Abstract
Saccharomyces cerevisiae is a petite-phenotype-positive (“petite-positive”) yeast, which can successfully grow in the absence of oxygen. On the other hand, Kluyveromyces lactis as well as many other yeasts are petite negative and cannot grow anaerobically. In this paper, we show that Saccharomyces kluyveri can grow under anaerobic conditions, but while it can generate respiration-deficient mutants, it cannot generate true petite mutants. From a phylogenetic point of view, S. kluyveri is apparently more closely related to S. cerevisiae than to K. lactis. These observations suggest that the progenitor of the modern Saccharomyces and Kluyveromyces yeasts, as well as other related genera, was a petite-negative and aerobic yeast. Upon separation of the K. lactis and S. kluyveri-S. cerevisiae lineages, the latter developed the ability to grow anaerobically. However, while the S. kluyveri lineage has remained petite negative, the lineage leading to the modern Saccharomyces sensu stricto and sensu lato yeasts has developed the petite-positive characteristic.
Cells of Saccharomyces cerevisiae constantly produce mutants that are stable during vegetative reproduction and are characterized by a reduced colony size, hence their name petite, on solid media in which a fermentable carbon source is the limiting factor (12). Petite mutants, which are a special class of respiration-deficient mutants, have been shown to have large deletions in their mitochondrial DNA (mtDNA) or to lack the mitochondrial genome entirely (for reviews, see references 9, 11, and 25). Yeasts can be divided into two groups depending on their ability to produce, spontaneously or when induced by interchelating dyes, petite mutants. One group, petite-phenotype-positive (“petite-positive”) yeasts, including several Saccharomyces yeasts, readily gives rise to petite mutants (26). The other group, petite-negative yeasts, which includes a majority of yeasts, like Schizosaccharomyces pombe and Kluyveromyces lactis, fails to yield these mutants (6, 9, 16). While large deletions and rearrangements have not been detected in mtDNA of petite-negative yeasts, curiously, for some of these yeasts, both nuclear lesions for respiratory function and point mutations or short deletions in mtDNA have been described (1, 15). mtDNA molecules, which are respiration deficient because of point mutations or short deletions, are called mit negative. Regarding the evolution of the petite-positive phenotype, it apparently originated independently at least twice during the evolutionary history of yeasts. It originated once in the lineage leading to the modern Saccharomyces species (26) and once in the lineage leading to the modern Brettanomyces/Dekkera species (9, 10). So far, the biochemical and physiological requirements for development of the petite-positive characteristic have been unclear. However, it is interesting to point out that both petite-positive yeast groups, Saccharomyces and Brettanomyces/Dekkera, can grow anaerobically (3, 27), while many other yeasts, which are petite negative, cannot grow in the absence of oxygen. For example, K. lactis, a close relative of Saccharomyces yeasts (19), is petite negative, and it cannot grow in the absence of oxygen (31). On the basis of these observations, it has been suggested that in yeast the petite-positive characteristic might coincide with the ability to grow anaerobically (2, 7).
In this paper, we analyze the ability of S. kluyveri to grow anaerobically and to generate respiration-deficient mutants. We show that S. kluyveri can grow at anaerobic conditions, but while it can generate respiration-deficient mutants, it cannot generate true petite mutants. Thus, upon separation of the K. lactis and Saccharomyces lineages, the latter developed the ability to grow anaerobically. However, while the S. kluyveri lineage remained petite negative, the other lineage, leading to Saccharomyces sensu stricto and sensu lato yeasts, developed the petite-positive characteristic.
MATERIALS AND METHODS
Yeast strains.
The following laboratory strains were used in the anaerobic batch cultivation experiments: S. kluyveri Y057 (type strain, NRRL Y-12651, originating from the National Center for Agricultural Utilization Research, Peoria, Ill.), S. kluyveri Y708 (MATa, prototrophic derivative of Y057), and K. lactis Y707 (CBS 2359, originating from the Centraal Bureau voor Schimmelcultures, Delft, The Netherlands). In the petite-mutation induction experiments, two parental haploid strains of S. kluyveri, Y090 (MATα thr) and Y091 (MATa his aux), which were provided by L. Marsh (Albert Einstein College of Medicine, Bronx, N.Y.), were used (aux is an unidentified auxotrophic marker). Two respiration-deficient mutants, Y176 and Y178, were derived from Y091, and one respiration-deficient mutant, Y182, originated from Y090. Y designations were used for strains from the laboratory collection.
