Abstract
Cattle can efficiently perform de novo generation of glucose through hepatic gluconeogenesis to meet post-weaning glucose demand. Substantial evidence points to cattle and non-ruminant animals being characterized by phylogenetic features in terms of their differing capacity for hepatic gluconeogenesis, a process that is highly efficient in cattle yet the underlying mechanism remains unclear. Here we used a variety of transcriptome data, as well as tissue and cell-based methods to uncover the mechanisms of high-efficiency hepatic gluconeogenesis in cattle. We showed that cattle can efficiently convert propionate into pyruvate, at least partly, via high expression of acyl-CoA synthetase short-chain family member 1 (ACSS1), propionyl-CoA carboxylase alpha chain (PCCA), methylmalonyl-CoA epimerase (MCEE), methylmalonyl-CoA mutase (MMUT), and succinate-CoA ligase (SUCLG2) genes in the liver (P < 0.01). Moreover, higher expression of the rate-limiting enzymes of gluconeogenesis, such as phosphoenolpyruvate carboxykinase (PCK) and fructose 1,6-bisphosphatase (FBP), ensures the efficient operation of hepatic gluconeogenesis in cattle (P < 0.01). Mechanistically, we found that cattle liver exhibits highly active mechanistic target of rapamycin complex 1 (mTORC1), and the expressions of PCCA, MMUT, SUCLG2, PCK, and FBP genes are regulated by the activation of mTORC1 (P < 0.001). Finally, our results showed that mTORC1 promotes hepatic gluconeogenesis in a peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) dependent manner. Collectively, our results not only revealed an important mechanism responsible for the quantitative differences in the efficiency of hepatic gluconeogenesis in cattle versus non-ruminant animals, but also established that mTORC1 is indeed involved in the regulation of hepatic gluconeogenesis through PGC-1α. These results provide a novel potential insight into promoting hepatic gluconeogenesis through activated mTORC1 in both ruminants and mammals.
Keywords: Hepatic gluconeogenesis, Cattle, mTORC1, Peroxisome-proliferator-activated receptor γ coactivator-1α
Graphical abstract
1. Introduction
Glucose, as an important energy pool, is not only the major nutrient for the survival and growth of cattle but also an important guarantee for lactose synthesis and milk production (Cant et al., 2002; Zhao et al., 2007). Differing from non-ruminant animals such as pigs and humans, whose glucose requirements are mainly provided by the digested non-fibrous carbohydrates from the small intestine (Woerle et al., 2003), cattle mainly obtain glucose produced by gluconeogenesis from ruminal volatile fatty acids (VFAs). Notably, de novo production of glucose from lactic acid, glycerol, or amino acids through hepatic gluconeogenesis only serves to compensate for the lack of an exogenous glucose supply (Aschenbach et al., 2010; Lemon et al., 1981). In comparison, propionate, valerate, and isobutyrate can be used as precursors for hepatic gluconeogenesis for net synthesis of glucose, and serve as a main glucose and energy resource in ruminants (Bergman, 1990; Donkin et al., 1995; Hostettler-Allen et al., 1994). Of those VFAs, propionate is the predominant substrate for hepatic gluconeogenesis in cattle, and its contribution rate can reach as high as 60%–74% (Larsen et al., 2009; Reynolds et al., 2003; Zebeli et al., 2015). These characteristics determined that more hepatic gluconeogenesis processes occurred in cattle than in non-ruminants, which may be induced by the higher efficiency of gluconeogenesis in cattle. Many steps of gluconeogenesis are the reverse reaction of glycolysis, wherein the respective rate-limiting steps catalysing glucose 6-phosphatase (G6PC), fructose 1,6-bisphosphatase (FBP), and phosphoenolpyruvate carboxykinase (PCK) reverse the glycolysis and promote gluconeogenesis (Westermeier et al., 2019). The expression of genes encoding G6PC, FBP, and PCK is a prerequisite for hepatic gluconeogenesis, thus playing a major role in endogenous glucose production to maintain lactose synthesis and milk production in cattle. However, whether cattle have a higher capacity to express those genes involved in gluconeogenesis is not yet fully understood, and the potential underlying mechanism that promotes higher gluconeogenesis in cattle remains unknown.
Transcriptional coactivator peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α (PGC-1α) controls gluconeogenesis to maintain energy homeostasis in response to nutrient and hormonal signals (Bhalla et al., 2014; Hu et al., 2017). An important component in the energy and nutrient pathways is the mechanistic target of rapamycin (mTOR), a kinase that regulates cell growth, size, survival, and metabolism (Liu et al., 2020). Substantial evidence points to the mechanistic target of rapamycin complex 1 (mTORC1) having a crucial role in regulating the metabolism of proteins, lipids, and nucleotides through the P70 ribosomal S6 kinase 1 (S6K1), sterol regulatory element-binding protein 1 (SREBP1), PPARγ, or hypoxia-inducible factor-1α (HIF1α), respectively (Sengupta et al., 2010; Wullschleger et al., 2006). Further, the mTOR-PGC-1α pathway was proven to mediate mitochondria biogenesis and control mitochondrial oxidative function (Cunningham et al., 2007; Wang et al., 2017). However, the specific participation of mTORC1 in hepatic gluconeogenesis has yet to be elucidated. To sum up, we hypothesised that the mTORC1-PGC-1α pathway may serve as a potential target to promote gluconeogenesis in cattle. Herein, we investigated the gene expression between cattle and pigs/humans to determine the factors that determine their hepatic gluconeogenesis. Furthermore, the potential roles of mTORC1-PGC-1α in promoting high hepatic gluconeogenesis were tested in both primary hepatocyte cells of cattle and human hepatic cell lines (LO2 cells).
2. Materials and methods
2.1. Liver transcriptome data sources and processing of cattle and non-ruminants
Liver transcriptome data of human (n = 20), pig (n = 20), and cattle (n = 20) were obtained from the public database (https://www.ebi.ac.uk/ena) (Alexandre et al., 2015; Song et al., 2019; Wang et al., 2008). The samples used in this study can be found in Table 1. By using the Salmon tool with a decoy-aware transcriptome index for the species reference genome, the abundances and effective transcript lengths were estimated for each sample. Then, we use the ‘tximport’ package in R to aggregate the transcript-level quantifications to the gene level for the gene-level differential expression analysis. In total, 13,389 1:1 orthologues genes across these 3 species were obtained through ENSEMBL BioMart (Ensembl Genes 103). Unless otherwise stated, only these 1:1 orthologues genes were used in the following analysis. We evaluated the quality of the transcriptome data using principal component analysis (PCA) and identified the differentially expressed genes (DEGs) across species using edgeR. The DEGs were defined as those genes with a |log 2 fold change| > 1 and adjusted P-value < 0.05. Boxplots were created using the R package ‘ggplot 2’. The Kyoto Encyclopaedia of Genes and Genomes (KEGG) (https://www.genome.jp/kegg/) was used to map the genes to the KEGG pathway.
Table 1.
The samples of human, cattle, and pig.
| Species | Sequence read runs (SRR) |
|---|---|
| Human | SRR10212253; SRR10212254; SRR10212255; SRR10212256; SRR10212265; SRR10212267; SRR10212268; SRR10212269; SRR10212270; SRR10212271; SRR10212272; SRR10212273; SRR10212276; SRR10212278; SRR10212283; SRR10212296; SRR10212299; SRR10212301; SRR10212305; SRR10212316 |
| Cattle | ERR789812; ERR789813; ERR789814; ERR789815; ERR789816; ERR789817; ERR789818; ERR789819; ERR789820; ERR789821; ERR789822; ERR789823; ERR789824; ERR789825; ERR789826; ERR789827; SRR3176171; SRR3176228; SRR3176229; SRR3176230 |
| Pig | SRR10799611; SRR10799613; SRR10799615; SRR10799617; SRR10799619; SRR10799620; SRR10799622; SRR10799624; SRR10799625; SRR10799626; SRR10799627; SRR10799628; SRR10799629; SRR10799630; SRR10799631; SRR10799632; SRR10799633; SRR10799634; SRR10799635; SRR10799636 |
ERR = experiment read runs.
