Abstract
We analyzed the thermodynamics of binding of cocaine and several cocaine metabolites to a humanized anti-cocaine mAb (h2E2), which is under development for the treatment of cocaine use disorders, using isothermal titration calorimetry. The calculated equilibrium dissociation (binding) constants were consistent with previous findings using other methods. All three ligands that display high affinity (nM) binding to the mAb (cocaine, cocaethylene, and benzoylecgonine) displayed similar enthalpically driven binding with substantial enthalpy-entropy compensation. The increased affinity of the cocaethylene metabolite compared to cocaine and benzoylecgonine is mostly attributable to a substantially less negative entropic binding component for cocaethylene, resulting in a more favorable binding energy, and thus, a higher affinity. The much lower affinity cocaine metabolites, norcocaine and ecgonine methyl ester, have much lower binding enthalpies than the high affinity ligands, and in contrast to the three high affinity ligands, have favorable (positive) entropic thermodynamic components of binding. Surprisingly, approximately 3.7 molecules of norcocaine are bound per mAb Fab site, as determined by isothermal titration calorimetry. This is in contrast to the three high affinity ligands, which bound with the expected stoichiometry of one drug molecule bound per one mAb Fab site. The results are discussed in relation to the previously published Fab:benzoylecgonine crystal structure for this h2E2 mAb, and compared to the isothermal titration calorimetry results published previously using an unrelated anti-cocaine mAb, mAb08.
Keywords: Monoclonal antibody, Cocaine and metabolite binding, Isothermal titration calorimetry, Thermodynamics, Ligand affinity
Highlights
-
•
ITC was used to measure the thermodynamics of ligand binding to anti-cocaine h2E2 mAb.
-
•
Binding of high and low affinity cocaine metabolites were compared to cocaine binding.
-
•
Low affinity metabolites have favorable/positive entropic binding components.
-
•
h2E2 mAb ITC results differ from those published for mAb08 anti-cocaine mAb.
-
•
Surprisingly, ≈3.7 molecules of low affinity norcocaine are bound per mAb Fab site.
Abbreviations
- mAb
monoclonal antibody
- h2E2
humanized anti-cocaine monoclonal antibody
- Fab
fragment antigen-binding
- ITC
isothermal titration calorimetry
- PBS
phosphate buffered saline
- CE
cocaethylene
- Coc
cocaine
- BE
benzoylecgonine
- NC
norcocaine
- EME
ecgonine methyl ester
- K
binding association constant
- Kd
binding dissociation constant
- HC
antibody heavy chain
- LC
antibody light chain
1. Introduction
As a therapy for cocaine use disorders, we have developed a high affinity anti-cocaine mAb, named h2E2, which also binds the pharmacologically active metabolite of cocaine, cocaethylene (CE), and the inactive metabolite, benzoylecgonine (BE), with high (nM) affinity [[1], [2], [3]]. As part of the characterization of this potential therapeutic antibody, we developed several assays to measure and assess the binding of cocaine and several cocaine metabolites to this h2E2 anti-cocaine mAb and its antibody fragments. Techniques utilized included quenching of intrinsic tyrosine and tryptophan mAb fluorescence [4] and differential scanning fluorimetry (DSF) using extrinsic fluorescent dyes [5]. We also used the fluorescent DASPMI rotor dye to assess high affinity cocaine and cocaine metabolite mAb binding by monitoring changes in both DASPMI absorbance [6] and fluorescence [7] upon ligand binding. In addition, using non-reducing SDS-PAGE, we demonstrated cocaine (ligand-induced) stabilization of the ligand binding protein domains of the intact mAb and its fragments [8], as well as mAb Fab fragment sub-domain ligand-induced stabilization [9]. Recently, we demonstrated that oxidation of tryptophan residues located in the h2E2 mAb drug binding site abolished high affinity binding [10].
