Abstract
Synthesis of cobalamin de novo by Salmonella enterica serovar Typhimurium strain LT2 and the absence of this ability in Escherichia coli present several problems. This large synthetic pathway is shared by virtually all salmonellae and must be maintained by selection, yet no conditions are known under which growth depends on endogenous B12. The cofactor is required for degradation of 1,2-propanediol and ethanolamine. However, cofactor synthesis occurs only anaerobically, and neither of these carbon sources supports anaerobic growth with any of the alternative electron acceptors tested thus far. This paradox is resolved by the electron acceptor tetrathionate, which allows Salmonella to grow anaerobically on ethanolamine or 1,2-propanediol by using endogenously synthesized B12. Tetrathionate provides the only known conditions under which simple cob mutants (unable to make B12) show a growth defect. Genes involved in this metabolism include the ttr operon, which encodes tetrathionate reductase. This operon is globally regulated by OxrA (Fnr) and induced anaerobically by a two-component system in response to tetrathionate. Salmonella reduces tetrathionate to thiosulfate, which it can further reduce to H2S, by using enzymes encoded by the genes phs and asr. The genes for 1,2-propanediol degradation (pdu) and B12 synthesis (cob), along with the genes for sulfur reduction (ttr, phs, and asr), constitute more than 1% of the Salmonella genome and are all absent from E. coli. In diverging from E. coli, Salmonella acquired some of these genes unilaterally and maintained others that are ancestral but have been lost from the E. coli lineage.
Virtually all Salmonella isolates synthesize B12 de novo under anaerobic conditions (27, 34, 43). The ability to synthesize and import B12 requires more than 35 known genes (48)—approaching 1% of the genome. However, mutations that eliminate B12 synthesis from otherwise wild-type strains cause no growth defect under the standard aerobic or anaerobic lab conditions used thus far. Since evolutionary maintenance of such a large fraction of the genome requires selection, it seems inescapable that natural conditions must exist under which endogenously synthesized B12 is important to growth of salmonellae. Salmonella enterica serovar Typhimurium makes B12 de novo only in the absence of oxygen (27). Degradation of ethanolamine or 1,2-propanediol requires B12 and provides a carbon and energy source, but growth on these compounds has been observed only under aerobic conditions requiring exogenous B12 (28, 46). These paradoxical aspects of B12 metabolism have been reviewed (47).
The B12 paradox may be resolved by the finding, described here, that the electron acceptor tetrathionate supports anaerobic use of ethanolamine or 1,2-propanediol as the sole carbon and energy source by using endogenously synthesized B12. Under anaerobic conditions, tetrathionate supports considerably better growth on these carbon sources than the other alternative electron acceptors tested. Tetrathionate plus either ethanolamine or 1,2-propanediol provides the only known conditions under which B12 synthesis is essential to the growth of wild-type Salmonella.
While tetrathionate metabolism has not been studied extensively, its reduction is likely to follow the pathway diagrammed in Fig. 1 (4, 36). The enzymes encoded by the ttr operon reduce tetrathionate to thiosulfate (9, 21), which can be reduced further to sulfite plus hydrogen sulfide by enzymes encoded by the phs operon (11, 20). Sulfite can be reduced to hydrogen sulfide by the dissimilatory anaerobic sulfite reductase encoded by the asr genes (17, 25, 26). Salmonella also has an assimilatory sulfite reductase (CysJI), which acts with or without oxygen (32). It is not clear where in nature tetrathionate might be encountered, but it seems likely to occur in bacterial communities that include sulfate-reducing bacteria; it has been detected in humid soils that support growth of such bacteria (52).
FIG. 1.
Reduction of tetrathionate to sulfide. The tetrathionate reductase (Ttr) described here performs the initial reduction to thiosulfate (S2O32−). This area of metabolism has been reviewed by Barrett and Clark (4).
Taken together, the three sulfur-reducing enzyme systems (encoded by ttr, phs, and asr) and the genes for 1,2-propanediol degradation (pdu) and de novo B12 synthesis (cbi) comprise more than 1% of the Salmonella genome and seem to act as part of a logical pattern of metabolism. All of these genes are characteristic of S. enterica and absent from Escherichia coli. This suggests that B12-dependent anaerobic catabolism of small molecules, supported by reduction of sulfur compounds, may be central to the life of salmonellae. A model is described for evolution of this system during the divergence of Salmonella and Escherichia coli. (Throughout the rest of this article, propanediol refers to 1,2-propanediol.)
MATERIALS AND METHODS
Bacterial strains and crosses.
All strains are derived from S. enterica serovar Typhimurium strain LT2. Key strains and their sources are listed in Table 1. The transposable elements TPOP1 and TPOP2 were described previously (44). All transductional crosses were mediated by the high-frequency transducing mutant of phage P22 (HT105, int) (51). Growth of phage and procedures for crosses have been described previously (7). Standard methods for cell culture have been described previously (1, 6).
TABLE 1.
Bacterial strains
| Strain | Genotypea | Source or referenceb |
|---|---|---|
| TT22362 | Wild-type S. enterica serovar Typhimurium strain LT2 | Lab collection |
| TT22336 | cbiD24::MudJ | 3 |
| TT22338 | oxrA::Tn10 | Charles Miller |
| TT22340 | phs-213::MudJ | Justin Tingey, unpublished data |
| TT22341 | eut-336 (eutS-K)::Tn10dTPOP1 | 31 |
| TT22342 | aceA112::MudJ | SGSC |
| TT22343 | aceB113::MudJ | SGSC |
| TT22344 | pdu-12::MudA | 60 |
| TT22474 | prp-54(prpR-E)::Cm(swap) | This study |
| TT20444 | phs208::Tn10dGn ttrB122::MudJ | N. Patrick Higgins |
| TT22347 | oxrA2::Tn10 ttrB123::MudJ | This study |
| TT22467 | oxrA2::Tn10 ttrB123::MudJ ttrS130::Tn10dCm | This study |
| TT22468 | oxrA2::Tn10 ttrS130::Tn10dCm | This study |
| TT22469 | ack-408::Tn10 | SGSC |
| TT18691 | ttrB122::MudJ | This study |
| TT18694 | ttrB123::MudJ | This study |
| TT18683 | ttrA121::MudJ | This study |
| TT20590 | ttrB122::MudJ arcA201::Tn10dTc | 1; this study |
| TT20464 | ttrB122::MudJ cya-961::Tn10 metE205 ara-9 | Peter Postma; this study |
| TT20469 | ttrB112::MudJ crp-773::Tn10 metE205 ara-9 | Peter Postma; this study |
| TT22372 | ttrS133(del)::Cm(swap) | This study |
| TT22373 | ttrS133(del)::Cmr, ttrB123::MudJ | This study |
| TT18665 | ttrS108::Tn10dTc | This study |
| TT20433 | ttrS108::Tn10dTc ttrB123::MudJ | This study |
| TT20369 | ttrS130::Tn10dCm | This study |
| TT22348 | ttrS130::Tn10dCm ttrB123::MudJ | This study |
| TT18675 | ttrS117::Tn10dTc | This study |
| TT20440 | ttrS117::Tn10dTc ttrB123::MudJ | This study |
| TT22349 | ttrS108::Tn10dTc ttrS130::Tn10dCm | This study |
| TT22350 | ttrS108::Tn10dTc ttrS130::Tn10dCm ttrB123::MudJ | This study |
| TT22351 | ttrS130::Tn10dCm ttrS117::Tn10dTc | This study |
| TT22352 | ttrS130::Tn10dCm ttrS117::Tn10dTc ttrB123::MudJ | This study |
| TT22353 | ttrP127::Tn10dTPOP2 | This study |
| TT22354 | ttrP127::Tn10dTPOP2 ttrA121::MudJ | This study |
| TT22355 | ttrP128::Tn10dTPOP2 | This study |
| TT22356 | ttrP128::Tn10dTPOP2 ttrA121::MudJ | This study |
| TT22470 | ttrR132(del)::Cm(swap) | This study |
| TT22471 | ttrR132(del)::Cm(swap) ttrB123::MudJ | This study |
| TT22472 | ttrR132::Cm(swap) ttrS126::Tn10dTPOP2 | This study |
| TT22473 | ttrR132::Cm(swap) ttrS126::Tn10dTPOP2 ttrA120::MudJ | This study |
| TT22359 | ttrB129::Tn10dTPOP2 | This study |
| TT22360 | ttrB129::Tn10dTPOP2 ttrA120::MudJ | This study |
| TT22339 | ttrC131::Tn10dTPOP2 | This study |
| TT22361 | ttrC131::Tn10dTPOP2 ttrA121::MudJ | This study |
Swap refers to a resistance determinant that replaces a substantial amount of the target gene sequence.
SGSC, Salmonella Genetic Stock Center.
Chemical reagents and growth media.
