Abstract
KRAS is the most commonly mutated oncogene in human cancers with limited therapeutic options, thus there is a critical need to identify novel targets and inhibiting agents. The 78-kDa glucose-regulated protein GRP78, which is upregulated in KRAS cancers, is an essential chaperone and the master regulator of the unfolded protein response (UPR). Following up on our recent discoveries that GRP78 haploinsufficiency suppresses both KRASG12D-driven pancreatic and lung tumorigenesis, we seek to determine the underlying mechanisms. Here, we report that knockdown of GRP78 via siRNA reduced oncogenic KRAS protein level in human lung, colon, and pancreatic cancer cells bearing various KRAS mutations. This effect was at the post-transcriptional level and is independent of proteasomal degradation or autophagy. Moreover, targeting GRP78 via small molecule inhibitors such as HA15 and YUM70 with anti-cancer activities while sparing normal cells significantly suppressed oncogenic KRAS expression in vitro and in vivo, associating with onset of apoptosis and loss of viability in cancer cells bearing various KRAS mutations. Collectively, our studies reveal that GRP78 is a previously unidentified regulator of oncogenic KRAS expression, and, as such, augments the other anti-cancer activities of GRP78 small molecule inhibitors to potentially achieve general, long-term suppression of mutant KRAS-driven tumorigenesis.
Keywords: GRP78, KRAS, Lung cancer, Colon cancer, Pancreatic cancer, Small molecule inhibitors
Abbreviations: KRAS, Kirsten rat sarcoma viral oncogene; ER, Endoplasmic Reticulum; GRP78, 78-kilodalton glucose-regulated protein; UPR, Unfolded Protein Response; PERK, protein kinase R(PKR)-like endoplasmic reticulum kinase; PDAC, Pancreatic ductal adenocarcinoma; LUAD, Lung adenocarcinoma; eIF2α, Eukaryotic translation initiation factor 2A; eIF4A, Eukaryotic initiation factor-4A; eIF5A, Eukaryotic translation initiation factor 5A; eIF4E, Eukaryotic translation initiation factor 4E; 4E-BP1, Eukaryotic translation initiation factor 4E-binding protein 1; 3-MA, 3-methyladenine
Introduction
The most frequently mutated oncogenic driver is Kirsten rat sarcoma viral oncogene homolog (KRAS), which accounts for about 15% to 30% of all human malignancies. KRAS mutations are particularly prevalent in pancreatic ductal adenocarcinoma, colorectal cancer and non-small cell lung cancer and have traditionally been regarded as “undruggable” [24]. Oncogenic KRAS alleles differ from the wild type by a single missense point mutation that results in an amino acid substitution typically at position 12,13 or 61, which impairs the rate of GTP hydrolysis and consequently de-regulates RAS signaling [23,32]. Attempts to target mutant KRAS or its downstream signaling have been mostly unsuccessful, largely due to the molecular structure of the KRAS protein as well as compensatory upregulation of alternative oncogenic pathways [8]. While the recent conditional approval of sortorasib which targets the KRAS G12C mutation represents a breakthrough for patients bearing this allele, there are multiple mutated forms of KRAS and with the likelihood of eventual development of resistance, it remains an urgent need to identify new therapeutic targets and agents to combat KRAS mutant cancers [16,22].
The 78-kDa glucose regulated protein (GRP78), also referred to as HSPA5/BiP, is a multifunctional protein that can impact a wide range of human diseases, including cancer, via diverse mechanisms [5,9,18,21,38]. GRP78 is traditionally regarded as a luminal endoplasmic reticulum (ER) chaperone with major functions in protein folding and maintenance of ER homeostasis as a key regulator of the unfolded protein response (UPR). However, under pathological stress, GRP78 can be relocated to compartments outside the ER, including the cell surface, the cytosol, the mitochondria, and the nucleus, and in some cell types, GRP78 can even be secreted and exert new effects on growth and signaling [13,18,36]. These discoveries change the paradigm on GRP78 functions, which continue to evolve and expand.
GRP78 upregulation is widely observed in aggressive tumors, including lung, pancreatic and colorectal cancer, and associates with poor prognosis [18]. Pancreatic ductal adenocarcinoma (PDAC) remains one of the deadliest diseases with limited therapeutic options, with KRAS mutated in more than 90% of PDAC [42]. Recently, we discovered that haploinsufficiency of a single cellular protein, GRP78, while having no harmful effect on the normal pancreas, is sufficient to suppress acinar-to-ductal metaplasia, ERK and AKT signaling and impede KRASG12D driven pancreatic tumorigenesis [30]. Non-small cell lung cancer accounts for the majority (85%) of all lung cancers and lung adenocarcinoma (LUAD) and is the most common type of lung cancer in the US. Activating mutants of KRAS are found in 25 to 50% of human LUAD cases [4]. Likewise, we observed that GRP78 haploinsufficiency can suppress KRASG12D-mediated lung tumor progression and prolong survival in mouse models of LUAD [25]. Furthermore, the same study showed that GRP78 knockdown in a human lung cancer cell line (A427) bearing the same KRAS G12D mutation led to activation of the UPR and apoptotic markers, which could represent one mechanism whereby GRP78 deficiency suppresses tumor growth. To explore other potential mechanisms, surprisingly we discovered that GRP78 knockdown also led to a loss of KRAS protein expression in the same cells, thus raising the intriguing question whether KRAS expression is regulated by GRP78. Here, we report that GRP78 knockdown reduces KRAS protein level in a panel of human LUAD, PDAC and colorectal cancer cell lines bearing various mutant KRAS alleles, and the regulation is at the post-transcriptional level. Recently, two specific small molecule inhibitors of GRP78 (HA15 and YUM70) have been identified which exhibit potent anti-cancer activities with minimal toxicity to normal cells and organs in cancer models [6,29]. Here we showed that these agents mimicked the effect of GRP78 knockdown and can suppress KRAS protein expression in vitro and in vivo, leading to loss of cancer cell viability. These findings reveal that targeting GRP78, in addition to activating the UPR, could also disrupt KRAS expression, thus providing a new mechanism for GRP78 inhibitors in combating KRAS-driven tumorigenesis.
Materials and methods
Cell culture
Human lung adenocarcinoma cell line A427 was cultured in Eagle's Minimum Essential Medium (EMEM) supplemented with 10% fetal bovine serum (FBS; GeminiBio, West Sacramento, CA) and 1% penicillin/streptomycin (Pen/Strep) (Corning Inc., Glendale, AZ). Human non-small cell lung carcinoma cell lines H460, H1299, H522, H1975 and normal human bronchial epithelial cell line BEAS-2B were cultured in RPMI-1640 medium supplemented with 10% FBS and 1% Pen/Strep. Human colorectal carcinoma cell lines HCT116, LS180, RKO, HT-29 and human pancreatic ductal carcinoma cell line PANC-1 and HEK-293T cell line were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS and 1% Pen/Strep. Human ductal pancreatic adenocarcinoma cell line CFPAC-1 was cultured in Iscove's Modified Dulbecco's Medium (IMDM) supplemented with 10% FBS and 1% Pen/Strep. All cell lines were cultured at 37°C in a humidified atmosphere of 95% air and 5% CO2. The A427 cell line was a gift from Dr. David Shackelford (UCLA), HCT116, LS180, RKO, and HT-29 cell lines were a gift from Dr. Chengyu Liang (The Wistar Institute), PANC-1 and CFPAC-1 cell lines were a gift from Dr. Ganging Liang (USC), H1299 cell line was a gift from Dr. Keigo Machida (USC), BEAS-2B and H522 cell lines were a gift from Dr. Ite Offringa (USC), H1975 cell line was a gift from Dr. Steven Dubinett (UCLA). The H460 cell line was purchased from the American Type Culture Collection (ATCC).
Transfection of siRNAs
The cells were seeded in triplicate on 6cm dishes at approximately 60-70% confluency and allowed to attached overnight. Next day, the cells were transfected with siRNAs targeting GRP78 (si78) or scrambled control (siCtrl) using the Lipofectamine RNAiMAX Transfection Reagent (ThermoFisher Scientific, Waltham, MA, Cat# 13778075) in accordance with manufacturer's recommendation for forward transfection. For reverse transfection, the cells were mixed with the transfection mix at the same time as seeding and the cells were incubated with the siRNA transfection mix for 48 hr before harvesting for analysis. The custom siRNAs for siCtrl, siGRP78, and siKRAS were purchased from GE Healthcare Dharmacon, Inc. (Chicago, IL) with the following sequences: siCtrl 5’- GAGAUCGUAUAGCAACGGU -3’, si78 5’- GGAGCGCAUUGAUACUAGA-3’ and siKRAS (ON-TARGETplus siRNA Cat# J-005069-08-0002). The custom siRNAs for HRAS and NRAS were purchased from Integrated DNA Technologies (IDT, Coralville, IA) with the following sequences: siHRAS 5’-UAAUUUACUGUGAUCCCAUCUG-3’, siNRAS 5’-UUCAGUUUCAUCUUUCUCCUGG-3’.
