Abstract
Context
DNA methylation in the diagnosis of gestational diabetes.
Objective
To assess the value of DNA methylation in the diagnosis of gestational diabetes (GDM) and in the prediction of maternal postpartum glucose disturbances.
Methods
Two-stage observational study performed between July 2006 and December 2010, at University Hospital. Forty-eight randomly selected pregnant women formed the discovery cohort (24 with GDM and 24 controls) and 252 pregnant women (94 with GDM and 158 controls) formed the replication cohort. GDM women were re-evaluated 4 years postpartum. The main outcome measures were GDM, type 2 diabetes or prediabetes at 4 years postpartum.
Results
We identified 3 CpG sites related to LINC00917, TRAPPC9, and LEF1 that were differentially methylated in women with GDM and abnormal glucose tolerance; and sites associated with LINC00917 and TRAPPC9 were independently associated with an abnormal glucose tolerance status 4 years postpartum after controlling for clinical variables. Moreover, the site associated with LINC00917 and the combination of the 3 sites had the highest predictive values.
Conclusion
Our results suggest that some of these sites may be implicated in the development of GDM and postpartum abnormal glucose tolerance.
Keywords: DNA methylation, gestational diabetes, postpartum glucose disturbance, epigenetic
Gestational diabetes (GDM) is a relatively common glucose tolerance disorder of pregnancy that is often associated with the later development of type 2 diabetes (1), metabolic syndrome (2-4), and cardiovascular disease (5). Its prevalence has increased in parallel with these metabolic disorders, suggesting a common pathogenic link between them. The decrease of insulin action (insulin resistance) that occurs during the second half of pregnancy places the β-cells of the pancreas under stress, which can lead to hyperglycemia and GDM when the β-cell secretory capacity is overwhelmed by tissue insulin resistance. The early identification of women at high risk for developing GDM is a main priority for healthcare systems worldwide, and drives the search for new biomarkers that can predict GDM before β-cell failure and loss of metabolic control occurs.
Genetic and environmental factors are known to influence the pathogenesis of GDM and, recently, epigenetic mechanisms have also been implicated (6). Epigenetics is described as the heritable changes in gene function that occur without changes to the DNA sequence, and can provide molecular mechanisms to explain the contributions of genetic and environmental factors in a disease process (7). Epigenetic modifications include DNA methylation (DNAm), histone modification, such as methylation and acetylation, and small noncoding RNAs (microRNAs). DNAm is one of the most well-studied epigenetic mechanisms associated with stable alterations of gene expression and involves the covalent addition of a methyl group to the fifth carbon position of a cytosine residue within a cytosine phosphate–guanine (CpG) dinucleotide. Methylation of repressor elements can trigger increased gene expression, whereas methylation of promoter or enhancer regions can repress gene expression. Also, methylation in the gene body can increase gene expression or influence alternative splicing (8).
DNAm has been proposed as a potential biomarker or predictor of disease (9, 10). In the context of metabolic control, several studies have reported an association between aberrant DNAm in insulin-sensitive tissues and type 2 diabetes, and these changes are also reflected in peripheral blood (reviewed in (11)), which would allow for the identification of global methylation regions as potent biomarkers of disease (12, 13). Indeed, specific changes in the epigenome have been associated with the onset and progression of diabetes and could be used as markers of type 2 diabetes risk (14, 15). In the same line, the comprehensive analysis of epigenetic modifications may contribute toward a better understanding of the underlying mechanisms of GDM, and aid in identifying mothers at risk of developing metabolic instability later in life.
Despite the important role of epigenetics in human disease, studies investigating DNAm alterations in blood from women with GDM are scarce, and most have focused on the study of epigenetic traits in placental tissue and cord blood (10) and on the identification of markers to explain the effect of the GDM environment on offspring. Relatively less is known about the potential role of DNAm in GDM pathogenesis per se or its utility as a biomarker in diagnosis, risk prediction, and prognosis in the mother. Several studies have assessed DNAm in the whole blood of patients with GDM during pregnancy, with conflicting results (16-22); most of these studies used limited sample sizes (16-20), were not designed to identify later adverse outcomes in mothers, and/or did not include a validation cohort.
The present study aimed to identify a specific DNAm signature in pregnant women with GDM and to test its predictive value for detecting glucose instability after pregnancy, including type 2 diabetes and/or prediabetes.
Patients and Methods
Participants, Recruitment, and Inclusion Criteria
Participants were recruited at the Hospital Universitari de Tarragona Joan XXIII, Spain, between July 2006 and December 2010. Pregnant women with and without GDM were included in a prebirth longitudinal study. Blood samples were obtained between 26 and 30 weeks of pregnancy and were stored in a biobank until the analysis was done. Data used for this analysis are from baseline examinations of the entire cohort. A sample of 124 women with GDM and 200 without GDM, with availability of blood samples in the bio-bank were selected (Pere Vigili Institute Biobank—national biobank Spanish Registry: C.0001725 https://biobancos.isciii.es/ListadoColecciones.aspx?id=C.0001725). From this sample, 6 patients with GDM and 18 control women were excluded due to difficulties in extracting sufficiently pure DNA. Women with GDM were invited for metabolic screening at 4 years postpartum.
During pregnancy, all women were screened for GDM at 24-28 weeks following the Spanish Diabetes and Pregnancy Group recommendations (23). Subjects with a 1-hour 50-g glucose challenge test ≥7.78 mmol/L (140 mg/dL) underwent a 3-hour 100-g oral glucose tolerance test (OGTT); those with 2 or more values above the threshold proposed by the National Diabetes Data Group (24) were considered to have GDM, whereas those with all values below the threshold were classified as controls. We included only women with a singleton pregnancy and with gestational age confirmed by ultrasound performed before 20 weeks. Women with pre-existing diabetes, inflammatory or chronic diseases, or on medication that is known to affect carbohydrate metabolism during pregnancy were excluded. The Ethics Committee of the center approved the experimental protocol (CEIC/1/12/2011) and all participants gave their written, informed consent.
From all the participants, we selected 2 nonoverlapping subcohorts. Forty-eight randomly selected women formed the discovery cohort (cohort 1), including 24 with GDM and 24 controls. The replication cohort (cohort 2) included 252 pregnant women (94 with GDM and 158 acting as controls). Baseline sociodemographic and clinical information was obtained by a predefined questionnaire. Height, weight, and clinical, analytical, and obstetrical outcomes were registered prospectively in the entire population. Weight, abdominal and waist circumference, and blood pressure were determined according to standard methods. Gestational weight gain (GWG) was the difference between the maternal weight recorded at the last antenatal visit and the prepregnancy weight. Body mass index (BMI) was calculated using the formula BMI = weight (kg)/height (m2). The change in BMI was calculated by the formula BMI gain = current BMI – pregravid BMI.
