Abstract
Background
The tumor microenvironment contains stromal cells, including endothelial cells and fibroblasts, that aid tumor growth and impair immune cell function. Many solid tumors remain difficult to cure because of tumor-promoting stromal cells, but current therapies targeting tumor stromal cells are constrained by modest efficacy and toxicities. TEM8 is a surface antigen selectively upregulated on tumor and tumor stromal cells, endothelial cells and fibroblasts that may be targeted with specific natural killer (NK) cell engagement.
Methods
A Tri-specific Killer Engager (TriKE) against TEM8—‘cam1615TEM8’—was generated using a mammalian expression system. Its function on NK cells was assessed by evaluation of degranulation, inflammatory cytokine production, and killing against tumor and stroma cell lines in standard co-culture and spheroid assays. cam1615TEM8-mediated proliferation and STAT5 phosphorylation in NK cells was tested and compared with T cells by flow cytometry. NK cell proliferation, tumor infiltration, and tumor and tumor-endothelium killing by cam1615TEM8 and interleukin-15 (IL-15) were assessed in NOD scid gamma (NSG) mice.
Results
cam1615TEM8 selectively stimulates NK cell degranulation and inflammatory cytokine production against TEM8-expressing tumor and stromal cell lines. The increased activation translated to superior NK cell killing of TEM8-expressing tumor spheroids. cam1615TEM8 selectively stimulated NK cell but not T cell proliferation in vitro and enhanced NK cell proliferation, survival, and tumor infiltration in vivo. Finally, cam1615TEM8 stimulated NK cell killing of tumor and tumor endothelial cells in vivo.
Conclusions
Our findings indicate that the cam1615TEM8 TriKE is a novel anti-tumor, anti-stroma, and anti-angiogenic cancer therapy for patients with solid tumors. This multifunctional molecule works by selectively targeting and activating NK cells by costimulation with IL-15, and then targeting that activity to TEM8+ tumor cells and TEM8+ tumor stroma.
Keywords: Killer Cells, Natural; Immunotherapy; Tumor Microenvironment
Background
Cancer immunotherapies for solid tumors need to overcome the cancer-promoting stromal cells within the tumor microenvironment, including fibroblasts, endothelial cells, pericytes, and immune cells, as they compose a large portion of the tumor mass in carcinomas.1 Tumor stromal cells abet cancer growth via growth factors and cause immunosuppression and immune cell dysfunction.2 Finally, the tumor vasculature, although abnormal, disorganized, and dilated, supplies cancer cells with the nutrients and oxygen necessary for tumor growth and dissemination.3
Current anti-angiogenic therapies include bevacizumab, sorafenib, and sunitinib, but these treatments have been plagued by toxicities and limited efficacy because the targeted pathways are important in normal adult angiogenesis and angiogenic signaling redundancies result in tumor resistance.4 Preclinical attempts to target other stromal cell types, such as cancer-associated fibroblasts, have similarly encountered on target, off tumor toxicities due to target protein expression in healthy tissue.5
Tumor Endothelial Marker 8 (TEM8, encoded by the ANTXR1 gene) is a highly conserved 80–85 kDa integrin-like adhesion molecule that was discovered on the endothelium of colorectal cancer, and subsequent studies identified its expression on multiple stromal cells—endothelial cells, fibroblasts, and pericytes—in the tumor microenvironment of diverse human cancer types.6–8 TEM8 expression has also been described on cancer cell lines and on breast cancer and pancreatic cancer stem cells.8–11 TEM8 is unique as it is not expressed on healthy tissue or during normal angiogenesis.6 8 Preclinical approaches targeting TEM8 with TEM8 blocking antibodies, TEM8-MMAE antibody-drug conjugates, and TEM8 CAR T cells have demonstrated remarkably low toxicities.7 8 12
Natural killer (NK) cells are a subset of lymphocytes with potent cytolytic activity against virally infected and malignantly transformed cells. They express activating and inhibitory receptors whose signals are integrated to determine NK cell killing.13 NK cell activity is also regulated by cytokines, particularly interleukin-2 (IL-2) and interleukin-15 (IL-15), that stimulate NK cell proliferation and activation.14 Numerous therapies have used NK cells to combat cancer, including monoclonal antibodies, allogeneic NK cells, and cytokine therapy with recombinant IL-15 and IL-15 superagonist complexes.15–18 Our group has developed a drug platform—Tri-specific Killer Engager (TriKE)—composed of three arms that form a cytolytic bridge between NK cell and target: a CD16-agonizing nanobody stimulating NK cell degranulation; an IL-15 moiety inducing NK proliferation and activation; and an antigen-specific single-chain variable fragment (scFv) mediating antigen-specific target cell recognition. In this study, we describe a novel TriKE—hereafter, named ‘cam1615TEM8’—targeting NK cells against TEM8-expressing tumor and tumor stroma.
Materials and methods
cam1615TEM8 plasmid and protein production
Refer to online supplemental table 1 for a list of reagents and equipment. A hybrid gene encoding the cam1615TEM8 TriKE was cloned into the Minicircle plasmid (SBI: MN502A-1). The hybrid gene encoded a camelid anti-CD16 nanobody,19 an IL-15 moiety, an anti-TEM8 scFv from a previously described anti-TEM8 monoclonal antibody,20 and a 10x histidine tag. DNA sequencing confirmed the final plasmid (Genomics Center, University of Minnesota, Minneapolis, Minnesota, USA). The cam1615TEM8-encoding plasmid was transfected into Expi293F cells. Protein was isolated with the AKTA Avant chromatography system. Protein purity and size was determined by polyacrylamide gel electrophoresis and densitometry using ImageJ1.21 An anti-TEM8 scFv without the anti-CD16 camelid nanobody and IL-15 moiety was produced similarly.
jitc-2022-004725supp001.pdf (530.9KB, pdf)
Peripheral blood mononuclear cell donors, NK cell enrichment, and NK cell expansion
Peripheral blood mononuclear cells (PBMCs) from the blood of deidentified healthy donors were obtained, after participants gave informed consent, from Memorial Blood Centers (Minneapolis, Minnesota, USA) and used in compliance with the Committee on the Use of Human Subjects in Research (IRB# 9709 M00134) and in accordance with the Declaration of Helsinki. Cells were isolated using density-gradient centrifugation with Ficoll-Paque Premium. Whole PBMCs were used directly (‘fresh’) or were cryopreserved and thawed 1 day before use. NK cells were enriched using kits from STEMCELL Technologies. For in vivo experiments, enriched NK cells were ex vivo expanded as described, with minor modifications.22 NK cells were expanded in G-REX 6-well plates. The irradiated K562 mIL21 41BBL cells were generated and used as described.23
Cell lines and cell culture
Cell lines and culture conditions are outlined in online supplemental table 2. ANTXR1 expression data were obtained from the DepMap 20Q1 Public dataset.24 A549s transfected with green fluorescent protein and luciferase—hereafter A549/GFP/Luc—were obtained from Dr. Dan Vallera at the University of Minnesota. NucLight Red-labeled tumor cell lines were generated using IncuCyte NucLight Red lentivirus reagent.
RT-qPCR for cell line ANTXR1 expression
Samples were lysed, homogenized using QIAShredder columns, and RNA was isolated using the RNEasy Mini Kit from Qiagen. Isolated RNA was treated with DNAse I and cDNA was produced using SuperScript IV reverse transcriptase. RT-qPCR was performed using TaqMan Fast Advanced Master Mix reagents in a QuantStudio 5 Real-Time PCR system. GAPDH was used as a housekeeping gene, and the relative expression of ANTXR1 was calculated using the delta-delta Ct method. The TaqMan probes used are listed in online supplemental table 1.
Human tissue ANTXR1 expression data
mRNA expression data for ANTXR1 in various human tissues were obtained from the GeneAtlas U133A, gcrma database, which was generated using Affymetrix U133A array to profile gene expression in diverse human tissues.25 ANTXR1 data from the 220092_s probe set were adapted to create custom graphs. Original data and graphs were available through the BioGPS website (http://biogps.org).