Anaerobic batch cultivations.
S. kluyveri was cultivated under anaerobic conditions in glucose minimal medium prepared as previously described (29). This medium was supplemented with ergosterol and unsaturated fatty acids in the form of Tween 80 (28), which is needed for the anaerobic growth of S. cerevisiae (3, 4). The final concentrations of the medium components were as follows: 20.0 g of glucose/liter, 5.0 g of ammonium sulfate/liter, 3.0 g of potassium dihydrogen phosphate/liter, 0.5 g of magnesium sulfate heptahydrate/liter, 15 mg of EDTA/liter, 4.5 mg of zinc sulfate heptahydrate/liter, 0.84 mg of manganese chloride dihydrate/liter, 0.30 mg of cobalt(II) chloride hexahydrate/liter, 0.30 mg of copper(II) sulfate pentahydrate/liter, 0.40 mg of disodium molybdenum dihydrate/liter, 4.5 mg of calcium chloride dihydrate/liter, 3.0 mg of iron sulfate heptahydrate/liter, 1.0 mg of boric acid/liter, 0.1 mg of potassium iodide/liter, 0.05 mg of d-(−)-biotin/liter, 1.0 mg of calcium d-(+)-panthotenate/liter, 1.0 mg of nicotinic acid/liter, 25.0 mg of myo-inositol/liter, 1.0 mg of thiamine chloride hydrochloride/liter, 1.0 mg of pyridoxol hydrochloride/liter, 0.2 mg of p-aminobenzoic acid/liter, 10 mg of ergosterol/liter, 420 mg of Tween 80/liter, and 50 μl of antifoam 289 (Sigma A-5551)/liter. Precultures were grown for 20 to 25 h, at 30°C and 75 rpm, in 500-ml cotton-stoppered shake flasks with 100 ml of medium. The medium used for precultures was the same as described for anaerobic batch cultivations, except that the concentration of ammonium sulfate was 7.5 g/liter, the concentration of potassium dihydrogen phosphate was 14.4 g/liter, ergosterol and Tween 80 were omitted, and the initial pH was set to 6.5. Anaerobic batch cultivations of S. kluyveri Y708 were performed in a bioreactor with a working volume of 4 liters at 30°C and a stirring rate of 500 rpm. pH was kept constant at 5.0 by addition of 2 M potassium hydroxide. The bioreactor was continuously flushed with N2 (containing less than 3 ppm O2) at a flow rate of 0.5 liter of N2/min (equivalent to 0.125 liter of N2 per liter of medium per min). The off-gas was led through a cooled condenser to a gas analyzer. In order to minimize the diffusion of oxygen into the bioreactor, Norprene tubing (Cole-Palmer) was used throughout the setup. The bioreactor was inoculated with an amount of preculture resulting in an initial biomass concentration of 1 mg (dry weight)/liter in the bioreactor. The assumption that anaerobic conditions prevailed was tested by performing the same experiment with the strictly aerobic yeast K. lactis CBS 2359. Anaerobic growth of S. kluyveri Y057 was tested in a 2-liter jacketed bioreactor (Applikon, Scheidam, The Netherlands) with a working volume of 1 liter. In these cultivations, there was no pH control and no samples were taken to make sure that no oxygen was introduced. The bioreactor was flushed with nitrogen at 1 liter/min (equivalent to 1 liter of nitrogen per liter of medium per min). The bioreactor was flushed with nitrogen for 24 h prior to inoculation.
Analysis of growth and product formation.