Standard aerobic cell culture was conducted in Difco nutrient broth supplemented with 0.1 mM NaCl (14). Minimal medium was the No-carbon-E (NCE) medium (13), supplemented with trace metals (0.3 μM CaCl2, 0.1 μM ZnSO4, 0.045 μM FeSO4, 0.2 μM Na2Se2O3, 0.2 μM Na2MoO4, 2 μM MnSO4, 0.1 μM CuSO4, 3 μM CoCl2, and 0.1 μM NiSO4). Unless otherwise indicated, carbon sources were provided at the following concentrations: glucose, 11 mM; glycerol, 43 mM; ethanolamine (Aldrich Chemical Co.), 25 mM in solid media, 10 mM in liquid; and 1,2-propanediol and potassium acetate (Aldrich Chemical Co.), 25 mM in solid media, 50 mM in liquid. Tetrathionate (Sigma Chemical Co.) was added at the final concentrations indicated in the figure legends and tables; for growth on plates and for assay of induction of the ttrBCA genes, it was added at a final concentration of 10 mM. For growth in liquid culture, it was added at 40 mM, since under these conditions a higher growth yield was obtained by adding the fourfold excess. Trimethylamine N-oxide, potassium nitrate, fumaric acid, and dimethyl sulfoxide were added at concentrations of 5, 10, 15, and 20 mM. Cyanocobalamin (Sigma Chemical Co.) was added at a final concentration of 200 nM unless otherwise specified. AdoB12 (5′-deoxyadenosylcobalamine) and Cbi (cobinamide dicyanide; Sigma Chemical Co.) were each added at a final concentration of 15 nM. Antibiotics were added to nutrient broth medium at the following concentrations: kanamycin, 50 μg/ml; tetracycline, 20 μg/ml; and chloramphenicol, 20 μg/ml. In minimal medium, tetracycline was added to a final concentration of 10 μg/ml.
Cell growth and enzyme assays.
Anaerobic conditions (37°C) for petri dishes were provided by an anaerobic chamber (Forma Anaerobic System Model 1024) with a gas mixture of CO2, H2, and N2 (5:6:89). For anaerobic liquid cultures, media were preincubated in the anaerobic chamber for 12 to 24 h. Cells for these cultures were pregrown aerobically to stationary phase in NCE glycerol, washed twice in NCE, and then diluted 10,000-fold into the liquid media inside the anaerobic chamber. The anaerobic culture tubes were then crimp capped, and the medium and headspace were flushed with nitrogen (7). Incubation was at 37o with shaking. Cultures that contained AdoB12 or Cbi were prepared and incubated in the dark. Turbidity was monitored in a spectrophotometer at 650 nm. Chlorate sensitivity was tested anaerobically on agar plates containing minimal E medium supplemented with 11 mM glucose with or without 0.2 mM potassium chlorate. β-Galactosidase activity from liquid cell cultures was assayed as described previously (37).
Sulfur assay.
Elemental sulfur was detected in cell cultures with a modified version of a method of cyanolysis (19). Soluble forms of sulfur were removed by pelleting cells and washing pellets with water. The cell pellet was dried, and elemental sulfur was dissolved in acetone by overnight incubation at 37°C with shaking. Sample dilutions were made in acetone. Elemental sulfur was detected by cyanolysis; thiocyanate derivatives were formed by adding 0.1 ml of 0.1 M KCN to a 1-ml sample at room temperature. To detect thiocyanates, 0.1 ml of an aqueous Fe(NO3)3 solution (Aldrich Chemical Co.) [0.25 M Fe(NO3)3 in 3 M HNO3] was added and the samples were read at 460 nm in a Beckman DU 640 spectrophotometer. Final sulfur concentrations were estimated by comparing absorbance to a standard curve prepared with elemental sulfur.
Isolation of insertion mutations in and near the ttr locus.
Insertion mutants unable to reduce tetrathionate were obtained by using MudJ (Knr-lac) or Tn10dTc; these mutants were identified by their failure to produce acid on MacConkey indicator medium (Difco) containing 10 mM tetrathionate. Mutants defective in synthesis of the molybdopterin cofactor (required for tetrathionate reductase) were identified on the basis of their resistance to chlorate and have been described previously (54). Of 18 ttr::MudJ insertions isolated (from 10,000 random insertion mutants), 4 expressed the lacZ gene from the ttr promoter. Other insertions of the MudJ (Knr-lac) element were obtained by screening a random insertion pool for clones whose β-galactosidase level was induced by tetrathionate (white colonies on nutrient broth–X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside] plates, blue colonies on nutrient broth–X-Gal tetrathionate plates).
Insertions of Tn10dCm linked to the ttr operon were obtained by transducing a ttr::MudJ insertion mutant with a P22 lysate grown on a pool of random Tn10dCm insertion mutants; chloramphenicol-resistant (Cmr) transductants were screened for those that had become Lac− by loss of the recipient MudJ insertion. These clones had inherited a donor fragment that carried both a wild-type ttr operon and a linked Tn10dCm element; the mutant ttr region of the recipient was replaced, leaving a ttr+ strain with a nearby Tn10dCm insertion. The same strategy was used to obtain insertions of the Tn10dTc (TPOP2) element (44) in and near the ttr operon. Additional TPOP insertions within the ttr coding sequence were obtained by transducing a strain carrying the distal MudJ insertion (ttrA120) with phage grown on a pool of random TPOP insertion mutants. Insertions upstream of the recipient MudJ insertion were identified because they allowed tetracycline (instead of tetrathionate) to induce expression of β-galactosidase.
Determination of the ttr operon sequence and insertion sites.
Starting with strains carrying multiple insertions in the ttr operon, fragments of the operon were amplified by PCR using primers complementary to known sequences at the ends of the particular inserted elements. The amplified fragments were sequenced according to the method of Sanger et al. (49) at the University of Utah Health Sciences DNA Sequence Facility. This information facilitated the design of primers that were used to amplify and sequence the regions between the ttr operon and nearby genes and to determine the insertion site of transposable elements.
Construction of ttrR and ttrS and prpRBCDE deletion-substitution (swap) mutations.
The linear transformation method was that of Murphy (38) and used cells expressing recombination genes of phage lambda. The ability of these lambda enzymes to support targeted recombination with very short homologous sequences has recently been demonstrated (12, 61). Methods were optimized for Salmonella by Eric Kofoid (personal communication). The transformation recipient was a wild-type Salmonella strain carrying plasmid pPT223 (TT22236), which includes the lam, bet, and exo genes of phage lambda expressed from a lac promoter; this plasmid was constructed and supplied by Poteete and Fenton (42). Cells were pregrown in Luria broth with isopropyl-β-d-thiogalactopyranoside (2 mM) to induce the plasmid recombination genes and washed three times in 10% glycerol prior to electroporation.
For the ttrR swap, the 5′ end of primer 1 included 40 bp just outside of the downstream end of the ttrR coding sequence, followed by a sequence adjacent to the promoter of the chloramphenicol resistance gene of pACYC184. Primer 2 had at its 5′ end 40 bases complementary to a sequence centered on the translational start codon of ttrR (which overlaps ttrS), followed by 20 bp homologous to the region immediately outside of the pACYC184 chloramphenicol resistance gene. These primers were used to amplify the chloramphenicol resistance gene (using Taq polymerase [Promega]), and the resulting linear fragment was electroporated into a recipient strain as described above. The resulting Cmr recombinants carried the resistance determinant in place of all of the ttrR gene except for the upstream 30 bp, which were left in place because they include the downstream end of the overlapping ttrS gene and its stop codon.
To construct the ttrS swap, all of the ttrS coding sequence was eliminated except for the distal 44 bases needed for initiation of the overlapping ttrR gene; the deletion was replaced by the intact Cmr gene from Tn10dCm plus the downstream end of the Tn10 element and 10 bases downstream of the ttrS130 insertion site. This was done to reconstruct any possible promoters associated with insertion (ttrS130::Tn10dCm). The first 40 bases of primer 3 are homologous to the 40 bases immediately outside the upstream end of the ttrS gene predicted by Hensel et al. (GenBank accession number AJ224978). The next 20 bases of this primer are the same Cmr sequence used in primer 1 described above. The first 40 bases of primer 4 correspond to the ttrS sequence immediately 5′ to the Shine-Dalgarno sequence of the ttrR gene. The next 20 bases of this primer were designed for the amplification (from ttrS130::Tn10dCm) of the entire Cmr marker, the adjacent Tn10 material, and 10 bases of downstream ttrS sequence. This same technique was used to replace the entire prp operon (23) (prpRBCDE) with a Cmr cassette from pACYC184.
Nucleotide sequence accession number.
The accession number for the ttr operon sequence and the location of insertions in this sequence are available from GenBank (accession number AF282268).
RESULTS
Fermentation of 1,2-propanediol and ethanolamine.