In addition, we determined the structure of the h2E2 mAb Fab fragment, both in the presence and absence of co-crystallized benzoylecgonine (BE) ligand [11]. The crystal structure obtained was consistent with previous data concerning the involvement of multiple tyrosine and tryptophan residues in the cocaine binding pocket. However, the structure containing co-crystallized benzoylecgonine did not explain the previously noted differences in the mAb affinities (Kds) for the three high affinity ligands (CE, cocaine, and BE), because the carboxylate group on the co-crystallized BE molecule was shown to have no direct contacts with any amino acid residues in the mAb Fab binding site. Thus, the molecular extensions (i.e., esterifications) of this carboxylate group which differentiate cocaine (a methyl ester) and cocaethylene (CE, an ethyl ester) from BE, are also predicted by the BE-mAb crystal structure to have no interactions with the Fab binding site, and therefore should not result in the experimentally observed increases in ligand affinities (decreased Kds) for the cocaine and CE ligands, relative to BE. However, we recently hypothesized that cocaine and cocaethylene may have slightly higher binding affinities to this h2E2 mAb due to additional contacts of their esterified carboxylates with a specific tyrosine residue located near the benzoylecgonine binding site on the mAb light chain (contacts which are not seen in the BE-mAb crystal). This was based on the observation that nitration of a specific tyrosine residue located near the carboxylate of the BE molecule in the crystal structure binding site (LC Tyr34) differentially affected the binding affinities of the three high affinity ligands, cocaine, cocaethylene, and benzoylecgonine [12].
Isothermal titration calorimetry (ITC) has not been used routinely to monitor the binding of antigens to antibodies, since most experimental and therapeutic mAbs bind large protein antigens rather than small molecules like cocaine or other drugs, making ITC thermodynamic binding measurements involving antibodies directed against large biomolecule antigens problematic. Nonetheless, one previous study examined the binding of methamphetamine to the antibody developed for its therapeutic treatment of methamphetamine abuse, using isothermal titration calorimetry (ITC) [13]. In addition, ITC was used to characterize the binding of cocaine to another anti-cocaine mAb (called mAb08 [14]). This mAb08 binds cocaine, BE, and CE with similar nM affinities as observed for the h2E2 mAb, however, the mAb08 anti-cocaine mAb binds cocaine with the highest affinity, followed by BE and then CE [14], in contrast to our anti-cocaine mAb, h2E2, which binds CE with the highest affinity, followed by cocaine, and then BE. This suggests a different mode of antigen/drug binding to the two mAbs, which could lead to different contributions of enthalpic and entropic components driving the drug binding affinities for the two anti-cocaine mAbs. Thus, in the present study we determined the thermodynamic components of binding of cocaine and cocaine metabolites to the h2E2 mAb and compared these results with previous findings using the mAb08 anti-cocaine mAb. In addition, we attempted to interpret the h2E2 mAb ITC binding thermodynamics to understand the reason for the rank order of h2E2 mAb ligand affinities for cocaine, CE, and BE, in light of the crystal structure of the Fab fragment of the h2E2 mAb determined with bound BE [11].
2. Materials and methods
2.1. Materials
The generation, production, and purification of the h2E2 anti-cocaine monoclonal antibody by the manufacturer, Catalent, was previously described [3]. The purity of this h2E2 mAb is estimated to be at least 98% by SDS-PAGE, with no other proteins detectable. Fab fragments of this mAb were generated by Endo-Lys-C digestion, and purified as previously described [4]. 10 mM ligand stock solutions of cocaine and cocaine metabolite ligands were made from solids as described [1]. Intact h2E2 mAb, Fab fragment, and the Fc fragment of h2E2 mAb were extensively dialyzed at 4 °C versus PBS buffer, diluted from a PBS buffer concentrate (10X) purchased from Cambrex (BioWhittaker, without calcium or magnesium, catalog number 17-517Q), prior to ITC analysis.
