Abstract
Nutritional pancreatic atrophy (NPA) is a classical Se/vitamin E deficiency disease of chicks. To reveal molecular mechanisms of its pathogenesis, we fed day-old chicks a practical, low-Se diet (14 μg Se/kg), and replicated the typical symptoms of NPA including vesiculated mitochondria, cytoplasmic vacuoles, and hyaline bodies in acinar cells of chicks as early as day 18. Target pathway analyses illustrated a > 90% depletion (P < 0.05) of glutathione peroxidase 4 (GPX4) protein and up-regulated apoptotic signaling (cytochrome C/caspase 9/caspase 3) in the pancreas and(or) acinar cells of Se deficient chicks compared with Se-adequate chicks. Subsequently, we overexpressed and suppressed GPX4 expression in the pancreatic acinar cells and observed an inverse (P < 0.05) relationship between the GPX4 production and apoptotic signaling and cell death. Applying pull down and mass spectrometry, we unveiled that GPX4 bound prothymosin alpha (ProTalpha) to inhibit formation of apoptosome in the pancreatic acinar cells. Destroying this novel protein-protein interaction by silencing either gene expression accelerated H2O2-induced apoptosis in the cells. In the end, we applied GPX4 shRNA to silence GPX4 expression in chick embryo and confirmed the physiological relevance of the GPX4 role and mechanism shown ex vivo and in the acinar cells. Altogether, our results indicated that GPX4 depletion in Se-deficient chicks acted as a major contributor to their development of NPA due to the lost binding of GPX4 to ProTalpha and its subsequent inhibition on the cytochrome c/caspase 9/caspase 3 cascade in the acinar cells. Our findings not only provide a novel molecular mechanism for explaining pathogenesis of NPA but also reveal a completely new cellular pathway in regulating apoptosis by selenoproteins.
Keywords: Apoptosis, Chick, Nutritional pancreatic atrophy, Prothymosin alpha, Selenium, Vitamin E
Graphical abstract
Highlights
-
•
Nutritional pancreatic atrophy (NPA) was replicated in Se-deficient chicks.
-
•
Pancreas of NPA chicks showed GPX4 depletion and apoptotic activation.
-
•
Altering GPX4 in acinar cells changed their susceptibility to apoptosis.
-
•
ProTα could bind GPX4 in acinar cells and modulate their fate of apoptosis.
-
•
GPX4 shRNA activated apoptotic pathway in embryonic pancreas of chicks.
Abbreviations
- ACTB
beta actin
- BD
basal diet;
- CASP 3
caspase 3
- CASP 9
caspase 9
- CYTC
Cytochrome c
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GPX1
2, 3, 4, glutathione peroxidase 1, 2, 3, 4
- NPA
nutritional pancreatic atrophy
- ProTalpha
prothymosin alpha
- Q-PCR
real-time quantitative PCR
- Se
selenium
- –Se
Se-deficient
- +Se
Se-adequate
- SELENOF
selenoprotein F
- SELENON
selenoprotein N
- SELENOP
selenoprotein P
- SELENOW
selenoprotein W
- SELENOU
selenoprotein U
- shGPX1
GPX1 short hairpin RNA
- shGPX4
GPX4 short hairpin RNA
- siGPX1
GPX1 small interfering RNA
- siGPX4
GPX4 small interfering RNA
- siProTalpha
ProTalpha small interfering RNA
- Vit. E
vitamin E
- –Vit. E
vitamin E deficient
- +Vit. E
vitamin E adequate
1. Introduction
Nutritional pancreatic atrophy (NPA) was first reported by Thompson and Scott as a classical selenium (Se)/vitamin E (Vit. E) deficiency disease in chicks [1]. Histologically, the atrophy starts with an intracellular vacuolation and hyaline body formation, followed by a loss of acinar zonation in the pancreas [2]. Many attempts [2,3] were made to elucidate the pathogenesis mechanism of NPA. However, the only consensus from those attempts was that Se acted as the primary nutritive factor, whereas Vit. E was only partially effective or not at all in preventing this type of disorders [[4], [5], [6]]. It has remained largely unknown if the role of Se is mediated directly by one or several of the 25 selenoproteints identified in chickens [[7], [8], [9]] and(or) indirectly by regulating expression and function of Se-independent proteins [10,11]. Digital gene expression (DGE) analyses can now be used to help provide a clue to address this question [12].
Previous work [13] indicated that the yield of mitochondria from Se-deficient pancreases was only 72% of that of Se-adequate controls. Whitacre and Combs [14] found that early-stage Se deficiency led to marked decreases in synthesis of RNA and protein in the pancreas. Subsequently, they suggested that the primary lesion in severe Se deficiency might be induced by the loss of mitochondrial integrity due to a diminished antioxidant protection via glutathione peroxidase (GPX) in cytosol and mitochondria1 matrix space [15]. However, others [16] showed that mitochondrial degeneration did not appear to be the initial lesion in NPA of Se-deficient chicks. Thus, it remains unclear if and how dietary Se deficiency impairs mitochondrial integrity to cause NPA in chicks.