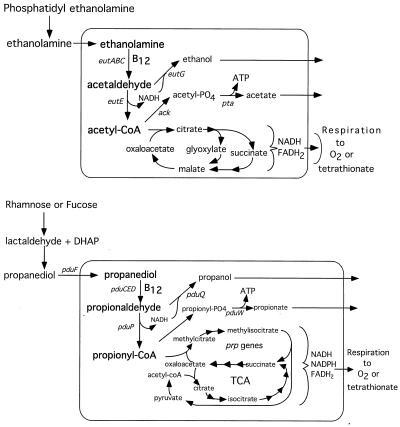
Ethanolamine is degraded by B12-dependent conversion to acetyl coenzyme A (acetyl-CoA), which can enter the tricarboxylic acid (TCA) cycle and the glyoxalate shunt; this is diagrammed in Fig. 2 and has been reviewed previously (47). Propanediol is converted to propionyl-CoA, joined to oxaloacetate (the 2-methyl-citrate pathway), and converted to succinate plus pyruvate, which can be converted to acetyl-CoA and enter the standard TCA cycle (23, 24, 56, 60). In the absence of any electron acceptor, ethanolamine and propanediol might be fermented, providing an ATP source by conversion of acetyl-CoA (or propionyl-CoA) to acetyl-PO4 (or propionyl-PO4) and thence to acetate (or propionate) plus ATP. The latter reactions could be performed by the Ack and Pta enzymes or by similar enzymes encoded by the eut and pdu operons (5, 31). Excess reducing equivalents generated by conversion of acetaldehyde (or propionaldehyde) to acetyl-CoA (or propionyl-CoA) could, in principle, be balanced by reducing some aldehyde to ethanol (or propanol) and excreting the alcohol. This scheme might allow ethanolamine and propanediol to provide an energy source without respiration (fermentative growth). When this was tested, Salmonella seemed unable to ferment either ethanolamine or propanediol for use as both a carbon and energy source (see below).
FIG. 2.
Metabolism of ethanolamine and of propanediol. The upper part of this diagram outlines the known metabolism of ethanolamine and indicates the proposed role of various proteins encoded by the ethanolamine (eut) operon (31). The lower part of the diagram outlines the known metabolism of propanediol as described in references 5 and 24.
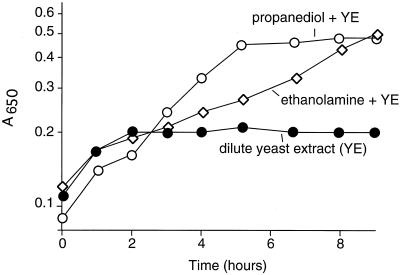
Fermentative growth on ethanolamine or propanediol envisioned above depends on excretion of carbon. Perhaps fermentation might provide energy if some additional source of carbon was provided. To test this, dilute yeast extract was provided at a concentration (0.2%) that could not support anaerobic growth alone. Growth with this added carbon source was stimulated by propanediol or ethanolamine (Fig. 3).
FIG. 3.
Stimulation of anaerobic growth by ethanolamine or propanediol. Cells of wild-type serovar Typhimurium, strain LT2, were grown anaerobically on minimal NCE medium supplemented with 0.2% yeast extract (YE) to provide a carbon source with or without 80 mM propanediol or 98 mM ethanolamine as an energy source.
The fermentative growth on ethanolamine or propanediol (facilitated by added carbon sources) was eliminated by mutations which block B12 synthesis (cob); addition of B12 overcame the effect of the cob mutation (data not shown). As can be seen in Fig. 3, the growth rate was lower on ethanolamine than on propanediol; this deficit was corrected by addition of B12 (data not shown). This B12 limitation is expected, since the B12 synthesis genes (cob) are induced by propanediol but not by ethanolamine (7, 10, 45). Thus, when ethanolamine alone is provided, the level of endogenous B12 may limit growth. Mutations in the eut operon eliminated the ethanolamine stimulation, and mutations in the pdu operon eliminated growth on propanediol. Thus, the inferred fermentative use of ethanolamine and propanediol as energy sources seems to involve the standard degradative pathways. (Data on use of endogenous B12 are given below.)
Qualitative tests of respiratory electron acceptors.
Salmonella is unable to use propanediol or ethanolamine as the sole carbon and energy source under anaerobic conditions even with the alternative respiratory electron acceptor fumarate, dimethyl sulfoxide, or trimethylamine N-oxide; very slight growth was seen with nitrate (data not shown). This result was seen both on solid media and when growth was scored qualitatively (as plus or minus) in anaerobic tubes. As expected, all of these electron acceptors allowed anaerobic growth on glycerol. However, only tetrathionate allowed strong anaerobic growth on all three carbon sources. No electron acceptor was required for growth on the fermentable carbon source glucose.
Growth of wild-type strains with tetrathionate.
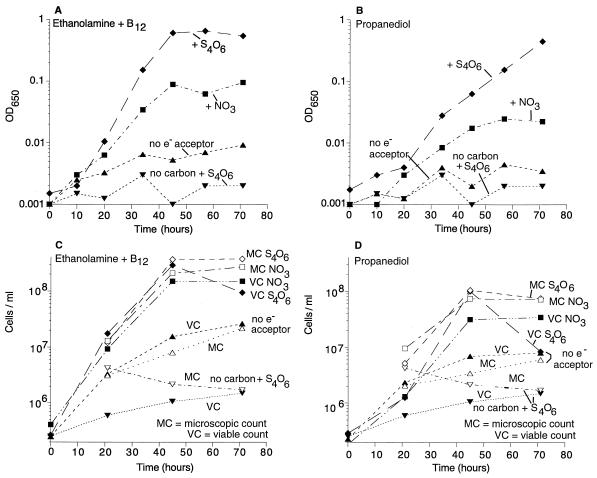
Growth was initially measured in anaerobic liquid cultures by monitoring the increase in optical density (OD) at 650 nm. Tetrathionate supported anaerobic growth on acetate, demonstrating that it is truly serving as a respiratory electron acceptor (Table 2). As can be seen in Fig. 4A and B, tetrathionate supported anaerobic growth on ethanolamine plus B12 and on propanediol. (Reasons for supplying B12 for ethanolamine tests are described below.) In the absence of an electron acceptor, none of these cultures reached an OD in excess of 0.01. Similarly no growth was seen on tetrathionate alone, in the absence of added carbon source. In agreement with previous qualitative results, some growth was seen with nitrate as an electron acceptor.
TABLE 2.
Effect of mutations on anaerobic growth of S. enterica serovar Typhimurium
| Straina | Relevant genotype | Relevant phenotype | Growth under indicated conditionsb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Glycerol
|
Acetate
|
Ethanolaminec
|
Propanediol
|
|||||||
| Solidd | Liquide | Solidd | Liquide | Solidd | Liquide | Solidd | Liquide | |||
| TT22362 | Wild type | Ttr+ Eut+ Pdu+ | + | 1.5 | + | 4.3 | + | 4.3 | + | 5.4 |
| TT22341 | eut-336 (eutS-K) | Deletion of eut operon | + | 1.7 | + | 5.5 | − | —f | + | 14 |
| TT22342 | aceA112::MudJ | Isocitrate lyase negative | + | 1.6 | − | 21 | − | 20 | + | 4.1 |
| TT22343 | aceB113::MudJ | Malate synthase A negative | + | 1.5 | − | 31 | − | 44 | + | 4.1 |
| TT22469 | ack-408::Tn10 | Acetate kinase negative | + | 1.5 | ± | 26 | ± | 10 | + | 5.4 |
| TT22344 | pdu-12::MudA | Propanediol negative | + | 1.5 | + | 4.8 | + | 4.9 | − | —f |
| TT22474 | prp-54(prpR-E) | Propionate negative | + | 1.5 | + | 4.0 | + | 3.8 | − | 83 |
| TT22359 | ttrB129::Tn10dTPOP2 | Tetrathionate reductase negative | − | 19 | − | 24 | − | 13 | − | 12 |
| TT22338 | oxrA::Tn10 | OxrA negative (Fnr−) | − | 10 | − | 10 | − | 17 | − | 17 |
| TT22340 | phs-213::MudJ | Thiosulfate reductase negative | + | 1.6 | + | 4.2 | + | 4.1 | + | 5.4 |
See Table 1.
The growth media were NCE minimal medium supplemented with tetrathionate and the indicated carbon and energy sources. Growth rates were estimated by monitoring optical density and microscopic counts. All strains were able to grow anaerobically on solid media with glucose; TT22362 (wild type), TT22359 (ttrB), and TT22338 (oxrA) were also grown in liquid cultures with glucose; in each case, the doubling time was measured to be 1.2 h.
Ethanolamine liquid cultures also contained 0.2 μM cyano-B12.
Anaerobic growth phenotype for strains grown on solid media after 72 h with 10 mM tetrathionate and the indicated carbon source as described in Materials and Methods. Symbols: +, wild-type growth; ±, slight growth; −, no visible growth.
Doubling times (in hours) calculated for the indicated strain grown anaerobically with 40 mM tetrathionate on the indicated carbon source in liquid culture as described in Materials and Methods.
A slight drop in optical density was seen with time.
FIG. 4.
Anaerobic growth on ethanolamine plus B12 or on propanediol with the electron acceptor tetrathionate (S4O6) or nitrate (NO3). Additions to NCE minimal medium were as follows: sodium tetrathionate (40 mM) or potassium nitrate (10 mM), ethanolamine (10 mM), and B12 (0.2 mM) (A and C) or propanediol (50 mM) (B and D). Growth was monitored on the basis of absorbance at 650 nm (A and B), viable cell counts (filled symbols in panels C and D), and microscopic counts (open symbols in panels C and D) using a Petroff-Hausser bacterial cell counter (C and D). The data for cells grown with S4O6 but no carbon source are replotted in graphs A and B (for turbidity) and C and D (for microscopic and viable cell counts).
Late in anaerobic growth on tetrathionate, a precipitate formed. To be sure that the OD measurements reflected cell growth and not this precipitate, we monitored the increase in viable cells (CFU) and the increase in visible cells (examined with a microscope). These measurements (Fig. 4C and D) generally reflected the OD measurements through most of the growth period but revealed some important differences. This precipitate was not observed during growth on nitrate.