Generation of TEM8 knockout A549 cell line
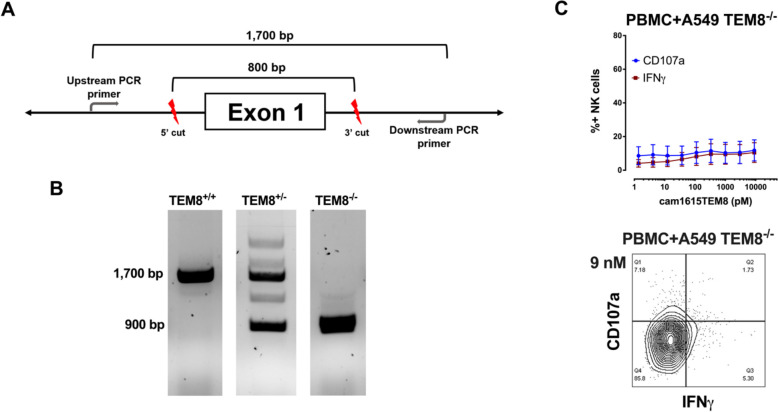
A CRISPR/Cas9 system was designed to knock out exon 1 of the ANTXR1 gene (NCBI Gene ID 84168) in the A549 cell line. Guide RNAs flanking the first exon were designed using the IDT Alt-R Custom Cas9 crRNA Design tool. crRNA, tracrRNA, and Alt-R HiFi Cas9 Nuclease V3 were transfected into A549 cells using Lipofectamine CRISPRMAX.
Antibodies and fluorescent stains
Refer to online supplemental table 3 for antibodies and other fluorescent markers.
NK cell degranulation and cytokine production assay
Fresh or frozen PBMCs were co-cultured for 5 hours with target cells (5:1 effector:target ratio) and cam1615TEM8 was serially diluted 1:3 from 9 nM to 1.37 pM (unless otherwise noted). NK cell degranulation was detected using an anti-CD107a reagent.26 GolgiPlug and GolgiStop were added for the last 4 hours of co-culture. Cells were stained with Live/Dead Fixable Near IR, anti-CD56, and anti-CD3; fixed; permeabilized; and stained with anti-interferon-γ (anti-IFNγ). All flow-based assays were run on an LSRII Flow Cytometer and analyzed with FlowJo software (Tree Star, Ashland, Orlando, USA). TEM8 blocking assays were modified by first incubating for 45 min in 1800 nM anti-TEM8 scFv before adding PBMCs and 9 nM cam1615TEM8.
Tumor spheroid killing assay
About 20,000 NucLight Red-expressing target cells were added per well to a round bottom ultra-low adhesion 96-well plate. Spheroids were allowed to form for 5 days before 100,000 NK cells enriched from fresh PBMCs were added with 3 nM cam1615TEM8 or 3 nM IL-15 for 6 days within an IncuCyte S3 (four separate donors in quintuplicate). The IncuCyte S3 software was used to calculate red fluorescence area as a measure of tumor size.
NK cell and T cell proliferation assay
PBMCs were stained with CellTrace Violet Cell Proliferation Kit and incubated for 7 days with IL-15 or cam1615TEM8 serially diluted as noted. PBMCs were then stained with Live/Dead Fixable Near IR, Annexin V, anti-CD56, and anti-CD3. ‘Proliferated’ cells divided at least once, while ‘highly proliferated’ cells divided at least twice.
NK cell and T cell pSTAT5 induction assay
PBMCs were incubated for 20 min with IL-15 or cam1615TEM8 serially diluted as noted. PBMCs were then stained with anti-CD56 and anti-CD3, fixed, permeabilized with True-Phos Perm Buffer, and stained with anti-STAT5/pY694. pSTAT5 levels in CD56+CD3 NK cells and CD56- CD3+ T cells were measured by flow cytometry.
NK-92 metabolic activity assay
NK-92s were grown in IL-2-free media for 24 hours before the start of the assay. At the start of the assay, 50,000 viable NK-92s or NK-92/CD16s were plated per well in a clear bottom 96-well plate in triplicate with IL-15 or cam1615TEM8 serially diluted 1:10 seven times to generate dose-response curves. The highest concentration of IL-15 was 150 nM, while the highest concentration of cam1615TEM8 was 1500 nM. After 48 hours, resazurin was added and incubation resumed for 4 hours, then resorufin (the fluorescent reduction product of resazurin) fluorescence at 590 nm was measured.27 A four-parameter dose-response curve of resorufin fluorescence versus log(treatment) was generated to determine the EC50 value for the IL-15-mediated metabolic activity.
A549 in vivo assay
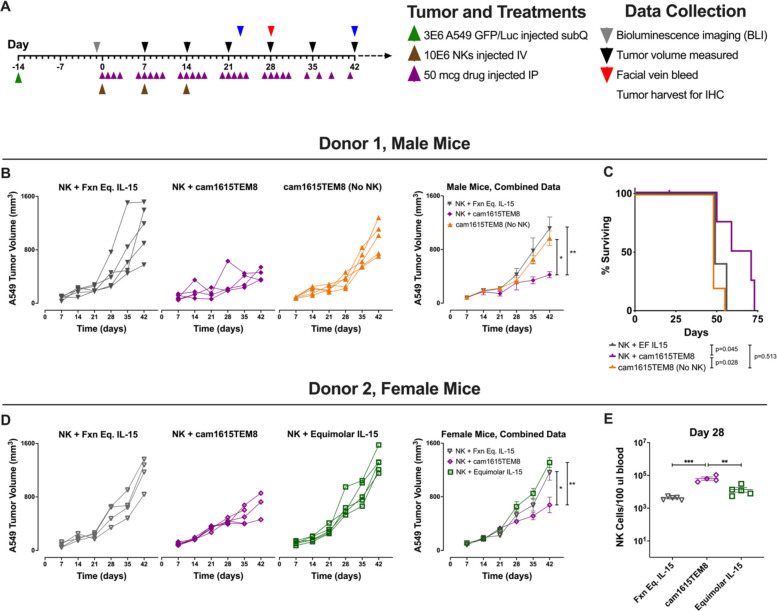
The in vivo study was conducted in accordance with the Institutional Animal Care and Use Committee at the University of Minnesota (IACUC# 1908-37330A). 3E6 A549/GFP/Luc in Matrigel diluted 3:2 with ice cold phosphate buffered saline (PBS) were injected subcutaneously over the left ribcage of 8–12 week old male or female NOD scid gamma (NSG) mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) (day −14). The tumors were allowed to engraft for 13 days, then initial tumor burden was assessed by bioluminescence imaging on day −1 with a Xenogen Ivis Spectrum imaging system following intraperitoneal luciferin injection to assign five male mice and/or five female mice to treatment groups with equal tumor burden. Once mice were assigned to groups, no mice were excluded from experiments; exclusions before this time were related to inappropriate tumor engraftment. Treatment procedures for each group were the same.
On days 0, 7, and 14, 10E6 ex vivo expanded NK cells were injected via tail vein intravenously. Fifty micrograms of cam1615TEM8 or functionally equivalent IL-15 was injected intraperitoneally five times per week for 4 weeks, then three times per week thereafter. Tumor length and width measurements were determined using a digital caliper. Tumor volume was calculated as length*width2/2. Mice that reached a tumor length of more than 2 cm or had an ulceration of the tumor were sacrificed and counted as dead for survival studies. On day 28, mice were bled and red blood cells (RBCs) from 100 μL of blood were lysed using ACK lysing buffer. Samples were stained with anti-CD56, anti-CD3, anti-human CD45, and anti-mouse CD45 and evaluated by flow cytometry to quantify human NK cells.
