Abstract
Background:
The circulating tumor DNA (ctDNA) diagnostic accuracy for detecting phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) mutations in breast cancer (BC) is under discussion. We aimed to compare plasma and tissue PIK3CA alterations, encompassing factors that could affect the results.
Methods:
Two reviewers selected studies from different databases until December 2020. We considered BC patients with matched tumor tissue and plasma ctDNA. We performed meta-regression and subgroup analyses to explore sources of heterogeneity concerning tumor burden, diagnostic technique, sample size, sampling time, biological subtype, and hotspot mutation. Pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and the related area under the curve (AUC) were elaborated for the overall population and each subgroup.
Results:
The pooled analysis was carried out on 25 cohorts for a total of 1966 patients. The overall ctDNA sensitivity and specificity were 0.73 (95% CI: 0.70–0.77) and 0.87 (95% CI: 0.85–0.89). The AUC was 0.93. Pooled concordance, negative predictive value and positive predictive value values were 0.87 (95% CI: 0.82–0.92), 0.86 (95% CI: 0.81–0.90), and 0.89 (95% CI: 0.81–0.95) with pooled PLR, NLR, and DOR of 7.94 (95% CI: 4.90–12.86), 0.33 (95% CI: 0.25–0.45), and 33.41 (95% CI: 17.23–64.79), respectively. The pooled results consistently favored next-generation sequencing (NGS)- over polymerase chain reaction-based methodologies. The best ctDNA performance in terms of sensitivity, specificity, and AUC (0.85, 0.99, and 0.94, respectively) was observed in the low-time sampling subgroup (⩽18 days between tissue and plasma collection). Meta-regression and subgroup analyses highlighted sampling time as a possible major cause of heterogeneity.
Conclusions:
These findings reliably estimate the high ctDNA accuracy for the detection of PIK3CA mutations. A ctDNA-first approach for the assessment of PIK3CA mutational status by NGS may accurately replace tissue tumor sampling, representing the preferable strategy at diagnosis of metastatic BC in patients who present with visceral involvement and at least two metastatic lesions, primarily given low clinical compliance or inaccessible metastatic sites.
Keywords: breast cancer, ctDNA, diagnostic accuracy, meta-analysis, PIK3CA
Background
The onset of somatic activating mutations of the phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) has been associated with acquired resistance to endocrine therapy (ET) in ~40% of advanced hormone-positive (H+) HER2-negative (HER2− ) breast cancer (BC) cases.1–4 Encouraging results regarding the use of the PIK3CA inhibitor alpelisib with ET for relapsed or progressed BC patients have been reported, confirming the predictive role of PIK3CA mutations in this setting.5–7 Although tissue biopsy is considered the gold standard for prognostic and predictive information, a high concordance rate between tissue and liquid biopsy has been reported in different histotypes.8–12 Several studies demonstrated that the detection of PIK3CA mutations using circulating tumor DNA (ctDNA) might represent a reliable option to suggest a better tailored therapeutic strategy. 2 In this regard, the Food and Drug Administration (FDA) has approved the liquid biopsy-based FoundationOne Liquid CDx test (Foundation Medicine, Inc., Cambridge, Massachusetts) as a companion diagnostic for alpelisib.
Nonetheless, the ctDNA diagnostic accuracy in detecting PIK3CA mutations is under discussion while not broadly endorsed by all the regulatory agencies. 13 Therefore, we performed a systematic review of the literature and an individual patient data meta-analysis that comprised studies evaluating the ctDNA diagnostic accuracy for detecting PIK3CA mutations compared to reference tissue biopsy. We aimed to provide a comparative analysis between plasma and tissue, discussing the pre-analytical and analytical factors that could affect the results.
Methods
Search strategy and study selection
We performed a systematic review of the literature reports on paired tumor tissue and blood samples to estimate ctDNA diagnostic accuracy in evaluating the PIK3CA mutational status in BC patients. We reviewed studies published up to 31 December 2020 through Medline (PubMed), EMBASE databases, and Cochrane Library using the following terms: ‘breast cancer’, ‘BC’, ‘breast’, ‘phosphoinositide 3-kinase’, ‘PIK3CA’, ‘tissue’, ‘liquid’, ‘blood’ (Supplemental Figure 1). Furthermore, we examined abstracts presented at the American Society of Clinical Oncology, the European Society for Medical Oncology, and the San Antonio Breast Cancer Symposium meetings. We searched for unpublished data reported on https://www.clinicaltrials.gov. Restriction for human studies and the English language was applied. We selected records meeting the following inclusion criteria: (1) patients with a histologically confirmed diagnosis of either early (stages I/II/III) or advanced (stage IV) BC; (2) studies detecting PIK3CA pathogenic variants in tissue and plasma samples; and (3) studies testing PIK3CA mutations by plasma ctDNA analysis. Studies not matching the inclusion criteria and ongoing clinical trials were excluded from the analysis. Only plasma ctDNA data from mixed plasma/serum cohorts were considered. When a study encompassed various follow-ups, we picked up the most recent one. The search protocol was registered in the PROSPERO 2021 database with the code: CRD42020222096.
Data extraction and assessment of the quality of the included studies
Two authors (L.C. and V.G.) independently assessed data extraction and examination. Disagreements were solved by consulting a third author (A.G.). Information retrieved from the included studies comprised: first author name, year of publication, study design, number of patients, biological subtype, study treatment, tumor burden (stage, number of metastatic lesions, and visceral and non-visceral disease), site (primitive or metastasis), diagnostic technique [polymerase chain reaction (PCR), digital droplet PCR (ddPCR), beads, emulsions, amplification, and magnetics (BEAMing), and next-generation sequencing (NGS)] with the limit of detection and PI3K reference range, ctDNA mutant allele fraction (MAF), sampling time, number of true-positives (TPs), true-negatives (TNs), false-positives (FPs), and false-negatives (FNs) (Supplemental Tables 1–7). The meta-analysis was designed according to the PRISMA guidelines (Supplemental Figure 1).14–17 Two authors (L.C. and V.G.) separately assessed the qualitative and quantitative analysis of the studies according to the QUAlity of Diagnostic Accuracy Studies 2 (QUADAS-2) tool, 18 considering four domains: patient selection, index test, reference standard, and flow and timing. The risk of selective outcome reporting bias was investigated, and divergences were overcome by consensus.
Statistical analysis
We extracted data considering the evaluation of PIK3CA mutational status on tissue as the gold standard and on ctDNA as the experimental procedure (Supplemental Table 2). The following rates were calculated: sensitivity, specificity, concordance, negative predictive value (NPV), positive predictive value (PPV), positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and the respective 95% CI (Supplemental Table 6). The random effect DerSimonian Laird model, evaluating the variance between studies, was used to pool PLR, NLR, and DOR. 19 A summary receiver operating characteristics (sROC) curve and the area under the curve (AUC) calculation were elaborated. Meta-regression and differing subgroup analyses were performed to explore heterogeneity concerning disease stage, diagnostic technique, sample size, sampling time, biological subtype [H+/Her2− versus HER2-positive (HER2+)], and hotspot mutations (E542/545X versus H1047X). We considered the median days between tissue and plasma collection to divide patients into low- and high-time subgroups. Fagan’s nomogram was produced to identify the association between pre-test probability, likelihood ratio, and post-test probability. 20 Spearman’s rank correlation coefficient between sensitivity and 1-specificity logit evaluated the bias connected to the threshold effect. A p-value <0.05 was considered a significant bias produced by the threshold effect. A p-value of Cochran’s Q test <0.05 and an index of inconsistency (I2) >50% were considered associated with significant heterogeneity within and between studies, respectively. We used STATA software (StataCorp. Stata statistical software: release 15. College Station, TX: StataCorp LLC, 2017) 21 to investigate publication bias producing Deek’s plot for asymmetry. All analyses were performed using the MetaDisc statistical software (version 1.4). 22
Results
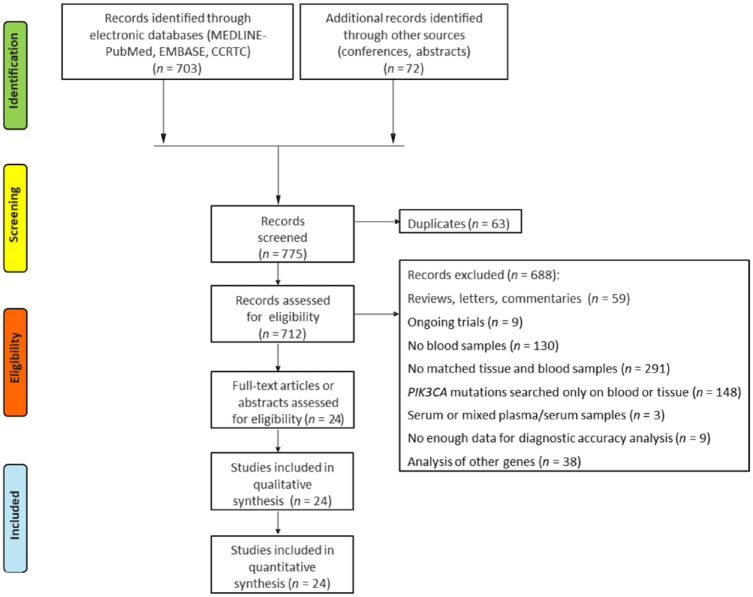
The systematic review of the literature provided 775 records. After screening and eligibility assessment, 24 studies met the inclusion criteria. Namely, one trial contained both prospective and retrospective cohorts: this was analyzed as two separate datasets. 23 The pooled analysis was finally carried out on 25 cohorts for a total of 1966 patients (Figure 1). The main features of selected studies are summarized in Table 1 and Supplemental Table 1.
