Abstract
IS1, the smallest active transposable element in bacteria, encodes a transposase that promotes inter- and intramolecular transposition. Host-encoded factors, e.g., histone-like proteins HU and integration host factor (IHF), are involved in the transposition reactions of some bacterial transposable elements. Host factors involved in the IS1 transposition reaction, however, are not known. We show that a plasmid with an IS1 derivative that efficiently produces transposase did not generate miniplasmids, the products of intramolecular transposition, in mutants deficient in a nucleoid-associated DNA-binding protein, H-NS, but did generate them in mutants deficient in histone-like proteins HU, IHF, Fis, and StpA. Nor did IS1 transpose intermolecularly to the target plasmid in the H-NS-deficient mutant. The hns mutation did not affect transcription from the indigenous promoter of IS1 for the expression of the transposase gene. These findings show that transpositional recombination mediated by IS1 requires H-NS but does not require the HU, IHF, Fis, or StpA protein in vivo. Gel retardation assays of restriction fragments of IS1-carrying plasmid DNA showed that no sites were bound preferentially by H-NS within the IS1 sequence. The central domain of H-NS, which is involved in dimerization and/or oligomerization of the H-NS protein, was important for the intramolecular transposition of IS1, but the N- and C-terminal domains, which are involved in the repression of certain genes and DNA binding, respectively, were not. The SOS response induced by the IS1 transposase was absent in the H-NS-deficient mutant strain but was present in the wild-type strain. We discuss the possibility that H-NS promotes the formation of an active IS1 DNA-transposase complex in which the IS1 ends are cleaved to initiate transpositional recombination through interaction with IS1 transposase.
IS1 is an insertion element present in chromosomes and plasmids of enteric bacteria (for a review, see reference 38). IS1 (768 bp long) carries imperfect terminal inverted repeats (IRL and IRR) that are about 30 bp long (17, 40). IS1 mediates the formation of cointegrates between the IS1-carrying plasmid and a target plasmid in which two copies of IS1 are duplicated at the two junctions in direct orientation (10, 39). This element encodes two open reading frames, insA and B′-insB (16, 32, 33). Transcription occurs from a promoter present in the left-terminal region (IRL) preceding insA (31). A translational frameshift occurs in the −1 direction at the AAAAAAC (A6C) sequence in the overlapping region between the two open reading frames, producing the InsA–B′-InsB transframe protein, IS1 transposase (7, 49). Unless frameshifting occurs, IS1 produces InsA protein from insA which can bind to the IRs (51, 74) and inhibits transposition (30, 75). An IS1 mutant (IS1-31) with a single adenine insertion at the frameshifting site efficiently produces transposase and therefore can frequently transpose and mediate cointegration (49). The plasmid with this IS1 mutant generates miniplasmids, the deletion products generated by intramolecular transposition (50, 51), as well as IS1 circles consisting of the entire IS1 sequence and a sequence 6 to 9 bp long which appears as a spacer between the IRL and IRR of IS1 (45).
The transposition and cointegration mediated by IS1 are believed to be initiated by the step in which each strand of the donor molecule is cut at the 3′ end of IS1, yielding a pair of 3′-OH termini which are transferred to the target molecule. IS1 transposase induction of the SOS response is dependent on the IS1 ends, indicating that SOS induction is caused by transposase-mediated DNA cleavages at the IS1 ends (26). IS1 circles are transposed to target plasmids at a very high frequency owing to the presence of transposase, and the SOS response is induced in cells containing IS1 circles (53). These IS1 circles appear to act as intermediates leading to simple insertion into the target DNA via the cleavage of the circles, thereby inducing the SOS response.
The transposition activity of transposable elements is mediated by various host factors. Histone-like proteins (or DNA chaperones), such as HU and integration host factor (IHF), function in the transposition reaction of some bacterial transposable elements in vivo, in vitro, or both (for reviews, see references 25 and 28). Of these histone-like proteins, IHF binds to the ends of IS1 and to sites within the major hot-spot region for the insertion of IS1 in the target plasmid pBR322 (11). Host factors required for IS1 transposition, however, have yet to be identified. We present findings which show that a nucleoid-associated protein, H-NS, is required for transposition of IS1 but that the other histone-like proteins, HU, IHF, Fis, and StpA, are not. H-NS appears to be important for transposase-induced cleavages at the IS1 ends. Its role in the transpositional recombination mediated by IS1 is discussed based on the results of the analysis of the functional domains of H-NS.
MATERIALS AND METHODS
Escherichia coli K-12 strains and plasmids.
The strains used are listed in Table 1. Plasmids used were pSEK117, pSEK131 (48), pSEK1831 (46), pKY6 (72), pSEK80 (47), pYS6, pYS7, pYS10T, pYS30, pSEK131ΔHS (see below), pMC1403 (2), pCM101 (30), and pSTV28 (Takara). pSEK131 and pYS10T have the pUC-type replication origin. pSEK80 has the R100-type replication origin. pSTV28 and its derivatives (pYS6 and pYS7) have the p15A-type replication origin.
TABLE 1.
E. coli K-12 strains used
| Strain | Genotypeb | Source or reference |
|---|---|---|
| YK1100 | W3110 trpC9941 | 72 |
| YK2920 | YK1100 trpC9941 himA hip | 72 |
| YK1130 | YK1100 trpC9941 hupA | 65 |
| YK1220 | YK1100 trpC9941 hupB | 65 |
| YK1340 | YK1100 trpC9941 hupA hupB | 72 |
| YK2741 | YK1100 trpC9941 hupA hupB himA | 72 |
| YK4122 | YK1100 | 72 |
| YK4124 | YK1100 hns-2 | 72 |
| YK4205 | YK1100 himA hip hns-2 | 22 |
| YK4137 | YK1100 hupA hupB hns-2 | 13 |
| YK5244 | YK1100 hns-2 stpA3::Spr | Y. Kano |
| YK5246 | YK1100 stpA3::Spr | Y. Kano |
| CSH26 | Δpro-lac ara thi | C. Ueguchi |
| CU211 | CSH26 Δhns::Kmr | C. Ueguchi |
| KT1008 | MC4100 | 60 |
| KT1004 | KT1008 rpoS(Am) | K. Tanaka |
| KT1008 hns | KT1008 Δhns::Kmr | K. Tanaka |
| KT1004 hns | KT1004 rpoS(Am) Δhns::Kmr | K. Tanaka |
| RZ4500 | MG1655 lacZΔ145 | W. Reznikoff (66) |
| MDW246 | RZ4500 fis-767 | W. Reznikoff (66) |
| CU241 | CSH26 λpCU45 | C. Ueguchi (63) |
| CU242 | CH241 Δhns::Kmr | C. Ueguchi (63) |
| HM12 | CU241 hns12 | C. Ueguchi (63) |
| HM52 | CU241 hns52 | C. Ueguchi (37, 63) |
| HM60 | CU241 hns60 | C. Ueguchi (37, 63) |
| JE6638 | F−purE trp lys proC leu thi lacZ xyl ara mtl mal man gal mel polA1 tonA tsx str rif nalA | National Institute of Genetics collection |
| CSH26 sulA::lacZ | CSH26 sulA::lacZ | T. Horiuchi |
| YS28a | CSH26 sulA::lacZ Δhns::Kmr | This study |
Strain YS28 is a derivative with Δhns::Kmr from CSH26 sulA::lacZ which was constructed by P1 transduction to kanamycin resistance using a P1 lysate grown on CU211.