GPX4 can reduce phospholipid hydroperoxides and act in conjunction with Vit. E to inhibit lipid peroxidation [17,18]. This monomeric selenoperoxidase is not only vital for development, but also indispensable for many cell types of adult animals [19]. The cardio-respiratory failure and perinatal death caused by missense mutation in selenocysteine synthase in mice can be compensated by Se-independent GPX4 [20]. Recently, GPX4 has been identified as a central regulator of ferroptosis [21]. In addition, GPX4 mediates the suppression of pyroptosis and ferroptosis in acute pancreatitis [22]. Although GPX4 is often lowly expressed and is resistant to Se deficiency in mammalian tissues [23], it is abundantly expressed in chicken tissues and readily affected by Se deficiency [[24], [25], [26]]. Because of its unique expression pattern in chickens and its potent role in cell death [[27], [28], [29]], GPX4 might serve as a key selenoprotein involved in the pathogenesis of NPA.
While GPX4 has been shown to protect against oxidative stress-induced cell death including apoptosis [19,28,30], the molecular linker for mediating this protection remains unknown. Prothymosin alpha (ProTalpha) is an acidic protein with a rare amino acid composition and serves as a key regulator at the gate-point of apoptotic signaling [31]. Specifically, ProTalpha can bind cytochrome c (CYTC) and then inhibit the assembly of apoptosome formed by CYTC, activated apoptotic protease activating factor 1 (Apaf-1) [32], and procaspase-9 (pro-CASP 9) in the presence of dATP [33], and block the activation of caspase-3(CASP 3) [34]. Apparently, the ability of ProTalpha to bind CYTC [35] renders it as a primary target in the search for modulators of cell death [[36], [37], [38], [39]]. However, it remain unknown if GPX4 interacts with ProTalpha to protect against apoptosis in NPA.
Therefore, we performed a series of experiments using hatching chicks and primary pancreatic acinar cells to determine: 1) if the classical Se deficiency disease NPA could be replicated in modern chickens by feeding a practical, low-Se corn-soy diet; 2) how the onset of NPA was correlated with the global expression of selenogenome and Se-independent genes related to cell death and tissue lesions; and 3) if and how GPX4 played a central role in apoptosis associated with NPA. In the end, we applied subgerminal cavity injection of a shRNA-expressing lentivirus to silence GPX4 expression in chick embryo for testing its in vivo function.
2. Materials and methods
2.1. Animal, diet, and experimental design
All animal experiments were performed according to guidelines provided by the Animal Welfare Act and Animal Welfare Ordinance. Our animal protocol was approved by the Animal Care and Use Committee of China Agriculture University (Beijing, China; Registration number: SYDW202001; allowed experimental animal number = 240). Day-old male broiler chicks (4 groups, n = 60/group; 5 replicates of 12 chicks/cage) were selected from DafaZhengda Poultry (Beijing, China). The basal diet (BD; 14 μg Se/kg; Supplementary Table 1) was composed of corn and soybean produced in the Se-deficient area of Sichuan, China, and was not supplemented with Se or Vit. E (–Se–Vit. E). Other three experimental diets were supplemented with rac-α-tocopheryl acetate at 50 mg/kg (–Se + Vit. E), Se (as sodium selenite) at 0.3 mg/kg (+Se–Vit. E), or both (+Se + Vit. E). The feeding trial lasted for 4 wk. Animal care and growth performance data collection were the same as described previously [24,40]. Incidences of NPA, based on the gross symptoms, were checked and recorded thrice daily.
2.2. Tissue sample collection, preparation, and biochemical analyses
On days 7, 14, 18, 21, and 28 of the study, blood and pancreas were collected from individual chicks (n = 8/group) as previously described [24]. After pancreas was immediately dissected on an ice-cold surface to measure weights and lengths, the tissue was washed with ice-cold isotonic saline. After a small fraction was taken for histology (day 21, see below), the rest of pancreas was minced, divided into aliquots, snap-frozen in liquid nitrogen, and stored at –80°C until use. Tissue concentrations of malondialdehyde, protein, and Se, total activities of GPX, catalase, and superoxide dismutase, and total antioxidant capacity were determined as previously described [[41], [42], [43]].
2.3. Histology, immunohistochemistry, and electron microscope
Morphology of pancreas: To preserve tissue morphology and retain the antigenicity of the target molecules, pancreas samples of chicks (n = 5/group) on day 21 were first perfused with formaldehyde fixative solution. Histologic changes were examined by two certified veterinary pathologists independently after the fixed pancreas were embedded in paraffin, sectioned at 6 mm, and stained with hematoxylin and eosin. Images were taken with a Zeiss AxioVert A1 inverted microscope (Carl Zeiss, Jena, Germany) [24].
Ultrastructural examination: For electron microscopy, pancreas tissue specimens were fixed with 2.5% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.2) for 3 h, washed in the same buffer for 1 h, and post fixed with 1% osmium tetroxide in sodium phosphate buffer for 1 h (all steps at 4°C). The tissues were then dehydrated in graded series of ethanol, starting at 50% with each step for 10 min after two changes in propylene oxide. The tissue specimens were embedded in araldite. Ultrathin sections were stained with Mg-uranyl acetate and lead citrate for transmission electron microscope (Hitachi-7650, Tokyo, Japan) evaluation.