2.2. Derivation of primary hepatocyte cells
The primary hepatocytes of calf were cultured in Roswell Park Memorial Institute (RPMI) 1640 (HyClone, UT, USA) supplemented with 5% fetal bovine serum (FBS, Gibco, Grand Island, USA) and 1% Penicillin-Streptomycin (Solarbio, Beijing, China) at 37 °C in 5% CO2. The isolation and culture of primary hepatocytes cells from calf were performed according to the previously published study by Song et al. (2016). The detailed methods are listed as follows: (1) separation: the thickest blood vessel was selected for perfusion, and the flow rate of perfusate A (140 mM NaCl [Sangon, Shanghai, China], 6.7 mM KCl [Sangon], 10 mM HEPES [Sangon], 2.5 mM glucose [Sangon], 0.5 mM EDTA [Sangon]) was maintained at 50 mL/min for 15 min. Then, perfusate B (140 mM NaCl, 6.7 mM KCl, 10 mM HEPES, 2.5 mM glucose, 5 mM CaCl2 [Sangon]) was perfused at a rate of 50 mL/min, lasting about 30 min. Perfusate C (0.2 g/L type IV collagenase [Solarbio, Beijing, China] dissolved in perfusate B) was perfused at a rate of 20 mL/min for 30 min; (2) cultivation: the digestion was stopped with medium containing 10% FBS, then centrifuged at 50 × g for 3 min at room temperature, after which the cells were suspended in RPMI 1640 and seeded on 10-cm dishes (NEST, Shanghai, China). Then, cells were transferred to a CO2 incubator (Thermo Fisher Scientific, Waltham, MA, USA), in which the medium was replaced every 24 h, and the morphology and growth of liver cells were observed under a microscope.
2.3. Cell culture, transient transfection, and drug treatment
Cells of the human normal hepatocyte line LO2 were purchased from American Type Culture Collection (ATCC) and cultured in RPMI 1640 supplemented with 10% FBS at 37 °C in 5% CO2.
LO2 cells with 1 × 105 per well of a 6-well plate were transfected with small interfering RNA (siRNA) oligonucleotides or Myc-Rheb Q64L by using Lipofectamine 2,000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Myc-Rheb Q64L, which directly targets mTORC1 for activation, was a gift from professor Wang Ping's laboratory at the Medical College of Tongji University, Shanghai. Non-specific control siRNA and siRNAs for rapamycin-sensitive adapter protein of mTOR (raptor, the key component of the mTOR pathway) were purchased from GenePharma (Shanghai, China). We examined the role of the mTOR pathway in regulating gluconeogenesis genes by knockdown raptor. The sequences of siRNAs raptor and the control siRNA were the following: si raptor: 5′-CGACTACTACATCTCCGTGTA-3′, and si NC: 5′-UUCUCCGAACGUGUCACGU-3΄. SiRNA and plasmid transfection of LO2 cells was performed according to the manufacturer's instructions.
We treated the primary hepatocytes of calf with rapamycin (100 nM)-mTORC1 inhibitor (V900930, Sigma Aldrich, MO, USA), or treated the LO2 cells with rapamycin (100 nM), MHY1485 (2 μM)-mTORC1 activator (S7811, Selleck, Shanghai, China), ZLN005 (20 μM)-PGC-1α activator (S7447, Selleck), and SR18292 (20 μM)-PGC-1α inhibitor (S8528, Selleck) for 24 h to examine whether the mRNA levels of gluconeogenesis genes were regulated by mTORC1.
2.4. RNA extraction and RT-qPCR
Total RNA was isolated from primary hepatocytes and LO2 by using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's directions. The NanoDrop ND-1,000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to detect RNA purity (OD260/OD280) and concentration of each sample. RNA integrity was confirmed by electrophoresis with a denaturing agarose gel. Real-time quantitative PCR (RT-qPCR) analysis was performed in technical triplicate, using the TB Green RT-qPCR kit (RR820A, Takara, Dalian, China) on a Roche LightCycler 96 RT-qPCR system (Roche, Basel, Switzerland). All data were generated using the cDNA from triplicate wells for each condition. The comparative Ct method was used to calculate the relative quantity of the target gene mRNA, which was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and relative to the calibrator and expressed as the fold change = 2−ΔΔCt (Livak et al., 2001). The following procedures were used for qPCR experiments: 30 s at 95 °C, followed by 40 cycles of 5 s at 95 °C and 30 s at 60 °C. The primer sequences used for RT-qPCR were designed using NCBI Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast) and listed in Table 2, Table 3.
Table 2.
Primer sequences for RT-qPCR of cattle.
| Gene | Forward primer sequence (5′-3′) | Reverse primer sequence (5′-3′) |
|---|---|---|
| ACSS1 | CTGGACGCCTACTTCGAGAC | GCAGCTTCTCCCTTGATGTC |
| PCCA | TGGGCCAACATTCTCCCATGA | TGGTGAGGATACGCACCTTGT |
| MCEE | GGAGTGTCCGTCGTTTTTGT | CTGGTTTTCCATGTGCTCCT |
| MMUT | ATGCAACTCGAGCAAGATGT | ACAAGAAGACGAGGTCTGCG |
| SUCLG2 | GCCTTTGAAAAACCAGGCTGC | CGGAATTCTGCGTTGTCATCA |
| FBP1 | TCCTGCCCTCACCGAGTATG | TCATACAGTAGTCTCAGCTTTCCA |
| FBP2 | AAAGAAGTTTCCTGAGGACGGC | CTGCTCGATGATGTAGGCCA |
| PCK1 | GACGGCCTCAACTACTCAGC | AGTGAGAGCCAACCAGCAGT |
| PCK2 | AGGCAAACCCTGGAAACCTG | ATACCAGGGGGACTCCTTTGG |
| GAPDH | GGGTCATCATCTCTGCACCT | GGTCATAAGTCCCTCCACGA |
ACSS1 = acyl-CoA synthetase short-chain family member 1; PCCA = propionyl-CoA carboxylase alpha chain; MCEE = methylmalonyl-CoA epimerase; MMUT = methylmalonyl-CoA mutase; SUCLG2 = succinate-CoA ligase; FBP = fructose 1,6-bisphosphatase; PCK = phosphoenolpyruvate carboxykinase; GAPDH = glyceraldehyde-3-phosphate dehydrogenase.
Table 3.
Primer sequences for RT-qPCR of human.
| Gene | Forward primer sequence (5′-3′) | Reverse primer sequence (5′-3′) |
|---|---|---|
| ACSS1 | CGAGAGCGTTGCTTTGATCT | GGGCATGTAGATGGCAACAC |
| PCCA | CGTGGAGTTCCTTGTGGACT | CAGCTTGTTTGTGCCTGAGA |
| MCEE | GGAGCACATGGAAAACCAGT | TGGAAGCAGTGAAGGACTCA |
| MMUT | GAGTGGAGCATATCGCCAGG | CACTTCACGAGGAGTCTGGAA |
| SUCLG2 | TCACAGCTGATCCTAAGGTTG | GCGACGTTCTCTTGGATACC |
| FBP1 | CCTGCCGTCACTGAGTACAT | CAGCAGTCTCAGCTTTCCAT |
| FBP2 | AAGAAATTCCCTGAGGATGGCAG | GCCACGGGATTGCATTCATAC |
| PCK1 | CCTGACCGCAGAGAGATCAT | CCGCCAGGTACTTCTTCTCA |
| PCK2 | CCACTGGCATTCGAGATTTT | CCCGCTGAGAAGGAGTTACA |
| GAPDH | CAACGAATTTGGCTACAGCA | AGGGGTCTACATGGCAACTG |
ACSS1 = acyl-CoA synthetase short-chain family member 1; PCCA = propionyl-CoA carboxylase alpha chain; MCEE = methylmalonyl-CoA epimerase; MMUT = methylmalonyl-CoA mutase; SUCLG2 = succinate-CoA ligase; FBP = fructose 1,6-bisphosphatase; PCK = phosphoenolpyruvate carboxykinase; GAPDH = glyceraldehyde-3-phosphate dehydrogenase.