For the hns mutant alleles, hns-2 and Δhns::Kmr have an insertion of a fragment carrying the hygromycin resistance gene and a substitution of a fragment carrying the kanamycin resistance gene within the hns gene, respectively, producing an H-NS protein with a truncation of the region downstream of the 37th amino acid; HM12 has a mutation, hns12, in the proximal region of hns producing an H-NS protein with a substitution of cysteine-12 for arginine in the N-terminal domain; HM52 and HM60 have an hns mutation in the distal region of hns producing H-NS proteins with a substitution of aspartate-113 for glycine in the C-terminal domain and a C-terminal truncation from amino acid residue 92, respectively.
Small-scale preparation of plasmid DNA was performed as described previously (14). The alkaline lysis method (43) was used to prepare the plasmid DNA for cloning and nucleotide sequencing.
Media.
L broth and L-rich broth (73) were used. The L-agar plates contained 1.5% (wt/vol) agar (Eiken) in L broth. Antibiotics were added to the L-agar plates when necessary, at 100 μg of ampicillin (Wako)/ml, 30 or 150 μg of chloramphenicol (Sigma)/ml, 30 μg of kanamycin (Sigma)/ml, or 20 μg of spectinomycin (Sigma)/ml.
Enzymes and H-NS protein.
Restriction endonucleases (BamHI, EcoRI, MluI, PstI, PvuII, and SalI from Takara and BsrGI, BspHI, BstEII, HindIII, NsiI, SapI, and XmnI from New England Biolabs), bacterial alkaline phosphatase, and T4 DNA ligase (Takara) were used in the buffers recommended by the suppliers. H-NS protein purified by the method of Tanaka et al. (61) was provided by T. Mizuno (Nagoya University).
Oligonucleotides.
The oligonucleotide primers used are listed in Table 2. These primers were synthesized chemically in an Oligo1000M DNA synthesizer (Beckman).
TABLE 2.
Primers used
| Primer | Sequence (5′→3′)a | Positionb |
|---|---|---|
| p1 | CGGGAAGCATGCATGCGGCCGCTCTCGAGAATGAGACGTTGA | 51–92 |
| p2 | TATCACTTATGGTGACCTCGTACGAGATCTTTAAGGGCA | 912–874 |
| p3 | gcgcgtcgacTGAGGTGCTCCAATGGC | 44–60 |
| p4 | cgGGTAATCATGACTAGTTACTGGTAGTGTTTTAT | 768–736 |
| p5 | AGAAACAGAAGCCACTGGAG | 70–51 |
| p6 | TGCATGACAAAGTCATCGGG | 704–723 |
| p7 | gggTGTACACTCGAGACGCTGGAAGAAATGCCGGAAAAATTAG | 921–961 |
| p8 | CAGGAAACAGCTATGAC | 1,769–1,753 |
Mutated or additional nucleotides used to introduce a restriction site are indicated by underlining or lowercase letters, respectively.
Primers p1 and p2 hybridize to the pHSG398 sequence with coordinates 1 to 2,228 (59); primers p3 to p6 hybridize to the IS1 sequence with coordinates 1 to 768 (40); primer p7 hybridizes to plasmid pYS6 with the hns gene sequence with coordinates 1 to 1,380 (42); primer p8 hybridizes to pYS6 with the vector pSTV28 sequence with coordinates 1 to 2,999 according to Takara.
PCR.
PCR was performed as described previously (45). For the analysis of the generation of miniplasmids, the total plasmid DNA was prepared from each E. coli strain harboring pSEK131 as described previously (14), and PCR was performed with the total plasmid DNA template (200 ng) and a pair of primers, p5 and p6 (50 pmol each; Table 2), in 50 μl of Cetus buffer with 2.5 U of Ex Taq DNA polymerase (Takara) under the following thermal cycling conditions: 94°C for 2 min followed by 25 cycles of 94°C for 1 min, 65°C for 1 min, and 72°C for 3 min. PCR products were electrophoresed in a 1% agarose gel.
Plasmid construction.
pYS6 carrying the hns gene was constructed by replacing the HindIII-BamHI segment in pSTV28 with the HindIII-BamHI fragment bearing the hns gene from plasmid pKY6. pYS7 carrying a mutated hns gene was constructed by replacing the BsrGI-BamHI segment of the hns gene in plasmid pYS6 with the BsrGI-BamHI fragment mutated by PCR using primers p7 and p8 (Table 2).
Two plasmids, pNS17 carrying mini-IS1 and pKEN200T carrying the IS1 transposase gene, were constructed first to obtain pYS10T. pNS17 was constructed by replacing the PstI-BstEII fragment from IS1 in pSEK17, a pUC18 derivative (48), with the NsiI-BstEII fragment bearing the chloramphenicol resistance (Cmr) gene, which was amplified by PCR with pHSG398, a pUC-based Cmr plasmid (59), as the template and with primers p1 and p2 (Table 2). pKEN200T was constructed by replacing the SalI-BspHI region in the polylinker sequence of pKEN100 (a p15A-based kanamycin resistance [Kmr] plasmid with the araC gene and promoter-operator region of the araBAD operon of Salmonella enterica serovar Typhimurium [47]) with the SalI-BspHI fragment bearing the transposase gene obtained by PCR with pSEK1451 (a pUC119 derivative carrying an IS1 mutant [IS1-37] with a guanine insertion in the A6C sequence and encoding a mutant transposase which promotes transposition as efficiently as the wild-type transposase [48]) as the template and with primers p3 and p4 (Table 2). Lastly, the HindIII fragment bearing both the transposase gene placed under the arabinose promoter and the araC gene of pKEN200T was inserted in the HindIII site of pNS17, yielding pYS10T.
pYS30 was constructed by insertion of the HindIII fragment bearing the IS1 transposase gene and the araC gene of pKEN200T into the HindIII site of pHSG398. pSEK131ΔHS was constructed by ligation of the pSEK131 DNA digested with HindIII and SapI.
Nucleotide sequencing.
Nucleotide sequences were determined by the dideoxynucleotide method (34, 44) as described previously (47).
Transposition assay.
Plasmid pYS10T was introduced by transformation into strains CSH26 and CU211, both already harboring pSEK80. The transformants were grown for 16 h at 37°C in L-rich broth containing 0.2% (wt/vol) arabinose or 0.2% (wt/vol) glucose. Plasmid DNAs were electroporated into JE6638(polA) cells in a Gene Pulser (Bio-Rad). Cells were plated on an L-agar plate containing 20 μg of spectinomycin/ml to obtain spectinomycin-resistant (Spr) transformants with pSEK80 and on an L-agar plate containing 20 μg of spectinomycin/ml and 150 μg of chloramphenicol/ml to obtain Spr Cmr transformants with pSEK80 that had an insertion of mini-IS1 or a cointegrate between pSEK80 and pYS10T. The transposition or cointegration frequency was calculated by dividing the number of Spr Cmr transformants by the number of Spr transformants.
Gel retardation assay.
Plasmid pSEK117 DNA (0.5 μg) digested with several restriction enzymes was incubated at room temperature for 20 min with an appropriate amount of H-NS in a reaction mixture containing 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 80 mM NaCl, 10 mM β-mercaptoethanol, and 4% (vol/vol) glycerol. Samples obtained were immediately electrophoresed in a 4% polyacrylamide gel. The gel was stained with ethidium bromide to detect protein-DNA complexes.
SOS induction assay.
E. coli strains CSH26 sulA::lacZ and YS28 were transformed with a pYS plasmid. The transformants were grown overnight at 37°C in L broth. The cultures were then diluted 100-fold with fresh medium without glucose and grown at 37°C to an optical density at 600 nm of 0.4. After the addition of arabinose (final concentration, 0.2% [wt/vol]), the cultures were grown for another 3 h. The specific activities of β-galactosidase were determined as described previously (36).
