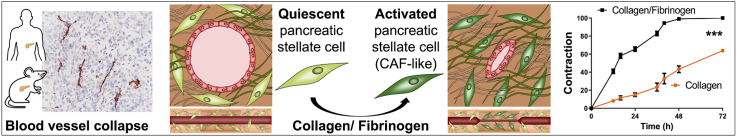
Abstract
Fibrotic tumors, such as pancreatic ductal adenocarcinoma (PDAC), are characterized for high desmoplastic reaction, which results in high intra-tumoral solid stress leading to the compression of blood vessels. These microarchitectural alterations cause loss of blood flow and poor intra-tumoral delivery of therapeutics. Currently, there is a lack of relevant in vitro models capable of replicating these mechanical characteristics and to test anti-desmoplastic compounds. Here, a multi-layered vascularized 3D PDAC model consisting of primary human pancreatic stellate cells (PSCs) embedded in collagen/fibrinogen (Col/Fib), mimicking tumor tissue within adjunct healthy tissue, is presented to study the fibrosis-induced compression of vasculature in PDAC. It is demonstrated how the mechanical and biological stimulation induce PSC activation, extracellular matrix production and eventually vessel compression. The clinical relevance is confirmed by correlating with patient transcriptomic data. Furthermore, the effects of gradual vessel compression on the fluid dynamics occurring within the channel is evaluated in silico. Finally, it is demonstrated how cancer-associated fibroblast (CAF)-modulatory therapeutics can inhibit the cell-mediated compression of blood vessels in PDAC in vitro, in silico and in vivo. It is envisioned that this 3D model is used to improve the understanding of mechanical characteristics in tumors and for evaluating novel anti-desmoplastic therapeutics.
Keywords: Pancreatic ductal adenocarcinoma, Cancer-associated fibroblasts, Collagen, Fibrinogen, Computational flow dynamics, 3D in vitro model
Graphical abstract
A 3D engineered in vitro model is presented to mimic the compression of vasculature in PDAC. The biomaterial-based activation of primary pancreatic stellate cells is investigated, inducing clinically relevant cell behavior and compression of vasculature. Furthermore, the effects of vasculature compression is demonstrated using computational simulation as well as the application of this model to evaluate novel TME-modulating therapeutics.
Highlights
-
•
A multi-layered model was generated including a central endothelialized channel mimicking pancreatic tumor stroma.
-
•
Pancreatic stellate cells were activated to cancer-associated fibroblasts (CAFs) by mechanical and biological stimuli.
-
•
CAF activation caused clinically-relevant compression of the central channel.
-
•
Computational flow simulation revealed high flow velocity and pressure in compressed channels.
-
•
Use of anti-fibrotic ITGA5 inhibitor reduced vascular compression which was validated in mouse tumor xenografts.
1. Introduction
The tumor microenvironment (TME) in fibrotic tumors such as pancreatic ductal adenocarcinoma (PDAC) is characterized by a high amount of desmoplasia [[1], [2], [3]]. In PDAC, cancer-associated fibroblasts (CAFs) form the most prevalent cell type accounting for around 80% of all cells in the TME and are the main driving factor of this desmoplasia as they produce excessive amounts of extracellular matrix (ECM) proteins such as collagen, fibronectin or hyaluronic acid [[4], [5], [6]]. Among several types of CAFs [7,8], myofibroblastic CAFs are highly contractile and ECM regulating CAF type which mainly originate from pancreatic stellate cells (PSCs). Based on the crosstalk with tumor cells and environmental stimuli the PSCs obtain an activated CAF-like state [[4], [5], [6]]. The abundance of CAFs and the resulting high amount of desmoplasia in PDAC creates a dense environment, which forms a nearly impenetrable barrier for therapeutics [1,3]. Additionally, this desmoplasia, accompanied by an excess deposition of ECM, creates a high intratumoral stress causing the compression of blood vessels in and surrounding the tumor, preventing therapeutics to reach the tumor site in the first place [3,9,10].
In recent years, novel treatment strategies have been developed that focus not on targeting the tumor cells directly, but on modulating the TME to reduce tumor progression and increase the efficacy of chemotherapy in combinational treatment approaches [[11], [12], [13]]. One strategy, for instance, is to directly reduce the high amount of ECM in PDAC using enzymatic therapeutics that directly degrade ECM proteins such as collagenase or hyaluronidase, which will eventually enable the penetration of other therapeutics [[14], [15], [16], [17]]. Differently, given the high amount of CAFs in the tumor, strategies to inhibit CAFs or the progenitor PSCs also form a promising strategy to inhibit the high desmoplasia in PDAC [18,19]. For example, we recently demonstrated that the modulation of CAFs using an integrin alpha5 binding peptide inhibits the activation of PSCs resulting in a lower deposition of collagen in the PDAC TME in patient-derived xenograft models in mice [20]. Interestingly, we found that the reduction of the PSC-driven deposition of collagen also re-opened the vasculature within the tumor increasing the efficacy of co-injected chemotherapy.
Although such strategies form a promising approach for altering the mechanical environment in PDAC and improving for instance drug penetration or facilitate re-opening of the vasculature, the exact mechanisms of such treatments and direct effects on the mechanical environment are not yet fully understood. In particular, the complex environment in animal models including the crosstalk of different cellular and acellular components in the TME, makes it challenging to fully identify the effect of treatments on the mechanical environment in PDAC. Therefore, there is an urgent need for suitable in vitro models that are able to mimic the complex interactions and mechanical alterations in the PDAC TME in a controlled and biologically relevant fashion.
In the recent years, several different biomaterials have been developed that are able to mimic the ECM found in PDAC to a great extent [[21], [22], [23]]. In particular, the combination of such biomaterials with different biofabrication techniques has facilitated the generation of complex in vitro tumor models for the evaluation of novel treatment strategies [3,24,25]. While most biomaterial approaches aim to mimic the exact composition of the ECM found in tumors, the biomaterial itself can also be used to mimic a diseased state based on mechanical or biological stimulation of embedded cells [[26], [27], [28]]. Based on this versatility, the use of a cell-stimulating 3D biomaterial forms a promising approach to mimic the CAF-driven mechanical environment in PDAC.
In this study, we demonstrate the feasibility of a 3D engineered multi-layered tissue consisting of PSCs embedded in a collagen and a collagen/fibrinogen (Col/Fib) matrix to replicate the complex mechanical environment in PDAC. We further present the inclusion of a central endothelialized channel to study the cell-mediated contraction of vasculature in PDAC in greater detail. First, we demonstrate the presence of compressed vasculature in not only clinical but also in pre-clinical conditions demonstrating the importance of this model for different stages in the development of novel therapeutics. Next, we study the activation of PSCs in our model based on mechanical and biological stimulation and demonstrate the biological relevance of our model by comparing the expression of PDAC specific markers in our model and in publicly available patient data. Next, we present the cell-mediated compression of an endothelialized channel in our model and demonstrate the effects on the blood flow inside the channel using computational fluid dynamics (CFD) simulations. Finally, we present the capability of our novel model to serve as a tool for the in vitro evaluation of novel therapeutics that aim to modulate the mechanical environment in PDAC by comparing the efficacy of PSC-mediating therapeutics in vitro, in silico and in vivo. The proposed technique is likely to advance the field of novel 3D in vitro models by offering relevant biomimetic characteristics in terms of biological and mechanical stimulation, which are not achievable with other culture systems. Most of all, our novel in vitro model is not only limited to mimic PDAC, but may be translated to other fibrotic cancer types and other diseases.
2. Results and discussion
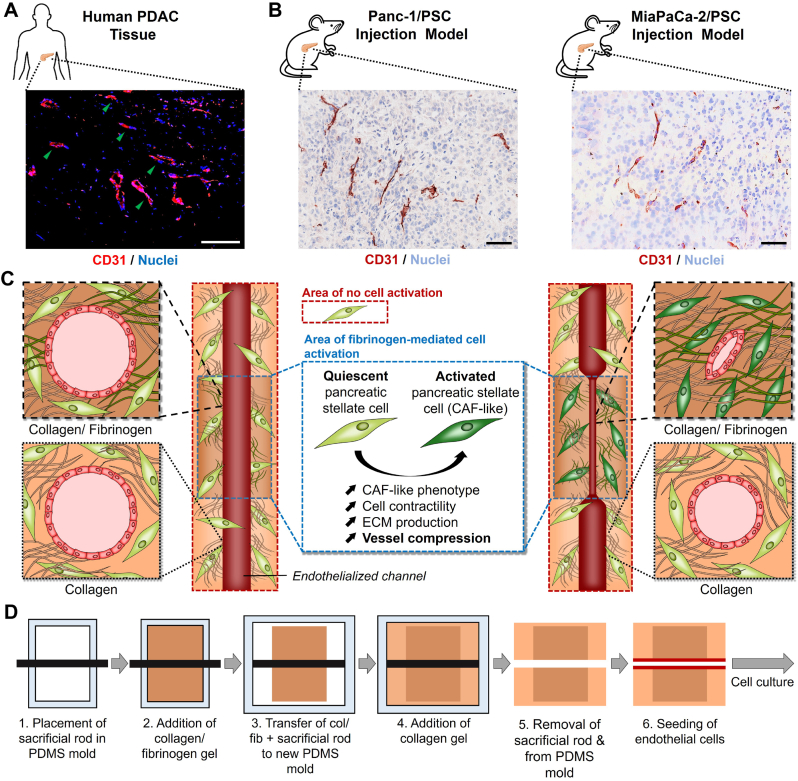
First, we wanted to investigate whether the compression of blood vessels is a phenomenon that is only present in human patients, which might reduce the efficacy of novel therapeutics in clinical studies, or also shared in pre-clinical animal models, which might already affect the development of novel therapeutics. Here we studied the expression of CD31, a common endothelial cell marker [29,30], in human PDAC sections as well as in sections obtained from subcutaneously co-injected Panc-1 pancreatic cancer cells with PSCs and MiaPaCa-2 pancreatic cancer cells with PSCs in mice (Fig. 1A and B). We were able to confirm the presence of compressed blood vessels in human pancreatic cancer patients (highlighted by green arrows) and also found similar compressed vessels in both, Panc-1/PSC and MiaPaCa-2/PSC, which demonstrates that even at pre-clinical stages blood vessel compression might play a crucial role in the efficacy of novel therapeutics.
Fig. 1.
Investigation of blood vessel compression in patients and mice and experimental setup. A) Immunofluorescent staining for CD31 (red, pseudocolor) and nuclei (blue) in tumor section obtained from PDAC patient. Scale bar = 100 μm. B) Immunohistochemical staining for CD31 (red) and nuclei (light blue) in tumor section obtained from mice co-injected with Panc-1/PSC (left) or MiaPaCa-2/PSC (right). Scale bar = 100 μm. C) Schematic representation of the 3D model design and experimental setup, highlighting the fibrinogen-mediated activation of pancreatic stellate cells (PSCs) in the center part of the model and the mechanisms causing compression of a central endothelialized channel while the PSCs in the collagen compartment remain quiescent not causing compression (CAF = cancer-associated fibroblast). D) Schematic representation of the process to generate multi-layered collagen/Col/Fib constructs (PDMS = Polydimethylsiloxane).
Based on this observation we designed a novel 3D engineered pancreatic tissue comprised of a central channel mimicking the vasculature in PDAC surrounded by different matrix compositions of collagen, which forms the most abundant ECM protein in PDAC (Fig. 1C and D). As it has been previously reported that low concentrations of fibrinogen in solution induce PSC activation towards a CAF-like phenotype [31], which display a higher contractility [20,32], we designed a central part in the model comprising of a collagen/fibrinogen (Col/Fib) mix including PSCs, which, based on PSC activation and contraction, causes the vessel to compress. This central area is surrounded by a collagen matrix similarly including PSCs, which, however, does not trigger PSC activation, hence remaining an open vessel. We chose to use a solid rod as sacrificial structure for the central channel to obtain an equally sized channel throughout the entire tissue.