Figure 1.
PRISMA flow diagram of the studies included in the quantitative synthesis.
Table 1.
Comparison of tissue versus ctDNA for detection of PIK3CA mutations according to technique and performance.
| Study | Sample | Methodology | Reference range (PIK3CA) | Mutation | Cross-tab analysis | Diagnostic accuracy | % | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Chung et al. 24 | Tissue | Hybrid capture-based NGS (Hi-Seq, Illumina) (Foundation Medicine) | NA | H1047L (1); H1047R (1); E545K (1) | Tissue + | Tissue − | Total | Sensitivity | 100 | |
| ctDNA+ | 3 | 0 | 3 | PPV | 100 | |||||
| Plasma | Hybrid capture-based NGS (Hi-Seq, Illumina) (FoundationACT ctDNA assay) | E542K; E545K; H1047R; H1047L | E545K (1); H1047L (1); H1047R (1); E726K (1) | ctDNA− | 0 | 11 | 11 | Specificity | 100 | |
| Total | 3 | 11 | 14 | NPV | 100 | |||||
| Concordance | 100 | |||||||||
| Baselga et al. 25 | Tissue | Sanger Sequencing | R88Q R93Q/W K111E/N G118D E365K C420R E542K E545G/K Q546K H1047R/K/Y |
NA | Tissue + | Tissue − | Total | Sensitivity | 71.2 | |
| CtDNA+ | 99 | 64 | 163 | PPV | 60.7 | |||||
| Plasma | BEAMing | NA | ctDNA− | 40 | 243 | 283 | Specificity | 79.2 | ||
| Total | 139 | 307 | 446 | NPV | 85.9 | |||||
| Concordance | 76.7 | |||||||||
| Chae et al. 26 | Tissue | NGS (Guardant360 and FoundationOne testing) | NA (indel/point mutation) | NA | Tissue + | Tissue − | Total | Sensitivity | 25 | |
| ctDNA+ | 3 | 2 | 5 | PPV | 60 | |||||
| Plasma | NA | ctDNA− | 9 | 31 | 40 | Specificity | 93.9 | |||
| Total | 12 | 33 | 45 | NPV | 77.5 | |||||
| Concordance | 75.6 | |||||||||
| Board et al. 27 | Tissue | RT-PCR (ARMS primers/Scorpion probes) | E542K; E545K; H1047R; H1047L | E542K (3); E545K (9); H1047R (10); H1047L (2) | Tissue + | Tissue − | Total | Sensitivity | 33.3 | |
| ctDNA+ | 8 | 1 | 9 | PPV | 88.9 | |||||
| Plasma | E542K (1); E545K (6); H1047R (4); H1047L (2) | ctDNA− | 16 | 46 | 62 | Specificity | 97.9 | |||
| Total | 24 | 47 | 71 | NPV | 74.2 | |||||
| Concordance | 76.1 | |||||||||
| Dawson et al. 28 | Tissue | NGS (HiSeq, llumina) (paired-end sequencing) | Selected regions (TAm-Seq) | E545K (6); H1047L (1); H1047R (4); E545K+ H1047R(1) | Tissue + | Tissue − | Total | Sensitivity | 100 | |
| ctDNA+ | 12 | 0 | 12 | PPV | 100 | |||||
| Plasma | dPCR; NGS (HiSeq, llumina) (paired-end sequencing) | NA; selected regions (TAm-Seq) | Exon 10 (6); Exon 21 (5); Exon 10 + Exon 21 (1) | ctDNA− | 0 | 18 | 18 | Specificity | 100 | |
| Total | 12 | 18 | 30 | NPV | 100 | |||||
| Concordance | 100 | |||||||||
| Higgins et al. 23 (p) | Tissue | PCR [Pyromark Q24 (Qiagen)], BEAMing (Inostics GmbH) | E542K; E545K; H1047R | E542K (2); E545K (2); H1047R (10) | Tissue + | Tissue − | Total | Sensitivity | 57.1 | |
| ctDNA+ | 8 | 8 | 16 | PPV | 50 | |||||
| Plasma | BEAMing (Inostics GmbH) | E542K; E545K; H1047R | E542K (3); E545K (2); H1047R (10); E545K + H1047R (2) | ctDNA− | 6 | 29 | 35 | Specificity | 78.4 | |
| Total | 14 | 37 | 51 | NPV | 82.9 | |||||
| Concordance | 72.5 | |||||||||
| Higgins et al. 23 (r) | Tissue | BEAMing | E545K; H1047R; H1047L | E545K (3); H1047R (10); H1047L (1) | Tissue + | Tissue − | Total | Sensitivity | 100 | |
| ctDNA+ | 14 | 0 | 14 | PPV | 100 | |||||
| Plasma | ctDNA− | 0 | 35 | 35 | Specificity | 100 | ||||
| Total | 14 | 35 | 49 | NPV | 100 | |||||
| Concordance | 100 | |||||||||
| Rothe et al. 29 | Tissue | NGS (Ion Torrent; Illumina) | NA (Ion AmpliSeq™ Cancer Hotspot Panel v2) | H1047R (1) H1047L (3) E453K (2) |
Tissue + | Tissue − | Total | Sensitivity | 75 | |
| ctDNA+ | 3 | 1 | 4 | PPV | 75 | |||||
| Plasma | H1047R (1) E453K (3) H1047L (1) |
ctDNA− | 1 | 12 | 13 | Specificity | 92.3 | |||
| Total | 4 | 13 | 17 | NPV | 92,3 | |||||
| Concordance | 88.2 | |||||||||
| García-Saenz et al. 30 | Tissue | RT-PCR (COBAS® PIK3CA Mutation Test; TaqMan assays on the QuantStudio® 3D Digital PCR System); ABI 3130 genetic analyzer | R88Q; N345K; C420R; E542K; E545X (E545A, E545D, E545G, and E545K); Q546X (Q546E, Q546K, Q546L, and Q546R); M1043I; H1047X (H1047L, H1047R, and H1047Y); G1049R | E542K (4); E545K (5); H1047R (11) | Tissue + | Tissue − | Total | Sensitivity | 100 | |
| ctDNA+ | 11 | 0 | 11 | PPV | 100 | |||||
| Plasma | dPCR (QuantStudio® 3D Digital PCR System) | E542K; E545K; H1047R | E542K (2); E545K (4); H1047R (5) | ctDNA− | 9 | 29 | 38 | Specificity | 100 | |
| Total | 20 | 29 | 49 | NPV | 76.3 | |||||
| Concordance | 81.6 | |||||||||
| Shatsky et al. 31 | Tissue | NGS (The FoundationOne genomic assay) | NA | H1047R (3); E542K (1); E545K (2); Q75E (1) | Tissue + | Tissue − | Total | Sensitivity | 77.8 | |
| ctDNA+ | 7 | 1 | 8 | PPV | 87.5 | |||||
| Plasma | NGS (The Guardant 360 assay) | NA | H1047R (5); Amplification (2); R108H (1); E542K(1); E545K (4); H1047Q + E545K (1); E81K (1); E453K (1) | ctDNA− | 2 | 28 | 30 | Specificity | 96.6 | |
| Total | 9 | 29 | 38 | NPV | 93.3 | |||||
| Concordance | 92.1 | |||||||||
| Spoerke et al. 32 | Tissue | RT-PCR BEAMing (OncoBEAM BC1 BEAMing Digital PCR panel) |
C420R, E542K, E545A/G/K, and H1047L/R/Y | H1047R (16); H1047R (8); H1047R + E545K (1); H1047L + E542K + E545K (1) | Tissue + | Tissue − | Total | Sensitivity | 78.1 | |
| ctDNA+ | 50 | 7 | 57 | PPV | 87.7 | |||||
| Plasma | C420R, E542K, E545K/G, Q546K, M1043I, and H1047R/L/Y | ctDNA− | 14 | 71 | 85 | Specificity | 91 | |||
| Total | 64 | 78 | 142 | NPV | 83.5 | |||||
| Concordance | 80.5 | |||||||||
| Tzanikou et al. 33 | Tissue | ddPCR | E545K; H1047R | E545K (2); H1047R (1); E545K + H1047R (7) | Tissue + | Tissue − | Total | Sensitivity | 38.5 | |
| ctDNA+ | 5 | 2 | 7 | PPV | 71.4 | |||||
| Plasma | E545K (1); E545K + H1047R (5) | ctDNA− | 8 | 1 | 9 | Specificity | 33.3 | |||
| Total | 13 | 3 | 16 | NPV | 11.1 | |||||
| Concordance | 37.7 | |||||||||