Apoptosis assay: The TUNEL test was performed using an apoptosis detection kit (Roche, Basel, Switzerland) according to the manufacturer's protocol. On each slide, apoptotic cells were counted from at least five different fields using the Image-proplus software (version 6.0 for windows, Media Cybernetics, Silver Spring, MD, USA) with the aid of a microscope (Carl Zeiss) at 400 × magnification.
Immunohistochemistry: Formalin-fixed, paraffin-embedded samples were cut at a thickness of 5 μm. Each tissue section was deparaffinized and rehydrated with graded ethanol concentrations. For antigen retrieval, slides were boiled in EDTA (1 mM, pH 8.0) for 15 min in a microwave oven. Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide for 10 min at room temperature. After rinsing with PBS, slides were incubated overnight at 4°C with a polyclonal antibody against GPX4 (1:100 dilution). The antigen-antibody binding was detected with an Envision Detection Kit, Peroxidase/DAB, Rabbit/Mouse (Gene Tech, Shanghai, China). Sections were counter stained with hematoxylin. For negative control, isotype-matched primary antibody replaced the anti-GPX4 antibody followed by the same secondary antibody used in the other sections. Positive areas stained with GPX4 were examined in all specimens using a microscope (Carl Zeiss). The anti-GPX4 (Santa Cruz Biotech, Santa Cruz, CA, USA) antibody was used for the immunohistochemical examination, and the secondary antibody was obtained from the ZSGB-Biotechnology (Beijing, China). Images were taken with a Zeiss AxioVert A1 inverted microscope (Carl Zeiss) [24]. The intensity of the dye color was graded as 0 (no color), 1 (light yellow), 2 (light brown), or 3 (brown), and the number of positive cells were graded as 0 (<5%), 1 (5-25%), 2 (25-50%), 3 (51-75%), or 4 (>75%). The two grades were added together and specimens were assigned to one of 4 levels: 0–1 score (−), 2 scores (+), 3–4 scores (++), more than 5 scores (+++). The positive expression rate was expressed as the percentage of the addition of (++) and (+++) to the total number.
2.4. Isolation, culture, and viability assay of pancreatic acinar cells
Pancreas was collected from male broiler poults (DafaZhengda Poultry Co., Ltd.) at 2 weeks of age and was digested with 1 mg/mL collagenase V (Sigma C-9263) for three sequential 5-min periods (with fresh collagenase each time) in shaking water bath (120 cycles/min, 37°C). The digested tissue was then gently pipetted, filtered through a nylon mesh (150 mesh), and centrifuged in buffer containing 4% bovine serum albumin and washed three times. Cells were then harvested and cultured on a sticky culture dish (Corning, Corning, NY, USA) in F-12K Nutrient Mixture (1 × ) medium (21127-022, Gibco, Gaithersburg, MD, USA) supplemented with 10% of fetal bovine serum (FBS, Gibco). The cells were cultured in the medium to allow attachment of fibroblasts to the culture dish. After 6-8 h of culture, floating cells were collected and re-plated on conventional culture dishes (Corning) in F-12K Nutrient Mixture (1 × ) medium supplemented with 20% FBS. The standard buffer in this work had the following composition (in mM): 118 NaCl, 4.7 KCl, 2.5 CaCl2, 1.13 MgCl2, 1.0 NaH2PO4, 5.5 d-glucose, 10 HEPES, 2.0 l-glutamine, and 2 g/L bovine serum albumin, 2% minimum essential medium amino acids mixture (Gibco), 0.1 g/L soybean trypsin inhibitor, pH adjusted to 7.4 with 4 M NaOH. The cell viability was determined using the Cell Titer-Glo luminescent cell viability assay kit (Promega, Madison, WI, USA), according to the kit instructions.
2.5. Gene expression and protein production analyses
Digital gene expression (DGE): Sequence tag preparation was performed with DGE Tag Profile Kit (Illumina, San Diego, California, USA), according to the manufacturer's instructions. Briefly, 1 μg of total RNA per sample (pancreases of 18-day old chicks, 4 groups, and each group pooled from 5 individual chicks) was incubated with oligo-dT beads to capture the polyadenlyated RNA fraction. The samples were sequenced according to the manufacturer's instructions at the gene pool. Image analysis and base-calling were performed using the illumine pipeline, where sequence tags were obtained after quality filtering. All tags were mapped to the in silico generated transcriptome of chicken (the most closely related, fully annotated genome available for the chicken). We used Hisat2 (v2.1.0) to align the tags to the chicken genome [44] and FeatureCounts (v1.6) to count the number of reads mapped to each gene [45]. Based on the transcriptome results obtained, we applied DESeq2 (1.32.0) to perform the DGE analysis using a P-value <0.05 and fold-change >2 [46]. Short tags generated by end nucleases from the 3ʼ ends of genes (the copy number of each tag indicates the expression level of the corresponding gene) were sequenced by the DGE analysis [46]. These analyses were performed to investigate the metabolism-related genes that were differentially expressed under the Se deficiency compared with Se adequacy in chicks. To take a more contextual view of the functions of these differentially expressed genes, we explored enrichment analyses of those genes into involved molecular mechanisms. The Gene Ontology (GO) database and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database were used to analyze the GO and pathway enrichment annotations, respectively. The results were defined as significant if the GO terms or pathways had a P value < 0.05 and a FDR <0.05 [47].