Tumors for histology were excised, placed in 10% formalin for 48 hours, and then placed in 70% ethanol. Tumor samples were embedded into paraffin and stained with H&E, anti-TEM8, anti-CD31, and anti-Granzyme B by the Biological Materials Procurement Network (BioNet) at the University of Minnesota. Whole slide digital scans were created with Zeiss Axio Scan.Z1 and analyzed using QuPath V.0.2.3.28 Day 23 stained samples were inspected for the seven areas of the highest Granzyme B+ cell density at ×240 magnification. Day 42 stained samples were inspected for the seven areas of the highest CD31+ cell density at ×120 magnification. Day 42 stained samples also were evaluated for TEM8 expression within an entire tumor section. A Granzyme B+ cell detection mask was generated with the QuPath Positive Cell Detection analysis tool. The % TEM8+ area and the % CD31+ area were quantified using image analysis with the QuPath software.
Data analysis, representation, and statistical analysis
GraphPad Prism (GraphPad Prism Software, La Jolla, California, USA) was used to generate graphs with error bars showing mean±SD (technical replicates) or SE of the mean (biological replicates) and to calculate statistical significance, indicated as *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001. GraphPad was used to calculate one-way analysis of variance with Tukey’s multiple comparison tests when comparing three or more means, t-tests (unpaired and multiple-paired) when comparing two means, log-rank test for survival, and four-parameter logistic curve to calculate the half maximal effective concentration (EC50).
Results
cam1615TEM8 Tri-specific Killer Engager (TriKE) design
The cam1615TEM8 TriKE is composed of three arms: a camelid nanobody that stimulates CD16 on NK cells, a wild-type IL-15 moiety that stimulates IL-15 signaling on NK cells, and a scFv that engages TEM8 to mediate antigen-specific target cell recognition (figure 1A). cam1615TEM8 was isolated from Expi293F supernatant by a C-terminal 10x histidine tag, resulting in a highly pure protein product of about 60 kDa (figure 1B).
Figure 1.
cam1615TEM8 Tri-specific Killer Engager (TriKE) layout, isolation, and induction of natural killer (NK) cell degranulation and cytokine production against TEM8+ tumor cell lines. (A) Schematic of cam1615TEM8 TriKE. (B) Representative polyacrylamide gel of cam1615TEM8 isolated from Expi293F supernatant and corresponding ImageJ densitometry plot used to calculate purity. Green arrows indicate desired product at approximately 60 kDa and maroon arrows indicate impurities. (C, D) cam1615TEM8-mediated NK cell degranulation and interferon-γ (IFNγ) production dose-response curves, and representative CD107a versus IFNγ plots of the highest concentration tested (9 nM), (C) without target cells (n=4), (D) against TEM8+ targets, A549 (n=7) and DU145 (n=5), and against TEM8- targets, HT-29 (n=5) and Raji (n=4). (E, F) Effect of TEM8 blocking with anti-TEM8 single-chain variable fragment (scFv) on NK cell degranulation (E) and IFNγ production (F) for TEM8+ and TEM8- tumor cell lines (n=4). PBMCs, peripheral blood mononuclear cells.
ANTXR1 expression in human tissues
TEM8 expression, encoded by the ANTXR1 gene, has been described on human tumor cells8–11 and tumor stromal cells—endothelial cells, fibroblasts, pericytes.6–8 It is seemingly absent from healthy human tissues and not expressed or required during normal angiogenesis.6–8 ANTXR1 expression data from human tissues were obtained from the GeneAtlas U133A, gcrma database, which revealed ANTXR1 expression highest in muscle cells—cardiomyocytes, smooth muscle cells, and skeletal muscle cells (online supplemental figure 1). ANTXR1 expression levels in most tissues were similar to the Raji (Burkitt’s lymphoma) cell line.
cam1615TEM8 induces dose-dependent NK cell function against TEM8+ tumor targets
PBMCs were incubated with target cells at a range of cam1615TEM8 concentrations to evaluate cam1615TEM8-mediated NK cell degranulation (CD107a) and inflammatory cytokine (IFNγ) production.26 RT-qPCR of various cancer cell lines revealed the A549 (non-small cell lung) and DU145 (prostate) cell lines highly expressed ANTXR1 and were designated TEM8+ cell lines; the HT-29 (colon) and Raji (Burkitt’s lymphoma) cell lines did not express detectable levels of ANTXR1 and were designated TEM8- cell lines (online supplemental figure 2). cam1615TEM8 induced no NK cell degranulation or IFNγ production in the absence of target cells (figure 1C). cam1615TEM8 induced strong NK cell degranulation and IFNγ production against A549 and DU145 TEM8+ tumor cell lines, but almost no NK cell degranulation or cytokine production against HT-29 or Raji TEM8- tumor cell lines (figure 1D), indicating that target cell TEM8 expression is necessary for cam1615TEM8-mediated NK cell function. TEM8 blockade, through pre-incubation with excess anti-TEM8 scFv, resulted in significantly less cam1615TEM8-induced NK cell degranulation and IFNγ production against TEM8+ lines, whereas no significant reduction in NK degranulation or IFNγ production was seen against TEM8- lines (figure 1E, F). Finally, an A549 TEM8 knockout cell line was created using a CRISPR/Cas9 system designed to knockout exon 1 of ANTXR1 (figure 2A). The knockout status of single cell clones was tested by PCR, and a homozygous knockout clone was identified—hereafter, A549 TEM8-/- (figure 2B). Knocking out TEM8 eliminated cam1615TEM8-mediated NK cell function against A549s (figure 2C), further demonstrating specificity.
Figure 2.
Knocking out ANTXR1 on A549s abolishes cam1615TEM8-mediated natural killer (NK) cell activity. (A) A CRISPR/Cas9 system was designed to knock out exon 1 of the ANTXR1 gene. (B) Knockout status of single cell clones was determined by PCR, where the wild-type ANTXR1 gene creates a product of ~1700 base pairs, while amplification of the exon 1 knockout creates a product of ~900 base pairs. Representative images of the wild-type homozygote (TEM8+/+), wild-type/knockout heterozygote (TEM8+/-), and knockout homozygote (TEM8-/-) PCR products. (C) cam1615TEM8-mediated NK cell degranulation and interferon-γ (IFNγ) production dose-response curves, and representative CD107a versus IFNγ plot of the highest concentration tested (9 nM), against A549 TEM8-/- (n=4). PBMCs, peripheral blood mononuclear cells.
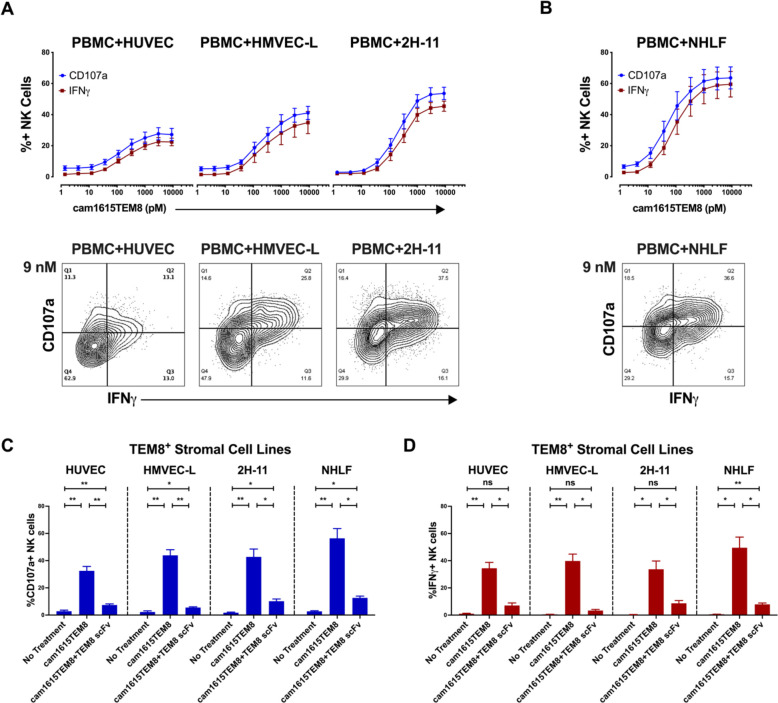
cam1615TEM8 induces NK cell function against TEM8+ endothelial cells and fibroblasts
TEM8 has been identified on multiple stromal cells within the tumor microenvironment of diverse human cancer types.6–8 For unknown reasons, TEM8 is upregulated on some stromal cell lines in vitro, thereby providing an opportunity to study NK cell-mediated toxicity against stromal cells that have been shown to express TEM8 within the tumor microenvironment. While NK cell cytotoxicity toward tumor cells is well described, their ability to target non-cancerous stromal cells is less clear. A previous study demonstrated that the human umbilical vein endothelial cell (HUVEC) line expresses low levels of TEM8.29 Consistent with this report, we detected ANTXR1 expression in HUVECs, but at lower levels than the TEM8+ tumor cell lines. Human microvascular endothelial cells from lung (HMVEC-L) expressed slightly lower levels of ANTXR1 (online supplemental figure 2). cam1615TEM8 stimulated modest NK cell degranulation and IFNγ production against HUVECs (figure 3A). cam1615TEM8 induced more robust NK cell degranulation and IFNγ production against HMVEC-L.