2.2. Methods
All isothermal titration calorimetry (ITC) experiments were performed at 20 °C using a MicroCal VP-ITC instrument. Twenty 14 μL injections of titrant (cocaine or cocaine metabolite) were performed for each experiment, adding to the 1.4227 ml cell sample containing h2E2 mAb or h2E2 mAb fragment or PBS buffer only, with all protein samples and ligand titrants dissolved in 0.22 μm filtered PBS buffer. The ITC sample cell was stirred at 329 rpm, and there was a delay of 240 s between each injection of ligand. Thermograms obtained by injection of the respective ligands into the buffer alone were subtracted from the mAb ITC data before fitting the corrected data to obtain the parameters reported in the Table. The data were analyzed and fit using the one binding site model incorporated into the Origin 7.0 software supplied with the instrument. Kd (dissociation constant) values were calculated by taking the reciprocal of the Origin best fitted ITC K (association constant) values, and ΔG values reported in the Table were calculated using the Origin fitted values of ΔH and program calculated values of ΔS, and the Gibbs free energy equation, ΔG = ΔH – TΔS. Approximately 100 μL of each diluted protein sample loaded into the ITC cell was used to determine the protein concentrations of all ITC samples, using the molar extinction coefficients of 219,500, 73,965, and 71,570 M−1cm−1 at 280 nm for the intact h2E2 mAb, the h2E2 Fab fragment, and the h2E2 Fc fragment, respectively. The ITC titration syringe and sample cell were extensively cleaned and washed with mild detergent and water after every run, as recommended by the instrument manufacturer. The experimental errors for the binding parameters reported in the Table are all standard deviations derived from 4 independent ITC experiments performed with each ligand.
3. Results
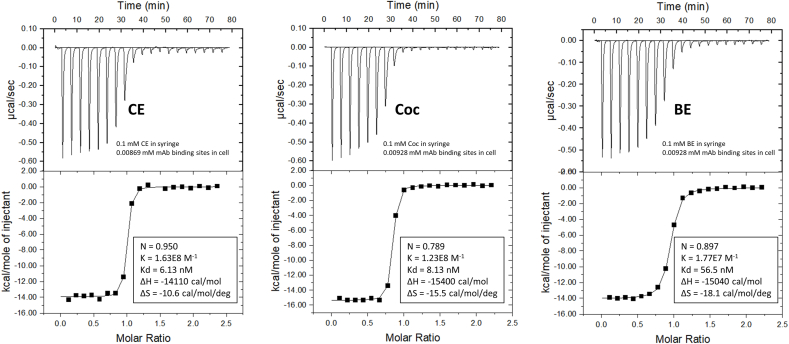
Isothermal titration calorimetry (ITC) was performed to analyze the thermodynamics of binding of cocaine and four cocaine metabolites to the h2E2 anti-cocaine mAb in PBS buffer at 20 °C, using appropriate concentrations of mAb and drug titrants. A summary of the results is given in Table 1, and representative ITC data and fitting of the data to a one site binding model are shown in Fig. 1 for the 3 high affinity (nM) ligands, cocaethylene (CE), cocaine (Coc), and benzoylecgonine (BE). These data are all well described by the one site binding model, with approximately one molecule of cocaine or cocaine metabolite binding per one Fab binding site present (see Fig. 1 and Table 1). The average Kd values obtained via ITC are similar, but slightly larger, to those obtained at the same temperature (20 °C) using the intrinsic tyrosine and tryptophan quenching method previously described [4], with ITC Kd values in PBS buffer of approximately 5, 8 and 54 nM for cocaethylene, cocaine, and benzoylecgonine, respectively (see Table 1), compared to approximately 1, 4, and 20 nM for cocaethylene, cocaine, and benzoylecgonine, respectively, using the intrinsic fluorescence quenching technique reported previously [4]. The previously published fluorescence quenching data were obtained using TBS buffer, not PBS buffer, possibly accounting for at least part of the small differences in measured affinities given by the two techniques. Also shown in Table 1 are ITC binding results obtained with 2 lower affinity cocaine metabolites, norcocaine (NC, Kd ≈ 1400 nM) and ecgonine methyl ester (EME, Kd ≈ 19000 nM). These lower (μM) affinity ligands both have much less favorable enthalpies of binding (ΔH) than the 3 high affinity ligands, by about 5–7 fold (Table 1). However, unlike the high affinity ligands, these lower affinity ligands have positive (energetically favorable) binding entropies (ΔS). In addition, unlike the high affinity ligands, these low affinity metabolites of cocaine also have N values greater than one, i.e., more than one ligand binds for each mAb Fab site present. Thus N ≈ 2 for ecgonine methyl ester, and N ≈ 3.7 for norcocaine (Table 1).
Table 1.
