIVATED PROTEIN KINASE6 and regulates immunity. Plant Physiol 167: 1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus PH, Ikura M (2002) Calmodulin in action: diversity in target recognition and activation mechanisms. Cell 6: 739–742 [DOI] [PubMed] [Google Scholar]
- Knight MR, Campbell AK, Smith SM, Trewavas AJ (1991) Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352: 524–526 [DOI] [PubMed] [Google Scholar]
- Knoblauch M, Froelich DR, Pickard WF, Peters WS (2014) Seorious business: structural proteins in sieve tubes and their involvement in sieve element occlusion. J Exp Bot 65: 1879–1893 [DOI] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N terminus of bacterial elongation factor tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Jiang S, Yu X, Cheng C, Chen S, Cheng Y, Yuan JS, Jiang D, He P, Shan L (2015) Phosphorylation of trihelix transcriptional repressor ASR3 by MAP KINASE4 negatively regulates Arabidopsis immunity. Plant Cell 27: 839–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, He X, Luo X, Xu L, Liu L, Min L, Jin L, Zhu L, Zhang X (2014) Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol 166: 2179–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YB, Han LB, Wang HY, Zhang J, Sun ST, Feng DQ, Yang CL, Sun YD, Zhong NQ, Xia GX (2016) The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in cotton. Plant Physiol 170: 2392–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Ding P, Lian K, Wang J, Ma M, Li L, Li L, Li M, Zhang X, Chen S, et al. (2016) Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. Elife 5: e13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S (2009) The CBL-CIPK network in plant calcium signaling. Trends Plant Sci 14: 37–42 [DOI] [PubMed] [Google Scholar]
- Luan S, Wang C (2021) Calcium signaling mechanisms across kingdoms. Annu Rev Cell Dev Biol 37: 311–340 [DOI] [PubMed] [Google Scholar]
- Ma Z, Song T, Lin Z, Ye W, Wang Y (2015) A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27: 2057–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack E, Braam J (2003) Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol 159: 585–598 [DOI] [PubMed] [Google Scholar]
- Mccormack E, Tsai YC, Braam J (2005) Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci 10: 383–389 [DOI] [PubMed] [Google Scholar]
- Moeder W, Urquhart W, Ung H, Yoshioka K (2011) The role of cyclic nucleotidegated ion channels in plant immunity. Mol Plant 4: 442–452 [DOI] [PubMed] [Google Scholar]
- Monaghan J, Matschi S, Shorinola O, Rovenich H, Matei A, Segonzac C, Malinovsky FG, Rathjen JP, MacLean D, Romeis T, et al. (2014) The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16: 605–615 [DOI] [PubMed] [Google Scholar]
- Mysore KS, Crasta OR, Tuori RP, Folkerts O, Swirsky PB, Martin GB (2002) Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. Tomato. Plant J 32: 299–315 [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Brunner F, Kemmerling B, Piater L (2010) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev 198: 249–266 [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Nennstiel D, Jabs T, Sacks WR, Scheel D (1994) High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell 78: 449–460 [DOI] [PubMed] [Google Scholar]
- Qin J, Wang K, Sun L, Xing H, Wang S, Li L, Chen S, Guo HS, Zhang J (2018) The plant-specific transcription factors CBP60g and SARD1 are targeted by a Verticillium secretory protein VdSCP41 to modulate immunity. Elife 7: e34902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Zhao J, Zuo K, Cao Y, Ling H, Sun X, Tang K (2004) Isolation and characterization of an ERF-like gene from Gossypium barbadense. Plant Sci 167: 1383–1389 [Google Scholar]
- Qutob D, Kemmerling B, Brunner F, Küfner I, Engehardt S, Gust AA, Luberacki B, Seitz HU, Stahl D, Rauhut T (2006) Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell 18: 3721–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VS, Ali GS, Reddy A (2002) Genes encoding calmodulin-binding proteins in the Arabidopsis genome. J Biol Chem 277: 9840–9852 [DOI] [PubMed] [Google Scholar]
- Ron M, Avni A (2004) The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaible LC, Cannon OS, Waddoups V (1951) Inheritance of resistance to Verticillium Wilt in a tomato cross. Phytopathology 41: 986–990 [Google Scholar]
- Sejalon-Delmas N, Mateos FV, Bottin A, Rickauer M, Dargent R, Esquerré-Tugayé MT (1997) Purification, elicitor activity, and cell wall localization of a glycoprotein from Phytophthora parasitica var. Nicotianae, a fungal pathogen of tobacco. Phytopathology 87: 899–909 [DOI] [PubMed] [Google Scholar]