Growth was monitored by measuring optical density at 600 nm with a Hitachi U-1100 spectrophotometer. The final biomass concentration was determined by measuring the culture dry weight as previously described (18). Glucose consumption and production of extracellular metabolites were monitored during the anaerobic batch cultivation by sampling for analysis of glucose, ethanol, glycerol, acetate, succinate, and pyruvate concentrations in the fermentation broth. Immediately after sampling, the fermentation broth was filtered through a 0.45-μm-pore-size cellulose acetate filter, and the filtrate was frozen at −20°C until analysis. The concentrations of the above-mentioned compounds were all determined by high-performance liquid chromatography analysis on an Aminex HPX-87H column (Bio-Rad), and the final ethanol and glycerol concentrations were verified by enzymatic assays (Roche). The concentration of CO2 in the off-gas was measured on-line with a 1308 acoustic gas analyzer (Bruël & Kjær, Nærum, Denmark).
Induction of respiration-deficient strains.
The parental haploid S. kluyveri strains Y090 and Y091 were grown in glucose-containing medium (YPD) that contained 20 g of glucose/liter, 10 g of yeast extract/liter, and 10 g of Bacto Peptone/liter at 28°C. In several independent experiments, overnight cultures of Y090 and Y091 were diluted 100 times with fresh YPD medium, and ethidium bromide (EtBr) was added to final concentrations of 0.1 to 50 μg/ml. The cultures were incubated for a couple of days until they were completely saturated. Then, the cells were pelleted, washed, and resuspended in sterile water and approximately 100 to 300 EtBr-mutagenized cells were spread on each petite-mutation detection plate (GGlyYP), which contained 20 g of glycerol/liter, 1 g of glucose/liter, 1 g of yeast extract/liter, and 10 g of Bacto Peptone/liter. The inoculated plates were incubated for a week and afterwards were examined for the presence of small colonies, putative respiration-deficient mutants. The obtained small colonies were transferred to YPD plates and afterwards were replica plated onto glycerol medium (GlyYP) containing 20 g of glycerol/liter, 1 g of yeast extract/liter, and 10 g of Bacto Peptone/liter. Growth on GlyYP medium requires respiration, since glycerol cannot be fermented.
Characterization of respiration-deficient strains.
A few colonies that did not grow on the GlyYP medium were then characterized by their mtDNA and behavior in genetic crosses. mtDNA from the wild-type strain and different mutants was prepared using zymolyase treatment followed by centrifugation in CsCl in the presence of bisbenzimide (24) and was analyzed with different restriction enzymes. Respiration-deficient strains were mated with the respiration-competent parental strains and among themselves by using the random mass-mating approach, and diploids were selected on minimal medium, which contained 20 g of glucose/liter and 6.7 g of Difco nitrogen base/liter. Several randomly chosen diploid colonies were analyzed for their growth on GlyYP medium. In addition, total cellular DNA was isolated from these diploid colonies and the mtDNA restriction pattern was analyzed as previously described (23).
RESULTS AND DISCUSION
Anaerobic growth of S. kluyveri.
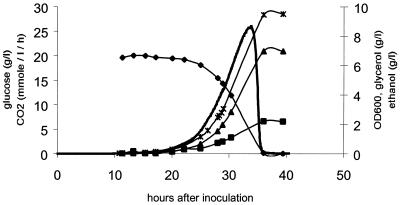
S. kluyveri and K. lactis were tested for anaerobic growth in batch cultures on glucose minimal medium supplemented with Tween 80 and ergosterol. During the cultivation of K. lactis, three or four generations of slow growth were observed during the first 24 h, and after that there was no further growth or sugar consumption. The initial growth was probably due to the aerobic inoculum that was used. The absence of sustained growth of K. lactis was taken as proof of anaerobic conditions in our experimental design, since this yeast can grow under severe oxygen limitation but not under anaerobic conditions (17). On the other hand, S. kluyveri Y708 was found to be capable of rapid anaerobic growth (μmax = 0.24 h−1) on glucose minimal medium supplemented with ergosterol and unsaturated fatty acids (Fig. 1; Table 1). S. cerevisiae has so far been the only yeast species for which good anaerobic growth has been described, and it seems that this property is very rare among yeasts (30). During anaerobic batch cultivation with S. kluyveri Y708, most glucose was converted to ethanol and carbon dioxide with a concomitant production of glycerol to reoxidize surplus NADH. Less than 10% (wt/wt) of the glucose was converted to biomass, and small amounts of various organic acids were also produced (Table 1). The ethanol yield given in Table 1 was probably underestimated due to evaporation of ethanol during the cultivation, which was also evident from the fact that only 95% of the carbon consumed during the cultivation could be accounted for in the measured products. The yields on glucose were almost identical to what has been found for anaerobic batch cultivation of a haploid S. cerevisiae strain (Table 1). To verify that S. kluyveri could grow anaerobically, another strain of S. kluyveri (Y057) was tested for anaerobic growth. In this experiment there was, furthermore, no pH control or sampling during the cultivation, and the N2 flow rate was higher in order to make the conditions more strictly anaerobic. Rapid anaerobic growth was also observed in this experiment, and the maximum specific growth rate was estimated from the CO2 signal to be 0.25 h−1. Thus, S. kluyveri, like S. cerevisiae, is capable of growth under anaerobic conditions where K. lactis cannot grow.