A summary of isothermal titration calorimetry (ITC) results in PBS at 20°C for cocaine and cocaine metabolite binding to the h2E2 anti-cocaine mAb. Thermograms obtained by injection of the respective ligands into the buffer alone were subtracted from the mAb ITC data before fitting the corrected data to obtain the parameters reported in the Table. The experimental errors for the binding parameters reported in the Table are all standard deviations derived from 4 independent ITC experiments performed with each ligand.
| Ligand Binding Parameter | Cocaethylene (CE) (n = 4) |
Cocaine (Coc) (n = 4) |
Benzoylecgonine (BE) (n = 4) |
Norcocaine (NC) (n = 4) |
Ecgonine Methyl Ester (EME) (n = 4) |
|---|---|---|---|---|---|
| N (# binding sites, per Fab present) | 0.922 ± 0.046 | 0.785 ± 0.032 | 0.901 ± 0.069 | 3.66 ± 0.07 | 1.90 ± 0.42 |
| K (association) (M−1) | 2.31E8 ± 1.3E8 | 1.33E8 ± 4.54E7 | 1.88E7 ± 3.26E6 | 704500 ± 36098 | 61375 ± 25024 |
| ΔH (cal/mol) | −13988 ± 279 | −15410 ± 368 | −15440 ± 833 | −3064 ± 150 | −2336 ± 452 |
| ΔS (cal/mol/deg) | −9.67 ± 1.98 | −15.4 ± 1.8 | −19.4 ± 2.5 | 16.3 ± 0.58 | 13.80 ± 2.33 |
| ΔG (cal/mol) | −11152 ± 302 | −10881 ± 187 | −9755 ± 106 | −7850 ± 27 | −6381 ± 247 |
| Kd (nM) | 5.29 ± 2.40 | 8.13 ± 2.37 | 54.4 ± 8.77 | 1422 ± 72 | 18708 ± 8313 |
| RBA (relative binding affinity, relative to the Kd for cocaine) | 0.65 | (1.00) | 6.7 | 175 | 2301 |
Fig. 1.
Representative ITC data for the binding of the high affinity ligands, cocaethylene (CE), cocaine (Coc), and benzoylecgonine (BE) to h2E2 mAb at 20°C in PBS buffer, pH = 7.4. The baseline-leveled raw ITC data is the upper panel, and the fitting to the resultant titration curve using a single binding site model is the lower panel for each ligand. The concentrations of ligands in the syringe and h2E2 mAb in the sample cell are given in the top panel, and the fitted parameters describing the binding for each ligand are given in the bottom panels.
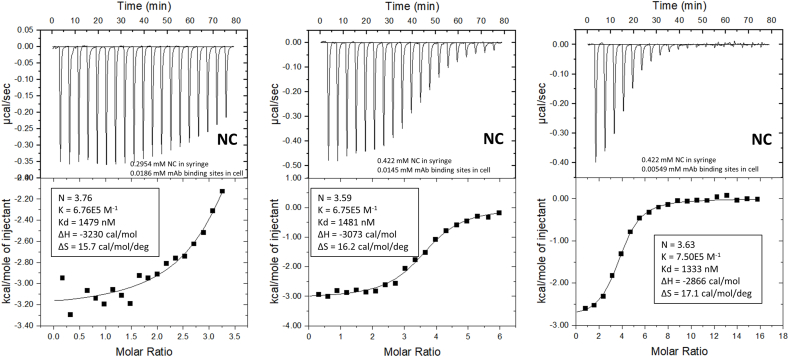
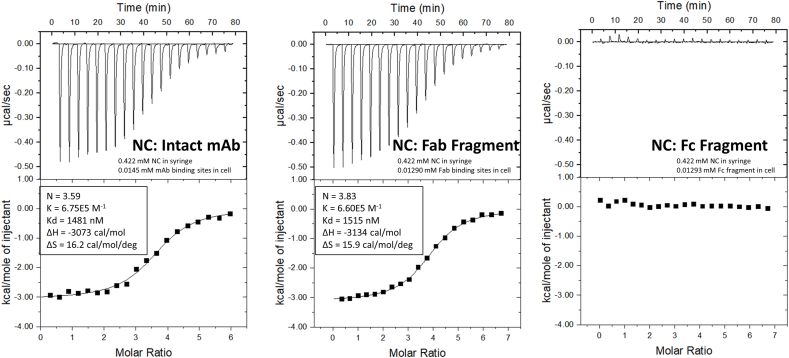
To further investigate this unexpectedly high number of norcocaine (NC) molecules binding to each h2E2 mAb, we analyzed norcocaine ITC data that spanned different portions of the ITC binding curve, by varying the concentration of norcocaine in the titrating syringe, and the concentration of mAb in the ITC sample cell. As can be seen from the binding fitting parameters and N values for norcocaine shown in Fig. 2, the N values (and other ITC binding parameters) obtained were not dependent on the concentrations of NC or mAb used in each experiment. To confirm this and assess which part(s) of the mAb bind norcocaine, we also analyzed the intact mAb, the Fab fragment, and the Fc fragment of the mAb using the same titrating concentration of NC, and very similar concentrations of Fab sites or Fc fragments present in the ITC cell. These results, shown in Fig. 3, clearly demonstrate that norcocaine is binding only to the Fab fragment, and confirm that there are ≈3.6–3.8 molecules of norcocaine binding to each Fab fragment of the mAb. Similar ITC experimental results were obtained using the EME metabolite, also demonstrating no EME binding to the Fc fragment (data not shown).