Cloning and production of lentivirus expressing inducible shRNA targeting GRP78
The lentiviral plasmid for inducible expression of shRNA Tet-pLKO-puro is a gift from Dr. Min Yu (USC). The Tet-pLKO-puro backbone was digested by EcoRI and AgeI (New England Biolabs, Ipswich, MA) and purified by agarose gel electrophoresis by ZymoClean Gel DNA Recovery Kit (Zymo Research, Irvine, CA, Cat#D4001). DNA oligos with sequences specifically targeting GRP78 were synthesized by Integrated DNA Technologies (Coralville, IA) and annealed in NEB Buffer 2. Annealed DNA oligos were ligated into the cut and purified Tet-pLKO-puro backbone above. The complete targeting vector was packaged into lentivirus by the Core Facilities at the School of Pharmacy of USC. H460 cells were transduced by lentivirus carrying shGRP78 and selected with 2μg/mL puromycin for 1 week. Then the transduced and selected cells were induced by 0.5-2μg/mL doxycycline for 48 hr. Cells were harvested for protein extraction and subsequent Western blot analysis. The shGRP78 targeting sequence is as follows: 5’-CTTGTTGGTGGCTCGACTCGA-3’.
Immunoblot analysis
Preparation of cell lysates and immunoblot analysis procedure have been described in detailed previously [14]. Total protein from whole cell lysates were electrophoresed in 10% or 12% SDS-PAGE gels, transferred to Supported Nitrocellulose membrane (Bio-Rad Laboratories, Cat#1620097, Hercules, CA) and probed with the following antibodies. The primary antibodies used in this study and their dilutions are as followed: mouse anti-BiP/GRP78 (1:1000, BD Biosciences, San Jose, CA, Cat#610979), mouse anti-KRAS (1:1000, Santa Cruz Biotechnology, Inc., Dallas, TX, sc-30), mouse anti-NRAS (1:1000, Santa Cruz Biotechnology, Inc., sc-31), rat anti-HRAS (1:1000, Santa Cruz Biotechnology, Inc., sc-35), mouse anti-GAPDH (1:1000, Santa Cruz Biotechnology, Inc., sc-32233), mouse anti-eIF4I/II (1:1000, Santa Cruz Biotechnology, Inc., sc-377315), mouse anti-eIF5A (1:1000, Santa Cruz Biotechnology, Inc., sc-390202), mouse anti-Sgk269/PEAK1 (1:1000, Santa Cruz Biotechnology, Inc., sc-100403), mouse anti-eIF4E (P2) (1:1000, Santa Cruz Biotechnology, Inc., sc-9976), mouse anti-4E-BP1 (P1) (1:1000, Santa Cruz Biotechnology, Inc., sc-9977), mouse anti-phospho-4E-BP1 (62.Ser 65) (1:1000, Santa Cruz Biotechnology, Inc., sc-293124), rabbit anti-peIF2α (1:1000, Cell Signaling, Danvers, MA, Cat#9721), rabbit anti-phospho-eIF4E (Ser209) (1:1000, Cell Signaling, Cat#9741), rabbit anti-Cleaved PARP (1:1000, Cell Signaling, Cat#5625), mouse anti-CHOP (L63F7) (1:1000, Cell Signaling, Cat#2895), rabbit anti-cleaved Caspase-7 (Asp198) (D6H1) (1:1000, Cell Signaling, Cat#8438), rabbit anti-PI3 Kinase p85 (1:1000, Cell Signaling, Cat#4292), rabbit anti-SRC (1:1000, Cell Signaling, Cat#2109). The secondary antibodies used in this study and their conditions are as followed: mouse IgG1 binding protein conjugated to HRP (1:1000, Santa Cruz Biotechnology, Inc., sc-525408), mouse anti-rabbit IgG conjugated to HRP (1:1000, Santa Cruz Biotechnology, Inc., sc-2357), goat anti-rat IgG HRP-conjugated (1:1000, Proteintech Group, Inc., Rosemont, IL, Cat#SA00001-15).
Treatment of small molecule inhibitors and chemical compounds
The design and synthesis of YUM70 has been previously described [29]. HA15 was purchased from MedChemExpress LLC (Monmouth Junction, NJ, Cat#HY-100437). 3-Methyladenine (3-MA) and Chloroquine was purchased from Millipore Sigma (Burlington, MA, Cat#M9281 and Cat#C6628). MG115 and MG132 was purchased from Cayman Chemical (Ann Arbor, MI, Cat#15413 and Cat#1002628). GSK2606414 was a gift from Dr. J. Alan Diehl (Case Western Reserve University). All compounds were dissolved in DMSO except for 3-MA and Chloroquine which were dissolved in sterile double distilled water. DMSO was used as vehicle control for all drug treatment experiments.
RNA extraction and reverse transcription quantitative real-time PCR
Detailed procedures for RNA extraction, reverse transcription and quantitative real-time PCR have been previously described [14]. The sequences for the primers used in this study are as followed: GRP78 5’- GTCAGGCGATTCTGGTCATT -3’ and 5’-GGTGAAAGACCCCTGACAAA-3’, GAPDH 5’- TGCACCACCAACTGCTTAGC -3’ and 5’- GGCATGGACTGTGGTCATGAG-3’, KRAS 5’-GACTGAATATAAACTTGTGGTAGTTGGA -3’ and 5’-CATATTCGTCCACAAAATGATTCTGA-3’.
WST-1 Cell viability assay
Cells were seeded in a 96-well plate at a density of 10,000 cells per well and allowed to attached overnight. Next day, the cells were either treated with DMSO vehicle control, HA15, or YUM70 at the indicated concentration and incubated for 48 hr. Cell viability was measured using the (4-[3-(4-Iodophenyl)-2-(4-nitro-phenyl)-2H-5-tetrazolio]-1,3-benzene sulfonate) WST-1 Cell Proliferation Assay Kit (Takara Bio USA, Inc., San Jose, CA, Cat#MK400) in accordance with manufacturer's recommendations. Colorimetric readout was detected at 450nm wavelength and quantified using a Model 680 Microplate Reader (Bio-Rad Laboratories, Hercules, CA).
Mouse xenograft tumor tissues
Human pancreatic cancer cell MIA PaCa-2 xenograft mouse model and treatment with YUM70 have been described previously [29]. All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee at the University of Michigan. Briefly, MIA PaCa-2 cells (2.0 × 106 in PBS, 100 µl) were inoculated subcutaneously into the dorsal flank of female NCr nude mice (Taconic Bioscience, NY, NY). YUM70 (25-30 mg/kg in 10% DMSO, 60% propylene glycol and 30% saline v/v, 100 µL) or vehicle control (solvent for YUM70) was administrated by intraperitoneal injection 5 days a week for 48 days. Tumor tissues were harvested at the end of treatment. To prepare tumor tissues for protein level analysis, tumor tissues were homogenized in cold RIPA buffer (50mM Tris-HCl, 150mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with Protease and Phosphatase inhibitor cocktail (ThermoFisher Scientific, Waltham, MA, Cat#78440). The homogenized lysates were incubated on ice for 30 minutes followed by centrifugation at 13,000 RPM for 15 minutes. The clarified supernatants containing soluble proteins were used in subsequent immunoblot.
Statistical Analysis
All pair-wise comparisons were analyzed by the two-tailed Student's t-test with unequal variance in Microsoft Excel. All graphs were presented as the mean ± Standard Deviation (S.D.). A P-value of ≤ 0.05 is signified by *, P-value of ≤ 0.01 by **, and P-value of ≤ 0.001 by ***, ns denotes not significant.