Postpartum Evaluation
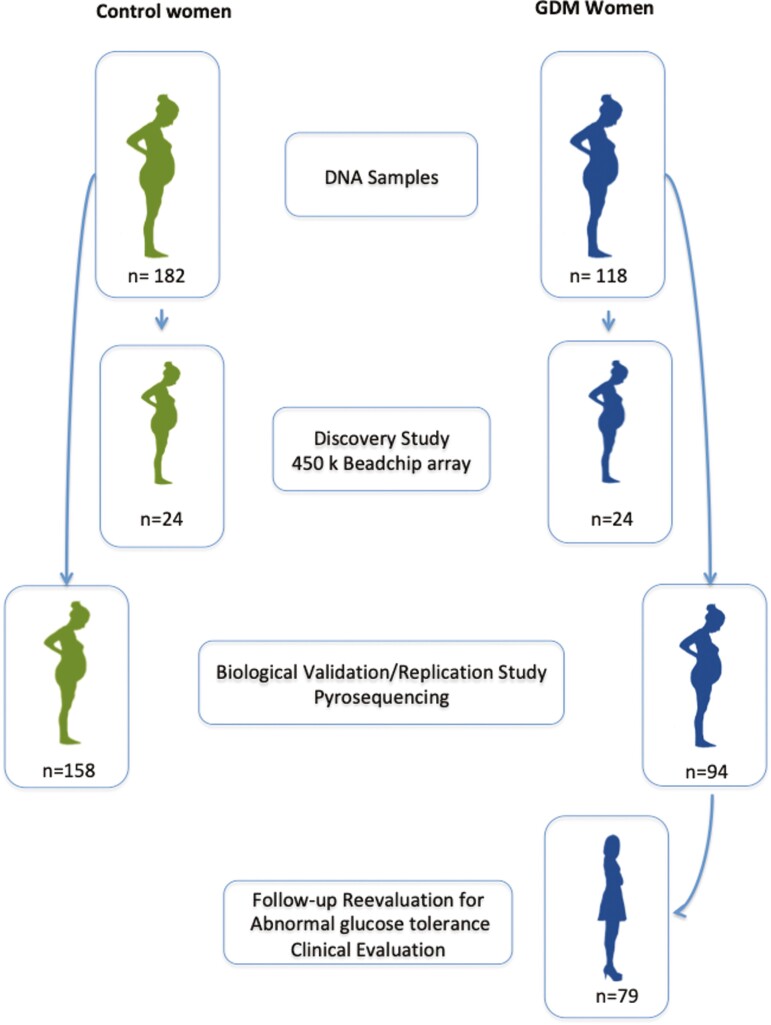
Women with GDM were invited to participate in a re-evaluation study at 4 years postpartum. At this time, we recorded any new event since the end of the index pregnancy. A 2-hour 75-g OGTT was performed in the morning after an overnight fast. Results of the OGTT were evaluated according to the criteria of the American Diabetes Association (25) and women were classified into 2 groups: normal glucose tolerant and abnormal glucose tolerant. Women with fasting glucose <100 mg/dL, glucose concentrations 2 hours after the OGTT <7.8 mmol/L (International Federation of Clinical Chemistry [IFCC]) (140 mg/dL), and HbA1c <39 mmol/mol (IFCC) (5.7%) were included in the normal glucose-tolerant group. Those with fasting plasma glucose ≥5.56 mmol/L (100 mg/dL) and/or glucose concentrations 2 hours after OGTT ≥7.8 mmol/L (140 mg/dL) and/or HbA1c ≥39 mmol/mol (IFCC) (5.7%) were included in the abnormal glucose tolerance group. This latter group included all the subjects with diabetes and prediabetes. The flowchart of the study is shown in Fig. 1.
Figure 1.
Flow chart of the population included in the study.
Analytical Methods
Glucose, HbA1c, insulin, and lipid concentrations were measured at fasting. Glucose levels were determined on an ADVIA 2400 autoanalyzer (Siemens AG, Munich, Germany) using standard enzymatic methods. Plasma insulin was determined by immunoassay on an ADVIA Centaur System (Siemens AG) (RRID:AB_2800499). The insulin assay showed cross-reactivity of 0.1% to intact human proinsulin or primary circulating split form (des 31,32 proinsulin). HbA1c was estimated by high-performance liquid chromatography–based ion exchange chromatography (Menarini/Akray ADAMS-A1c HA-8160; Menarini Diagnostics, Florence, Italy) standardized and traceable to the Diabetes Control and Complications Trial assay (26). The intra-assay coefficients for people without and with diabetes are 1.5 and 0.5%, respectively
DNA Methylation Profiling
DNA was extracted from peripheral whole blood samples collected in 10-mL EDTA tubes using the Gentra Puregene Blood kit (Qiagen, Valencia, CA). DNA concentration was quantified using the PicoGreen assay (Thermo Scientific, Waltham, MA, USA) and the quality of the samples was assessed by absorbance measurements. DNAm profiling was performed in the discovery cohort using the Infinium Human Methylation 450 BeadChip (Illumina, San Diego, CA), which enables the simultaneous quantitative measurement of the methylation status at 485 577 CpG sites, covering 99% of RefSeq Genes and intergenic regions (www.Illumina.com). The arrays were scanned with the Illumina HiScan Scanner. Data on methylation levels at each CpG as beta values (1 corresponds to complete methylation and 0 to no methylation) were processed with minfi v1.7.15, a R-bioconductor package (27). All controls were checked for inconsistencies in each measured plate. Each 1 of the 4 microchips used contained samples from participants with and without GDM and were randomly distributed. Probes with a signal detection P > .01 (unable to differentiate from background) were excluded from the analysis. In the final analysis, we excluded CpG sites associated with single nucleotide polymorphisms, cross-reactive probes, and CpG sites located on sex chromosomes, leaving 361 922 CpG sites for analysis.
Differential methylated regions identified in the discovery cohort were studied by sequencing on bisulfite-converted genomic DNA in the replication cohort, amplifying 400 to 600 bp in 2 to 3 fragments by region. MethPrimer software was used to design the primers for directional region amplification (http://www.urogene.org/ methprimer/index1.html). We analyzed a maximum of 2 amplicons per region and each amplification reaction was functionally validated with the QIAxcel System (Qiagen). Libraries compatible with Illumina were generated by polymerase chain reaction using 2 rounds of polymerase chain reaction (first for specific amplification and inclusion of a tail for the second step; second polymerase chain reaction was used to include the barcodes and remaining sequences required for Illumina sequencing). Products were verified on the QIAxcel System and were cleaned up for next-generation sequencing. Amplicons were sequenced on an Illumina MiSeq system and amplicon analysis was performed with our own pipelines inspired by BWA-meth (https://github.com/brentp/bwa-meth), where reads were mapped to an in silico bisulfite reference and the percentage of Cs and Ts was calculated from the variant call format file (28).
