Abstract
Despite the broad array of roles for epigenetic mechanisms on regulating diverse processes in eukaryotes, no experimental system is currently available in plants for the direct assessment of histone function. In this work, we present the development of a genetic strategy in Arabidopsis (Arabidopsis thaliana) whereby modified histone H4 transgenes can completely replace the expression of endogenous histone H4 genes. Accordingly, we established a collection of plants expressing different H4 point mutants targeting residues that may be post-translationally modified in vivo. To demonstrate its utility, we screened this new H4 mutant collection to uncover substitutions in H4 that alter flowering time. We identified different mutations in the H4 tail (H4R17A) and the H4 globular domain (H4R36A, H4R39K, H4R39A, and H4K44A) that strongly accelerate the floral transition. Furthermore, we identified a conserved regulatory relationship between H4R17 and the ISWI chromatin remodeling complex in plants: As with other biological systems, H4R17 regulates nucleosome spacing via ISWI. Overall, this work provides a large set of H4 mutants to the plant epigenetics community that can be used to systematically assess histone H4 function in Arabidopsis and a roadmap to replicate this strategy for studying other histone proteins in plants.
A systematic histone H4 replacement system reveals that H4R17 regulates flowering time by mediating ISWI-dependent nucleosome spacing in plants.
In a Nutshell.
Introduction
In eukaryotic cells, genomic DNA is organized into chromatin, whose basic unit is the nucleosome, which consists of 147 bp of DNA wrapped around one histone octamer made up of two copies of the histone proteins H2A, H2B, H3, and H4 (Luger et al., 1997). Histones play a significant role in regulatory processes operating at the chromatin level, such as transcription and DNA replication, and can therefore exert widespread effects on organismal growth, development, and fitness (Kouzarides, 2007). One mechanism by which histones contribute to these processes is through the post-translational modifications (PTMs) of histone residues. Traditionally, the functional significance of histone PTMs has primarily been deduced through the analysis of phenotypes resulting from the mutation of histone-modifying enzymes or histone-reading proteins. However, while this method has been successful at identifying functions for many histone PTMs, there are several limitations to this approach. For example, this strategy presents difficulties if there are many redundant proteins writing or reading the same PTM, or if the writers or readers of a specific histone PTM have not been identified. Moreover, histone-modifying enzymes often target non-histone substrates in addition to histones, complicating the analysis of mutant phenotypes (Glozak et al., 2005).
To circumvent these obstacles, one effective strategy to study the functions of histone PTMs is to mutate the acceptor histone residue to a nonmodifiable residue and then assess the resulting phenotype(s). One inherent advantage of this strategy is that it can be applied to investigate the roles of modifiable and nonmodifiable residues of histones. In addition, this histone replacement strategy can be used in biological backgrounds expressing wild-type histone genes, or in backgrounds where expression of endogenous histone genes is partially or completely eliminated. A major advantage of removing endogenous histones in this strategy is that it increases the likelihood of detecting phenotypes associated with the expression of genes encoding histone mutants, which may otherwise be masked if competing wild-type histones are also present. Systematic mutagenesis experiments with core histones were initially conducted in budding yeast (Saccharomyces cerevisiae) and revealed many new insights into histone function (Dai et al., 2008; Nakanishi et al., 2008; Govin et al., 2010; Fu et al., 2021). These experiments utilized histone shuffle systems to either provide an episomal plasmid expressing histone mutant genes in a background from which the endogenous histone genes had been deleted, or to directly mutate the endogenous histone gene copies using homologous recombination. The most recent system developed in yeast utilized an efficient clustered regularly interspaced short palindromic repeats (CRISPR)-associated nuclease 9 (Cas9)-based histone shuffle strategy that allows for the rapid development of multiplex histone mutations (Fu et al., 2021). In multicellular eukaryotes, Drosophila (Drosophila melanogaster) was the first organism in which systematic histone mutagenesis was performed, by using site-specific transgenesis to replace the endogenous histone coding region with that of a modified histone gene or histone array (Hodl and Basler, 2009, 2012; Gunesdogan et al., 2010; McKay et al., 2015). Additionally, high-throughput screens of histones H3 and H4 were recently conducted in Drosophila using a CRISPR/Cas9-mediated knock-in technology for histone gene replacement at the endogenous histone locus (Zhang et al., 2019).
In contrast to the aforementioned biological model systems, plant systems present additional challenges to implementing complete histone gene replacement. While all replication-dependent histone genes are clustered at a single genomic locus in Drosophila (Lifton et al., 1978), and yeast contains only two copies of each core histone gene at the haploid cell stage (Fu et al., 2021), there are 47 genes, dispersed throughout the genome, that encode H2A, H2B, H3, and H4 in Arabidopsis (Arabidopsis thaliana) (Tenea et al., 2009). Because the histone genes are not clustered together in plants, the establishment of complex histone deletion mutants necessary for partially or completely replacing endogenous histone genes with modified histone genes is more challenging. Histone replacement was however recently reported in Arabidopsis, using a combination of the traditional crossing of histone mutants and artificial microRNAs to generate backgrounds largely depleted of wild-type histone H3.1 (Jiang et al., 2017). Notably, this strategy is relatively time-consuming and may not completely eliminate endogenous histones. While the earliest strategies used to implement histone gene replacement in both yeast and Drosophila were not applicable to plants due to their reliance on either the plasmid shuffle strategy and/or site-specific recombination systems, some aspects of the newest histone replacement strategies in other systems should facilitate the establishment of complete histone gene replacement in plants. For example, recent advancements in the deployment of multiplex CRISPR/Cas9-based technologies in plants make it possible to create mutations in large gene families like those of histones.
While histones contribute to diverse processes in plants, a system enabling histone gene replacement would allow plant researchers to further elucidate the biological roles of histones in a more high-throughput manner. One of the most important developmental decisions during the plant life cycle is the transition from vegetative growth to reproductive development (Andres and Coupland, 2012; Song et al., 2015). Thus, the ways in which epigenetic mechanisms regulate the floral transition is a major area of research that could benefit from the application of histone replacement strategies. Diverse histone PTMs, including histone H3 lysine 4 (H3K4) methylation, H3K36 di- and tri-methylation, H3K9 methylation, H3K27 methylation, and H3 acetylation have been shown to regulate the expression of key flowering time regulatory genes such as FLOWERING LOCUS C (FLC) and FLOWERING LOCUS T (FT) (Bastow et al., 2004; He et al., 2004; Kim et al., 2005; Deng et al., 2007; Jiang et al., 2008; Pien et al., 2008; Xu et al., 2008; He, 2009; Yu et al., 2011; Bu et al., 2014; Crevillen et al., 2014, 2019; Cui et al., 2016; Pajoro et al., 2017; Ning et al., 2019; Zheng et al., 2019). However, compared with H3, the role of H4 in regulating the floral transition has been characterized to a much lesser extent.
Here, we present the establishment of a CRISPR-based histone mutagenesis platform in the plant model system Arabidopsis that allows for complete histone replacement. As a proof-of-concept, we targeted histone H4, which is encoded by the largest number of endogenous genes (eight genes) among functionally distinct plant histone proteins (Okada et al., 2005; Wierzbicki and Jerzmanowski, 2005; Tenea et al., 2009), for a systematic assessment of the roles of modifiable residues on this protein. After in vivo validation of our histone replacement strategy, we generated a large population of H4 point mutants to study the role(s) of 38 histone H4 residues. Using this H4 population, we identified a role for H4R17 in the regulation of flowering time. Furthermore, we demonstrated the functional relationship between H4R17 and an imitation switch (ISWI) chromatin-remodeling complex. Overall, this study demonstrates the utility of implementing histone replacement strategies in plants and provides a new resource that the plant community can use to probe for H4 functions in various aspects of plant growth and development.
Results
Generation of an Arabidopsis mutant expressing a single histone H4 gene
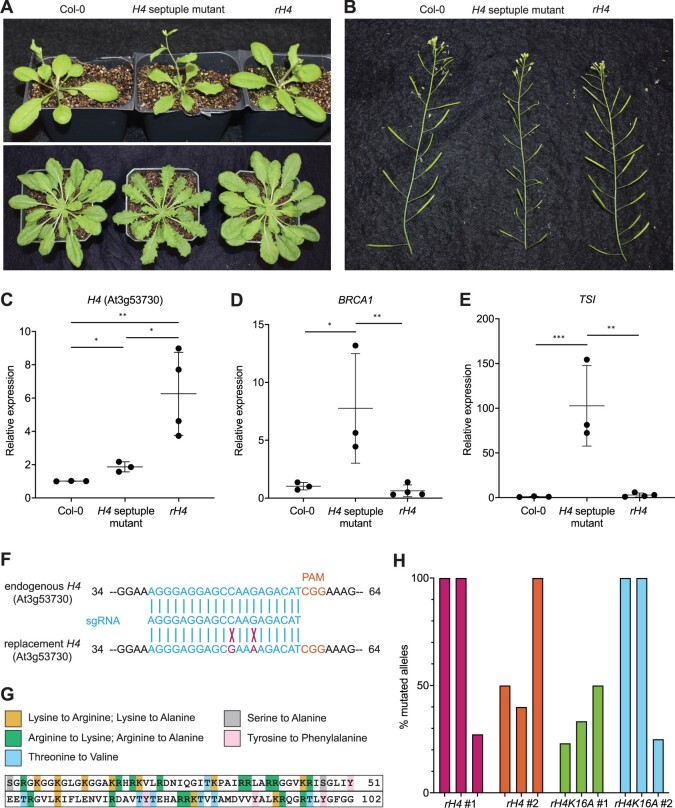
To create a library of Arabidopsis plants with replacement of endogenous histone H4 with H4 point mutants, we first generated a histone H4-depleted background using multiplex CRISPR/Cas9. The eight histone H4 genes in Arabidopsis (Columbia-0 [Col-0] accession) all code for the same histone H4 protein, which is 98% identical to human histone H4 (100/102 identical amino acids [aa]; with conservative substitutions at aa 60 and 77) (Supplemental Figure S1A). We designed three single-guide RNAs (sgRNAs) that can target Cas9 to seven of the eight endogenous H4 genes (Supplemental Figure S1B). We then transformed Col-0 plants via Agrobacterium (Agrobacterium tumefaciens) using a multiplex Cas9/sgRNA construct containing the three sgRNAs against the H4 genes, selected primary transformants (T1), and exposed these T1 plants to repeated heat stress treatments at 37°C for 30 h to increase the efficiency of targeted mutagenesis by Cas9 (LeBlanc et al., 2017). We assessed CRISPR/Cas9 activity at all seven H4 genes via PCR and sequencing in T1 plants, leading to the identification in the T2 generation of one plant with homozygous loss-of-function mutations in all seven targeted H4 genes (hereafter referred to as the H4 septuple mutant) (Supplemental Figure S1B). Morphological and molecular characterization of the H4 septuple mutant plants showed that they are slightly smaller than wild-type Col-0 plants and display a serrated leaf phenotype (Figure 1A). In addition, fertility was much lower in the H4 septuple mutant compared with Col-0 plants (Figure 1B). We determined that the transcript levels of the remaining endogenous H4 gene (At3g53730) is upregulated approximately two-fold in the H4 septuple mutant relative to Col-0, likely to compensate for H4 depletion due to the loss of function mutations in the other seven H4 genes (Figure 1C). The H4 septuple mutant exhibited misregulation of markers of genomic and epigenomic instability, including upregulation of the DNA damage response gene BREAST CANCER SUSCEPTIBILITY1 (BRCA1) and transcriptional derepression of the heterochromatic DNA repeat TRANSCRIPTIONNALLY SILENT INFORMATION (TSI) (Figure 1, D and E). Additionally, we observed that the H4 septuple mutant displays similar morphological phenotypes (small serrated leaves, abnormal silique phyllotaxy, and partial sterility) and a high overlap of differentially expressed genes (DEGs) with a mutant (fas1-4 in FASCIATA1) lacking the histone chaperone chromatin assembly factor-1 (CAF-1) that loads histones H3.1/H4 during replication (e.g. 62% of downregulated DEGs and 55% of upregulated DEGs in the H4 septuple mutant are shared with fas1-4) (Supplemental Figure S2). These findings strongly suggest that reduced H4 levels result in issues with replication due to an insufficient amount of histones for replication forks to proceed normally. Overall, our results demonstrate that multiplex CRISPR/Cas9 can be used to rapidly create an Arabidopsis mutant background containing a minimal number of functional genes coding for a specific histone.
Figure 1.