Fig. 2.
ITC data for the binding of norcocaine (NC) to the intact h2E2 mAb at 20°C in PBS buffer, pH = 7.4. The concentrations of NC in the titration syringe and the concentration of the h2E2 mAb in the ITC cell were varied to generate fitted curves covering varying portions of the total binding curve. The baseline-leveled raw ITC data is the upper panel, and the fitting to the resultant titration curve using a single binding site model is the lower panel for each condition. The concentrations of ligands in the syringe and h2E2 mAb in the sample cell are given in the top panel, and the fitted parameters describing the binding for each condition are given in the bottom panels.
Fig. 3.
ITC data for the binding of norcocaine (NC) to the intact h2E2 mAb, h2E2 Fab fragment, and h2E2 Fc fragment at 20°C in PBS buffer, pH = 7.4. The concentration of NC in the titration syringe was kept constant, and the concentration ligand binding sites for the intact mAb and the Fab fragment were very similar, as shown in the figure. Very similar NC binding parameters were found for the intact mAb and the Fab fragment (left and center panels), while no NC binding was detected for the Fc fragment (right panel). The baseline-leveled raw ITC data is the upper panel, and the fitting to the resultant titration curve using a single binding site model is the lower panel for each protein analyzed. The concentrations of ligands in the syringe and h2E2 mAb in the sample cell are given in the top panel, and the fitted parameters describing the binding for each condition are given in the bottom panels.
To further characterize the binding of cocaine and the cocaine metabolites that bind to the h2E2 mAb with high affinity, we also performed ITC binding experiments using these three ligands and the Fab fragment of the h2E2 mAb. Very similar binding parameters, stoichiometries, and binding thermodynamics were obtained using the Fab fragments as were obtained using the intact mAb, demonstrating the lack of effect on the binding of the high affinity ligands to the mAb due to the Fc region of the intact antibody (data not shown).
4. Discussion
There are only a few published reports characterizing mAb antigen binding thermodynamics using isothermal titration calorimetry (ITC). This is likely because most antibodies recognize proteins and larger biomolecules, making binding to them less amenable to thermodynamic binding analysis by ITC. A total of only about two dozen mAb drug binding studies have utilized ITC, with several analyzing the binding of peptides to antibodies [15,16]. In addition, several studies have involved the binding of drugs by mAbs, including a methamphetamine binding antibody [13], and a different anti-cocaine mAb than the h2E2 mAb investigated in this study, called mAb08 [14]. Despite the paucity of such studies, the determination and analysis of ligand binding thermodynamics is often important in drug discovery and drug development [17].