Gene Ontology and Functional Enrichment Analysis
The PANTHER GO-Slim database (version 15.0, based on GO release 2018-07-03, released February 14, 2020), including 3219 total terms of 2137 biological process, 548 cellular components, and 534 molecular functions, was used for functional enrichment analysis. GO terms of identified differentially methylated genes included categories such as cellular component, biological process and molecular function. The significance value was set as P < .05 using the Bonferroni correction for multiple analyses.
The interactive relationships between differentially methylated genes were identified using the STRING database (29) (https://string-db.org/). A combined score of >0.7 (high) of only experimentally validated interactions was considered to be statistically significant. The enriched pathways were identified using Kyoto Encyclopedia of Genes and Genomes (30) and Reactome (31) analysis.
Statistical Analysis
Each array and sample passed quality control assessment based on the performance of internal array controls. Initial processing, probe type correction and assessment of array data were conducted with minfi package using Swan normalization (32), a within-array normalization correction for the technical differences between the Type I and Type II array designs.
In subcohort 1 (discovery cohort processed with Illumina’s 450K methylation chip), we investigated the association of whole blood DNAm with GDM at 361 922 CpG sites. As DNAm levels quantified by beta values at most of the CpG sites are not normally distributed, we used nonparametric median regression models, in which the dependent variables were the beta values at CpG sites and the independent variable was “GDM at recruitment yes or no”. We used raw P values and selected only those CpG sites with P < .001.
In subcohort 2 (replication cohort, CpG validation using methyl-seq of amplicons), we examined the association of whole blood DNAm with GDM for the top 7 CpG sites of which the association in subcohort 1 was more significant. Data are presented as percentages for categorical variables, mean (SD) for normally distributed continuous variables, and median (interquartile range [IQR]) for non-normally distributed variables. Data normality was tested with the Kolmogorov–Smirnov test. For comparisons of proportions, differences between groups were analyzed using the chi-square test, while for comparisons of normally and non-normally distributed quantitative variables an unpaired t-test or Mann–Whitney U test was applied. After applying the Kolmogorov–Smirnov test to identify distributions, we used Student’s t-test or the Wilcoxon test to evaluate differences of quantitative variables between 2 groups, and the chi-square test. Subsequently, the general linear model was used to adjust the differences between the GDM and control groups for confounding variables (pregestational BMI, GWG, and maternal age), when required. Effect size measures, Cohen’s d and h, were calculated using the online calculator Psychometrica (https://www.psychometrica.de/effect_size.html). To assess the potential predictive ability of DNAm for abnormal glucose metabolism at 4 years postpartum in women with GDM, we used logistic regression analysis to assess the independence of the associations observed between the differentially methylated sites and abnormal glucose tolerance (type 2 diabetes or prediabetes). Subsequently, the area under the curve of the receiver operator characteristic (ROC) curves was computed using the predicted probability of abnormal glucose tolerance (type 2 diabetes or prediabetes) and the true status of abnormal glucose tolerance for each participant and the paired-sample design was used to test differences between the curves. A 2-sided P < .05 was considered as significant. Data were analyzed with SPSS software v20.0 (IBM, Armonk, NY)
Results
Maternal and Birth Characteristics of the Entire Cohort
Table 1 shows the characteristics of all the study participants. Women with GDM were on average 1 year older than controls, were heavier, gained less weight during pregnancy, had GDM in a previous pregnancy, were less frequently nulliparous, and had higher fasting glucose levels.
Table 1.
Clinical and analytical characteristics of the population studied
| Control (n = 182) | GDM (n = 118) | P value | Effect size | |
|---|---|---|---|---|
| Maternal | ||||
| Maternal age (years) | 31.2 ± 4.4 | 32.6 ± 4.5 | .007 | 0.315 |
| BMI (kg/m2) | 22.9 (20.9–26.8) | 24.5 (21.5–30.1) | .018 | 0.280 |
| Gestational weight gain (kg) | 11.8 ± 6.0 | 8.2 ± 5.0 | <.001 | –0.674 |
| Gestational age at enrollment (weeks) | 27 (26–28) | 27.5 (26–29) | .250 | 0.132 |
| Smoker | 33 (18.1) | 21 (17.8) | .941 | 0.008 |
| Family history of diabetes | 83 (45.6) | 61 (51.7) | .240 | |
| GDM in previous pregnancy | 7 (3.8) | 22 (18.6) | <.001 | 0.502 |
| Nulliparous | 80 (44) | 35 (29.7) | .013 | |
| Fasting glucose (mg/dL) | 80.8 ± 7.3 | 84.1 ± 10.6 | .002 | 0.377 |
| Glucose 60 min after 100 g OGTT | 154.7 ± 23.9 | 209.9 ± 28.6 | <.001 | 2.136 |
| Glucose 120 min after 100 g OGTT | 120.7 ± 23.3 | 185.3 ± 19.8 | <.001 | 2.937 |
| Glucose 180 min after 100 g OGTT | 96.5 ± 25.9 | 135.8 ± 38.5 | <.001 | 1.249 |
| Cholesterol (mg/dL) | 259 ± 45 | 258 ± 56 | .926 | –0.020 |
| HDL cholesterol (mg/dL) | 73 ± 13 | 70 ± 13 | .149 | –0.231 |
| Triglycerides (mg/dL) | 167 (138–202) | 176 (146–221) | <.001 | 0.196 |
| HOMA-IR | 1.51 (1.13–2.52) | 1.84 (1.35–2.71) | .061 | 0.235 |
| Offspring | ||||
| Gestational age at delivery (weeks) | 39 (38–40) | 39 (38–40) | .140 | 0.168 |
| Birth weight (g) | 3241 ± 484 | 3194 ± 500 | .422 | –0.096 |
| Length (cm) | 49.4 ± 2.1 | 49.1 ± 2.2 | .178 | –0.140 |
| Male sex | 92 (50.5) | 59 (50) | .926 | 0.009 |
| Caesarean delivery | 49 (26.9) | 40 (33.9) | .063 | 0.120 |
Data are presented as n (%), mean ± SD, or median (IQR).
Abbreviations: BMI, body mass index; DM, diabetes; GDM, gestational diabetes mellitus; HDL, high-density lipoprotein; HOMA-IR, homeostasis model assessment-estimated insulin resistance; OGTT, oral glucose tolerance test.
The clinical characteristics of the discovery subcohort are shown elsewhere (Table S1 (33)). No differences were observed between groups with the exception of higher glucose levels in the GDM group than in the control group.