| Bianchini et al. 34 | Tissue | NGS (AmpliSeq HD, Oncomine Pan-Cancer) | NA | NA | Tissue + | Tissue − | Total | Sensitivity | 46.6 | |
| ctDNA+ | 27 | 2 | 29 | PPV | 93 | |||||
| Plasma | ctDNA− | 31 | 84 | 115 | Specificity | 97.7 | ||||
| Total | 58 | 86 | 144 | NPV | 73 | |||||
| Concordance | 77.1 | |||||||||
| Oliveira et al. 35 | Tissue | Amplicon NGS based (MiSeq Illumina) | NA (59 cancer panel genes) | NA | Tissue + | Tissue − | Total | Sensitivity | 76.2 | |
| ctDNA+ | 16 | 0 | 16 | PPV | 100 | |||||
| Plasma | Amplicon NGS based (MiSeq Illumina) | NA | NA | ctDNA− | 5 | 1 | 6 | Specificity | 100 | |
| Total | 21 | 1 | 22 | NPV | 16.7 | |||||
| Concordance | 77.3 | |||||||||
| Di Leo et al. 36 | Tissue | COBAS | NA (PIK3CA assay covering exons 7, 9, and 20) | NA | Tissue + | Tissue − | Total | Sensitivity | 80.5 | |
| ctDNA+ | 70 | 21 | 91 | PPV | 76.9 | |||||
| Plasma | BEAMing | E542K E545G/KQ546K M1043I H1047L/R/Y |
NA | ctDNA− | 17 | 142 | 159 | Specificity | 87.1 | |
| Total | 87 | 163 | 250 | NPV | 89.3 | |||||
| Concordance | 84,8 | |||||||||
| Blackwell et al. 37 | Tissue | Hybridization-captured NGS based | Foundation Medicine, Inc. | N345 K (2), C420R (2) E542 K (2), E545 K (1), Q546 K (1) H1047R (10), H1047L (2) | Tissue + | Tissue − | Total | Sensitivity | 94.4 | |
| ctDNA+ | 17 | 1 | 18 | PPV | 94.4 | |||||
| Plasma | BEAMing | E542K, E545K, E545G, Q546K; M1043I; H1047L; c.3139C>T_p.H1047R; c.3140A>G_p.H1047R | N345 K (1), C420R (1) E542 K (2), E545 K (1), Q546 K (1) H1047R (10), H1047L (2) | ctDNA− | 1 | 12 | 13 | Specificity | 92.3 | |
| Total | 18 | 13 | 31 | NPV | 92.3 | |||||
| Concordance | 93,5 | |||||||||
| Moynahan et al. 38 | Tissue | NGS (HiSeq, Illumina) | NA | NA | Tissue + | Tissue − | Total | Sensitivity | 73.3 | |
| ctDNA+ | 63 | 50 | 113 | PPV | 55.8 | |||||
| Plasma | ddPCR | E542K; E545K; H1047R | E542K (39); E545K (61); H1047R (138): multiplea: (4) aThree E545K/E542 |
ctDNA− | 23 | 111 | 134 | Specificity | 68.9 | |
| Total | 86 | 161 | 247 | NPV | 82.9 | |||||
| Concordance | 70.4 | |||||||||
| Moreno et al. 39 | Tissue | NGS (Ion Torrent; Illumina) | NA (a customized panel of 54 genes) | H1047R (7) A511T (1) V344G (1) Va46G (1) A668C (1) G106V (1) T462A (1) G451_D54del (1) C420R (1) |
Tissue + | Tissue − | Total | Sensitivity | 72.7 | |
| CtDNA+ | 8 | 0 | 8 | PPV | 100 | |||||
| Plasma | NGS (Ion Torrent; Illumina) | NA (a customized panel of 54 genes) | H1047R (7) A511T (1) V344G (1) V346G (1) A668C (1) G106V (1) T462A (1) C420R (1) |
ctDNA− | 3 | 27 | 30 | Specificity | 100 | |
| Total | 11 | 27 | 38 | NPV | 90 | |||||
| Concordance | 92.1 | |||||||||
| Takano et al. 40 | Tissue | ddPCR | E542K, E545K, H1047R | E542K (2); E545K (1); H1047R (10) | Tissue + | Tissue − | Total | Sensitivity | 60 | |
| ctDNA+ | 6 | 0 | PPV | 100 | ||||||
| Plasma | H1047R (8) | ctDNA− | 4 | 16 | 20 | PPV | 100 | |||
| Total | 10 | 16 | 26 | Specificity | 100 | |||||
| NPV | 80 | |||||||||
| Concordance | 84.7 | |||||||||
| Slembrouck et al. 41 | Tissue | NGS | NA | E542K (1); E545K (6); H1047R (1); No hotspot mutation (12) | Tissue + | Tissue − | Total | Sensitivity | 100 | |
| ctDNA+ | 8 | 0 | 8 | PPV | 100 | |||||
| Plasma | ddPCR | E542K, E545K, H1047R, H1047L | E542K (3); E545K (6); H1047R (1); No hotspot mutation (12) | ctDNA− | 0 | 12 | 12 | Specificity | 100 | |
| Total | 8 | 12 | 20 | NPV | 100 | |||||
| Concordance | 100 | |||||||||
| Rudolph et al. 42 | Tissue | NGS | Mutations, deletions, amplifications (FoundationOne® T5a panel) | NA | Tissue + | Tissue − | Total | Sensitivity | 100 | |
| ctDNA+ | 13 | 0 | 13 | PPV | 100 | |||||
| Plasma | BEAMing | E542K, E545K, H1047R, H1047L | NA | ctDNA− | 0 | 37 | 37 | Specificity | 100 | |
| Total | 13 | 37 | 50 | NPV | 100 | |||||
| Concordance | 100 | |||||||||
| Perkins et al. 43 | Tissue | PCR; MALDI-TOF (OncoCarta panel) | NA | H1047R (4) | Tissue + | Tissue − | Total | Sensitivity | 75 | |
| ctDNA + | 3 | 0 | 3 | PPV | 100 | |||||
| Plasma | MALDI-TOF (OncoCarta panel) | NA | H1047R (3) | ctDNA − | 1 | 15 | 16 | Specificity | 100 | |
| Total | 4 | 15 | 19 | NPV | 93.8 | |||||
| Concordance | 100 | |||||||||
| Ma et al. 44 | Tissue | NGS | NA | NA | Tissue + | Tissue − | Total | Sensitivity | 50 | |
| ctDNA+ | 3 | 0 | 3 | PPV | 100 | |||||
| Plasma | ctDNA− | 3 | 6 | 9 | Specificity | 100 | ||||
| Total | 6 | 6 | 12 | NPV | 66.7 | |||||
| Concordance | 75 | |||||||||
| Kim et al. 45 | Tissue | RT-PCR | C420R; E542K; E545A/G/K; H1047L/R/Y |
Tissue + |
Tissue - |
Total | Sensitivity | 100 | ||
| ctDNA+ | 54 | 0 | 54 | PPV | 100 | |||||
| Plasma | RT-PCR; NGS (Foundat ion One) | R88Q; N345K; C420R; E542X; E545X; Q546X; M1043I; H1047X; G1049R; AKT1 | E542K (2); G1049R (1); H1047L (2); H1047R (10); M1043I (1); N345K (1); Q546K (1) | ctDNA− | 0 | 18 | 18 | Specificity | 100 | |
| Total | 54 | 18 | 72 | NPV | 100 | |||||
| Concordance | 100 | |||||||||
| Beaver et al. 46 | Tissue | Sanger Sequencing ddPCR (Custom TaqMan probes) | E545K; H1047R | E545K (3); H1047R (7) E545K (4); H1047R (10) |
Sensitivity | 92,9 | ||||
| ctDNA+ | 13 | 0 | 13 | PPV | 100 | |||||
| Plasma | ctDNA− | 1 | 15 | 16 | Specificity | 100 | ||||
| Total | 14 | 15 | 29 | NPV | 93,8 | |||||
| Concordance | 96,6 | |||||||||
ARMS, amplification-refractory mutation system; BEAMing, beads, emulsions, amplification, and magnetics; ctDNA, circulating tumor DNA; ddPCR, digital droplet polymerase chain reaction; dPCR, digital polymerase chain reaction; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; NA, not available; NGS, next-generation sequencing; NPV, negative predictive value; PCR, polymerase chain reaction; PIK3CA, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha; PPV, positive predictive value; RT-PCR, real-time polymerase chain reaction.