2.5. Western blot
The protein of fresh liver tissue samples of cattle (n = 5) and pig (n = 5) collected from local slaughterhouses was extracted by RIPA buffer, and protein content was quantified using the BCA Protein Assay Kit (Tiangen, Beijing, China). Further, the protein of cells was extracted as previously described (Liu et al., 2010). Then, a Western blot of all isolated protein samples was performed according to Liu et al. (2010). Briefly, the protein samples were resolved by SDS-PAGE gel electrophoreses and transferred onto a nitrocellulose filter membrane (0.45 μm, GE). The membranes were blocked with 5% non-fat milk powder in phosphate buffer saline with 0.1% Tween-20 (PBST, Sangon) and subsequently incubated overnight with the primary antibody at 4 °C. After being washed with PBST, the membranes were incubated with appropriate 1:100,000 dilutions of horseradish peroxidase (HRP)-conjugated secondary antibody at room temperature for 2 h. The membranes were washed again in PBST and the proteins were detected using an imaging system (Bio-Rad, Hercules, CA, USA).
The antibodies against pT389-S6K1 (9234 S/L), p-S6 (4858 S), S6K1 (9202 S), S6 (2217 S), raptor (2280), pS473-AKT (9271), AKT (9272) were obtained from Cell Signaling Technology, and the Anti-c-Myc (9E10 sc-40) antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The secondary antibodies were obtained from Sigma Aldrich. The primary antibodies were diluted at 1:1,000 to 1:2,000 and the secondary antibodies were diluted at 1:5,000.
2.6. Statistical analyses
The analysis was done by using Student's t-test or two-way Analysis of variance (ANOVA) using SPSS 21.0 software with replicates as experimental units and differences were considered to be statistically significant at P < 0.05. If a significant treatment effect was observed by two-way ANOVA, the significance of the differences between treatments was determined using Sidak's multiple comparisons test.
3. Results
3.1. Differently expressed genes in cattle and non-ruminants
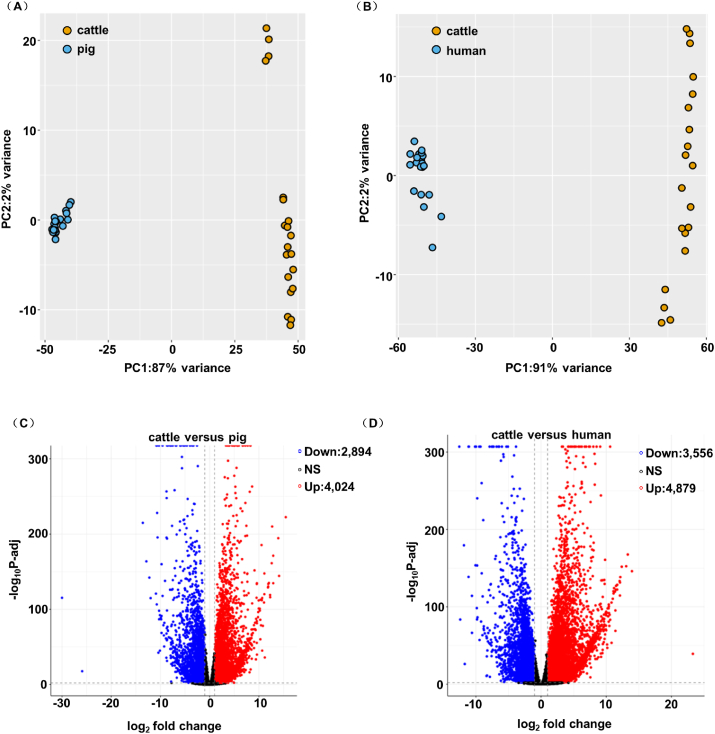
To explore the mechanism conferring such a different capacity of hepatic gluconeogenesis in cattle versus non-ruminants, we downloaded liver transcriptome data of human, pig, and cattle from a public database (https://www.ebi.ac.uk/ena) based on sequencing depth and species consistency (Table 1). The global relationships of liver transcriptome data among human, pig, and cattle samples can be explored through PCA, which revealed a clear separation with no overlap between human/pig and cattle samples, thus confirming that the transcriptome profiles of the liver in human/pig and cattle are distinct (Fig. 1A and B). The results of hierarchical clustering analysis confirmed a large separation between the samples of the three species. According to the DESeq2 package for differential analysis of count data (DESeq2), there are 6,923 DEGs in the comparison of cattle and pigs, of which 4,024 were up-regulated and 2,894 were down-regulated in cattle. Similarly, there are 8,435 DEGs with 4,879 of them up-regulated and 3,556 down-regulated in cattle compared with humans (Fig. 1C and D).
Fig. 1.
Differently expressed genes in cattle and non-ruminants. (A and B) Principal component analysis (PCA) plots from the whole transcriptome of cattle versus pig or human liver tissues. Different colors correspond to different species. Yellow PCA plots represent the cattle liver transcriptome data (n = 20). Blue PCA plots represent the pig or human livers' transcriptome data (n = 20). (C and D) Volcano plot for the differentially expressed genes (DEGs) of liver of cattle compared with that of pig or human. Red points are up-regulated DEGs. Blue points are down-regulated DEGs. Black points are those genes neither significantly up- nor down-regulated. The cut-off used to designate a DEG was an adjusted P < 0.01.
3.2. Propionate metabolism and gluconeogenesis genes are highly expressed in cattle
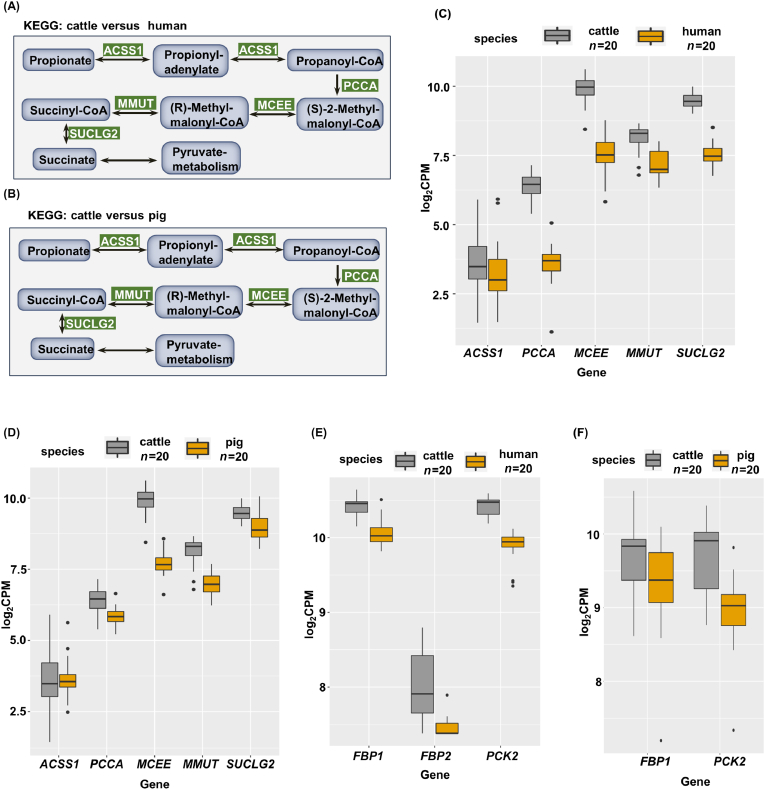
In the pathway of propionate metabolism, several categories of enzymes, including acyl-CoA synthetase short-chain family member 1 (ACSS1), propionyl-CoA carboxylase alpha chain (PCCA), methylmalonyl-CoA epimerase (MCEE), methylmalonyl-CoA mutase (MMUT), and succinate-CoA ligase (SUCLG2), figure prominently in regulating the conversion of propionate to pyruvate. The KEGG pathway of propionate metabolism through transcriptional changes in the liver of human/pig versus cattle showed that the expression levels of the main enzymes converting propionate to pyruvate were increased to different extents (Fig. 2A–D).
Fig. 2.