Quantitative RT-PCR: Total RNA was isolated from pancreas tissue and pancreatic acinar cells using Tri-reagent (Invitrogen) and then purified using QiagenRNAEasy Mini Kit according to the manufacturer's instructions. For qRT-PCR analysis, cDNA was synthesized from 1.0 μg of RNA by using a cDNA synthesis kit (Invitrogen). Real time PCR was done in a 25 μL of reaction using SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA), chicken specific oligo nucleotides (Supplementary Table 2), and Step One Plus Real-Time PCR System (Applied Biosystems). Relative fold differences between an experimental and calibrator sample were calculated by using comparative Ct (2−ΔΔCt) method. β-actin (ACTB) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used as the reference genes [48].
Western blot: Tissues and cells were homogenized with the cell lysis buffers for Western and(or) immune-precipitation (Catalog no. P0013, Beyotime Institute of Biotechnology, Beijing, China) and centrifuged at 12,000 g for 10 min at 4°C. The resulting supernatants of homogenates (10–40 μg protein/lane) were loaded onto a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 12.5%), transferred to polyvinylidene difluoride membranes, and incubated with appropriate antibodies (Supplementary Table 3) as described previously [49]. The signal of the protein bands was detected using a chemiluminescence analysis system (Amersham Imager 600, Amersham Biosciences, Piscataway, NJ, USA).
2.6. siRNA preparation and transfection
The siRNA corresponding to the GPX4 gene was designed and synthesized by Shanghai Biolino Acid Technology Co., Ltd. The sequences (siGPX4-1: sense 5′-GGUGCUUCGUCUGAAUCAU-3′, anti-sense 5′-AUGAUUCAGACGAAGCACC-3'; siGPX4-2: sense 5′-GCGCAGAUCAAAGCCUUUG-3′, anti-sense 5′-CAAAGGCUUUGAUCUGCGC-3′) were as the GPX4 siRNA sequences. A random siRNA sequence (sense 5′-CGGUCGUGGACACCGACGCCCUAAA-3'; anti-sense 5' -UUUAGGGCGUCGGUGUCCACGACCG-3′) was used as a negative control and had no homology with any genes. Pancreatic acinar cells were seeded into six-well plates at 70-80% confluence and transfected with 3 μL of 20 μM siGPX4-1 and siGPX4-2 and 3μL of Lipofect AMINE 2000 transfection reagent (Invitrogen) in 2 mL of Opti-MEM (Invitrogen), following the manufacturer's instructions. The pancreatic acinar cells were transfected with siRNA using Lipofect AMINE 2000 for 12 h, 24 h, 48 h, and 72 h for the GPX4 mRNA analysis, respectively. After transfection for 42 h, the cells were treated with 50 μM or 100 μM H2O2 in differentiation medium for 6 h, and then were harvested for analysis.
2.7. Cloning and expression of chicken GPX4
Total RNA was extracted from chicken pancreas samples (50 mg) using Trizol reagent (Invitrogen). The first strand cDNA was synthesized using oligo dT primers and superscript II reverse transcriptase (Invitrogen), according to the manufacturer's instructions. Unless stated otherwise, the resulting cDNA pool was used directly for PCR amplification. Using the chicken sequence from GenBank (accession no. XM_003642871.2), primers were designed to amplify chicken GPX4 (sense 5′- TCTAAGCTT CGGAGAATGTGCGCTCA -3'; anti-sense 5′-CGGAATTCGACAGCAGATACTACATTAGGGC-3′). The PCR amplifications were performed using Taq DNA polymerase (Fermentas), and the PCR product was separated on a 1% agarose gel. The amplified DNA fragments were then purified from the agarose gels and subcloned into the pMD-18T (TaKaRa, Dalian, China) plasmid vector. The DNA fragment that contained the cDNA was released from the modified pMD-18T plasmid by digestion with EcoRI and Hind III and was then sub-cloned into the mammalian expression vector pcDNA3.1(+) (Invitrogen), yielding pcDNA3.1/GPX4. The fragment insert was then sequenced (Huada, Beijing, China). Fragments containing the predicted GPX4 binding (restriction) sites were amplified by PCR using primers: sense5′-GCAAGCTTCGATGTGCGCTCAGGCGGACGA-3'; anti-sense5′- ATGGTACCCAGGTAGGCGGGCAGATCCT-3'. The amplified fragment was cloned downstream of EGFP CDS (coding sequence) with stop codons in pcDNA3.1(+)/EGFP at the Hind III and KpnI sites. And the resultant constructs were named pcDNA3.1/GPX4-EGFP.
Transfections were performed using the Entranster-H DNA transfection reagent (Engreen, Beijing, China), following the manufacturer's instructions. A 3 μg of pcDNA3.1/GPX4-EGFP that encoded chicken GPX4 was added to 200 μL of Opti-MEM (Invitrogen) before mixing with 3 μL of Entranster-H DNA transfection reagent. The plasmid DNA/Entranster-H mixture was incubated at room temperature for 25 min and then added into each well of the 6-well plates. After transfection for 48 h, the cells were treated with 50 μM H2O2 in the differentiation medium for 6 h and then harvested for analysis.