Overall diagnostic accuracy analysis
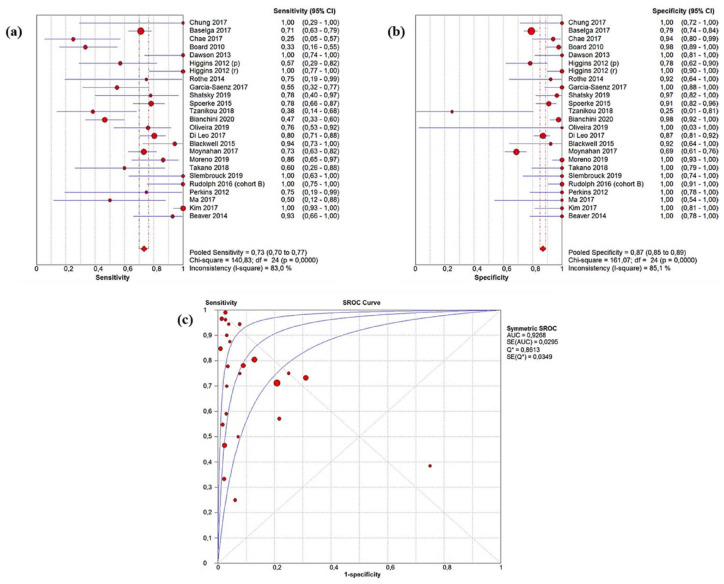
Across the included studies, sensitivity ranged from 25 to 100%, specificity from 69 to 100%, and concordance from 37 to 100% with lower rates being associated with early BC. 33 The pooled ctDNA sensitivity and specificity of ctDNA were 0.73 (95% CI: 0.70–0.77) and 0.87 (95% CI: 0.85–0.89) (Figure 2(a) and (b)). The AUC resulting from the sROC curve was 0.93 (Figure 2(c)). According to Youden’s index, the best pooled cut-off able to minimize the FP was 0.6. 47 We obtained pooled concordance, NPV, and PPV equal to 0.87 (95% CI: 0.82–0.92), 0.86 (95% CI: 0.81–0.90), and 0.89 (95% CI: 0.81–0.95), respectively. Pooled PLR, NLR, and DOR were 7.94 (95% CI: 4.90–12.86), 0.33 (95% CI: 0.25–0.45), and 33.41 (95% CI: 17.23–64.79) (Table 2). Assuming a pre-test probability of 37%, Fagan’s plot showed that detecting a ctDNA PIK3CA mutation would raise the post-test chance to diagnose a tissue PIK3CA mutation to 77%, whereas the missed identification would decrease the post-test probability to 15% (Supplemental Figure 2).
Figure 2.
Pooled ctDNA sensitivity (a), specificity (b), and sROC curve related to the overall population (c).
ctDNA, circulating tumor DNA; sROC, summary receiver operating characteristics.
Table 2.
Meta-analysis results.
| No of patients | Sensitivity (95% CI) | Specificity (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) | AUC | |
|---|---|---|---|---|---|---|---|
| Overall | 1966 | 0.73 (0.70–0.77) | 0.87 (0.85–0.89) | 7.94 (4.90–12.86) | 0.33 (0.25–0.45) | 33.41 (17.23–64.79) | 0.93 |
| Tumor burden | |||||||
| Early | 55 | 0.76 (0.57–0.90) | 1.00 (0.87–1.00) | 8.47 (0.97–73.91) | 0.21 (0.02–2.55) | 45.17 (1.13–1810.10) | 1.00 |
| Advanced | 1836 | 0.77 (0.73–0.80) | 0.86 (0.84–0.88) | 8.16 (4.98–13.37) | 0.29 (0.22–0.39) | 40.53 (20.32–80.82) | 0.92 |
| Sample size | |||||||
| Low | 274 | 0.78 (0.70–0.85) | 0.96 (0.91–0.98) | 10.6 (2.5–45.9) | 0.27 (0.15–0.46) | 48.4 (11.38–205.9) | 0.90 |
| High | 1698 | 0.72 (0.68–0.75) | 0.85 (0.83–0.87) | 7.2 (4.2–12.3) | 0.36 (0.25–0.51) | 27.11 (12.75–57.6) | 0.87 |
| Diagnostic technique | |||||||
| NGS | 307 | 0.83 (0.75–0.89) | 0.98 (0.94–0.99) | 11.65 (5.43–24.99) | 0.23 (0.09–0.62) | 59.80 (14.29–250.23) | 0.98 |
| ddPCR/BEAMing | 1485 | 0.74 (0.70–0.78) | 0.84 (0.82–0.86) | 6.63 (3.97–11.08) | 0.31 (0.22–0.43) | 28.84 (13.45–61.86) | 0.92 |
| PCR | 174 | 0.51 (0.39–0.64) | 0.96 (0.91–0.99) | 9.30 (0.64–136.17) | 0.54 (0.31–0.96) | 20.61 (1.57–270.46) | 0.77 |
| Sampling time | |||||||
| Low-time | 219 | 0.85 (0.75–0.92) | 0.99 (0.96–1.00) | 16.24 (6.23–42.31) | 0.21 (0.1–0.47) | 101.50 (23.22–443.62) | 0.94 |
| High-time | 679 | 0.66 (0.59–0.73) | 0.83 (0.80–0.87) | 4.63 (2.46–8.73) | 0.47 (0.31–0.70) | 11.81 (5.15–27.10) | 0.89 |
| Biological subtype | |||||||
| H+/HER2− | 1357 | 0.73 (0.69–0.77) | 0.83 (0.80–0.86) | 5.97 (3.58–10.00) | 0.32 (0.24–0.45) | 22.94 (11.18–47.07) | 0.87 |
| HER2+ | 52 | 0.57 (0.35–0.77) | 1.00 (0.88–1.00) | 5.65 (1.69–18.95) | 0.55 (0.37–0.82) | 14.94 (3.00–74.54) | 0.86 |
| Hotspot mutation | |||||||
| E542/545X | 421 | 0.70 (0.58–0.81) | 0.95 (0.92–0.97) | 8.74 (3.47–22.02) | 0.36 (0.16–0.82) | 29.65 (7.55–116.41) | 0.88 |
| H1047X | 520 | 0.74 (0.65–0.82) | 0.98 (0.96–0.99) | 18.57 (6.19–55.72) | 0.30 (0.17–0.54) | 83.38 (17.64–394.06) | 0.93 |
AUC = area under the curve; BEAMing = beads, emulsions, amplification, and magnetics; CI, confidence interval; ddPCR = digital droplet polymerase chain reaction; DOR, diagnostic odds ratio HER2 = human epidermal growth factor receptor 2; HR = hormone receptor; NGS, next-generation sequencing; NLR, negative likelihood ratio; PLR, positive likelihood ratio.