We have used multiple techniques to analyze and quantitate aspects of cocaine and cocaine metabolite binding to our h2E2 anti-cocaine mAb under development for the treatment of cocaine use disorders [[4], [5], [6], [7], [8]]. However, none of these methods permitted estimation or calculation of the thermodynamics of drug and drug metabolite binding to the h2E2 mAb. Therefore, we employed ITC to answer questions regarding the enthalpic vs entropic contributions of binding for cocaine and several cocaine metabolites that bind with similar (nM) affinities to cocaine (cocaethylene (CE) and benzoylecgonine (BE)), as well as cocaine metabolites having much lower affinities than cocaine (i.e., μM affinities, including norcocaine (NC) and ecgonine methyl ester (EME)). Not surprisingly, as seen in Table 1, the binding of the high affinity binding drugs (CE, Coc, and BE) is enthalpically driven. However, the rank order of binding affinities of these high affinity drugs are partly determined by their entropic components, with CE having the highest affinity, due to a much lower negative value (less unfavorable value) for the entropy change (ΔS), when, in fact, the enthalpic component (ΔH) is slightly less favorable for CE compared to cocaine and BE, i.e., ≈-14,000 cal/mol for CE, compared to ≈ -15,400 for cocaine and benzoylecgonine (see Table 1). We speculate this may be explained by the ethyl ester which is added to the carboxyl group of BE to form CE leading to no additional, enthalpically favorable binding interactions with the mAb, but resulting in a less entropically unfavorable ligand (CE) ethyl ester solvation/desolvation effect. This hypothesis is consistent with inspection of the h2E2 mAb BE:Fab co-crystallized structure, showing that this portion of the BE molecule, which is esterified to form CE, is not in contact with the mAb, sticking up from the mAb binding site [11]. Therefore, according to the BE:Fab published structure [11], the mAb binding site is not predicted to make enthalpically favorable interactions with the esterified carboxyl group present in cocaine (a methyl ester) and CE (an ethyl ester). However, the nitration of a nearby light chain tyrosine residue (LC Y34) does decrease the affinity of the mAb for CE and Coc, and not for BE, possibly indicating an additional interaction with that tyrosine might exist for CE and Coc, which is not seen in the published BE crystal. Regardless, the Kds determined by ITC for these nM affinity ligands are all similar, but slightly larger, than the Kds determined for these same ligands at the same temperature using other techniques [1,4], possibly due to differences in the experimental buffers used or in the ligand solvation/desolvation effects measured by ITC but not the other methods. Nonetheless, the relative binding affinities obtained with these different techniques are similar, and the rank order of the affinities for the h2E2 anti-cocaine mAb are the same as previously determined using other techniques (Table 1).
In contrast to the high affinity drugs, the lower affinity ligands norcocaine (NC) and ecgonine methyl ester (EME), are far less enthalpically driven (i.e., have much smaller negative ΔH values), and display favorable entropic components for their binding (i.e., positive ΔS values, see Table 1). Interestingly and unexpectedly, both lower affinity cocaine metabolites also bind with a greater than 1:1 stoichiometry to Fab sites present. Very unusual and unexplained is the binding of ≈3.7 molecules of norcocaine per Fab. Norcocaine differs from cocaine by the deletion of a single methyl group on the tropane ring of cocaine. However, this part of the molecule is intimately involved in interacting with the mAb binding site, according to the published BE:Fab h2E2 mAb crystal structure [11]. Thus, the methyl group on the tropane ring of cocaine (absent in norcocaine) is in contact with both HC W33 and LC W91 tryptophan mAb residues, the oxidation of which abolished high affinity binding of cocaine to the h2E2 mAb [10]. The present ITC results indicate that this demethylation of cocaine to form norcocaine completely changes the way that norcocaine binds to the mAb, and apparently gives rise to NC binding to multiple, low affinity sites on the Fab portion of the h2E2 mAb.