Propionate metabolism and gluconeogenesis genes are highly expressed in cattle. (A and B) Differential expression of propionate metabolism genes in the liver of cattle versus that of human or pig in the Kyoto Encyclopaedia of Genes and Genomes (KEGG) propionate pathway. Green indicates the genes found up-regulated in cattle liver tissue, P < 0.01. (C and D) The expression of acyl-CoA synthetase short-chain family member 1 (ACSS1), propionyl-CoA carboxylase alpha chain (PCCA), methylmalonyl-CoA epimerase (MCEE), methylmalonyl-CoA mutase (MMUT), and succinate-CoA ligase (SUCLG2) in the liver of cattle compared with that of human and pig. Grey indicates cattle, and yellow indicates human or pig (n = 20), P < 0.01. (E and F) Differential expression of fructose 1,6-bisphosphatase 1 (FBP1), FBP2, and phosphoenolpyruvate carboxykinase (PCK2) in the liver of cattle compared with that of human or pig. Grey indicates cattle, and yellow indicates human or pig (n = 20), P < 0.01.
The transcription of rate-limiting enzyme genes, such as those of PCK and FBP, is an important step toward activating hepatic gluconeogenesis (Westermeier et al., 2019), particularly in cattle. To determine the differences in the expression of PCK and FBP in cattle and non-ruminant animals, we analysed the liver transcriptome data of pigs/humans and cattle, and found that the cattle liver can powerfully stimulate the expression of PCK as well as FBP (Fig. 2E and F).
3.3. The activation of mTORC1 is strongly induced in liver of cattle
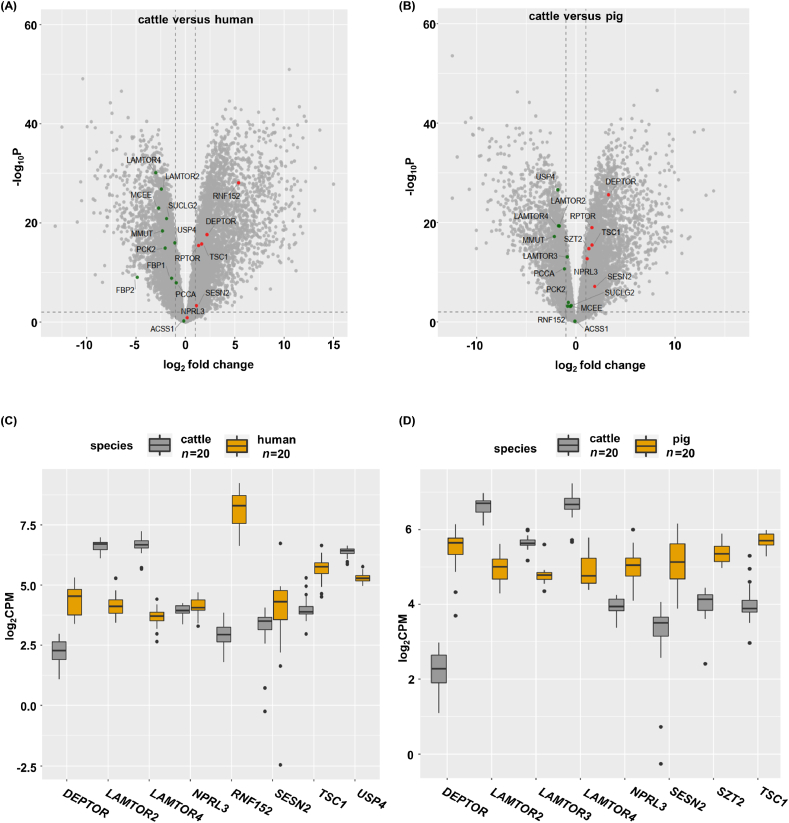
Considering the crucial role of the mTORC1 pathway in glucose metabolism, to further explore the transcriptional regulation mechanism of PCK and FBP, we analysed the transcriptional differences in the major components of the mTORC1 pathway in the liver transcriptomics of pig/human and cattle. We identified genes with significant changes in expression between cattle and non-ruminants, finding an increase in positive regulators in the mTORC1 pathway (LAMTOR2, LAMTOR3, and LAMTOR4) in cattle liver, while the mRNA levels of negative regulators (DEP-domain-containing mTOR-interacting protein [DEPTOR], nitrogen permease related-like 3 [NPRL3], SESN2, SZT2, and tuberous sclerosis complex 1 [TSC1]) were consistently reduced in cattle liver (Fig. 3A–D). We next examined the expression of ring finger protein 152 (RNF152) and ubiquitin specific protease 4 (USP4) in the cattle liver, which also indicated the activation of the mTORC1 pathway. Indeed, the transcriptome data identified the markedly reduced and enhanced abundance of mRNAs that encode RNF152 and USP4, respectively, in the liver tissue of cattle (Fig. 3A–D).
Fig. 3.
The activation of mechanistic target of rapamycin complex 1 (mTORC1) is strongly induced in the liver of cattle. (A and B) Volcano plot for the differentially expressed genes (DEGs) in the liver of cattle compared with that of human or pig. Red points are down-regulated DEGs. Green points are up-regulated DEGs. The cut-off used to designate a DEG was an adjusted P < 0.01. (C and D) DEGs of mTORC1 signaling in the liver of cattle, human, and pig. Grey indicates cattle, and yellow indicates human or pig (n = 20), P < 0.01.
3.4. Increased gluconeogenesis genes are regulated by mTORC1 pathway in cattle primary hepatic cells
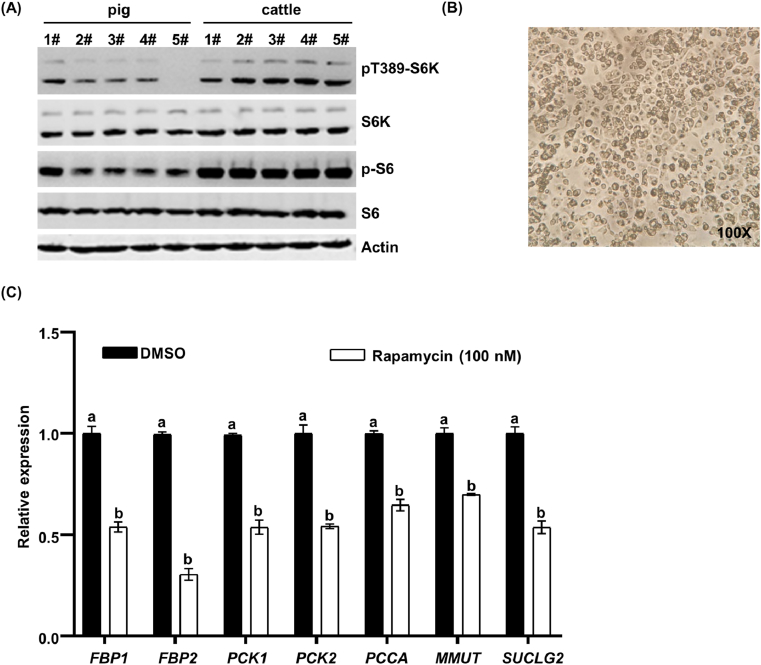
To further elucidate the mechanism by which mTORC1 regulates the expression of gluconeogenesis genes, we were particularly interested in examining the extent of mTORC1 activation in the liver of pigs versus cattle. The activation of mTORC1 was monitored by measuring the phosphorylation of S6K1 at Thr389, which is a well-characterized mTORC1-dependent phosphorylation site (Ruvinsky et al., 2006). Our data showed that the activation of mTORC1 was significantly enhanced in cattle liver, as indicated by the pT389-S6K1 (Fig. 4A). In addition, the phosphorylation of S6, which is a target of S6K1, was strongly correlated with the phosphorylation status of S6K1 (Fig. 4A), indicating that mTORC1 activity is promoted in the liver of cattle. Moreover, to directly examine whether the mRNA levels of gluconeogenesis genes are regulated by mTORC1, we isolated primary hepatocyte cells of calf (Fig. 4B) and treated the cells with the mTORC1 inhibitor rapamycin (Chen et al., 2020). There was a significant decrease in FBP, PCK, PCCA, MMUT, and SUCLG2 gene expression in response to the rapamycin treatment (Fig. 4C). The primer quality was evaluated by melting curve (Supplementary Figs. S1–S2).