Quality analysis and publication bias
Based on the QUADAS-2 results, the records were overall affected by a low risk of bias, increasing the strength of scientific evidence of the study. Only one study (Perkins et al. 43 ) presented a high risk of bias in the patient selection task since the authors did not include patients tested with negative tissue results (Supplemental Figure 3). The presence of publication bias was explored through Deek’s funnel plot, showing a potential risk (p = 0.04) (Supplemental Figure 2).
Threshold effect and heterogeneity
Spearman’s rank correlation coefficient was −0.276 (p-value = 0.181), thus not significantly associated with bias. Considering the positive publication bias, we performed meta-regression and subgroup analysis to explore sources of heterogeneity not linked to the threshold effect. The meta-regression demonstrated that sampling time was significantly associated with heterogeneity (Supplemental Table 1b).
Subgroup analysis
Furthermore, as a means of investigating heterogeneous results while answering specific clinical questions, we split participant data into subgroups according to tumor burden, sample size, diagnostic technique, sampling time, biological subtype, and hotspot mutation (Table 2).
Tumor burden
Extracting data from cohorts singly evaluating different disease stages, 4 and 23 cohorts were finally assigned to early and advanced subgroups for a total of 55 and 1836 patients, respectively (Supplemental Table 3).23–46 Regarding the advanced setting, we observed an AUC of 0.92, which showed an excellent discrimination ability between mutated and wild-type patients (Supplemental Figure 4 and Table 2). Furthermore, even if not evaluated in terms of diagnostic accuracy due to missing data, we investigated both the disease distribution and the number of metastatic lesions from nine and eight cohorts, respectively.23,25,28–30,32,34–36,38,43,44 Most of the examined population had a visceral involvement and at least two metastatic lesions (Supplemental Table 5). Likewise, we found comparable pooled diagnostic values for the early subgroup, even if arising from a very limited sample size (Supplemental Figure 4 and Table 2). We observed lower absolute sensitivity rates in the earlier stages, 26 however, showing similar pooled diagnostic values compared to the advanced setting (Table 2).
Sample size
According to the median number of included patients (45 individuals), 12 and 13 studies were collected in the low- and high-size subgroups, showing the highest ctDNA performance in low-size studies according to the diagnostic values (Supplemental Figure 7a and b). Noteworthy, smaller studies added compelling insights in terms of pooled specificity and DOR compared to the heterogeneity of larger samples (0.96 and 40.42 versus 0.85 and 27.11, respectively) (Supplemental Figure 4).
Diagnostic technique
The most used techniques were ddPCR/BEAMing (12 cohorts, 1485 patients), followed by NGS (9 cohorts, 307 patients) and PCR (5 cohorts, 174 patients) (Supplemental Table 3). The ctDNA PIK3CA MAF was reported as the median and/or media of all mutated cases or calculated by extracting data from supplementary (7/25 studies) (Supplemental Table 7).24,28,29,31,32,39,46 Namely, NGS seemed to outperform ddPCR/BEAMing and PCR in terms of sensitivity (0.83 versus 0.74 and 0.51, respectively) (Supplemental Figure 6 and Table 2). The ddPCR/BEAMing subgroup reported a lower pooled specificity (0.84) than NGS (0.98) and PCR (0.96). Furthermore, NGS outclassed PCR-based assays in terms of detection sensitivity, specificity, and AUC (0.98), not eventually leading to heterogeneity for specificity (Supplemental Figure 6a) while showing compelling PLR, NLR, and DOR rates that favored NGS over PCR-based methodologies (Table 2).
Sampling time
Among 20 studies, tissue biopsies were mainly performed on the primary site, with four studies carrying out tissue biopsies on metastatic lesions (Supplemental Table 5). According to data available for 13 cohorts, the time between tissue and plasma sampling was variable, ranging from 0 day to over 15 years23–26,29–31,35,39,43,44,46 (Supplemental Table 7d). Patients were assigned into low- and high-time subgroups, respectively (⩽ and >18 days), according to the median time between tissue and plasma collection. The best ctDNA performance in terms of sensitivity, specificity, and AUC (0.85, 0.99, and 0.94, respectively) was observed in the low-time subgroup, showing compelling findings for PLR, NLR, and DOR rates (16.24, 0.21, and 101.50, respectively) with acceptable heterogeneity (Supplemental Figure 7 and Table 2).
Biological subtype
The H+/HER2− and HER2+ subgroups were included in 5 and 10 studies (Supplemental Table 7)25,32,34,36–38,40,44,46 with very few data being available on triple-negative BCs.28,29,45 We found a comparable ctDNA performance for AUC (0.87 and 0.86, respectively) and other diagnostic rates, however observing higher ctDNA sensitivity favoring the H+/HER2− over the HER2+ subgroup (0.73 versus 0.57, respectively) (Supplemental Figure 7 and Table 2).
Hotspot mutation
Considering the most involved PIK3CA mutations within exons 9 and 20, 12 and 10 studies were pooled for the H1047X and E542/545X subgroups (520 and 421 patients, respectively) (Supplemental Table 4).48–58 Specifically, ctDNA assays revealed a slightly more accurate trend in detecting H1047X than E542/545X in terms of sensitivity, specificity, and AUC (0.74, 0.98, and 0.93 versus 0.70, 0.95, and 0.88, respectively) (Supplemental Figure 7c–d and Table 2).
Discussion
In BC clinical practice, the tissue from primary lesions is typically available for diagnosis and biomarker testing in the basal setting. On the other hand, re-biopsies to obtain metastatic specimens of adequate quality and quantity may not always be feasible due to the location of the metastatic sites or patients’ comorbidities. A growing body of evidence demonstrated that ctDNA represents a promising tool for predicting response to targeted treatment in solid tumors.11,59 The choice of tumor tissue or liquid genotyping should be individualized in the clinical setting based on patient and disease characteristics, primarily considering that a reflex tumor tissue biopsy, if feasible, should be performed in the case of a ctDNA negative result to prevent FN results. With regard to BC, BELLE-2, BELLE-3, and SOLAR-1 were the first trials to include a survival analysis in ctDNA PIK3CA-positive patients. In this scenario, however, there is a lack of well-established data on sensitivity and specificity rates and concordance with tissue genotyping.