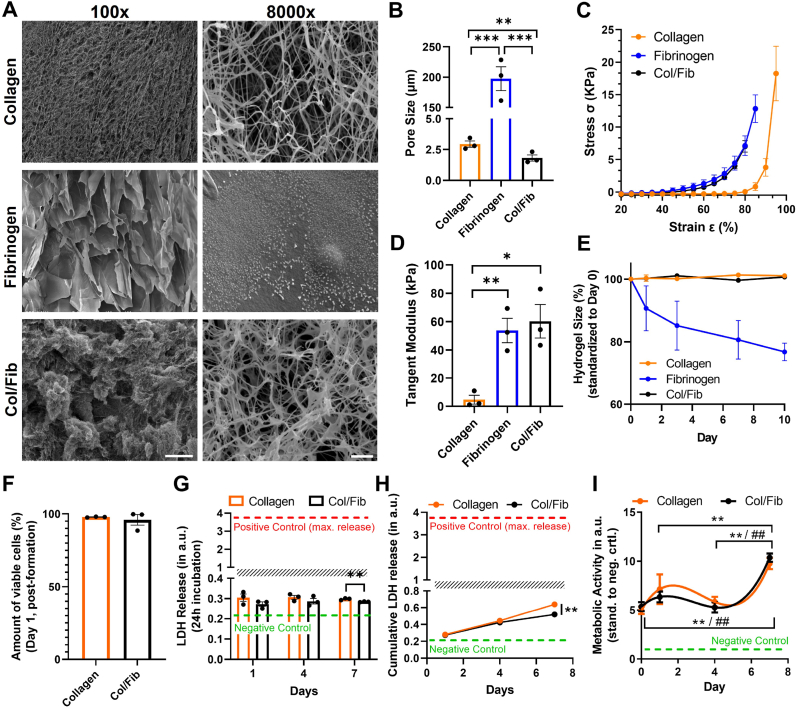
As it has been reported that the structural and mechanical properties of a hydrogel matrix might also induce the activation of PSCs [33], we first investigated the morphology and pore size of the different matrices using scanning electron microscopy (SEM) (Fig. 2A and Fig. S1A, Supporting Information). We observed that collagen presented a homogenous surface at lower magnification, while displaying a fibrous network at higher magnification with an average diameter of 2.94 ± 0.44 μm between the pores (Fig. 2B and Fig. S1B, Supporting Information). Fibrinogen, on the other hand, displayed a more heterogeneous structure with large pores of 197.49 ± 33.9 μm at lower magnification, while smooth surfaces at higher magnification. It has been shown that the morphology of fibrinogen gels depends on the ratio and concentration of fibrinogen to thrombin, which might explain this more macroscale structure compared to collagen [34]. Interestingly, the combination of Col/Fib appeared to comprise both, the more heterogeneous architecture of fibrinogen at lower magnification while presenting a fibrous network at higher magnification with pore size of 1.81 ± 0.43 μm, demonstrating the successful combination of the two matrices. It has to be noted, that while the pore size in collagen and Col/Fib are small, cell migration should not be hampered as the cells should still be able to stretch between the fibers.
Fig. 2.
Mechanical characterization of collagen, fibrinogen and Col/Fib gels as well as PSCactivity and viability. A) Scanning electron microscopy (SEM) images for collagen, fibrinogen and collagen/fibrinogen (Col/Fib) hydrogel at 100x magnification, scale bar = 200 μm, and 8000x magnification, scale bar = 2 μm. B) Quantification of the average respective pore size, n = 3, 20 pores/independent sample. C) Stress – strain curve for collagen, fibrinogen and Col/Fib hydrogels based on compression analysis, n = 3. D) Tangent modulus for collagen, fibrinogen and Col/Fib hydrogels between 75% and 80% strain based on compression analysis, n = 3. E) Stability of collagen, fibrinogen and Col/Fib hydrogels based on the hydrogel size for a total duration of 10 days, n = 3. F) Quantification of viable PSCs in collagen and Col/Fib gels on day 1 post-tissue formation based on LIVE/DEAD staining (Fig. S2, Supporting Information), n = 3. G) Release of lactate dehydrogenase (LDH) into culture medium based on 24 h incubation of collagen or Col/Fib gels, negative control = full culture medium, positive control = LDH released on mechanical destruction of tissues, n = 4. H) Cumulative LDH release in culture medium for collagen and Col/Fib gels for 7 days of culture, n = 3. Negative control = full culture medium, positive control = LDH released on mechanical destruction of tissues. I) Metabolic activity (based on AlamarBlue assay) of PSCs in collagen and Col/Fib gels on days 1, 4 and 7 post-preparation, n = 3. Significance indicated for * Col/Fib and # collagen. Mean + SEM, *p < 0.05, ##/**p < 0.01, ***p < 0.001.
Next, we investigated the mechanical properties of the different gels based on compression testing. We found that fibrinogen and Col/Fib share a highly similar strain-stress relation reaching a tangent modulus (calculated between 75% and 80% strain) of 53.78 ± 14.96 kPa and 60.25 ± 20.45 kPa, respectively, while collagen displayed a lower stiffness in general with a tangent modulus of 4.73 ± 5.43 kPa (Fig. 2C and D). These differences might be based on the enzymatic crosslinking of fibrinogen, creating a mechanically stiffer structure while collagen is based on thermal solidification of the gel. Interestingly, collagen also displayed a very low stress from 20% to 80% strain, which might be based on the alignment of collagen fibers and leakage of water from the construct at lower strains. Finally, we observed that collagen and Col/Fib gels displayed a high stability over a duration of 10 days, while fibrinogen consistently decreased in size (Fig. 2E). As a result, we continued the experiments with collagen or Col/Fib only, presenting the more stable gels.
After investigating the mechanical and structural properties of the empty gels we generated PSC-laden collagen and Col/Fib gels and first investigated whether the generation of gels as well as the materials affect the viability and metabolic activity of PSCs. We observed a very high amount of viable cells for both, PSCs in collagen and Col/Fib 1 days post-preparation of 97.84 ± 0.28% and 99.63 ± 0.14%, respectively, and no significant difference between the two gels, demonstrating that both gels and the fabrication technique do not harm the PSCs (Fig. 2F and Fig. S2, Supporting Information). Moreover, PSCs displayed a clearly elongated shape indicating proper cell attachment to the matrices facilitating migration and contraction. Furthermore, the cells remained highly viable for at least 7 days post-preparation demonstrating the suitability of the hydrogels for long-term cell culture (Fig. S2, Supporting Information). However, based on a higher contraction of the both gels at later time-points and resulting clustering of cells individual quantification of the amount of viable cells was not possible.
To further confirm the high viability of cells we measured the release of lactate dehydrogenase (LDH) into the culture as indicator of cytotoxicity. Here, we first investigated the release of LDH after incubation for 24 h in fresh culture medium (Fig. 2G and Fig. S3, Supporting Information). We found that while PSCs in collagen show a slightly higher release of LDH, indicating higher toxicity, the overall measured values are very low being roughly 10x lower compared to the positive control displaying the maximum release of LDH possible. This indicates an overall high viability of PSCs in both, collagen and Col/Fib, despites small differences. Additionally we investigated the cumulative release of LDH for a total duration of 7 days while only refreshing parts of the medium (Fig. 2H and Fig. S3, Supporting Information). We found that after 7 days the concentration of LDH in the medium doubled, however, still remained significantly low compared to the positive control again indicating an overall high viability of PSCs for the culture duration. Interestingly, also here we observed a slightly higher release of LDH for collagen gels at day 7 post-preparation which for both conditions might be based on the contraction of the hydrogels entrapping parts of the LDH.
To confirm that PSCs in our model are not only viable but also metabolically active, we performed an AlamarBlue assay. In both, collagen and Col/Fib, PSCs display a high metabolic activity for a total duration of 7 days (Fig. 2I). Interestingly, we observed that from days 1–4, a small decrease in activity could be seen, which might be based on the cells adapting to the new environment.
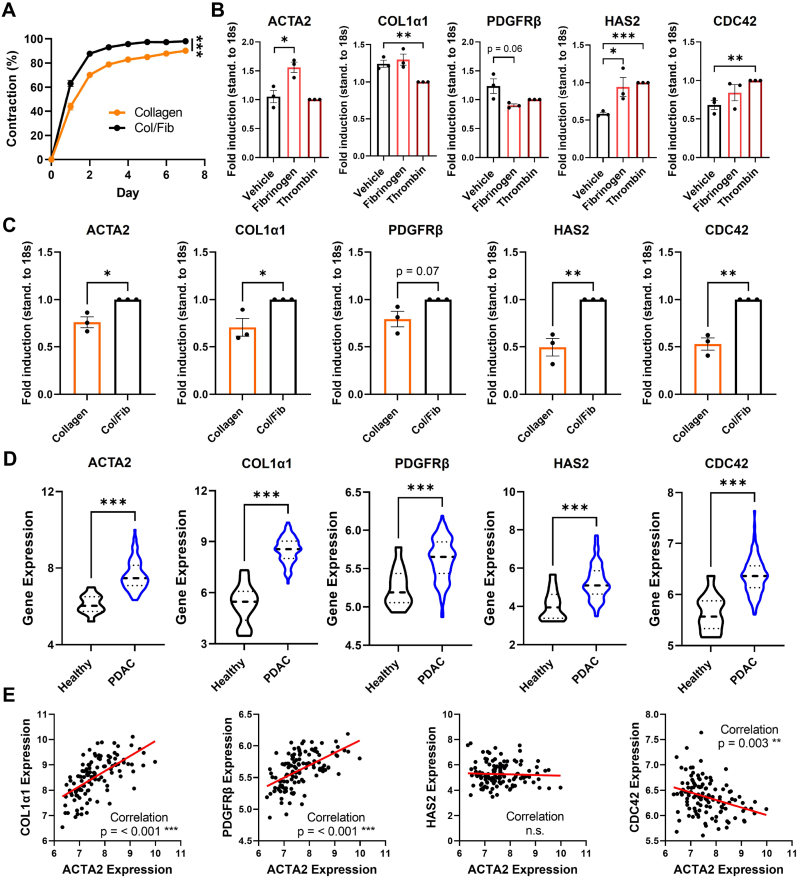
As we already observed a contraction of hydrogels in our viability assays, we next we investigated whether the presence of fibrinogen is significantly increasing this contraction. We found that, while both PSC-laden collagen and Col/Fib gels displayed a contraction, Col/Fib were significantly more contracted compared to collagen (Fig. 3A and Fig. S4, Supporting Information). Already on day 1 post-preparation, Col/Fib gels demonstrated a 1.44 times higher contraction than collagen gels (63.16 ± 5.21% and 43.84 ± 5.83%, respectively), while maintaining this difference throughout the experiment displaying a 1.15 times higher contraction on day 5 when the Col/Fib gels reach a plateau with a contraction of 97.40 ± 1.33% for Col/Fib and 84.97 ± 2.41% for collagen. That PSCs in collagen also displayed contraction might be based on autocrine activation of PSCs when cultured in higher concentrations; however, it is clear that fibrinogen does indeed have a beneficial effect on contraction. Interestingly, the observed contraction period overlapped with the initial decrease in metabolic activity (Fig. 2I), which indicates that cells present a certain adapt-or-grow principle favoring either adaption or proliferation.
Fig. 3.
Characterization of PSC-laden collagen and Col/Fib hydrogels and transcriptomic comparison in human PDAC patients. A) Contraction of PSC-laden collagen and Col/Fib gels for a total duration of 7 days post-preparation standardized to the initial hydrogel size, n = 3. B) Gene expression analysis on 2D cultured PSCs treated with either vehicle (DPBS), 2 mg/mL fibrinogen or 1U/mL thrombin, n = 3. C) Gene expression analysis in 3D cultured PSC-laden collagen and Col/Fib gels, n = 3. D) Transcriptomic analysis in human cohort from public database GEO (GSE62165), 13 healthy vs. 118 PDAC patients. E) Correlation between the expression of ACTA2 and other genes found upregulated in patients. Mean + SEM, *p < 0.05, **p < 0.01, ***p < 0.001.