Fig. 4.
Increased gluconeogenesis genes are regulated by mechanistic target of rapamycin complex 1 (mTORC1) signaling in cattle. (A) The activation of mTORC1 in calf and pig liver tissues was monitored by measuring the phosphorylation of P70 ribosomal S6 kinase 1 (S6K1) and S6. (B) Primary hepatocyte cells of cattle isolated and cultured for 4 h were observed under a microscope (OLYMPUS CKX53, Japan). (C) The primary hepatocyte cells of calf were treated with rapamycin for 24 h, and endogenous expression of FBP1, FBP2, PCK1, PCK2, PCCA, MMUT, and SUCLG2 genes was examined by RT-qPCR. Statistical significance was determined as the mean ± SEM. a, b Bars with a different letter mean a significant difference (P < 0.001). DMSO = dimethyl sulfoxide.
3.5. Activated mTORC1 pathway could increase the gluconeogenesis in human LO2 cells
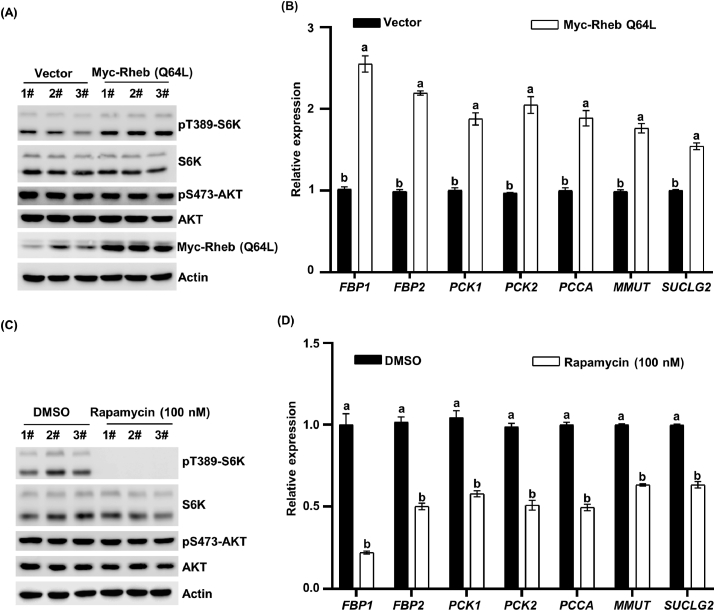
Using the ectopically expressed Myc-Rheb Q64L, which serves as the constitutively active mutant of Rheb, not only strongly induced the higher level of pT389-S6K1 (Fig. 5A), but also significantly increased the gluconeogenesis gene expression of FBP, PCK, PCCA, MMUT, and SUCLG2 (Fig. 5B), indicating that mTORC1 activation plays a critical role in the expression of gluconeogenesis genes.
Fig. 5.
Activated mechanistic target of rapamycin complex 1 (mTORC1) pathway could increase the gluconeogenesis in human hepatic cell lines (LO2 cells). (A) Overexpression of Myc-Rheb Q64L in LO2 cells and the indicated protein was detected using a Western blot. (B) Overexpression of Myc-Rheb Q64L in LO2 cells, and endogenous expression of FBP1, FBP2, PCK1, PCK2, PCCA, MMUT, and SUCLG2 genes was examined by RT-qPCR. Statistical significance was determined as the mean ± SEM. a, b Bars with a different letter mean a significant difference (P < 0.001). (C) LO2 cells were treated with rapamycin for 24 h, and the indicated protein was detected using a Western blot. (D) LO2 cells were treated with rapamycin for 24 h, and endogenous expression of FBP1, FBP2, PCK1, PCK2, PCCA, MMUT, and SUCLG2 genes was examined by RT-qPCR. Statistical significance was determined as the mean ± SEM. a, b Bars with a different letter mean a significant difference (P < 0.001). DMSO = dimethyl sulfoxide.
Furthermore, our results again confirmed that treatment with rapamycin also resulted in a remarkable decrease in FBP, PCK, PCCA, MMUT, and SUCLG2 gene expression in human hepatocyte LO2 cells (Fig. 5D). Phosphorylation of S6K1 at Thr389 was used as the positive control, and as expected, the phosphorylation level was significantly inhibited in response to rapamycin treatment (Fig. 5C).
3.6. mTORC1 pathway promotes gluconeogenesis genes through PGC-1α
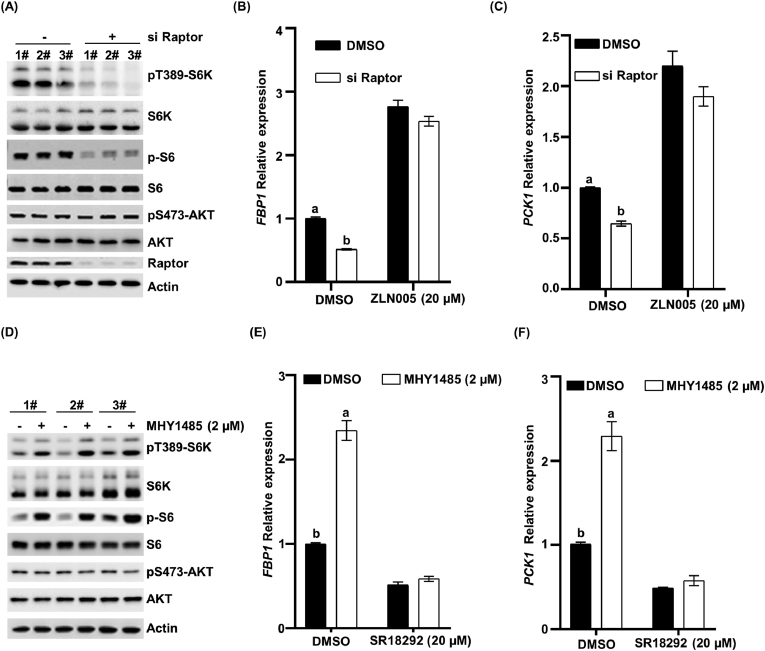
As an important transcription factor, PGC-1α can regulate PCK and FBP gene expression in hepatocytes. We next sought to determine whether PGC-1α has a regulatory effect on mTORC1-mediated gluconeogenesis gene expression in LO2 cells. To this end, we assessed the abundance of mRNAs that encode FBP and PCK in LO2 cells with the treatment of ZLN005 (the activator of PGC-1α) alone or in combination with raptor knockdown. Notably, our result showed that the mRNA levels of FBP and PCK were increased with the ZLN005 treatment (Fig. 6B and C). Endogenous raptor (an essential protein for mTORC1 lysosomal localization and the scaffold protein that recruits substrates to the complex) was depleted in LO2 cells using specific siRNAs, and knockdown efficiency was detected by using a Western blot (Fig. 6A). We found that the depletion of raptor significantly decreased S6K1 phosphorylation at Thr389 (Fig. 6A). Consistent with the effects of mTORC1 on gluconeogenesis genes, the downregulation of FBP and PCK expression was observed in raptor deficient cells (Fig. 6B and C). Importantly, we found that co-treatment of cells with ZLN005 markedly restored the raptor knockdown-induced downregulation of FBP and PCK expression (Fig. 6 B and C). Similarly, we found that SR18292, which inhibits the activity of PGC-1α, drastically inhibited the gluconeogenesis gene expression of FBP, PCK (Fig. 6E and F). Moreover, the mTORC1 activator MHY1485 elevated the level of pT389-S6K1 (Fig. 6D) and significantly increased the levels of PCK and FBP transcripts (Fig. 6E and F). Furthermore, the effect of MHY1485 was eliminated by SR18292 treatment (Fig. 6E and F). In contrast, there was no significant change in mTORC2 activity as determined by the phosphorylation of AKT at Ser 473. Our data, therefore, indicate that the mTORC1 pathway promotes gluconeogenesis genes through PGC-1α.
Fig. 6.