Results
Effect of GRP78 deficiency on KRAS protein levels in human cancer cell lines
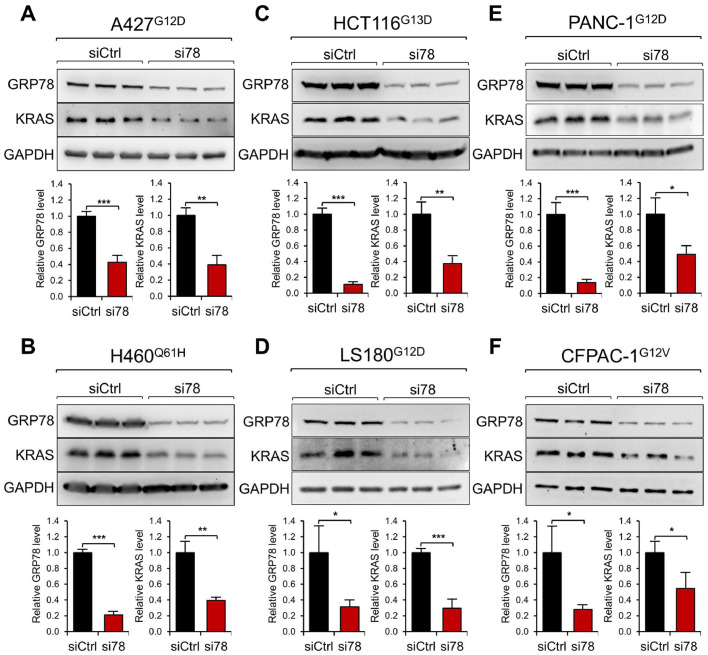
Lung (A427, H460), colon (HCT116, LS180), and pancreatic (PANC-1, CFPAC-1) cancer cell lines bearing various mutant KRAS alleles (Supp. Table S1) were transfected with scramble control siRNA (siCtrl) or siRNA targeting GRP78 (si78). Western blot analysis of the cell lysates harvested 48 hr after siRNA treatment revealed that while GRP78 protein levels were reduced by 60-90% by si78 as expected, significant reductions in KRAS protein levels in all six cancer cell lines ranging from 40-70% in the si78-treated cells were detected (Fig. 1A-F). Validation of the anti-Ras antibodies used in this study was performed through the use of siRNA molecules specifically targeting each Ras protein, correlating with reduction of the specific RAS protein band in Western blot (Suppl. Fig. S1A). Further, using a doxycycline-inducible shRNA targeting a different coding region of GRP78, we observed dose-dependent decrease in GRP78 accompanied by a concomitant decrease in KRAS protein level in H460 cells (Supp. Fig. S1B). In contrast, knockdown of GRP78 in a normal human bronchial epithelial cell line (BEAS-2B) bearing WT KRAS alleles yielded no reduction in KRAS protein level, which was efficiently knockdown by siRNA against KRAS as expected (Supp. Fig. S2A). Knockdown of GRP78 in lung (H522, H1975) and colon (RKO, HT-29) cancer cell lines harboring only wild-type KRAS alleles also showed no effect on KRAS protein levels (Supp. Table S1, Supp. Fig. S2B, C). These results indicate that in a panel of pancreatic, lung and colon cancer cells being examined, GRP78 deficiency can reduce oncogenic KRAS protein level irrespective of the type of KRAS mutant alleles.
Fig. 1.
Knockdown of GRP78 via siRNA reduces KRAS protein level in human cancer cell lines with various KRAS mutations. Human lung cancer A427 (A) and H460 (B), colon cancer HCT116 (C) and LS180 (D), and pancreatic cancer PANC-1 (E) and CFPAC-1 (F) cells were used. The superscript indicates the KRAS mutation for each cell line. The cells were transfected with control siRNA (siCtrl) or siRNA targeting GRP78 (si78) for 48 hr. Whole cell lysate was subjected to Western blot analysis for GRP78 and KRAS protein levels with GAPDH serving as loading control. The quantitation of the relative protein levels of GRP78 and KRAS after normalization against GAPDH levels is shown in the graphs below. Data are presented as mean ± S.D. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001 (Student's t test).
GRP78 deficiency-mediated reduction of KRAS protein is at the post-transcriptional level
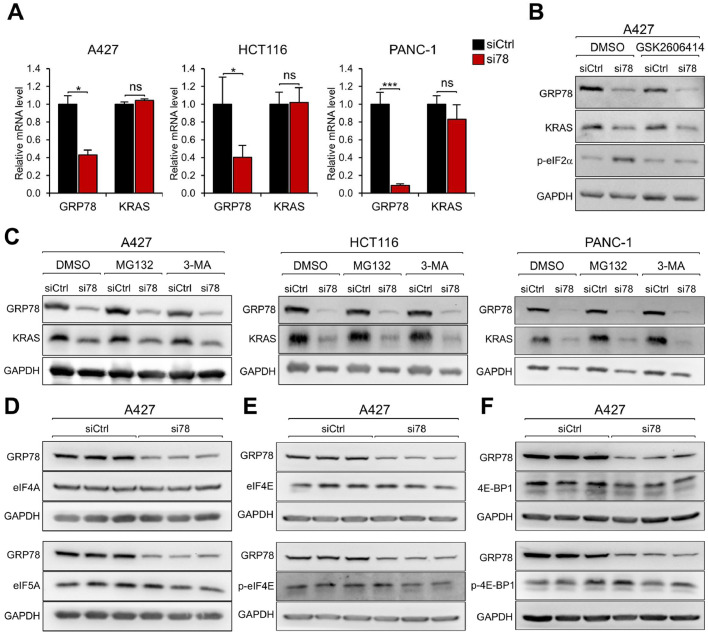
To investigate the mechanism for the reduction of the KRAS protein level, A427, HCT116, and PANC-1 cells were treated with siCtrl and si78 and the KRAS mRNA level was measured by RT-qPCR. Knockdown of GRP78 markedly reduced GRP78 mRNA level as expected while there was no significant decrease in KRAS mRNA level in all three cell lines (Fig. 2A). These results indicate that the reduction in KRAS expression is not at the transcript level.
Fig. 2.
GRP78 deficiency reduces KRAS protein at the post-transcriptional level. (A) A427, HCT116, and PANC-1 cells were transfected with siCtrl or si78 for 48 hr. Total RNA was extracted and subjected to reverse transcription and real-time PCR to measure the mRNA levels of GRP78 and KRAS with GAPDH serving as control. The quantitation of the relative mRNA levels of GRP78 and KRAS after normalization against GAPDH levels is shown. Data are presented as mean ± S.D. * P ≤ 0.05, *** P ≤ 0.001, ns: not significant (Student's t test). (B) A427 cell was transfected with siCtrl or si78 for 36 hr followed by treatment of DMSO or GSK2606414 (1μM) for 12 hr. Whole cell lysate (WCL) was subjected to Western blot analysis for GRP78, KRAS, phosphorylated-eIF2α (p-eIF2α) protein levels with GAPDH serving as loading control. (C) A427, HCT116, and PANC-1 cells were transfected with siCtrl or si78 for 36 hr followed by treatment of DMSO, MG132 (10μM), or 3-MA (10mM) for 12 hr. WCL was subjected to Western blot analysis for GRP78 and KRAS protein levels with GAPDH serving as loading control. (D) A427 cells were transfected with siCtrl or si78 for 48 hr and WCL was analyzed by Western blot for GRP78, eIF4A, and eIF5A protein levels with GAPDH serving as loading control. (E) Same as in (D) except eIF4E and phosphorylated eIF4E (p-eIF4E) protein levels were analyzed. (F) Same as in (D) except 4E-BP1 and phosphorylated 4E-BP1 (p-4E-BP1) protein levels were analyzed.
As the master regulator of the UPR, GRP78 depletion can trigger ER stress and activate the UPR leading to attenuation of protein synthesis via the PERK/eIF2α pathway [15,40]. To test this, we utilized the PERK inhibitor GSK2606414 to block eIF2α phosphorylation. A427 cells were transfected with siCtrl or si78 for 36 hr followed by GSK2606414 treatment. GRP78 knockdown reduced KRAS protein level and increased phosphorylated eIF2α (Serine 51) level as expected, however treatment with GSK2606414, while successfully blocking GRP78 knockdown mediated increase in eIF2α phosphorylation, failed to restore KRAS protein level in the si78 treated cells (Fig. 2B). This result suggests that protein translational arrest due to eIF2α phosphorylation is unlikely to be a mechanism for the reduction of KRAS protein expression.