A CRISPR-based genetic system for expression of H4 point mutant constructs in Arabidopsis. A, Morphological phenotypes of Col-0, H4 septuple mutant, and rH4 plants grown in long-day conditions for 24 days (top) and short-day conditions for 7 weeks (bottom). B, Siliques of Col-0, H4 septuple mutant, and rH4 plants grown in long-day conditions for 4 weeks. C–E, RT-qPCR analysis of (C) H4 (At3g53730), (D) BRCA1, and (E) TSI in Col-0, the H4 septuple mutant, and four independent rH4 T1 lines. Three biological replicates were included for Col-0 and H4 septuple mutant plants. Horizontal bars indicate the mean. Error bars show standard deviation. *P < 0.05; **P < 0.005; ***P < 0.0005, as determined by unpaired Student’s t test. F, Design of the sgRNA targeting the remaining endogenous H4 gene (At3g53730) in the H4 septuple mutant. Mismatches of the replacement H4 gene with the sgRNA shown with an X. G, Schematic of point mutations in the H4 replacement plasmid library. H, Percentage of mutated alleles in six rH4 plants and six rH4K16A plants. Each plant assessed was from the T2 generation; three plants from the same T1 parent were used in this experiment (i.e. two independent T2 lines per genotype).
Establishment of a histone H4 replacement system in Arabidopsis
To set up a complete histone H4 replacement in plants, we designed an H4 replacement plasmid that contains (1) a sgRNA targeting the last remaining endogenous H4 gene (At3g53730) and (2) a Cas9-resistant H4 gene allowing for expression of At3g53730 under its native promoter (i.e. H4 replacement gene). Our strategy was to transform the H4 septuple mutant, which already expresses Cas9, with the H4 replacement plasmid and select T1 plants that contain mutations at the endogenous At3g53730 gene. To prevent Cas9 from targeting the replacement H4 gene, we introduced two silent mutations in the transgene that prevent recognition from the sgRNA targeting the endogenous At3g53730 gene (Figure 1F). After transformation of the H4 septuple mutant with the H4 replacement plasmid, we recovered many T1 transformants expressing the replacement H4 gene (hereafter referred to as rH4 plants); in contrast to the H4 septuple mutant, all rH4 plants were normal in size, did not exhibit serrated leaves, and showed normal fertility (Figure 1, A and B). Moreover, the relative transcript levels of BRCA1 and TSI in rH4 plants were comparable to those of Col-0 (Figure 1, D and E). The expression of At3g53730 in first-generation rH4 plants was upregulated approximately four- to nine-fold relative to Col-0 (Figure 1C). These results indicate that high expression levels of the replacement H4 gene in rH4 plants are responsible for suppressing the morphological phenotypes of the H4 septuple mutant.
We then used site-directed mutagenesis to create a large library of H4 replacement plasmids carrying different point mutations in the H4 replacement gene. We generated mutations covering every amino acid (i.e. lysine, arginine, threonine, serine, and tyrosine) in the encoded H4 that could theoretically be post-translationally modified in vivo. To this end, we mutated each modifiable amino acid to a residue that cannot be post-translationally modified (i.e. alanine, valine, or phenylalanine). We also changed lysine and arginine residues to residues with similar biochemical properties (i.e. arginine and lysine, respectively). In total, we modified 38 amino acid residues of H4 to generate 63 different H4 replacement genes containing a specific point mutation (Figure 1G). We subcloned these H4 mutant genes into the H4 replacement plasmid and individually transformed them into the H4 septuple mutant. We selected two independent transgenic lines for each H4 mutant, except for plants expressing the H4 replacement genes H4R40A, H4R45A, H4K59A, H4R78A, H4K79R, and H4R92K due to lethality induced by these mutations (Supplemental Figure S3A). Again, we exposed all T1 plants to repeated heat stress treatments to maximize the efficiency of targeted mutagenesis of the remaining endogenous H4 gene by Cas9. To estimate the frequency of mutations at the remaining endogenous H4 gene in the plants expressing each H4 replacement gene, we genotyped three plants each from two independent rH4 lines and two independent rH4K16A lines at the T2 generation stage. We amplified the remaining endogenous H4 gene (At3g53730) from these T2 plants, cloned the resulting PCR products and sequenced at least 10 individual clones corresponding to each plant, and calculated the percentage of mutated alleles. Approximately half of the plants were characterized by a complete elimination of the wild-type At3g53730 allele, while the other plants varied from 50% to 75% wild-type alleles remaining (Figure 1H). Taking into account that expression of the H4 replacement gene was either equivalent or much higher compared with the remaining endogenous H4 gene (Figure 1C), these results suggest that the chromatin of most T2 plants in our H4 replacement collection contains large amounts of H4 point mutants. Overall, these results show that our CRISPR/Cas9 strategy was successful in creating a large collection of Arabidopsis plants expressing different H4 point mutants replacing wild-type H4 proteins.
Differential regulation of flowering time in plants expressing histone H4 mutants
To demonstrate the utility of the H4 replacement collection in identifying pathways regulated by H4 in Arabidopsis, we initiated a screen of the plants expressing H4 mutants for defects in flowering time. The transition between vegetative and reproductive development is sensitive to various chromatin disruptions in Arabidopsis, but most of the findings in this field have focused on the roles of PTMs on histone H3 (He and Amasino, 2005; He, 2009; Srikanth and Schmid, 2011; Yaish et al., 2011; Berry and Dean, 2015).
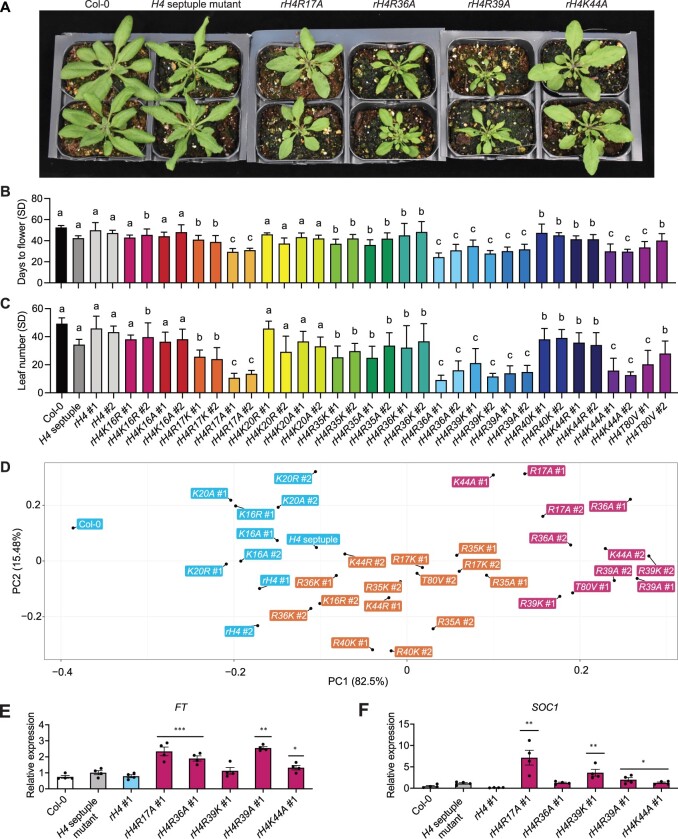
We grew our collection of H4 mutants using plants from two independent T2 lines for each H4 mutant and measured flowering time (as days to flowering and leaf number) when grown under both short-day conditions (8-h light, 16-h dark) and long-day conditions (16-h light, 8-h dark). We observed many morphological and developmental phenotypes at the vegetative stage of growth in T2 plants expressing the different H4 mutants (Figure 2A and Supplemental Figure S4A), which demonstrates that our H4 replacement strategy can be used to reveal various developmental phenotypes associated with mutations on histone H4. In regard to flowering time, plants expressing H4 point mutants were not associated with a consistent and significant late flowering-time phenotype compared with rH4 plants (i.e. replacement with the intact H4 gene) for both transgenic lines corresponding to the same mutation in either long-day or short-day conditions. By contrast, we identified 16 rH4 mutants with early flowering phenotype using the same criteria (Figure 2, B and C, Supplemental Figures S4, B and C and S5). Many rH4 mutant lines exhibited early flowering in both long-day and short-day conditions, with the rH4R17A, rH4R36A, rH4R39K, rH4R39A, and rH4K44A mutants exhibiting the most consistent and drastic acceleration of flowering time (Supplemental Figure S3B). To reduce the dimensionality of the data, we performed a principal component analysis of the mean values for the four flowering time variables measured in our analyses: number of days in long-day conditions, leaf number in long-day conditions, number of days in short-day conditions, and leaf number in short-day conditions (Figure 2D). We performed k-means clustering on principle component 1 (PC1) and PC2, which together explained 98% of the standing variance (Supplemental Figure S4D) and revealed three clusters in the data. Cluster a, corresponding to a flowering response most similar to that of wild-type plants, contained Col-0, the H4 septuple mutant, rH4, rH4K16A, rH4K20R, and rH4K20A. Cluster b, corresponding to a moderately early flowering time phenotype, contained rH4R17K, rH4R35K, rH4R35A, rH4R36K, rH4R40K, and rH4K44R. Cluster c, corresponding to a drastically early flowering time phenotype, contained rH4R17A, rH4R36A, rH4R39K, rH4R39A, and rH4K44A. The two rH4K16R lines were split between Cluster a and Cluster b, and the two rH4T80V lines were split between Cluster b and Cluster c. While the rH4K16R, rH4K16A, rH4K20R, and rH4K20A mutants appeared slightly early flowering relative to rH4 plants (Figure 2, B and C and Supplemental Figure S4, B and C), all of these mutant lines, except for a single rH4K16R line, clustered within the wild-type cluster (Cluster a) through these analyses. Due to the differences in the flowering time responses of the two independent transgenic lines expressing the rH4K16R and rH4K20R mutants, we performed RT-qPCR on H4 (At3g53730) and established that the early flowering lines (rH4K16R #1 and rH4K20R #2) also display higher H4 expression (Supplemental Figure S4E), thus supporting the hypothesis that a higher expression of the mutant H4 transgene causes earlier flowering for these mutants. We also performed RT-qPCR analyses on the key genes FT and SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1) regulating flowering time in selected rH4 mutants and observed their upregulation, consistent with the early flowering behavior of rH4 mutants from Cluster c (Figure 2, E and F). Taken together, our histone H4 replacement system enables the assessment of expressing histone H4 mutants on flowering time regulation, thus demonstrating the usefulness of the system for probing histone H4 function in plants.
Figure 2.
Mutations in specific residues of histone H4 generate early flowering phenotypes in Arabidopsis. A, Rosette phenotype of Col-0, the H4 septuple mutant, rH4R17A, rH4R36A, rH4R39A, and rH4K44A mutant lines grown in long-day conditions for 3 weeks. For the rH4 plants, individual T2 plants (top and bottom) from independent T1 parents are shown. B and C, Mean days to flower (B) and rosette leaf number (C) at flowering in short-day (SD) conditions for Col-0, the H4 septuple mutant, and various H4 replacement backgrounds (two independent T2 transgenic lines each). Error bars show standard deviation (n ≥ 7). Lowercase letters indicate cluster identified by k-means clustering. D, Principal component analysis for flowering time data along the first two principal components PC1 and PC2. Variance explained by each principal component is indicated on the respective axis. The three clusters produced by k-means clustering are represented in blue (cluster a), orange (cluster b), and pink (cluster c). E and F, RT-qPCR analysis of FT (E) and SOC1 (F) in Col-0, the H4 septuple mutant, rH4 #1, rH4R17A #1, rH4R36A #1, rH4R39K #1, rH4R39A #1, and rH4K44A #1 plants. Error bars show standard deviation. *P < 0.05; **P < 0.005; ***P < 0.0005, as determined by unpaired Student’s t test. Bar colors represent cluster assignment from (D).