This individual patient data meta-analysis aimed to outline the diagnostic accuracy of ctDNA in evaluating the PIK3CA mutational status compared to tissue biopsy. Zhou et al. 60 have previously reported pooled optimal values of diagnostic performance of plasma ctDNA for prediction of PIK3CA mutation for sensitivity (0.86), specificity (0.98), AUC (0.99), PLR (42.8), and NLR (0.14). However, these results should be cautiously interpreted for the small sample size (247 patients from seven publications). 60 We found a highly accurate ctDNA performance in terms of sensitivity, specificity, and concordance with tissue testing from a larger sample size. The AUC curve supported these findings. Translating these overall pooled results in the clinic, the three-quarters of patients with a PIK3CA-positive tissue biopsy would test positive on ctDNA while only failing to be detected on plasma in the remaining cases. Furthermore, as shown by the NLR in Fagan’s plot, a negative result of PIK3CA on plasma would lead to a three-fold decreased risk of finding a positive PI3KCA mutation on tissue. Nonetheless, the wide variability of the selected population in terms of several clinical, methodological, and technical conditions must be considered. While the meta-regression technique highlighted the sampling time as the main reason for heterogeneity, stratified subgroup analyses were performed to investigate the impact of specific variables on the diagnostic accuracy performance. Our meta-analysis, including more than 1800 patients with advanced PIK3CA-positive BC, provided a reliable estimation of the high ctDNA diagnostic accuracy in the metastatic setting, showing an AUC > 0.9, which is considered very accurate in clinical practice. We observed that most patients presented with visceral involvement and at least two metastatic lesions, thus including those tumors shedding high enough ctDNA that would eventually avoid FN results. However, albeit showing comparable diagnostic values in early-stage BC, the controversial influence of PIK3CA mutations on survival outcomes in this subset of patients should be considered. In this regard, the scarce sample size (55 patients) along with the lower sensitivity rates critically affected the clinical utility of ctDNA which is to date already limited in early-stage BC, requiring further studies in the adjuvant setting before drawing any conclusions.
Considering the molecular diagnostic techniques, these pooled results consistently favored NGS over PCR-based methodologies. Overall, we found that NGS panels covered a broader spectrum of PI3KCA mutations, far beyond the FDA-approved detection of 11 activating mutations. These results were consistent with the exploratory analysis of the SOLAR-1 trial, revealing the ability of NGS testing to detect 60 different mutations across multiple exons and select PI3KCA-mutated patients who also benefited from alpelisib.61,62 Considering the FDA-approved therascreen® RGQ PCR Kit (QIAGEN GmbH) ability to detect only hotspot mutations across three exons together with the eventual risk of generating FP results as highlighted by the ongoing market surveillance process, these findings would support the implementation of broader NGS panels either on tissue or plasma to screen for uncommon PIK3CA activating mutations that, however, remain to be further validated in clinical trials. Regarding the sampling time, remarkably, identifying a ctDNA PIK3CA mutation within 18 days from the tissue sampling would suggest a highly accurate concordance with histological genotyping, supporting the reliable use of a plasma-first approach that would likely allow overcoming the issue of intra-tumor heterogeneity. Referring to biological subtypes and common PIK3CA hotspot mutations, the ctDNA comparable performance between subgroups advised a similar impact on clinical decisions, even if the difference in both magnitude and different detection methods must be considered. Indeed, most of the patients were H+/Her2− and tested with PCR-based methodologies. Despite thoroughly encompassing all the publicly available data for detecting ctDNA PIK3CA mutations, some limitations of this meta-analysis should be considered. First, some of the included studies had missing data, affecting subgroup analyses. Second, our pooled results came from retrospective and prospective trials with different design conceptions that did neither aim to directly evaluate the prognostic/predictive role of PI3KCA mutations nor the correlation between the clearance of PI3KCA mutated allelic frequency and the radiologic response, although emerging data seemed to further validate the dynamic role of PI3KCA detected on ctDNA in the real-time longitudinal monitoring of BC. 63 Third, as partially discussed above, the heterogeneity of analyzed studies, including different disease stages and distribution, dissimilar sample sizes, the different prevalence of testing platforms, and timing for tissue and plasma sample collection, could have negatively affected the overall results. Notwithstanding, electronic databases, meeting proceedings, and other sources of gray literature research guarantee the systematicity of the literature review suggesting the high heterogeneity of the included studies is responsible for bias. Interestingly, subgroup analyses and meta-regression highlighted the sampling time as a possible cause of heterogeneity, reflecting the wide range between tissue and plasma sampling (0 and 15 years). Such heterogeneity should not affect the overall results, stating the ctDNA clinical utility for the PIK3CA mutational status evaluation.
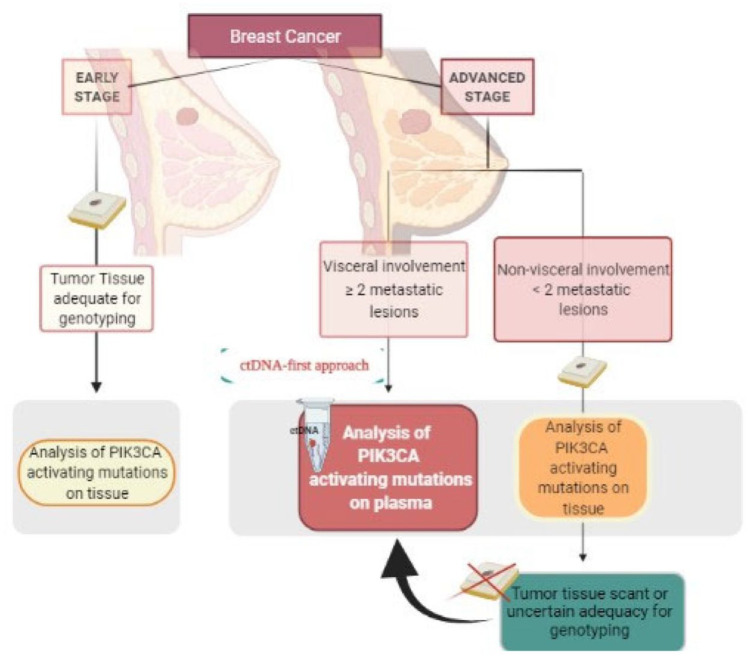
In conclusion, these findings reliably estimate the ctDNA accuracy for detecting PIK3CA mutations, validating the role of liquid biopsy in the management of advanced BC. Considering the highest ctDNA accuracy in the metastatic setting, using highly sensitive NGS panels and when plasma is evaluated within 18 days from the tissue sampling, a ctDNA-first approach for the assessment of PIK3CA mutational status by NGS may accurately replace tissue tumor sampling, representing the preferable strategy at diagnosis of metastatic BC in patients who present with visceral involvement and at least two metastatic lesions, primarily given low clinical compliance or inaccessible metastatic sites (Figure 3). Larger clinical trials are warranted to further define the clinical utility of ctDNA accuracy for the detection of PIK3CA mutations in the early-stage BC setting.
Figure 3.
Algorithm depicting the role of ctDNA for the assessment of PIK3CA mutations in BC patients.
BC, breast cancer; ctDNA, circulating tumor DNA; PIK3CA, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha.
Supplemental Material
Supplemental material, sj-docx-1-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-10-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-11-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-12-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-13-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-14-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-2-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-3-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-4-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-5-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-6-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-7-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-8-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-9-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Acknowledgments
V.G. and M.L. contributed to the current work under the Doctoral Program in Experimental Oncology and Surgery, University of Palermo. The authors thank Dr. Chiara Drago for the English language revision.
Footnotes
ORCID iDs: Lorena Incorvaia  https://orcid.org/0000-0002-1199-7286
https://orcid.org/0000-0002-1199-7286
Antonio Russo  https://orcid.org/0000-0002-4370-2008
https://orcid.org/0000-0002-4370-2008
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Antonio Galvano, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Luisa Castellana, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Valerio Gristina, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Maria La Mantia, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Lavinia Insalaco, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Nadia Barraco, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Alessandro Perez, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Sofia Cutaia, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Valentina Calò, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Tancredi Didier Bazan Russo, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Edoardo Francini, Department of Experimental and Clinical Medicine, University of Florence, Florence, Italy.
Lorena Incorvaia, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Mario Giuseppe Mirisola, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Salvatore Vieni, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Christian Rolfo, Center for Thoracic Oncology, Tisch Cancer Institute, Mount Sinai Medical System & Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Viviana Bazan, Department of Experimental Biomedicine and Clinical Neurosciences, School of Medicine, University of Palermo, Palermo, Italy.
Antonio Russo, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Via del Vespro 129, Palermo 90127, Italy.
Declarations
Ethics approval and consent to participate: Not Applicable.
Consent for publication: Not Applicable.
Author contribution(s): Antonio Galvano: Conceptualization; Data curation; Formal analysis; Methodology; Software; Supervision; Writing – review & editing.
Luisa Castellana: Conceptualization; Data curation; Formal analysis; Investigation; Writing – original draft.