Proteasome and autophagy are two major pathways for the cell to degrade proteins. To test whether these processes are involved, A427, HCT116, and PANC-1 cells were transfected with siCtrl or si78 followed by treatment of DMSO, proteasome inhibitors (MG132) or autophagy inhibitor (3-MA). GRP78 knockdown reduced KRAS protein in all three cell lines. However, combined treatment with MG132 or 3-MA failed to restore KRAS protein level (Fig. 2C). Similar results were obtained in H460 cells treated with DMSO, MG115, MG132, 3-MA or chloroquine (Supp. Fig. S1C). Thus, KRAS protein reduction is unlikely due to proteasome, lysosome or autophagy-mediated protein degradation in the cells tested, suggesting that the regulation could be at the translational level.
Previous studies showed that eukaryotic translation initiation factors eIF4A and eIF5A and the PEAK1 kinase regulate the translation of the KRAS oncoprotein [11,33,34]. However, we observed that A427, HCT116, and PANC-1 cells treated with si78 exhibited no reduction in the protein levels of eIF4A, eIF5A and PEAK1 compared to siCtrl (Fig. 2D and Supp. Fig. S3A-C), implying that GRP78 deficiency does not alter the availability of these factors implicated in KRAS translational control. Recent studies also suggest that phosphorylation of eIF4E and 4E-BP1 regulates translation of stress or oncogenic proteins [28]. Our results showed that si78 treatment had no effect the phosphorylated or total level of eIF4E and 4E-BP1 (Fig. 2E, F). Collectively, these results indicate that GRP78-deficiency mediated reduction of KRAS protein is at the post-transcriptional level and may involve some novel mechanisms which remain to be identified.
Targeting GRP78 via small molecule inhibitors reduces oncogenic KRAS protein level
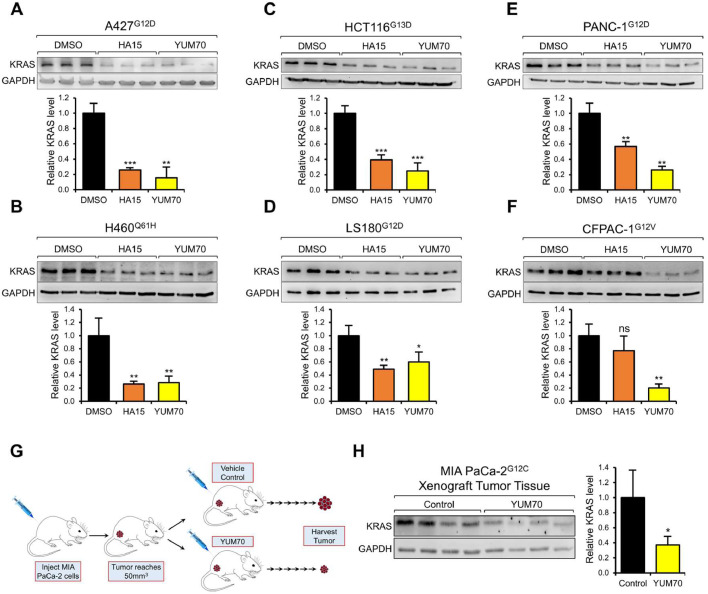
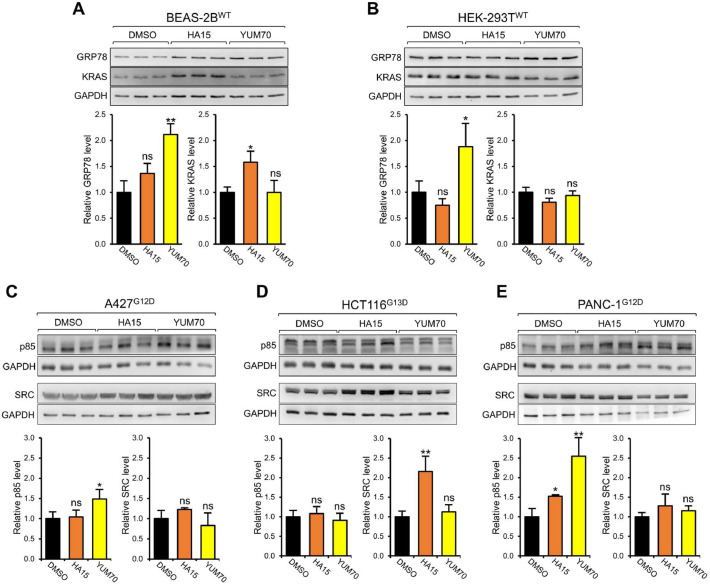
Next, taking advantage of the two GRP78 small molecule inhibitors HA15 and YUM70 which specifically bind GRP78 and inhibit its ATPase activity, we tested the effect of these clinically relevant agents on KRAS expression. We observed that in A427, HCT116, PANC-1, H460, LS180, and CFPAC-1 cells, treatment with either HA15 or YUM70 for 24 hr resulted in significant decrease of KRAS protein level (50 to 90%) in all six cell lines, except for the relatively modest effect of HA15 (20%) in CFPAC-1 cells (Fig. 3A-F). In contrast, both HA15 and YUM70 treatment did not reduce wild type KRAS levels in the normal human bronchial epithelial cell line (BEAS-2B), as well as in normal human embryonic kidney epithelial cell line (293T) (Fig. 4A-B). Furthermore, both HA15 and YUM70 treatment did not reduce the expression of two non-related oncoproteins SRC kinase and p85, with reported half-lives of 5 to 8 hr [17,44] (Fig. 4C-E). These results indicate that GRP78 inhibitors mimicked the effect of GRP78 knockdown on reducing mutant KRAS protein levels, while other key oncoproteins were not affected. Recently, we demonstrated that YUM70 suppressed tumor growth in a human pancreatic cancer xenograft model utilizing MIA PaCa-2 cells, however the mechanisms remained to be further explored [29]. Examination of the tumor tissues from this xenograft model treated with either YUM70 or vehicle control revealed a significant reduction of KRAS protein level in the YUM70-treated tumor tissues compared to vehicle control (Fig. 3G, H). For this analysis, Western blots were performed to detect the KRAS protein levels due to issues raised towards the selectivity and sensitivity of commercially available anti-KRAS antibodies for immunofluorescence or immunohistochemical analyses [39]. Collectively, our results showed that GRP78 inhibitors can efficiently suppress oncogenic KRAS expression in human cancer cells in both in vitro and in vivo settings.
Fig. 3.
GRP78 small molecule inhibitors reduce KRAS protein level in vitro and in vivo. A427 (A), H460 (B), HCT116 (C), LS180 (D), PANC-1 (E), and CFPAC-1 (F) cells were treated with either DMSO, HA15 (10μM) or YUM70 (10μM) for 24 hr. The superscript indicates the KRAS mutation for each cell line. Whole cell lysate was subjected to Western blot analysis for KRAS protein level with GAPDH serving as loading control. The quantitation of the relative KRAS protein level after normalization to GADPH levels is shown in the graphs below. (G) Schematic illustration of the pancreatic cancer xenograft experiment and treatment conditions. (H) MIA PaCa-2 xenograft tumor tissues were harvested from the mice at the end of the treatment and subjected to Western blot analysis for KRAS protein level with GAPDH serving as loading control. The quantitation of the relative KRAS protein level after normalization to the GAPDH levels is shown in the graph on the right. Data are presented as mean ± S.D. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, ns: not significant (Student's t test).
Fig. 4.
Effect of GRP78 small molecule inhibitors of GRP78 on normal human cell lines and other oncogenic proteins. BEAS-2B (A), and HEK-293T (B) cells were treated with either DMSO, HA15 (10μM) or YUM70 (10μM) for 24 hr and whole cell lysate (WCL) was subjected to Western blot analysis for GRP78 and KRAS protein levels with GAPDH serving as loading control. The quantitation of the relative protein levels of GRP78 and KRAS after normalization to the GADPH levels is shown in the graphs below. A427 (C), HCT116 (D), and PANC-1 (E) cells were treated with either DMSO, HA15 (10μM) or YUM70 (10μM) for 24 hr and WCL was subjected to Western blot analysis for SRC and p85 protein levels with GAPDH serving as loading control. The quantitation of the relative protein levels of SRC and p85 after normalization to the GADPH levels is shown in the graphs below. Data are presented as mean ± S.D. *P ≤ 0.05, ** P ≤ 0.01, ns: not significant (Student's t test). The superscript indicates the KRAS mutation for each cell line.