In vivo modulation of PRMT7 activity does not replicate the early flowering phenotype of rH4R17A plants
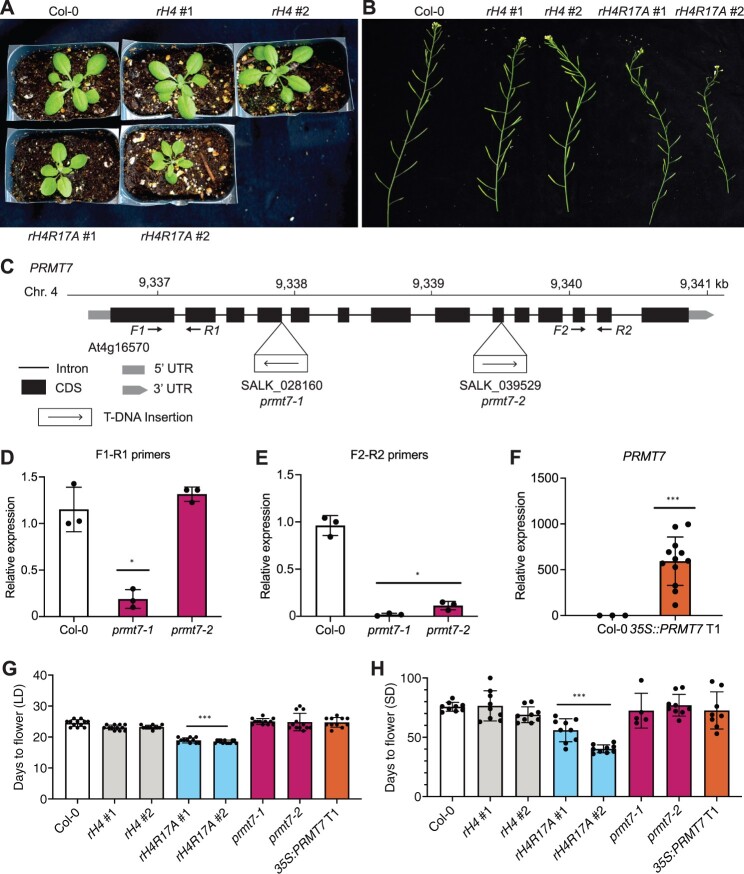
Of the five H4 mutations (H4R17A, H4R36A, H4R39K, H4R39A, and H4K44A) identified in our screen that cause the strongest effect on flowering time, only one (H4R17A) mapped to the N-terminal tail (aa 1-20) of H4, where most histone PTMs are made. Mutations in the unstructured N-terminal tail of H4 are less likely to affect flowering time by disrupting histone H4 folding and/or nucleosome structure than mutations in the histone-fold domain. Therefore, we focused our subsequent analyses on trying to elucidate the mechanism by which the H4R17A mutation affects the timing of the transition to reproductive development. For this work, we used H4 replacement plants for which there was a complete replacement of the endogenous histone H4 with the H4R17A mutant (Supplemental Figure S6A). In addition to a significantly earlier floral transition, we observed that the H4R17A mutation also causes other developmental phenotypes, including smaller, upwardly curled leaves, and reduced fertility compared with wild-type plants (Figure 3, A and B). When we introduced the H4R17A replacement construct in wild-type Col-0 instead of the H4 septuple mutant background, we noticed an attenuation of the effects of H4R17A on plant development when the normal H4 protein content of Arabidopsis competes with the H4 point mutant for insertion on chromatin (Supplemental Figure S6, B and C).
Figure 3.
PRMT7 does not regulate the floral transition in Arabidopsis. A, Rosette phenotype of Col-0, rH4 #1, rH4 #2, rH4R17A #1, and rH4R17A #2 plants grown in long-day conditions for 3 weeks. B, Silique phenotype of Col-0, rH4 #1, rH4 #2, rH4R17A #1, and rH4R17A #2 plants grown in long-day conditions for 4 weeks. C, Schematic diagram of the PRMT7 locus. The location of the T-DNA insertions and the primers (F1–R1 and F2–R2) used for gene expression analyses are shown. D–F, RT-qPCR showing relative PRMT7 transcript levels in Col-0, prmt7-1, and prmt7-2 plants (D and E), and Col-0 and 35S:PRMT7 T1 plants (F). The average of three biological replicates and standard deviation are shown for Col-0 and prmt7 mutants. For the 35S:PRMT7 plants, individual data points represent independent T1 plants. *P < 0.05; **P < 0.005; ***P < 0.0005, as determined by unpaired Student’s t test (sample versus Col-0). E and F, Mean days to flower in long-day (E) or short-day conditions (F) for Col-0, rH4 #1, rH4 #2, rH4R17A #1 and rH4R17A #2, prmt7-1, prmt7-2, and 35S:PRMT7 T1 plants. Error bars show standard deviation. *P < 0.05; **P < 0.005; ***P < 0.0005, as determined by one-way ANOVA with Tukey’s honestly significant difference (HSD) post hoc test. n ≥ 11 for long days, n ≥ 5 for short days.
One hypothesis regarding the mechanism by which the H4R17A mutation causes early flowering is that the encoded variant H4 protein prevents deposition of a PTM on H4R17. PROTEIN ARGININE METHYLTRANSFERASE 7 (PRMT7) is the only known histone-modifying enzyme that targets H4R17 in eukaryotes, as it has been shown to mono-methylate H4R17 in mammals (Feng et al., 2013, 2014; Jain and Clarke, 2019). The Arabidopsis genome contains a single orthologous gene for PRMT7 (At4g16570), which has not been functionally characterized. To assess a potential role for PRMT7 in regulating flowering time via methylation of H4R17, we measured flowering time in prmt7 mutants (SALK_028160 and SALK_039529) and in plants overexpressing the PRMT7 gene (i.e. 35S:PRMT7). We confirmed by RT-qPCR that both T-DNA alleles used in these experiments prevent the expression of a full-length PRMT7 transcript and that PRMT7 is overexpressed in the 35S:PRMT7 plants generated here (Figure 3, C–F). Importantly, neither prmt7 mutants nor PRMT7 overexpressing plants displayed altered flowering time in either long-day or short-day conditions (Figure 3, G and H and Supplemental Figure S7). In addition, we observed none of the other vegetative or reproductive phenotypes characteristic of rH4R17A plants in plants lacking or overexpressing PRMT7. These results strongly suggest that replacement of H4 with H4R17A does not affect development in Arabidopsis by interfering with PRMT7 activity on histone H4.
H4R17A interferes with the binding of the catalytic subunits of Arabidopsis ISWI to H4
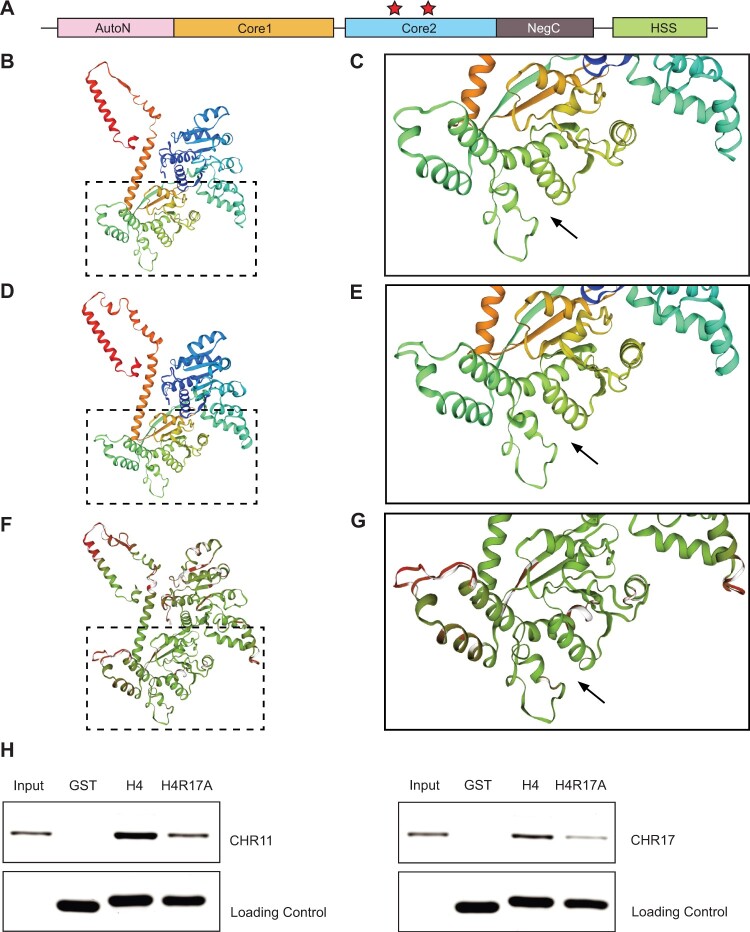
In addition to affecting the deposition of PTMs, mutation of histone residues can prevent binding of proteins to chromatin (Hyland et al., 2005; Norris et al., 2008). Therefore, we next investigated the possibility that replacement of histone H4 with H4R17A affects plant development by negatively affecting the function of plant ISWI chromatin-remodeling complexes. In yeast and animals, R17 of H4 has been shown to directly interact with ISWI to regulate nucleosome remodeling activity in vitro and in vivo (Hamiche et al., 2001; Clapier et al., 2002; Fazzio et al., 2005; Yan et al., 2016; Dann et al., 2017). Replacement of H4 with H4R17A was shown to severely reduce the ability of ISWI from the fungus Myceliophthora thermophila to interact with H4 in isothermal titration calorimetry assays (Yan et al., 2016). Comparative analysis of the protein sequence of the ISWI catalytic subunits in Arabidopsis (CHROMATIN-REMODELING PROTEIN 11 [CHR11] and CHR17) revealed a strict conservation of the two amino acids (E474 and D524 in CHR11; E479 and D529 in CHR17) directly involved in making contacts with H4R17 in the ISWI orthologs from other species, in addition to high protein sequence identity across the H4R17-interacting domain (71%, 167/235 CHR11/CHR17 versus M. thermophila ISWI) (Figure 4A and Supplemental Figure S8). Therefore, we hypothesized that the H4R17A mutation might prevent binding of histone H4 to Arabidopsis ISWI enzymes (Yan et al., 2016, 2019). To test this hypothesis, we first assessed homology models of the structure of CHR11, which indicated conservation of the H4R17-binding pocket in the plant ISWI protein (Figure 4, B–G). We then performed in vitro binding assays with recombinant CHR11 or CHR17 and histone H4 and determined that plant ISWI subunits bind to intact histone H4, while we detected lower binding for the H4R17A mutant protein (Figure 4H). Taken together, these results suggest that expression of H4R17A in plants interferes with the function of the ISWI chromatin-remodeling complexes.
Figure 4.
The H4R17A mutation prevents binding by Arabidopsis ISWI enzymes. A, Domain architecture of ISWI. HSS, HAND–SAND–SLIDE; core1, first RecA-like domain; core2, second RecA-like domain. Red stars indicate the residues implicated in binding H4R17 on the second RecA-like ATPase core domain (core2) identified in M. thermophila (Yan et al., 2016) and S. cerevisiae (Yan et al., 2019). B and C, Homology model of Arabidopsis CHR11 amino acids (aa) 176–706. D and E, Reference structure of M. thermophila ISWI (5JXR) aa 173–718. F and G, Superposition of Arabidopsis CHR11 and M. thermophila ISWI structures with consistency color scheme (green indicates more consistent and red indicates less consistent). Black arrow denotes the predicted (Arabidopsis) or validated (M. thermophila) binding pocket of histone H4 arginine 17 (Yan et al., 2016). The boxed regions are enlarged in (C), (E), and (G). H, In vitro pull-down assay with recombinant GST or GST-tagged histone proteins (H4 or H4R17A) and CHR11 (left) or CHR17 (right).