Epigenome-wide DNA Methylation Analysis
We measured epigenome-wide DNAm in the peripheral blood of women in the discovery cohort to examine for changes in DNAm between GDM and control pregnancies. In the regression-fitted analysis, we identified 50 differentially methylated CpG sites with raw P values between 10–4 and 10–5 (Table S2 (33)). In total, 31 sites were annotated to 28 unique genes, and 3 genes were each associated with 2 CpG sites. The first pair (cg05090351 and cg23000734) mapped to CTBP2 and both located in the transcription start site (TSS); the second pair (cg23000734 and cg11504805) mapped to RASA3 and were located in the gene body; and the third pair (cg10436026 and cg00454719) mapped to SMAD9 and were also located in the gene body. In total, 15 CpG sites were located within a CpG island, 8 at a CpG island “shore” (up to 2 kb away from the island boundary), 8 within a CpG island “shelf” (2.5-5 kb away from the island), and the remaining 19 were in “open sea” regions. In total, of the 50 CpGs, 14 were in the gene body, 14 in the TSS1500 (within 1500 bp of the TSS), and 4 in TSS200 (within 2000 bp).
After correcting for multiple testing using a false discovery rate of 5%, none of the CpG sites identified were significantly different between the groups. However, in an effort to avoid Type II errors (false negative), we selected the top 7 differentially methylated CpG sites with the lowest uncorrected P values to look for potential candidates in this preliminary screening and as targets for bisulfite sequencing (Table 2). Three CpG sites were hypomethylated in GDM cases compared with controls, whereas the other 4 were hypermethylated. Each of the genomic locations of the 7 CpG sites was annotated with 7 unique genes (Table 2). Two CpGs were located within a CpG island, 2 at a CpG island shore, 2 within a CpG island shelf, and 1 in the open sea region. Of the 7 CpGs, 2 (28.6%) were in the body and 2 (28.6%) were in the TSS1500.
Table 2.
The top 7 significantly differentially methylated CpG sites between GDM and control
| Probe ID | Methylation | Chr | Nearest gene | Located | P value |
|---|---|---|---|---|---|
| cg13773818 | Increased | 9 | ZNF79 | N_Shore | 3.67 ×10–5 |
| cg05090351 | Increased | 10 | CTBP2 | Island | 5.94 ×10–5 |
| cg19403586 | Decreased | 8 | MRPS28 | N_Shelf | 8.78 ×10–5 |
| cg06341779 | Decreased | 4 | LEF1 | Island | 1.31 ×10–4 |
| cg10758618 | Decreased | 8 | TRAPPC9 | N_Shelf | 1.39 ×10–4 |
| cg23519572 | Increased | 13 | RASA3 | S_Shore | 1.77 ×10–4 |
| cg03554962 | Increased | 16 | LINC00917 | Open_sea | 2.00 ×10–4 |
Pathway Enrichment Analysis
Genes related to the top 7 differentially methylated CpG sites were analyzed using Panther GO-Slim classification analysis. The detailed results of the enrichment analysis of the molecular function, biological processes, and cellular components are shown elsewhere (Table S3 (33)). Based on molecular function analysis, 2 genes (LEF1 and ZNF79) were related to protein- and DNA-binding function; for biological process, LEF1 was associated with canonical and noncanonical Wnt signaling; and in cellular components, 4 genes were involved in protein secretion and membrane-bounded organelles. Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis identified the Wnt signaling pathway that included LEF1 and CTBP2 (P = .0152), and analysis of Reactome pathways highlighted repression of Wnt target genes involving the same genes (P < .001) and of diseases of signal transduction, which included RASA3 and CTBP2 (P = .0487).
Confirmation of the Differentially Methylated CpGs in the Replication Cohort
We performed pyrosequencing of the 7 CpG sites selected from the discovery cohort, as well as those in the vicinity, and from independent samples of the replication cohort (94 women with GDM and 158 controls). The clinical and analytical characteristics of the replication cohort are shown elsewhere (Table S4 (33)).
Differences in DNAm between the GDM and control groups were assessed after adjusting for age, BMI, and GWG (Table S5 (33)). Results showed that methylation at Cg03554962 (LINC00917) was higher in the GDM group than in the control group and the same trend was observed for the 5 adjacent positions studied, reaching statistical significance for 4. CpG cg05090351 (CTBP2) was also hypermethylated in the replication cohort and the adjacent sites showed the same trend, but only 1 of them reached statistical significance. While no differences were found for the differential methylation for the Cg13773818 (ZNF79) site, 2 of the 7 adjacent positions studied were hypermethylated.
By contrast, Cg06341779 (LEF1), which was hypomethylated in the GDM group in the discovery study, was significantly hypermethylated in the replication cohort, and in 4 of the 5 adjacent sites studied. No differences in DNAm were observed in the CpG sites related to Cg23519572 (RASA3), Cg10758618 (TRAPP9) and Cg19403586 (MRSP28) or in the majority of the adjacent sites studied. These findings support the need to verify array data.
Development of Abnormal Glucose Tolerance (Diabetes or Prediabetes) at 4 Years Postpartum
Seventy-nine of the 97 women with GDM (81.4%) included in the validation analysis returned for re-evaluation at 4 years postpartum, and glucose tolerance was assessed. Based on the results of this re-evaluation, we divided the cohort into 2 groups, normal glucose tolerance (n = 54) and abnormal glucose tolerance (n = 25). The clinical characteristics of this population are shown in Table 3.
Table 3.
Clinical and analytical characteristics of the GDM women at the 4-year postpartum evaluation
| Normal glucose tolerance (n = 54) | Abnormal glucose tolerance (n = 25) | P value | Effect size | |
|---|---|---|---|---|
| Maternal age (years) | 33.08 ± 4.16 | 33.48 ± 3.33 | .667 | 0.102 |
| Pregestational BMI (kg/m2) | 25.09 ± 5.20 | 29.47 ± 6.48 | .002 | 0.778 |
| Gestational weight gain (kg) | 8.53 ± 5.03 | 7.46 ± 5.49 | .402 | -0.207 |
| Birth weight (g) | 3.250 ± 443 | 3.091 ± 557 | .184 | -0.330 |
| Fasting glucose (mmol/L) | 4.63 ± 0.56 | 5.0 ± 0.73 | .018 | 0.596 |
| 60-min glucose after 100-g OGTT (mmol/L) | 11.50 ± 1.61 | 11.52 ± 1.58 | .954 | 0.014 |
| 120-min glucose after 100 g OGTT (mmol/L) | 10.25 ± 1.10 | 10.43 ± 1.04 | .519 | 0.159 |
| 180-min glucose after 100-g OGTT (mmol/L) | 7.67 ± 2.10 | 7.98 ± 2.66 | .580 | 0.137 |
| Cholesterol (mmol/L) | 6.79 ± 1.27 | 6.01 ± 1.42 | .040 | 0.327 |
| HDL cholesterol (mmol/L) | 1.84 ± 0.36 | 1.74 ± 0.39 | .360 | -0.572 |
| Triglycerides (mmol/L) | 2.16 ± 0.59 | 2.41 ± 0.86 | .246 | -0.268 |
| BMI at 4 years postpartum (kg/m2) | 24.59 ± 5.05 | 31.39 ± 7.36 | <.001 | 1,150 |
| Weight gain at 4 years postpartum (kg/m2) | -0.59 ± 5.16 | 4.77 ± 6.67 | <.001 | 0.940 |
| BMI gain at 4 years postpartum (kg/m2) | -0.24 ± 2.04 | 1.92 ± 2.85 | <.001 | 0.924 |
Abbreviations: BMI, body mass index; GDM, gestational diabetes mellitus; HDL, high-density lipoprotein; OGTT, oral glucose tolerance test.