Next, we examined the effect of HA15 and YUM70 on HRAS and NRAS protein levels in the same panel of human cancer cells bearing various mutant KRAS alleles but wild type alleles for HRAS and NRAS. Here we noted that the effects varied in a cell-context dependent manner. For example, while the inhibitors decreased HRAS and NRAS protein levels in A427, H460, and HCT116 cells, only modest effects were observed for LS180, PANC-1, and CFPAC-1 cells (Supp. Fig. S4). Interestingly, in the lung cancer cell line H1299 harboring mutant NRASQ61K, HA15 or YUM70 substantially reduced the mutant NRAS protein level (Supp. Table S1, Supp. Fig. S5). Overall, our results showed that HA15 and YUM70 consistently reduced oncogenic KRAS protein level across the cell lines tested while their suppressive effects on HRAS and NRAS are cell-context dependent, with no compensatory upregulation of these proteins in any of the cell lines tested (Supp. Fig. S6).
GRP78 small molecule inhibitors trigger apoptosis and reduce viability of cancer cells bearing various KRAS mutations
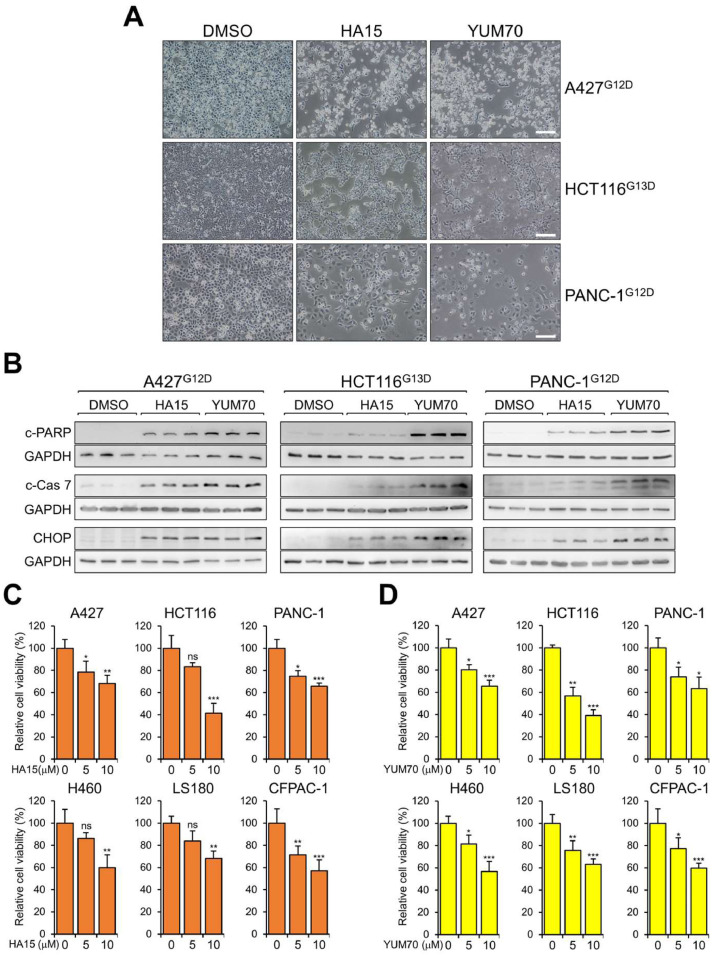
We then investigated the effect of HA15 and YUM70 treatment on the survival of the panel of lung, colon and pancreatic cancer cell lines bearing various KRAS mutations. Upon treatment with either HA15 or YUM70 for 48 hr, alterations in cell morphologies including membrane blebbing and rounding of cell shape, as well as loss of cell number, were evident (Fig. 5A). Consistent of cell death, Western blot analysis showed induction of the cleaved form of PARP, an apoptotic marker in cells treated with either GRP78 inhibitors, with YUM70 exhibiting a stronger effect than HA15 at the same drug concentration being tested (Fig. 5B). The more potent effect of YUM70 correlated with increased cleavage of Caspase-7, a caspase localized at the outer ER membrane whose activation is regulated by GRP78 [27], as well as higher induction of CHOP, an ER stress apoptotic marker [18]. In WST-1 assays, we observed a significant and dose-dependent decrease in cell viability in cells treated with HA15 or YUM70 (Fig. 5C, D). In agreement with previous studies that HA15 and YUM70 at the concentrations used did not affect the viability of normal cells [6,29], both HA15 and YUM70 treatment showed minimal toxicity in the normal human bronchial epithelial cell line BEAS-2B (Supp. Fig. S2D). Collectively, our results demonstrated that GRP78 small molecule inhibitors HA15 and YUM70 can induce apoptosis and reduce viability of cancer cell lines bearing various KRAS mutations.
Fig. 5.
Small molecule inhibitors of GRP78 induce apoptosis and reduce cancer cell viability bearing mutant KRAS alleles. (A) A427, HCT116, and PANC-1 cells were treated with either DMSO, HA15 (10μM) or YUM70 (10μM) for 48 hr. Images from phase contrast microscopy are shown. Scale bars represent 20μM. (B) Same as in (A) except the cells were harvested and whole cell lysate was subjected to Western blot analysis for cleaved PARP (c-PARP), cleaved Caspase-7 (c-Cas 7), and CHOP protein levels with GAPDH serving as loading control. (C) A427, HCT116, PANC-1, H460, LS180, and CFPAC-1 cells were treated with either DMSO or HA15 (10μM) for 48 hr and cell viability was measured by WST-1 assay. The experiment was repeated 4 times for each cell line. The relative percentage of cell viability was quantified in the graphs. (D) Same as in (C) except the cells were treated with DMSO or YUM70 (10μM). Data are presented as mean ± S.D. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, ns: not significant (Student's t test).
Discussion
While it is well established that GRP78 is a key regulator of the UPR, such that its depletion or inhibition of its catalytic activity will trigger ER stress, leading to cancer cell death by concomitant induction of autophagic and apoptotic mechanisms [6,18,29], evidence is emerging that GRP78 deficiency may also lead to disruption of key oncogenic drivers located outside the ER. For example, in genetically engineered mouse models of PTEN-driven cancer, GRP78 haploinsufficiency suppressed tumor progression and AKT activation and this led to the discovery that GRP78 is a binding partner of PI3K on the cell surface and required for its downstream signaling activity [10,19,41,45]. Prompted by the recent observations that GRP78 is critical for KRAS-driven pancreatic and lung tumorigenesis in mouse models, YUM70 which specifically binds and inhibits GRP78 exhibits potent activity against pancreatic cancer, as well as other drugs that reduce GRP78 expression suppressed pancreatic cancer chemoresistance and lung cancer metastasis [12,14,20,25,29,30], we investigated whether targeting GRP78 may alter KRAS expression in human cancers bearing various oncogenic KRAS alleles. Our studies reveal several new findings, which expand on the role of GRP78 as a regulator of key oncogenic drivers, affecting processes beyond the ER.
First, in a panel of human lung, colorectal and pancreatic cancer cell lines bearing various KRAS mutations, knockdown of GRP78 consistently reduced KRAS protein but not mRNA levels, thus the regulation is at the post-transcriptional level. Intriguingly, blockage of eIF2α phosphorylation failed to restore KRAS protein levels, suggesting that it is not due to the general translational shutdown resulting from ER stress. Moreover, GRP78 knockdown has no effect on the levels of eIF4A, eIF5A and PEAK1 reported to regulate KRAS translation [11,33,34]. GRP78 knockdown also has no effect on the phosphorylation of eIF4E or 4E-BP1 reported to regulate translation of stress or oncogenic proteins including Myc, which is inhibited by acute metabolic stress [28]. The failure of proteasome and autophagy inhibitors to restore KRAS protein levels further implied that protein degradation is unlikely to be the mechanism, while future studies are required to dissect the precise regulatory steps involved.