Mechanistic target of rapamycin complex 1 (mTORC1) pathway promotes gluconeogenesis gene expression through peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α (PGC-1α). (A) Knockdown raptor in hepatic cell lines (LO2 cells) and the indicated protein was detected via Western blot. (B and C) The LO2 cells with or without knockdown raptor were treated with ZLN005 (20 μM) for 24 h, and the endogenous expression of FBP1 and PCK1 was examined by RT-qPCR. Statistical significance was determined as the mean ± SEM. a, b Bars with a different letter mean a significant difference (P < 0.001). (D) The LO2 cells were treated with MHY1485 (2 μM) for 24 h, and the indicated protein was detected via Western blot. (E and F) The LO2 cells were treated with SR18292 (20 μM) alone or in combination with MHY1485 (2 μM) for 24 h. The endogenous expression of FBP1, PCK1 was examined by RT-qPCR. Statistical significance was determined as the mean ± SEM. a, b Bars with a different letter mean a significant difference (P < 0.001). DMSO = dimethyl sulfoxide.
4. Discussion
Cattle and non-ruminant animals are characterized by phylogenetic features and differing capacity for hepatic gluconeogenesis, with the propionate originating from ruminal metabolism being the major substrate for hepatic gluconeogenesis in cattle (Reynolds et al., 2003; Young, 1977). However, the mechanism enabling cattle to attain their high capacity for hepatic gluconeogenesis from propionate remains largely unknown. Compared with non-ruminant animals, hepatic gluconeogenesis is more essential for supporting glucose (lactose) synthesis and milk production during the transition from pregnancy to lactation in cattle. Therefore, the mechanism that regulates hepatic gluconeogenesis in cattle and non-ruminant animals must differ between them (Aschenbach et al., 2010; Young, 1977).
Our data from the transcriptome, liver tissue, and hepatocyte cell sources revealed that ACSS1, PCCA, MCEE, MMUT, and SUCLG2, which are key genes in propionate-pyruvate metabolism (Reynolds et al., 2003), were highly expressed in cattle liver. But these enzyme-mediated processes are all reversible in the propionate metabolism pathway, so how do those highly expressed enzymes promote the conversion of propionate to pyruvate? We further speculate that the concentration of propionate may play a decisive role in the metabolism pathway, and the enzymatic cascade reaction will promote the conversion of propionate to pyruvate in response to increased propionate. Coincidentally, the rumen of cattle can produce a large amount of propionate, which would shift the propionate metabolism pathway to proceed in the direction of producing pyruvate. Moreover, our data show that the treatment of mTORC1 inhibitors can limit the expression of PCCA, MMUT, and SUCLG2. Whether mTORC1 plays an important role in the regulation of the expression of PCCA, MMUT, and SUCLG2, and whether it participates in the regulation of these genes’ expression directly or indirectly, requires further research. More importantly, as an important VFA, propionate can function like an amino acid — not only as a substrate for protein synthesis, but also as a signal for activating the mTORC1 signaling pathway (Hu et al., 2020) — in addition to serving as a hepatic gluconeogenesis substrate. Still, whether propionate can regulate the signaling transduction involved and promote the expression of these enzymes also warrants further investigation.
The liver plays a major role in maintaining glucose homeostasis in mammals. Although many steps of gluconeogenesis are the reverse reaction of glycolysis, the respective rate-limiting steps catalyzed by G6PC, FBP, and PCK reverse the glycolysis and promote gluconeogenesis (Lebigot et al., 2015; Westermeier et al., 2019). Yet, the distinguishing physiological characteristic of cattle is their high-efficiency hepatic gluconeogenesis. Whether cattle are highly capable of expressing the genes responsible for gluconeogenesis remains not fully understood. In this study, we found that mRNA abundance of PCK and FBP is augmented in cattle liver, indicating that the high efficiency of hepatic gluconeogenesis is a unique feature of cattle. Similarly, a previous study reported direct evidence of the capability of propionate to control the expression of mRNA like PCK for key enzymes for gluconeogenesis in cattle liver (Zhang et al., 2016). Interestingly, other research also found enhanced mRNA levels of PCK2 and FBP1 in bovine intestinal epithelial cells induced by propionate (Zhan et al., 2020). However, we only detected the expression of the gluconeogenesis genes, but did not measure the activity of enzymes, which requires further research. Nevertheless, the regulatory mechanism of PCK and FBP expression is still unclear.
According to previous research (Puigserver et al., 2003; Yoon et al., 2001), the transcription factor PGC-1α could bind to the promoters of FBP and PCK and initiate the expression of these two genes. However, the PGC-1α transcripts were not highly expressed in the transcriptome data. We speculate the functioning of the transcription factor not only depends on its quantity but is also determined by its activity and intracellular location. Similarly, even though it is well recognized that PPAR has an important role in the adaptation of the liver to the transition period in dairy cows, the gene or protein expression of its isotypes such as PPARA (the gene symbol of PGC-1α) and PPARG had no significant changes during the transition period (Bionaz et al., 2015; Gao et al., 2021; Schaff et al., 2012). Therefore, although the transcription level of PGC-1α has not been improved, its activation and location may be regulated by post-translational modification, which may involve the regulation of protein kinase mTORC1 (Cunningham et al., 2007).
The mTORC1 signaling pathway has several components, namely mTOR, which confers to the complex its kinase activity, the raptor (Kim et al., 2002), mammalian lethal with SEC13 protein 8 (mLST8) (Kim et al., 2003); proline-rich AKT substrate 40 kDa (PRAS40), and DEPTOR (Peterson et al., 2009; Vander Haar et al., 2007). Classical theory holds that the activation of mTORC1 is tightly coupled to lysosomal localization and the activation of Rheb. To be specific, the lysosomal positioning is necessary for mTORC1 activation, which is mainly mediated by multiple factors, such as SESN2 (Chantranupong et al., 2014; Saxton et al., 2016; Wolfson et al., 2016), KICSTOR/SZT2 (Bar-Peled et al., 2013; Shen et al., 2018; Wolfson et al., 2017), GATOR2-GATOR1 (comprising NPRL2, NPRL3, and DEP domain-containing 5 [DEPDC5]) (Bar-Peled et al., 2013), Ragulator (comprising LAMTOR1, LAMTOR2, LAMTOR3, LAMTOR4, and LAMTOR5) (Bar-Peled et al., 2012), and RagGTPase (Chantranupong et al., 2016; Sancak et al., 2010; Wolfson et al., 2017). The lysosomal-localized mTORC1 needs to be activated by the AKT-TSC-Rheb signal axis (Dibble et al., 2015; Inoki et al., 2003; Tee et al., 2003). In addition, our previous studies revealed that the lysosome-anchored E3 ligase RNF152 catalyzes the ubiquitination of Rheb and RagA, resulting in the mTORC1 pathway's inactivation; conversely, the deubiquitinase USP4 promotes the activation of the mTORC1 pathway by removing the ubiquitin chain from Rheb (Deng et al., 2019; Deng et al., 2015). Coincidentally, our results showed that the above-mentioned positive regulators in the mTORC1 pathway were all up-regulated and negative regulators were all down-regulated in cattle when compared with non-ruminant animals. In addition, we found significantly enhanced pT389-S6K1 and p-S6, indicating mTORC1 activity is promoted in the liver of cattle. Based on the expression of positive and negative regulators and key indicators in the mTORC1 pathway from the tissue and cell samples, we showed that cattle livers had a much higher mTORC1 activity when compared with non-ruminant animals.
Compelling evidence points to mTORC1 playing a crucial role in regulating the metabolism of proteins, lipids, and nucleotides (Hu et al., 2020; Liu et al., 2020). However, the functioning of mTORC1 in hepatic gluconeogenesis has not been revealed. In this study, our results show that the expression of PCCA, MMUT, SUCLG2, PCK, and FBP gluconeogenesis genes is controlled by the activation of the mTORC1 pathway, which points to the regulation of the mTORC1 being of central importance in the control of gluconeogenesis in cattle liver. Other research has shown increased expression of PCK mRNA in the liver of post-partum dairy cows to support the increasing importance of gluconeogenesis (Gao et al., 2021). Similarly, the expression of PCK mRNA and protein in dairy cattle is also affected by feed intake and dietary forage-to-concentrate ratios which influenced the amounts of substrates for gluconeogenesis (Greenfield et al., 2000; Zhang et al., 2019). So, the high fiber utilization of ruminants and the high concentration of VFAs also determine that cattle should have high gluconeogenesis capacity. Combined with our data, whether physiological stages, dietary composition, and/or feed intake do more to promote the expression of PCK mRNA by regulating the activation of mTORC1 is an unresolved issue that deserves in-depth study. Further study with more replication of animals that are fed with different kinds of diets could be considered to determine the relationship between differential diets, VFA concentrations, mTOR, and hepatic gluconeogenesis.