As we have shown above that Col/Fib presents a higher stiffness compared to collagen, which might directly activate PSCs, we were interested to analyze the activating effects of the fibrinogen component on PSCs in greater detail. For this, we treated PSCs cultured in a 2D monolayer with either fibrinogen or thrombin, the two components added to the commonly shared collagen matrix, and analyzed the expression of common PSC activation markers (alpha smooth muscle actin (ACTA2) and platelet-derived growth factor receptor (PDGFRβ) [18,35,36]), markers for the production of ECM proteins directly involved in the potential compression of blood vessels in PDAC (Collagen type 1 alpha 1 (COL1α1) and hyaluronic acid synthase 2 (HAS2) [1,4,37]) and a marker related to cell-ECM attachment (cell division cycle 42 (CDC42) [38,39]) (Fig. 3B). We found that fibrinogen was mainly inducing the expression of ACTA2, indicating PSC activation, and HAS2, while thrombin induced HAS2 and CDC42, displaying that both components play a crucial role in our model. Interestingly, we did not observe significant increase in the expression of COL1α1 when treated with fibrinogen and even a decrease when treated with thrombin, while PDGFRβ was reduced in both conditions, indicating that the expression of these markers might be induced by other growth factors or stimuli. Next, we investigated the expression of these markers in the 3D collagen and Col/Fib gels (Fig. 3C). Remarkably, we found that nearly all markers except PDGFRβ (p = 0.07) are significantly upregulated in Col/Fib gels compared to collagen gels alone indicating a clear activation of PSCs in the model as well as induced contraction and ECM production of PSCs. In particular ACTA2 displayed a 1.3 times upregulation, indicating the activation of PSCs. Furthermore, HAS2, a gene involved in the production of hyaluronic acid, which is a protein that forms a crucial part in vessel compression in PDAC due to its capability to swell when in aqueous solution [9], and CDC42, a gene directly related to cell-ECM attachment and related migration/contraction [38,39], displayed a 2 and 1.9 times upregulation, respectively, which might be crucial for the potential contraction of blood vessel in our model. While the upregulation of ACTA2, HAS2 and CDC42 might be related to pure presence of fibrinogen and thrombin in this culture, COL1α1 and PDGFRβ might be induced by the increased mechanical stiffness of the material. Eventually, the combination of both, mechanical stiffness and presence of fibrinogen/thrombin, is most likely to induce the activation of PSCs, resulting in the induced contraction of the Col/Fib gels.
To confirm that the genes upregulated in the Col/Fib gels are of clinical relevance for PDAC patients, we performed a transcriptomic analysis of publicly available data [40]. Data from 118 PDAC patients versus 13 control patients was analyzed to know the upregulation of aforementioned genes. Intriguingly, we found that all analyzed genes were significant upregulated in PDAC patients compared to the control, which shows the importance of these genes in PDAC as well as demonstrates the clinical relevance of our model (Fig. 3D). Remarkably, we also found that high expressions of COL1α1 and PDGFRβ can be significantly correlated to a higher expression of ACTA2 (Fig. 3E), indicating that activated PSCs are the main contributors to the expression of these genes. Surprisingly, we did not find such significant correlation in HAS2 and even an opposite trend in CDC42, where a higher ACTA2 expression was significantly correlated with a lower CDC42 expression. This might be based on the presence of other components in the patient TME that are actively involved in ECM remodeling and the mechanical environment in the TME such as macrophages [41,42].
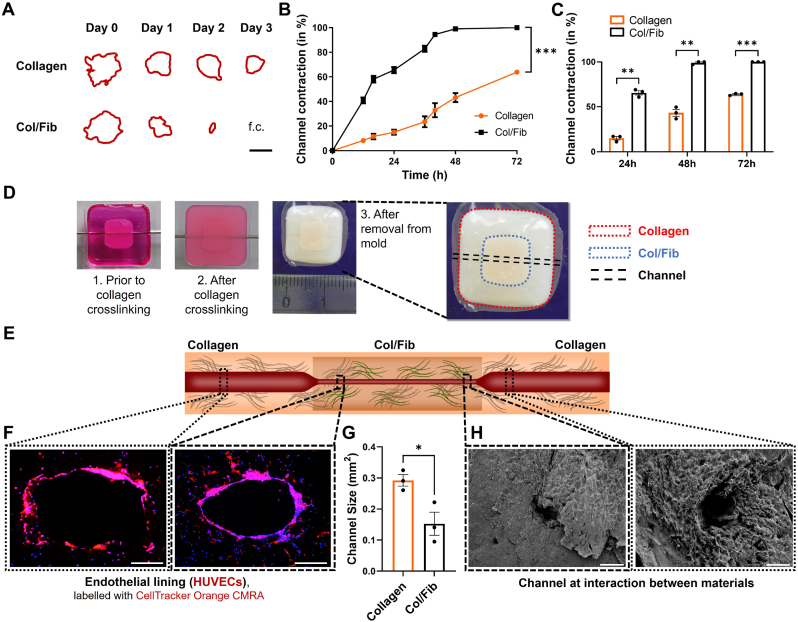
After studying the behavior of PSCs in collagen and Col/Fib gels in greater detail and demonstrating that Col/Fib indeed induced the activation of PSCs and promoted the contraction of the hydrogels, we wanted to investigate if this contractile behavior is also able to cause a blood vessel to compress. For this we first studied the contraction of a channel in collagen and Col/Fib gels using a custom-designed contraction assay based on a Ø6mm and 2 mm high disk of the gels including a Ø1mm channel in the middle (Fig. S5, Supporting Information). The design of this assay allowed us to track the contraction of the same channel on daily basis using conventional brightfield microscopy, despite the lack of optical clearance in collagen and Col/Fib gels (Fig. 4A). Remarkably, we found that Col/Fib gels display a significantly higher channel contraction compared to the collagen gel alone (Fig. 4B). While channels in collagen depicted a contraction of 15 ± 3.4%, 43.2 ± 6.4% and 63.8 ± 1.2% on day 1, 2 and 3, respectively, Col/Fib gels displayed a contraction of 65.5 ± 4.3%, 99 ± 1.6% on day 1 and 2 and were fully contracted on day 3 (Fig. 4C). Even despite the fact that channels in Col/Fib gels displayed a slightly bigger size in the beginning on day 0, these channels completely closed while collagen channels remained open demonstrating the enhancing effect of fibrinogen presence on the contraction in gels (Fig. S6, Supporting Information). As we found a high upregulation of markers for PSC activation and contraction, cell contraction might form a major reason for the contraction of this channel, however, the induction of COL1α1 and HAS2 also indicates the production of ECM proteins might play an additional role in the contraction of the channel in our model.
Fig. 4.
Contraction ofacentral channelincollagen and Col/Fib gels. A) Outline of channels in PSC-laden collagen and Col/Fib gels for a total duration of 3 days (f.c. = fully closed) based on a custom-made contraction assay (Fig. S5, Supporting Information). Scale bar = 500 μm. B) Contraction of channels in PSC-laden collagen and Col/Fib gels followed for a total duration of 3 days and standardized to the initial days, n = 3. C) Highlighted standardized contraction for 24 h, 48 h and 96 h post-preparation, n = 3. D) Photographic images of the full model (1) prior to crosslinking of the surrounding collagen gel highlighting the central, already crosslinked, Col/Fib gel, (2) after the crosslinking of collagen and (3) after removal of the central sacrificial rod and from the mold, highlighting the different parts of the model. For better visualization of the central Col/Fib gel, a top layer of collagen was not applied for these photographic images. E) Schematic representation of a contracted central endothelialized channel in multi-layered collagen/Col/Fib gels. F) Sections of central endothelialized channel highlighting the monolayer of CellTracker-labelled HUVECs (red) and nuclei (blue) in the collagen and Col/Fib part of a multi-layered collagen/Col/Fib construct 3 days post-formation. Scale bar = 200 μm. G) Quantification of the size of the central channel in the collagen and Col/Fib part of a multi-layered collagen/Col/Fib construct, n = 3. H) SEM images at the intersection of the collagen and Col/Fib part of a multi-layered collagen/Col/Fib construct. Scale bar = 500 μm. Mean + SEM, *p < 0.05, **p < 0.01, ***p < 0.001.
Next, we were interested to investigate if the found contraction in our custom-made model can also be seen in the full multi-layer tissue model consisting of areas with pure collagen and with Col/Fib and a central endothelialized channel. First, we found that, based on the proposed technique to generate these multi-layered tissues (Fig. 1C), we were able to generate tissues that depict a central channel in the middle of the whole tissue (Fig. 4D and E), which could successfully be endothelialized using primary human umbilical vein endothelial cells (HUVECs) (Fig. 4F and Fig. S7, Supporting Information). Interestingly, we saw that HUVECs seem to form a complete monolayer in the Col/Fib gels while we clearly observed gaps in the layer of HUVECs in collagen gels. This shows that fibrinogen in the gels also promotes HUVECs adhesion and growth to form a monolayer as also reported in literature [[43], [44], [45], [46]]. Furthermore, we found that the channels in collagen depicted a significantly larger size of 0.292 ± 0.03 mm2, while channels in Col/Fib showed a ∼50% smaller size of 0.152 ± 0.06 mm2 (Fig. 4G). This difference could further be demonstrated by SEM focusing on the intersection between collagen and Col/Fib (Fig. 4H). These results demonstrate that our model was successful to mimic the compression of blood vessels found in PDAC in a single construct replicating the realistic situation in vivo. Although we did not observe a full vessel compression in our model, which might be attributed to an overall larger size of this model (Ø8mm, ∼8 mm height) that would require a larger number of cells or longer culture duration to successfully contract the channel, we were able to generate a model that allowed to study cell-mediated contraction of blood vessels in a biologically relevant environment.
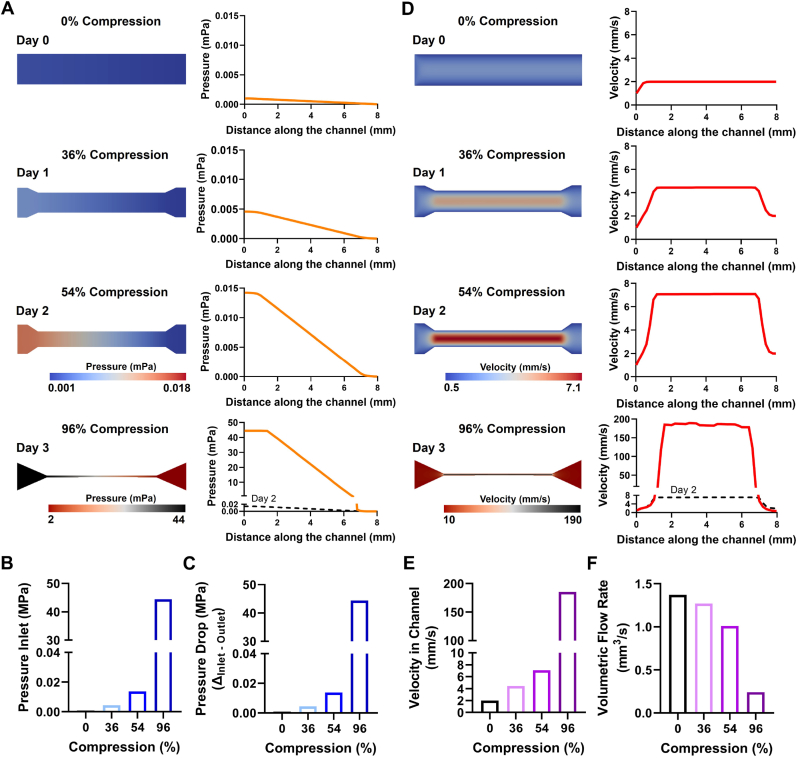
To further analyze what effects such compressed vessels have in PDAC, in particular on the efficacy of intravenously administered therapeutics, we investigated the blood flow in channels of different levels of compression in greater detail using CFD simulations. Based on the observed contraction on different days throughout the experiments we designed a blood vessel with 0%, 36%, 54% and 96% compression representing days 0, 1, 2 and 3 of the experiments (Fig. S8, Supporting Information). Using CFD, we computed the flow right before, inside and right after the compressed channel and investigated the differences in pressure and flow velocity based on the different rate of compression. First, we found that with increasing compression of the channel, the pressure at the inlet of the constriction continuously increases at lower stages of compression from 0.000895 mPa (mPa) for 0%, 0.004359 mPa for 36% to 0.01375 mPa for 54%, while the pressure displays a drastic increase when reaching 96% compression to 44.1 mPa (Fig. 5A and B). Furthermore, the higher pressure at the inlets of the constriction also created an increasing pressure drop (Δ Pressure Inlet – Pressure Outlet) with higher levels of compression in the channels, which was nearly equal to the pressure at the inlet due to a nearly negligible pressure at the outlet (Fig. 5C). The increasing pressure at the inlets of the compressed channels demonstrated a clear obstruction of the blood flow, causing potential therapeutics or nanomedicines to accumulate before the compressed channel and not reaching their target. Furthermore, the high pressure might also cause early extravasation of the therapeutics, which, depending on the drug, might cause severe side-effects in non-target tissues. Additionally, the obstruction does not only cause therapeutics to accumulate but also cells, such as erythrocytes or immune cells, and proteins in the blood stream are accumulating at the inlet of the compressed vessel which might further obstruct the blood flow reducing nutrient and oxygen supply creating an increasing hypoxic environment. Furthermore, immune cells are not able to reach the tumor area, causing PDAC to escape immune response. Besides the clear differences in pressure, we also found that with increasing channel compression the velocity in the channels drastically increases from 1.99 mm/s, 4.43 mm/s, 7.08 mm/s for 0%, 36% and 54% compression, respectively, up to 185 mm/s for channels of 96% compression (Fig. 5D and E). This indicates that therapeutics or immune cells that are able to enter the compressed channel are potentially passing the channel without a chance to extravasate as the flow velocity is too high within the channel, which drastically limits the effects of intravenously injected therapeutics. Although the flow rate in the channels was significantly increasing with higher levels of compression, the volumetric flow rate (Q) was continuously decreasing with higher levels of compression from 1.37 mm3/s, 1.27 mm3/s and 1.01 mm3/s for 0%, 36% and 54% of compression, respectively, down to 0.24 mm3/s for a compression 96% (Fig. 5F). This shows that in compressed channels the same amount of blood flows at a much higher velocity. To further analyze the flow dynamics in the compressed channels we calculated the Reynolds number and Péclet number for each compression stage (Table S1, Supporting Information). We found very low Reynolds numbers for all compression stages ranging from 2.0937 for 0% compression up to 8.1568 for 96% compression indicating a laminar flow in the channels. Furthermore we calculated the Péclet number to investigate the diffusion properties of molecules in the channels. As no drug or small molecule was simulated within the flow, here we used the self-diffusion coefficient of water at 25 °C. We found a Péclet number of 3479 at 0% compression already indicating that mass transfer is mainly advection based which is continuously increasing with increasing channel compression up to 323168 at 96% compression. This demonstrates that the high flow velocities in the channel do not allow for diffusion-based transfer of molecules in the flow.