The present ITC results utilizing our h2E2 anti-cocaine mAb are very different from those reported previously for the mAb08 anti-cocaine mAb [14]. The mAb08 antibody exhibited a similar range of affinities for Coc, CE and BE as the h2E2 mAb, but in a different rank order (Kd values for mAb08 determined by ITC for Coc, BE, and CE were 2.5, 18.6, and 34.4 nM, respectively [14]). The binding of these 3 high affinity ligands is more enthalpically driven for the mAb08 mAb than the h2E2 mAb, with ΔH values ranging from -21 to -28 kcal/mol [14], compared with -14.0 to -15.4 kcal/mol for the h2E2 mAb investigated in the present study (see Table 1). However, the ΔG values, and thus the Kds, are similar for the two antibodies, due to the much larger unfavorable entropic component of binding observed for the mAb08 antibody, i.e., -37 to -55 cal/mol/°K for mAb08 [14], compared to -9.7 to -19.4 cal/mol/°K for the h2E2 anti-cocaine mAb (Table 1). This indicates a very different drug binding modality for the two anti-cocaine mAbs. The crystal structure of the mAb08 mAb has not been published, but the crystal structure of the Fab fragment complexed with the cocaine metabolite, BE, has been [11]. This crystal structure indicates that the tropane ring is one of the central components of the mAb binding site, consistent with the present findings that loss of the single methyl group on that ring in the norcocaine metabolite greatly lowers binding affinity and completely changes the binding thermodynamics for norcocaine, as well as the stoichiometry of binding (Table 1, and Fig. 2, Fig. 3). Unfortunately, norcocaine and ecgonine methyl ester binding were not reported for mAb08. The reason for the favorable (positive) entropic component of binding to the h2E2 mAb for the low affinity ligands, norcocaine and ecgonine methyl ester (Table 1), is not clear at present. In general, the entropic contribution to binding energies consists of three components [18]: solvent entropy changes resulting from solvent release upon binding; conformational entropy changes occurring due to changes in conformational freedom of both the protein and ligand; and entropic changes due to the loss of translational and rotational degrees of freedom following complex formation (which contributes unfavorably to the binding entropy). Thus, we speculate that it may be favorable solvent entropy changes resulting from solvent release upon binding of the low affinity ligands to multiple sites on the h2E2 mAb protein that result in the observed favorable entropic component of binding (Table 1).
Although the present ITC study does not explain on a molecular interaction level why the binding affinities for the h2E2 mAb decrease in the order of CE > Coc > BE, it does show that CE is substantially different from both cocaine and BE in its thermodynamics of binding, having a smaller unfavorable entropic binding component, and thus less “enthalpy-entropy compensation” (EEC [17,19]), which is commonly observed with the binding of many drug molecules. This enthalpy-entropy compensation decreases the favorable Gibbs free energy associated with binding, and thus the binding affinity. We hypothesize that the smaller EEC for CE, as compared to cocaine, and especially benzoylecgonine, measured for the h2E2 mAb may be due to less unfavorable solvation/desolvation entropic effects mediated due to the ethyl ester extension of the carboxylic acid of benzoylecgonine to form cocaethylene.
Financial disclosure statement
Dr. Norman is named as a co-inventor on a portfolio of patents for the matter and use of the h2E2 humanized anti-cocaine monoclonal antibody.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Andrew B. Norman has patent issued to University of Cincinnati. None.
Acknowledgments
This work was supported in part by the National Institutes of Health National Institute on Drug Abuse Grant U01DA050330. We are grateful to Catalent PharmaSolutions, Inc. (Madison, WI) for providing the recombinant humanized h2E2 anti-cocaine mAb protein expressed using their GPex® technology. We thank Dr. Rhett Koval in the Department of Molecular Genetics, Biochemistry and Microbiology at the University of Cincinnati College of Medicine for the use of the MicroCal VP-ITC instrument, and Dr. Zhenyu Yuan in Dr. Koval's laboratory for instruction in the use of the ITC instrument, and the fitting of the ITC data.