Consistently, in our study, we revealed that mTORC1 promotes the expression of gluconeogenesis genes in a PGC-1α dependent manner. Given that hepatic gluconeogenesis plays a major role in blood glucose homeostasis, targeting the mTORC1-PGC-1α pathway might be used as a strategy to improve blood glucose control. However, the mechanism by which mTORC1 regulates the transcriptional activity of PGC-1α requires further investigation.
5. Conclusions
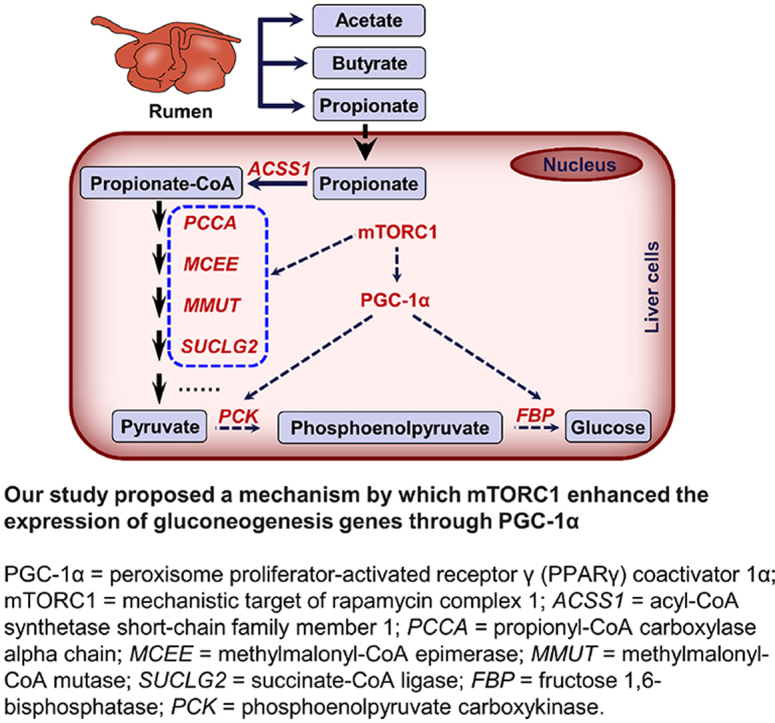
Our study firstly reveals that the hyperactivation of mTORC1 mediated the expression of downstream genes, including PCCA, MMUT, SUCLG2, PCK, and FBP, to promote high-efficiency hepatic gluconeogenesis in cattle (Fig. 6). Our results not only serve as an important mechanism for quantitative and qualitative differences in the efficiency of hepatic gluconeogenesis between cattle and non-ruminants, but also provide a theoretical basis for improving glucose (lactose) synthesis and milk production during lactation. More importantly, our results also revealed that mTORC1 controlled hepatic gluconeogenesis through PGC-1α (Fig. 6), which offers new insight into the molecular mechanisms of mTORC1 on glucose homeostasis, indicating the mTORC1-PGC-1α pathway was positively correlated with glucose parameters, and may act as a potential therapeutic target for metabolic disease.
Author contributions
Guoyan Wang: Conceptualization, Methodology, Data curation, Formal analysis, Writing – original draft. Jun Zhang: Conceptualization, Methodology, Data curation, Formal analysis, Writing – original draft. Shengru Wu: Conceptualization, Methodology, Data curation, Formal analysis, Writing – original draft. Senlin Qin: Methodology and Investigation. Yining Zheng: Methodology and Investigation. Chao Xia: Methodology and Investigation. Huijun Geng: Investigation. Junhu Yao: Conceptualization, Resources, Supervision, Funding acquisition, Project administration, Writing – review & editing. Lu Deng: Conceptualization, Resources, Supervision, Validation, Funding acquisition, Project administration, Writing – review & editing. All authors read and approved the final manuscript.
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China, China (grant numbers 32070782, 32072761).
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2022.07.010.
Contributor Information
Junhu Yao, Email: yaojunhu2004@sohu.com.
Lu Deng, Email: denglu128128@nwafu.edu.cn.
Appendix. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
References
- Alexandre P.A., Kogelman L.J.A., Santana M.H.A., Passarelli D., Pulz L.H., Fantinato-Neto P., et al. Liver transcriptomic networks reveal main biological processes associated with feed efficiency in beef cattle. BMC Genom. 2015;16:1073. doi: 10.1186/s12864-015-2292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschenbach J.R., Kristensen N.B., Donkin S.S., Hammon H.M., Penner G.B. Gluconeogenesis in dairy cows: the secret of making sweet milk from sour dough. IUBMB Life. 2010;62:869–877. doi: 10.1002/iub.400. [DOI] [PubMed] [Google Scholar]
- Bar-Peled L., Chantranupong L., Cherniack A.D., Chen W.W., Ottina K.A., Grabiner B.C., et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L., Schweitzer L.D., Zoncu R., Sabatini D.M. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- Bhalla K., Liu W.-J., Thompson K., Anders L., Devarakonda S., Dewi R., et al. Cyclin D1 represses gluconeogenesis via inhibition of the transcriptional coactivator PGC1α. Diabetes. 2014;63:3266–3278. doi: 10.2337/db13-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bionaz M., Osorio J., Loor J.J. Triennial LACTATION SYMPOSIUM: Nutrigenomics in dairy cows: nutrients, transcription factors, and techniques. J Anim Sci. 2015;93:5531–5553. doi: 10.2527/jas.2015-9192. [DOI] [PubMed] [Google Scholar]
- Cant J.P., Trout D.R., Qiao F., Purdie N.G. Milk synthetic response of the bovine mammary gland to an increase in the local concentration of arterial glucose. J Dairy Sci. 2002;85:494–503. doi: 10.3168/jds.S0022-0302(02)74100-3. [DOI] [PubMed] [Google Scholar]
- Chantranupong L., Sabatini D.M. Cell biology: the TORC1 pathway to protein destruction. Nature. 2016;536:155–156. doi: 10.1038/nature18919. [DOI] [PubMed] [Google Scholar]
- Chantranupong L., Wolfson R.L., Orozco J.M., Saxton R.A., Scaria S.M., Bar-Peled L., et al. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep. 2014;9:1–8. doi: 10.1016/j.celrep.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhou X. Research progress of mTOR inhibitors. Eur J Med Chem. 2020;208:112820. doi: 10.1016/j.ejmech.2020.112820. [DOI] [PubMed] [Google Scholar]
- Cunningham J.T., Rodgers J.T., Arlow D.H., Vazquez F., Mootha V.K., Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- Deng L., Chen L., Zhao L., Xu Y., Peng X., Wang X., et al. Ubiquitination of Rheb governs growth factor-induced mTORC1 activation. Cell Res. 2019;29:136–150. doi: 10.1038/s41422-018-0120-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Jiang C., Chen L., Jin J., Wei J., Zhao L., et al. The ubiquitination of rag A GTPase by RNF152 negatively regulates mTORC1 activation. Mol Cell. 2015;58:804–818. doi: 10.1016/j.molcel.2015.03.033. [DOI] [PubMed] [Google Scholar]
- Dibble C.C., Cantley L.C. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015;25:545–555. doi: 10.1016/j.tcb.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkin S.S., Armentano L.E. Insulin and glucagon regulation of gluconeogenesis in preruminating and ruminating bovine. J Anim Sci. 1995;73:546–551. doi: 10.2527/1995.732546x. [DOI] [PubMed] [Google Scholar]
- Gao S.T., Girma D.D., Bionaz M., Ma L., Bu D.P. Hepatic transcriptomic adaptation from prepartum to postpartum in dairy cows. J Dairy Sci. 2021;104:1053–1072. doi: 10.3168/jds.2020-19101. [DOI] [PubMed] [Google Scholar]
- Greenfield R.B., Cecava M.J., Donkin S.S. Changes in mRNA expression for gluconeogenic enzymes in liver of dairy cattle during the transition to lactation. J Dairy Sci. 2000;83:1228–1236. doi: 10.3168/jds.S0022-0302(00)74989-7. [DOI] [PubMed] [Google Scholar]
- Hostettler-Allen R.L., Tappy L., Blum J.W. Insulin resistance, hyperglycemia, and glucosuria in intensively milk-fed calves. J Anim Sci. 1994;72:160–173. doi: 10.2527/1994.721160x. [DOI] [PubMed] [Google Scholar]
- Hu X., Guo F. Amino acid sensing in metabolic homeostasis and health. Endocr Rev. 2020;42:56–76. doi: 10.1210/endrev/bnaa026. [DOI] [PubMed] [Google Scholar]