Fig. 5.
Computational fluid dynamics (CFD) simulation for different compression stages. A) CFD simulations of channels with a compression of 0, 36, 54 and 96% depicting the pressure inside of the channel. B) Pressure at the inlet of the channel for 0, 36, 54 and 96% compression. C) Pressure drop along the channel for 0, 36, 54 and 96% compression. D) CFD simulations of channels with a compression of 0, 36, 54 and 96% depicting the flow velocity inside of the channel. E) Average velocity inside of the channel for 0, 36, 54 and 96% compression. F) Volumetric flow rate for channel with 0, 36, 54 and 96% compression.
In summary, we found that not only the full obstruction of a channel can have direct consequences on the blood flow as the pressure nearly increased 5 and 3 times and the flow velocity almost doubled for earlier stages of compression (0%–36% to 54% compression) before experiencing a drastic increase of 3000 times higher pressure and 26 times higher flow velocity from 54% to 96% compression. Altogether, the high pressure right before the entrance of the constriction, demonstrating an obstruction of flow, the high velocity but low volumetric flow rate combined, not only prevent sufficient amounts of therapeutics and immune cells from reaching the target tissue and might cause early extravasation but even therapeutics and cells that enter compressed channels experience a high flow velocity and a high pressure drop which might not give sufficient time and support to extravasate. Furthermore, mass transfer within the flow is mainly advection driven, based on the high flow velocities in the compressed channels.
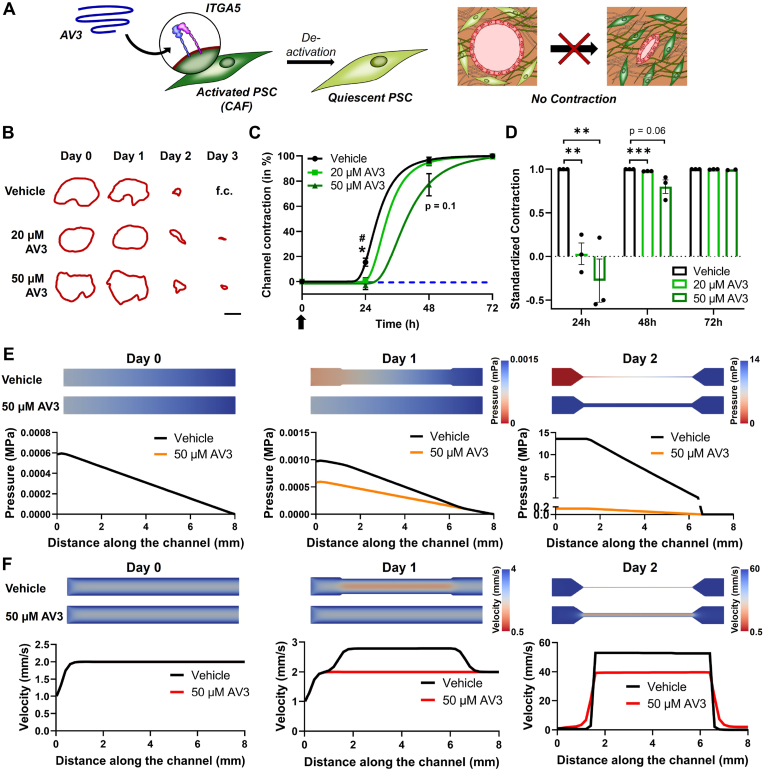
After demonstrating that our novel model is successfully able to replicate the blood vessel compression found in clinical and pre-clinical situations as well as after analyzing what effects such compressed vessels have on the blood flow and subsequently on therapeutics or immune cells in the blood stream, we wanted to demonstrate that our model can also be used to investigate the efficacy of therapeutics able to re-open blood vessels. Here we studied the efficacy of AV3, an integrin alpha 5 (ITGA5) antagonist that was developed in our group and that has recently demonstrated to inhibit CAFs in PDAC causing a re-opening of the vasculature in vivo (Fig. 6A) [20]. Furthermore, ITGA5 is known to bind to fibrinogen in the ECM [43,44]. The blocking of ITGA5 should inhibit binding of PSCs to fibrinogen and therefore prevent contraction. First, we confirmed the increased expression of ITGA5 in 2D cultured PSCs (Fig. S9A, Supporting Information), being mainly driven by the presence of fibrinogen and not thrombin going along with the activation of PSCs as previously shown based on the expression of ACTA2 (Fig. 3B). Similarly, we found an upregulation of ITGA5 in Col/Fib 3D gels compared to collagen gels, which might be based on the activation of PSCs in the environment as well as excess presence of fibrinogen (Fig. S9B, Supporting Information). Furthermore, we found a significant upregulation of ITGA5 in PDAC patients based on the transcriptomic dataset, which can be attributed to the activation of PSCs demonstrated by the significant correlation of a high ACTA2 expression and ITGA5 expression (Fig. S9C, Supporting Information). Based on the correlation of ACTA2 and ITGA5, we next investigated if AV3 is indeed able to inhibit PSC activation. We incubated PSC-laden Col/Fib gels with either DMSO, serving as vehicle, or AV3 and found a significant inhibition of ACTA2 expression based on the treatment, which goes along with our previously published data (Fig. S9D, Supporting Information) [20]. Next, we investigated if the treatment with AV3 is also able to inhibit the compression of blood vessels in our custom-made channel contraction assay. We found that treating PSC-laden Col/Fib gels with 20 and 50 μM on the day of preparation (day 0) we can significantly delay the contraction of the channel in Col/Fib gels (Fig. 6B–D). Especially 1 day after gel preparation, AV3-treated gels hardly displayed any contraction with only 0.63 ± 4.05% for 20 μM and −2.93 ± 5.98% for 50 μM, displaying even a further relaxation of the channel, compared to 15.33 ± 5.6% for the vehicle group. Although the gels treated with vehicle and 20 μM AV3 displayed a nearly 100% contraction on day 2 with 96.79 ± 4.07% and 94.62 ± 3.90%, respectively, the gels treated with 50 μM AV3 still displayed a contraction of only 77.09 ± 15.2%, a clear inhibition compared to the control group. After 3 days, all gels presented a nearly complete contraction, although both AV3-treated groups still displayed a slightly open channel. Nonetheless, we clearly observed a significant inhibition on the contraction in the initial days, demonstrating that the treatment with AV3 was indeed able to prevent blood vessel compression, which goes along with our previous findings. Furthermore, refreshing the treatment with AV3 on for instance day 1 or day 2 might further delay the compression of the channels.
Fig. 6.
Evaluation of the ITGA5 antagonist AV3 in vitro and in silico. A) Schematic representation demonstrating the de-activation of PSCs to their quiescent phenotype by binding of AV3 to ITGA5 on activated PSCs resulting in the prevention of contraction. B) Outline of channels in PSC-laden Col/Fib gels treated with vehicle (DMSO), 20 μM or 50 μM AV3 followed for a total duration of 3 days (f.c. = fully closed). Scale bar = 500 μm. C) Contraction of channels in PSC-laden Col/Fib gels treated with vehicle (DMSO), 20 μM or 50 μM AV3 followed for a total duration of 3 days and standardized to the initial days, n = 3. Arrow indicates treatment with AV3. *indicates significant between vehicle and 50 μm AV3, #indicates significance between vehicle and 20 μm AV3. D) Contraction of channels in PSC-laden Col/Fib gels treated with vehicle (DMSO), 20 μM or 50 μM AV3 followed for a total duration of 3 days and standardized to the vehicle control, n = 3. E) Computational fluid dynamics (CFD) simulations of channels after treatment with vehicle or 50 μM AV3 depicting the pressure inside of the channel. F) CFD simulations of channels after treatment with vehicle or 50 μM AV3 depicting the flow velocity inside of the channel. Mean + SEM, */#p < 0.05, **p < 0.01, ***p < 0.001.
Next, we investigated the flow within the channels using CFD simulations to further demonstrate the differences of AV3 treatment on the pressure and flow velocity right before, inside and right after the compressed channel on days 0, 1 and 2 post-treatment. First, we found that with increasing compression of the channels the pressure at the inlet continuously increased, confirming the results observed before. Furthermore, we found that the treatment with 50 μM AV3 can drastically reduce this pressure at the inlet, up to 86x lower pressure on day 2, where we detected a pressure at the inlet of the channel of 13.6 mPa in the vehicle-treated group compared to a pressure of 0.15 mPa in the AV3-treated group (Fig. 6E and Fig. S10A, Supporting Information). Similarly, we found a drastic reduction in the pressure drop along the channel (Fig. S10B, Supporting Information). These findings demonstrate that an inhibition of ∼20% compared to a nearly fully contracted vessel can cause significant differences in the pressure inside of this channel, indicating drastic differences in the blockage found in the vessel and hence in the therapeutics reaching the tumor site. Similarly, we investigated the flow in the channels based on the treatment with vehicle compared to 50 μM AV3. We found that treatment with AV3 can reduce the flow velocity in the channel up to 1.35x on 2 post-treatment, presenting a flow velocity of 53.44 mm/s compared to 39.54 mm/s for the vehicle and AV3-treated group, respectively (Fig. 6F and Fig. S10C, Supporting Information). Although the flow velocity in the AV3-treated group is still comparably high, we found that the standardized volumetric flow rate in the AV3-treated is 5.88x higher compared to the volumetric flow rate in the vehicle-treated group, indicating that in the vehicle group the same amount of blood flows at much higher velocity compared to the AV3-treated group (Figs. S10D and E, Supporting Information).