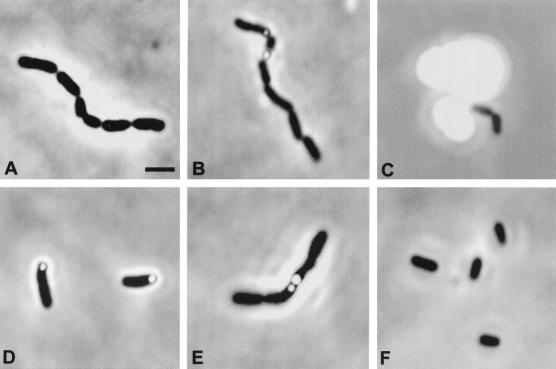
On ethanolamine, the OD increase for the first 40 h on tetrathionate reflected the increase in cell number or viable counts; thereafter, OD increased without a parallel increase in cell number. This late discrepancy was not seen during growth on NO3. Some of the late OD increase was due to a precipitate that was visualized with a microscope both as smaller refractile granules associated with 5 to 10% of the cells in the culture (Fig. 5B) and as larger, extracellular, granules (Fig. 5C). However, some of the OD increase reflected the accumulation of biomass, since cells grown on tetrathionate form long chains which are counted as single cells in both the microscopic and viable-cell enumerations (Fig. 5A and B). During growth on ethanolamine plus tetrathionate, 10 to 20% of the cell units are present as chains; these are short at the earlier stages of growth and increase to an average length of 10 cells (range, 6 to 14) as cells enter stationary phase. Cells in these chains appear more rounded and distinct than in typical rod-like cell filaments (with or without septa) formed by Salmonella during SOS induction. Thus, OD measurements of growth on ethanolamine with tetrathionate appear to overestimate cell number (because of the precipitate), and the cell counts underestimate biomass accumulation due to the presence of chains. The precipitate that is seen late in the growth experiment appears to be elemental sulfur (see below).
FIG. 5.
Cell chain and granule formation during growth of wild-type serovar Typhimurium on ethanolamine or propanediol plus tetrathionate. Cells were viewed on a Zeiss Axioplan phase-contrast microscope. The scale bar in panel A represents 2 μm, and all photos are at the magnification indicated in panel A. Photographs are from stationary-phase cultures grown as described in the legend to Fig. 4. Panels A to C show cells (A and B) and refractile granules (B and C) observed in an ethanolamine-tetrathionate culture after 44 h of incubation at 37°C. (D and E) Cells and refractile granules observed in propanediol-tetrathionate culture after 67 h of incubation at 37°C. (F) Cells grown on propanediol plus NO3. Neither cell chains nor refractile granules were seen in either ethanolamine- or propanediol-grown cultures when nitrate was the electron acceptor.
On propanediol, roughly 90% of the OD increase that is observed after 40 h of growth is not paralleled by an increase in cell number. The increase seems to be due to changes in absorption or refractivity of cells that are forming short chains (four to six cells long) and accumulating cell-associated refractile granules (see below and Fig. 5D and E). Approximately 80% of the cells in these cultures had associated refractile granules.
Neither cell chains nor refractile granules were seen during growth on ethanolamine or propanediol when nitrate was used as an electron acceptor. Cells grown on propanediol and nitrate are shown in Fig. 5F.
A surprising aspect of the data shown in Fig. 4 is the observation that cells growing with tetrathionate are dying (but not lysing) late in the growth period. This loss of viability was seen for all carbon sources whenever tetrathionate was used as the electron acceptor; it was not seen during growth on nitrate. The loss of viability is temporally associated with appearance of the small cell-associated refractile granules that appear to be within cells but that could be on their surface (Fig. 5B, D, and E).
In Fig. 4D it can be seen that tetrathionate provides only about a 10-fold increase in viable cell number on propanediol. This growth yield is increased severalfold when glutamate is added (data not shown). We propose that accumulated intermediates in the methyl-citrate pathway (for propanediol degradation) inhibit the TCA cycle, limiting synthesis of glutamate and related amino acids.
Nature of the granules appearing in tetrathionate-grown cultures.
A variety of bacteria are known to produce elemental sulfur granules during growth involving oxidation or reduction of sulfur compounds (36, 50). This suggested that Salmonella might deposit sulfur to form the granules that appear within and outside of cells during growth on tetrathionate. To test this, we assayed elemental sulfur by the method of Hazeu et al. (19) as described in Materials and Methods. The method detects sulfur that can be removed by centrifugation (inside or outside of cells); it does not detect dissolved sulfides. The assays revealed that, late during anaerobic growth on tetrathionate, elemental sulfur was accumulating in cells and as precipitable material to a concentration of about 1 mM (based on the volume of the culture).
Anaerobic respiration of ethanolamine and propanediol requires endogenous cobalamin.
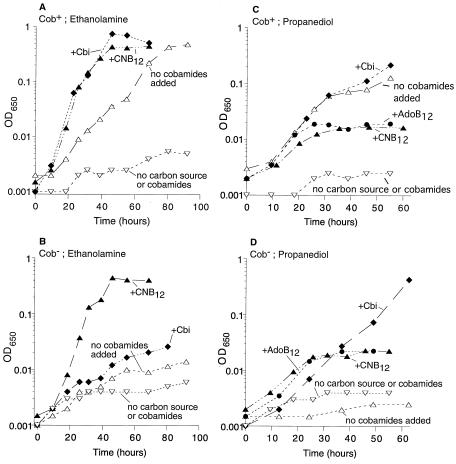
Tetrathionate appears to provide conditions that allow S. enterica serovar Typhimurium to use ethanolamine and propanediol under anaerobic conditions, the conditions under which cobalamin is synthesized. Mutants blocked in B12 synthesis were used to show that anaerobic growth on these carbon sources did, in fact, require endogenous B12. These experiments revealed some unexpected things.
Wild-type cells (Fig. 6A) grow anaerobically on ethanolamine plus tetrathionate without any added cobalamin. Growth rate is stimulated twofold by added cyano-B12 or by the intermediate cobinamide (Cbi). The limited growth on endogenous B12 may reflect the fact that the B12 synthetic operon is induced by propanediol but not by ethanolamine; thus, during growth on ethanolamine, B12 synthesis relies on a repressed cob operon. A mutant blocked prior to Cbi in B12 synthesis did not grow without added cobamides (Fig. 6B). Growth was stimulated by cyano-B12 but not by the intermediate Cbi. This probably reflects the fact that ethanolamine does not induce the cob operon (see below), and the cob mutation used (cbiD24) is a polar insertion that reduces the expression of genes required for conversion of Cbi to AdoB12. These growth experiments used OD650 to monitor growth; turbidity increases in these experiments, corresponded closely to microscopic cell counts.
FIG. 6.
Effect of cobamides on anaerobic growth on ethanolamine or propanediol. Wild-type serovar Typhimurium (A and C) and a mutant with an insertion in cbiD (Cob−) (B and D) were grown anaerobically on ethanolamine plus tetrathionate (A and B) or propanediol plus tetrathionate (C and D). Additions were as follows: cyano-B12 (0.2 μM in panels A, B, and D; 15 nM in panel C), cobinamide dicyanide (15 nM), and AdoB12 (15 nM).
On propanediol, which induces the cob operon, wild-type cells grew best without added B12 or with added Cbi (Fig. 6C). Surprisingly, their growth yield was inhibited by exogenous cyano-B12. Growth was monitored with cyano-B12 added to make final concentrations of 2, 15, and 200 nM. Inhibition is seen even at cyano-B12 levels (2 and 15 nM) insufficient to repress the cob operon (2), making it unlikely that B12 represses a function encoded by the cob operon and essential to propanediol catabolism (Fig. 6C). It seemed possible that excess added cyano-B12 might be inhibiting propanediol dehydratase (as seen in vitro), but added AdoB12 also reduced growth yield (Fig. 6C). A mutant blocked in cobalamin synthesis showed no growth on propanediol without cobalamin (Fig. 6D), and growth was fully restored by adding Cbi. Added cyano-B12 or AdoB12 corrected growth only to the level seen for inhibited wild-type cells (Fig. 6D).
All of the inhibitory effects noted above involve addition of forms of B12 with dimethyl benzimidazole (DMB) as the lower ligand. Recently, Keck and Renz reported that Salmonella cannot synthesize DMB anaerobically and makes pseudo-B12 (adenine as lower ligand) under these conditions (29). Based on this finding, the best growth seen here is supported by endogenous pseudo-B12. The growth inhibition data suggest that anaerobic propanediol utilization is inhibited by DMB-containing cofactors. Consistent with this possibility, addition of DMB inhibits anaerobic growth of wild-type cells on propanediol plus tetrathionate (data not shown). The strong stimulation by Cbi may reflect its conversion to pseudo-B12. This conversion may be less sensitive to the polarity effects that impaired use of Cbi during ethanolamine growth.
Defining the pathways of ethanolamine and propanediol respiration by mutant phenotypes.