Valerio Gristina: Data curation; Project administration; Supervision; Validation; Visualization; Writing – original draft; Writing – review & editing.
Maria La Mantia: Data curation; Visualization; Writing – review & editing.
Lavinia Insalaco: Validation.
Nadia Barraco: Resources; Supervision.
Alessandro Perez: Validation; Visualization.
Sofia Cutaia: Investigation.
Valentina Calò: Data curation.
Tancredi Didier Bazan Russo: Validation.
Edoardo Francini: Data curation; Project administration; Supervision; Validation; Visualization; Writing – review & editing.
Lorena Incorvaia: Resources.
Mario Giuseppe Mirisola: Validation; Visualization.
Salvatore Vieni: Validation; Visualization.
Christian Rolfo: Project administration; Resources; Software; Supervision; Validation; Visualization.
Viviana Bazan: Project administration; Resources; Software; Supervision; Validation; Visualization.
Antonio Russo: Project administration; Resources; Software; Supervision; Validation; Visualization.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
Competing interests: C.R. is a speaker for Merck Sharp and Dohme, AstraZeneca and has research collaborations with Guardant Health; advisory board activity: Archer, Inivata and MD Serono, Novartis, and BMS; non-financial support from Guardant Health; and research grant from LCRF-Pfizer. A.R. reported personal fees from Bristol, Pfizer, Bayer, Kyowa Kirin, Ambrosetti for advisory board activity; speaker honorarium from Roche Diagnostics. The remaining authors declare no potential conflicts of interest.
Availability of data and materials: The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1. Araki K, Miyoshi Y. Mechanism of resistance to endocrine therapy in breast cancer: the important role of PI3K/Akt/mTOR in estrogen receptor-positive, HER2-negative breast cancer. Breast Cancer 2018; 25: 392–401. [DOI] [PubMed] [Google Scholar]
- 2. Del Re M, Crucitta S, Lorenzini G, et al. PI3K mutations detected in liquid biopsy are associated to reduced sensitivity to CDK4/6 inhibitors in metastatic breast cancer patients. Pharmacol Res 2021; 163: 105241. [DOI] [PubMed] [Google Scholar]
- 3. Ma CX, Crowder RJ, Ellis MJ. Importance of PI3-kinase pathway in response/resistance to aromatase inhibitors. Steroids 2011; 76: 750–752. [DOI] [PubMed] [Google Scholar]
- 4. Martínez-Sáez O, Chic N, Pascual T, et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res 2020; 22: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. FDA approves alpelisib for metastatic breast cancer | FDA [Internet], https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-alpelisib-metastatic-breast-cancer (accessed 27 April 2021).
- 6. Rugo HS, Lerebours F, Ciruelos E, et al. Alpelisib plus fulvestrant in PIK3CA-mutated, hormone receptor-positive advanced breast cancer after a CDK4/6 inhibitor (BYLieve): one cohort of a phase 2, multicentre, open-label, non-comparative study. Lancet Oncol 2021; 22: 489–498. [DOI] [PubMed] [Google Scholar]
- 7. Study finds alpelisib effective after CDK4/6 inhibition in advanced breast cancer | The ASCO Post [Internet], https://ascopost.com/issues/june-25-2020/study-finds-alpelisib-effective-after-cdk46-inhibition-in-advanced (accessed 27 April 2021).
- 8. Passiglia F, Rizzo S, Di Maio M, et al. The diagnostic accuracy of circulating tumor DNA for the detection of EGFR-T790M mutation in NSCLC: a systematic review and meta-analysis. Sci Rep 2018; 8: 13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galvano A, Taverna S, Badalamenti G, et al. Detection of RAS mutations in circulating tumor DNA: a new weapon in an old war against colorectal cancer: a systematic review of literature and meta-analysis. Ther Adv Med Oncol 2019; 11: 1758835919874653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liang DH, Ensor JE, Liu Z-B, et al. Cell-free DNA as a molecular tool for monitoring disease progression and response to therapy in breast cancer patients. Breast Cancer Res Treat 2016; 155: 139–149. [DOI] [PubMed] [Google Scholar]
- 11. Nacchio M, Sgariglia R, Gristina V, et al. KRAS mutations testing in non-small cell lung cancer: the role of liquid biopsy in the basal setting. J Thorac Dis 2020; 12: 3836–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pisapia P, Pepe F, Gristina V, et al. A narrative review on the implementation of liquid biopsy as a diagnostic tool in thoracic tumors during the COVID-19 pandemic. Mediastinum 2021; 5: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Russo A, Incorvaia L, Del Re M, et al. The molecular profiling of solid tumors by liquid biopsy: a position paper of the AIOM-SIAPEC-IAP-SIBioC-SIC-SIF Italian Scientific Societies. ESMO Open 2021; 6: 100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moher D, Liberati A, Tetzlaff J, et al.; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med 2009; 151: 264–W64. [DOI] [PubMed] [Google Scholar]
- 15. Moher D, Shamseer L, Clarke M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev 2015; 4: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mantia ML, Koyyala VPB. The war against coronavirus disease 19 through the eyes of cancer physician: an Italian and Indian young medical oncologist’s perspective. Indian J Med Paediatr Oncol 2020; 41: 305–307. [Google Scholar]
- 17. Passiglia F, Galvano A, Gristina V, et al. Is there any place for PD-1/CTLA-4 inhibitors combination in the first-line treatment of advanced NSCLC? A trial-level meta-analysis in PD-L1 selected subgroups. Transl Lung Cancer Res 2021; 10: 3106–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011; 155: 529–536. [DOI] [PubMed] [Google Scholar]
- 19. Borenstein M, Hedges LV, Higgins JPT, et al. Introduction to meta-analysis [Internet], www.wiley.com (2009, accessed 27 April 2021).
- 20. Fagan TJ. Letter: nomogram for Bayes theorem. New Engl J Med 1975; 293: 257. [DOI] [PubMed] [Google Scholar]
- 21. StataCorp. Stata statistical software: release 15. College Station, TX: StataCorp LLC., 2017. [Google Scholar]
- 22. Zamora J, Abraira V, Muriel A, et al. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol 2006; 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Higgins MJ, Jelovac D, Barnathan E, et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res 2012; 18: 3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chung JH, Pavlick D, Hartmaier R, et al. Hybrid capture-based genomic profiling of circulating tumor DNA from patients with estrogen receptor-positive metastatic breast cancer. Ann Oncol 2017; 28: 2866–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baselga J, Im SA, Iwata H, et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2017; 18: 904–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chae YK, Davis AA, Jain S, et al. Concordance of genomic alterations by next-generation sequencing in tumor tissue versus circulating tumor DNA in breast cancer. Mol Cancer Ther 2017; 16: 1412–1420. [DOI] [PubMed] [Google Scholar]
- 27. Board RE, Wardley AM, Dixon JM, et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat 2010; 120: 461–467. [DOI] [PubMed] [Google Scholar]
- 28. Dawson S-J, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. New Engl J Med 2013; 368: 1199–1209. [DOI] [PubMed] [Google Scholar]
- 29. Rothé F, Laes JF, Lambrechts D, et al. Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann Oncol 2014; 25: 1959–1965. [DOI] [PubMed] [Google Scholar]
- 30. García-Saenz JA, Ayllón P, Laig M, et al. Tumor burden monitoring using cell-free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging. BMC Cancer 2017; 17: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shatsky R, Parker BA, Bui NQ, et al. Next-generation sequencing of tissue and circulating tumor DNA: the UC San Diego moores center for personalized cancer therapy experience with breast malignancies. Mol Cancer Ther 2019; 18: 1001–1011. [DOI] [PubMed] [Google Scholar]
- 32. Spoerke JM, Gendreau S, Walter K, et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun 2016; 7: 11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tzanikou E, Markou A, Politaki E, et al. PIK3CA hotspot mutations in circulating tumor cells and paired circulating tumor DNA in breast cancer: a direct comparison study. Mol Oncol 2019; 13: 2515–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bianchini G, De Laurentiis M, Arpino G, et al. 11P BioItaLEE: Comparative biomarker analysis of liquid biopsies and paired tissue samples of patients treated with ribociclib and letrozole as first-line therapy for advanced breast cancer (aBC). Ann Oncol 2020; 31: S20. [Google Scholar]