FIG. 1.
Shown are glucose (⧫), ethanol (▴), and glycerol (■) concentrations, the optical density at 600 nm (OD600) (∗), and carbon dioxide evolution (solid line) during anaerobic batch cultivation of S. kluyveri Y708.
TABLE 1.
Growth parameters for anaerobic batch cultivation of S. kluyveri Y708 and S. cerevisiae TN1
| Growth parametera | S. kluyveri | S. cerevisiaeb |
|---|---|---|
| Growth rate (μmax [h−1]) | 0.24 | 0.41 |
| Yield | ||
| Biomass (YSX [g/g]) | 0.089 | 0.092 |
| Ethanol (YSE [g/g]) | 0.350 | 0.376 |
| Glycerol (YSGly [g/g]) | 0.109 | 0.107 |
| Carbon dioxide (YSC [g/g]) | 0.389 | 0.397 |
| Acetate (YSA [g/g]) | 0.003 | 0.004 |
| Succinate (YSSuc [g/g]) | 0.004 | 0.003 |
| Pyruvate (YSPyr [g/g]) | 0.004 | 0.004 |
Cultures were grown on glucose at 30°C and pH 5.0. The maximum specific growth rate (μmax) was calculated from the first 13 OD600 measurements shown in Fig. 1. The yield coefficients (Y) are given as grams of biomass, ethanol, glycerol, carbon dioxide, acetate, succinate, and pyruvate, respectively, formed per gram of glucose consumed.
Data were taken from another source (22).
Respiration-deficient mutants.
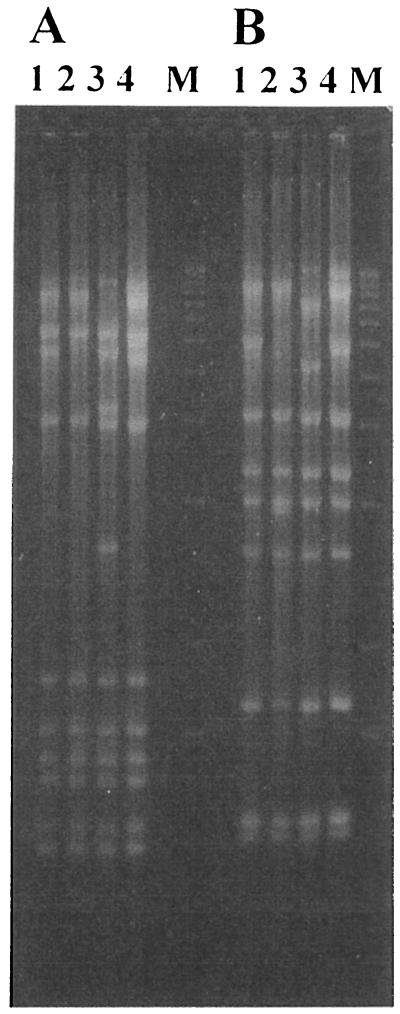
Approximately 50,000 S. kluyveri colonies originating from nonmutated and EtBr-mutated cells of Y090 and Y091 were plated on petite-mutation detection plates, and after several days of growth, the plates were examined for the presence of small colonies. While small colonies were not detected among nonmutated cells, the EtBr-treated cultures yielded approximately 100 small colonies. Note that in the case of S. cerevisiae under similar conditions, all cells would have turned into petite mutants (11, 26). When the small S. kluyveri colonies were transferred to YPD medium and then checked again for growth on GlyYP and GGlyYP media, a great majority were shown to be respiration competent. However, 10 putative mutants could not grow with glycerol as the sole carbon source. The respiratory defects of these mutants could be due to mitochondrial or nuclear mutations. The obtained respiration-deficient strains were then examined for the structure of their mitochondrial genome and behavior in genetic crosses. All examined strains contained mtDNA, but in the case of Y176 and Y178, the mtDNA restriction pattern differed slightly from the wild-type one (Fig. 2). While a majority of restriction fragments could still be observed, apparently a limited deletion or rearrangement also took place in these two strains (Fig. 2). It was likely that Y176 and Y178 were mit-negative-like mutants. To confirm that the respiration deficiency had an extrachromosomal origin, genetic crosses were performed. Respiration-deficient S. kluyveri strains were crossed to the wild-type parental strains, and the