Figure 3.
cam1615TEM8 induction of natural killer (NK) cell function against TEM8+ endothelial and fibroblast lines. (A, B) cam1615TEM8-mediated NK cell degranulation and interferon-γ (IFNγ) production dose-response curves, and representative CD107a versus IFNγ plots of the highest concentration tested (9 nM), against (A) TEM8+ endothelial cell lines: human umbilical vein endothelial cells (HUVECs) (n=7), human microvascular endothelial cells from lung (HMVEC-Ls) (n=4), and the immortalized mouse endothelial line 2 H-11 (n=4); and (B) TEM8+ fibroblasts: normal human lung fibroblasts (NHLFs) (n=5). (C, D) Effect of TEM8 blocking with anti-TEM8 single-chain variable fragment (scFv) on NK cell degranulation (C) and IFNγ production (D) for TEM8+ endothelial and fibroblast lines (n=4). PBMCs, peripheral blood mononuclear cells.
Mouse TEM8 is highly homologous to human TEM8, and the TEM8-engaging scFv sequence within cam1615TEM8 was reported to bind both human and mouse.8 30 2H-11 cells are a murine model for tumor endothelial cells because they display activity in endothelial function assays and express murine homologs for human endothelial markers, including TEM8.31 2H-11s expressed low levels of ANTXR1, even compared with HUVECs (online supplemental figure 2). cam1615TEM8 still stimulated strong NK cell degranulation and IFNγ production against 2H-11s (figure 3A), indicating that activity against murine tumor stroma could be targeted in a xenogeneic model.
TEM8 is also expressed by cancer-associated fibroblasts, which contribute to tumor growth and impair immune activity.2 8 Normal human lung fibroblasts (NHLFs) expressed higher levels of ANTXR1 than the endothelial cell lines tested (online supplemental figure 2). cam1615TEM8 potently induced NK cell degranulation and IFNγ production against NHLFs (figure 3B), indicating that cam1615TEM8 can target NK cells against multiple TEM8+ stromal cell types present within the tumor microenvironment. TEM8 blocking resulted in significantly less cam1615TEM8-induced NK cell function against these cell lines (figure 3C, D).
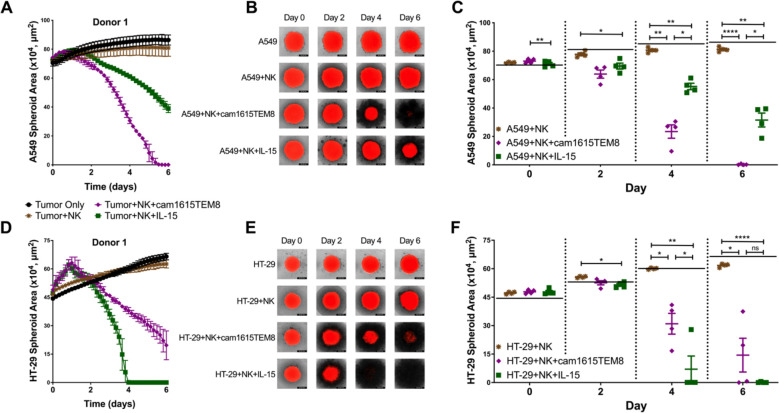
cam1615TEM8 stimulates NK cell killing of TEM8+ tumor spheroids
Since cam1615TEM8 preferentially induces NK cell function against TEM8+ tumor cell lines, it was evaluated if this would translate to killing within the more stringent and physiologic spheroid model. NucLight Red-expressing A549 (TEM8+) spheroids were formed and treated with NK cells alone or in the presence of cam1615TEM8 or IL-15. cam1615TEM8 resulted in more rapid and complete A549 spheroid elimination than IL-15 (figure 4A–C). In contrast, cam1615TEM8 did not improve NK cell-mediated killing of NucLight Red-expressing HT-29 (TEM8-) spheroids compared with IL-15 (figure 4D–F).
Figure 4.
cam1615TEM8 stimulates superior natural killer (NK) cell-mediated killing of TEM8+ tumor spheroids. (A, D) NK cells from a representative donor killing A549 and HT-29 NucLight Red-expressing spheroids alone or in the presence of 3 nM cam1615TEM8 or interleukin-15 (IL-15). (B, E) Overlaid brightfield and red fluorescence images of A549 and HT-29 spheroids co-cultured with the representative donor for each treatment. (C, F) Pooled data of A549 and HT-29 tumor spheroid areas on days 0, 2, 4, and 6. The horizontal black bar at each timepoint represents the average spheroid size of A549 or HT-29 only wells.
The camelid anti-CD16 nanobody within cam1615TEM8 TriKE enhances IL-15 signaling specifically in NK cells
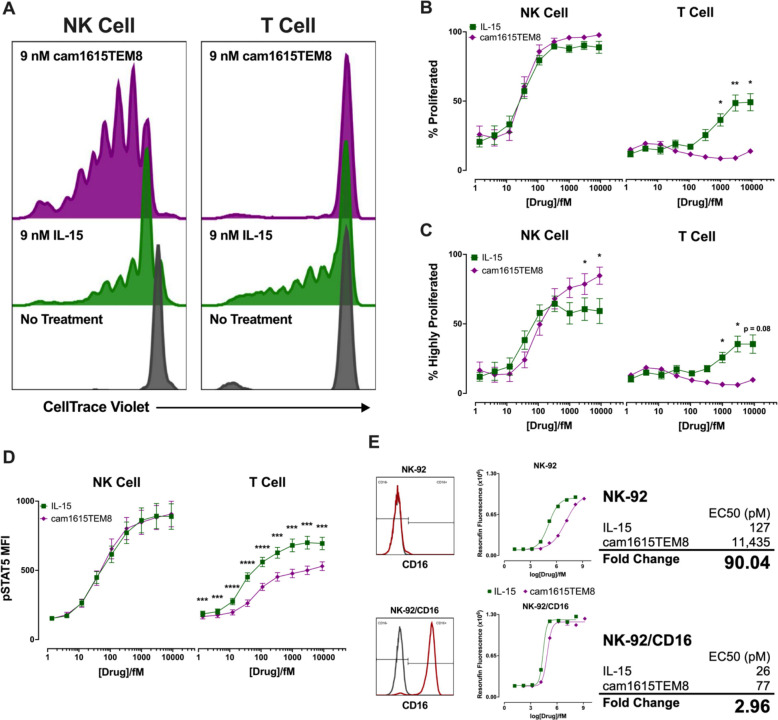
IL-15 stimulates NK cell activation and survival by signaling through the IL-15 receptor complex.14 The cam1615TEM8 IL-15 moiety was evaluated in a proliferation assay. Briefly, whole PBMCs were stained with CellTrace Violet and incubated with IL-15 or cam1615TEM8 for 7 days, at which time proliferation of live NK and T cells was assessed by flow cytometry (figure 5A). NK cell proliferation was similar between IL-15 and cam1615TEM8 at all concentrations, with both treatments inducing strong, dose-dependent NK proliferation. However, cam1615TEM8 stimulated substantially less T cell proliferation when compared with IL-15 (figure 5B). The cam1615TEM8 induced more ‘highly proliferated’ NK cells—defined as two or more cell divisions—than IL-15, while IL-15 induced more highly proliferated T cells than cam1615TEM8 (figure 5C). To further understand the differential in signaling from cam1615TEM8, STAT5 phosphorylation was evaluated. IL-15 and cam1615TEM8 induced similar levels of pSTAT5 in NK cells, whereas IL-15 stimulated higher levels of pSTAT5 than cam1615TEM8 in T cells (figure 5D).