In examining the effect of GRP78 deficiency on various RAS protein expression and cell viability, we noted that wild type KRAS protein levels and viability of a normal human lung epithelial cell line were not affected, consistent with the observations that at dosages exhibiting strong anti-cancer activity, YUM70 did not affect viability of normal pancreatic tissue-derived cells (HPNE) and HA15 did not show toxicity towards primary human melanocytes and fibroblasts, and no adverse effects were was observed in normal organs in vivo [6,29]. Likewise, in several lung and colon cancer cell lines expressing wild type KRAS, its level was not affected by GRP78 knockdown. On the other hand, wild type HRAS and NRAS protein levels in cancer cell lines harboring mutant KRAS alleles were either not affected or reduced by GRP78 inhibition. This raises the interesting question why when both wild type and mutant KRAS alleles are present in the same cancer cells, how might GRP78 targeting affects both forms of KRAS proteins? While the precise mechanisms remain to be determined, cellular stress is reported to function in synergistic cooperation with oncogenic mutations including mutant RAS to drive cancer progression by enhancing the ability of cells to tolerate these stresses through multiple mechanisms [26]. The expression of GRP78, a well-established stress-responding chaperone inside tumor cells, likely contributes majorly to the homeostasis of mutant KRAS-driven cancer cells [18]. Thus, GRP78 deficiency could trigger a non-discriminative response suppressing both oncogenic and wild type KRAS expression in the same cells. Additionally, this could also involve feedback regulation between oncogenic KRAS and the wild-type RAS present in the same cells [43]. On the other hand, while mutant NRAS expression may also be reduced by HA15 and YUM70, the levels of SRC kinase and p85, both are potent drivers of tumorigenesis, are not affected by these agents in the cell lines examined. Thus, it is tempting to speculate that GRP78 deficiency or inhibition may directly or indirectly impact some unique features of mutant RAS translational machineries or stress-adaptive microRNAs [7] which warrant vigorous future investigations.
Despite the recent breakthrough advance of KRAS specific inhibitors, for downregulation of KRAS and its oncogenic effects, it has been suggested that physical loss or degradation may provide a more robust and durable anti-tumor effect. Advances has been made towards manipulation of KRAS protein stability, however, there appears to be different modes of KRAS degradation attributed to different inducers of KRAS degradation in different cells, adding complexity to this approach [31]. Regarding physical loss of the oncoprotein, stable suppression of oncogenic KRAS tumorigenicity by virus-mediated RNA interference against specific mutant KRAS allele in pancreatic cancer has been reported [3]. Nonetheless, the success of this approach requires the development of an efficient delivery system that can affect most of the tumor cells. Here in our studies, we have demonstrated that the anti-GRP78 small molecule inhibitors HA15 and YUM70 can suppress multiple types of oncogenic KRAS protein expression in PDAC, LUAD and colorectal cancer cell lines, leading to apoptosis and loss of cell viability. Other agents capable of suppressing or inhibiting GRP78 have been reported and are in clinical development or use [1,2,9,14,18]. Importantly, they can be easily administered to achieve efficacy in curbing tumor growth.
In contrast to pancreatic cancer, studies in non-small cell lung cancer showed that knockdown of mutant KRAS by itself may not be sufficient treatment due to compensatory oncogenic pathways, however this may offer opportunities to couple with other targeted therapeutic approaches [35]. Consistent with this view, a highly desirable target for anti-cancer therapy is a cellular moiety which controls multiple functions required for the cancer cells to proliferate at a high rate. In addition to its role as a key regulator of the UPR, as a multifunctional protein and a key chaperone, GRP78 can interact directly or indirectly with a wide variety of client proteins, impacting their downstream pathways [13,18]. GRP78 deficiency has been shown to suppress PI3K, AKT, TGF-β and CD44 signaling among many other signaling pathways, as well as EGFR expression, in various human cancer cell lines and cancer mouse models [10,30,36,37,41,45]. Upon GRP78 knockdown or inhibition, the simultaneous blockade of these pathways, coupled with the onset of ER stress-induced apoptosis and autophagy will provide robust defense against the development of resistance by the cancer cells prior to their elimination [6,18,21,29]. In summary, our studies raise the possibility that targeting GRP78 via knockdown or small molecule inhibitors may offer a new strategy to suppress oncogenic KRAS expression, and as such, augment the other anti-cancer activities mediated by these agents to achieve stable, long-term suppression of mutant KRAS-driven tumorigenesis.
Author Contribution Statement
D.P.H. and A.S.L. conceptualization; D.P.H., B.H., H.W., D.F.R., R.V.K. and S.S. investigation and data acquisition; D.P.H., B.H., Z.L., and A.S.L. formal analysis; D.P.H and A.S.L. writing manuscript; D.P.H., B.H., Z.L., N. N. and A.S.L. review manuscript; N.N. and A.S.L. funding acquisition.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declaration of competing interests
N. Neamati reports a patent for GRP78 degraders pending. No disclosures were reported by the other authors.
Acknowledgments
Funding Information
National Institutes of Health, Grant/Award Number: R01 CA238029 and R01 CA027607 (to A.S.L.) and R01 CA188252 (to N.N.)
Acknowledgements
We thank Drs. David Shackelford, Chengyu Liang, Ganging Liang, Keigo Machida, Steven Dubinett, and Ite Offringa for the gifts of cell lines, J. Alan Diehl and Min Yu for compound and plasmid and Wei Li for helpful discussions. We thank the Core Facilities of the School of Pharmacy of USC for production of lentivirus. This work was supported in part by National Institutes of Health grants R01 CA238029 and CA027607 to A.S.L and R01 CA188252 to N.N. The University of Southern California Norris Comprehensive Cancer Center Molecular Genomics Core which provided primer service was supported by NIH grant P30 CA014089.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2022.100837.
Appendix. Supplementary materials
Supp. Fig. S1. Validation of RAS antibodies and effect of inducible GRP78 knockdown and protein degradation inhibitors on KRAS protein levels in H460 cells. (A) HCT116 cells were transfected with siCtrl, siKRAS, siNRAS, or siHRAS for 48 hr. Whole cell lysate (WCL) was subjected to Western blot analysis for KRAS, NRAS, or HRAS protein levels. Molecular size markers were shown on the left and the antibodies used for each blot were listed at the bottom. The red arrow indicates the band for the respective RAS protein knockdown by its specific siRNA. (B) H460 cells bearing the Q61H KRAS mutant alleles were transduced by lentivirus carrying an inducible shRNA construct targeting GRP78. The cells were selected with puromycin for 1 week and induced with the indicated dose of doxycycline (Dox) for 48 hr. WCL was subjected to Western blot analysis for GRP78 and KRAS protein level with GAPDH serving as loading control. The quantitation of the relative protein levels after normalization to the GADPH levels is shown in the graph on the right. (C) H460 cells were transfected with siCtrl or si78 for 36 hr followed by treatment of DMSO, MG115(10μM), MG132 (10μM), 3-MA (10mM), or Chloroquine (20μM) for 12 hr. WCL was subjected to Western blot analysis for GRP78 and KRAS protein levels with GAPDH serving as loading control.
Supp. Fig. S2. Effect of GRP78 knockdown on cell lines expressing wild type KRAS and small molecule GRP78 inhibitors on the viability of a normal human bronchial epithelial cell line. (A) BEAS-2B cells were transfected with control siCtrl, si78 or siKRAS for 48 hr. Whole cell lysate (WCL) was subjected to Western blot analysis for GRP78 and KRAS protein levels with GAPDH serving as loading control. The quantitation of the relative protein level after normalization to the GADPH levels is shown in the graph below. (B) Human lung cancer cell lines H522 or H1975 were transfected with siCtrl or si78 for 48 hr. WCL was subjected to Western blot analysis for GRP78 and KRAS protein levels with GAPDH serving as loading control. (C) Same as in (B) except human colon cancer cell lines RKO and HT-29 were used. The quantitation of the relative protein level is shown in the graph below. (D) BEAS-2B cells were treated with either DMSO, HA15 (10μM) or YUM70 (10μM) for 48 hr and cell viability was measured by WST-1 assay and the relative percentage of cell viability was quantified in the graphs on the right. Data are presented as mean ± S.D. *P ≤ 0.05, ** P ≤ 0.01, ns: not significant (Student's t test). The superscript indicates the wild type (WT) KRAS allele status for each cell line.
Supp. Fig. S3. Knockdown of GRP78 reduced KRAS but not PEAK1, eIF4A, or eIF5A protein levels. (A) A427 cells bearing the G12D KRAS mutant allele were transfected with siCtrl or si78 for 48 hr. Whole cell lysate (WCL) was subjected to Western blot analysis for GRP78, KRAS, and PEAK1 protein levels with GAPDH serving as loading control. The quantitation of the relative protein levels after normalization to the GADPH levels is shown in the graph on the right. (B) HCT116 cells were transfected with siCtrl or si78 for 48hr and WCL was subjected to Western blot analysis for GRP78, eIF4A, and eIF5A protein levels with GAPDH serving as loading control. (C) Same as in (B) except PANC-1 cells were used.