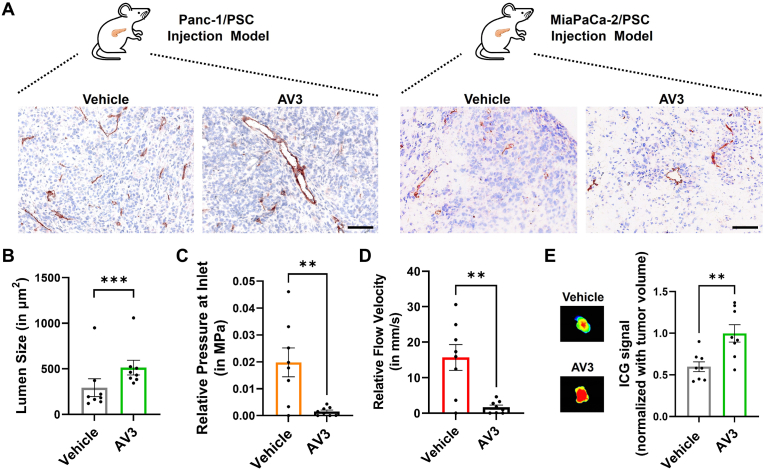
Finally, we wanted to confirm that the results we obtained from our in vitro and in silico model indeed represent the functioning of AV3 in vivo. Here, we first investigated the potential of AV3 to open the vasculature in Panc-1/PSC and MiaPaCa-2/PSC co-injection tumors in mice. Hereby Panc-1/PSC and MiaPaCa-2/PSC tumors were combined to mimic patient heterogeneity in the clinics. Interestingly, we found that based on immunohistochemical staining of CD31 AV3-treated mice indeed presented more open blood vessels with a significantly larger lumen size on average, confirming the functioning of AV3 in vivo as predicted by our engineered in vitro model (Fig. 7A and B, outliers have been identified using Z-score statistical analysis and have not been included in statistical analysis (Fig. S11, Supporting Information)). Strikingly we found that vessels in the vehicle group depict a compression of 42.9% compared to the vessel in AV3-treated group, considering the average lumen size in AV3-treated mice as “most open”, hence 0% compression. To further understand the effects of this opening of vasculature in vivo, we investigated the relative pressure at the inlet of the compressed vessel and the relative velocity inside of the compressed channel. For this we created standard curves based on the obtained CFD simulation data from Fig. 5, Fig. 6, and studied the effects of the relative vessel contraction in the vehicle group compared to the AV3-treated mice (Fig. 7C,D and Fig. S12, Supporting Information). Intriguingly, we found that the vehicle group displayed a 12.8x higher pressure at the inlet of the compressed vessel with 0.02 ± 0.015 MPa compared to 0.0015 ± 0.001 MPa for the vehicle and AV3-treated group, respectively (Fig. 7C). Furthermore, the velocity inside of the compressed vessel was 9.3x higher in the vehicle treated group with 15.67 ± 10.25 mm/s compared 1.68 ± 1.73 mm/s in the AV3-treated group, respectively (Fig. 7D). To confirm if the overall compression of vessels in the vehicle group, combined with the theoretical higher pressure at the inlet and higher flow velocity inside of the compressed channel have effect on the delivery of drugs to the tumor region, we investigated the distribution of indocyanine green (ICG) dye using near-infrared imaging and found a significantly higher accumulation of the dye in tumors of AV3-treated compared to vehicle treated mice (Fig. 7E). While arguably other factors might further increase the higher accumulation of ICG in tumors, such as for instance reduction of tumor ECM density itself, as previously shown [20], the results from our in vivo experiments confirmed the potential of our novel 3D engineered model in combination with CFD simulation to successfully predict the effects of novel TME-modulating therapeutics on the vasculature in vivo and additionally give an indication of the potential efficacy of intravenously injected therapeutics to reach and accumulate at the tumor site.
Fig. 7.
Evaluation of the ITGA5 antagonist AV3 in vivo. A) Immunohistochemical staining for CD31 (red) and nuclei (blue) in Panc-1/PSC and MiaPaca-2/PSC tumors treated with either vehicle or AV3. Scale bar = 100 μm. B) Quantification of the average lumen size based on CD31 staining including tumors from Panc-1/PSC and MiaPaCa-2/PSC (combined data, n = 4/cell type model, n = 8 total). Outliers have been identified using Z-score statistical analysis and have not been included in statistical analysis (Fig. S11, Supporting Information). C) Theoretical relative pressure at the inlet of compressed vessels in Panc-1/PSC and MiaPaCa-2/PSC tumors in mice based on previously established CFD simulations of different compression levels, based on n = 8 animals. D) Theoretical relative flow velocity in compressed vessels in Panc-1/PSC and MiaPaCa-2/PSC tumors in mice based on previously established CFD simulations of different compression levels, based on n = 8 animals. E) Accumulation of ICG dye in Panc-1/PSC and MiaPaCa-2/PSC tumors in mice treated with either vehicle or AV3. Isolated tumors were imaged using near-infrared animal imager, combined data, n = 4/cell type model, n = 8 total. Mean + SEM, **p < 0.01, ***p < 0.001.
Altogether, in our model we found that AV3 treatment can drastically decrease the pressure in the vessels while also reducing the flow velocity in the channel and increasing the volumetric flow rate, eventually facilitating the entrance of other therapeutics or immune cells into the channel allowing them to reach the tumor site as well as prevent a rapid clearance. Using CFD simulation in combination with our in vitro model allowed us to not only test the inhibitory potential of novel therapeutics on the contraction of the vasculature in PDAC, but also aided in predicting how such modulation of the vasculature can affect the pressure and velocity in the channel in vivo, eventually helping to design novel combinational therapeutics. Furthermore, the use of CFD simulations to further predict the flow in the channels can aid in the smart design of novel treatment strategies in vivo based on observations made in vitro and in silico, such as determining the effects of simultaneous injection of a, for instance, TME-modulating and chemotherapy, compared to a pre- or post-treatment of different therapeutics to increase their efficacy.
We envision that by adding ECM modulating cells such as macrophages to our model in the future, we might be able to replicate the TME in vivo in an even more biologically relevant fashion. As macrophages would actively degrade ECM, while CAFs would be inhibited using TME-modulating therapeutics, using our model we could further analyze the mechanistic changes in TME in greater detail. Furthermore, while increasing the complexity in terms of cellular content, one might change the ECM composition to represent PDAC in a more complex fashion or directly alter the mechanical properties of the matrix. Currently our novel model mainly relies on cell-own ability to produce more PDAC specific ECM as well as alter the mechanics of the tissues. While arguable two different areas of stiffness could be achieved in the center and surrounding of our model based on the covalent crosslinking of fibrinogen, collagen mainly relies on thermal gelation. By for instance introducing crosslinkable components to the collagen, one might be able to achieve different zones of stiffness immediately which would help to study cellular reaction to PDAC mechanics in greater detail [47,48]. Furthermore introducing more components in such fashion, like for instance fibronectin or other materials might result in a more tunable hydrogel matrix that is eventually mimicking the mechanics of PDAC to a greater extent [49]. In such way, one might also be able to study the diffusion of a drug, when extravasating, into the tissue based on different stiffness or morphology which might have a direct effect on drug performance and availability. Being able to control the exact mechanical properties of the PDAC surrounding combined with a more complex cellular composition and supporting computational simulations might help to understand the performance of novel therapeutics in much greater fashion than conventional culture models or animal experiments.
3. Conclusion
To our best knowledge, this is the first proof-of-concept study presenting a 3D engineered vascularized model that mimics the compression of vasculature in PDAC displaying the biomaterial-driven controlled activation of PSCs. Furthermore, the combination of observations from this novel in vitro model with in silico CFD simulation, facilitated the assessment of the blood flow during different compression stages, which allows to predict the behavior of intravenously administered therapeutics in vivo. Moreover, this model can be used to specifically investigate the effects of CAF-modulatory therapeutics on the compression and fluid dynamics of tumor vasculature in a simple and rapid fashion, which might eventually facilitate the generation of tailored therapeutics that are aimed to re-open the tumor vasculature to induce tumor delivery, hence the efficacy of anti-tumor agents. By implementing additional cell types that are known to actively remodel the ECM, such as macrophages, one might be able to study the effects of cellular crosstalk on the mechanical environment in PDAC in greater detail. Furthermore, this model is not solely limited to PDAC, as other fibrotic tumors also display the presence of compressed blood vessels. One may use the same proposed model to study cell-mediated changes in the mechanical microenvironment in different tumor types. Altogether, this novel model allowed to study the compression of vasculature, which so far was only observed in vivo or in clinical conditions, in a controlled fashion. We envision that this model might find broad application in the evaluation of novel treatment strategies that modulate the TME, which will eventually help reducing and refining animal experiments as well as designing optimized combinational therapies.
In summary, the realistic biomimetic characteristics of our 3D in vitro vascularized PDAC model has paved the way for novel models to study the mechanical environment in fibrotic tumors in a controlled fashion, where the future development of more complex models for the rapid and effective screening of (combinational) therapies is envisioned.
4. Experimental section
Immunofluorescent staining on human PDAC sections. Anonymous paraffin-embedded human PDAC sections were obtained from Laboratory Pathology East Netherlands (LabPON; Hengelo, The Netherlands). Sections were deparaffinized in xylene and rehydrated in a serious of ethanol followed by MilliQ water. Antigen retrieval was performed by heat induction at 95 °C in citrate buffer at pH 6.0 (Dako, Glostrup, Denmark). The sections were incubated with the primary antibody against CD31 (Mouse monoclonal antibody, 1:50, Cell Marque, Rocklin, CA, USA) and in phosphate-buffered saline (PBS) overnight at 4 °C. After washing in PBS, the sections were incubated with secondary fluorescent antibody (Alexa Fluor 488, Donkey anti-mouse IgG, 1:100, Thermofisher Scientific, Waltham, MA, USA) for 30 min at room temperature, before being washed in PBS and mounted with Fluoroshield™ with DAPI (Sigma-Aldrich, St. Louis, MO, USA) and imaged using Nanozoomer-RS (Hamamatsu Photonics, Hamamatsu, Japan).
Cell Culture (PSCs, Panc-1 & MiaPaCa-2). Primary human patient-derived pancreatic stellate cells (PSCs, ScienCell, Carlsbad, CA, USA) were cultured in stellate cell medium supplemented with 2 v/v % fetal bovine serum (FBS), 100 U/mL penicillin/100 μg/mL streptomycin (Pen/Strep) and 1 vol% stellate cell growth supplement (SteCGS) according to manufacturer's instruction (all products from ScienCell). Panc-1 pancreatic cancer cells and MiaPaCa-2 pancreatic cancer cells (both from the American Type Culture Collection (ATCC), Manassas, VA, USA) were cultured in DMEM - High Glucose HyClone medium (GE Healthcare Life Sciences, Chicago, IL, USA) containing 10 vol% FBS, 100 U/mL penicillin/100 μg/mL streptomycin (Thermofisher Scientific) and 2 mM l-glutamine (Thermofisher Scientific). The cells were maintained at 37 °C in a humidified 5% CO2 atmosphere, and passed at 80% confluence. PSCs were used between passages 4–10 according to the manufacturer's suggestion.
Animal experiments and ethics statement. All the animal experiments in this study were performed according to the Institutional Animal Care and Use Committees guidelines and approved by the animal ethical committee of Utrecht University (2014.III.02.022), The Netherlands, and Central Animal Welfare (AVD1100020174305).
Co-injection model of Panc-1/PSC and MiaPaCa-2/PSC. Six-week old male CB17 severe combined immunodeficiency (SCID) mice (Janvier Labs, Le Genest-Saint-Isle, France) were subcutaneously co-injected with either Panc-1 cancer cells or MiaPaCa-2 cancer cells (2 × 106 cells) and PSCs (4 × 106 cells). At the end of the experiment (≈45 days post-injection), animals were sacrificed under anesthesia, after which tumors were harvested, embedded in cryomatrix (Thermofisher Scientific) and immediately snap-frozen in ice-cold 2-methyl butane (Thermofisher Scientific). All samples were stored at −80 °C until further analysis.
Immunohistochemical staining on section from Panc-1/PSC and MiaPaCa-2/PSC co-injection model. Snap-frozen tumors were cryosectioned into 4 μm thick sections, fixed in acetone for 10min, air-dried and stained for CD31 expression. In brief, the air-dried sections were surrounded with a hydrophobic PAP pen (Sigma-Aldrich) before being rehydrate in phosphate buffered saline (PBS). The rehydrated sections were incubated with primary antibody against CD31 (Rat anti-mouse, 1:50, SouthernBiotech, Birmingham, AL, USA) overnight at 4 °C. Next the sections were incubated with hydrogen peroxide (H2O2, Sigma-Aldrich) to block endogenous peroxidases, washed with PBS, before being incubated with secondary antibody (Rabbit anti-rat IgG/HRP, 1:100, Dako, Denmark) in PBS containing 5% normal human serum (Sigma-Aldrich) for 1 h at room temperature. Afterwards the sections were washed again and incubated with tertiary antibody (Goat anti-rabbit IgG/HRP, 1:100, Dako) in PBS containing 5% normal human serum for 1 h, before stained with AEC solution (Sigma-Aldrich) for 15 min, washed with MilliQ water, counterstained with hematoxylin solution for 1 min and rinsed in tapwater for 10 min. Finally the sections were mounted using Aquatex® aqueous mounting agent for microscopy (Sigma-Aldrich) and imaged using Nanozoomer-RS.
Generation of collagen, fibrinogen and collagen/fibrinogen (Col/Fib) gels. Collagen gels were prepared according to the manufacturer's instructions. In brief, to prepare 1 mL of gel solution, 45 μl of 1 M NaOH (Sigma-Aldrich), 50 μl of 1 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, Sigma-Aldrich) buffer, 5 μl of sterile water, 100 μl of 10x Medium (Sigma-Aldrich) were mixed with 800 μl of collagen solution (0.5%, type 1 from bovine calfskin, Matrix Biosciences, Mörlenbach, Germany) to obtain a final collagen concentration of 4 mg/mL. The hydrogel solution was further diluted with sterile Dulbecco's phosphate buffered saline (DPBS, Sigma-Aldrich) to achieve a final concentration of 3.5 mg/mL. The gels were transferred to a custom-made PDMS mold (7 mm × 7 mm x 5 mm (length x width x height)) and allowed to fully solidify at 37 °C in a humidified atmosphere.