Assay for promoter activity specified by IS1.
CSH26 and CU211 were transformed by pMC1403 (a derivative of pBR322 with a lacZ gene lacking its promoter) or pCM101 (a pMC1403 derivative having the 5′ portion of IS1 [the 1-to-208 region based on the coordinates given to IS1] cloned in frame with the promoter-less lacZ gene). Promoter activities were measured as described previously (53).
RESULTS
Intramolecular IS1 transposition in mutants deficient in various histone-like proteins.
IS1-31, an IS1 derivative with an insertion of 1 bp in the translational frameshifting site, efficiently mediates the production of miniplasmids, which are deletion products generated by intramolecular transposition (50, 51). The necessity of the histone-like proteins HU and IHF for intramolecular transposition was examined using pSEK131, a pUC119 derivative with IS1-31. HU is composed of two homologous polypeptides, HU-1 and HU-2, encoded by the hupB and hupA genes, respectively (21, 23), and IHF is composed of two components, IHFα and IHFβ, encoded by the himA and hip genes, respectively (9, 35). Miniplasmids were generated from pSEK131 in a hupA hupB double mutant deficient in HU and in a himA hip double mutant deficient in IHF as well as in the isogenic wild-type strain (Fig. 1A, lanes 1 to 3). For cell growth, IHF is reported to compensate for the absence of HU, indicating that there is functional redundancy between IHF and HU (20). However, miniplasmids were generated in a hupB hupA himA triple mutant (Fig. 1A, lane 4), which indicates that neither HU nor IHF is required for intramolecular IS1 transposition.
FIG. 1.
Generation of miniplasmids from pSEK131. (A) Ethidium bromide-stained agarose gel (0.7%) showing miniplasmids in various E. coli strains harboring pSEK131. Lane 1, YK1100 (wild type [wt]); lane 2, YK1340 (hupA hupB); lane 3, YK2920 (himA hip); lane 4, YK2741 (himA hupA hupB); lane 5, YK4122 (wt); lane 6, YK4124 (hns); lane 7, YK4137 (hns hupA hupB); lane 8, YK4205 (hns himA hip); lane 9, YK5246 (stpA); lane 10, YK5244 (stpA hns); lane 11, RZ4500 (wt); lane 12, MDW246 (fis); lane 13, CSH26 (wt); lane 14, CU211 (hns); lane 15, KT1008 (wt); lane 16, KT1004 (rpoS); lane 17, KT1008 hns (hns); lane 18, KT1004 hns (rpoS hns). Positions of pSEK131 (monomers [M] and dimers [D]) and several kinds of miniplasmids are indicated. ccc and oc indicate covalently closed circular and open circular DNA molecules, respectively. Note that miniplasmids of pSEK131 were generated in the hns wild-type strains (+) but not in the hns mutant strains (−). (B) Ethidium bromide-stained agarose gels (1%) showing PCR-amplified fragments from miniplasmids of pSEK131. PCR was performed with (a) the total plasmid DNA prepared from the same E. coli strains as those used above (lanes 1 to 18); (b) the total plasmid DNA prepared from strains CU241 (wt; lane 1) and CU242 (Δhns::Kmr; lane 2) and from the CU241 derivatives with an hns mutation in the N- or C-terminal domain, HM12 (hns12; lane 3), HM52 (hns52; lane 4), and HM60 (hns60; lane 5); and (c) the total plasmid DNA prepared from the H-NS-deficient strain YK4124 harboring plasmid pYS6 with the wild-type hns gene (lane 2), plasmid pYS7 with the hns gene mutated in the central domain (lane 3), or pSTV28 without the hns gene (lane 1). The DNA fragments marked with α indicate PCR fragments from pSEK131, and small fragments marked with β indicate those from the most abundant miniplasmids of pSEK131. Molecular sizes of linear marker DNA fragments are shown on the side of the gel. Note that PCR fragments from miniplasmids were generated in the hns mutant strain (−∗) with a mutation in the N- or C-terminal domain but not in the one with a mutation in the central domain.
Miniplasmids were generated in a Fis-deficient mutant as well as in the isogenic wild-type strain (Fig. 1A, lanes 11 and 12), indicating that Fis is not required for the intramolecular transposition reaction. Miniplasmids, however, were not generated in an H-NS-deficient mutant, YK4124 (hns-2), but were generated in the wild-type strain (Fig. 1A, lanes 5 and 6). Moreover, miniplasmids were not generated in a mutant [CU211(Δhns::Kmr)] with a mutation in another hns allele (Fig. 1A, lane 14). Furthermore, no miniplasmids were generated in a mutant deficient in both H-NS and HU or in a mutant deficient in both H-NS and IHF (Fig. 1A, lanes 7 and 8). These findings indicate that H-NS is required for the IS1-mediated intramolecular transposition reaction.
IS3 is a representative member of the IS3 family, which is distinct from the family to which IS1 belongs (38). IS3 encodes orfA and orfB and produces three proteins, OrfA, OrfB, and OrfAB. The OrfAB transframe protein is transposase, which is produced by −1 translational frameshifting between orfA and orfB (46). IS3-1, an IS3 derivative with a guanine insertion in the translational frameshift site leading to the in-frame alignment of orfA and orfB, generates miniplasmids at a high frequency (46). Miniplasmids were generated from plasmid pSEK1831, a pUC118 derivative with IS3-1, in mutants deficient in HU, IHF, or Fis and even in the mutant deficient in H-NS (data not shown). This shows that these proteins are not important for the IS3-mediated intramolecular transposition reaction.
The presence or absence of miniplasmids in the total plasmid DNA prepared from the various pSEK131-carrying strains used above was also examined by PCR with a pair of primers that hybridize to the IRL and the IRR and prime DNA synthesis toward the outside of IS1. The PCR yielded a 3.3-kb fragment representing the parental plasmid pSEK131 and several small fragments representing miniplasmids, of which those generating a 1.3-kb fragment appeared to be the most abundant (Fig. 1B, panel a). Such small fragments were not amplified by PCR with the total plasmid DNA prepared from the H-NS-deficient mutants but were amplified by PCR with the total plasmid DNA from the other mutants (Fig. 1B, panel a). This confirms the results above, showing that H-NS is required for the intramolecular IS1 transposition reaction.
Further evidence of the involvement of H-NS in IS1-mediated intramolecular transposition.
As stated above, no miniplasmids were generated in the H-NS-deficient mutant. H-NS has been shown to modulate the transcription of many genes (for reviews, see references 1 and 64), but the hns mutation did not affect transcription from the indigenous promoter of IS1 that is located in the left-terminal region and is responsible for the expression of the transposase gene (data not shown). This shows that H-NS does not mediate expression of the transposase gene in IS1-31 and indicates that H-NS is directly involved in the transposition reaction.
The E. coli StpA protein is an intraspecies homologue of H-NS and substitutes for H-NS in some biological reactions (52, 69, 76). Miniplasmids were generated in an stpA mutant (Fig. 1A and B, lanes 9) but not in an stpA hns double mutant (Fig. 1A and B, lanes 10). This shows that StpA is not required for intramolecular transposition but that H-NS is.
The stationary phase-specific sigma factor ςs encoded by the rpoS gene accumulates even in the logarithmic-growth phase in H-NS-deficient mutant cells (71). This accumulation of ςs may be responsible for the lack of generation of miniplasmids from pSEK131 in the H-NS-deficient mutant. However, miniplasmids were not generated from pSEK131 in an rpoS hns double mutant or in an hns single mutant (Fig. 1A and B, lanes 17 and 18), but they were generated in an rpoS single mutant (Fig. 1A and B, lanes 16). This is evidence that the unusual accumulation of ςs is not responsible for the failure of intramolecular transposition in the H-NS-deficient mutant.