Supp. Fig. S4. Effect of GRP78 small molecule inhibitors on NRAS and HRAS protein levels in cancer cells harboring various KRAS mutations. A427 (A), H460 (B), HCT116 (C), LS180 (D), PANC-1 (E), and CFPAC-1 (F) cells were treated with either DMSO, HA15 (10μM) or YUM70 (10μM) for 24 hr and whole cell lysate was subjected to Western blot analysis for NRAS and HRAS protein levels with GAPDH serving as loading control. The quantitation of the relative protein levels of NRAS and HRAS after normalization to GAPDH level is shown in the graphs below. Data are presented as mean ± S.D. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, ns: not significant (Student's t test). The superscript indicates the KRAS mutation for each cell line.
Supp. Fig. S5. GRP78 small molecule inhibitors reduce mutant NRAS protein level. Human non-small cell lung carcinoma H1299 cells bearing the NRAS Q61K mutant allele were treated with either DMSO, HA15 (10μM) or YUM70 (10μM) for 24 hr and whole cell lysate was subjected to Western blot analysis for NRAS protein level with GAPDH serving as loading control. The quantitation of the relative NRAS protein level after normalization to GAPDH level is shown in the graph below. Data are presented as mean ± S.D. *** P ≤ 0.001 (Student's t test).
Supp. Fig. S6. Summary of the effects of GRP78 small molecule inhibitors on the protein levels of KRAS, NRAS, and HRAS. The relative protein levels of KRAS, NRAS, and HRAS (from Fig. 3 and Supp. Fig. 5) are combined into one graph for each cell line. Data are presented as mean ± S.D. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, ns: not significant (Student's t test).
References
- 1.Bakewell SJ, Rangel DF, Ha DP, et al. Suppression of stress induction of the 78-kilodalton glucose regulated protein (GRP78) in cancer by IT-139, an anti-tumor ruthenium small molecule inhibitor. Oncotarget. 2018;9(51):29698–29714. doi: 10.18632/oncotarget.25679. Published 2018 Jul 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Booth L, Roberts JL, Cash DR, et al. GRP78/BiP/HSPA5/Dna K is a universal therapeutic target for human disease. J Cell Physiol. 2015;230(7):1661–1676. doi: 10.1002/jcp.24919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2(3):243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma [published correction appears in Nature. 2014 Oct 9;514(7521):262. Rogers, K [corrected to Rodgers, K]] [published correction appears in Nature. 2018 Jul;559(7715):E12] Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casas C. GRP78 at the Centre of the Stage in Cancer and Neuroprotection. Front Neurosci. 2017;11:177. doi: 10.3389/fnins.2017.00177. Published 2017 Apr 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerezo M, Lehraiki A, Millet A, et al. Compounds Triggering ER Stress Exert Anti-Melanoma Effects and Overcome BRAF Inhibitor Resistance [published correction appears in Cancer Cell. 2016 Jul 11;30(1):183] Cancer Cell. 2016;29(6):805–819. doi: 10.1016/j.ccell.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 7.Chitnis N, Pytel D, Diehl JA. UPR-inducible miRNAs contribute to stressful situations. Trends Biochem Sci. 2013;38(9):447–452. doi: 10.1016/j.tibs.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dang CV, Reddy EP, Shokat KM. Soucek L. Drugging the ‘undruggable’ cancer targets. Nat Rev Cancer. 2017;17(8):502–508. doi: 10.1038/nrc.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elfiky AA, Baghdady AM, Ali SA, Ahmed MI. GRP78 targeting: Hitting two birds with a stone. Life Sci. 2020;260 doi: 10.1016/j.lfs.2020.118317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y, Wey S, Wang M, et al. Pten null prostate tumorigenesis and AKT activation are blocked by targeted knockout of ER chaperone GRP78/BiP in prostate epithelium. Proc Natl Acad Sci U S A. 2008;105(49):19444–19449. doi: 10.1073/pnas.0807691105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujimura K, Wang H, Watson F, Klemke RL. KRAS oncoprotein expression is regulated by a self-governing eIF5A-PEAK1 feed-forward regulatory loop. Cancer Res. 2018;78(6):1444–1456. doi: 10.1158/0008-5472.CAN-17-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gifford JB, Huang W, Zeleniak AE, et al. Expression of GRP78, master regulator of the unfolded protein response, increases chemoresistance in pancreatic ductal adenocarcinoma. Mol Cancer Ther. 2016;15(5):1043–1052. doi: 10.1158/1535-7163.MCT-15-0774. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Gronow M, Selim MA, Papalas J, Pizzo SV. GRP78: a multifunctional receptor on the cell surface. Antioxid Redox Signal. 2009;11(9):2299–2306. doi: 10.1089/ARS.2009.2568. [DOI] [PubMed] [Google Scholar]
- 14.Ha DP, Tsai YL, Lee AS. Suppression of ER-stress induction of GRP78 as an anti-neoplastic mechanism of the cardiac glycoside Lanatoside C in pancreatic cancer: Lanatoside C suppresses GRP78 stress induction. Neoplasia. 2021;23(12):1213–1226. doi: 10.1016/j.neo.2021.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hetz C, Zhang K, Kaufman RJ. Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Biol. 2020;21(8):421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong DS, Fakih MG, Strickler JH, et al. KRASG12C inhibition with sotorasib in advanced solid tumors. N Engl J Med. 2020;383(13):1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko HR, Kim CK, Lee SB, et al. P42 Ebp1 regulates the proteasomal degradation of the p85 regulatory subunit of PI3K by recruiting a chaperone-E3 ligase complex HSP70/CHIP. Cell Death Dis. 2014;5(3):e1131. doi: 10.1038/cddis.2014.79. Published 2014 Mar 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee AS. Glucose-regulated proteins in cancer: molecular mechanisms and therapeutic potential. Nat Rev Cancer. 2014;14(4):263–276. doi: 10.1038/nrc3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu R, Li X, Gao W, et al. Monoclonal antibody against cell surface GRP78 as a novel agent in suppressing PI3K/AKT signaling, tumor growth, and metastasis. Clin Cancer Res. 2013;19(24):6802–6811. doi: 10.1158/1078-0432.CCR-13-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lizardo MM, Morrow JJ, Miller TE, et al. Upregulation of glucose-regulated protein 78 in metastatic cancer cells is necessary for lung metastasis progression. Neoplasia. 2016;18(11):699–710. doi: 10.1016/j.neo.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene. 2013;32(7):805–818. doi: 10.1038/onc.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagasaka M, Potugari B, Nguyen A, Sukari A, Azmi AS, Ou SI. KRAS Inhibitors- yes but what next? Direct targeting of KRAS- vaccines, adoptive T cell therapy and beyond. Cancer Treat Rev. 2021;101 doi: 10.1016/j.ctrv.2021.102309. [DOI] [PubMed] [Google Scholar]
- 23.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72(10):2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11(11):761–774. doi: 10.1038/nrc3106. Published 2011 Oct 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rangel DF, Dubeau L, Park R, et al. Endoplasmic reticulum chaperone GRP78/BiP is critical for mutant Kras-driven lung tumorigenesis. Oncogene. 2021;40(20):3624–3632. doi: 10.1038/s41388-021-01791-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redding A, Aplin AE, Grabocka E. RAS-mediated tumor stress adaptation and the targeting opportunities it presents. Dis Model Mech. 2022;15(2) doi: 10.1242/dmm.049280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, Lee AS. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J Biol Chem. 2003;278(23):20915–20924. doi: 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- 28.Ruan H, Li X, Xu X, et al. eIF4E S209 phosphorylation licenses myc- and stress-driven oncogenesis. Elife. 2020;9:e60151. doi: 10.7554/eLife.60151. Published 2020 Nov 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samanta S, Yang S, Debnath B, et al. The Hydroxyquinoline Analogue YUM70 Inhibits GRP78 to Induce ER Stress-Mediated Apoptosis in Pancreatic Cancer. Cancer Res. 2021;81(7):1883–1895. doi: 10.1158/0008-5472.CAN-20-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen J, Ha DP, Zhu G, et al. GRP78 haploinsufficiency suppresses acinar-to-ductal metaplasia, signaling, and mutant kras-driven pancreatic tumorigenesis in mice. Proc Natl Acad Sci U S A. 2017;114(20):E4020–E4029. doi: 10.1073/pnas.1616060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shukla S, Allam US, Ahsan A, et al. KRAS protein stability is regulated through SMURF2: UBCH5 complex-mediated β-TrCP1 degradation. Neoplasia. 