Functional relationship between H4R17 and ISWI in the regulation of flowering time
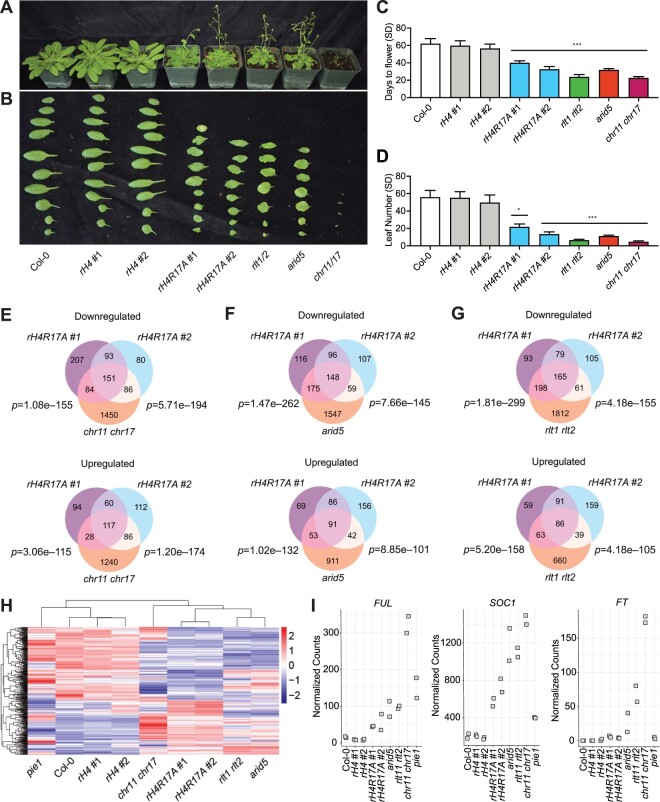
We next examined morphological phenotypes induced by mutations in genes encoding different Arabidopsis ISWI subunits (CHR11, CHR17, HOMEOBOX-1/RINGLET 1 [RLT1], RLT2 and AT-RICH INTERACTING DOMAIN5 [ARID5]). We observed that mutants alleles in these genes result in plants with similar phenotypes as rH4R17A mutant plants: early flowering, upwardly curled leaves, reduced fertility, and a small size relative to wild-type plants (Figure 5, A–D and Supplemental Figure S9; Li et al., 2012). Defects in the timing of the floral transition and other developmental aspects seen in the rH4R17A plants were more similar to those of the single mutant arid5 defective in the ISWI accessory subunit or in the double mutant rlt1 rlt2 (RLT1 and RLT2 were shown to act redundantly [Li et al., 2012]) than to mutations in the ISWI catalytic subunit genes CHR11 and CHR17 (also shown to act redundantly [Li et al., 2012]), which cause more severe developmental phenotypes (Figure 5, A–D and Supplemental Figure S9; Li et al., 2012). The increased severity of the phenotypes displayed by the chr11 chr17 double mutant may be caused by the joint disruption of the ISWI and SWR1 chromatin-remodeling complexes, which both contain CHR11 and CHR17 (Luo et al., 2020). By contrast, ARID5 and RLT1/RLT2 are present in ISWI, but not in SWR1. In addition, RLT1 and RLT2 are only 2 of 12 DNA binding homeobox and Different Transcription (DDT) factors-domain proteins in Arabidopsis, and different ISWI complexes associate with different DDT-domain proteins in vivo (Dong et al., 2013; Tan et al., 2020).
Figure 5.
H4R17 and ISWI are functionally related and involved in the regulation of gene expression and plant development. A, Morphological phenotypes of Col-0, rH4 #1, rH4 #2, rH4R17A #1 and rH4R17A #2, rlt1 rlt2, arid5, and chr11 chr17 plants grown in short-day conditions for 7 weeks. B, Rosette leaf phenotype of Col-0, rH4 #1, rH4 #2, rH4R17A #1 and rH4R17A #2, rlt1 rlt2, arid5, and chr11 chr17 plants. Rosette leaves were cut from plants shortly after bolting in long-day conditions. C and D, Mean days to flower (C) and rosette leaf number (D) at flowering in short-day conditions for Col-0, rH4 #1, rH4 #2, rH4R17A #1 and rH4R17A #2, rlt1 rlt2, arid5, and chr11 chr17 plants. Error bars show standard deviation. *P < 0.01; **P < 0.001; ***P < 0.0001, as determined by one-way ANOVA (genotype versus Col-0) with Tukey’s HSD post hoc test. n = 12. E–G, Venn diagrams showing DEGs (relative to Col-0) identified by RNA-seq in rH4R17A and chr11 chr17 (E), rH4R17A and arid5 (F), and rH4R17A and rlt1 rlt2 plants (G). Statistical analyses were performed using the hypergeometric test, assessing overlap between rH4R17A #1 or rH4R17A #2 and chr11 chr17, arid5, or rlt1 rlt2 mutants, respectively (left P-value represents rH4R17A #1 versus ISWI subunit mutant and right P-value represents rH4R17A #2 versus ISWI subunit mutant). H, Heatmap representation of relative expression levels of shared DEGs identified in the rH4R17A #1 and rH4R17A #2 lines. Legend represents scaled Z-score of normalized read counts. Clustering of rows and columns was calculated using Euclidean distance. I, Normalized read counts for FUL, SOC1, and FT in Col-0, rH4 #1, rH4 #2, rH4R17A #1 and rH4R17A #2, arid5, rlt1 rlt2, chr11 chr17, and pie1 plants.
To further investigate the interplay in plants between H4R17 and ISWI, we performed transcriptome deep sequencing (RNA-seq) analysis on the rH4R17A, arid5, rlt1 rlt2, chr11 chr17, and pie1 (defective in the catalytic subunit of the SWR1 complex PIE1 [PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1]) mutants grown in short-day conditions. We identified 1,771 downregulated genes and 1,471 upregulated genes in the chr11 chr17 double mutant (3,242 DEGs in total), while there were only 535 downregulated genes and 299 upregulated genes in the rH4R17A #1 mutant line (834 DEGs), and 410 downregulated genes and 375 upregulated genes in the rH4R17A #2 mutant line (785 DEGs), relative to wild-type plants (Figure 5E). In spite of the large difference in the total amount of DEGs between chr11 chr17 mutants and the rH4R17A plants, we observed a high overlap between the DEGs in the chr11 chr17 and rH4R17A #1 mutants (45.6%, 380/834), as well as between the DEGs in the chr11 chr17 and rH4R17A #2 mutants (56.0%, 440/785) (Figure 5E). Additionally, we observed a high overlap of DEGs (average 50.0% rH4R17A versus arid5; average 52.5% rH4R17A versus rlt1 rlt2) when comparing the rH4R17A lines to the ISWI subunit mutants arid5 and rlt1 rlt2 (Figure 5, F and G). Furthermore, in the rH4R17A, chr11 chr17, arid5, and rlt1 rlt2 mutants, we detected a similar pattern of RNA expression not shared by rH4 or Col-0 plants, as indicated by the clustering of both rH4R17A lines and all ISWI subunit mutants together (Figure 5H). As ARID5 was previously demonstrated to recognize the histone modification H3K4me3 (Tan et al., 2020), we investigated the relationship between DEGs in rH4R17A mutants and H3K4me3-enriched genes (Chica et al., 2013) and detected a significant overlap (64.7%, 540/834 for rH4R17A #1; 73.0%, 573/785 for rH4R17A #2) (Supplemental Figure S10A), further supporting a functional relationship between H4R17 and ARID5. By contrast, we observed a weaker extent of overlap (average 25.8%) between the DEGs identified in pie1 mutants and the DEGs of rH4R17A mutants and we failed to observe similar patterns of RNA expression between pie and the rH4R17A lines (Figure 5H and Supplemental Figure S10B). Principal component analysis (PC1 and PC2 together explaining 60% of the variance) also indicated similarities between rH4R17A and ISWI subunit mutants, while Col-0 and rH4 plants clustered separately (Supplemental Figure S10C). We then investigated the expression of key flowering time regulatory genes and found that the flowering promoter genes FRUITFULL (FUL), SOC1, and FT are all co-upregulated in the rH4R17A, rlt1 rlt2, arid5, and chr11 chr17 backgrounds (Figure 5I and Supplemental Figure S10D). We did not observe these patterns of co-expression when comparing rlt1 rlt2, arid5, and chr11 chr17 mutants to rH4 plants (Figure 5, H and I and Supplemental Figure S10D). We also performed Gene Ontology (GO) term enrichment analysis and identified multiple additional biological pathways co-regulated by H4R17 and ISWI, including flavonoid metabolism and biosynthesis, pattern specification, specification of symmetry, morphogenesis, the ultraviolet (UV) response pathway, and the regulation of cell death (Supplemental Figure S11 and Supplemental Data Set S1). The shared developmental phenotypes and transcriptional profiles of the rH4R17A, rlt1 rlt2, arid5, and chr11 chr17 mutants suggest that H4R17 plays an important role in plants as in other eukaryotes in regulating the activity of ISWI on chromatin.
Effects of the H4R17A mutation on global nucleosome positioning
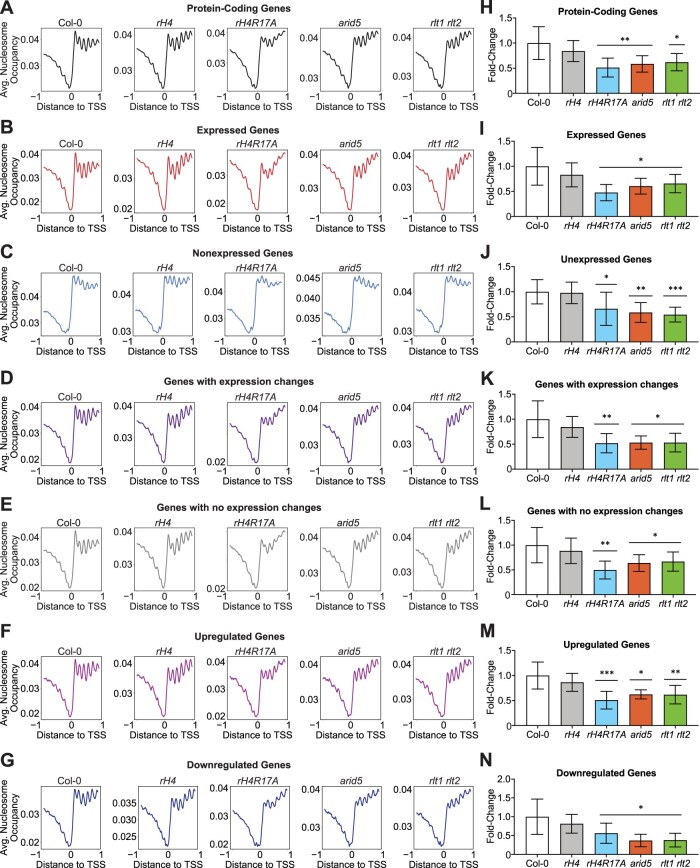
ISWI functions as a chromatin remodeling complex that properly organizes nucleosome spacing at transcriptionally active genes in eukaryotes (Clapier and Cairns, 2009; Gkikopoulos et al., 2011; Yadon and Tsukiyama, 2011; Li et al., 2014). Due to the similarity in the phenotypes and transcriptional profiles between rH4R17A plants and mutants in the Arabidopsis ISWI complex, we hypothesized that the expression of H4R17A interferes with nucleosome spacing in plants. To address this hypothesis, we assessed global nucleosome positioning in rH4R17A mutants using micrococcal nuclease digestion followed by deep sequencing (MNase-seq). Consistent with previous results, we detected a relatively lower nucleosome density in the 1-kb region upstream of the transcription start site (TSS) of protein-coding genes, and a relatively high, evenly spaced distribution of nucleosomes in the 1-kb region downstream of the TSS for Col-0 plants (Figure 6A and Supplemental Figure S12A; Li et al., 2014). Moreover, we observed that expressed protein-coding genes generally displayed more highly phased nucleosome arrays in the gene body and a sharper peak of nucleosome-free DNA in the promoter when compared with nonexpressed protein-coding genes, in line with previous studies (Figure 6, B and C and Supplemental Figure S12B; Li et al., 2014; Zhang et al., 2015). In terms of the different genotypes analyzed, rH4 plants displayed highly similar nucleosome positioning patterns to those of Col-0, as expected. By contrast, while rH4R17A, arid5, and rlt1 rlt2 mutants showed the same general pattern of lower nucleosome density upstream of the TSS and high nucleosome density downstream of the TSS, these genotypes all exhibited a reduction of evenly spaced nucleosome distributions in the gene body (Figure 6A and Supplemental Figure S12A), similar to the pattern reported for the chr11 chr17 mutant (Li et al., 2014). Additionally, we analyzed the nucleosome distribution patterns at genes with expression changes in rH4R17A, chr11 chr17, rlt1 rlt2, and/or arid5 mutants as well as genes without expression changes in these mutants. We determined that the nucleosome distribution patterns at DEGs and non-DEGs are both affected in rH4R17A, arid5, and rlt1 rlt2 mutants (Figure 6, D and E and Supplemental Figure S12C and D), in line with previously published MNase-seq results for the chr11 chr17 mutant (Li et al., 2014). Additionally, nucleosome distribution patterns at DEGs were affected in rH4R17A, arid5, and rlt1 rlt2 mutants regardless of whether the expression of these genes was upregulated or downregulated (Figure 6, F and G). To provide a more quantitative assessment of nucleosome spacing in our assays, we calculated the average change in nucleosome occupancy at the +2 through +6 nucleosome peaks as a measure of nucleosome phasing. This analysis confirmed that the rH4R17A, arid5, and rlt1 rlt2 mutations cause a significant reduction in regular nucleosome phasing in gene bodies (Figure 6, H–N). Taken together, these results indicate that H4R17 positively regulates the action of the ISWI complex to establish nucleosome arrays in protein-coding genes.
Figure 6.