- Hu Y., Shin D.-J., Pan H., Lin Z., Dreyfuss J.M., Camargo F.D., et al. YAP suppresses gluconeogenic gene expression through PGC1α. Hepatology. 2017;66:2029–2041. doi: 10.1002/hep.29373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K., Li Y., Xu T., Guan K.L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Sarbassov D.D., Ali S.M., King J.E., Latek R.R., Erdjument-Bromage H., et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Sarbassov D.D., Ali S.M., Latek R.R., Guntur K.V., Erdjument-Bromage H., et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- Larsen M., Kristensen N.B. Effect of abomasal glucose infusion on splanchnic amino acid metabolism in periparturient dairy cows. J Dairy Sci. 2009;92:3306–3318. doi: 10.3168/jds.2008-1889. [DOI] [PubMed] [Google Scholar]
- Lebigot E., Brassier A., Zater M., Imanci D., Feillet F., Therond P., et al. Fructose 1,6-bisphosphatase deficiency: clinical, biochemical and genetic features in French patients. J Inherit Metab Dis. 2015;38:881–887. doi: 10.1007/s10545-014-9804-6. [DOI] [PubMed] [Google Scholar]
- Lemon P.W., Nagle F.J. Effects of exercise on protein and amino acid metabolism. Med Sci Sports Exerc. 1981;13:141–149. [PubMed] [Google Scholar]
- Liu G.Y., Sabatini D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol. 2020;21:183–203. doi: 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Li H., Li S., Shen M., Xiao N., Chen Y., et al. The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J Biol Chem. 2010;285:18858–18867. doi: 10.1074/jbc.M109.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Peterson T.R., Laplante M., Thoreen C.C., Sancak Y., Kang S.A., Kuehl W.M., et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P., Rhee J., Donovan J., Walkey C.J., Yoon J.C., Oriente F., et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- Reynolds C.K., Aikman P.C., Lupoli B., Humphries D.J., Beever D.E. Splanchnic metabolism of dairy cows during the transition from late gestation through early lactation. J Dairy Sci. 2003;86:1201–1217. doi: 10.3168/jds.S0022-0302(03)73704-7. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I., Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci. 2006;31:342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., Sabatini D.M. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R.A., Knockenhauer K.E., Wolfson R.L., Chantranupong L., Pacold M.E., Wang T., et al. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science. 2016;351:53–58. doi: 10.1126/science.aad2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaff C., Borner S., Hacke S., Kautzsch U., Albrecht D., Hammon H.M., et al. Increased anaplerosis, TCA cycling, and oxidative phosphorylation in the liver of dairy cows with intensive body fat mobilization during early lactation. J Proteome Res. 2012;11:5503–5514. doi: 10.1021/pr300732n. [DOI] [PubMed] [Google Scholar]
- Shen K., Huang R.K., Brignole E.J., Condon K.J., Valenstein M.L., Chantranupong L., et al. Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes. Nature. 2018;556:64–69. doi: 10.1038/nature26158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Sabunciyan S., Yang G., Florea L. A multi-sample approach increases the accuracy of transcript assembly. Nat Commun. 2019;10:5000. doi: 10.1038/s41467-019-12990-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Li N., Gu J., Fu S., Peng Z., Zhao C., et al. beta-Hydroxybutyrate induces bovine hepatocyte apoptosis via an ROS-p38 signaling pathway. J Dairy Sci. 2016;99:9184–9198. doi: 10.3168/jds.2016-11219. [DOI] [PubMed] [Google Scholar]
- Tee A.R., Manning B.D., Roux P.P., Cantley L.C., Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- Vander Haar E., Lee S.I., Bandhakavi S., Griffin T.J., Kim D.H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Wang J., Chen L., Li D., Yin Y., Wang X., Li P., et al. Intrauterine growth restriction affects the proteomes of the small intestine, liver, and skeletal muscle in newborn pigs. J Nutr. 2008;138:60–66. doi: 10.1093/jn/138.1.60. [DOI] [PubMed] [Google Scholar]
- Wang X., Huang N., Yang M., Wei D., Tai H., Han X., et al. FTO is required for myogenesis by positively regulating mTOR-PGC-1α pathway-mediated mitochondria biogenesis. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermeier F., Holyoak T., Asenjo J.L., Gatica R., Nualart F., Burbulis I., et al. Gluconeogenic enzymes in beta-cells: Pharmacological targets for improving Insulin secretion. Trends Endocrinol Metab. 2019;30:520–531. doi: 10.1016/j.tem.2019.05.004. [DOI] [PubMed] [Google Scholar]
- Woerle H.J., Meyer C., Dostou J.M., Gosmanov N.R., Islam N., Popa E., et al. Pathways for glucose disposal after meal ingestion in humans. Am J Physiol Endocrinol Metab. 2003;284:E716–E725. doi: 10.1152/ajpendo.00365.2002. [DOI] [PubMed] [Google Scholar]
- Wolfson R.L., Chantranupong L., Saxton R.A., Shen K., Scaria S.M., Cantor J.R., et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351:43–48. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson R.L., Chantranupong L., Wyant G.A., Gu X., Orozco J.M., Shen K., et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature. 2017;543:438–442. doi: 10.1038/nature21423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson R.L., Sabatini D.M. The dawn of the age of amino acid sensors for the mTORC1 pathway. Cell Metab. 2017;26:301–309. doi: 10.1016/j.cmet.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J.C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- Young J.W. Gluconeogenesis in cattle: significance and methodology. J Dairy Sci. 1977;60:1–15. doi: 10.3168/jds.S0022-0302(77)83821-6. [DOI] [PubMed] [Google Scholar]
- Zebeli Q., Ghareeb K., Humer E., Metzler-Zebeli B.U., Besenfelder U. Nutrition, rumen health and inflammation in the transition period and their role on overall health and fertility in dairy cows. Res Vet Sci. 2015;103:126–136. doi: 10.1016/j.rvsc.2015.09.020. [DOI] [PubMed] [Google Scholar]
- Zhan K., Yang T.Y., Chen Y., Jiang M.C., Zhao G.Q. Propionate enhances the expression of key genes involved in the gluconeogenic pathway in bovine intestinal epithelial cells. J Dairy Sci. 2020;103:5514–5524. doi: 10.3168/jds.2019-17309. [DOI] [PubMed] [Google Scholar]
- Zhang J., Shi H., Li S., Cao Z., Yang H., Wang Y. Integrative hepatic metabolomics and proteomics reveal insights into the mechanism of different feed efficiency with high or low dietary forage levels in Holstein heifers. J Proteomics. 2019;194:1–13. doi: 10.1016/j.jprot.2018.12.026. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Koser S.L., Donkin S.S. Propionate induces mRNA expression of gluconeogenic genes in bovine calf hepatocytes. J Dairy Sci. 2016;99:3908–3915. doi: 10.3168/jds.2015-10312. [DOI] [PubMed] [Google Scholar]
- Zhao F.Q., Keating A.F. Expression and regulation of glucose transporters in the bovine mammary gland. J Dairy Sci. 2007;90(Suppl 1):E76–E86. doi: 10.3168/jds.2006-470. [DOI] [PubMed] [Google Scholar]