It is known that hns mutants tend to accumulate mutations (29). To disprove the possibility that a second-site mutation(s) which may have occurred in the hns mutant is responsible for the failure of intramolecular IS1 transposition, plasmid pYS6, which carries the wild-type hns gene, was introduced into the H-NS-deficient mutant and the generation of miniplasmids from pSEK131 was examined by PCR. Miniplasmids were generated in the H-NS-deficient mutant harboring both pSEK131 and pYS6 (Fig. 1B, panel c, lane 2) but not in the mutant harboring both pSEK131 and the vector plasmid pSTV28 with no hns gene (Fig. 1B, panel c, lane 1). This shows that the hns gene carried by pYS6 complemented the mutated hns gene, resulting in the generation of miniplasmids, and that any possible second-site mutations are not responsible for the failure of intramolecular IS1 transposition in the H-NS-deficient mutant.
H-NS requirement in IS1-mediated intermolecular transposition.
The necessity for H-NS in IS1-mediated intermolecular transposition was examined using plasmid pYS10T with mini-IS1, in which the transposase-coding region was replaced with the Cmr gene as the donor. In this plasmid, the transposase gene was placed in the region downstream of the arabinose-inducible promoter (Fig. 2A). It was introduced into the wild-type strain (CSH26) or the H-NS-deficient mutant (CU211) that harbored the target plasmid pSEK80 with the spectinomycin resistance (Spr) gene. The plasmid DNA prepared from cells bearing both the donor and target plasmids was electroporated into cells of strain JE6638 (polA1), in which the donor plasmid pYS10T, a pUC18 derivative, cannot replicate and the target plasmid pSEK80, a derivative of plasmid R100, can. Cmr Spr transformants were obtained at a frequency of 1.4 × 10−4 per target plasmid molecule upon electroporation of the plasmid DNA prepared from the wild-type strain grown in the presence of arabinose (Table 3). Gel electrophoresis and cleavage by relevant restriction enzymes showed that the plasmids from 77% of the Cmr Spr transformants were target plasmids, with an insertion of mini-IS1, and that those from the rest of the transformants were cointegrates, with two copies of mini-IS1 at the junctions between the donor and target plasmid sequences. No Cmr Spr transformants were obtained upon electroporation of the plasmid DNA from the H-NS-deficient strain, even when grown in the presence of arabinose. (The frequency [<1.4 × 10−6 per target plasmid molecule] was at least 100-fold lower than the frequency for the wild-type strain [Table 3].) This shows that H-NS is required not only for intramolecular transposition but for the intermolecular transposition of IS1 as well.
FIG. 2.
Structures of pYS10T (A) and pYS30 (B). pYS10T carries mini-IS1, which has an IRL and an IRR (closed triangles) and the chloramphenicol resistance gene (Cmr) in place of the transposase gene. The transposase gene was placed in the region downstream of the promoter of the araBAD operon (Para; open triangle), whose expression is controlled by the araC gene product. Apr, ampicillin resistance gene. pYS30 carries the Para-driven transposase gene. The origin of replication of the pUC plasmid is shown by a closed circle.
TABLE 3.
Transposition frequency of mini-IS1 on plasmid pYS10T
| Host | Presence of arabinosea | Transposition frequencyb |
|---|---|---|
| CSH26 (wt)c | + | 1.4 × 10−4 |
| − | 5.3 × 10−7 | |
| CU211 (hns) | + | <1.4 × 10−6 |
| − | <2.6 × 10−7 |
Plasmid DNA was prepared from cells grown in the presence (+) or absence (−) of arabinose.
See Materials and Methods.
wt, wild type.
Identification of sites preferentially bound by H-NS in the IS1-carrying plasmid.
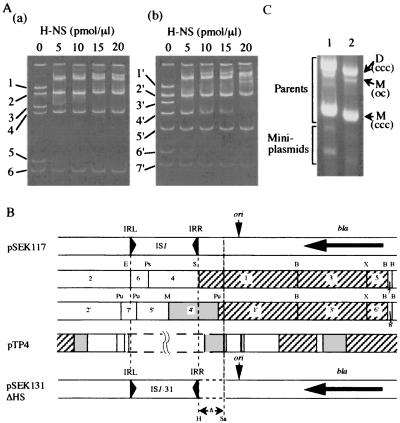
The site(s) bound by H-NS in pSEK117 was examined using a gel retardation assay. pSEK117, the parent of pSEK131, carries wild-type IS1 instead of the mutant IS1-31 in pSEK131. pSEK117 was used in this assay because it does not generate miniplasmids, which complicate the assay. The gel retardation assay using the pSEK117 plasmid DNA digested with a set of restriction enzymes showed that H-NS preferentially bound to three fragments (those labeled 1, 3, and 5 in Fig. 3) but did not bind to any others, including the two fragments with the IS1 sequence (those labeled 4 and 6 in Fig. 3), indicating that IS1 has no H-NS-binding site within its sequence. (Note that an excess amount of H-NS protein caused retardation of all the DNA fragment bands due to the nonspecific DNA-binding ability of H-NS [data not shown].) The gel retardation assay using the pSEK117 plasmid DNA digested with another set of restriction enzymes showed that four fragments (those labeled 1′, 3′, 4′, and 6′ in Fig. 3) were preferentially bound by H-NS. Of these retarded fragments, one (4′) has the IRR region of the IS1 sequence as well as the sequence neighboring it. This means that at least one H-NS-binding site is present in the sequence neighboring the IRR of IS1, because the IS1 sequence has no H-NS-binding site.
FIG. 3.
(A) Ethidium bromide-stained acrylamide gels (4%) showing the results of the gel retardation assay with H-NS and restriction fragments derived from pSEK117. (a) Restriction fragments digested with BspHI, EcoRI, PstI, SalI, and XmnI and (b) those digested with BspHI, MluI, PvuII, and XmnI. Fragments were incubated with H-NS protein at the indicated concentrations (pmol/μl). Numbers refer to fragments whose positions are shown in panel B. (B) Restriction maps of pSEK117 and pTP4 (a derivative of pUC119 with a cloned fragment [18]). The restriction fragments from pSEK117 are numbered in order of size. Restriction sites: B, BspHI; E, EcoRI; H, HindIII; M, MluI; Ps, PstI; Pu, PvuII; S, SalI; Sa, SapI; X, XmnI. The bla gene is shown with a thick arrow. ori is the replication origin of pUC119. The region in pTP4 which differs from that in pSEK117 is shown by broken lines. An HaeIII restriction map of pTP4 is depicted. The restriction fragments of pSEK117 and pTP4 preferentially bound by H-NS are indicated by hatched or dark boxes. Restriction fragments indicated by the dark boxes required a greater amount of H-NS protein for retardation than those indicated by the hatched boxes. pSEK131ΔHS has a deletion of the HindIII-SapI fragment (dotted lines) which borders the IRR sequence in pSEK117. (C) An ethidium bromide-stained agarose gel (0.7%) showing the production of miniplasmids from pSEK131 (lane 1) and pSEK131ΔHS (lane 2) in YK4122. For further details, see the legend for Fig. 1.
Plasmid pSEK117 is a derivative of pUC119. Jordi et al. (18) performed a gel retardation assay with plasmid pTP4, a pUC119 derivative with a cloned fragment, and showed that some restriction fragments were bound by H-NS. Retarded fragments with the pUC119 sequence could be identified, as shown in Fig. 3B. This, together with the above results, allowed for the identification of the DNA segments with an H-NS-binding site(s), one of which is located in the region neighboring the IRR of IS1 (Fig. 3B).