2014;16(2):115–128. doi: 10.1593/neo.14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170(1):17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh K, Lin J, Lecomte N, et al. Targeting eIF4A-dependent translation of KRAS signaling molecules. Cancer Res. 2021;81(8):2002–2014. doi: 10.1158/0008-5472.CAN-20-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strnadel J, Choi S, Fujimura K, et al. eIF5A-PEAK1 signaling regulates YAP1/TAZ protein expression and pancreatic cancer cell growth. Cancer Res. 2017;77(8):1997–2007. doi: 10.1158/0008-5472.CAN-16-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sunaga N, Shames DS, Girard L, et al. Knockdown of oncogenic KRAS in non-small cell lung cancers suppresses tumor growth and sensitizes tumor cells to targeted therapy. Mol Cancer Ther. 2011;10(2):336–346. doi: 10.1158/1535-7163.MCT-10-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai YL, Ha DP, Zhao H, et al. Endoplasmic reticulum stress activates SRC, relocating chaperones to the cell surface where GRP78/CD109 blocks TGF-β signaling. Proc Natl Acad Sci U S A. 2018;115(18):E4245–E4254. doi: 10.1073/pnas.1714866115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tseng CC, Stanciauskas R, Zhang P, et al. GRP78 regulates CD44v membrane homeostasis and cell spreading in tamoxifen-resistant breast cancer. Life Sci Alliance. 2019;2(4) doi: 10.26508/lsa.201900377. Published 2019 Aug 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang M, Wey S, Zhang Y, Ye R, Lee AS. Role of the unfolded protein response regulator GRP78/BiP in development, cancer, and neurological disorders. Antioxid Redox Signal. 2009;11(9):2307–2316. doi: 10.1089/ars.2009.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waters AM, Ozkan-Dagliyan I, Vaseva AV, et al. Evaluation of the selectivity and sensitivity of isoform- and mutation-specific RAS antibodies. Sci Signal. 2017;10(498):eaao3332. doi: 10.1126/scisignal.aao3332. Published 2017 Sep 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wek RC, Cavener DR. Translational control and the unfolded protein response. Antioxid Redox Signal. 2007;9(12):2357–2371. doi: 10.1089/ars.2007.1764. [DOI] [PubMed] [Google Scholar]
- 41.Wey S, Luo B, Tseng CC, et al. Inducible knockout of GRP78/BiP in the hematopoietic system suppresses Pten-null leukemogenesis and AKT oncogenic signaling. Blood. 2012;119(3):817–825. doi: 10.1182/blood-2011-06-357384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ying H, Dey P, Yao W, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016;30(4):355–385. doi: 10.1101/gad.275776.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young A, Lou D, McCormick F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 2013;3(1):112–123. doi: 10.1158/2159-8290.CD-12-0231. [DOI] [PubMed] [Google Scholar]
- 44.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3’-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18(3):1379–1387. doi: 10.1128/MCB.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Tseng CC, Tsai YL, Fu X, Schiff R, Lee AS. Cancer cells resistant to therapy promote cell surface relocalization of GRP78 which complexes with PI3K and enhances PI(3,4,5)P3 production. PloS One. 2013;8(11):e80071. doi: 10.1371/journal.pone.0080071. Published 2013 Nov 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp. Fig. S1. Validation of RAS antibodies and effect of inducible GRP78 knockdown and protein degradation inhibitors on KRAS protein levels in H460 cells. (A) HCT116 cells were transfected with siCtrl, siKRAS, siNRAS, or siHRAS for 48 hr. Whole cell lysate (WCL) was subjected to Western blot analysis for KRAS, NRAS, or HRAS protein levels. Molecular size markers were shown on the left and the antibodies used for each blot were listed at the bottom. The red arrow indicates the band for the respective RAS protein knockdown by its specific siRNA. (B) H460 cells bearing the Q61H KRAS mutant alleles were transduced by lentivirus carrying an inducible shRNA construct targeting GRP78. The cells were selected with puromycin for 1 week and induced with the indicated dose of doxycycline (Dox) for 48 hr. WCL was subjected to Western blot analysis for GRP78 and KRAS protein level with GAPDH serving as loading control. The quantitation of the relative protein levels after normalization to the GADPH levels is shown in the graph on the right. (C) H460 cells were transfected with siCtrl or si78 for 36 hr followed by treatment of DMSO, MG115(10μM), MG132 (10μM), 3-MA (10mM), or Chloroquine (20μM) for 12 hr. WCL was subjected to Western blot analysis for GRP78 and KRAS protein levels with GAPDH serving as loading control.
Supp. Fig. S2. Effect of GRP78 knockdown on cell lines expressing wild type KRAS and small molecule GRP78 inhibitors on the viability of a normal human bronchial epithelial cell line. (A) BEAS-2B cells were transfected with control siCtrl, si78 or siKRAS for 48 hr. Whole cell lysate (WCL) was subjected to Western blot analysis for GRP78 and KRAS protein levels with GAPDH serving as loading control. The quantitation of the relative protein level after normalization to the GADPH levels is shown in the graph below. (B) Human lung cancer cell lines H522 or H1975 were transfected with siCtrl or si78 for 48 hr. WCL was subjected to Western blot analysis for GRP78 and KRAS protein levels with GAPDH serving as loading control. (C) Same as in (B) except human colon cancer cell lines RKO and HT-29 were used. The quantitation of the relative protein level is shown in the graph below. (D) BEAS-2B cells were treated with either DMSO, HA15 (10μM) or YUM70 (10μM) for 48 hr and cell viability was measured by WST-1 assay and the relative percentage of cell viability was quantified in the graphs on the right. Data are presented as mean ± S.D. *P ≤ 0.05, ** P ≤ 0.01, ns: not significant (Student's t test). The superscript indicates the wild type (WT) KRAS allele status for each cell line.
Supp. Fig. S3. Knockdown of GRP78 reduced KRAS but not PEAK1, eIF4A, or eIF5A protein levels. (A) A427 cells bearing the G12D KRAS mutant allele were transfected with siCtrl or si78 for 48 hr. Whole cell lysate (WCL) was subjected to Western blot analysis for GRP78, KRAS, and PEAK1 protein levels with GAPDH serving as loading control. The quantitation of the relative protein levels after normalization to the GADPH levels is shown in the graph on the right. (B) HCT116 cells were transfected with siCtrl or si78 for 48hr and WCL was subjected to Western blot analysis for GRP78, eIF4A, and eIF5A protein levels with GAPDH serving as loading control. (C) Same as in (B) except PANC-1 cells were used.
Supp. Fig. S4. Effect of GRP78 small molecule inhibitors on NRAS and HRAS protein levels in cancer cells harboring various KRAS mutations. A427 (A), H460 (B), HCT116 (C), LS180 (D), PANC-1 (E), and CFPAC-1 (F) cells were treated with either DMSO, HA15 (10μM) or YUM70 (10μM) for 24 hr and whole cell lysate was subjected to Western blot analysis for NRAS and HRAS protein levels with GAPDH serving as loading control. The quantitation of the relative protein levels of NRAS and HRAS after normalization to GAPDH level is shown in the graphs below. Data are presented as mean ± S.D. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, ns: not significant (Student's t test). The superscript indicates the KRAS mutation for each cell line.
Supp. Fig. S5. GRP78 small molecule inhibitors reduce mutant NRAS protein level. Human non-small cell lung carcinoma H1299 cells bearing the NRAS Q61K mutant allele were treated with either DMSO, HA15 (10μM) or YUM70 (10μM) for 24 hr and whole cell lysate was subjected to Western blot analysis for NRAS protein level with GAPDH serving as loading control. The quantitation of the relative NRAS protein level after normalization to GAPDH level is shown in the graph below. Data are presented as mean ± S.D. *** P ≤ 0.001 (Student's t test).
Supp. Fig. S6. Summary of the effects of GRP78 small molecule inhibitors on the protein levels of KRAS, NRAS, and HRAS. The relative protein levels of KRAS, NRAS, and HRAS (from Fig. 3 and Supp. Fig. 5) are combined into one graph for each cell line. Data are presented as mean ± S.D. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, ns: not significant (Student's t test).
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.