Fibrinogen gels were prepared by dissolving fibrinogen (from bovine plasma, Type I–S, 65–85% protein (≥75% of clottable protein, Sigma-Aldrich) in warm DPBS to obtain a final concentration of 80 mg/mL. Fibrinogen was allowed to fully dissolve by incubation at 37 °C for 1 h before preparing aliquots and stored at −20 °C for further use. Similarly, thrombin (from bovine plasma, lyophilized powder, ≥ 2000 NIH units/mg protein, Sigma-Aldrich) was dissolved in DPBS to obtain a final concentration of 100U/mL. Thrombin aliquots were prepared in small volumes and stored at −20 °C. Thrombin was always thawed immediately prior to use to avoid loss of enzymatic activity and never re-frozen. To prepare fibrinogen gels, fibrinogen (80 mg/mL) was diluted to a final concentration of 10 mg/mL in warm DPBS. Next, thrombin was added in a ratio of 1:100 to achieve a thrombin centration of 1U/mL before homogenous mixing of the two components. The gel solution was transferred to custom-made PDMS molds (7 mm × 7 mm x 5 mm) and allowed to fully solidify at 37 °C in a humidified atmosphere.
Collagen/fibrinogen (Col/Fib) hybrid gels were prepared by first mixing the collagen stock solution (4 mg/mL, prepared as aforementioned) with fibrinogen stock solution (80 mg/mL) in a ratio of 7:1 to achieve a final concentration of 3.5 mg/mL collagen and 10 mg/mL fibrinogen. Next, thrombin was added in a ratio of 1:100 to achieve a final concentration of 1U/mL, the solution was homogenously mixed and transferred to custom made PDMS molds (7 mm × 7 mm x 5 mm) and allowed to solidify at 37 °C in a humidified atmosphere.
Scanning electron microscopy of collagen, fibrinogen and Col/Fib gels. All gels were prepared as aforementioned and allowed to soak in DPBS at 37 °C for 48 h. To prepare the samples for SEM, all gels were fixed with 2.5 v/v% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA, USA) for 1 h at room temperature and at 4 °C overnight. The fixed gels were washed three times with MilliQ water before being frozen in liquid nitrogen. Afterwards the gels were lyophilized (TFD5503 Freeze Dryer, ilShin BioBase Europe, Ede, The Netherlands), gold-sputtered (Sputter Coater 108 Auto, Cressington Scientific Instruments, Watford, UK) and imaged using a scanning electron microscope (JSM-IT100, JEOL, Tokyo, Japan) at an accelerating voltage of 5 kV and a probe current of 35. The pore size for each gel was determined using ImageJ (Public, developed by Wayne Rasband (NIH)) for 18 pores/sample while measuring the largest diameter for each pore.
Mechanical characterization of collagen, fibrinogen and Col/Fib gels. The compression modulus of the gels was measured using a digital mechanical analyzer (DMA850, TA Instruments, New Castle, Delaware, USA). First, cubes of 7 mm × 7 mm x 5 mm were prepared as mentioned above and soaked in DPBS at 37 °C for 48 h. The samples were transferred to the DMA, pre-loaded with 0.00001 N, allowed to equilibrate for 5min at a temperature of 20 °C and measured using a displacement of 0.1 mm/min until the samples broke or slipped out of the measurement clamps considered as the maximum achievable displacement.
Stability of collagen, fibrinogen and Col/Fib gels. For the stability of the different gels, disks of Ø6mm and 2 mm height were prepared as aforementioned and kept in DPBS at 37 °C for a total duration of duration of 10 days. The size of the constructs was photographically imaged on daily basis and the size was determined using ImageJ.
Generation of PSC-laden collagen and Col/Fib gels. PSCs were cultured as aforementioned, treated with 0.25% trypsin/ethylenediaminetetraacetic acid (EDTA) (Gibco, Thermofisher Scientific) for 5min at 37 °C, neutralized with 10-fold full PSC culture medium, counted and centrifuged for 5min at 300 g to achieve a cell pallet. The PSC pallet was resuspended in collagen or collagen/fibrinogen gels (prepared as aforementioned) at a concentration of 2 × 106 cells/mL. The PSC-laden gels were transferred to custom-made PDMS molds (dimensions specified for each experiment), allowed to solidify at 37 °C in a humidified atmosphere, before being transferred to full PSC culture medium for further culture.
Viability of PSCs in collagen and Col/Fib gels. 1, 4 and 7 days post-preparation the viability of the cells was determined by incubating the PSC-laden gels with 2 μM calcein AM (LIVE) and 4 μM ethidium homodimer 1 (DEAD, both from Thermofisher Scientific) in DPBS for 20min before immediately imaged with a fluorescent microscope (EVOS Cell Imaging System, Thermofisher Scientific). Quantification was performed by counting alive and dead cells using ImageJ. The direct quantification of viable cells based on LIVE/DEAD on later days (4 and 7) was not possible due to the high contraction of the gels, limiting optical clearance and causing cells to cluster not allowing individual counting.
Metabolic activity of PSCs in collagen and Col/Fib gels. The metabolic activity of cells was measured on day 1, 4 and 7 post-preparation. To monitor the activity, AlamarBlue dye (10x, Invitrogen, Carlsbad, USA) was diluted in full culture medium to a final concentration 1x. The gels were incubated in AlamarBlue-containing medium for 4 h, before transferring the supernatant to a black Nunc™ 96 well plate (Thermofisher Scientific). The fluorescent signal was measured using a VIKTOR™ plate reader (PerkinElmer, Waltham, MA, USA).
Release of lactate dehydrogenase (LDH) from collagen and Col/Fib gels. The release of LDH was measured on day 1, 4 and 7 post-preparation in two different ways: i) Incubating the gels with fresh culture medium on day 0, 3 and 6 and collecting the supernatant after 24 h of incubation or ii) continuously culture the medium from day 0 and only refreshing 50% of the medium on day 1, 4 and 7 to determine the accumulative release of LDH in the medium. For both conditions 1 mL of culture medium was collected, 150 μL of Poly(ethylene glycol) (PEG)-400 (Sigma-Aldrich) was added as cryoprotectant and the samples were stored at – 20 °C until measurement. A negative control was prepared in a similar fashion by collecting fresh cell culture medium, adding PEG-400 and freezing at – 20 °C. The positive control was prepared as follows: The cultured gels were collected on day 7 in the respective culture media and mechanically disrupted using an Ultra Turrax (IKA T18 basic, IKA®-Werke GmbH & Co. KG, Staufen, Germany) until complete homogenization of the gels inducing cell death while keeping the sample on ice to avoid heat-induced denaturation of LDH. Then the homogenized gels and cells were centrifuged at 1000×g for 5 min. Next, the supernatant was collected, PEG-400 was added and the samples were stored at – 20 °C. To measure the LDH content, all samples were thawed at room temperature for 45 min and the concentration of LDH was determined using Cytotoxicity detection kitPLUS (Roche #04744926001, Basel, Switzerland) according to the manufacturer's instructions. In brief, the collected supernatant was incubated with a provided freshly prepared reaction mix for 30 min under constant agitation at 150 RPM before the reaction was stopped with the provided stop solution. Next the absorbance was measured at λ = 490 nm using a Tecan plate reader (Tecan Group Ltd., Männedorf, Switzerland).
Contraction of PSC-laden collagen and Col/Fib gels. PSC-laden gels were prepared as previously described. The gels were cultured in full culture medium for a duration of 7 days while being imaged on daily basis. The size of the gels was determined using ImageJ and the contraction was determined by standardizing the observed size for each day to the initial size on day 0.
Gene expression in 2D cultured PSCs. PSCs were seeded at a concentration of 40.000 cells/well in a conventional 12 well plate (tissue culture treated, Greiner Bio-One, Kremsmünster, Austria) and allowed to attach for 24 h. Next, the PSCs were treated with either 2 mg/mL fibrinogen or 1U/mL thrombin diluted in full culture medium for 24 h before being lysed for RNA isolation. The RNA was isolated using the GenElute™ Mammalian Total RNA Miniprep Kit (Sigma-Aldrich) and the RNA amount was measured using a Nanodrop® ND-1000 spectrophotometer (Thermofisher Scientific). Subsequently, cDNA was synthesized with iScript™ cDNA synthesis Kit (BioRad, Venendaal, Netherlands). 10 ng cDNA were used for each PCR reaction. The real-time PCR primers (Table S2, Supporting Information) were purchased from Sigma Aldrich. Quantitative real time PCR was performed using 2x SensiMix SYBR and Fluorescein Kit (Bioline GmbH, Luckenwalde, Germany) using a BioRad CFX384 Real-Time PCR detection system (BioRad). Gene levels were normalized to the expression of the housekeeping gene ribosomal protein S18 (RPS18).
Gene expression in PSCs cultured in collagen and Col/Fib gels. PSC-laden gels were prepared as aforementioned and cultured for 24 h. Next, the collagen and collagen/fibrinogen matrix was enzymatically degraded using a mix of 0.5 mg/mL collagenase (from Clostridium Histolyticum, activity ≥800 units/mg, Sigma-Adrich) and 2 mg/mL dispase II (Sigma-Aldrich) in DPBS for 30min at 37 °C to isolate the PSCs. RNA isolation, cDNA synthesis and real-time PCR were performed as previously described. Gene levels were normalized to the expression of the house-keeping gene RPS18.
Transcriptomic expression analysis in human cohort from public database. A PDAC gene expression from the Expression Omnibus Database (GEO) was selected and downloaded. GSE62165 comprises of data from 118 PDAC patients versus 13 control samples from the healthy pancreas [40]. To access the expression of mRNA in PDAC versus control samples, GEO2R was used and the resulted normalized gene expression was plotted as well as the correlation between ACTA2 and different genes.
Channel contraction in PSC-laden collagen or Col/Fib disks. PSC-laden gels were prepared as aforementioned using a custom-made PDMS mold of a Ø6mm and 2 mm height (Fig. S5, Supporting Information). Each disk was punctured using an Accupunch (Electron Microscopy Sciences) of a size of Ø1mm to achieve a channel in the hydrogel of the same size. The disks were cultured for a duration of 3 days while being imaged on daily basis. The channel size was determined using ImageJ and the contraction was determined by standardizing the observed size for each day to the initial size on day 0.
Cell culture (HUVEC). Human umbilical vein endothelial cells (HUVECs, Lonza) were cultured according to the manufacturer's instructions. In brief, HUVECs were cultured in EGM-2 endothelial cell growth medium (Lonza) supplemented with 2% FBS, 0.04% hydrocortisone, 0.4% hFGF-B, 0.1% VEGF, 0.1% R3-IGF, 0.1% ascorbic acid, 0.1% hEGF, 0.1% GA-1000, 0.1% heparin (all concentrations in v/v%, all products from Lonza). The cells were maintained at 37 °C in a humidified 5% CO2 atmosphere, and passed at 80% confluence. HUVECs were used between passages 1–4 according to the manufacturer's suggestion.
Generation of vascularized collagen + Col/Fib multi-layered constructs. Multi-layered constructs were prepared using a sandwich method. First, a PSC-laden collagen/fibrinogen construct was prepared using a custom-made PDMS mold (Ø8mm, 4 mm height) including a blunt needle (Ø1mm) in the middle serving as sacrificial structure for the central channel. After initial solidification where the fibrinogen component is already crosslinked while the collagen is still in a semi-solidified state, the PSC-laden Col/Fib gels including the needle were rapidly transferred to another custom-made PDMS mold (Ø8mm, 10 mm height) before being surrounded by a PSC-laden collagen solution (Fig. 1C and D). The hybrid construct was allowed to fully solidify at 37 °C in a humidified atmosphere. Next, the needle was carefully removed from the construct to obtain a final channel of ≈ Ø1mm. In the meantime, HUVECs were labelled with CellTracker Orange CMRA (Thermofisher Scientific, 20 μM in FBS-free culture medium for 30min). Afterwards, labelled HUVECs (5 × 106 cells/mL) were seeded into the channel and allowed to attach for 6 h before not-attached HUVECs were washed out and the multi-layered construct was cultured as previously described.