To see whether the H-NS-binding site(s) in the sequence neighboring the IRR of IS1 is important for the intramolecular IS1 transposition, plasmid pSEK131ΔHS, a pSEK131 derivative with a deletion of the segment with the binding site(s) (Fig. 3B), was examined for the generation of miniplasmids. pSEK131ΔHS was able to generate miniplasmids (Fig. 3C), showing that the H-NS-binding site(s) in the region neighboring IS1 is not important for the intramolecular transposition reaction.
Analysis of the functional domain of H-NS that affects IS1 transposition.
The previous mutational analysis of hns indicated that H-NS consists of three distinct domains, the N-terminal, central, and C-terminal regions, which are responsible for transcriptional repression, dimerization and/or oligomerization, and DNA-binding, respectively (62, 63). To determine which domain of H-NS is important for IS1 transposition, the generation of miniplasmids from pSEK131 in three mutants with a mutation in the hns gene on the chromosome was examined by PCR. Mutant strain HM12 has a missense mutation in the proximal end region of hns which produces an H-NS protein with a mutated N-terminal domain, causing loss of the ability to repress the transcription of proV but retaining the DNA-binding activity (63). Miniplasmids were generated in HM12 harboring pSEK131 as well as in the isogenic wild-type strain CU241 harboring pSEK131 (Fig. 1B, panel b, lanes 1 and 3), showing that an amino acid substitution in the N-terminal domain of H-NS does not affect IS1 transposition. This may support the idea that the N-terminal domain of H-NS is not important for IS1 transposition. Mutant strains HM52 and HM60 have missense and amber mutations, respectively, in the distal end region of hns which produce an H-NS protein with a mutated or truncated C-terminal domain, causing loss of the DNA-binding activity (63). Miniplasmids were generated in each mutant strain harboring pSEK131 (Fig. 1B, panel b, lanes 4 and 5), showing that neither an amino acid substitution nor truncation in the C-terminal domain of H-NS affects IS1 transposition. This indicates that the C-terminal domain is not important for IS1 transposition.
The mutants with a mutation in the N- or C-terminal domain of H-NS which were used in the above experiments have been shown not to impair dimer formation of H-NS (63). Therefore, to determine whether the central domain of H-NS responsible for dimer formation is important for IS1 transposition, the generation of miniplasmids from pSEK131 in the H-NS-deficient mutant YK4124 harboring plasmid pYS7 with a mutated hns gene in the central domain was examined by PCR. The mutated hns gene has a missense mutation (the 30th codon, CUG for leucine, to CCG for proline) causing loss of the dimer-forming activity of the H-NS protein as well as the repression of transcription from the proV and bgl promoters (62). Miniplasmids were not generated in YK4124 harboring both pSEK131 and pYS7 (Fig. 1B, panel c, lane 3) or in YK4124 harboring both pSEK131 and the vector plasmid pSTV28 with no hns gene (Fig. 1B, panel c, lane 1), but they were generated in YK4124 harboring both pSEK131 and pYS6 with the hns gene (Fig. 1B, panel c, lane 2). This shows that the central domain of H-NS is important for IS1 transposition.
Lack of induction of the SOS response by IS1 transposase in the H-NS-deficient mutant.
IS1 transposase induces the SOS response by way of transposase-induced cleavages at the IS1 ends (26). The SOS response in cells harboring pYS plasmids with mini-IS1 was therefore examined using an E. coli strain with an SOS gene, sulA, fused with the lacZ gene. β-Galactosidase activity in the H-NS-deficient and wild-type strains that harbored no plasmid and were grown in the presence of mitomycin C, a drug known to induce the SOS response, was significantly increased compared to that in the strains grown in the absence of mitomycin C (Table 4). β-Galactosidase activity in the H-NS-deficient strain harboring pYS10T with the arabinose-inducible transposase gene (Fig. 2A) was not increased in the presence of arabinose, whereas it was increased in the wild-type strain harboring pYS10T (Table 4). These findings suggest that the ends of IS1 are not cleaved in the H-NS-deficient strain and therefore there is no induction of the SOS response. β-Galactosidase activity was not increased in the presence of arabinose in the H-NS-deficient strain harboring pYS30 (Table 4), which has the arabinose-inducible transposase gene but lacks IR sequences in mini-IS1 (Fig. 2B), as was also the case for the H-NS-deficient or wild-type strain harboring no plasmid (Table 4), supporting the argument described above. β-Galactosidase activity, however, was increased to some extent in the presence of arabinose in the wild-type strain harboring pYS30. This increase may be the result of transposase action on chromosomal copies of IS1.
TABLE 4.
SOS responses induced by transposase and mini-IS1
| Plasmida | Hostb | Presence of arabinosec | LacZd(U) | Net increasee |
|---|---|---|---|---|
| pYS10T | wt | + | 68 | 33 |
| − | 35 | |||
| hns | + | 39 | −1 | |
| − | 40 | |||
| pYS30 | wt | + | 39 | 11 |
| − | 28 | |||
| hns | + | 35 | 1 | |
| − | 34 | |||
| None | wt | + | 58 | −2 |
| − | 60 | |||
| hns | + | 52 | −2 | |
| − | 54 | |||
| wt | +f | 68 | 37 | |
| − | 31 | |||
| hns | +f | 106 | 73 | |
| − | 33 |
For the structures of pYS10T and pYS30, see Fig. 2.
Host strains were CSH26 (wt) and YS28 (hns), both of which carry sulA::lacZ. wt, wild type.
Cells were grown in the presence (+) or absence (−) of arabinose.
β-Galactosidase (LacZ) activity in the cell lysate. Each value is the mean of those obtained from three independent experiments; standard errors in all cases were less than 4.7%.
Net increase in β-galactosidase activity.
Mitomycin C (final concentration, 5 ng/ml) was added instead of arabinose.
DISCUSSION
We showed that in vivo the intramolecular transposition mediated by IS1 does not require HU, IHF, or Fis and that the intramolecular transposition mediated by IS3, which is distinct from IS1, does not require these host proteins. Histone-like proteins (or DNA chaperones) are involved in the transposition of certain transposons as well as in many site-specific recombination systems. The HU protein is required in an early step in phage Mu transposition in vitro (4, 5, 56) and in vivo (15, 19). HU is thought to promote the formation of a tight DNA bend in the end region which facilitates the communication of Mu A monomers during complex assembly by binding specifically to sites within the region between two Mu A-binding sites in the Mu left end (27). IHF stimulates the Mu DNA strand transfer reaction in vitro by binding at the transpositional enhancer (57, 58). IHF modulates Tn10 transposition in vivo and in vitro; IHF stimulates Tn10 excision and forces transposon end-target DNA interactions into a constrained pathway that yields only unknotted intratransposon inversion circles (3, 54). IHF binds to the ends of γδ, a Tn3 family element, cooperatively with transposase and stimulates transpositional immunity of γδ (67, 68). Fis is required for various site-specific recombination systems, and HU and IHF act in some of these systems as well (for a review, see reference 12). These proteins are thought to promote the formation of the nucleoprotein structures which facilitate synapsis of the DNA strands required for site-specific recombination. IS1 and IS3, which were shown here to require none of the histone-like proteins (HU, IHF, and Fis) for their transposition in vivo, are thought to form a nucleoprotein complex that is different from the complexes of the other elements discussed.