To examine for relationships between the methylation profile and the development of abnormal glucose tolerance between the 2 groups, we assessed the differences in the percentage of DNAm at the studied sites and adjacent positions. We observed differences in 3 positions, each 1 related to a different gene: TRAPPC9 (normal glucose tolerance 95.49% [IQR 94.60-96.93] vs abnormal glucose tolerance 94.43% [IQR 95.68-97.50]; P = .039), LEF1 (1.05% [IQR 0.61-1.82] vs 1.71% [IQR 1.09-2.46]; P = .039), and LINC00917 (92.07% [IQR 88.50-94.99] vs 94.57% [IQR 93.31-96.25]; P = .014) (Table S6 (33)). To assess the independence of these associations, we performed a logistic regression analysis adjusting for pregestational BMI, maternal age, and GWG and the association between any type of glucose disturbance. Results showed that the methylation percentage at the positions related to TRAPPC9 and LINC00917 persisted (Table S7) (33)).
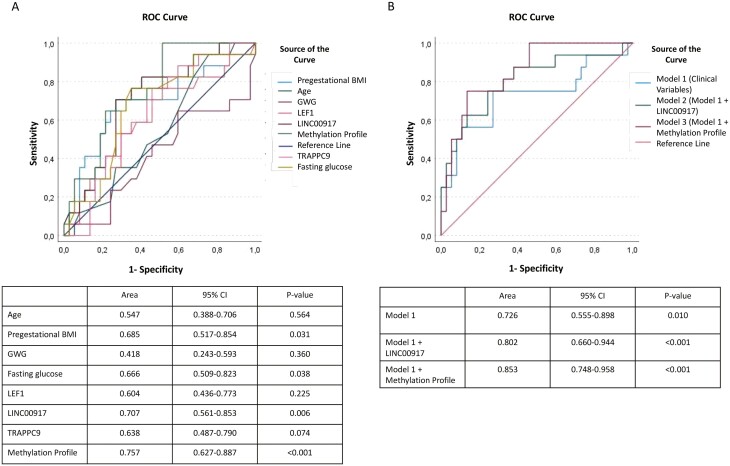
To explore the predictive ability of conventional risk factors and the methylation profile, we performed ROC curve analysis (Fig. 2). The predictive value of LINC00917 was greater than that of prepregnancy BMI and fasting glucose, but the difference did not reach statistical significance. The optimal cut-off point for LINC00917 for predicting any kind of glucose disturbance was 93.83%, with sensitivity of 70.6 and specificity of 73.0, and with a positive predictive value of 54.5 and negative predictive value of 86.6. The combination of the 3 sites identified (referred to as the methylation profile) showed a higher area under the curve (AUC), but the differences were not significantly different (Fig. 2A). We further analyzed prediction models incorporating clinical and biochemical risk factors. In Model 1, the conventional risk factors prepregnancy BMI, age, fasting plasma glucose, gestational age, and family history of diabetes during pregnancy were included. The AUC for the conventional model was 0.760 (95% CI 0.555-0.898). Introducing LINC00917 into the model increased the AUC to 0.802 (95% CI 0.660-0.944; P < .001); finally, the addition of the methylation profile to Model 1 increased the prediction model, with an AUC of 0.853 (95% CI 0.748-0.958; P < .001) (Fig. 2B). However, when comparing the areas under the ROC curves, the differences observed with respect to the clinical model (0.076 and 0.127, respectively) were not statistically significant (P = .196 and P = .083, respectively).
Figure 2.
Receiver operating characteristic (ROC) curves showing area under the curve (AUC), 95% CI, and P value in assessing clinical parameters. LEF1, LINC 00917, and TRAPP9 methylation and the combination of the 3 methylated sites (methylation profile) as predictors of abnormal glucose tolerance at 4 years postpartum (A) and assessing the combination of clinical parameters (Model 1), Model 1 + LINC00917 (Model 2) and Model 1 + methylation profile (Model 3) as a predictors of abnormal glucose tolerance (B).
Discussion
In this longitudinal study, we provide new data on epigenomics in women with GDM and identify maternal DNAm during pregnancy as a potential biomarker to detect risk for developing glucose abnormalities later in life.
Global DNAm is altered during hyperglycemia (34, 35), and increased global methylation in placental tissue has been associated with GDM (36, 37). However, there are limited data describing DNAm in the peripheral blood of patients with GDM, and the results are inconclusive (16-19, 21, 22, 38, 39). Small sample sizes and the absence of reproducibility or confirmatory cohorts have been the main weaknesses in most studies (16-18, 38).
In a recent study on Black South African women, global DNAm in peripheral blood was found not to be associated with GDM, which was contrary to the authors’ hypothesis (20). Two studies with larger cohorts have been recently published using the Infinium MethylationEPIC BeadChip (21, 22), which covers 850 000 CpG sites. Fragoso-Bargas et al (22) identified 4 CpG sites associated with BMI and hyperglycemia in peripheral blood leukocytes of a subcohort of the EPI-PREG study; however, the data were not validated in an independent cohort. Canouil et al (21) used the same array to assess maternal methylation in a group of the FinnGeDi prospective multicenter cohort, but failed to find any significant differentially methylated positions associated with GDM (after false discovery rate adjustment) in an epigenome-wide association study. In our exploratory study, we identified 7 putative genes associated with both hyper- and hypomethylated sites, and we validated these in a larger population, finding the same trend in 2 CpG sites: Cg03554962 (LINC00917) and Cg05090351 (CTBP2). Of note, some of the studied genes have been linked to alterations of metabolic homeostasis (40-43).
We found a hypermethylated site related to LINC00917, a long noncoding RNA, in the GDM group. While these types of molecules lack a complete functional reading frame and rarely encode a functional protein, recent studies have associated some long noncoding RNAs with human disease. Of note, LINC00917 has been associated with obesity in children (40), a lipid-lowering statin response (41), and diabetic retinopathy (44), suggesting a relationship with metabolic control. Our findings during pregnancy and at 4 years postpartum support this hypothesis.