References
- 1.Wetzel H.N., Webster R.P., Saeed F.O., Kirley T.L., Ball W.J., Norman A.B. Characterization of a recombinant humanized anti-cocaine monoclonal antibody produced from multiple clones for the selection of a master cell bank candidate. Biochem. Biophys. Res. commun. 2017;487:690–694. doi: 10.1016/j.bbrc.2017.04.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wetzel H.N., Tabet M.R., Ball W.J., Norman A.B. The effects of a humanized recombinant anti-cocaine monoclonal antibody on the disposition of cocaethylene in mice. Int. Immunopharm. 2014;23:387–390. doi: 10.1016/j.intimp.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Norman A.B., Gooden F.C., Tabet M.R., Ball W.J. A recombinant humanized anti-cocaine monoclonal antibody inhibits the distribution of cocaine to the brain in rats. Drug Metab. Dispos.: Bio. Fate Chem. 2014;42:1125–1131. doi: 10.1124/dmd.114.057034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirley T.L., Norman A.B. Characterization of a recombinant humanized anti-cocaine monoclonal antibody and its Fab fragment. Hum. Vaccines Immunother. 2015;11:458–467. doi: 10.4161/21645515.2014.990856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirley T.L., Norman A.B., Wetzel H.N. A novel differential scanning fluorimetry analysis of a humanized anti-cocaine mAb and its ligand binding characteristics. J. Immunol. Methods. 2020;476 doi: 10.1016/j.jim.2019.112676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirley T.L., Norman A.B. Ligand binding to a humanized anti-cocaine mAb measured by dye absorption spectroscopy. Biochem. Biophys. Res. commun. 2021;535:93–98. doi: 10.1016/j.bbrc.2020.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirley T.L., Norman A.B. Cocaine binding to the Fab fragment of a humanized anti-cocaine mAb quantitated by dye absorption and fluorescence spectroscopy. J. Immunol. Methods. 2021;496 doi: 10.1016/j.jim.2021.113103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirley T.L., Norman A.B. Ligand binding to a humanized anti-cocaine mAb detected by non-reducing SDS-PAGE. Biochem. Biophy. Rep. 2020;23 doi: 10.1016/j.bbrep.2020.100795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirley T.L., Norman A.B. Multi-domain unfolding of the Fab fragment of a humanized anti-cocaine mAb characterized by non-reducing SDS-PAGE. Biochem. Biophys. Res. commun. 2020;533:580–585. doi: 10.1016/j.bbrc.2020.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirley T.L., Norman A.B., Greis K.D. Oxidation of specific tryptophan residues inhibits high-affinity binding of cocaine and its metabolites to a humanized anticocaine mAb. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.101689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan K., Zhou M., Ahrendt A.J., Duke N.E.C., Tabaja N., Ball W.J., Kirley T.L., Norman A.B., Joachimiak A., Schiffer M., Wilton R., Pokkuluri P.R. Structural analysis of free and liganded forms of the Fab fragment of a high-affinity anti-cocaine antibody, h2E2, Acta crystallographica. Section F. Struct. Biol. Commun. 2019;75:697–706. doi: 10.1107/S2053230X19013608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirley T.L., Greis K.D., Norman A.B. Tyrosine nitration of a humanized anti-cocaine mAb differentially affects ligand binding of cocaine and its metabolites. Biochem. Biophy. Rep. 2022;30 doi: 10.1016/j.bbrep.2022.101278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevens M.W., Tawney R.L., West C.M., Kight A.D., Henry R.L., Owens S.M., Gentry W.B. Preclinical characterization of an anti-methamphetamine monoclonal antibody for human use. mAbs. 2014;6:547–555. doi: 10.4161/mabs.27620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramakrishnan M., Alves De Melo F., Kinsey B.M., Ladbury J.E., Kosten T.R., Orson F.M. Probing cocaine-antibody interactions in buffer and human serum. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brockhaus M., Ganz P., Huber W., Bohrmann B., Loetscher H.R., Seelig J. Thermodynamic studies on the interaction of antibodies with beta-amyloid peptide. J. Phys. Chem. B. 2007;111:1238–1243. doi: 10.1021/jp0664059. [DOI] [PubMed] [Google Scholar]
- 16.Reese H.R., Xiao X., Shanahan C.C., Chu W., Van Den Driessche G.A., Fourches D., Carbonell R.G., Hall C.K., Menegatti S. Novel peptide ligands for antibody purification provide superior clearance of host cell protein impurities. J. Chromatogr. 2020;A 1625 doi: 10.1016/j.chroma.2020.461237. [DOI] [PubMed] [Google Scholar]
- 17.Geschwindner S., Ulander J., Johansson P. Ligand binding thermodynamics in drug discovery: still a hot tip? J. Med. Chem. 2015;58:6321–6335. doi: 10.1021/jm501511f. [DOI] [PubMed] [Google Scholar]
- 18.Du X., Li Y., Xia Y.L., Ai S.M., Liang J., Sang P., Ji X.L., Liu S.Q. Insights into protein-ligand interactions: mechanisms, models, and methods. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lumry R., Rajender S. Enthalpy-entropy compensation phenomena in water solutions of proteins and small molecules: a ubiquitous property of water. Biopolymers. 1970;9:1125–1227. doi: 10.1002/bip.1970.360091002. [DOI] [PubMed] [Google Scholar]