We demonstrated that the intra- and intermolecular transposition mediated by IS1 requires H-NS, a protein abundant in E. coli which has a molecular mass of approximately 15.6 kDa (55). H-NS binds to DNA in a relatively nonspecific fashion but preferentially binds to curved double-stranded DNA (41, 70, 77). H-NS constrains negative supercoils, thereby affecting DNA topology, and modulates the transcription of many genes (for reviews, see references 1 and 64). Mutations in the hns gene caused an increase in transcription of the Mu transposase gene, leading to an increase in the Mu transposition rate, and the depletion of H-NS probably caused destabilization of the Mu repressor-DNA complex which keeps the transposase gene transcriptionally inactive (8). To our knowledge, no other cases in which H-NS positively regulates the DNA recombination reaction have been reported. The rate of spontaneous chromosomal deletions and an inversion of a DNA element with the fimA promoter are stimulated in hns mutants (24, 29), but in these cases H-NS is supposed to negatively regulate the DNA recombination reaction. Note that the positive role of H-NS in the transpositional recombination mediated by IS1 is the only example of a histone-like protein that is essential for the transposition of the transposable element in vivo, except for HU, which is involved in Mu transposition. The role of H-NS in IS1 transposition may therefore shed light on a novel function of H-NS.
We showed that the N-terminal and C-terminal domains of H-NS, which are responsible for the repression of the proV promoter and DNA binding, respectively, are dispensable for intramolecular IS1 transposition. Dispensability of the DNA-binding domain may also be supported by the fact that the IS1 sequence is not preferentially bound by H-NS. We then showed that the central domain of H-NS, which is responsible for the dimerization of the H-NS protein (62), is indispensable for intramolecular IS1 transposition. These results lead us to conclude that the ability of H-NS to form a dimer is important for IS1 transposition. The transposition reaction of IS1 should require the formation of an IS1 DNA-transposase complex in which both IRs are held closely together by transposase for subsequent reactions such as end cleavage and strand transfer. We postulate that H-NS interacts with IS1 transposase to form an active DNA-transposase complex for IS1 transposition. In this connection, it is noteworthy that the fimA promoter inversion and the bgl gene silencing for which the C-terminal DNA-binding domain of H-NS is dispensable are thought to be regulated through protein-protein interactions with other molecules at the fimA invertible element and the bgl promoter (6, 63). Although H-NS regulates DNA recombination positively in the IS1 system but negatively in the fimA system, H-NS may be involved in these systems by a common protein-protein interaction mechanism.
We showed here that the SOS response was not induced in the H-NS-deficient strain, which is strong evidence that H-NS has an important role in DNA cleavage at the IRs owing to the action of transposase. An IS1 circle with a spacer sequence of 6 to 9 bp has been shown to transpose to target plasmids at a very high frequency because of transposase (53). This transposition was accompanied by removal of the spacer sequence in the IS1 circle and by duplication of a sequence at the target site. IS1 circles appear to act as intermediates leading to simple insertion into the target DNA via IS1 circle cleavage, which efficiently induces the SOS response. Interestingly, IS1 circle transposition did not require H-NS (53), although IS1 transposition did. We assume that when the IS1 circle, in which both IRs are intrinsically closely located, serves as the donor, an active DNA-transposase complex is formed even in the absence of H-NS but that when IS1 serves as the donor, H-NS is essential for the formation of the active complex in which DNA cleavage occurs at the IRs.
We found that as in the wild-type strain, IS1 circles transpose preferentially to AT-rich regions in the H-NS-deficient strains. In the H-NS-deficient strain, however, IS1 circles transpose even to non-AT-rich regions (53). These observations led us to assume that H-NS participates in the step involving the target DNA molecule. The interaction of H-NS with the IS1 transposase protein may also be important for target capture and/or formation of a nucleoprotein complex consisting of IR DNA, target DNA, and transposase.
ACKNOWLEDGMENTS
This research was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports, and Culture of Japan.
We thank K. Tanaka, T. Horiuchi, W. Reznikoff, and C. Ueguchi for kindly providing us with the E. coli strains used in this study. We also thank T. Mizuno for the gift of purified H-NS protein.
REFERENCES
- 1.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 2.Casadaban M J, Chou J, Cohen S N. In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980;143:971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chalmers R, Guhathakurta A, Benjamin H, Kleckner N. IHF modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. Cell. 1998;93:897–908. doi: 10.1016/s0092-8674(00)81449-x. [DOI] [PubMed] [Google Scholar]
- 4.Craigie R, Arndt-Jovin D J, Mizuuchi K. A defined system for the DNA strand-transfer reaction at the initiation of bacteriophage Mu transposition: protein and DNA substrate requirements. Proc Natl Acad Sci USA. 1985;82:7570–7574. doi: 10.1073/pnas.82.22.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craigie R, Mizuuchi K. Transposition of Mu DNA: joining of Mu to target DNA can be uncoupled from cleavage at the ends of Mu. Cell. 1987;51:493–501. doi: 10.1016/0092-8674(87)90645-3. [DOI] [PubMed] [Google Scholar]
- 6.Donato G M, Kawula T H. Phenotypic analysis of random hns mutations differentiate DNA-binding activity from properties of fimA promoter inversion modulation and bacterial motility. J Bacteriol. 1999;181:941–948. doi: 10.1128/jb.181.3.941-948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escoubas J M, Prère M F, Fayet O, Salvignol I, Galas D, Zerbib D, Chandler M. Translational control of transposition activity of the bacterial insertion sequence IS1. EMBO J. 1991;10:705–712. doi: 10.1002/j.1460-2075.1991.tb08000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falconi M, McGoven V, Gualerzi C, Hillyard D, Higgins N P. Mutations altering chromosomal protein H-NS induce mini-Mu transposition. New Biol. 1991;3:615–625. [PubMed] [Google Scholar]
- 9.Flamm E L, Weisberg R A. Primary structure of the hip gene of Escherichia coli and of its product, the beta subunit of integration host factor. J Mol Biol. 1985;183:117–128. doi: 10.1016/0022-2836(85)90206-2. [DOI] [PubMed] [Google Scholar]
- 10.Galas D J, Chandler M. Structure and stability of Tn9-mediated cointegrates. Evidence for two pathways of transposition. J Mol Biol. 1982;154:245–272. doi: 10.1016/0022-2836(82)90063-8. [DOI] [PubMed] [Google Scholar]
- 11.Gamas P, Chandler M, Prentki P, Galas D. E. coli integration host factor binds specifically to the ends of the insertion sequence IS1 and to its major insertional hot-spot in pBR322. J Mol Biol. 1987;195:261–272. doi: 10.1016/0022-2836(87)90648-6. [DOI] [PubMed] [Google Scholar]