abilities of progeny to grow on GlyYP medium and the mtDNA restriction patterns of the progeny were analyzed. When Y176 and Y178 were crossed with Y090, a fraction of the daughter cells produced were respiration deficient (Table 2), demonstrating the extrachromosomal characteristic of the respiratory defect. Apparently, the wild-type mtDNA was transmitted to the progeny preferentially over the mutant mtDNA molecule. A similar transmission pattern has been reported previously for petite mutants, as well as for respiration-competent mitochondrial deletion mutants of S. cerevisiae (reviewed in reference 25). On the other hand, in the rest of the respiration-deficient mutants, including Y182, mtDNAs exhibited the wild-type restriction pattern. However, when Y182 was crossed with Y178, which was also respiration deficient, the progeny consisted of both respiration-competent and -deficient cells (Table 2). It could be that the two mitochondrial genomes recombined and generated a respiration-competent mtDNA molecule. Thus, it is likely that the respiration-deficient phenotype observed for Y182 was due to a point mutation in the mtDNA molecule and not to a chromosomal defect.
FIG. 2.
mtDNA isolated from different S. kluyveri strains: lanes 1, Y091; lanes 2, Y176; lanes 3, Y178; lanes 4, Y182. CsCl-purified mtDNA molecules were digested with HaeIII (A) or MspI (B). Note that the digestion patterns of the respiration-deficient strains differ only slightly from that of the wild-type strain. Lane M, 1-kb DNA ladder (Gibco BRL/Life Technologies).
TABLE 2.
Genetic crosses with S. kluyveri respiration-deficient mutantsa
| Cross | No. of daughter colonies
|
||
|---|---|---|---|
| Total | Gly+ | Gly− | |
| Y090 × Y091 | 50 | 50 | 0 |
| Y090 × Y176 | 39 | 30 | 9 |
| Y090 × Y178 | 39 | 37 | 2 |
| Y091 × Y182 | 40 | 40 | 0 |
| Y176 × Y182 | 24 | 24 | 0 |
| Y178 × Y182 | 42 | 34 | 8 |
Note that Y090 and Y091 are respiration competent, while Y176, Y178, and Y182 are respiration deficient. A number of zygotic clones were tested for growth on GlyYP medium. Gly+ denotes daughter colonies that could grow on GlyYP medium. Gly− denotes daughter colonies that could not grow on GlyYP medium. A majority of Gly− clones from the Y090 × Y176 and Y090 × Y178 crosses had the same restriction pattern of mtDNA as Y176 and Y178, respectively.
In short, respiration deficiency mutations can be generated in S. kluyveri cells. However, while a fraction of these mutants have the extrachromosomal characteristic, only limited deletions and rearrangements could be observed within the mtDNA molecule. So far, true petite mutants, characterized by extensive deletions within the mtDNA molecule, could not be generated. Thus, S. kluyveri behaves, with regard to the petite phenotype, like K. lactis.