Our results in GDM also show differential methylation around genes related to the Wnt pathway (LEF1 and CTBP2), which is involved in a broad range of physiological processes. Indeed, Wnt derangement is associated with type 2 diabetes and metabolic syndrome (45-47). CtBP2 belongs to the CtBP family of transcriptional corepressors (48, 49). CtBP2 is essential in the regulation of stem cell differentiation (42) and adipogenesis (43), and its inhibition promotes adipogenesis. CtBP2 also seems to protect against insulin resistance induced by lipids. In mice, differential methylation of ctbp2 has been observed in the pancreas of offspring of GDM mothers with a downregulation of gene expression (50), which is consistent with previous data showing a downregulation of CTBP2 in the offspring of diabetic mothers (51). LEF1 is a member of the T-cell Factor/LEF1 family of high-mobility group transcription factors and is a downstream mediator of the Wnt/β-catenin signaling pathway. LEF1 is essential for stem cell maintenance and organ development, but has also been related to type 1 diabetes (52) and adipogenesis (53).
We observed that CpGs annotated to the CTBP2 promoter were hypermethylated in women with GDM, while those annotated to the body area of LEF1 were hypomethylated. Different methylation patterns are known to influence gene transcription differentially. The methylation patterns found in the discovery cohort could act synergistically to downregulate Wnt signaling. Unfortunately, these changes were not confirmed in the replication cohort, and LEF1-related CpG positions were hypermethylated in women with GDM.
Other methylation patterns observed in the discovery cohort were not confirmed in the replication cohort. One of the top hypermethylated genes was RASA3, which was also identified in a GDM study by Dias et al (38). RASA3 is a member of the family of GTPase-activating proteins that functions as a negative regulator of Ras signaling and insulin receptor signaling. RASA3 is localized on chromosome 13, close to insulin receptor substrate 2 (IRS2), and deletions in this chromosome have been associated with diabetes (54). RASA3 methylation is also implicated in diabetic embryopathy (55). Unfortunately, we failed to confirm the changes to RASA3 methylation in the replication cohort. Similarly, the CpG sites related to MRPS28 and TRAPPC9 could not be validated.
Two markers (LINC00917 and TRAPPC9) in the replication cohort were uncovered as potentially useful indicators of early glucose metabolism alterations in GDM 4 years after delivery, with the caveat that findings of epigenetic markers do not imply causality related to the outcome. It should be noted that the addition of a specific DNAm signature link to a specific LINC00917 position or the DNAm profiling, increased the predictive capacity of the clinical model. Although, this increase was not statistically significant, probably in part due to the limited sample size, however, we believe that it is a good point to be considered in future studies of new cohorts. LINC00917 was hypermethylated both in the discovery and replication cohorts, but this could not be confirmed for TRAPPC9. TRAPPC9 encodes Trafficking protein particle complex 9, which functions as an activator of nuclear factor kappa B, and mutations in this gene are associated with cognitive disability (56). In animal models of GDM, hypermethylation of this gene has been associated with its upregulation in pancreas cells of offspring mice (50), but no clear relationship with type 2 diabetes mellitus has been described.
Our study has several limitations. The number of women who developed type 2 diabetes was low. Also, the replication cohort was not matched for age and BMI, and to avoid this limitation we adjusted for these confounding factors. Lastly, we performed methylation analysis in whole blood, without considering the cell type and count, assuming a similar cell type distribution. Nevertheless, our study has several strengths, including a relatively large sample size, information on long-term follow-up of the women with GDM, and replication in an independent cohort of pregnant women. Also, the data on the metabolic status of women with GDM at 4 years postpartum has allowed us to evaluate the predictive capacity of DNAm in these patients. Finally, although we were unable to validate all the CpG sites identified in the microarray-based analysis, we confirmed hypermethylation of a new unique gene related site, LINC00917, and a trend in a second site related to CTBP2.
We provide evidence of new differential methylated sites related to GDM in peripheral blood of pregnant women, some of them with potential pathophysiological significance in the development of GDM and abnormal glucose tolerance. However, we could not establish a unique biomarker to identify women with GDM and/or at risk of development of abnormal glucose tolerance postpartum. While future studies will be needed to determine the true value of DNAm in the pathophysiology of GDM and in the prediction of diabetes or other metabolic alterations in the long term, our study is encouraging in terms of its applicability, especially to study the impact of GDM on future health.
Acknowledgments
We acknowledge the patients and volunteers involved in this study for their collaboration. We also acknowledge the BioBank IISPV (PT17/0015/0029) integrated into the Spanish National Biobanks Network. We also wish to highlight the essential contribution of the midwives and the Gynecology and Obstetrics Department of the Hospital Universitari Joan XXIII from Tarragona.
Glossary
Abbreviations
- AUC
area under the curve
- BMI
body mass index
- CpG
cytosine phosphate–guanine
- DNAm
DNA methylation
- GDM
gestational diabetes
- GWG
gestational weight gain
- IFCC
International Federation of Clinical Chemistry
- IQR
interquartile range
- OGTT
oral glucose tolerance test
- ROC
receiver operating characteristic
- TSS
transcription start site
Contributor Information
Mónica Ballesteros, Department of Medicine and Surgery, Rovira i Virgili University, Tarragona, Spain; Department of Obstetrics and Gynecology. University Hospital of Tarragona Joan XXIII, Institut d’Investigació Sanitària Pere Virgili (IISPV), Tarragona, Spain; CIBER de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM)-Instituto de Salud Carlos III, Madrid, Spain.
Pilar Gil-Lluís, Department of Endocrinology and Nutrition, University Hospital of Tortosa Verge de la Cinta, Tarragona, Spain.
Miriam Ejarque, CIBER de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM)-Instituto de Salud Carlos III, Madrid, Spain; Department of Endocrinology and Nutrition. Research Unit. University Hospital of Tarragona Joan XXIII-Institut d´Investigació Sanitària Pere Virgili (IISPV), Tarragona, Spain.
Cristina Diaz-Perdigones, CIBER de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM)-Instituto de Salud Carlos III, Madrid, Spain; Department of Endocrinology and Nutrition. Research Unit. University Hospital of Tarragona Joan XXIII-Institut d´Investigació Sanitària Pere Virgili (IISPV), Tarragona, Spain.
Laia Martinez-Guasch, Department of Medicine and Surgery, Rovira i Virgili University, Tarragona, Spain; CIBER de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM)-Instituto de Salud Carlos III, Madrid, Spain; Department of Endocrinology and Nutrition. Research Unit. University Hospital of Tarragona Joan XXIII-Institut d´Investigació Sanitària Pere Virgili (IISPV), Tarragona, Spain.
Sonia Fernández-Veledo, CIBER de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM)-Instituto de Salud Carlos III, Madrid, Spain; Department of Endocrinology and Nutrition. Research Unit. University Hospital of Tarragona Joan XXIII-Institut d´Investigació Sanitària Pere Virgili (IISPV), Tarragona, Spain.