- 35. Oliveira M, Ruiz-Pace F, Matito J, et al. Determinants of concordance in clinically relevant genes (CRG) from synchronously acquired tumor biopsies (tBx) and ctDNA in metastatic breast cancer (MBC). J Clin Oncol 2019; 37: 1075–1075. [Google Scholar]
- 36. Di Leo A, Johnston S, Lee KS, et al. Buparlisib plus fulvestrant in postmenopausal women with hormone-receptor-positive, HER2-negative, advanced breast cancer progressing on or after mTOR inhibition (BELLE-3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2018; 19: 87–100. [DOI] [PubMed] [Google Scholar]
- 37. Blackwell K, Burris H, Gomez P, et al. Phase I/II dose-escalation study of PI3K inhibitors pilaralisib or voxtalisib in combination with letrozole in patients with hormone-receptor-positive and HER2-negative metastatic breast cancer refractory to a non-steroidal aromatase inhibitor. Breast Cancer Res Treat 2015; 154: 287–297. [DOI] [PubMed] [Google Scholar]
- 38. Moynahan ME, Chen D, He W, et al. Correlation between PIK3CA mutations in cell-free DNA and everolimus efficacy in HR(+), HER2(−) advanced breast cancer: results from BOLERO-2. Br J Cancer 2017; 116: 726–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moreno F, Gayarre J, López-Tarruella S, et al. Concordance of genomic variants in matched primary breast cancer, metastatic tumor, and circulating tumor DNA: the MIRROR study. JCO Precis Oncol 2019; 3: 1–16. [DOI] [PubMed] [Google Scholar]
- 40. Takano T, Tsurutani J, Takahashi M, et al. A randomized phase II trial of trastuzumab plus capecitabine versus lapatinib plus capecitabine in patients with HER2-positive metastatic breast cancer previously treated with trastuzumab and taxanes: WJOG6110B/ELTOP. Breast 2018; 40: 67–75. [DOI] [PubMed] [Google Scholar]
- 41. Slembrouck L, Renders D, Borght SV, et al. Abstract P5-06-28: Optimization and validation of PIK3CA mutation detection with droplet digital PCR in liquid biopsies of patients with metastatic breast cancer. Cancer Res 2020; 80: P5-06-28. [Google Scholar]
- 42. Rudolph M, Anzeneder T, Schulz A, et al. AKT1 (E17K) mutation profiling in breast cancer: prevalence, concurrent oncogenic alterations, and blood-based detection. BMC Cancer 2016; 16: 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Perkins G, Yap TA, Pope L, et al. Multi-purpose utility of circulating plasma DNA testing in patients with advanced cancers. PLoS One 2012; 7: e47020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ma F, Li Q, Chen S, et al. Phase I study and biomarker analysis of pyrotinib, a novel irreversible Pan-ErbB receptor tyrosine kinase inhibitor, in patients with human epidermal growth factor receptor 2–Positive metastatic breast cancer. J Clin Oncol 2017; 35: 3105–3112. [DOI] [PubMed] [Google Scholar]
- 45. Kim S-B, Dent R, Im S-A, et al. Ipatasertib plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer (LOTUS): a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol 2017; 18: 1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Beaver JA, Jelovac D, Balukrishna S, et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res 2014; 20: 2643–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Youden WJ. Index for rating diagnostic tests. Cancer 1950; 3: 32–35. [DOI] [PubMed] [Google Scholar]
- 48. Saura C, Oliveira M, Feng Y-H, et al. Neratinib plus capecitabine versus lapatinib plus capecitabine in HER2-Positive metastatic breast cancer previously treated with ⩾2 HER2-Directed regimens: phase III NALA trial. J Clin Oncol 2020; 38: 3138–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gray R, Bhattacharya S, Bowden C, et al. Independent review of E2100: a phase III trial of bevacizumab plus paclitaxel versus paclitaxel in women with metastatic breast cancer. J Clin Oncol 2009; 27: 4966–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Slamon DJ, Neven P, Chia S, et al. Overall survival with ribociclib plus fulvestrant in advanced breast cancer. New Engl J Med 2020; 382: 514–524. [DOI] [PubMed] [Google Scholar]
- 51. Litton JK, Scoggins ME, Hess KR, et al. Neoadjuvant talazoparib for patients with operable breast cancer with a germline BRCA pathogenic variant. J Clin Oncol 2020; 38: 388–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Olaparib as adjuvant treatment in patients with germline BRCA mutated high risk HER2 negative primary breast cancer – Full Text View – ClinicalTrials.gov [Internet], https://clinicaltrials.gov/ct2/show/NCT02032823 (accessed 15 Sptember 2020).
- 53. Adjuvant treatment for high-risk triple negative breast cancer patients with the anti-PD-l1 antibody avelumab – Full Text View – ClinicalTrials.gov [Internet], https://clinicaltrials.gov/ct2/show/NCT02926196 (accessed 15 September 2020).
- 54. Patel HK, Bihani T. Selective estrogen receptor modulators (SERMs) and selective estrogen receptor degraders (SERDs) in cancer treatment. Pharmacol Ther 2018; 186: 1–24. [DOI] [PubMed] [Google Scholar]
- 55. Pagani O, Francis PA, Fleming GF, et al. Absolute improvements in freedom from distant recurrence to tailor adjuvant endocrine therapies for premenopausal women: results from TEXT and SOFT. J Clin Oncol 2020; 38: 1293–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cardoso F, Senkus E, Costa A, et al. 4th ESO–ESMO international consensus guidelines for advanced breast cancer (ABC 4). Ann Oncol 2018; 29: 1634–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Augereau P, Patsouris A, Bourbouloux E, et al. Hormonoresistance in advanced breast cancer: a new revolution in endocrine therapy. Ther Adv Med Oncol 2017; 9: 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Turner NC, Slamon DJ, Ro J, et al. Overall survival with palbociclib and fulvestrant in advanced breast cancer. New Engl J Med 2018; 379: 1926–1936. [DOI] [PubMed] [Google Scholar]
- 59. Massihnia D, Galvano A, Fanale D, et al. Triple negative breast cancer: shedding light onto the role of pi3k/akt/mtor pathway. Oncotarget 2016; 7: 60712–60722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhou Y, Wang C, Zhu H, et al. Diagnostic accuracy of PIK3CA mutation detection by circulating free DNA in breast cancer: a meta-analysis of diagnostic test accuracy. PLoS One 2016; 11: e0158143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Juric D, Andre F, Singer CF, et al. Abstract P4-10-04: clinical outcomes of alpelisib in hormone receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer by next-generation sequencing-detected PIK3CA alteration status and phosphatase and tensin homolog loss: biomarker analysis from the SOLAR-1 study. Cancer Res 2020; 80: P4-10-04. [Google Scholar]
- 62. Pisapia P, Pepe F, Baggi A, et al. Next generation diagnostic algorithm in non-small cell lung cancer predictive molecular pathology: the KWAY Italian multicenter cost evaluation study. Crit Rev Oncol Hematol 2022; 169: 103525. [DOI] [PubMed] [Google Scholar]
- 63. Kodahl AR, Ehmsen S, Pallisgaard N, et al. Correlation between circulating cell-free PIK3CA tumor DNA levels and treatment response in patients with PIK3CA-mutated metastatic breast cancer. Mol Oncol 2018; 12: 925–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-10-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-11-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-12-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-13-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-14-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-2-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-3-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-4-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-5-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-6-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-7-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-8-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-9-tam-10.1177_17588359221110162 for The diagnostic accuracy of PIK3CA mutations by circulating tumor DNA in breast cancer: an individual patient data meta-analysis by Antonio Galvano, Luisa Castellana, Valerio Gristina, Maria La Mantia, Lavinia Insalaco, Nadia Barraco, Alessandro Perez, Sofia Cutaia, Valentina Calò, Tancredi Didier Bazan Russo, Edoardo Francini, Lorena Incorvaia, Mario Giuseppe Mirisola, Salvatore Vieni, Christian Rolfo, Viviana Bazan and Antonio Russo in Therapeutic Advances in Medical Oncology