Determination of H4R17 function on regulating nucleosome positioning. A–G, Average nucleosome occupancy relative to the TSS (in kb) of all protein-coding genes (A), expressed protein-coding genes (B), non-expressed protein-coding genes (C), genes with expression changes in rH4R17A, arid5, rlt1 rlt2, and/or chr11 chr17 mutants (D), genes with no expression changes in rH4R17A, arid5, rlt1 rlt2, and/or chr11 chr17 mutants (E), upregulated genes (F) and downregulated genes (G) in rH4R17A, arid5, rlt1 rlt2, and/or chr11 chr17 mutants. The MNase-seq results were generated from two independent biological replicates and RNA-seq data were obtained from the same tissues used for MNase-seq. Cutoffs were defined as follows: Expressed ≥0.5 TPM; nonexpressed <0.5 TPM. Genes with expression changes were defined as >±1.5-fold versus Col-0 and genes with no expression changes were defined as <±1.1-fold versus Col-0. H–N, Fold-change in Δnucleosome occupancy of +2 through +6 nucleosome peaks relative to Col-0 corresponding to all protein-coding genes (H), expressed protein-coding genes (I), unexpressed protein-coding genes (J), genes with expression changes in rH4R17A, arid5, rlt1 rlt2, and/or chr11 chr17 mutants (K), genes with no expression changes in rH4R17A, arid5, rlt1 rlt2, and/or chr11 chr17 mutants (L), upregulated genes (M) and downregulated genes (N) in rH4R17A, arid5, rlt1 rlt2, and/or chr11 chr17 mutants. Error bars show standard deviation. *P < 0.05; **P < 0.005; ***P < 0.0005, as determined by paired Student’s t test.
Discussion
A novel system for studying histone function in plants
In this study, we present a new histone replacement system that facilitates the analysis of histone H4 functions in plants. Our results serve as a proof-of-concept that complete histone replacement systems can be rapidly established in Arabidopsis. In the future, this approach may be applied to generate similar systems to study the functions of different histones or histone variants. The histone replacement system developed in this study for histone H4 will supplement already existing systems in yeast and Drosophila to offer new biological insights into the roles of H4 in plants. Our methodology provides extensive coverage of H4 mutants in a multicellular eukaryote, as the histone replacement system generated in Drosophila has only been used to generate 14 H4 point mutants (Zhang et al., 2019), compared with the 63 H4 point mutants generated with our system, which have been made available through the Arabidopsis Biological Resource Center (ABRC).
This collection of H4 point mutants has revealed a multitude of roles for H4 residues in plants. For example, six separate H4 point mutations located in the globular domain appeared to cause lethality. Many of the equivalent mutations also cause lethality or reduced sporulation efficiency in yeast (Dai et al., 2008; Govin et al., 2010), revealing insights into the effect of these mutations on chromatin. One possibility is that the H4R40A and H4R45A substitutions disrupt the nucleosome entry site, which is highly sensitive to mutations that affect DNA wrapping around the nucleosome (Zhou et al., 2019). Additionally, H4R78A and H4K79A reside on the low sporulation patch identified in yeast (Govin et al., 2010), a region that lies in close proximity to contact sites with the DNA and has been demonstrated to affect meiosis. Similar functions could also result in the early flowering phenotypes observed for certain H4 globular domain mutants (e.g. H4R35, H4R36, H4R39, and H4K44, whose corresponding amino acid positions reside on the nucleosome entry site while that of H4T80 is located on the low sporulation patch). In this way, experiments in plants build on knowledge gathered from other model systems to elucidate how histone residues regulate chromatin function.
Our CRISPR-based strategy to replace endogenous histones offers several advantages over other methods that can potentially be used to achieve complete histone replacement. For example, the successful generation of the H4 septuple mutant in two generations in this work demonstrates that multiplex CRISPR/Cas9 can be used to efficiently inactivate many histone genes in plants (LeBlanc et al., 2017). Using CRISPR/Cas9 greatly reduces the amount of time and resources required to generate a histone depletion background, especially when compared with crossing individual histone mutants. The presence of tandem duplicated copies of histone genes (e.g. the H3.1 genes At5g10390 and At5g10400) can also preclude using traditional crossing schemes to generate backgrounds lacking a specific histone or histone variant. In addition, deploying multiplex CRISPR/Cas9 to inactivate endogenous histones will allow researchers to rapidly re-establish histone replacement systems in a particular mutant background, for example, to screen for point mutations in histones that enhance or suppress a phenotype of interest. Another advantage of our histone H4 replacement strategy is that we consistently observed high expression of the H4 replacement gene, which rescues the morphological phenotype of the H4 septuple background in all of our T1 plants (Figure 1, A–C). Several factors could contribute to this phenomenon. While T-DNA integration into the Arabidopsis genome occurs randomly, a requirement for minimal H4 expression appears to shift the recovery of T-DNA insertions into more transcriptionally active chromatin regions (Koncz et al., 1989; Brunaud et al., 2002; Szabados et al., 2002; Alonso et al., 2003; Kim et al., 2007). Moreover, dosage compensation mechanisms acting to upregulate the expression of the endogenous histone H4 gene At3g53730, as seen in this study (Figure 1C), may also act on the histone H4 replacement gene, as the expression of both of these genes is driven by the At3g53730 promoter. Histone dosage compensation has also been observed in the histone replacement systems implemented in the multicellular eukaryote Drosophila (McKay et al., 2015; Zhang et al., 2019). These histone dosage compensation mechanisms may be related to the recently described process of transcriptional adaptation, in which mutant mRNA decay causes the upregulation of related genes (El-Brolosy et al., 2019; Serobyan et al., 2020). For both of the above reasons, transgenic plants lacking endogenous H4 proteins are observed, and therefore, rH4 plants expressing exclusively mutant histones can predictably be obtained using our strategy.
Several changes may be implemented to improve future histone replacement systems in Arabidopsis and other plants. To control for differential effects caused by random T-DNA integration (Gelvin, 2017), in this study we characterized two independent transgenic lines expressing each H4 replacement construct. Ideally, gene targeting would be utilized to introduce the H4 mutations directly at an endogenous histone H4 locus. While gene targeting technologies in plants relying on homologous recombination currently have very low efficiency compared with yeast and animals, as additional improvements in gene targeting are developed, in situ histone replacement systems in plants analogous to platforms already existing in yeast and Drosophila may also become feasible. Additionally, more precise control over the dosage of the replacement histone could also serve to improve this method. It was recently shown that while yeast histone replacement systems utilizing single-copy integrated histone genes expressing certain mutant histones cannot survive, the addition of a second copy of the mutant histone gene rescues this lethality (Jiang et al., 2017). In the system described here, we utilized a single endogenous histone H4 gene as the H4 replacement gene, rather than generating eight histone H4 replacement constructs corresponding to each endogenous histone H4 gene present in the Arabidopsis genome. While we observed that our rH4 plants appear morphologically wild type due to high H4 expression, it may be important to study the function of the other endogenous H4 genes, or the requirement for Arabidopsis to have eight copies of the H4 genes in its genome. Although labor-intensive, future strategies simultaneously using multiple endogenous H4 genes as H4 replacement genes could therefore be more reflective of the H4 supply available to wild-type plants.
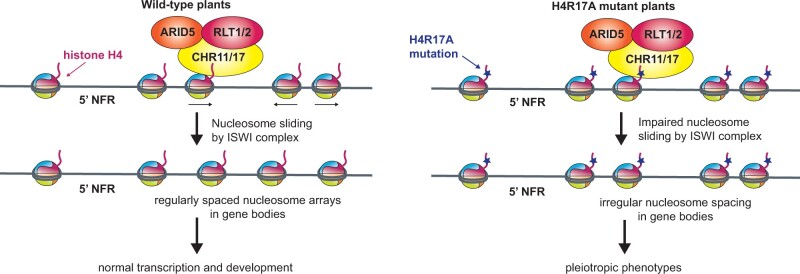
H4R17 regulates nucleosome remodeling and developmental processes in plants
This study uncovered a role for H4R17 in regulating multiple developmental processes in Arabidopsis, including leaf development, fruit development, and flowering. Our findings suggest that this role for H4R17 is not mediated via PTM of this residue. Based on our results, we propose a model similar to that of animal systems where H4R17 regulates developmental processes in plants through its regulation of the ISWI complex (Figure 7; Hamiche et al., 2001; Clapier et al., 2001, 2002; Fazzio et al., 2005; Dann et al., 2017). In wild-type plants, H4R17 positively regulates the ISWI complex to slide nucleosomes and adequately establish the nucleosome positioning patterns in the gene bodies of protein-coding genes (Figure 7). In rH4R17A mutant plants however, the positive regulation of ISWI by histone H4 is impaired so that evenly spaced nucleosome distributions are no longer observed in gene bodies. The altered nucleosome positioning patterns in gene bodies and the large-scale transcriptional changes in turn cause the observed pleiotropic developmental phenotypes. Interestingly, while transcription and nucleosome positioning have been shown to be highly interconnected (Workman and Kingston, 1998; Jiang and Pugh, 2009; Hughes et al., 2012; Struhl and Segal, 2013), we and others have observed that mutations in H4R17 and plant ISWI complex subunits affect nucleosome positioning patterns in both differentially and non-DEGs (Li et al., 2014). Our results support previous work demonstrating that it is unlikely that the nucleosome positioning defects in ISWI mutants are caused by the transcriptional changes observed in these backgrounds (Li et al., 2014; Luo et al., 2020). Moreover, our results are consistent with the idea that many factors on top of nucleosome positioning in gene bodies affect the transcription levels of a gene, and thus in some cases, altered genic nucleosome positioning appears to majorly affect transcription, while in others, little change is observed (Jiang and Pugh, 2009; Bai and Morozov, 2010). Additionally, processes related to genetic robustness may also serve to counteract transcriptional fluctuations due to perturbations of nucleosome positioning (Masel and Siegal, 2009). For these reasons, we observed independence between the nucleosome positioning and transcriptional phenotypes of rH4R17A and mutants in ISWI components. While we cannot exclude the possibility that H4R17 interacts with other chromatin-associated factors to induce certain phenotypes when mutated, our genetic and biochemical evidence strongly supports our proposed model.
Figure 7.
Model for the role of H4R17 in plants. Proposed model for the role of histone H4 arginine 17 in the regulation of ISWI complexes in Arabidopsis. 5′-NFR, 5′-nucleosome-free region.
ISWI chromatin-remodeling complexes contain between two and four subunits in eukaryotes, consisting of a conserved ATPase catalytic subunit and at least one accessory subunit (Corona and Tamkun, 2004; Clapier and Cairns, 2009; Aydin et al., 2014). Multiple types of ISWI complexes have been identified in animals, and the different accessory subunits in these complexes have been proposed to modulate the activity of the shared catalytic subunit as well as the specificity and target recognition of the complex (Lusser et al., 2005; Aydin et al., 2014; Toto et al., 2014). In plants, three types of ISWI complexes have been identified: the plant-specific CHR11/CHR17-RLT1/RLT2-ARID5 (CRA)-type complex, the CHR11/CHR17-DDP1/2/3-MSI3 (CDM)-type complex, and the CHR11/CHR17-DDR1/3/4/5-DDW1 (CDD)-type complex (Tan et al., 2020). In addition, the shared ISWI catalytic subunits CHR11 and CHR17 were also recently demonstrated to act as accessory subunits of the SWR1 chromatin remodeling complex in plants (Luo et al., 2020). We observed that rH4R17A mutants exhibited less severe transcriptional defects than arid5, rlt1 rlt2, or chr11 chr17 mutants. One explanation for this result is that the rH4R17A mutation could affect the action of all types of ISWI complexes in plants without completely abolishing their function, while the arid5 and rlt1 rlt2 mutations could have a more severe influence on only one specific type of ISWI complex (i.e. the CRA-type complex). For example, as ARID5 recognizes H3K4me3 (Tan et al., 2020), it may play a role in directing the ISWI complex to a specific subset of genes, and thus mutation of ARID5 could cause the mislocalization of the ISWI complex. These different mechanisms of action of the rH4R17A and ISWI accessory subunit mutations could induce the differing gene expression and nucleosome positioning phenotypes that we observed. Further characterization of the different ISWI complexes in plants, including their different targeting specificities to chromatin loci and the impact of the other identified CDM-type and CDD-type complexes on the regulation of global transcription and nucleosome positioning, will contribute to elucidating their specific consequences on chromatin regulation.