Joan Vendrell, Department of Medicine and Surgery, Rovira i Virgili University, Tarragona, Spain; CIBER de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM)-Instituto de Salud Carlos III, Madrid, Spain; Department of Endocrinology and Nutrition. Research Unit. University Hospital of Tarragona Joan XXIII-Institut d´Investigació Sanitària Pere Virgili (IISPV), Tarragona, Spain.
Ana Megía, Department of Medicine and Surgery, Rovira i Virgili University, Tarragona, Spain; CIBER de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM)-Instituto de Salud Carlos III, Madrid, Spain; Department of Endocrinology and Nutrition. Research Unit. University Hospital of Tarragona Joan XXIII-Institut d´Investigació Sanitària Pere Virgili (IISPV), Tarragona, Spain.
Funding
This study was supported by grants from the Associació Catalana de Diabetis (ajuts recerca clinica to P.G.-L.) and from the Spanish Ministry of Economy and Competitiveness (PI 12/00717; PI 15/01562, and PI 18/00516 to A.M., RTI2018-093919-B-I00 to S.F.-V., PI14/00228 and PI17/01503 to J.V.) cofinanced by the European Regional Development Fund (ERDF) “A way to make Europe/Investing in your future.” The Spanish Biomedical Research Center in Diabetes and Associated Metabolic Disorders (CIBERDEM) (CB07708/0012) is an initiative of the Instituto de Salud Carlos III. S.F.V. acknowledges the Miguel Servet tenure-track program (CP10/00438 and CPII16/00008) from the Fondo de Investigación Sanitaria, cofinanced by the ERDF.
Disclosures
The authors report no conflict of interest.
Ethics Approval and Consent to Participate
The study was conducted according to the guidelines of the Declaration of Helsinki. Ethical approval was obtained by the Institutional Review Board of the “Institut d’Investigacio Sanitaria Pere Virgili” (243/C/2016). Informed consent was obtained from all subjects involved in the study.
Consent for publication
Not applicable.
Data Availability
Some or all datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
References
- 1. Bellamy L, Casas JP, Hingorani AD, Williams D. Type 2 diabetes mellitus after gestational diabetes: a systematic review and meta-analysis. Lancet 2009;373(9677):1773-1779. [DOI] [PubMed] [Google Scholar]
- 2. Noctor E, Crowe C, Carmody LA, et al. ATLANTIC-DIP: prevalence of metabolic syndrome and insulin resistance in women with previous gestational diabetes mellitus by International Association of Diabetes in Pregnancy Study Groups criteria. Acta Diabetol. 2015;52(1):153-160. Doi: 10.1007/s00592-014-0621-z [DOI] [PubMed] [Google Scholar]
- 3. Puhkala J, Raitanen J, Kolu P, Tuominen P, Husu P, Luoto R. Metabolic syndrome in Finnish women 7 years after a gestational diabetes prevention trial. BMJ Open 2017;7(3):e014565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu Y, Shen S, Sun L, Yang H, Jin B, Cao X. Metabolic syndrome risk after gestational diabetes: a systematic review and meta-analysis. PLoS One. 2014;9(1):e87863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carr DB, Utzschneider KM, Hull RL, et al. Gestational diabetes mellitus increases the risk of cardiovascular disease in women with a family history of type 2 diabetes. Diabetes Care. 2006;29(9):2078-2083. [DOI] [PubMed] [Google Scholar]
- 6. Michalczyk AA, Dunbar JA, Janus ED, et al. Epigenetic markers to predict conversion from gestational diabetes to type 2 diabetes. J Clin Endocrinol Metab. 2016;101(6):2396-2404. [DOI] [PubMed] [Google Scholar]
- 7. Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6-21. [DOI] [PubMed] [Google Scholar]
- 8. Hernando-Herraez I, Garcia-Perez R, Sharp AJ, Marques-Bonet T. DNA methylation: insights into human evolution. PLoS Genet. 2015;11(12):e1005661e1005661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gillberg L, Ling C. The potential use of DNA methylation biomarkers to identify risk and progression of type 2 diabetes. Front Endocrinol (Lausanne) 2015;6:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Elliott HR, Sharp GC, Relton CL, Lawlor DA. Epigenetics and gestational diabetes: a review of epigenetic epidemiology studies and their use to explore epigenetic mediation and improve prediction. Diabetologia. 2019;62(12):2171-2178. Doi: 10.1007/s00125-019-05011-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Willmer T, Johnson R, Louw J, Pheiffer C. Blood-based DNA methylation biomarkers for type 2 diabetes: potential for clinical applications. Front Endocrinol (Lausanne) 2018;9:744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13(10):679-692. [DOI] [PubMed] [Google Scholar]
- 13. Bacos K, Gillberg L, Volkov P, et al. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat Commun. 2016;7:11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Mello VDF, Pulkkinen L, Lalli M, Kolehmainen M, Pihlajamäki J, Uusitupa M. DNA methylation in obesity and type 2 diabetes. Ann Med. 2014;46(3):103-113. [DOI] [PubMed] [Google Scholar]
- 15. Pirola L, Balcerczyk A, Okabe J, El-Osta A. Epigenetic phenomena linked to diabetic complications. Nat Rev Endocrinol. 2010;6(12):665-675. [DOI] [PubMed] [Google Scholar]
- 16. Kang J, Lee C-N, Li H-Y, Hsu K-H, Lin S-Y. Genome-wide DNA methylation variation in maternal and cord blood of gestational diabetes population. Diabetes Res Clin Pract. 2017;132:127-136. [DOI] [PubMed] [Google Scholar]
- 17. Kang J, Lee C-N, Li H-Y, Hsu K-H, Wang S-H, Lin S-Y. Association of interleukin-10 methylation levels with gestational diabetes in a Taiwanese population. Front Genet. 2018;9:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu P, Farrell WE, Haworth KE, et al. Maternal genome-wide DNA methylation profiling in gestational diabetes shows distinctive disease-associated changes relative to matched healthy pregnancies. Epigenetics 2018;13(2):122-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Enquobahrie DA, Moore A, Muhie S, Tadesse MG, Lin S, Williams MA. Early pregnancy maternal blood DNA methylation in repeat pregnancies and change in gestational diabetes mellitus status—a pilot study. Reprod Sci. 2015;22(7):904-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dias S, Adam S, Van Wyk N, Rheeder P, Louw J, Pheiffer C. Global DNA methylation profiling in peripheral blood cells of South African women with gestational diabetes mellitus. Biomarkers 2019;24(3):225-231. [DOI] [PubMed] [Google Scholar]
- 21. Canouil M, Khamis A, Keikkala E, et al. Epigenome-wide association study reveals methylation loci associated with offspring gestational diabetes mellitus exposure and maternal methylome. Diabetes Care. 2021;44(9):1992-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fragoso-Bargas N, Opsahl JO, Kiryushchenko N, et al. Cohort profile: Epigenetics in Pregnancy (EPIPREG) – population-based sample of European and South Asian pregnant women with epigenome-wide DNA methylation (850k) in peripheral blood leukocytes. PLoS One. 2021;16(8):e0256158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grupo Español de Diabetes y Embarazo (GEDE), Grupo Españñol de Diabetes y Embarazo. Asistencia a la gestante con diabetes. Guía de práctica clínica actualizada en 2014. Av en Diabetol. 2015;31(2):45-59. [Google Scholar]