The origin of the petite-positive and anaerobic characteristics.
A majority of ascomycetous yeasts are strictly aerobic, and these yeasts cannot be propagated at low oxygen levels. However, several aerobic yeasts, like K. lactis, can provide energy for growth by fermentation. Thus, oxygen is not absolutely necessary for the energy metabolism. The oxygen dependence is at least partially due to the dependence, directly or indirectly, of several biochemical pathways, like biosynthesis of sterols, pyrimidines, and deoxyribonucleotides (3, 8, 21), on the presence of molecular oxygen. For example, the fourth step of the de novo pyrimidine biosynthetic pathway, catalyzed by dihydroorotate dehydrogenase, in S. pombe (an aerobic yeast) is mitochondrial and dependent on the integrity of the respiratory chain (21). On the other hand, S. cerevisiae has DHOdehase (dihydroorotate dehydrogenase), which is cytoplasmic and is not dependent on a functional respiratory chain and thus can make pyrimidines in the absence of oxygen (21). However, while S. cerevisiae is generally considered as being capable of anaerobic growth (an anaerobic yeast), it is not absolutely independent of oxygen. At least one of the essential metabolic reactions, de novo generation of deoxyribonucleotides catalyzed by ribonucleotide reductase, is dependent on the presence of microconcentrations of oxygen (8, 14).
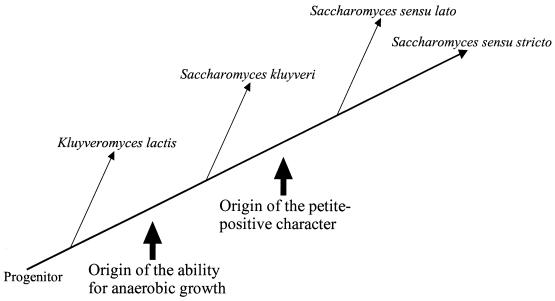
Based on a comparison of the modern yeast genera which are closely related to Saccharomyces, it is likely that the progenitor of these yeasts was fully dependent on the presence of oxygen and the integrity of the respiratory chain. However, upon diversifying, some of the lineages progressively decreased their dependence on oxygen. Especially notable is that the origin of the “fermentative lifestyle” greatly reduced the need for oxygen during proliferation. A decreasing dependence on oxygen-requiring reactions was the basis of, and a necessity for, the development of the petite-positive phenotype. In the present work, we have tried to study the connection between the origin of the anaerobic phenotype and the ability to generate and tolerate petite mutations. Previously, it has been shown that the anaerobic phenotype found in S. cerevisiae originated after the separation of the S. cerevisiae and K. lactis lineages (19, 31). Apparently, our results suggest that the anaerobic phenotype evolved just after the separation of the S. cerevisiae-S. kluyveri lineage from the K. lactis lineage (Fig. 3). It is likely that the metabolic change in the progenitor of the S. cerevisiae-S. kluyveri lineage was accompanied by a large genome rearrangement, including duplication of several genes but also loss of several other genes (13, 20). However, while S. kluyveri cells could grow under the anaerobic conditions established in our experiments, this yeast cannot tolerate the presence of the mitochondrial petite mutation. Thus, it seems that further modifications in the yeast metabolism were necessary for development of the petite-positive characteristic, and apparently they took place after the S. cerevisiae and S. kluyveri lineages separated (Fig. 3).
FIG. 3.
Simplified model to explain the evolution of anaerobic growth and the petite mutation in Saccharomyces yeasts. The relative phylogenetic relationships were adapted from another source (19).
ACKNOWLEDGMENTS
This work was supported by the Danish Research Council and the Novo Nordisk Foundation.
Wolfgang Knecht is acknowledged for his comments on the manuscript, and Jeanne Hvidtfeldt is acknowledged for technical assistance on the S. kluyveri crosses.