Figure 5.
cam1615TEM8 selectively stimulates proliferation and pSTAT5 induction in natural killer (NK) cells rather than T cells. (A) Representative proliferation graphs of CellTrace Violet stained NK cells and T cells in response to 7-day incubation without treatment, in 9 nM interleukin-15 (IL-15), and in 9 nM cam1615TEM8. (B, C) IL-15 and cam1615TEM8-mediated NK and T cell proliferation dose-response curves. (B) ‘Proliferated’ cells were defined as having proliferated at least once. (C) ‘Highly proliferated’ cells were defined as having proliferated at least twice. (D) pSTAT5 induction in NK and T cells following incubation in IL-15 or cam1615TEM8. (E) NK-92 metabolic assays in NK-92s and NK-92s expressing CD16 (NK-92/CD16). NK-92 metabolic activity was measured by resorufin fluorescence. Also shown are flow cytometry graphs of CD16 expression (red) versus control (gray) for NK-92 and NK-92/CD16. A four-parameter logistic curve was generated to calculate the EC50s of IL-15- and cam1615TEM8-mediated metabolic activity in NK-92 and NK-92/CD16. The ‘fold change’ of cam1615TEM8 versus IL-15-stimulated metabolic activity was calculated as cam1615TEM8 EC50/IL-15 EC50.
To understand the differential in signaling between the NK cells and T cells, we wanted to determine if: (1) the IL-15 moiety in the TriKE is weaker (due to steric hindrance within the TriKE scaffold) resulting in less T cell proliferation, and (2) if the anti-CD16 nanobody improved IL-15 signaling in CD16-expressing cells through compensation of the steric deficits. To reduce confounding variables, a system measuring IL-15 stimulated metabolic activity in the NK-92 cell line was employed. NK-92s are an NK cell-like cell line that signal through IL-15, but the native line does not express CD16.32 Native NK-92s were treated with a range of concentrations of IL-15 or cam1615TEM8, and after 2 days’ metabolic activity, measured by resorufin fluorescence, was determined to generate a dose-response curve for both treatments. The EC50 value of cam1615TEM8-stimulated metabolic activity in native NK-92s was ~90 times higher than IL-15, confirming that the nanobody and scFv surrounding the IL-15 moiety hindered its activity (figure 5E). Therefore, a concentration of monomeric IL-15 1/90th that of cam1615TEM8 has an equal amount of IL-15 function—this ratio is designated as ‘functionally equivalent’ IL-15 (Fxn Eq. IL-15). When tested in NK-92s transfected to stably express CD16 (NK-92/CD16),32 the EC50 of cam1615TEM8-stimulated metabolic activity was only ~3 times higher than IL-15, indicating that the disparity in signaling between IL-15 and cam1615TEM8 is greatly reduced when cells express CD16, thus explaining the differential between NK and T cell signaling (figure 5E).
cam1615TEM8 stimulates in vivo NK cell control of A549 tumors
Based on the TEM8-specific NK cell targeting of tumor and stroma in vitro, cam1615TEM8’s ability to stimulate NK cell targeting of TEM8-expressing tumor and tumor endothelium in vivo was evaluated. Of critical importance for these studies, mouse TEM8 is highly homologous to human TEM8, absent or very lowly expressed on normal mouse tissues, and upregulated on the vasculature of human tumor xenografts in mice.30 Moreover, figure 3A shows cam1615TEM8 activates NK cells against murine endothelial cells, indicating that targeting of endothelium can be modeled in the xenograft system despite absence of human stroma.
A549/GFP/Luc tumors (TEM8+) were engrafted in NSG mice, followed by injection of expanded NK cells 2 weeks later, and treatment with cam1615TEM8 or functionally equivalent IL-15. Mice were assessed for a number of parameters during the course of several studies with this setup (figure 6A). On day 42, tumors from mice treated with NK cells and cam1615TEM8 were significantly smaller than tumors treated with NK cells and functionally equivalent IL-15 (figure 6B, D). cam1615TEM8 was also tested in the absence of NK cells to control for NK cell-independent effects, as TEM8 blocking antibodies have been shown to reduce human tumor xenograft growth in previous studies.7 NK cells and cam1615TEM8 resulted in significantly smaller tumors than cam1615TEM8 alone, demonstrating that the reduction in tumor volume was NK cell dependent (figure 6B). Mice treated with NK cells and cam1615TEM8 also survived longer than mice treated with NK cells and functionally equivalent IL-15 or with cam1615TEM8 in the absence of NK cells (figure 6C). Equimolar IL-15 was also tested to determine whether NK cells with higher doses of IL-15 demonstrated improved tumor control, but NK cells and cam1615TEM8 was still superior and resulted in significantly smaller tumors compared with NK cells and equimolar IL-15 (figure 6D).
Figure 6.
cam1615TEM8 slows tumor growth and enhances natural killer (NK) cell expansion compared with functionally equivalent interleukin-15 (IL-15). (A) Treatment schema. (B, D) Left: Tumor growth curves from day 7 to day 42 for individual mice from two trials. Right: Combined tumor volume data depicting the mean tumor volume for all mice treated in each treatment group of the respective trial. Tumor volumes were compared at day 42. (C) Kaplan-Meier survival curves for mice receiving each treatment. One mouse in the ‘NK+cam1615TEM8’ treatment group was censored for a death unrelated to tumor burden. (E) Human NK cell count in 100 μL of mouse blood on day 28 for Donor 2. ‘Fxn Eq. IL-15’ indicates ‘functionally equivalent IL-15’.
Circulating human NK cells within mouse peripheral blood were quantified on day 28. Mice treated with cam1615TEM8 had greater NK cell numbers than mice treated with functionally equivalent IL-15 in two separate experiments. cam1615TEM8 mediated increased NK cell numbers compared with equimolar IL-15, while the two IL-15 doses did not differ from each other (figure 6E). In summary, cam1615TEM8 stimulated greater in vivo NK cell number than IL-15 and superior, NK cell-dependent tumor control than IL-15 or cam1615TEM8 in the absence of NK cells.
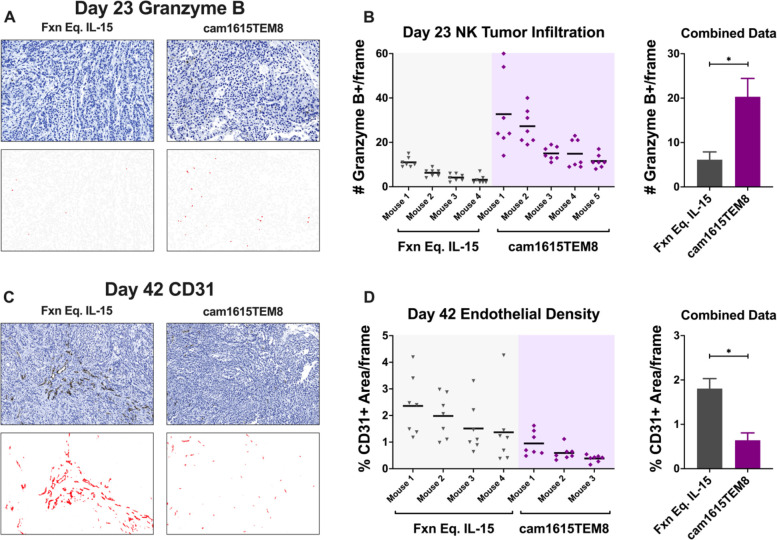
cam1615TEM8 stimulates NK cell infiltration and reduction of endothelial density in A549 tumors
To determine whether cam1615TEM8 would also stimulate enhanced NK cell tumor infiltration in the A549 xenograft model, day 23 tumors were harvested and analyzed by immunohistochemistry for human Granzyme B to detect human NK cells (the only cells containing this human marker within this model). A positive cell mask was developed using QuPath to quantify tumor-infiltrating human NK cells in response to cam1615TEM8 or functionally equivalent IL-15 (figure 7A). cam1615TEM8 treatment resulted in increased NK cell tumor infiltration compared with functionally equivalent IL-15 (figure 7B).