- 12.Glasgow A C, Hughes K T, Simon M I. Bacterial DNA inversion systems. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 637–659. [Google Scholar]
- 13.Goshima N, Kano Y, Tanaka H, Kohno K, Iwaki T, Imamoto F. IHF suppresses the inhibitory effect of H-NS on HU function in the hin inversion system. Gene. 1994;141:17–23. doi: 10.1016/0378-1119(94)90122-8. [DOI] [PubMed] [Google Scholar]
- 14.He M, Wilde A, Kaderbhai M A. A simple single-step procedure for small-scale preparation of Escherichia coli plasmid. Nucleic Acids Res. 1990;18:1660. doi: 10.1093/nar/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huisman O, Faelen M, Girard D, Jaffè A, Toussaint A, Rouvière-Yaniv J. Multiple defects in Escherichia coli mutants lacking HU protein. J Bacteriol. 1989;171:3704–3712. doi: 10.1128/jb.171.7.3704-3712.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jakowec M, Prentki P, Chandler M, Galas D J. Mutational analysis of the open reading frames in the transposable element IS1. Genetics. 1988;120:47–55. doi: 10.1093/genetics/120.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnsrud L. DNA sequence of the transposable element IS1. Mol Gen Genet. 1979;169:213–218. doi: 10.1007/BF00271673. [DOI] [PubMed] [Google Scholar]
- 18.Jordi B J M, Fielder A, Burns C M, Hinton J C D, Dover N, Ussery D W, Higgins F. DNA binding is not sufficient for H-NS-mediated repression of proU expression. J Biol Chem. 1997;272:12083–12090. doi: 10.1074/jbc.272.18.12083. [DOI] [PubMed] [Google Scholar]
- 19.Kano Y, Goshima N, Wada M, Imamoto F. Participation of hup product in replicative transposition of Mu phage in E. coli. Gene. 1989;76:353–358. doi: 10.1016/0378-1119(89)90175-3. [DOI] [PubMed] [Google Scholar]
- 20.Kano Y, Imamoto F. Requirement of integration host factor (IHF) for growth of Escherichia coli deficient in HU protein. Gene. 1990;89:133–137. doi: 10.1016/0378-1119(90)90216-e. [DOI] [PubMed] [Google Scholar]
- 21.Kano Y, Osato K, Wada M, Imamoto F. Cloning and sequencing of the HU-2 gene of Escherichia coli. Mol Gen Genet. 1987;209:408–410. doi: 10.1007/BF00329674. [DOI] [PubMed] [Google Scholar]
- 22.Kano Y, Yasuzawa K, Tanaka H, Imamoto F. Propagation of Mu in IHF-deficient Escherichia coli in the absence of the H-NS histone-like protein. Gene. 1993;126:93–97. doi: 10.1016/0378-1119(93)90594-s. [DOI] [PubMed] [Google Scholar]
- 23.Kano Y, Yoshino S, Wada M, Yokoyama K, Nobuhara M, Imamoto F. Molecular cloning and nucleotide sequencing of the HU-1 gene of Escherichia coli. Mol Gen Genet. 1985;201:360–362. doi: 10.1007/BF00425687. [DOI] [PubMed] [Google Scholar]
- 24.Kawula T H, Orndorff P E. Rapid site-specific DNA inversion in Escherichia coli mutants lacking the histone-like protein H-NS. J Bacteriol. 1991;173:4116–4123. doi: 10.1128/jb.173.13.4116-4123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleckner N, Chalmers R, Kwon D, Sakai J, Bolland S. Tn10 and IS10 transposition and chromosome rearrangements: mechanism and regulation in vivo and in vitro. Curr Top Microbiol Immunol. 1996;204:49–82. doi: 10.1007/978-3-642-79795-8_3. [DOI] [PubMed] [Google Scholar]
- 26.Lane D, Cavaillé J, Chandler M. Induction of SOS response by IS1 transposase. J Mol Biol. 1994;242:339–350. doi: 10.1006/jmbi.1994.1585. [DOI] [PubMed] [Google Scholar]
- 27.Lavoie B D, Chaconas G. Site-specific HU binding in the Mu transpososome: conversion of a sequence-independent DNA-binding protein into a chemical nuclease. Genes Dev. 1993;7:2510–2519. doi: 10.1101/gad.7.12b.2510. [DOI] [PubMed] [Google Scholar]
- 28.Lavoie B D, Chaconas G. Transposition of phage Mu DNA. Curr Top Microbiol Immunol. 1996;204:83–99. doi: 10.1007/978-3-642-79795-8_4. [DOI] [PubMed] [Google Scholar]
- 29.Lejeune P, Danchin A. Mutations in the bglY gene increase the frequency of spontaneous deletions in Escherichia coli K-12. Proc Natl Acad Sci USA. 1990;87:360–363. doi: 10.1073/pnas.87.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Machida C, Machida Y. Regulation of IS1 transposition by the insA gene product. J Mol Biol. 1989;208:567–574. doi: 10.1016/0022-2836(89)90148-4. [DOI] [PubMed] [Google Scholar]
- 31.Machida C, Machida Y, Ohtsubo H, Ohtsubo E. Both inverted repeat sequences located at the ends of IS1 provide promoter functions. J Mol Biol. 1984;177:247–267. doi: 10.1016/0022-2836(84)90455-8. [DOI] [PubMed] [Google Scholar]
- 32.Machida Y, Machida C, Ohtsubo H, Ohtsubo E. Factors determining frequency of plasmid cointegration mediated by insertion sequence IS1. Proc Natl Acad Sci USA. 1982;79:277–281. doi: 10.1073/pnas.79.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machida Y, Machida C, Ohtsubo H, Ohtsubo E. Insertion element IS1 encodes two structural genes required for its transposition. J Mol Biol. 1984;177:229–245. doi: 10.1016/0022-2836(84)90454-6. [DOI] [PubMed] [Google Scholar]
- 34.Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- 35.Miller H I. Primary structure of the himA gene of Escherichia coli: homology with DNA-binding protein HU and association with the phenylalanyl-tRNA synthetase operon. Cold Spring Harbor Symp Quant Biol. 1984;49:691–698. doi: 10.1101/sqb.1984.049.01.078. [DOI] [PubMed] [Google Scholar]
- 36.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- 37.Ohta T, Ueguchi C, Mizuno T. rpoS function is essential for bgl silencing caused by C-terminally truncated H-NS in Escherichia coli. J Bacteriol. 1999;181:6278–6283. doi: 10.1128/jb.181.20.6278-6283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohtsubo E, Sekine Y. Bacterial insertion sequences. Curr Top Microbiol Immunol. 1996;204:1–26. doi: 10.1007/978-3-642-79795-8_1. [DOI] [PubMed] [Google Scholar]
- 39.Ohtsubo E, Zenilman M, Ohtsubo H. Plasmids containing insertion elements are potential transposons. Proc Natl Acad Sci USA. 1980;77:750–754. doi: 10.1073/pnas.77.2.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohtsubo H, Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci USA. 1978;75:615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Owen-Hughes T A, Pavitt G D, Santos D S, Sidebotham J M, Hulton C S J, Hinton J C D, Higgins C F. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell. 1992;71:255–265. doi: 10.1016/0092-8674(92)90354-f. [DOI] [PubMed] [Google Scholar]
- 42.Pon C L, Calogero R A, Gualerzi C O. Identification, cloning, nucleotide sequence and chromosomal map location of hns, the structural gene for Escherichia coli DNA-binding protein H-NS. Mol Gen Genet. 1988;212:199–202. doi: 10.1007/BF00334684. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sekine Y, Eisaki N, Kobayashi K, Ohtsubo E. Isolation characterization of IS1 circles. Gene. 1997;191:183–190. doi: 10.1016/s0378-1119(97)00057-7. [DOI] [PubMed] [Google Scholar]
- 46.Sekine Y, Eisaki N, Ohtsubo E. Translational control in production of transposase and in transposition of insertion sequence IS3. J Mol Biol. 1994;235:1406–1420. doi: 10.1006/jmbi.1994.1097. [DOI] [PubMed] [Google Scholar]
- 47.Sekine Y, Izumi K, Mizuno T, Ohtsubo E. Inhibition of transpositional recombination by OrfA and OrfB proteins encoded by insertion sequence IS3. Genes Cells. 1997;2:547–557. doi: 10.1046/j.1365-2443.1997.1440342.x. [DOI] [PubMed] [Google Scholar]