The present view of the aerobic degradative pathways for ethanolamine and propanediol is outlined in Fig. 2. However, the anaerobic respiration of these compounds has not been investigated. Table 2 presents effects of various mutations on anaerobic growth using tetrathionate as an electron acceptor in solid and liquid media. The results on solid media are extremely clear—strong growth on tetrathionate and no visible growth seen for conditions indicated by a minus sign. In liquid medium, some increase in turbidity and cell number was measured for conditions that produced no growth on plates. The difference may reflect the observation that cells are dividing to form smaller and smaller cells under these starvation conditions and may do so with very little increase in biomass (dividing down). This increases turbidity and cell number but is not apparent on the plates.
Anaerobic growth on ethanolamine requires genes of the eut operon and the glyoxalate shunt (aceAB); this is also true for growth under aerobic conditions (Tom Fazzio, personal communication). The ack and pta genes (converting acetyl-CoA to acetate and producing ATP) are required for aerobic use of ethanolamine (Tom Fazzio, personal communication). However under anaerobic conditions with tetrathionate, an ack mutation caused only a partial loss of growth ability (Table 2), suggesting that the ack and pta genes are not absolutely required anaerobically.
Anaerobic growth on propanediol requires enzymes encoded by the pdu operon (which convert propanediol to propionyl-CoA) and some proteins from the prp operon, presumably those that convert propionyl-CoA to succinate plus pyruvate (23, 24, 57). A deletion of the entire prp operon (prp-54) eliminated growth on propanediol but made cells sensitive to growth inhibition by propionate on other carbon sources (J. Tittensor, unpublished results); we suspect that this is due to accumulated propionyl-CoA, which has previously been seen to inhibit growth (59). Thus, the failure of prp mutants to use propanediol could be due to lack of the prp pathway or to inhibition by accumulated propionyl-CoA. The glyoxalate shunt (aceA and aceB genes) is not required for use of propanediol.
Mutants unable to reduce tetrathionate (ttr [described below]) cannot use tetrathionate to support growth on any of the tested carbon sources whose utilization requires an electron acceptor (Table 2). Growth is also eliminated by oxrA (fnr) mutations, since this regulator is required for induction of the ttr operon (see below) (21). The pathway for tetrathionate reduction, illustrated in Fig. 1, suggested a requirement for subsequent steps in sulfur reduction (phs, asr). These subsequent steps are not required under the conditions tested here, since a phs insertion mutant grows normally.
Mutations (ttr) causing a defect in tetrathionate reduction.
The results below confirm and support those of Hensel et al. (21, 22), which were reported while this work was in progress. A large set of mutants unable to reduce tetrathionate was isolated by using MudJ, Tn10dTc, Tn10dCm, and TPOP elements (see Materials and Methods). These mutations all affected a single region (ttr) whose chromosomal position was confirmed by transductional linkage to markers near the previously determined position of ttr mutants (9). The region includes an operon of three structural genes for enzymes, all of which are required for tetrathionate reduction, and two genes that are essential to expression of the three-gene operon. These regulatory proteins are homologous with proteins that are part of known two-component regulatory systems (21). Our mutant set included insertions in all genes except the small ttrR gene, for which a deletion was constructed as described below. All sequenced insertion mutations are described in GenBank under accession number AF282268. Insertions near the ttr operon were isolated to obtain mutations in immediately adjacent open reading frames; the ttr+ phenotypes of the rkh and nth mutants ensured that these adjacent genes are not essential to tetrathionate reduction.
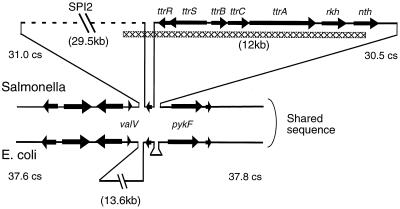
In Fig. 7, a map of the Salmonella ttr region is compared to the analogous region of the E. coli chromosome, which lacks ttr. It should be noted that the Salmonella ttr operon is part of a larger block of genes absent from E. coli and is near another such region (SPI2) that is unique to Salmonella. A small open reading frame shared by both S. enterica and E. coli is located between these two regions, suggesting that distinct genetic events account for the presence of the flanking sequence blocks.
FIG. 7.
Map of the Sallmonella ttr region and analogous region of the E. coli chromosome. Regions present in only one genome are represented as raised triangles. The map is not to scale; the sizes of various fragments, in kilobases, are shown in parentheses. The SPI2 region is represented by a dashed line to indicate its foreign evolutionary origin. Genes are represented by arrows that point in the direction of transcription. The hatched box represents the region of the Salmonella chromosome that was sequenced during the course of this work.
Global regulation of the ttr operon.
By use of fusions to the lac operon formed by ttr::MudJ insertions, conditions for induction of the operon were tested. Results in Tables 3 and 4 show that, under aerobic conditions, tetrathionate causes a slight induction (5- to 10-fold), but its major inductive effect (100- to 900-fold) occurs anaerobically. The requirement for anaerobic conditions is mediated by the OxrA protein, homologous to Fnr of E. coli (16). An insertion in the oxrA gene eliminated anaerobic induction of a ttr::MudJ fusion by tetrathionate (Table 3) and prevented anaerobic growth on ethanolamine and propanediol (Table 2). Neither the ArcA global regulator (also responding to anaerobic conditions) nor the Crp-cyclic AMP system (responding to carbon starvation) affected ttr operon induction.
TABLE 3.
Global control of ttr expression
| Strain | Relevant mutationa | β-Galactosidase levels in ttr-lac fusion strains grown as indicatedb
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Aerobic
|
Anaerobic
|
||||||||||
| Glucose
|
Glycerol
|
Glucose
|
Glycerol
|
||||||||
| Alone | + tetrathionate | Alone | + tetrathionate | Alone | + nitrate | + tetrathionate | + nitrate and tetrathionate | + nitrate | + nitrate and tetrathionate | ||
| TT18691 | ttrB1221::MudJ | 2 | 20 | 4 | 34 | 7 | 4 | 1,729 | 310 | 5 | 1,010 |
| TT20444 | phs-213::MudJ | 2 | 25 | 4 | 63 | 9 | 5 | 2,505 | 559 | 4 | 1,298 |
| TT20590 | arcA201::Tn10dTc | 3 | 24 | 3 | 46 | 4 | 3 | 1,530 | 240 | 3 | 628 |
| TT20464 | cya-961::Tn10 | 3 | 20 | 4 | 42 | 6 | 6 | 1,578 | 414 | 4 | 952 |
| TT20469 | crp-773::Tn10 | 3 | 21 | 4 | 45 | 5 | 5 | 1,496 | 573 | 4 | 1,152 |
| TT18694 | ttrB123::Mud-lac | 1 | 20 | 4 | 44 | 6 | 3 | 1,775 | 376 | 5 | 863 |
| TT22347 | oxrA2::Tn10 (fnr) | 2 | 24 | 3 | 37 | 4 | 6 | 52 | 46 | NTc | NT |
The first six strains carry the ttrB122::MudJ insertion, which fuses transcription of lac operon to the ttr promoter. The last two strains carry ttrB123::MudJ; this insertion lies 123 bases downstream of ttrB122::MudJ and also fuses transcription of the lac operon to the ttr promoter.
The level of β-galactosidase activity is presented in units defined by Miller (37). Carbon source concentrations and growth conditions are as described in Materials and Methods; nitrate was used at 20 mM, and tetrathionate was used at 10 mM.
NT, not tested.
TABLE 4.
Inducers of ttr operon transcription
| Growth conditions | β-Galactosidase levels in a ttr-lac fusion strain grown in minimal medium with the following additiona
|
|||||
|---|---|---|---|---|---|---|
| None | Sulfite (1 mM) | Sulfite (10 mM) | Thiosulfate (6 mM) | Thiosulfate (18 mM) | Tetrathionate (10 mM) | |
| Glucose + O2 | 2 | 2 | 2 | 4 | 5 | 20 |
| Glycerol + O2 | 3 | 3 | NT | 3 | 25 | 34 |
| Glucose − O2 | 5 | 7 | 92 | 55 | 15 | 1,729 |
The level of β-galactosidase activity is presented in units defined by Miller (37). The strain tested (TT18691) carries the insertion ttr-122::MudJ, which fuses lac operon transcription to the ttr promoter. Sulfite, thiosulfate, and tetrathionate were used as sodium salts at the concentrations indicated. NT, not tested.
When Salmonella grows in anaerobic nitrate medium, synthesis of other anaerobic respiratory enzymes is transcriptionally down-regulated by a pair of two-component sensor-response regulatory systems—NarX-NarL and NarQ-NarP. This regulation has been studied extensively and is reviewed in references 53, 54, and 55. Nitrate reduced ttr operon expression three- to fivefold (Table 3). For other promoters regulated by NarL-NarP, variable numbers of binding sites [the heptad repeat: TAC(c/t)N(a/c)T] are found within the first 200 bases upstream of the transcriptional start sites (55, 58). A search of the ttr control region (between the divergent ttrS and ttrB genes) revealed none of these sequence elements. Several poor matches are located near the region of overlap between the regulatory genes ttrR and ttrS, a region not thought to have promoters (see below). It is unclear how ttr operon transcription is down-regulated by nitrate. The small effect of nitrate on ttr expression may suggest that tetrathionate serves as an electron acceptor whose quality is comparable to that of nitrate.
Proximal inducers of the ttr operon.