2.8. Co-immunoprecipitation and confocal microscopy
Screening for the GPX4 interacting proteins: To identify candidate proteins interacting with GPX4, we performed GST pull-down and mass spectrometric (MS) screening. GST-GPX4 (TGA was mutated to TCA) protein was produced by inserting the GPX4 gene into an expression vector pET28a (Novagen, Madison, WI, USA) and expressing the plasmid in E. coli (BL21). The expressed protein was purified using the GST-tag (GE Healthcare, Piscataway, NJ, USA), immobilized on glutathione sepharose beads (GE Healthcare), and incubated with homogenate of chicken pancreatic acinar cells (chick, 2-wk old) for 12 h. The binding complex was pulled down by centrifugation, eluted, and subjected to mass spectrometry (MS). Peptide mass information was obtained using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF/TOF) MS (Autoflex II, Bruker Daltonik, Bremen, Germany). Proteins were identified using MASCOT software (Matrix Science, Boston, MA, USA) and the NCBI data base for chicken proteome to search for peptide mass finger printing and peptide sequence tagging. Candidates with more than 2 matched peptides were selected for subsequent tests [48].
Co-immunoprecipitation: To determine if candidate proteins selected above actually bound GPX4, we performed co-immunoprecipitation experiments. Supernatants from pancreatic acinar cell or pancreas lysates (70 μg of protein) were incubated with 1 μg of the anti-GPX4 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 2 h at 4°C with agitation. Thereafter, the protein-antibody complex was precipitated by centrifugation at 15,000×g for 2 min and washed four times with RIPA buffer and once with 50 mM Tris-HCl (pH 7.5). The collected beads were used for SDS-PAGE analysis and immunoblotting as described previously [48]. To determine if ProTalpha bound GPX4 in pancreas and if the binding was affected by dietary Se deficiency, we homogenized pancreas from chicks fed the –Se + Vit. E and +Se + Vit. E diet (18-day old) in lysis buffer. Then, 200 μL of the lysates (70 μg protein) were mixed with 1 μg of anti-GPX4 antibody (Santa Cruz). After the pull down by dynabeads Protein G, the collected beads were used for SDS-PAGE analysis and immunoblotting against the anti-ProTalpha antibody (Santa Cruz).
Confocal microscopy: To determine if GPX4 co-localized or bound ProTalpha in pancreatic aciner cells, we labeled anti-ProTalpha antibody with a fluorescent dye DyLight™ 594 with a 1:5 (mol/mol) molar excess according to the manufacturer's instruction. The DyLight™ 594-labeled ProTalpha antibody (DyLight™ 594- ProTalpha) was then purified through a fluorescent dye removal column (Pierce, Rockford, IL, USA). The DyLight™ 594-ProTalpha antibody was added to the cultured GPX4-enhanced green fluorescent protein (EGFP) overexpressing acinar cells at a final concentration of 100 mg/mL. After 48 h of trasnfection, the cells were fixed in 4% paraformaldehyde. The GPX4-EGFP overexpressing acinar cell nucleus were labeled with 4 mg/mL Hoechst 33258. Fluorescent images were captured using a Zeiss LSM 710 confocal laser scanning microscope (Carl Zeiss) with a Plan-Apochromat × 63/1.40 M27 glycerol immersion objective. The same gain, pin hole, and laser power settings were employed for imaging each treatment to achieve comparative results. The laser lines at 405 nm, 594 nm, and 488 nm were used for excitation of Hoechst 33258, DyLight™ 594, and EGFP, respectively. The emission wave length range for each dye was selected according to the ZEN 2009 SP1 software (Carl Zeiss) and corresponding single optical sections were collected using an image format of 1024 × 1024 pixels.
2.9. Biochemical impacts of GPX4 and ProTalpha interaction in acinar cells
Altering GPX4 expression on the binding of GPX4 and ProTalpha: To determine how GPX4 overexpression or silence affected protein production of ProTalpha and its binding to GPX4 in pancreatic acinar cells, we transfected the cells with control, GPX4 (OE), or the siGPX4 constructs, respectively. After 48 h transfection, cells (2 x 106) were harvested and lysed to obtain 200 μL aliquots of lysates (70 μg of protein) that were incubated with 1 μg of anti-ProTalpha antibody (Santa Cruz) for 2 h at 4 °C with agitation. After pulling down by dynabeads Protein G, the collected beads were used for SDS-PAGE analysis and immunoblotting against anti-GPX4 or anti-ProTalpha antibody (Santa Cruz).
Silencing ProTalpha on cell viability: To test if silencing ProTalpha led the gene deficiency and thereby induced apoptosis, we selected two regions of siRNA (siProTalpha-1 and siProTalpha-2) targeting to knock down the expression of ProTalpha in the pancreatic acinar cells of chicks. After 12 h, 24 h, 48 h, and 72 h transfections with control, negative ccontrol (Nc), siProTalpha-1, and siProTalpha-2, respectively, cells (2 x 106) were harvested for detecting mRNA and protein (48 h) levels of ProTalpha. Subsequently, we transfected the cells with siProTalpha-2 to determine effects of ProTalpha deficiency on cell viability. After 42 h of transfection, the cells (1.3 x 105) were treated with 50 μM or 100 μM H2O2 in differentiation medium for 6 h, and harvested for cell viability detection (Cell Titer-Glo luminescent cell viability assay kit, Promega).