Materials and methods
Plant materials
All Arabidopsis (A. thaliana) plants were derived from the Columbia-0 (Col-0) accession and were grown in Pro-Mix BX Mycorrhizae soil under cool‐white fluorescent lights (approximately 100 μmol m−2 s−1). Seeds were surface-sterilized with a 70% (v/v) ethanol, 0.1% (v/v) Triton X-100 solution for 5 min, and then with 95% (v/v) ethanol for 1 min. Seeds were spread on sterilized paper, air-dried, and plated on half-strength Murashige–Skoog (MS) plates. Seeds were stratified in the dark at 4°C for 2–4 days, transferred to the growth chamber for 5 days, and then transplanted to soil. Plants were grown in long‐day conditions (16-h light/8-h dark) or short-day conditions (8-h light/16-h dark), as indicated.
The chr11 (GK-424F01) chr17 (GK-424F04) double mutant was described previously (Li et al., 2012). The fas1-4 (SAIL_662_D10), arid5 (SALK_111627), prmt7-1 (SALK_028160), and prmt7-2 (SALK_039529) T-DNA insertion mutants were obtained from the ABRC. The pie1 T-DNA insertion mutants were initially obtained from ABRC (SALK_096434) and the pie1 mutants used in this study were seeds collected from homozygous pie1 plants. The rlt1 (SALK_099250) rlt2 (SALK_132828) double mutant was generated by crossing. Due to severely reduced fertility, chr11 chr17 and arid5 mutants were maintained in a heterozygous state. All newly generated mutant lines are described in Supplemental Data Set S2.
Generation of transgenic Arabidopsis plants
Binary vectors were transformed into Agrobacterium (A. tumefaciens) strain GV3101, using heat shock; plants were transformed using the floral dip procedure as described previously (Clough and Bent, 1998). Transgenic plants for generation of the H4 septuple mutant were selected on half-strength MS plates containing 1% (w/v) sucrose, carbenicillin (200 μg mL−1), and kanamycin (100 μg mL−1). Transgenic rH4 plants were selected on half-strength MS plates containing 1% (w/v) sucrose, carbenicillin (200 μg mL−1), and glufosinate ammonium (25 μg mL−1). Plants were subjected to heat stress treatments as described previously (LeBlanc et al., 2017). The plants were grown continuously at 22°C thereafter.
Scoring of flowering time, rosette leaf number, and rosette size
Days to flower were measured when a 1-cm bolting stem was visible. The number of rosette leaves was determined at the day of flowering. Rosette area was measured using the ARADEEPOPSIS workflow (Huther et al., 2020).
Dimensionality reduction and clustering
For flowering time data, principal component analysis of four variables (day number in long days, leaf number in long days, day number in short days, and leaf number in short days) was performed. We centered variables at mean 0 and set the standard deviation to 1. k-Means clustering was performed 40 times with random initializations on the first two principal components to identify three clusters. For RNA-seq data, principal component analysis of the 200 most variable DEGs in rH4R17A #1 and rH4R17A #2 lines was performed as described above. Analyses were executed in RStudio with R version 3.6.1 (R Development Core Team, 2018).
Plasmid construction
CRISPR constructs used to generate the H4 septuple mutant were inserted into the pYAO-Cas9-SK vector as described previously (Yan et al., 2015).
The H4 replacement plasmid was made by amplifying the promoter (from 967 bp upstream of the start codon to the start codon), gene body, and terminator (up to 503 bp downstream of the stop codon) of H4 (At3g53730) into pENTR/D (ThermoFisher Scientific, Waltham, MA, USA). Site-directed mutagenesis of H4pro:H4 in pENTR/D was first performed using QuikChange II XL (Agilent Technologies, Santa Clara, CA, USA) to create plasmids with 10 silent mutations in the H4 coding sequence. These silent mutations were engineered to test the resistance of the H4 replacement gene against multiple sgRNAs. Additional site-directed mutagenesis of this vector was performed to generate a library of 63 H4 point mutant genes.
Each H4pro:H4 sequence was then transferred into the binary vector pB7WG, containing the H4 sgRNA, using Gateway Technology. The binary vector pB7WG containing the H4 sgRNA was generated as follows: The AtU6‐26‐sgRNA vector containing the sgRNA targeting H4 (At3g53730) was first digested with the restriction enzymes SpeI and NheI, and the digestion products were run on a 1% (w/v) agarose gel. The band containing the H4 sgRNA was then cut out and ligated into the binary vector pB7WG, which had been digested with the restriction enzyme SpeI.
The PRMT7 overexpression construct was created by cloning the PRMT7 genomic coding region (from ATG to stop codon, including introns) into pDONR207, and then subcloning the gene into pMDC32 (Curtis and Grossniklaus, 2003).
Constructs for the production of recombinant protein were generated as follows: Using the NdeI and BamHI restriction sites, the coding sequences of H4 (aa 1–30) were cloned into pET28-Mff(1-61)-PP-GST (Addgene plasmid #73042; provided by D. Chan) and fused with a C-terminal glutathione-S-transferase (GST) tag (Davarinejad et al., 2022). Site-directed mutagenesis was used to create the H4R17A mutation (QuikChange II XL, Agilent Technologies). The coding sequences of CHR11 (encoding fragment aa 409–750) and CHR17 (encoding fragment aa 414–753) were cloned into the pET32a vector using the BamHI and SalI restriction sites.
DNA extraction, PCR, and sequencing analyses
Genomic DNA was extracted from Arabidopsis plants by grinding one leaf in 500 μL of extraction buffer (200 mM Tris–HCl pH 8.0, 250 mM NaCl, 25 mM ethylenediaminetetraacetic acid [EDTA] and 1% [w/v] SDS). Phenol/chloroform (50 μL) was added and tubes were vortexed, followed by centrifugation for 10 min at 3,220 × g at room temperature. The supernatant was transferred to a new tube and 70 μL of isopropanol was added, followed by centrifugation for 10 min at 3,200 × g at room temperature. The supernatant was removed and the DNA pellets were resuspended in 100 μL water.
PCR products were sequenced and analyzed using Sequencher 5.4.6 (Gene Codes Corporation, Ann Arbor, MI, USA) to identify CRISPR‐induced mutations. To assess the rate of mutation of the remaining endogenous H4 gene (At3g53730) in rH4 plants by the sgRNA in the H4 replacement plasmid, endogenous H4 PCR products were cloned into TOPO TA cloning vectors (Invitrogen, Carlsbad, CA, USA). Ten to sixteen individual clones corresponding to each plant were sequenced.
RT-qPCR
Total RNA was extracted from 18-day-old leaves from plants grown in long-day conditions with TRIzol (Invitrogen) and DNase I-treated using RQ1 RNase-Free DNase (Promega, Madison, WI, USA). Three biological replicates (different plants sampled simultaneously) were assessed. SuperScript II Reverse Transcriptase (Invitrogen) was used to generate first-strand cDNA from 1 µg of DNAse I-treated total RNA. Reverse transcription was initiated using random hexamers (Applied Biosystems, Foster City, CA, USA). Quantification of cDNA was done by qPCR using a CFX96 Real‐Time PCR Detection System (Bio‐Rad, Hercules, CA, USA) with KAPA SYBR FAST qPCR Master Mix (2×) Kit (Kapa Biosystems, Wilmington, MA, USA). Relative transcript levels were determined by using the comparative Ct method as follows: Relative quantity = 2(−((CtGOI unknown −Ctnormalizer unknown) − (CtGOI calibrator −Ctnormalizer calibrator))), where GOI is the gene of interest (Livak and Schmittgen, 2001). Actin was used as reference transcript for normalization.
Recombinant protein production
To produce the recombinant proteins AtCHR11, AtCHR17, and the histone-GST fusion proteins, the Rosetta (DE3) Escherichia coli strain (#70954, Sigma, St. Louis, MO, USA) was used. The bacteria were grown in LB medium and protein production was induced with 1 mM IPTG (final concentration). To purify recombinant CHR11 and CHR17, with each protein harboring an N-terminal Trx-His-S tag and a C-terminal His tag, the cell pellets were first resuspended in NPI-10 buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8) containing 1 mM PMSF and then sonicated. After centrifugation, the supernatant was passed through a Ni-NTA agarose column. The column was then washed with NPI-20 buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) and the recombinant proteins were eluted with NPI-250 buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0). For purification of histone-GST proteins, the cell pellets were resuspended in 1× phosphate-buffered saline (PBS, 137 mM NaCl, 10 mM phosphate buffer pH 7.4, 2.7 mM KCl) with 1 mM PMSF before sonification and centrifugation. The supernatant was passed over a glutathione Sepharose 4B column and the bound proteins were washed with 1× PBS before being eluted with EB buffer (50 mM Tris–HCl pH 8.0, 50 mM NaCl, 30 mM reduced l-glutathione, 10% glycerol). The proteins were divided into aliquots and kept at –80°C.
In vitro binding assays
Four micrograms of recombinant CHR11 or CHR17 was combined with 4 μg of GST or GST-tagged histone proteins in 400 μL of binding buffer (25 mM Tris–HCl pH 8.0, 250 mM NaCl, 0.05% [v/v] NP-40) and incubated at 4°C for 12 h for the binding assay. Each tube received 20 μL of pre-washed glutathione Sepharose 4B agarose beads, which were incubated for 30 min to draw down the GST-tagged histone proteins. The beads were washed twice with 1 mL of binding buffer, with rotation for 5 min at 4°C between washes. After the final wash, 15 μL of 2× SDS loading buffer was added to each tube and the proteins were eluted by boiling at 95°C for 5 min. A 10% (w/v) SDS–PAGE gel was used to separate the samples. The GST or GST-tagged histone N-terminal tail proteins were visualized on the gel using Coomassie staining.
Preparation of sequencing libraries
RNA-seq and MNase-seq libraries were prepared at the Yale Center for Genome Analysis (YCGA). For the analysis of the H4 septuple mutant and fas1-4 plants, unopened flowers from 6-week-old plants grown in long-day conditions were frozen in liquid nitrogen, ground with a pestle, and then RNA was extracted using a Direct-zol RNA Miniprep Plus Kit (Zymo Research, Irvine, CA, USA). For each biological replicate, pooled tissue collected simultaneously from three different plants was used. RNA quality was assessed with an Agilent Bioanalyzer 5300 using a DNF-471 RNA kit (Agilent) (Supplemental Figure S13A). For library preparation, mRNA was purified from 500 ng of total RNA with oligodT beads and sheared by incubation at 94°C in the presence of magnesium (KAPA mRNA HyperPrep). Following first-strand cDNA synthesis with random primers, second strand synthesis and A-tailing were performed with dUTP for generating strand-specific sequencing libraries. Adapter ligation with 3′ dTMP overhangs was ligated to library insert fragments. The library was then amplified to obtain fragments carrying the appropriate adapter sequences at both ends. Strands marked with dUTP were not amplified. Indexed libraries that met appropriate cut-offs for both for titer and quality were quantified by qPCR using a commercially available kit (KAPA Biosystems); insert size distribution was determined with LabChip GX or Agilent Bioanalyzer.
For the analysis of rH4 #1, rH4 #2, rH4R17A #1, rH4R17A #2, arid5, rlt1 rlt2, chr11 chr17, and pie1 plants, leaves of 4-week-old plants grown in short-day conditions were frozen in liquid nitrogen, ground with a pestle, and then total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). For each biological replicate, pooled leaf tissue collected simultaneously from three different plants was used. RNA quality was confirmed through analysis on an Agilent Bioanalyzer 2100 with an RNA Nano chip (Supplemental Figure S14). Library preparation was performed using an Illumina TruSeq Stranded Total RNA kit with Ribo-Zero for plant in which samples were normalized with a total RNA input of 1 µg and library amplification was carried out with eight PCR cycles.
MNase-digested DNA was collected as described previously (Pajoro et al., 2018) with the following modifications: 2 g of leaves from 4-week-old plants grown in short-day conditions was ground in liquid nitrogen and resuspended in 20 mL of lysis buffer for 15 min at 4°C. The resulting slurry was filtered through a 40-μm cell strainer into a 50-mL tube. Samples were centrifuged for 20 min at 3,200 ×g at 4°C. The resulting pellets were resuspended in 10 mL of HBB buffer (25 mM Tris–HCl, pH 7.6, 0.44 M sucrose, 10 mM MgCl2, 0.1% [v/v] Triton-X, and 10 mM β-mercaptoethanol) and centrifuged for 10 min at 1,500 × g at 4°C. Pellets were successively washed in 5 mL wash buffer and 5 mL reaction buffer. MNase-seq library preparation was performed using the KAPA Hyper Library Preparation kit (KAPA Biosystems, Part#KK8504). For each biological replicate, pooled leaf tissue collected simultaneously from three different plants was used. Libraries were validated using Agilent Bioanalyzer 2100 Hisense DNA assay and quantified using the KAPA Library Quantification Kit for Illumina Platforms kit. Sequencing was done on an Illumina NovaSeq 6000 using the S4 XP workflow.