Tetrathionate was the most effective inducer of the compounds tested (Table 4). The inducer seems to be tetrathionate per se, rather than its reduction products, because the strains used in these tests carry a ttr::MudJ insertion and are unable to reduce tetrathionate. Furthermore, mutations in the phs genes, which prevent further reduction of thiosulfate (Fig. 1), had little effect on induction by tetrathionate (Table 3). Sulfite (SO32−) and thiosulfate (S2O32−) caused very little induction, even at high concentrations (Table 4); the small effects seen could reflect internal production of tetrathionate.
Mechanism of proximal ttr control.
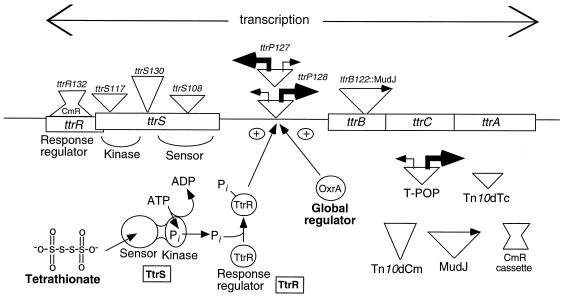
The two regulatory genes (ttrS and ttrR), which overlap by 26 bases (see Fig. 8), encode proteins similar to those of two-component regulatory systems (40) and are responsible for the tetrathionate-specific regulation of the ttr operon (21). The TtrS protein resembles sensor kinases, which (in other systems) act to phosphorylate another protein in response to a regulatory stimulus. The TtrR protein resembles these responsive regulatory proteins. Alignment of the TtrS sequence with that of known sensor kinases reveals two functional domains. The C-terminal sensory kinase domain (residue 325 to the end) includes all of the motifs required for autokinase activity (N box, G1 box, F box, and G2 box). The N-terminal domain of TtrS (residues 1 to 358) shows little homology to any gene in GenBank but contains the H box (autophosphorylation site) and is most likely involved in sensing tetrathionate. To examine the role of these proteins, we assayed the effects of mutations in the ttrR, ttrS, and promoter regions on transcription of the structural genes (ttrBCA). Results are shown in Table 5; normal regulation is shown in lines 1 and 10. These data support the model presented by Hensel et al. (21) and outlined in Fig. 8.
FIG. 8.
Proximal regulation of the ttrBCA operon. The key to inserted elements is shown on the lower right. In the model presented below the map, tetrathionate is sensed by the TtrS protein, which autophosphorylates and then transfers the phosphate group to activate TtrR. Activated TtrR cooperates with the global regulator OxrA (Fnr) to positively regulate expression of the ttrBCA operon.
TABLE 5.
Mutations that affect proximal regulation of the ttr operon
| Strain | β-Galactosidase level in cells grown as indicatedb
|
|||||
|---|---|---|---|---|---|---|
| Relevant genotypea
|
Aerobic
|
Anaerobic
|
||||
| ttr mutation testedd | lac fusion | Medium alone
|
Medium plus tetrathionate
|
Medium alone
|
Medium plus tetrathionate
|
|
| − Tc (+Tc) | − Tc (+Tc) | − Tc (+Tc) | − Tc (+Tc) | |||
| TT18694 | ttrB123::MudJ | A | 1 | 23 | 3 | 1,775 |
| TT22471 | ttrR132(del)::Cm(swap) | A | 3 | 3 | 3 | 4 |
| TT20440 | ttrS117::Tn10dTc | A | 1 | 2 | 6 | 12 |
| TT20433 | ttrS108::Tn10dTc | A | 1 | 2 | 7 | 7 |
| TT22348 | ttrS130::Tn10dCm | A | 2 | 2 | 815 | 297 |
| TT22350 | ttrS130::Tn10dCm ttrS108::Tn10dTc | A | 2 | 2 | 325 | 106 |
| TT22352 | ttrS130::Tn10dCm ttrS117::Tn10dTc | A | 2 | 2 | 5 | 5 |
| TT22467 | ttrS130::Tn10dCm oxrA2::Tn10 | A | 3 | 3 | 19 | 11 |
| TT22373 | ttrS133(del)::Cm(swap)c | A | 4 | 4 | 1,545 | 141 |
| TT18683 | ttrA121::MudJ | B | 3 | 25 | 13 | 1,734 |
| TT22356 | ttrP128::Tn10dTPOP2 | B | 34 (1,438) | 10 (1,455) | 17 (724) | 12 (1,676) |
| TT22354 | ttrP127::Tn10dTPOP2 | B | 9 (216) | 6 (139) | 10 (51) | 8 (163) |
All strains carried a ttr::MudJ insertion that fuses lac transcription to the ttrBCA promoter. Strains designated as A carry ttrB123::MudJ; those designated B carry ttrA121::MudJ.
The level of β-galactosidase activity is presented in units defined by Miller (37). Enzyme levels in parentheses were observed in cells grown in the presence of the antibiotic tetracycline (Tc), which induces promoters within the TPOP transposon.
All of the coding sequence unique to ttrS was removed and replaced by the chloramphenicol resistance gene and distal Tn10dCm transposon, followed by the 10 bases adjacent to the distal end of this Tn10dCm element in strain ttrS130::Tn10dCm.
Each of the ttr control mutations was tested for its effect on anaerobic acid production and growth in strains carrying a wild-type ttrBCA operon. The wild-type strain (LT2), as well as TT20369 (ttrS130::Tn10dCm), TT22349 (ttrS108::Tn10dTc ttrS130::Tn10dCm), and TT22372 [ttrS133(del)::Cm(swap)], formed red patches (produced acid) when grown on MacConkey-tetrathionate (10 mM) medium. All other strains formed white patches. Strains with an insertion in the promoter region (ttrP128::Tn10dTPOP2 or ttrP127::Tn10dTPOP2) formed red patches when grown on this medium in the presence of the antibiotic tetracycline (Tc), which induces promoters within the TPOP transposon. Growth was tested anaerobically on solid medium with propanediol or ethanolamine as the sole carbon source and 10 mM tetrathionate; LT2, as well as TT20369 (ttrS130::Tn10dCm), TT22349 (ttrS108::Tn10dTc, ttrS130::Tn10dCm), and TT22372 [ttrS133(del)::Cm(swap)], produced visible patches on this media. All other strains failed to grow under these conditions. One of the strains with an insertion in the promoter region (ttrP128::Tn10dTPOP) produced a visible patch when grown on this medium in the presence of tetracycline.
Since no ttrR mutants emerged during our search for mutants, a deletion was constructed which removes almost all of the ttrR gene but retains all ttrS sequences (see Materials and Methods). This mutant is unable to induce operon expression, suggesting that the response regulator works as a positive effector for transcription (Table 5, line 2). A similar conclusion was reached by Hensel et al. on the basis of very different experiments (21).
Insertions of Tn10dTc at either of two sites within the ttrS gene (Fig. 8) prevent operon induction and cause a complete Ttr− growth phenotype (Table 5, lines 3 and 4 and footnote d). One sensor kinase insertion (ttrS117) (Table 5, line 3) disrupts the downstream kinase domain (close to the conserved G2 box); the other (ttrS108) (Table 5, line 4) is located in the upstream sensor domain (Fig. 8).
Surprisingly, a Tn10dCm insertion (ttrS130) in the middle of the ttrS gene caused constitutive operon expression (independent of tetrathionate), and cells remained Ttr+ (Table 5, line 5 and footnote d; Fig. 8). This insertion is upstream of the kinase domain; it appears to cause tetrathionate-independent induction of the ttrBCA operon via TtrR, since expression requires anaerobic conditions and OxrA protein (Table 5, line 8). The constitutive phenotype is independent of the sensor domain since expression is unaffected by adding the upstream sensor insertion (ttrS108 in Table 5, line 6). A double mutant with the constitutive Tn10dCm element and the downstream Tn10dTc insertion, in the kinase domain, showed no operon expression (Table 5, line 7).
These results could be explained by a promoter within the Tn10dCm element that expresses a shorter kinase domain, which can activate TtrR without tetrathionate; alternatively, this promoter might transcribe ttrR, and the excess TtrR protein could be nonspecifically phosphorylated or could induce transcription without phosphorylation. Consistent with the idea of a promoter within the inserted material, the chloramphenicol resistance gene within the element is transcribed toward ttrR, and outward transcription from Tn10dCm elements has been seen in several operons. To determine whether the kinase domain of TtrS is essential for this constitutive phenotype, the entire ttrS gene was deleted and replaced with the Tn10dCm sequences likely to include a promoter. The insertion included the chloramphenicol resistance determinant, the downstream region of the Tn10dCm element, and 10 bases from the ttrS gene immediately distal to insertion ttrS130. This inserted material (derived from ttrS130::Tn10dCm) resulted in a better constitutive phenotype without the ttrS (kinase domain) than that seen for the parent ttrS130::Tn10dCm element in the presence of this domain (Table 5, compare lines 5 and 9). This rules out dependence on the kinase domain and supports the idea that constitutive expression is due to overproduction of the TtrR protein. It is not understood why tetrathionate reduces expression of the ttrBCA operon in all of the strains carrying insertion ttrS130::Tn10dCm or parts thereof. The effect does not seem to depend on the sensor domain of TtrS, since it is seen in the swap strain lacking almost all ttrS coding sequence and in strains with an upstream ttrS insertion.