Silencing ProTalpha on apoptotic signaling and cell death: To determine the ProTalpha deficiency activated apoptotic signaling, we transfected the pancreatic acinar cells with siProTalpha-2 for 42 h as described above. The transfected cells (2 x 106) were treated with 50 μM H2O2 in differentiation medium for 6 h, and then were harvested for Western blot analyses of ProTalpha, CYTC, pro-CASP 9, cleaved-CASP 9, pro-CASP 9, and cleaved-CASP 9. To determine the role and interaction of GPX4 and ProTalpha on cell apoptosis, we transfected the cells with siGPX4, siProTalpha-2, or both. After 42 h of transfection, cells (1.3 x 105) were treated with 0 μM or 50 μM H2O2 in differentiation medium for 6 h and then were harvested for cell apoptosis detection (Catalog no. C1065S, Beyotime).
2.10. Impacts of subgerminal cavity microinjection of lentiviral GPX4 vector
Construct preparation and subgerminal cavity injection: To determine the role of GPX4 in the embryonic development of chicks, we constructed short-hairpin RNA (shRNA) of GPX4 vectors using the pLL3.7 lentivirus-based vector that efficiently expresses shRNAs under the control of the U6 promoter with downstream HpaI and XhoI restriction sites (Supplemental Fig. 1A). The GPX4 shRNA sequence(5′-ATCGTTAACGGTGCTTCGTCTGAATCATCG AAATGATTCAGACGAAGCACCTTTTCTCGAGCAT-3′) and GPX1 shRNA sequence (5'-ATCGTTAACACGGCTTCACCCAACTTCACGAATGAAGTTGGGTGAAGCCGTTTTTCT CGAGCAT-3′) were designed, synthesized by the Sangon Biotech Co. (Beijing, China), and subcloned into the pLL3.7 vector. The recombinant vector sequence was confirmed by PCR, restriction enzyme digestion, and sequence analysis, and the resulting plasmids were named pLL3.7-GPX4-shRNA and pLL3.7-GPX1-shRNA, respectively. Plasmids used for transfection were purified using the Endofree Plasmid Maxi kit (Qiagen, Hilden, Germany). Lentiviral production was performed by Sangon Biotech Co. (Beijing, China). Briefly, 293FT cells were plated on 60 mm dishes and transfected with the pLL3.7-GPX4-shRNA or pLL3.7-GPX1-shRNAand packaging vectors, including pMDLg/pRRE, pRSV-Rev and pMD2.G. The resulting supernatant was collected and passed through a 0.22 μm filter after 48 h. Virus particles were concentrated via ultracentrifugation for 90 min at 25,000 g. The virus was re-suspended in PBS for 4 h and then stored at –80 °C. Cultures of 293FT cells were infected with serial dilutions of concentrated lentivirus to determine viral titres. The ratio of total cells to cells expressing EGFP was determined at 72 h after infection via microscopy.
A total of 6 x103 chicken embryonic stem (chES) cells derived from the area pellucida of the stage x chicken blastoderm were infected with 1 μL of the lentiviral vector. In subgerminal cavity injection groups, freshly laid, sterilized eggs were positioned horizontally for 4–6 h. A window (3 × 3 mm2) in the shell was opened using a dental drill and tweezers. One microliter (1 × 109 TU/mL) of pLL3.7-GPX4-shRNA, pLL3.7-GPX1-shRNA or control of recombinant vectors was injected into the subgerminal cavity of the blastoderm of stage X blastodermal embryos (Supplemental Fig. 1). The window was then sealed with Parafilm® and the eggs incubated until hatching. Eggs injected with the empty vector were used as controls.
Metabolic impacts of shGPX4 in chicken embryo: To determine the silence of GPX4 expression and its biochemical and metabolic impacts, we collected fresh embryos on day 8 to determine EGFP expression using a fluorescenece microscope (Carl Zeiss) and embryonic pancreas samples on day 18 for histological and anatomical measures. Relative mRNA and(or) protein levels of GPX4 and apoptotic signal proteins (ProTalpha, CYTC, pro-CASP 9, cleaved-CASP 9, pro-CASP 9, and cleaved-CASP 9) were also detected in the day 18 embryonic pancreas. Hatching rates of eggs transfected with shGPX4 and shGPX1 were compared with those transfected with the vector only.
2.11. Data analysis
Data from the four groups of the animal experiment were analyzed as 2 × 2 factorial arrangement of treatments using two-way ANOVA. The rest of data were analyzed by one-way ANOVA or Student t-test. Means were compared by Tukey honestly significant difference test. Data were presented as mean ± SE, and significance level was set at P < 0.05. All data were analyzed using SPSS for Windows (version 13; SPSS Inc, Chicago, IL, USA).