Figure 7.
cam1615TEM8 increases natural killer (NK) tumor infiltration and reduces endothelial density compared with functionally equivalent interleukin-15 (IL-15). A549 tumor was engrafted as outlined and mice were treated with NK cells and cam1615TEM8 or functionally equivalent IL-15. (A) Top: Representative anti-human Granzyme B stained tumor on day 23 at ×240 magnification. Bottom: The Granzyme B mask used for NK cell identification generated with QuPath Positive Cell Detection software. (B) Left: Dot plots of Granzyme B+ cell counts from seven frames in individual tumors. Right: Combined data for tumors from all mice. (C, D) Analogous data to A and B but with % anti-mouse CD31+ staining area at ×120 magnification on day 42 to evaluate endothelial density. ‘Fxn Eq. IL-15’ indicates ‘functionally equivalent IL-15’.
Day 42 tumors were harvested and analyzed by immunohistochemistry for human and mouse TEM8. Tumors treated with NK cells and functionally equivalent IL-15 and NK cells and cam1615TEM8 demonstrated diffuse TEM8 expression, with select regions of high TEM8 density (online supplemental figure 3A). Unfortunately, since A549 tumor cells express TEM8, it was difficult to discern TEM8 expression in tumor versus stroma. QuPath was used to quantify the percent TEM8+ in digital images of sections of whole tumor. The percent TEM8+ area within tumor sections did not differ between tumors treated with NK cells and cam1716TEM8 or with NK cells and functionally equivalent IL-15 (online supplemental figure 3B), indicating that the NK cell-mediated reduction in tumor growth with cam1615TEM8 did not affect global TEM8 expression, indicating that antigen escape is not operant in this model system.
To determine whether cam1615TEM8 would stimulate an NK cell-mediated reduction in tumor endothelial density, indicating that NK cells were being directed to the mouse-derived TEM8 expressed on tumor endothelium,7 day 42 tumors were harvested and analyzed by immunohistochemistry for mouse CD31. QuPath was used to quantify the percent CD31+ area in digital images of the areas with the highest endothelial density (figure 7C). Tumors treated with NK cells and cam1615TEM8 had significantly lower endothelial density compared with tumors treated with NK cells and functionally equivalent IL-15 (figure 7D). These data indicate that cam1615TEM8 decreased endothelial density by about threefold, suggesting that TEM8-targeted NK cells can kill both cancer and the cancerized stroma, a dual mechanism felt to be important in solid tumors.
Discussion
In this study, we describe a novel TriKE—cam1615TEM8—that targets NK cells to TEM8, an emerging cancer target that has been described on tumor and tumor stromal cells. We previously reported TriKEs created against CD19 for acute lymphoblastic leukemia (ALL) and chronic lymphocytic leukemia (CLL); CD33 and CLEC12A for AML; and CD133, EpCAM, B7-H3, and HER2 for various solid tumors.33–39 TriKEs differ from therapeutic antibodies that can mediate ADCC in that they mediate ADCC but also trigger priming and expansion through specific targeted delivery of IL-15 to NK cells. The priming not only enhances ADCC but also induces better natural cytotoxicity to amplify tumor antigen independent killing. The expansion enhances the number of effector NK cells capable of attacking the tumor. Therapeutic antibodies do not trigger proliferation of NK cells. Moreover, previous work has demonstrated that directly engaging CD16 on NK cells as with a CD16 targeting scFv can be more potent than CD16 engagement of Fc fragments from monoclonal antibodies.40 We previously demonstrated that the camelid CD16 engaging moiety within cam1615TEM8 provides further potency than CD16 engaging scFv.19 cam1615TEM8 specifically stimulated NK cell function better than IL-15 against TEM8-expressing A549 and DU145 cell lines and induced rapid killing of TEM8+ A549 tumor spheroids, which better mimic features of solid tumors.41 The TriKE also induced better A549 tumor control in a xenogeneic mouse model. Therefore, CD16 agonism and TEM8 engagement by cam1615TEM8 stimulates NK cell killing beyond the cytokine-mediated NK cell activation by IL-15.
Previously described TriKEs only targeted NK cells to tumor cells, but TEM8 has been described on tumor endothelial cells and other tumor stromal cells.6–8 NK cell cancer immunotherapies for solid tumors need to overcome the cancer-promoting stromal cells within the tumor microenvironment, including cancer-associated fibroblasts, endothelial cells, pericytes, and immune cells.1 2 42 43 Previous studies, in melanoma and breast cancer, have implicated tumor stromal cells like fibroblasts in intratumoral NK cell dysfunction.43 44 Therefore, targeting tumor-promoting stromal cells may promote NK cell function rather than dysfunction within the tumor microenvironment. cam1615TEM8-induced NK cells function against TEM8+ endothelial cells and fibroblasts, showing NK cell immunotherapy can target tumor stroma. The exact function of TEM8 on tumor or tumor stroma is unknown. It is similarly unclear why normal stromal cell lines such as HUVECs or NHLFs express this molecule in vitro. Some previous research has demonstrated that TEM8 is involved in in vitro endothelial cell migration.29 This may suggest that TEM8 is involved in endothelial migration into tumors during angiogenesis and tumor cell metastasis in vivo, a theory supported by previous studies that demonstrated impaired tumor growth and reduced tumor endothelial density within tumors grown in TEM8 knockout mice.7 Nevertheless, further research is needed to elucidate the function of this adhesion molecule within tumors.
Despite the limited clinical efficacy and toxicities of currently approved anti-angiogenic agents, this approach remains of interest because (1) angiogenesis is necessary for solid tumor growth, (2) fibroblasts and pericytes are present in many solid tumors so anti-stromal therapies may be efficacious for diverse cancer histotypes, (3) eliminating the tumor endothelium may have a significant ‘domino effect’ that kills many more cancer and tumor stromal cells deprived of oxygen and nutrients, and (4) tumor stromal cells are more genomically stable and less likely to undergo antigen loss and drug resistance than cancer cells.45 46 cam1615TEM8 targeted NK cells to both TEM8+ tumor and tumor endothelium in an A549 xenograft model, resulting in smaller tumors and decreased tumor endothelium compared with treatment with functionally equivalent IL-15. The striking decrease in endothelial density with cam1615TEM8 treatment was likely a driving factor to the reduction in tumor growth. Importantly, while previous studies have demonstrated that TEM8 blocking antibodies alone reduced tumor growth,7 cam1615TEM8 in the absence of human NK cells did not decrease tumor growth—that is, NK cells were necessary for the anti-tumor effect.
Tumors in mice treated with cam1615TEM8 had significantly more tumor-infiltrating NK cells than tumors in mice treated with functionally equivalent IL-15. Immune infiltration is generally considered a positive prognostic marker for immunotherapy outcomes.47 Most of the research of tumor-infiltrating lymphocytes has focused on tumor-infiltrating T cells, and the importance of tumor-infiltrating NK cells is poorly understood. Multiple studies have shown that a greater NK cell tumor infiltration correlated with a better prognosis in diverse cancer types, but the effect of tumor-infiltrating NK cells likely depends on NK cell expression of activating receptors, cancer cell expression of NK cell activating ligands, and the presence or absence of local immunomodulatory cells and molecules.48 Accordingly, cam1615TEM8 is an alluring option to increase tumor immunoreactivity by increasing the number of tumor-infiltrating NK cells, activating the infiltrating NK cells through the anti-CD16 nanobody and IL-15 moiety, and stimulating NK cell killing of immunosuppressive stromal cell types within the microenvironment.