REFERENCES
- 1.Ahne A, Muller-Derlich J, Merlos-Lange A M, Kanbay F, Wolf K, Lang B F. Two distinct mechanisms for deletion in mitochondrial DNA of Schizosaccharomyces pombe mutator strains: slipped mispairing mediated by direct repeats and erroneous intron splicing. J Mol Biol. 1988;202:725–734. doi: 10.1016/0022-2836(88)90553-0. [DOI] [PubMed] [Google Scholar]
- 2.Alexander M A, Jefries T W. Respiratory efficiency and metabolite partitioning as regulatory phenomena in yeasts. Enzyme Microb Technol. 1990;12:2–19. [Google Scholar]
- 3.Andreasen A A, Stier T J B. Anaerobic nutrition of Saccharomyces cerevisiae. I. Ergosterol requirement for the growth in a defined medium. J Cell Comp Physiol. 1953;41:23–36. doi: 10.1002/jcp.1030410103. [DOI] [PubMed] [Google Scholar]
- 4.Andreasen A A, Stier T J B. Anaerobic nutrition of Saccharomyces cerevisiae. II. Unsaturated fatty acid requirement for the growth in a defined medium. J Cell Comp Physiol. 1954;41:23–36. doi: 10.1002/jcp.1030430303. [DOI] [PubMed] [Google Scholar]
- 5.Barnett J A. The taxonomy of the genus Saccharomyces Meyen ex Reess: a short review for non-taxonomists. Yeast. 1992;8:1–23. [Google Scholar]
- 6.Bulder C J E A. Induction of petite mutation and inhibition of synthesis of respiratory enzymes in various yeasts. Antonie Leeuwenhoek. 1964;30:1–9. doi: 10.1007/BF02046695. [DOI] [PubMed] [Google Scholar]
- 7.Bulder C J E A. Lethality of the petite mutation in petite negative yeasts. Antonie Leeuwenhoek. 1964;30:442–454. doi: 10.1007/BF02046758. [DOI] [PubMed] [Google Scholar]
- 8.Chabes A, Domkin V, Larsson G, Liu A, Gräslund A, Wijmenga S, Thelander L. Yeast ribonucleotide reductase has a heterodimeric iron-radical-containing subunit. Proc Natl Acad Sci USA. 2000;97:2474–2479. doi: 10.1073/pnas.97.6.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X J, Clark-Walker G D. The petite mutation in yeasts: 50 years on. Int Rev Cytol. 2000;194:197–238. doi: 10.1016/s0074-7696(08)62397-9. [DOI] [PubMed] [Google Scholar]
- 10.Clark-Walker G D, McArthur C R, Daley D J. Does mitochondrial DNA length influence the frequency of spontaneous petite mutations in yeasts? Curr Genet. 1981;4:7–12. doi: 10.1007/BF00376779. [DOI] [PubMed] [Google Scholar]
- 11.Dujon B. Mitochondrial genetics and functions. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces. Life cycle and inheritance. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. pp. 505–635. [Google Scholar]
- 12.Ephrussi B, Hottinguer H, Chimenes A M. Action de l'acriflavine sur les levures. I. La mutation “petite colonie.”. Ann Inst Pasteur. 1949;76:351–357. [Google Scholar]
- 13.Gojkovic Z, Jahnke K, Schnackerz K D, Piškur J. PYD2 encodes 5,6-dihydropyrimidine amidohydrolase, which participates in a novel fungal catabolic pathway. J Mol Biol. 2000;295:1073–1087. doi: 10.1006/jmbi.1999.3393. [DOI] [PubMed] [Google Scholar]
- 14.Harder J, Follmann H. Identification of a free radical and oxygen dependence of ribonucleotide reductase in yeast. Free Radic Res Commun. 1990;10:281–286. doi: 10.3109/10715769009149896. [DOI] [PubMed] [Google Scholar]
- 15.Hardy C M, Galeotti C L, Clark-Walker G D. Deletions and rearrangements in Kluyveromyces lactis mitochondrial DNA. Curr Genet. 1989;16:419–427. doi: 10.1007/BF00340721. [DOI] [PubMed] [Google Scholar]