- 24. Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15(7):539-53. [DOI] [PubMed] [Google Scholar]
- 25. American Diabetes Association. . Standards of medical Care in diabetes. Diabetes Care. 2013;36(Suppl 1):S11-S66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoelzel W, Weykamp C, Jeppsson J-O, et al. IFCC reference system for measurement of hemoglobin A1c in human blood and the national standardization schemes in the United States, Japan, and Sweden: a method-comparison study. Clin Chem. 2004;50(1):166-174. [DOI] [PubMed] [Google Scholar]
- 27. Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014;30(10):1363-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perez-Gil D, Lendinez-Tortajada V, Sanchis-Juan A, et al. DPMAS: pipeline for 450K top table replication with MiSeq amplicons. Paper presented at: 4th Annual Infinium Human Methylation 450 Array Analysis Workshop; May 21, 2015; London, UK. Accessed June 24, 2019: https://figshare.com/articles/DPMAS_pipeline_for_450K_top_table_replication_with_MiSeq_amplicons/1418262 [Google Scholar]
- 29. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362-D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fabregat A, Jupe S, Matthews L, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018;46(D1):D649-D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maksimovic J, Gordon L, Oshlack A. SWAN: subset-quantile within array normalization for Illumina Infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13(6):R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ballesteros M, Gil-Lluis P, Ejarque M, et al. DNA methylation in gestational diabetes and its predictive value for postpartum glucose disturbances. Cora.RDR 2022. Deposited 24 May 2022. 10.34810/data191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matsha TE, Pheiffer C, Humphries SE, Gamieldien J, Erasmus RT, Kengne AP. Genome-wide DNA methylation in mixed ancestry individuals with diabetes and prediabetes from South Africa. Int J Endocrinol 2016;2016:3172093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pinzón-Cortés JA, Perna-Chaux A, Rojas-Villamizar NS, et al. Effect of diabetes status and hyperglycemia on global DNA methylation and hydroxymethylation. Endocr Connect 2017;6(8): 708-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nomura Y, Lambertini L, Rialdi A, et al. Global methylation in the placenta and umbilical cord blood from pregnancies with maternal gestational diabetes, preeclampsia, and obesity. Reprod Sci. 2014;21(1):131-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reichetzeder C, Dwi Putra SE, Pfab T, et al. Increased global placental DNA methylation levels are associated with gestational diabetes. Clin Epigenetics 2016;8:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dias S, Adam S, Rheeder P, Louw J, Phei C. Altered genome-wide DNA methylation in peripheral blood of South African women with gestational diabetes mellitus. Int J Mol Sci . 2019;20:5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Steyn A, Crowther NJ, Norris SA, Rabionet R, Estivill X, Ramsay M. Epigenetic modification of the pentose phosphate pathway and the IGF-axis in women with gestational diabetes mellitus. Epigenomics 2019;11(12):1371-1385. [DOI] [PubMed] [Google Scholar]
- 40. Comuzzie AG, Cole SA, Laston SL, et al. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One. 2012;7(12):e51954e51954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barber MJ, Mangravite LM, Hyde CL, et al. Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS One. 2010;5(3):e9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim TW, Kwak S, Shin J, et al. Ctbp2-mediated β-catenin regulation is required for exit from pluripotency. Exp Mol Med. 2017;49(10):e385-e385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang L, Xu L, Xu M, et al. Obesity-associated MiR-342-3p promotes adipogenesis of mesenchymal stem cells by suppressing CtBP2 and releasing C/EBPα from CtBP2 binding. Cell Physiol Biochem. 2015;35(6):2285-2298. [DOI] [PubMed] [Google Scholar]
- 44. Grassi MA, Tikhomirov A, Ramalingam S, Below JE, Cox NJ, Nicolae DL. Genome-wide meta-analysis for severe diabetic retinopathy. Hum Mol Genet. 2011;20(12):2472-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jin T. The WNT signalling pathway and diabetes mellitus. Diabetologia. 2008;51(10):1771-1780 [DOI] [PubMed] [Google Scholar]
- 46. Abou Ziki MD, Mani A. The interplay of canonical and noncanonical Wnt signaling in metabolic syndrome. Nutr Res. 2019;70:18-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arnold AC, Robertson D. Defective Wnt signaling: a potential contributor to cardiometabolic disease? Diabetes. 2015;64(10):3342-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell. 2002;9(2):213-224. [DOI] [PubMed] [Google Scholar]
- 49. Zhao L-J, Subramanian T, Vijayalingam S, Chinnadurai G. CtBP2 proteome: Role of CtBP in E2F7-mediated repression and cell proliferation. Genes Cancer 2014;5(1-2):31-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhuangli Z, Xiongfeng C, Yiqing X, et al. Gestational diabetes mellitus alters DNA methylation profiles in pancreas of the offspring mice. J Diabetes Complications. 2019;33(1): 15-22. [DOI] [PubMed] [Google Scholar]
- 51. Pavlinkova G, Salbaum JM, Kappen C. Maternal diabetes alters transcriptional programs in the developing embryo. BMC Genomics. 2009;10(1):274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Anjos SM, Tessier M-C, Polychronakos C. Association of the cytotoxic T lymphocyte-associated antigen 4 gene with type 1 diabetes: evidence for independent effects of two polymorphisms on the same haplotype block. J Clin Endocrinol Metab. 2004;89(12): 6257-6265. [DOI] [PubMed] [Google Scholar]
- 53. Yang X, Jansson P-A, Nagaev I, et al. Evidence of impaired adipogenesis in insulin resistance. Biochem Biophys Res Commun. 2004;317(4):1045-1051. [DOI] [PubMed] [Google Scholar]
- 54. Babaya N, Noso S, Hiromine Y, et al. Early-onset diabetes mellitus in a patient with a chromosome 13q34qter microdeletion including IRS2. J Endocr Soc. 2018;2(10):1207-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schulze KV, Bhatt A, Azamian MS, et al. Aberrant DNA methylation as a diagnostic biomarker of diabetic embryopathy. Genet Med. 2019;21(11):2453-2461. [DOI] [PubMed] [Google Scholar]
- 56. Marangi G, Leuzzi V, Manti F, et al. TRAPPC9-related autosomal recessive intellectual disability: report of a new mutation and clinical phenotype. Eur J Hum Genet. 2013;21(2):229-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.