Novel cancer immunotherapies must consider the off-tumor and systemic effects of immune activation and the resultant toxicities. Trials evaluating the systemic administration of unmodified cytokines, although modestly effective in a subset of patients, were limited by toxicities.49 Accordingly, selective delivery of immune activating cytokines may permit larger, more effective doses that result in greater tumor rejection and less systemic inflammation. The CD16 nanobody within cam1615TEM8 specifically enhanced signaling of the IL-15 moiety in CD16-expressing NK cells, resulting in a dose-dependent stimulation of NK cell proliferation without T cell proliferation. The exact mechanism of this preferential NK cell proliferation is unclear, but the preferential IL-15 stimulation of NK cells and relative absence of IL-15 stimulation of T cells may be an effective method of reducing dose-limiting toxicities.50
Most preclinical approaches targeting TEM8 with TEM8 blocking antibodies, TEM8-MMAE antibody-drug conjugates, and TEM8 CAR T cells have been well tolerated.7 8 12 This is consistent with studies that have demonstrated TEM8 is not expressed on healthy tissue or during normal angiogenesis.6 8 Using a publicly available gene expression database, we found the TEM8-expressing gene ANTXR1 to be expressed at very low levels in all tissues tested. The tissues with the highest ANTXR1 expression like heart muscle and skeletal muscle only slightly surpassed the ANTXR1 expression in the Burkitt’s lymphoma Raji cell line. Raji’s TEM8 levels were so low that cam1615TEM8 did not stimulate NK cell mediated degranulation and inflammatory cytokine production against them. Therefore, TEM8 expression by healthy tissues may be sufficiently low that cam1615TEM8 may not stimulate off-tumor toxicities. Importantly, while most preclinical approached targeting TEM8 have not encountered toxicities, one study of TEM8 targeting CAR T cells found that TEM8 targeting CAR T cells rapidly disappeared from circulation, causing lung and spleen inflammation and death in their in vivo model.51 Reasons for why TEM8 targeting CAR T cells have markedly disparate toxicity profiles in vivo are unclear, but it demonstrates that clinical translation of any TEM8 targeting therapy like cam1615TEM8 would require careful monitoring for toxicities. Notably, cam1615TEM8 did not result in fatal toxicities in our in vivo model, rather human NK cells and cam1615TEM8 prolonged survival compared with mice treated with NK cells and functionally equivalent IL-15.
In summary, this study demonstrates that cam1615TEM8 can mediate NK cell targeting of tumor and its stromal support system and architecture. cam1615TEM8 selectively stimulated IL-15 signaling in and proliferation of NK cells, which may reduce toxicities compared with current anti-tumor and anti-angiogenic therapies. These data suggest that cam1615TEM8 may be a valuable and novel anti-tumor, anti-stroma, and anti-angiogenic cancer therapy for patients with solid tumors.
Acknowledgments
This research received immunohistochemistry assistance from the University of Minnesota’s Biorepository and Laboratory Services program and was supported by the National Institutes of Health’s National Center for Advancing Translational Sciences, grant UL1TR002494. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health’s National Center for Advancing Translational Sciences.
Footnotes
Contributors: Wrote and revised manuscript: MFK, JSM, and MF. Conceived and designed studies and analysis: MFK, TRL, MAG, JSM, and MF. Collected data: MFK, LB, RH, MK, BK, YS, PH, and JTW. MF is the guarantor for the study.
Funding: This work was supported by NIH/NCI grants P01-CA111412 and R35 CA197292.
Competing interests: MF and JSM receive research support, consulting honoraria, and stock and, with the University of Minnesota, are shared owners of the TriKE technology licensed by the University to GT Biopharma, Inc. This relationship has been reviewed and managed by the University of Minnesota in accordance with its conflict of interest policies. No GTB funds were used in this research study.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by Committee on the Use of Human Subjects in Research at the University of Minnesota. IRB# 9709M00134. Note that all samples used were obtained deidentified from a local blood bank but usage was still approved under our IRB. Participants gave informed consent to participate in the study before taking part.
References
- 1.Dvorak HF. Tumors: wounds that do not heal. New England Journal of Medicine 1986;315:1650–9. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer 2016;16:582–98. 10.1038/nrc.2016.73 [DOI] [PubMed] [Google Scholar]
- 3.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature 2000;407:249–57. 10.1038/35025220 [DOI] [PubMed] [Google Scholar]
- 4.Chen HX, Cleck JN. Adverse effects of anticancer agents that target the VEGF pathway. Nat Rev Clin Oncol 2009;6:465–77. 10.1038/nrclinonc.2009.94 [DOI] [PubMed] [Google Scholar]
- 5.Tran E, Chinnasamy D, Yu Z, et al. Immune targeting of fibroblast activation protein triggers recognition of multipotent bone marrow stromal cells and cachexia. J Exp Med 2013;210:1125–35. 10.1084/jem.20130110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.St Croix B, Rago C, Velculescu V, et al. Genes expressed in human tumor endothelium. Science 2000;289:1197–202. 10.1126/science.289.5482.1197 [DOI] [PubMed] [Google Scholar]
- 7.Chaudhary A, Hilton MB, Seaman S, et al. TEM8/ANTXR1 blockade inhibits pathological angiogenesis and potentiates tumoricidal responses against multiple cancer types. Cancer Cell 2012;21:212–26. 10.1016/j.ccr.2012.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szot C, Saha S, Zhang XM, et al. Tumor stroma-targeted antibody-drug conjugate triggers localized anticancer drug release. J Clin Invest 2018;128:2927–43. 10.1172/JCI120481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen D, Bhat-Nakshatri P, Goswami C, et al. ANTXR1, a stem cell-enriched functional biomarker, connects collagen signaling to cancer stem-like cells and metastasis in breast cancer. Cancer Res 2013;73:5821–33. 10.1158/0008-5472.CAN-13-1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies G, Rmali KA, Watkins G, et al. Elevated levels of tumour endothelial marker-8 in human breast cancer and its clinical significance. Int J Oncol 2006;29:1311. [PubMed] [Google Scholar]
- 11.Alcalá S, Martinelli P, Hermann PC, et al. The anthrax toxin receptor 1 (ANTXR1) is enriched in pancreatic cancer stem cells derived from primary tumor cultures. Stem Cells Int 2019;2019:1–13. 10.1155/2019/1378639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byrd TT, Fousek K, Pignata A, et al. TEM8/ANTXR1-Specific CAR T cells as a targeted therapy for triple-negative breast cancer. Cancer Res 2018;78:489. 10.1158/0008-5472.CAN-16-1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanier LL. NK cell recognition. Annu Rev Immunol 2005;23:225–74. 10.1146/annurev.immunol.23.021704.115526 [DOI] [PubMed] [Google Scholar]
- 14.Meazza R, Azzarone B, Orengo AM, et al. Role of common-gamma chain cytokines in NK cell development and function: perspectives for immunotherapy. J Biomed Biotechnol 2011;2011:861920. 10.1155/2011/861920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis ZB, Felices M, Verneris MR, et al. Natural killer cell adoptive transfer therapy: exploiting the first line of defense against cancer. Cancer J 2015;21:486–91. 10.1097/PPO.0000000000000156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alderson KL, Sondel PM. Clinical cancer therapy by NK cells via antibody-dependent cell-mediated cytotoxicity. J Biomed Biotechnol 2011;2011:1–7. 10.1155/2011/379123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conlon KC, Lugli E, Welles HC, et al. Redistribution, hyperproliferation, activation of natural killer cells and CD8 T cells, and cytokine production during first-in-human clinical trial of recombinant human interleukin-15 in patients with cancer. J Clin Oncol 2015;33:74–82. 10.1200/JCO.2014.57.3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wrangle JM, Velcheti V, Patel MR, et al. ALT-803, an IL-15 superagonist, in combination with nivolumab in patients with metastatic non-small cell lung cancer: a non-randomised, open-label, phase 1B trial. Lancet Oncol 2018;19:694–704. 10.1016/S1470-2045(18)30148-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felices M, Lenvik TR, Kodal B, et al. Potent cytolytic activity and specific IL15 delivery in a second-generation trispecific killer Engager. Cancer Immunol Res 2020;8:1139. 10.1158/2326-6066.CIR-19-0837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dimitrov D, Zhu Z, B SC. 2016. United States of America patent 9765142 2016.