- 48.Sekine Y, Nagasawa H, Ohtsubo E. Identification of the site of translational frameshifting required for production of the transposase encoded by insertion sequence IS1. Mol Gen Genet. 1992;253:317–324. doi: 10.1007/BF00279376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sekine Y, Ohtsubo E. Frameshifting is required for production of the transposase encoded by insertion sequence 1. Proc Natl Acad Sci USA. 1989;86:4609–4613. doi: 10.1073/pnas.86.12.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sekine Y, Ohtsubo E. Translational frameshifting in IS elements and other genetic systems. In: Kimura M, Takahata N, editors. New aspects of the genetics of molecular evolution. Tokyo, Japan: Japan Scientific Societies Press; 1991. pp. 243–261. [Google Scholar]
- 51.Sekino N, Sekine Y, Ohtsubo E. IS1-encoded proteins, InsA and InsA-B′-InsB transframe protein (transposase): functions deduced from their DNA-binding ability. Adv Biophys. 1995;31:209–222. doi: 10.1016/0065-227x(95)99393-4. [DOI] [PubMed] [Google Scholar]
- 52.Shi X, Bennett G N. Plasmids bearing hfq and the hns-like gene stpA complement hns mutants in modulating arginine decarboxylase gene expression in Escherichia coli. J Bacteriol. 1994;176:6769–6775. doi: 10.1128/jb.176.21.6769-6775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiga Y, Sekine Y, Ohtsubo E. Transposition of IS1 circles. Genes Cells. 1999;4:551–559. doi: 10.1046/j.1365-2443.1999.00281.x. [DOI] [PubMed] [Google Scholar]
- 54.Signon L, Kleckner N. Negative and positive regulation of Tn10/IS10-promoted recombination by IHF: two distinguishable processes inhibit transposition off of multicopy plasmid replicons and active chromosomal events that favor evolution of new transposons. Genes Dev. 1995;9:1123–1136. doi: 10.1101/gad.9.9.1123. [DOI] [PubMed] [Google Scholar]
- 55.Spassky A, Rimsky S, Garreau H, Buc H. H1a, an E. coli DNA-binding protein which accumulates in stationary phase, strongly compacts DNA in vitro. Nucleic Acids Res. 1984;12:5321–5340. doi: 10.1093/nar/12.13.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Surette M G, Buch S J, Chaconas G. Transpososomes: stable protein-DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell. 1987;49:253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]
- 57.Surette M G, Chaconas G. A protein factor which reduces the negative supercoiling requirement in the Mu DNA strand transfer reaction is Escherichia coli integration host factor. J Biol Chem. 1989;264:3028–3034. [PubMed] [Google Scholar]
- 58.Surette M G, Lavoie B D, Chaconas G. Action at a distance in Mu DNA transposition: an enhancer-like element is the site of action of supercoiling relief activity by integration host factor (IHF) EMBO J. 1989;8:3483–3489. doi: 10.1002/j.1460-2075.1989.tb08513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZα-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka K, Handel K, Loewen P C, Takahashi H. Identification and analysis of the rpoS-dependent promoter of katE, encoding catalase HPII in Escherichia coli. Biochim Biophys Acta. 1997;1352:161–166. doi: 10.1016/s0167-4781(97)00044-4. [DOI] [PubMed] [Google Scholar]
- 61.Tanaka K, Yamada H, Yoshida T, Mizuno T. Overproduction and rapid purification of the Escherichia coli histone-like protein, H-NS. Agric Biol Chem. 1991;55:3139–3141. [Google Scholar]
- 62.Ueguchi C, Seto C, Suzuki T, Mizuno T. Clarification of the dimerization domain and its functional significance for the Escherichia coli nucleoid protein H-NS. J Mol Biol. 1997;274:145–151. doi: 10.1006/jmbi.1997.1381. [DOI] [PubMed] [Google Scholar]
- 63.Ueguchi C, Suzuki T, Yoshida T, Tanaka K, Mizuno T. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J Mol Biol. 1996;263:149–162. doi: 10.1006/jmbi.1996.0566. [DOI] [PubMed] [Google Scholar]
- 64.Ussery D W, Hinton J C D, Jordi B J A M, Granum P E, Seirafi A, Stephen R J, Tupper A E, Berridge G, Sidebotham J M, Higgins C F. The chromatin-associated protein: H-NS. Biochimie. 1994;76:968–980. doi: 10.1016/0300-9084(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 65.Wada M, Kano Y, Ogawa T, Imamoto F. Construction and characterization of the deletion mutant of hupA and hupB genes in Escherichia coli. J Mol Biol. 1988;204:581–591. doi: 10.1016/0022-2836(88)90357-9. [DOI] [PubMed] [Google Scholar]
- 66.Weinreich M D, Reznikoff W S. Fis plays role in Tn5 and IS50 transposition. J Bacteriol. 1992;174:4530–4537. doi: 10.1128/jb.174.14.4530-4537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wiater L A, Grindley N D F. γδ transposase and integration host factor bind cooperatively at both ends of γδ. EMBO J. 1988;7:1907–1911. doi: 10.1002/j.1460-2075.1988.tb03024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wiater L A, Grindley N D F. Integration host factor increases the transpositional immunity conferred by the γδ ends. J Bacteriol. 1990;172:4951–4958. doi: 10.1128/jb.172.9.4951-4958.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams R M, Rimsky S, Buc H. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant-negative derivatives. J Bacteriol. 1996;178:4335–4343. doi: 10.1128/jb.178.15.4335-4343.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamada H, Muramatsu S, Mizuno T. An Escherichia coli protein that preferentially binds to sharply curved DNA. J Biochem. 1990;108:420–425. doi: 10.1093/oxfordjournals.jbchem.a123216. [DOI] [PubMed] [Google Scholar]
- 71.Yamashino T, Ueguchi C, Mizuno T. Quantitative control of the stationary phase-specific sigma factor, ςs, in Escherichia coli: involvement of the nucleoid protein H-NS. EMBO J. 1995;14:594–602. doi: 10.1002/j.1460-2075.1995.tb07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yasuzawa K, Hayashi N, Goshima N, Kohno K, Imamoto F, Kano Y. Histone-like proteins are required for cell growth and constraint of supercoils in DNA. Gene. 1992;122:9–15. doi: 10.1016/0378-1119(92)90026-l. [DOI] [PubMed] [Google Scholar]
- 73.Yoshioka Y, Ohtsubo H, Ohtsubo E. Repressor gene finO in plasmids R100 and F: constitutive transfer of plasmid F is caused by insertion of IS3 into finO. J Bacteriol. 1987;169:619–623. doi: 10.1128/jb.169.2.619-623.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zerbib D, Jakowec M, Prentki M, Galas D J, Chandler M. Expression of proteins essential for IS1 transposition: specific binding of InsA to the ends of IS1. EMBO J. 1987;6:3163–3169. doi: 10.1002/j.1460-2075.1987.tb02627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zerbib D, Polard P, Escoubas J M, Galas D, Chandler M. The regulatory role of the IS1-encoded InsA protein in transposition. Mol Microbiol. 1990;4:471–477. doi: 10.1111/j.1365-2958.1990.tb00613.x. [DOI] [PubMed] [Google Scholar]
- 76.Zhang A, Rimsky S, Buc H, Belfort M. Escherichia coli protein analogs StpA and H-NS: regulatory networks, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]
- 77.Zuber F, Rimsky S, Buc H. Modulated expression of promoters containing upstream curved DNA sequences by the Escherichia coli nucleoid protein H-NS. Mol Microbiol. 1994;12:231–240. doi: 10.1111/j.1365-2958.1994.tb01012.x. [DOI] [PubMed] [Google Scholar]