Two TPOP2 insertions within the regulatory region were found to be in opposite orientations at the same site 20 bp upstream of the ttrBCA operon transcriptional start site. The two ends of TPOP have outward-directed, tetracycline-inducible promoters of different strengths (44). The insertion that directs the stronger (tetA) promoter toward the structural genes, when induced by tetracycline, can provide a Ttr+ phenotype as judged by both the acid production (on MacConkey medium) and anaerobic growth tests (Table 5, line 11 and footnote d). Induction of the other insertion, which directs the 10-fold-weaker (tetR) promoter across the ttr operon, provides acid production from tetrathionate but not anaerobic growth on ethanolamine or propanediol (Table 5, line 12 and footnote d). As expected, expression of ttrBCA in these two insertion mutants requires neither the global regulatory input (OxrA) nor a proximal inducer; induction is seen both aerobically and anaerobically. This demonstrates that tetrathionate use requires only expression of the ttrBCA genes; the two-component regulatory system does not appear to activate any unlinked genes required for use of tetrathionate.
DISCUSSION
The alternative electron acceptor tetrathionate allows Salmonella to grow anaerobically on ethanolamine or propanediol by using endogenously synthesized B12. These are the only conditions we know under which wild-type Salmonella requires B12 for growth. Almost 2% of the Salmonella genome (88 genes) is dedicated to the metabolism discussed here. Synthesis and import of B12 requires at least 30 genes; the eut, the propanediol (pdu), and the proprionate (prp) operons contain 17, 23, and 5 genes, respectively. Operons for sulfur reduction are ttr (five genes), phs (five genes), and asr (three genes). We presume that the natural environment of Salmonella must frequently include anaerobic conditions with tetrathionate, ethanolamine, and/or propanediol; these conditions select for maintenance of the B12 synthesis (cob) genes. The importance of B12 to propanediol use is supported by the fact that propanediol induces the cob operon (7, 45). This complex of abilities is found in virtually every Salmonella isolate; most are absent from E. coli. Aspects of this complex are used to enrich for and identify salmonellae in natural isolates and distinguish them from E. coli (8, 21, 39, 41, 43). The metabolism described here appears to be an important aspect of a Salmonella-specific lifestyle.
The mechanism of tetrathionate reduction has not been studied extensively in Salmonella (4, 15). By analogy with similar systems in other bacteria, we presume that reduction of tetrathionate can support electron transport and generate a proton gradient. Salmonella can grow anaerobically on tetrathionate plus glycerol, and use of glycerol as a carbon source is known to require electron transport (15). Furthermore, tetrathionate allows anaerobic growth on acetate, the catabolism of which provides no means of substrate-linked phosphorylation. Surprisingly, tetrathionate can serve as an electron acceptor even in strains with mutations in synthesis of both ubiquinone and menaquinone (Tom Fazzio, personal communication), suggesting that tetrathionate reductase may accept electrons directly from NADH or FADH2. It is not clear why tetrathionate should be superior to nitrate in supporting anaerobic growth on ethanolamine or propanediol or acetate.
Cell chains and lethality were noted at late stages of growth on tetrathionate. We suspect that these phenomena reflect toxic effects of thiosulfate, sulfite, or sulfide in the medium or sulfur accumulation within cells. Salmonella possesses the phs and asr systems, which can reduce thiosulfate completely to sulfide. A mutational block in the first step (phs) neither relieves nor exacerbates the toxicity. However, the effects of such mutants may be masked by the assimilatory thiosulfate reductase (CysM), which converts thiosulfate to sulfite, or by rhodanese, which can, in principle, convert thiosulfate to sulfite plus sulfide. While little work has been done on rhodanese in Salmonella, the activity has been reported in E. coli (18), and genomes of both E. coli and serovar Typhimurium include rhodanese homologues. Under natural conditions, toxic accumulations might be diluted more than was possible in the growth tubes used for these experiments.
A surprising aspect of anaerobic growth on tetrathionate was the appearance of sulfur granules. We presume that the sulfur granules are generated by the chemical reaction of sulfide ions with tetrathionate, which has been described previously (30). It is not known whether Salmonella enzymes contribute to this process. However, serovar Typhimurium (but not E. coli) can reduce mineral sulfur, and Salmonella mutants are known which fail in this (K. Nealson and D. Lies, personal communication; M. Price-Carter, unpublished results).
Most of the activities mentioned here are found in Salmonella but not in E. coli. (The ethanolamine operon is shared by both species.) We suggest that the Salmonella pattern evolved by acquisition of foreign genes and mutational loss of ancestral genes. The ability to synthesize B12 and catabolize propanediol was acquired by Salmonella (but not E. coli) as a single DNA fragment from an organism having a guanosine-plus-cytosine content and codon usage atypical for Salmonella (47). This occurred about 70 million years ago, perhaps 30 million years after the divergence of salmonellae and E. coli (33–35). In contrast, the ttr operon was probably carried by the common ancestor of E. coli and Salmonella and unilaterally lost from the E. coli lineage in the course of their divergence. The guanosine-plus-cytosine content and codon usage of ttr are typical of ancestral genes shared by Salmonella and E. coli (21). Tetrathionate reduction is found in many other enteric bacteria, suggesting that it was present in the common ancestor of enteric lineages (11). The phs and asr genes also seem likely to be ancestral genes lost from the E. coli lineage. The pathogenicity island SPI2, close to the ttr operon on the Salmonella chromosome (Fig. 7), is clearly of foreign origin and appears to have been added to the genome of Salmonella but not to that of E. coli (22). The evolution of the genes described here exemplifies the divergence of Salmonella and E. coli by genomic flux—lineage-specific events of gene loss and acquisition (35).
We thank Eric Kofoid for insightful comments and for constructing the ttrR deletion, Edward King for taking the pictures of the bacterial cell chains and granules, David Blair for generously sharing his phase-contrast microscope, and Renee Dawson for carefully reading the manuscript.
ACKNOWLEDGMENTS
This work was supported in part by National Institutes of Health grants GM34804 and GM59486.
REFERENCES
- 1.Ailion M, Bobik T A, Roth J R. Two global regulatory systems (Crp and Arc) control the cobalamin/propanediol regulon of Salmonella typhimurium. J Bacteriol. 1993;175:7200–7208. doi: 10.1128/jb.175.22.7200-7208.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ailion M, Roth J R. Repression of the cob operon of Salmonella typhimurium by adenosylcobalamin is influenced by mutations in the pdu operon. J Bacteriol. 1997;179:6084–6091. doi: 10.1128/jb.179.19.6084-6091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson D I, Roth J R. Mutations affecting regulation of cobinamide biosynthesis in Salmonella typhimurium. J Bacteriol. 1989;171:6726–6733. doi: 10.1128/jb.171.12.6726-6733.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett E, Clark M. Tetrathionate reduction and production of hydrogen sulfide from thiosulfate. Microbiol Rev. 1987;51:192–205. doi: 10.1128/mr.51.2.192-205.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobik T, Havemann G, Busch R, Williams D, Aldrich H. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bobik T, Xu Y, Jeter R, Otto K, Roth J R. Propanediol utilization genes (pdu) of Salmonella typhimurium: three genes for the propanediol dehydratase. J Bacteriol. 1997;179:6633–6639. doi: 10.1128/jb.179.21.6633-6639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bobik T A, Ailion M, Roth J R. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J Bacteriol. 1992;174:2253–2266. doi: 10.1128/jb.174.7.2253-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bopp C A, Brenner F W, Wells J G, Strockbine N A. : P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. Salmonella: description of the genus pp. 467–474. [Google Scholar]
- 9.Casse F M, Pascal M-C, Chippaux M. A mutant of Salmonella typhimurium deficient in tetrathionate reductase activity. Mol Gen Genet. 1972;119:71–74. doi: 10.1007/BF00270446. [DOI] [PubMed] [Google Scholar]
- 10.Chen P, Ailion M, Bobik T, Stormo G, Roth J. Five promoters integrate control of the cob/pdu regulon in Salmonella typhimurium. J Bacteriol. 1995;177:5401–5410. doi: 10.1128/jb.177.19.5401-5410.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark M, Barrett E. The phs gene and hydrogen sulfide production by Salmonella typhimurium. J Bacteriol. 1987;169:2391–2397. doi: 10.1128/jb.169.6.2391-2397.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko K A, Wanner B L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis R W, Botstein D, Roth J R. A manual for genetic engineering. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 14.Difco Laboratories. Difco manual of dehydrated culture media and reagents for microbiological and clinical laboratory procedures. Detroit, Mich: Difco Laboratories; 1965. [Google Scholar]