- 21.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods 2012;9:671–5. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Somanchi SS, Senyukov VV, Denman CJ, et al. Expansion, purification, and functional assessment of human peripheral blood NK cells. J Vis Exp 2011;48:e2540. 10.3791/2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu H, Blum RH, Bjordahl R, et al. Pluripotent stem cell-derived NK cells with high-affinity noncleavable CD16a mediate improved antitumor activity. Blood 2020;135:399–410. 10.1182/blood.2019000621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghandi M, Huang FW, Jané-Valbuena J, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature 2019;569:503–8. 10.1038/s41586-019-1186-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su AI, Wiltshire T, Batalov S, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A 2004;101:6062–7. 10.1073/pnas.0400782101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bryceson YT, March ME, Barber DF, et al. Cytolytic granule polarization and degranulation controlled by different receptors in resting NK cells. J Exp Med 2005;202:1001–12. 10.1084/jem.20051143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borra RC, Lotufo MA, Gagioti SM, et al. A simple method to measure cell viability in proliferation and cytotoxicity assays. Braz Oral Res 2009;23:255–62. 10.1590/s1806-83242009000300006 [DOI] [PubMed] [Google Scholar]
- 28.Bankhead P, Loughrey MB, Fernández JA, et al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017;7:16878. 10.1038/s41598-017-17204-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hotchkiss KA, Basile CM, Spring SC, et al. TEM8 expression stimulates endothelial cell adhesion and migration by regulating cell-matrix interactions on collagen. Exp Cell Res 2005;305:133–44. 10.1016/j.yexcr.2004.12.025 [DOI] [PubMed] [Google Scholar]
- 30.Carson-Walter EB, Watkins DN, Nanda A, et al. Cell surface tumor endothelial markers are conserved in mice and humans. Cancer Res 2001;61:6649. [PubMed] [Google Scholar]
- 31.Walter-Yohrling J, Morgenbesser S, Rouleau C, et al. Murine endothelial cell lines as models of tumor endothelial cells. Clin Cancer Res 2004;10:2179. 10.1158/1078-0432.ccr-03-1013 [DOI] [PubMed] [Google Scholar]
- 32.Snyder KM, Hullsiek R, Mishra HK, et al. Expression of a recombinant high affinity IgG Fc receptor by engineered NK cells as a docking platform for therapeutic mAbs to target cancer cells. Front Immunol 2018;9:2873–73. 10.3389/fimmu.2018.02873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felices M, Kodal B, Hinderlie P, et al. Novel CD19-targeted TriKE restores NK cell function and proliferative capacity in CLL. Blood Adv 2019;3:897–907. 10.1182/bloodadvances.2018029371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vallera DA, Felices M, McElmurry R, et al. IL15 trispecific killer Engagers (TriKE) make natural killer cells specific to CD33+ targets while also inducing persistence, in vivo expansion, and enhanced function. Clin Cancer Res 2016;22:3440. 10.1158/1078-0432.CCR-15-2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arvindam US, van Hauten PMM, Schirm D, et al. A trispecific killer engager molecule against CLEC12A effectively induces NK-cell mediated killing of AML cells. Leukemia 2021;35:1586–96. 10.1038/s41375-020-01065-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmohl JU, Felices M, Oh F, et al. Engineering of Anti-CD133 trispecific molecule capable of inducing NK expansion and driving antibody-dependent cell-mediated cytotoxicity. Cancer Res Treat 2017;49:1140–52. 10.4143/crt.2016.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmohl JU, Felices M, Taras E, et al. Enhanced ADCC and NK cell activation of an anticarcinoma bispecific antibody by genetic insertion of a modified IL-15 cross-linker. Mol Ther 2016;24:1312–22. 10.1038/mt.2016.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vallera DA, Ferrone S, Kodal B, et al. NK-cell-mediated targeting of various solid tumors using a B7-H3 tri-specific killer engager in vitro and in vivo. Cancers 2020;12:2659. 10.3390/cancers12092659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallera DA, Oh F, Kodal B, et al. A HER2 Tri-Specific NK cell Engager mediates efficient targeting of human ovarian cancer. Cancers 2021;13. 10.3390/cancers13163994. [Epub ahead of print: 08 08 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson S, Burke S, Huang L, et al. Effector cell recruitment with novel Fv-based dual-affinity re-targeting protein leads to potent tumor cytolysis and in vivo B-cell depletion. J Mol Biol 2010;399:436–49. 10.1016/j.jmb.2010.04.001 [DOI] [PubMed] [Google Scholar]
- 41.Nunes AS, Barros AS, Costa EC, et al. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol Bioeng 2019;116:206–26. 10.1002/bit.26845 [DOI] [PubMed] [Google Scholar]
- 42.Platonova S, Cherfils-Vicini J, Damotte D, et al. Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res 2011;71:5412. 10.1158/0008-5472.CAN-10-4179 [DOI] [PubMed] [Google Scholar]
- 43.Mamessier E, Sylvain A, Thibult M-L, et al. Human breast cancer cells enhance self tolerance by promoting evasion from NK cell antitumor immunity. J Clin Invest 2011;121:3609–22. 10.1172/JCI45816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balsamo M, Scordamaglia F, Pietra G, et al. Melanoma-associated fibroblasts modulate NK cell phenotype and antitumor cytotoxicity. Proc Natl Acad Sci U S A 2009;106:20847. 10.1073/pnas.0906481106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kerbel RS. Inhibition of tumor angiogenesis as a strategy to circumvent acquired resistance to anti-cancer therapeutic agents. Bioessays 1991;13:31–6. 10.1002/bies.950130106 [DOI] [PubMed] [Google Scholar]
- 46.Boehm T, Folkman J, Browder T, et al. Antiangiogenic therapy of experimental cancer does not induce acquired drug resistance. Nature 1997;390:404–7. 10.1038/37126 [DOI] [PubMed] [Google Scholar]
- 47.Ochoa de Olza M, Navarro Rodrigo B, Zimmermann S, et al. Turning up the heat on non-immunoreactive tumours: opportunities for clinical development. Lancet Oncol 2020;21:e419–30. 10.1016/S1470-2045(20)30234-5 [DOI] [PubMed] [Google Scholar]
- 48.Guillerey C. NK Cells in the Tumor Microenvironment. In: Birbrair A, ed. Tumor microenvironment: hematopoietic cells – Part B. Cham: Springer International Publishing, 2020: 69–90. [Google Scholar]
- 49.Berraondo P, Sanmamed MF, Ochoa MC, et al. Cytokines in clinical cancer immunotherapy. Br J Cancer 2019;120:6–15. 10.1038/s41416-018-0328-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kennedy LB, Salama AKS. A review of cancer immunotherapy toxicity. CA Cancer J Clin 2020;70:86–104. 10.3322/caac.21596 [DOI] [PubMed] [Google Scholar]
- 51.Petrovic K, Robinson J, Whitworth K, et al. TEM8/ANTXR1-specific CAR T cells mediate toxicity in vivo. PLoS One 2019;14:e0224015. 10.1371/journal.pone.0224015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-004725supp001.pdf (530.9KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information.