3. Results
3.1. Chicks fed low-Se diets developed classical symptoms of NPA
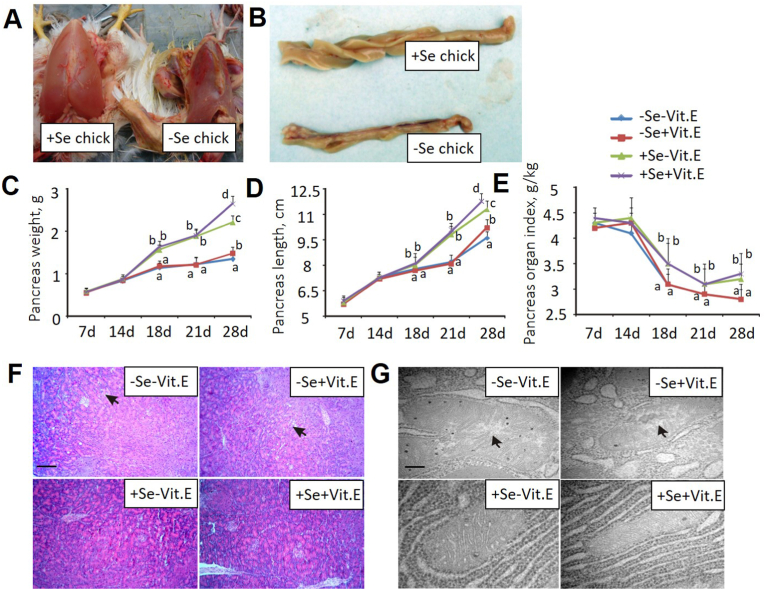
Chicks fed the two Se-deficient diets developed NPA, along with poor growth, poor feathering, and mortality as early as on day 18 (Fig. 1A). By day 21, NPA was diagnosed in 15 chicks in the –Se–Vit. E group and 2 chicks in the –Se + Vit. E group. In contrast, no single incidence of NPA appeared in those fed the +Se diets throughout. Compared with the +Se chicks, the -Se chicks had much smaller pancreas (Fig. 1B), with 27-42% decreases in weight (Fig. 1C), 4-18% decreases in length (Fig. 1D), and 6-15% decreases in pancreas organ index (Fig, 1E) on days 18–28, respectively. Compared with the –Vit. E chicks, the +Vit. E chicks had pancreas that was 14% higher in weight (Fig. 1C) and 6% increase in length (Fig. 1D) on day 28, respectively. By day 18, chicks fed the –Se diet showed histological signs of pancreatic atrophy: pronounced intracellular vacuolation and hyaline body formation (Fig. 1F).Transmission electron microscopy of severely disrupted acinar mitochondria in those chicks revealed crest broken and vesiculated mitochondria (Fig. 1G). Compared with the +Se chicks, the –Se chicks had lower (P < 0.05) concentrations of pancreatic Se (40-52%), activities of pancreatic GPX (45-62%), catalase (12-75%), and superoxide dismutase (22%), and total antioxidant capacity (53%), but higher pancreatic malondialdehyde contents (1.3-3.2 fold) at various time-points (Table 1).
Fig. 1.
Nutritional pancreatic atrophy induced by Se deficiency in chicks.
(A) Autopsy showing subcutaneous hemorrhage and a greenish, gelatinous edema under the skin of dependent portions of the body as well as thin (paper-thin) pectoral muscles of –Se + Vit. E vs + Se + Vit. E chicks on day 21.
(B) Pancreatic atrophy in the –Se + Vit. E verse the +Se + Vit. E chicks on day 21.
(C), (D), and (E) Effects of dietary Se and Vit. E concentrations on pancreas weights, length, and organ index on days 7–28. Data are means ± SE, n = 8. Values within a given time differ (P < 0.05) without sharing a common superscript letter.
(E) Hematoxylin and eosin (H&E) staining of chick pancreas on 28 days. Arrows indicate cytoplasmic vacuoles and hyaline bodies in acinar cells. Images were taken with 400 × magnification. The image was a representative of five sets of data.
(F) Ultrastructural images of the pancreatic acinar cells of chicks on day 28. Arrows indicate crest broken and vesiculated mitochondria in the cells. Images were taken with 15000 × magnification. The image was a representative of five sets of data.
Abbreviations: Se, selenium deficient; +Se, selenium adequate; –Vit. E, vitamin E deficient; +Vit. E, vitamin E adequate.
Table 1.
Effects of dietary Se and vitamin E concentrations on biochemical indicators in the pancreas of chicks.