- 16.Heslot H, Louis C, Goffeau A. Segregational respiratory-deficient mutants of a “petite negative” yeast Schizosaccharomyces pombe 972h−. J Bacteriol. 1970;104:482–491. doi: 10.1128/jb.104.1.482-491.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiers J, Zeeman A M, Luttik M, Thiele C, Castrillo J I, Steensma H Y, van Dijken J P, Pronk J T. Regulation of alcoholic fermentation in batch and chemostat cultures of Kluyveromyces lactis CBS 2359. Yeast. 1998;14:459–469. doi: 10.1002/(SICI)1097-0061(19980330)14:5<459::AID-YEA248>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 18.Klein C J L, Olsson L, Rønnow B, Mikkelsen J D, Nielsen J. Alleviation of glucose repression of maltose metabolism by MIG1 disruption in Saccharomyces cerevisiae. Appl Environ Microbiol. 1996;62:4441–4449. doi: 10.1128/aem.62.12.4441-4449.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurtzman C P, Robnett C J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek. 1998;73:331–371. doi: 10.1023/a:1001761008817. [DOI] [PubMed] [Google Scholar]
- 20.Langkjær R B, Nielsen M L, Daugaard P R, Liu W, Piškur J. Yeast chromosomes have been significantly reshaped during their evolutionary history. J Mol Biol. 2000;304:271–288. doi: 10.1006/jmbi.2000.4209. [DOI] [PubMed] [Google Scholar]
- 21.Nagy M, Lacroute F, Thomas D. Divergent evolution of pyrimidine biosynthesis between anaerobic and aerobic yeasts. Proc Natl Acad Sci USA. 1992;89:8966–8970. doi: 10.1073/pnas.89.19.8966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nissen T L, Hamann C W, Kielland-Brandt M C, Nielsen J, Villadsen J. Anaerobic and aerobic batch cultivations of Saccharomyces cerevisiae mutants impaired in glycerol synthesis. Yeast. 2000;16:463–474. doi: 10.1002/(SICI)1097-0061(20000330)16:5<463::AID-YEA535>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 23.Piškur J. Transmission of yeast mitochondrial loci to progeny is reduced when nearby intergenic regions containing ori/rep sequences are deleted. Mol Gen Genet. 1988;214:425–432. doi: 10.1007/BF00330476. [DOI] [PubMed] [Google Scholar]
- 24.Piškur J. Respiratory-competent yeast mitochondrial DNAs generated by deleting intergenic regions. Gene. 1989;81:165–168. doi: 10.1016/0378-1119(89)90347-8. [DOI] [PubMed] [Google Scholar]
- 25.Piškur J. Inheritance of the yeast mitochondrial genome. Plasmid. 1994;31:229–241. doi: 10.1006/plas.1994.1025. [DOI] [PubMed] [Google Scholar]
- 26.Piškur J, Smole S, Groth C, Petersen R F, Petersen M B. Structure and genetic stability of mitochondrial genomes vary among yeasts of the genus Saccharomyces. Int J Syst Bacteriol. 1998;48:1015–1024. doi: 10.1099/00207713-48-3-1015. [DOI] [PubMed] [Google Scholar]
- 27.Subik J, Kolarov J, Kovac L. Anaerobic growth and formation of respiration-deficient mutants of various species of yeasts. FEBS Lett. 1974;45:263–266. doi: 10.1016/0014-5793(74)80858-6. [DOI] [PubMed] [Google Scholar]
- 28.Verduyn C, Postma E, Scheffers W A, van Dijken J P. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J Gen Microbiol. 1990;136:395–403. doi: 10.1099/00221287-136-3-395. [DOI] [PubMed] [Google Scholar]
- 29.Verduyn C, Postma E, Scheffers W A, van Dijken J P. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 30.Visser W, Scheffers W A, Batenburg-van der Vegte W H, van Dijken J P. Oxygen requirements of yeasts. Appl Environ Microbiol. 1990;56:3785–3792. doi: 10.1128/aem.56.12.3785-3792.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wésolowski-Louvel M, Breunig K D, Fukuhara H. Kluyveromyces lactis. In: Wolf K, editor. Nonconvential yeasts in biotechnology. Berlin-Heidelberg, Germany: Springer Verlag; 1996. pp. 139–202. [Google Scholar]