RNA-seq processing and analysis
Two independent biological replicates for Col-0, rH4 #1, rH4 #2, rH4R17A #1, rH4R17A #2, arid5, rlt1 rlt2, chr11 chr17, and pie1 were sequenced. Paired-end reads were filtered and trimmed using Trim Galore! (version 0.5.0) with default options for quality (https://github.com/FelixKrueger/TrimGalore). The resulting datasets were aligned to the Araport11 genome (Cheng et al., 2017) using STAR (version 2.7.2a) allowing two mismatches (–outFilterMismatchNmax 2) (Dobin et al., 2013). Statistics for mapping and coverage of the RNA-seq data are provided in Supplemental Data Set S3. Protein-coding genes were defined as described in the Araport11 genome annotation (Cheng et al., 2017). The program featureCounts (version 1.6.4) (-M –fraction) (Liao et al., 2014) was used to count the paired-end fragments overlapping with the annotated protein-coding genes. Differential expression analysis of protein-coding genes was performed using DESeq2 version 1.26 (Love et al., 2014) on raw read counts to obtain normalized fold-changes and Padj-values for each gene. Genes were considered to be differentially expressed if they showed >±2-fold-change and Padj-value <0.05. DEGs are described in Supplemental Data Set S4. Venn diagrams, correlation plots, and correlation matrices were plotted using RStudio with R version 3.6.1 (R Development Core Team, 2018). Heatmaps were plotted with the pheatmap package (version 1.0.12) in RStudio using default clustering parameters on rows and columns. Consistency between biological replicates was assessed by Spearman correlation using deepTools2 (version 2.7.15) (Supplemental Figures S13B and S15; Ramirez et al., 2016). deepTools2 was used to generate bam coverage profiles for visualization with Integrative Genomics Viewer version 2.8.9 (Robinson et al., 2011). The enrichGO method of clusterProfiler (version 3.14.3) was used for GO term enrichment analysis (ont = “BP,” P-value cut-off = 0.05, q-value cut-off = 0.10). Hypergeometric tests were performed using the phyper function of R. H3K4me3-enriched genes were defined from a previously published ChIP-seq dataset gathered from wild-type plants grown in short-day conditions at the 17-leaf stage (Chica et al., 2013)
MNase-seq processing and analysis
Two independent biological replicates for Col-0, rH4 #2, rH4R17A #1, arid5, and rlt1 rlt2 were sequenced. Paired-end reads were filtered and trimmed using Trim Galore! (version 0.5.0) with default options for quality (https://github.com/FelixKrueger/TrimGalore). Bowtie2 version 2.4.2 (Langmead and Salzberg, 2012) was used to align the reads to the Araport11 genome (Cheng et al., 2017) with the –very-sensitive parameter. Statistics for mapping and coverage of the MNase-seq data are provided in Supplemental Data Set S3. Duplicate reads were removed using Picard toolkit version 2.9.0 (Toolkit, 2019) (MarkDuplicates with REMOVE_DUPLICATES=true) and the insertion size was filtered to be between 140 bp and 160 bp using SAMtools version 1.11 (Li et al., 2009). The average nucleosome occupancy corresponding to the regions 1 kb upstream and downstream of the TSS of all protein-coding genes was calculated using the bamCoverage (–MNase parameter specified) and computeMatrix functions of deepTools2 version 2.7.15 (Ramirez et al., 2016). Normalization was performed by scaling with the effective library size calculated by the calcNormFactors function using edgeR version 3.28.1 (Robinson et al., 2010). Consistency between biological replicates was confirmed by Spearman correlation using deepTools2 (Supplemental Figure S16). Fold-change in ΔNucleosome Occupancy of +2 through +6 nucleosome peaks relative to Col-0 was calculated with a custom Python script (https://github.com/etc27/MNaseseq-workflow/analysis/peak_height) as follows: ΔNucleosome Occupancy = peak maximum – (5′ peak minimum + 3′ peak minimum)/2.
Model building
The homology model for Arabidopsis CHR11 (aa 176-706) was built with Swiss-Model against the M. thermophila ISWI reference structure (5JXR) (Biasini et al., 2014; Yan et al., 2016).
Primers
All primers used in this study are listed in Supplemental Data Set S5 (Wu et al., 2008; Richter et al., 2019; Dong et al., 2021).
Statistical analyses
Statistical analysis data are provided in Supplemental Data Set S6. In general, we used unpaired t test and one-way ANOVA with Tukey’s HSD post hoc test for statistical analyses.
Accession numbers
Accession numbers of genes reported in this study are: At3g53730 (H4), At1g07660 (H4), At1g07820 (H4), At2g28740 (H4), At3g45930 (H4), At5g59690 (H4), At3g46320 (H4), At5g59970 (H4), At4g21070 (BRCA1), At1g65470 (FAS1), At1g65480 (FT), At2g45660 (SOC1), At4g16570 (PRMT7), At3g06400 (CHR11), AT5g18620 (CHR17), At3g43240 (ARID5), At1g28420 (RLT1), At5g44180 (RLT2), At3g12810 (PIE1), At5g60910 (FUL), and At5g09810 (ACTIN7). Raw and processed RNA-seq and MNase-seq data have been deposited in the Gene Expression Omnibus database with the accession code GSE190317.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. CRISPR/Cas9-induced mutations in the H4 septuple mutant of Arabidopsis.
Supplemental Figure S2. Co-regulation of biological processes in H4 septuple and fas1-4 mutants.
Supplemental Figure S3. Mapping of H4 globular domain mutations to the nucleosome structure.
Supplemental Figure S4. Impact of histone H4 mutations on the floral transition in Arabidopsis.
Supplemental Figure S5. Phenotypes of early flowering histone H4 mutants.
Supplemental Figure S6. Expression of H4R17A in wild-type Col-0 attenuates mutant phenotypes.
Supplemental Figure S7. Phenotypes of rH4R17A plants and prmt7 mutants.
Supplemental Figure S8. Conservation of ISWI proteins.
Supplemental Figure S9. The effect of ISWI and rH4R17A mutations on the floral transition and development.
Supplemental Figure S10. Co-regulation of gene expression observed between rH4R17A and ISWI mutants.
Supplemental Figure S11. GO term enrichment analysis for rH4R17A and ISWI mutants.
Supplemental Figure S12. H4R17 and the ISWI complex regulate nucleosome positioning in plants.
Supplemental Figure S13. Quality control analysis of RNA-seq replicates.
Supplemental Figure S14. Bioanalyzer electropherograms of RNA-seq replicates.
Supplemental Figure S15. Spearman correlation of RNA-seq replicates.
Supplemental Figure S16. Spearman correlation of MNase-seq replicates.
Supplemental Data Set S1. GO term enrichment analysis.
Supplemental Data Set S2. Genetic materials module.
Supplemental Data Set S3. Statistics for mapping and coverage of sequencing data.
Supplemental Data Set S4. DEGs identified in RNA-seq.
Supplemental Data Set S5. Cloning and PCR primers.
Supplemental Data Set S6. Statistical analysis data.
Supplementary Material
Acknowledgments
We thank all current and former members of our laboratory for discussions and advice during the course of this work. We especially acknowledge the contributions of Axel Poulet, Gonzalo Villarino, Benoit Mermaz, and Anisa Iqbal. We also acknowledge Christopher Bolick and his staff at Yale for help with plant growth and maintenance and thank the Yale Science Building Facilities Staff for maintenance of the laboratory facilities. We also thank Franziska Bleichert from Yale University for her assistance with structural analyses, Paja Sijacic and Roger Deal from Emory University for their generous gift of pie1 seeds, Lin Xu and Wu Liu from the National Laboratory of Plant Molecular Genetics, Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, China, for their generous gift of chr11 chr17 mutant seeds. The authors declare that they have no competing interests.
Funding
This work was supported by grant #R35GM128661 from the National Institutes of Health to Y.J. E.T.C. was supported by a Yale University Gruber Science Fellowship, the NIH Predoctoral Program in Cellular and Molecular Biology Training Grant T32GM007233, and the National Science Foundation Graduate Research Fellowship #2139841. U.V.P. was supported by a grant from the National Institutes of Health R35GM125003.
Conflict of interest statement. None declared.
Contributor Information
Emma Tung Corcoran, Faculty of Arts and Sciences, Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06511, USA.
Chantal LeBlanc, Faculty of Arts and Sciences, Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06511, USA.
Yi-Chun Huang, Faculty of Arts and Sciences, Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06511, USA.
Mia Arias Tsang, Faculty of Arts and Sciences, Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06511, USA.
Anthony Sarkiss, Faculty of Arts and Sciences, Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06511, USA.
Yuzhao Hu, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724, USA.
Ullas V Pedmale, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724, USA.
Yannick Jacob, Faculty of Arts and Sciences, Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06511, USA.
Y.J. and E.T.C. designed the experiments and wrote the article with contributions from C.L. Constructs were generated by E.T.C. and Y.-C.H. Plant transformations were performed by E.T.C., A.S., C.L., and Y.J. Genotyping was performed by E.T.C., A.S., M.A.T., C.L., and Y.J. RNA extractions and RT-qPCR were done by E.T.C., C.L., and Y.J. Flowering time measurements were obtained by E.T.C., M.A.T., and C.L. Plant pictures were taken by E.T.C., M.A.T., C.L., and Y.J. Some of the mutants used in this work were generated by Y.H. and U.V.P. Y.-C.H. performed the in vitro binding assays. C.L. performed the RNA-seq and MNase-seq experiments. E.T.C. did the bioinformatics analyses of all RNA-seq and MNase-seq experiments.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Yannick Jacob (yannick.jacob@yale.edu).