- 15.Gennis R B, Stewart V. Respiration. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichi coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 217–261. [Google Scholar]
- 16.Green J, Guest J R. The citric acid cycle and oxygen-regulated gene expression in Escherichia coli. In: Busby S J, Thomas C M, Brown N L, editors. Molecular microbiology. Berlin, Germany: Springer-Verlag; 1998. pp. 28–37. [Google Scholar]
- 17.Hallenbeck P, Clark M, Barrett E. Characterization of anaerobic sulfite reduction by Salmonella typhimurium and purification of the anaerobically induced sulfite reductase. J Bacteriol. 1989;171:3008–3015. doi: 10.1128/jb.171.6.3008-3015.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hama H, Kayahara T, Ogawa W, Tsuda M, Tsuchiya T. Enhancement of serine-sensitivity by a gene encoding rhodanese-like protein in Escherichia coli. J Biochem (Tokyo) 1994;115:1135–1140. doi: 10.1093/oxfordjournals.jbchem.a124469. [DOI] [PubMed] [Google Scholar]
- 19.Hazeu W, Bijleveld W, Grotenhuis J T, Kakes E, Kuenen J G. Kinetics and energetics of reduced sulfur oxidation by chemostat cultures of Thiobacillus ferrooxidans. Antonie Leeuwenhoek. 1986;52:507–518. doi: 10.1007/BF00423411. [DOI] [PubMed] [Google Scholar]
- 20.Heinzinger N, Fujimoto S, Clark M, Moreno M, Barrett E. Sequence analysis of the phs operon in Salmonella typhimurium and the contribution of thiosulfate reduction to anaerobic energy metabolism. J Bacteriol. 1995;177:2813–2820. doi: 10.1128/jb.177.10.2813-2820.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hensel M, Hinsley A, Nikolaus T, Sawers G, Berks B. The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol Microbiol. 1999;32:275–287. doi: 10.1046/j.1365-2958.1999.01345.x. [DOI] [PubMed] [Google Scholar]
- 22.Hensel M, Nikolaus T, Egelseer C. Molecular and functional analysis indicates a mosaic structure of Salmonella pathogenicity island 2. Mol Microbiol. 1999;31:489–498. doi: 10.1046/j.1365-2958.1999.01190.x. [DOI] [PubMed] [Google Scholar]
- 23.Horswill A, Escalante-Semerena J. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J Bacteriol. 1997;179:928–940. doi: 10.1128/jb.179.3.928-940.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horswill A, Escalante-Semerena J. Salmonella typhimurium LT2 catabolizes propionate via the 2-methyl citric acid cycle. J Bacteriol. 1999;181:5615–5623. doi: 10.1128/jb.181.18.5615-5623.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang C, Barrett E. Identification and cloning of genes involved in anaerobic sulfite reduction by Salmonella typhimurium. J Bacteriol. 1990;172:4100–4102. doi: 10.1128/jb.172.7.4100-4102.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C, Barrett E. Sequence analysis and expression of the Salmonella typhimurium asr operon encoding production of hydrogen sulfide from sulfite. J Bacteriol. 1991;173:1544–1553. doi: 10.1128/jb.173.4.1544-1553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeter R, Olivera B M, Roth J R. Salmonella typhimurium synthesizes cobalamin (vitamin B12) de novo under anaerobic growth conditions. J Bacteriol. 1984;159:206–216. doi: 10.1128/jb.159.1.206-213.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeter R M. Cobalamin dependent 1,2-propanediol utilization by Salmonella typhimurium. J Gen Microbiol. 1990;136:887–896. doi: 10.1099/00221287-136-5-887. [DOI] [PubMed] [Google Scholar]
- 29.Keck B, Renz P. Salmonella typhimurium forms adenylcobamide and 2-methyladenylcobamide, but no detectable cobalamin during strictly anaerobic growth. Arch Microbiol. 2000;173:76–77. doi: 10.1007/s002030050011. [DOI] [PubMed] [Google Scholar]
- 30.Klimmek O, Kroeger A, Steudel R, Holdt G. Growth of Wolinella succinogenes with polysulfide as terminal acceptor of phosphorylative electron transport. Arch Microbiol. 1991;155:177–182. [Google Scholar]
- 31.Kofoid E, Rappleye C, Stojiljkovic I, Roth J R. The seventeen-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologs of carboxysome shell proteins. J Bacteriol. 1999;181:5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kredich N. Biosynthesis of cysteine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 514–527. [Google Scholar]
- 33.Lawrence J G, Roth J R. The cobalamin (coenzyme B12) biosynthetic genes of Escherichia coli. J Bacteriol. 1995;177:6371–6380. doi: 10.1128/jb.177.22.6371-6380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrence J G, Roth J R. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and reacquisition of a multigene complex. Genetics. 1995;142:11–24. doi: 10.1093/genetics/142.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawrence J G, Roth J R. Genomic flux: genome evolution by gene loss and acquisition. In: Charlebois R L, editor. Organization of the prokaryotic genome. Washington, D.C.: American Society for Microbiology; 1999. pp. 263–289. [Google Scholar]
- 36.Le Faou A, Rajagopal B, Daniels L, Fauque G. Thiosulfate, polythionates and elemental sulfur assimilation and reduction in the bacterial world. FEMS Microbiol Rev. 1990;75:351–382. doi: 10.1111/j.1574-6968.1990.tb04107.x. [DOI] [PubMed] [Google Scholar]
- 37.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 38.Murphy K. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papavassiliou J, Samaraki-Lyberopoulou V, Piperakis G. Production of tetrathionate reductase by Salmonella. Can J Microbiol. 1969;15:238–240. doi: 10.1139/m69-041. [DOI] [PubMed] [Google Scholar]
- 40.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 41.Patil D M, Parhad N M. Growth of salmonellas in different enrichment media. J Appl Bacteriol. 1986;61:19–24. doi: 10.1111/j.1365-2672.1986.tb03754.x. [DOI] [PubMed] [Google Scholar]
- 42.Poteete A, Fenton A. Lambda rec-dependent growth and recombination of phage P22. Virology. 1984;134:161–167. doi: 10.1016/0042-6822(84)90281-2. [DOI] [PubMed] [Google Scholar]
- 43.Rambach A. New plate medium for facilitated differentiation of Salmonella spp. from Proteus spp. and other enteric bacteria. Appl Environ Microbiol. 1990;56:301–303. doi: 10.1128/aem.56.1.301-303.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rappleye C, Roth J R. A Tn10 derivative (T-POP) for isolation of insertions with conditional (tetracycline-dependent) phenotypes. J Bacteriol. 1997;179:5827–5834. doi: 10.1128/jb.179.18.5827-5834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rondon R M, Escalante-Semerena J C. The poc locus is required for 1,2-propanediol-dependent transcription of the cobalamin biosynthetic (cob) and propanediol utilization (pdu) genes of Salmonella typhimurium. J Bacteriol. 1992;174:2267–2272. doi: 10.1128/jb.174.7.2267-2272.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roof D M, Roth J R. Ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1988;170:3855–3863. doi: 10.1128/jb.170.9.3855-3863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roth J R, Lawrence J G, Bobik T A. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 48.Roth J R, Lawrence J G, Rubenfield M, Kieffer-Higgins S, Church G M. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J Bacteriol. 1993;175:3303–3316. doi: 10.1128/jb.175.11.3303-3316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schlegel H. General microbiology. 7th ed. Cambridge, England: Cambridge University Press; 1992. [Google Scholar]
- 51.Schmieger H. A method for detection of phage mutants with altered transducing ability. Mol Gen Genet. 1971;110:378–381. doi: 10.1007/BF00438281. [DOI] [PubMed] [Google Scholar]
- 52.Starkey R. Relations of microorganisms to transformations of sulfur in soils. Soil Sci. 1950;70:55–65. [Google Scholar]
- 53.Stewart V. Bacterial two-component regulatory systems. NATO ASI (Adv Sci Inst) Ser Ser H Cell Biol. 1998;103:141–158. [Google Scholar]
- 54.Stewart V. Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli. Mol Microbiol. 1993;9:425–434. doi: 10.1111/j.1365-2958.1993.tb01704.x. [DOI] [PubMed] [Google Scholar]
- 55.Stewart V, Rabin R S. Dual sensors and dual response regulators interact to control nitrate- and nitrite-responsive gene expression in Escherichia coli. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington D.C.: American Society for Microbiology; 1995. pp. 233–252. [Google Scholar]
- 56.Textor S, Wendisch V, De Graff A, Müller U, Linder M, Linder D, Buckel W. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch Microbiol. 1997;168:428–436. doi: 10.1007/s002030050518. [DOI] [PubMed] [Google Scholar]
- 57.Tsang A W, Horswill A R, Escalante-Semerena J C. Studies of regulation of expression of the propionate (prpBCDE) operon provide insights into how Salmonella typhimurium LT2 integrates its 1,2-propanediol and propionate catabolic pathways. J Bacteriol. 1998;180:6511–6518. doi: 10.1128/jb.180.24.6511-6518.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tyson K L, Bell A I, Cole J A, Busby S J W. Definition of nitrite and nitrate response elements at the anaerobically inducible Escherichia coli nirB promoter: interactions between FNR and NarL. Mol Microbiol. 1993;7:151–157. doi: 10.1111/j.1365-2958.1993.tb01106.x. [DOI] [PubMed] [Google Scholar]
- 59.Van Dyk T, LaRossa R. Involvement of ack-pta operon products in α-ketobutyrate toxicity in Salmonella typhimurium. Mol Gen Genet. 1987;207:435–440. doi: 10.1007/BF00331612. [DOI] [PubMed] [Google Scholar]
- 60.Walter D, Ailion M, Roth J R. Genetic characterization of the pdu operon: use of 1,2 propanediol in Salmonella typhimurium. J Bacteriol. 1996;179:1013–1022. doi: 10.1128/jb.179.4.1013-1022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu D, Ellis H M, Lee E C, Jenkins N A, Copeland N G, Court D L. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]