Immunostaining of collagen + Col/Fib multi-layered constructs. Multi-layered constructs were cultured for a total duration of 3 days, before being fixed with 4% formaldehyde (Sigma-Aldrich) for 30 min at room temperature and incubated with an increasing concentration of sucrose (Sigma-Aldrich) from 5%, 10% and 20% in PBS for 1 h each at room temperature and 30% in PBS overnight at 4 °C to replace the water content in the gels. Afterwards, the gels were embedded in cryomatrix and snap-frozen using ice-cold 2-methyl butane. Snap-frozen gels were cryosectioned into 8 μm thick sections protected from light, immediately fixed with 4% formaldehyde for 15 min at room temperature, mounted using Fluoroshield™ with DAPI and imaged using Nanozoomer-RS.
Scanning electron microscopy of collagen + Col/Fib multi-layered constructs. Multi-layered constructs were prepared and cultured as previously described. After 3 days, the constructs were fixed with 2.5 v/v% glutaraldehyde for 1 h at room temperature and at 4 °C overnight. The fixed gels were washed three times with MilliQ water before being frozen in liquid nitrogen. Afterwards the gels were lyophilized, broken at the intersection of collagen and Col/Fib, gold-sputtered and imaged using a scanning electron microscope at an accelerating voltage of 5 kV and a probe current of 35.
Flow simulation in different degrees of stenosis. Channels with different degrees of stenosis were designed in Solidworks (Dassault Systémes, Vélizy-Villacoublay, France) representing the level of channel contraction on day 0, 1, 2 and 3 based on experimental observations (Fig. S8, Supporting Information). The range of the channels’ diameters varied between 0.936 mm and 0.016 mm. The length of all the channels was set to 8 mm. Simulations were conducted using the end-to-end parallel simulation toolchain contained within the adaptable poly engineering simulator (APES) framework [50]. The designs of the channels were translated into the discretized form with a dx = 4 μm, based on the previous studies of stenotic flow [51]. The time step was set to dt = 0.3 μs Volume meshes were generated using Seeder which is a part of the APES framework. The resulting number of cells in the volume meshes ranged from 9 million to 84 million, depending on the fluid volume occupied by each constricted geometry. Conventionally, the left side of the channel was considered the inlet and the right side the outlet. The flow inside each channel was approximated with the physical properties of water. The flow rate at the inlet of each channel was set to 1000 mm³/h. At the outlets, a boundary condition of zero pressure was maintained. The walls of each channel were represented by a higher order boundary condition and a no slip boundary condition was prescribed at the impermeable walls. Simulations were conducted using the Musubi LBM solver available as open-source package for simulations on massively parallel supercomputers [52]. One physical second was simulated for each case using 1920 cores of the Cartesius system (national supercomputer in the Netherlands). Depending on the number of cells, simulations required between 2 and 19 h of execution time. Flow properties were analyzed at the end of the first second when the flow attained a steady state.
Calculation of Reynolds andPécletNumber. Based on the CFD simulation results for the flow velocity within the channel of different compression and the general channel properties the Reynolds number was calculated using equation (1):
| (1) |
where ρ = fluid density of water at 25 °C, v = flow velocity, d = diameter of the contracted channel and μ = dynamic fluid viscosity of water at 25 °C. The Péclet number was calculated using equation (2):
| (2) |
where L = length of contracted channel (betwenn 2 mm and 6 mm of the complete channel length), v = flow velocity and D = self-diffusion coefficient of water at 25 °C.
Integrin alpha 5 (ITGA5) expression in 2D-cultured PSCs, PSC-laden collagen and Col/Fib gels and human PDAC patients. The expression of integrin alpha 5 (ITGA5) in 2D cultured and 3D PSC-laden collagen and Col/Fib gels was determined using real-time PCR as previously described. The expression of ITGA5 was normalized to the expression of the house-keeping gene RPS18. The expression of ITGA5 in PDAC patients was determined in the human cohort GSE62165 as aforementioned.
Treatment of PSC-laden Col/Fib gels with ITGA5-antagonist AV3. For investigating the inhibition of ACTA2 based on the treatment with AV3, PSC-laden Col/Fib cubes of 7 mm × 7 mm x 5 mm were prepared as previously described. Immediately after formation the gels were incubated with ether 50 μM AV3, a peptide previously developed in our group, or DMSO (vehicle) for 24 h before being prepared for RNA isolation. Enzymatic degradation, RNA isolation, cDNA synthesis and real-time PCR were performed as previously described for the expression of ACTA2. The expression of ACTA2 was normalized to the expression of the house-keeping gene RPS18.
For investigating the effects of AV3 on the channel contraction in Col/Fib gels, PSC-laden Col/Fib disks (Ø6mm, 2 mm height) were prepared and punctured to achieve a ≈ Ø 1 mm channel in the disk. Immediately after preparation, the disks were incubated with either 20 μM, 50 μM or DMSO (vehicle) and the contraction of the channel was followed for a total duration of 3 days while being imaged on daily basis as previously described.
CFD simulation for AV3-treated and vehicle-treated channels. Computational flow dynamics simulation was performed as previously described. In brief, channels were designed using Solidworks based on the experimental data obtained from the in vitro experiments. Simulations were conducted using the end-to-end parallel simulation toolchain contained within the adaptable poly engineering simulator (APES) framework. Time step and flow rate at the inlet were set to dt = 0.3 μs and 1000 mm3/h as aforementioned. Simulations were conducted using the Musubi LBM solver available as open-source package for simulations on massively parallel supercomputers [52]. One physical second was simulated for each case using 1920 cores of the Cartesius system (national supercomputer in the Netherlands). Flow properties were analyzed at the end of the first second when the flow attained a steady state.
Treatment of animals (Panc-1/PSC & MiaPaca-2/PSC co-injection model) with AV3. As described above, six week old male CB17 SCID mice were subcutaneously co-injected with Panc-1 cancer cells (2 × 106 cells) and PSCs (4 × 106 cells) or MiaPaCa-2 cancer cells (2 × 106 cells) and PSCs (4 × 106 cells). Four tumor-bearing mice per treatment were taken and injected intraperitoneally with either vehicle or AV3 (20 mg/kg). Initially mice were injected three times a week. Later both groups treatment was reduced twice a week.
For immunohistochemical analysis of the vasculature, tumors were collected, sectioned and stained as described above.
To investigate the accumulation of indocyanine green dye (ICG, LICOR, Lincoln, NE, USA) in tumors, mice were injected with 5 mg/kg ICG intravenously via the tail vein. 24 h post-injection, tumors were isolated and imaged using a small animal imager (Pearl Imager, LICOR).
Graphs & Statistical Analysis. All graphs were made using GraphPad Prism Vol.9 (GraphPad Software Inc., San Diego, CA) based on calculations using Microsoft Excel. Schematics were made using Inkscape (Open-source vector graphics editor). Flow diagrams were made using ParaView (Open-source visualization toolkit by Sandia National Laboratories, Kitware Inc., Los Alamos National Laboratory). All values are expressed as mean ± standard error of the mean (SEM). Statistical significance of the results was performed by either two-tailed unpaired student's t-test for comparison between two treatment groups or two-way ANOVA for comparing multiple treatment groups. Significant difference was determined for a p-value of *p < 0.05, **p < 0.01 and ***p < 0.001, respectively.
CRediT authorship contribution statement
Marcel Alexander Heinrich: contributed equally to this work as primary author, conceived the study, designed experiments, and co-wrote the manuscript, and, performed the in vitro experiments. Irene Uboldi: contributed equally to this work as primary author, performed the in vitro experiments. Praneeth Reddy Kuninty: performed the animal studies and the immunohistochemical staining of tissue sections. Marc J.K. Ankone: performed the mechanical analysis of the hydrogels. Joop van Baarlen: provided the sections from human PDAC patients. Yu Shrike Zhang: helped in discussions on the project and reviewed the manuscript. Kartik Jain: performed the CFD simulations and supervised the fluid mechanical aspects. Jai Prakash: conceived the study, designed experiments, and co-wrote the manuscript. All authors reviewed the manuscript. Correspondence and requests for materials should be addressed to Jai Prakash. The authors acknowledge funding from the Dutch Technology Foundation and the Dutch Cancer Society (STW/ KWF; project no. 15204 to J.P.). Compute resources on the Dutch national supercomputer Cartesius were provided by SURFSara through NWO grant 2019/ENW/00768083.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Jai Prakash reports financial support was provided by Dutch Research Council. Kartik Jain reports financial support was provided by Dutch Research Council. Jai Prakash reports a relationship with ScarTec Therapeutics that includes: equity or stocks. Jai Prakash has patent #AU2016340763B2 issued to University of Twente/ScarTec Therapeutics. Jai Prakash has patent #EP3365353A1 pending to University of Twente/ScarTec Therapeutics. Other authors declare no conflict of interest.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2022.09.015.
Appendix ASupplementary data
The following is the Supplementary data to this article:
References
- 1.Ho W.J., Jaffee E.M., Zheng L. The tumour microenvironment in pancreatic cancer - clinical challenges and opportunities. Nat. Rev. Clin. Oncol. 2020;17:527–540. doi: 10.1038/s41571-020-0363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murakami T., Hiroshima Y., Matsuyama R., Homma Y., Hoffman R.M., Endo I. Role of the tumor microenvironment in pancreatic cancer. Ann. Gastroenterol. Surg. 2019;3:130–137. doi: 10.1002/ags3.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinrich M.A., Mostafa A.M.R.H., Morton J.P., Hawinkels L.J.A.C., Prakash J. Translating complexity and heterogeneity of pancreatic tumor: 3D in vitro to in vivo models. Adv. Drug Deliv. Rev. 2021;174:265–293. doi: 10.1016/j.addr.2021.04.018. [DOI] [PubMed] [Google Scholar]
- 4.Roife D., Sarcar B., Fleming J.B. Stellate cells in the tumor microenvironment. Adv. Exp. Med. Biol. 2020;1263:67–84. doi: 10.1007/978-3-030-44518-8_6. [DOI] [PubMed] [Google Scholar]
- 5.Vonlaufen A., Joshi S., Qu C., Phillips P.A., Xu Z., Parker N.R., Toi C.S., Pirola R.C., Wilson J.S., Goldstein D., Apte M.V. Pancreatic stellate cells: partners in crime with pancreatic cancer cells. Cancer Res. 2008;68:2085–2093. doi: 10.1158/0008-5472.CAN-07-2477. [DOI] [PubMed] [Google Scholar]