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andres F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Aydin OZ, Vermeulen W, Lans H (2014) ISWI chromatin remodeling complexes in the DNA damage response. Cell Cycle 13: 3016–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Morozov AV (2010) Gene regulation by nucleosome positioning. Trends Genet 26: 476–483 [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Berry S, Dean C (2015) Environmental perception and epigenetic memory: mechanistic insight through FLC. Plant J 83: 133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, et al. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42: W252–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunaud V, Balzergue S, Dubreucq B, Aubourg S, Samson F, Chauvin S, Bechtold N, Cruaud C, DeRose R, Pelletier G, et al. (2002) T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep 3: 1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Z, Yu Y, Li Z, Liu Y, Jiang W, Huang Y, Dong AW (2014) Regulation of Arabidopsis flowering by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression. PLoS Genet 10: e1004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD (2017) Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89: 789–804 [DOI] [PubMed] [Google Scholar]
- Chica C, Szarzynska B, Chen-Min-Tao R, Duvernois-Berthet E, Kassam M, Colot V, Roudier F (2013) Profiling spatial enrichment of chromatin marks suggests an additional epigenomic dimension in gene regulation. Front Life Sci 7: 80–87 [Google Scholar]
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Langst G, Corona DF, Becker PB, Nightingale KP (2001) Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol Cell Biol 21: 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Nightingale KP, Becker PB (2002) A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res 30: 649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corona DF, Tamkun JW (2004) Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta 1677: 113–119 [DOI] [PubMed] [Google Scholar]
- Crevillen P, Gomez-Zambrano A, Lopez JA, Vazquez J, Pineiro M, Jarillo JA (2019) Arabidopsis YAF9 histone readers modulate flowering time through NuA4-complex-dependent H4 and H2A.Z histone acetylation at FLC chromatin. New Phytol 222: 1893–1908 [DOI] [PubMed] [Google Scholar]
- Crevillen P, Yang H, Cui X, Greeff C, Trick M, Qiu Q, Cao X, Dean C (2014) Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515: 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Lu F, Qiu Q, Zhou B, Gu L, Zhang S, Kang Y, Cui X, Ma X, Yao Q, et al. (2016) REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat Genet 48: 694–699 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Hyland EM, Yuan DS, Huang H, Bader JS, Boeke JD (2008) Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell 134: 1066–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann GP, Liszczak GP, Bagert JD, Muller MM, Nguyen UTT, Wojcik F, Brown ZZ, Bos J, Panchenko T, Pihl R, et al. (2017) ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 548: 607–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davarinejad H, Huang YC, Mermaz B, LeBlanc C, Poulet A, Thomson G, Joly V, Munoz M, Arvanitis-Vigneault A, Valsakumar D, et al. (2022) The histone H3.1 variant regulates TONSOKU-mediated DNA repair during replication. Science 375: 1281–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ (2002) Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol 319: 1097–1113 [DOI] [PubMed] [Google Scholar]
- Deng W, Liu C, Pei Y, Deng X, Niu L, Cao X (2007) Involvement of the histone acetyltransferase AtHAC1 in the regulation of flowering time via repression of FLOWERING LOCUS C in Arabidopsis. Plant Physiol 143: 1660–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Gao Z, Liu S, Li G, Yang Z, Huang H, Xu L (2013) SLIDE, the protein interacting domain of Imitation Switch remodelers, binds DDT-domain proteins of different subfamilies in chromatin remodeling complexes. J Integr Plant Biol 55: 928–937 [DOI] [PubMed] [Google Scholar]
- Dong J, LeBlanc C, Poulet A, Mermaz B, Villarino G, Webb KM, Joly V, Mendez J, Voigt P, Jacob Y (2021) H3.1K27me1 maintains transcriptional silencing and genome stability by preventing GCN5-mediated histone acetylation. Plant Cell 33: 961–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Gunther S, Fukuda N, Kikhi K, Boezio GLM, Takacs CM, Lai SL, et al. (2019) Genetic compensation triggered by mutant mRNA degradation. Nature 568: 193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Gelbart ME, Tsukiyama T (2005) Two distinct mechanisms of chromatin interaction by the Isw2 chromatin remodeling complex in vivo. Mol Cell Biol 25: 9165–9174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Hadjikyriacou A, Clarke SG (2014) Substrate specificity of human protein arginine methyltransferase 7 (PRMT7): the importance of acidic residues in the double E loop. J Biol Chem 289: 32604–32616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Maity R, Whitelegge JP, Hadjikyriacou A, Li Z, Zurita-Lopez C, Al-Hadid Q, Clark AT, Bedford MT, Masson JY, et al. (2013) Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions. J Biol Chem 288: 37010–37025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Zhu Z, Meng G, Zhang R, Zhang Y (2021). A CRISPR-Cas9 based shuffle system for endogenous histone H3 and H4 combinatorial mutagenesis. Sci Rep 11: 3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin SB (2017) Integration of Agrobacterium T-DNA into the plant genome. Annu Rev Genet 51: 195–217 [DOI] [PubMed] [Google Scholar]
- Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, Workman JL, Barton GJ, Owen-Hughes T (2011) A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science 333: 1758–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glozak MA, Sengupta N, Zhang X, Seto E (2005) Acetylation and deacetylation of non-histone proteins. Gene 363: 15–23 [DOI] [PubMed] [Google Scholar]
- Govin J, Dorsey J, Gaucher J, Rousseaux S, Khochbin S, Berger SL (2010) Systematic screen reveals new functional dynamics of histones H3 and H4 during gametogenesis. Genes Dev 24: 1772–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunesdogan U, Jackle H, Herzig A (2010) A genetic system to assess in vivo the functions of histones and histone modifications in higher eukaryotes. EMBO Rep 11: 772–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiche A, Kang JG, Dennis C, Xiao H, Wu C (2001) Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc Natl Acad Sci USA 98: 14316–14321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y (2009) Control of the transition to flowering by chromatin modifications. Mol Plant 2: 554–564 [DOI] [PubMed] [Google Scholar]
- He Y, Amasino RM (2005) Role of chromatin modification in flowering-time control. Trends Plant Sci 10: 30–35 [DOI] [PubMed] [Google Scholar]
- He Y, Doyle MR, Amasino RM (2004) PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev 18: 2774–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodl M, Basler K (2009) Transcription in the absence of histone H3.3. Curr Biol 19: 1221–1226 [DOI] [PubMed] [Google Scholar]
- Hodl M, Basler K (2012) Transcription in the absence of histone H3.2 and H3K4 methylation. Curr Biol 22: 2253–2257 [DOI] [PubMed] [Google Scholar]
- Hughes AL, Jin Y, Rando OJ, Struhl K (2012) A functional evolutionary approach to identify determinants of nucleosome positioning: a unifying model for establishing the genome-wide pattern. Mol Cell 48: 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huther P, Schandry N, Jandrasits K, Bezrukov I, Becker C (2020) ARADEEPOPSIS, an automated workflow for top-view plant phenomics using semantic segmentation of leaf states. Plant Cell 32: 3674–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland EM, Cosgrove MS, Molina H, Wang D, Pandey A, Cottee RJ, Boeke JD (2005) Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol Cell Biol 25: 10060–10070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K, Clarke SG (2019) PRMT7 as a unique member of the protein arginine methyltransferase family: a review. Arch Biochem Biophys 665: 36–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF (2009) Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 10: 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Wang Y, Wang Y, He Y (2008) Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Liu Y, Wang A, Qin Y, Luo M, Wu Q, Boeke JD, Dai J (2017) Construction of comprehensive dosage-matching core histone mutant libraries for Saccharomyces cerevisiae. Genetics 207: 1263–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SI, Gelvin SB (2007) Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J 51: 779–791 [DOI] [PubMed] [Google Scholar]
- Kim SY, He Y, Jacob Y, Noh YS, Michaels S, Amasino R (2005) Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17: 3301–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Martini N, Mayerhofer R, Koncz-Kalman Z, Korber H, Redei GP, Schell J (1989) High-frequency T-DNA-mediated gene tagging in plants. Proc Natl Acad Sci USA 86: 8467–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- LeBlanc C, Zhang F, Mendez J, Lozano Y, Chatpar K, Irish V, Jacob Y (2017) Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J 93: 377–386 [DOI] [PubMed] [Google Scholar]
- Li G, Liu S, Wang J, He J, Huang H, Zhang Y, Xu L (2014) ISWI proteins participate in the genome-wide nucleosome distribution in Arabidopsis. Plant J 78: 706–714 [DOI] [PubMed] [Google Scholar]
- Li G, Zhang J, Li J, Yang Z, Huang H, Xu L (2012) Imitation Switch chromatin remodeling factors and their interacting RINGLET proteins act together in controlling the plant vegetative phase in Arabidopsis. Plant J 72: 261–270 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Lifton RP, Goldberg ML, Karp RW, Hogness DS (1978) The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harb Symp Quant Biol 42: 1047–1051 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Luo YX, Hou XM, Zhang CJ, Tan LM, Shao CR, Lin RN, Su YN, Cai XW, Li L, Chen S, et al. (2020) A plant-specific SWR1 chromatin-remodeling complex couples histone H2A.Z deposition with nucleosome sliding. EMBO J 39: e102008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser A, Urwin DL, Kadonaga JT (2005) Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol 12: 160–166 [DOI] [PubMed] [Google Scholar]
- Masel J, Siegal ML (2009) Robustness: mechanisms and consequences. Trends Genet 25: 395–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DJ, Klusza S, Penke TJR, Meers MP, Curry KP, McDaniel SL, Malek PY, Cooper SW, Tatomer DC, Lieb JD, et al. (2015) Interrogating the function of metazoan histones using engineered gene clusters. Dev Cell 32: 373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A (2008) A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol 15: 881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning YQ, Chen Q, Lin RN, Li YQ, Li L, Chen S, He XJ (2019) The HDA19 histone deacetylase complex is involved in the regulation of flowering time in a photoperiod-dependent manner. Plant J 98: 448–464 [DOI] [PubMed] [Google Scholar]
- Norris A, Bianchet MA, Boeke JD (2008) Compensatory interactions between Sir3p and the nucleosomal LRS surface imply their direct interaction. PLoS Genet 4: e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Endo M, Singh MB, Bhalla PL (2005) Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J 44: 557–568 [DOI] [PubMed] [Google Scholar]
- Pajoro A, Muino JM, Angenent GC, Kaufmann K (2018) Profiling nucleosome occupancy by MNase-seq: experimental protocol and computational analysis. Methods Mol Biol 1675: 167–181 [DOI] [PubMed] [Google Scholar]
- Pajoro A, Severing E, Angenent GC, Immink RGH (2017) Histone H3 lysine 36 methylation affects temperature-induced alternative splicing and flowering in plants. Genome Biol 18: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard Toolkit (2019) Broad Institute, GitHub Repository. https://broadinstitute.github.io/picard/. [Google Scholar]
- Pien S, Fleury D, Mylne JS, Crevillen P, Inze D, Avramova Z, Dean C, Grossniklaus U (2008) ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20: 580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. (2018) R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T (2016) deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Kinoshita A, Vincent C, Martinez-Gallegos R, Gao H, van Driel AD, Hyun Y, Mateos JL, Coupland G (2019) Floral regulators FLC and SOC1 directly regulate expression of the B3-type transcription factor TARGET OF FLC AND SVP 1 at the Arabidopsis shoot apex via antagonistic chromatin modifications. PLoS Genet 15: e1008065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serobyan V, Kontarakis Z, El-Brolosy MA, Welker JM, Tolstenkov O, Saadeldein AM, Retzer N, Gottschalk A, Wehman AM, Stainier DY (2020) Transcriptional adaptation in Caenorhabditis elegans. eLife 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T (2015) Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol 66: 441–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68: 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K, Segal E (2013) Determinants of nucleosome positioning. Nat Struct Mol Biol 20: 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados L, Kovacs I, Oberschall A, Abraham E, Kerekes I, Zsigmond L, Nagy R, Alvarado M, Krasovskaja I, Gal M, et al. (2002) Distribution of 1000 sequenced T-DNA tags in the Arabidopsis genome. Plant J 32: 233–242 [DOI] [PubMed] [Google Scholar]
- Tan LM, Liu R, Gu BW, Zhang CJ, Luo J, Guo J, Wang Y, Chen L, Du X, Li S, et al. (2020) Dual recognition of H3K4me3 and DNA by the ISWI component ARID5 regulates the floral transition in Arabidopsis. Plant Cell 32: 2178–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenea GN, Spantzel J, Lee LY, Zhu Y, Lin K, Johnson SJ, Gelvin SB (2009) Overexpression of several Arabidopsis histone genes increases agrobacterium-mediated transformation and transgene expression in plants. Plant Cell 21: 3350–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toto M, D’Angelo G, Corona DF (2014) Regulation of ISWI chromatin remodelling activity. Chromosoma 123: 91–102 [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Jerzmanowski A (2005) Suppression of histone H1 genes in Arabidopsis results in heritable developmental defects and stochastic changes in DNA methylation. Genetics 169: 997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman JL, Kingston RE (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem 67: 545–579 [DOI] [PubMed] [Google Scholar]
- Wu JF, Wang Y, Wu SH (2008) Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol 148: 948–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen WH (2008) Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol Cell Biol 28: 1348–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadon AN, Tsukiyama T (2011) SnapShot: chromatin remodeling: ISWI. Cell 144: 453–453.e451 [DOI] [PubMed] [Google Scholar]
- Yaish MW, Colasanti J, Rothstein SJ (2011) The role of epigenetic processes in controlling flowering time in plants exposed to stress. J Exp Bot 62: 3727–3735 [DOI] [PubMed] [Google Scholar]
- Yan L, Wang L, Tian Y, Xia X, Chen Z (2016) Structure and regulation of the chromatin remodeller ISWI. Nature 540: 466–469 [DOI] [PubMed] [Google Scholar]
- Yan L, Wei S, Wu Y, Hu R, Li H, Yang W, Xie Q (2015) High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant 8: 1820–1823 [DOI] [PubMed] [Google Scholar]
- Yan L, Wu H, Li X, Gao N, Chen Z (2019) Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat Struct Mol Biol 26: 258–266 [DOI] [PubMed] [Google Scholar]
- Yu CW, Liu X, Luo M, Chen C, Lin X, Tian G, Lu Q, Cui Y, Wu K (2011) HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol 156: 173–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhang W, Jiang J (2015) Genome-wide nucleosome occupancy and positioning and their impact on gene expression and evolution in plants. Plant Physiol 168: 1406–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhang X, Xue Z, Li Y, Ma Q, Ren X, Zhang J, Yang S, Yang L, Wu M, et al. (2019) Probing the function of metazoan histones with a systematic library of H3 and H4 mutants. Dev Cell 48: 406–419.e405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Hu H, Ren H, Yang Z, Qiu Q, Qi W, Liu X, Chen X, Cui X, Li S, et al. (2019) The Arabidopsis H3K27me3 demethylase JUMONJI 13 is a temperature and photoperiod dependent flowering repressor. Nat Commun 10: 1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Gaullier G, Luger K (2019) Nucleosome structure and dynamics are coming of age. Nat Struct Mol Biol 26: 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.