- Sistrunk ML, Antosiewicz DM, Purugganan MM, Braam J (1994) Arabidopsis TCH3 encodes a novel Ca2+ binding protein and shows environmentally induced and tissue-specific regulation. Plant Cell 6: 1553–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhang J (2020) Regulatory role of receptor-like cytoplasmic kinases in early immune signaling events in plants. FEMS Microbiol Rev. 44: 845–856 [DOI] [PubMed] [Google Scholar]
- Sun L, Qin J, Wang K, Zhang J (2017) Expansion of pathogen recognition specificity in plants using pattern recognition receptors and artificially designed decoys. Sci China Life Sci 60: 797–805 [DOI] [PubMed] [Google Scholar]
- Sun L, Zhu L, Xu L, Yuan D, Min L, Zhang X (2014) Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat Commun 5: 5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborski J, Krasileva KV (2020) Evolution of plant NLRs: from natural history to precise modifications. Annu Rev Plant Biol 71: 355–378 [DOI] [PubMed] [Google Scholar]
- Tian W, Hou C, Ren Z, Wang C, Zhao F, Dahlbeck D, Hu S, Zhang L, Niu Q, Li L, et al. (2019) A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572: 131–135 [DOI] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Sato M, Cohen JD, Katagiri F, Glazebrook J (2009) Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PloS Pathog 5: e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Truman W, Sato M, Nguyen LV, Katagiri F, Glazebrook J (2011) CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J 67: 1029–1041 [DOI] [PubMed] [Google Scholar]
- Wei ZM, Laby RJ, Zumoff CH, Bauer DW, He SY, Collmer A, Beer SV (1992) Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 257: 85–88 [DOI] [PubMed] [Google Scholar]
- Wernimont AK, Artz JD, Finerty P, Lin YH, Amani M, Allali-Hassani A, Senisterra G, Vedadi M, Tempel W, Mackenzie F, et al. (2010) Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat Struct Mol Biol 17: 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa H, Mitsuhara I, Ito N, Seo S, Kamada H (2001) Transcriptionally and post-transcriptionally regulated response of 13 calmodulin genes to tobacco mosaic virus-induced cell death and wounding in tobacco plant. Eur J Biochem 268: 3916–3929 [DOI] [PubMed] [Google Scholar]
- Yang CL, Shan L, Wang HY, Han LB, Wang FX, Cheng HQ, Wu XM (2015) Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dahliae. Mol Plant 8: 399–411 [DOI] [PubMed] [Google Scholar]
- Yu IC, Fengler KA, Clough SJ, Bent AF (2000) Identification of Arabidopsis mutants exhibiting an altered hypersensitive response in gene-for-gene disease resistance. Mol Plant Microbe Interact 13: 277–286 [DOI] [PubMed] [Google Scholar]
- Yu IC, Parker JE, Bent A (1998) Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci USA 95: 7819–7824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Jauregui E, Du L, Tanaka K, Poovaiah BW (2017) Calcium signatures and signaling events orchestrate plant-microbe interactions. Curr Opin Plant Biol 38: 173–183 [DOI] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010a). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zhang L, Kars I, Essenstam B, Liebrand T, Wagemakers L, Elberse J, Tagkalaki P, Tjoitang D, Van D, Kan JV (2014) Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol 164: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu S, Ding P, Wang D, Cheng YT, Jing H, Gao M, Xu F, Yan L, Zhu Z, et al. (2010b). Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci USA 107: 18220–18225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Chai J (2008) Plant pathogenic bacterial type III effectors subdue host responses. Curr Opin Microbiol 11: 179–185 [DOI] [PubMed] [Google Scholar]
- Zhou JM, Zhang Y (2020) Plant immunity: danger perception and signaling. Cell 181: 978–989 [DOI] [PubMed] [Google Scholar]
- Zhou J, Wang X, He Y, Sang T, Wang P, Dai S, Zhang S, Meng X (2020) Differential phosphorylation of the transcription factor WRKY33 by the protein kinases CPK5/CPK6 and MPK3/MPK6 cooperatively regulates camalexin biosynthesis in Arabidopsis. Plant Cell 32: 2621–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Oldroyd GED (2017) Plant signalling in symbiosis and immunity. Nature 543: 328–336 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones J, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zulawski M, Braginets R, Schulze WX (2013) PhosPhAt goes kinases—searchable protein kinase target information in the plant phosphorylation site database PhosPhAt. Nucleic Acids Res 41: 1176–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.