- 6.Sperb N., Tsesmelis M., Wirth T. Crosstalk between tumor and stromal cells in pancreatic ductal adenocarcinoma. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21155486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biffi G., Oni T.E., Spielman B., Hao Y., Elyada E., Park Y., Preall J., Tuveson D.A. Cancer Discov; 2018. IL1-Induced JAK/STAT Signaling Is Antagonized by TGFbeta to Shape CAF Heterogeneity in Pancreatic Ductal Adenocarcinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohlund D., Handly-Santana A., Biffi G., Elyada E., Almeida A.S., Ponz-Sarvise M., Corbo V., Oni T.E., Hearn S.A., Lee E.J., Chio, Hwang C.I., Tiriac H., Baker L.A., Engle D.D., Feig C., Kultti A., Egeblad M., Fearon D.T., Crawford J.M., Clevers H., Park Y., Tuveson D.A. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017;214:579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chauhan V.P., Boucher Y., Ferrone C.R., Roberge S., Martin J.D., Stylianopoulos T., Bardeesy N., DePinho R.A., Padera T.P., Munn L.L., Jain R.K. Compression of pancreatic tumor blood vessels by hyaluronan is caused by solid stress and not interstitial fluid pressure. Cancer Cell. 2014;26:14–15. doi: 10.1016/j.ccr.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katsuta E., Qi Q., Peng X., Hochwald S.N., Yan L., Takabe K. Pancreatic adenocarcinomas with mature blood vessels have better overall survival. Sci. Rep. 2019;9:1310. doi: 10.1038/s41598-018-37909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramanathan R.K., McDonough S.L., Philip P.A., Hingorani S.R., Lacy J., Kortmansky J.S., Thumar J., Chiorean E.G., Shields A.F., Behl D., Mehan P.T., Gaur R., Seery T., Guthrie K.A., Hochster H.S. Phase IB/II randomized study of FOLFIRINOX plus pegylated recombinant human hyaluronidase versus FOLFIRINOX alone in patients with metastatic pancreatic adenocarcinoma: SWOG S1313. J. Clin. Oncol. 2019;37:1062–1069. doi: 10.1200/JCO.18.01295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hingorani S.R., Zheng L., Bullock A.J., Seery T.E., Harris W.P., Sigal D.S., Braiteh F., Ritch P.S., Zalupski M.M., Bahary N., Oberstein P.E., Wang-Gillam A., Wu W., Chondros D., Jiang P., Khelifa S., Pu J., Aldrich C., Hendifar A.E. Halo 202: randomized phase II study of PEGPH20 plus nab-paclitaxel/gemcitabine versus nab-paclitaxel/gemcitabine in patients with untreated, metastatic pancreatic ductal adenocarcinoma. J. Clin. Oncol. 2018;36:359–366. doi: 10.1200/JCO.2017.74.9564. [DOI] [PubMed] [Google Scholar]
- 13.Schnittert J., Heinrich M.A., Kuninty P.R., Storm G., Prakash J. Reprogramming tumor stroma using an endogenous lipid lipoxin A4 to treat pancreatic cancer. Cancer Lett. 2018;420:247–258. doi: 10.1016/j.canlet.2018.01.072. [DOI] [PubMed] [Google Scholar]
- 14.McKee T.D., Grandi P., Mok W., Alexandrakis G., Insin N., Zimmer J.P., Bawendi M.G., Boucher Y., Breakefield X.O., Jain R.K. Degradation of fibrillar collagen in a human melanoma xenograft improves the efficacy of an oncolytic herpes simplex virus vector. Cancer Res. 2006;66:2509–2513. doi: 10.1158/0008-5472.CAN-05-2242. [DOI] [PubMed] [Google Scholar]
- 15.Stern R. Hyaluronidases in cancer biology. Semin. Cancer Biol. 2008;18:275–280. doi: 10.1016/j.semcancer.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 16.Eikenes L., Tari M., Tufto I., Bruland O.S., de Lange Davies C. Hyaluronidase induces a transcapillary pressure gradient and improves the distribution and uptake of liposomal doxorubicin (Caelyx) in human osteosarcoma xenografts. Br. J. Cancer. 2005;93:81–88. doi: 10.1038/sj.bjc.6602626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuriyama N., Kuriyama H., Julin C.M., Lamborn K., Israel M.A. Pretreatment with protease is a useful experimental strategy for enhancing adenovirus-mediated cancer gene therapy. Hum. Gene Ther. 2000;11:2219–2230. doi: 10.1089/104303400750035744. [DOI] [PubMed] [Google Scholar]
- 18.Pereira B.A., Vennin C., Papanicolaou M., Chambers C.R., Herrmann D., Morton J.P., Cox T.R., Timpson P. CAF subpopulations: a new reservoir of stromal targets in pancreatic cancer. Trends Cancer. 2019;5:724–741. doi: 10.1016/j.trecan.2019.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Norton J., Foster D., Chinta M., Titan A., Longaker M. Pancreatic cancer associated fibroblasts (CAF): under-explored target for pancreatic cancer treatment. Cancers. 2020;12:1347. doi: 10.3390/cancers12051347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuninty P.R., Bansal R., De Geus S.W.L., Mardhian D.F., Schnittert J., van Baarlen J., Storm G., Bijlsma M.F., van Laarhoven H.W., Metselaar J.M., Kuppen P.J.K., Vahrmeijer A.L., Östman A., Sier C.F.M., Prakash J. ITGA5 inhibition in pancreatic stellate cells attenuates desmoplasia and potentiates efficacy of chemotherapy in pancreatic cancer. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aax2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z., Vunjak-Novakovic G. Modeling tumor microenvironments using custom-designed biomaterial scaffolds. Curr. Opin. Chem. Eng. 2016;11:94–105. doi: 10.1016/j.coche.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Brien F.J. Biomaterials & scaffolds for tissue engineering, Mater. Today Off. 2011;14:88–95. doi: 10.1016/S1369-7021(11)70058-X. [DOI] [Google Scholar]
- 23.Brancato V., Olveira J.M., Correlo V.M., Reis R.L., Kundu S.C. Could 3D models of cancer enhance drug screening? Biomaterials. 2020;232 doi: 10.1016/j.biomaterials.2019.119744. [DOI] [PubMed] [Google Scholar]
- 24.Li J., Parra-Cantu C., Wang Z., Zhang Y.S. Improving bioprinted volumetric tumor microenvironments in vitro. Trends Cancer. 2020;6:745–756. doi: 10.1016/j.trecan.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albritton J.L., Miller J.S. 3D bioprinting: improving in vitro models of metastasis with heterogeneous tumor microenvironments. Dis. Model. Mech. 2017;10:3–14. doi: 10.1242/dmm.025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J., Weihs D., Vermolen F.J. Model Mechanobiol; 2019. Computational Modeling of Therapy on Pancreatic Cancer in its Early Stages, Biomech. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang X., Yang N., Fiore V.F., Barker T.H., Sun Y., Morris S.W., Ding Q., Thannickal V.J., Zhou Y. Matrix stiffness–induced myofibroblast differentiation is mediated by intrinsic mechanotransduction. Am. J. Respir. Cell Mol. Biol. 2012;47:340–348. doi: 10.1165/rcmb.2012-0050OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niu L., Jia Y., Wu M., Liu H., Feng Y., Hu Y., Zhang X., Gao D., Xu F., Huang G. Matrix stiffness controls cardiac fibroblast activation through regulating YAP via AT1R. J. Cell. Physiol. 2020;235:8345–8357. doi: 10.1002/jcp.29678. [DOI] [PubMed] [Google Scholar]
- 29.Katsuta E., Qi Q., Peng X., Hochwald S.N., Yan L., Takabe K. Pancreatic adenocarcinomas with mature blood vessels have better overall survival. Sci. Rep. 2019;9:1310. doi: 10.1038/s41598-018-37909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L., Shi G.-P. CD31: beyond a marker for endothelial cells. Cardiovasc. Res. 2012;94:3–5. doi: 10.1093/cvr/cvs108. [DOI] [PubMed] [Google Scholar]
- 31.Masamune A., Kikuta K., Watanabe T., Satoh K., Hirota M., Hamada S., Shimosegawa T. Fibrinogen induces cytokine and collagen production in pancreatic stellate cells. Gut. 2009;58:550–559. doi: 10.1136/gut.2008.154401. [DOI] [PubMed] [Google Scholar]
- 32.Mardhian D.F., Vrynas A., Storm G., Bansal R., Prakash J. FGF2 engineered SPIONs attenuate tumor stroma and potentiate the effect of chemotherapy in 3D heterospheroidal model of pancreatic tumor. Nanotheranostics. 2020;4:26–39. doi: 10.7150/ntno.38092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice A.J., Cortes E., Lachowski D., Cheung B.C.H., Karim S.A., Morton J.P., del Río Hernández A. Matrix stiffness induces epithelial–mesenchymal transition and promotes chemoresistance in pancreatic cancer cells. Oncogenesis. 2017;6:e352. doi: 10.1038/oncsis.2017.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vadival G.N., Sukmana I. Effect of mechanical and biophysical properties of fibrin gel on microvascual development in a three-dimensonal angiogenesis assay. Int. J. Automot. Eng. 2014;9:1662–1668. doi: 10.15282/ijame.9.2013.16.0138. [DOI] [Google Scholar]
- 35.Öhlund D., Handly-Santana A., Biffi G., Elyada E., Almeida A.S., Ponz-Sarvise M., Corbo V., Oni T., Hearn S.A., Lee E.J., Chio I.I.C., Hwang C.I., Tiriac H., Baker L.A., Engle D.D., Feig C., Kultti A., Egeblad M., Fearon D.T., Crawford J.M., Clevers H., Park Y., Tuveson D.A. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017;214:579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Östman A. PDGF receptors in tumor stroma: biological effects and associations with prognosis and response to treatment. Adv. Drug Deliv. Rev. 2017;121:117–123. doi: 10.1016/j.addr.2017.09.022. [DOI] [PubMed] [Google Scholar]
- 37.Weniger M., Honselmann K.C., Liss A.S. The extracellular matrix and pancreatic cancer: a complex relationship. Cancers. 2018;10:316. doi: 10.3390/cancers10090316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monypenny J., Zicha D., Higashida C., Oceguera-Yanez F., Narumiya S., Watanabe N. Cdc42 and Rac family GTPases regulate mode and speed but not direction of primary fibroblast migration during platelet-derived growth factor-dependent chemotaxis. Mol. Cell Biol. 2009;29:2730–2747. doi: 10.1128/MCB.01285-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sipes N.S., Feng Y., Guo F., Lee H.-O., Chou F.-S., Cheng J., Mulloy J., Zheng Y. Cdc42 regulates extracellular matrix remodeling in three dimensions. J. Biol. Chem. 2011;286:36469–36477. doi: 10.1074/jbc.M111.283176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janky R., Binda M.M., Allemeersch J., Van den Broeck A., Govaere O., Swinnen J., Roskams T., Aerts S., Topal B. Prognostic relevance of molecular subtypes and master regulators in pancreatic ductal adenocarcinoma. BMC Cancer. 2016;16 doi: 10.1186/s12885-016-2540-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valentin J.E., Stewart-Akers A.M., Gilbert T.W., Badylak S.F. Macrophage participation in the degradation and remodeling of extracellular matrix scaffolds. Tissue Eng. 2019;15:1687–1694. doi: 10.1089/ten.tea.2008.0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li K.Y., Yuan J.L., Trafton D., Wang J.X., Niu N., Yuan C.H., Liu X.B., Zheng L. Pancreatic ductal adenocarcinoma immune microenvironment and immunotherapy prospects. Chronic Dis. Transl. Med. 2020;6:6–17. doi: 10.1016/j.cdtm.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suehiro K., Mizuguchi J., Nishiyama K., Iwanaga S., Farrell D.H., Ohtaki S. Fibrinogen binds to integrin alpha(5)beta(1) via the carboxyl-terminal RGD site of the Aalpha-chain. J. Biochem. 2000;128:705–710. doi: 10.1093/oxfordjournals.jbchem.a022804. [DOI] [PubMed] [Google Scholar]
- 44.Yokoyama K., Zhang X.P., Medved L., Takada Y. Specific binding of integrin alpha v beta 3 to the fibrinogen gamma and alpha E chain C-terminal domains. Biochemistry. 1999;38:5872–5877. doi: 10.1021/bi9827619. [DOI] [PubMed] [Google Scholar]
- 45.Suehiro K., Gailit J., Plow E.F. Fibrinogen is a ligand for integrin α5β1 on endothelial cells. J. Biol. Chem. 1997;272:5360–5366. doi: 10.1074/jbc.272.8.5360. [DOI] [PubMed] [Google Scholar]
- 46.Dejana E., Lampugnani M.G., Giorgi M., Gaboli M., Marchisio P.C. Fibrinogen induces endothelial cell adhesion and spreading via the release of endogenous matrix proteins and the recruitment of more than one integrin receptor. Blood. 1990;75:1509–1517. doi: 10.1182/blood.V75.7.1509.1509. [DOI] [PubMed] [Google Scholar]
- 47.Kong B., Sun L., Liu R., Chen Y., Shang Y., Tana H., Zhao Y., Sun L. Recombinant human collagen hydrogels with hierarchically ordered microstructures for corneal stroma regeneration. Chem. Eng. J. 2022;428 doi: 10.1016/j.cej.2021.131012. [DOI] [Google Scholar]
- 48.Zhang H., Liu Y., Chen G., Wang H., Chen C., Li M., Lu P., Zhao Y. Immunotherapeutic silk inverse opal particles for post-surgical tumor treatment. Sci. Bull. 2020;65:380–388. doi: 10.1016/j.scib.2019.10.023. [DOI] [PubMed] [Google Scholar]
- 49.de la Peña D.O., Trabulo S.M.D., Collin E., Liu Y., Sharma S., Tatari M., Behrens D., Erkan M., Lawlor R.T., Scarpa A., Heeschen C., Mata A., Loessner D. Bioengineered 3D models of human pancreatic cancer recapitulate in vivo tumour biology. Nat. Commun. 2021;12:5623. doi: 10.1038/s41467-021-25921-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50.Klimach H., Jain K., Roller S. In: Parallel Computing: Accelerating Computational Science and Engineering (CSE), IOS Press. Bader M., Bode A., Bungartz H.J., Gerndt M., Joubert G.R., Peters F., editors. 2014. End-to-end parallel simulations with APES. [Google Scholar]
- [51.Jain K. Transition to turbulence in an oscillatory flow through stenosis. Biomech. Model. Mechanobiol. 2020;19:113–131. doi: 10.1007/s10237-019-01199-1. [DOI] [PubMed] [Google Scholar]
- [52.Hasert M., Masilamani K., Zimny S., Klimach H., Qi J., Bernsdorf J., Roller S. Complex fluid simulations with the parallel tree-based lattice Boltzmann solver Musubi. J. Comput. Sci. 2014;5:784–794. doi: 10.1016/j.jocs.2013.11.001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.