| Se, mg/kg | 0 | 0 | 0.3 | 0.3 |
|---|---|---|---|---|
| Vit.E, mg/kg | 0 | 50 | 0 | 50 |
| Se, ng/g tissue | ||||
| 7d | 192 ± 18.2a | 196 ± 13.2a | 363 ± 11.0b | 376 ± 34.2b |
| 14d | 157 ± 14.2a | 160 ± 13.4a | 334 ± 28.4b | 335 ± 24.9b |
| 21d | 132 ± 12.3a | 135 ± 12.3a | 302 ± 23.0b | 310 ± 30.4b |
| 28d | 118 ± 8.6a | 120 ± 9.5a | 299 ± 20.7b | 303 ± 24.2b |
| Glutathione peroxidase, unit/mg protein | ||||
| 7d | 20.3 ± 2.10 | 21.1 ± 3.30 | 22.0 ± 3.00 | 23.2 ± 2.20 |
| 14d | 14.7 ± 2.40a | 15.0 ± 2.20a | 23.0 ± 2.10b | 25.2 ± 2.40b |
| 21d | 12.3 ± 2.20a | 14.0 ± 3.10a | 25.0 ± 2.30b | 26.3 ± 2.10b |
| 28d | 11.4 ± 2.0a | 12.0 ± 1.90a | 24.5 ± 1.70b | 27.4 ± 2.30b |
| Catalase, unit/mg protein | ||||
| 14d | 1.4 ± 0.3 | 1.5 ± 0.3 | 1.6 ± 0.3 | 1.7 ± 0.3 |
| 18d | 0.6 ± 0.1a | 0.7 ± 0.3a | 1.5 ± 0.1b | 1.4 ± 0.1b |
| 28d | 0.3 ± 0.0a | 0.3 ± 0.0a | 1.1 ± 0.2b | 1.2 ± 0.3b |
| Superoxide dismutase, unit/mg protein | ||||
| 14d | 117 ± 6.70 | 116 ± 7.10 | 114 ± 7.50 | 116 ± 7.40 |
| 18d | 101 ± 8.60a | 107 ± 7.40a | 121 ± 9.20b | 120 ± 8.70b |
| 28d | 98.0 ± 6.40a | 96.4 ± 7.20a | 7 ± 8.10b | 4 ± 9.20b |
| Malondialdehyde,μmol/mg protein | ||||
| 14d | 4.2 ± 0.30b | 4.4 ± 0.20b | 1.9 ± 0.30a | 1.8 ± 0.20a |
| 18d | 6.2 ± 0.30b | 6.4 ± 0.20b | 1.6 ± 0.20a | 1.7 ± 0.20a |
| 28d | 7.4 ± 0.60b | 7.6 ± 0.70b | 1.8 ± 0.20a | 1.7 ± 0.20a |
| Total antioxidant capacity, mmol/g protein | ||||
| 14d | 2.6 ± 0.20a | 2.8 ± 0.30a | 4.7 ± 0.30b | 4.5 ± 0.30b |
| 18d | 2.0 ± 0.20a | 1.9 ± 0.20a | 4.3 ± 0.40b | 4.2 ± 0.30b |
| 28d | 1.8 ± 0.20a | 1.7 ± 0.20a | 3.7 ± 0.30b | 3.8 ± 0.20b |
Values are means ± SE, n = 5. Means in row without sharing a common letter differ, P < 0.05.
3.2. Gene expressions of Se metabolism and apoptotic pathway were altered in chicks with NPA
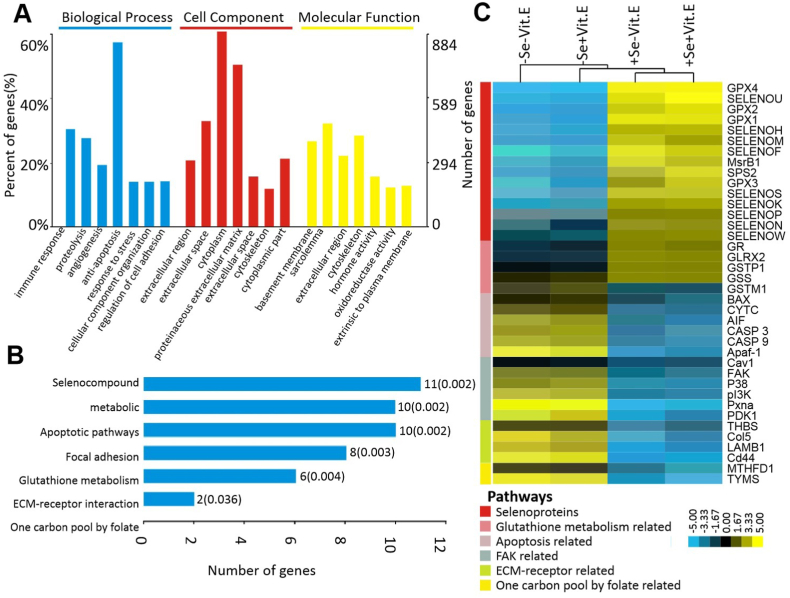
Dietary Se deficiency increased and decreased the expression of 360 and 524 unigenes, respectively, in the pancreas of chicks on day 18. The GO enrichment analysis outcomes are presented in Fig. 2A. Of the 884 differentially expressed unigenes, 530 were annotated and 65 were classified into metabolism pathways. KEGG analysis indicated that these genes were significantly enriched in the functions of selenocompound metabolism, apoptotic pathways, focal adhesion, glutathione metabolism, ECM-receptor interaction, and one carbon pool by folate (Fig. 2B). Expression changes of specific genes related to those functions among the four dietary treatment groups are listed in Fig. 2C. Compared with the +Se diets, the –Se diets decreased expression of 15 selenoprotein genes and glutathione metabolism-related genes (GR, GSTP1, and GSS), but up-regulated expression of apoptosis-related genes (AIF, Apaf-1, BAX, CASP 3, CASP 9, and CYTC), FAK-related genes (Cav 5, FAK, p38, PDX1, PI3K, andPxna), ECM-receptor related genes (Cd44, Col 5, LAMB1, and THBS), and one carbon pool/folate-related genes (MTHFD1 and TYMS). The quality of the DGE analyses was checked by Q-PCR analysis of four key genes (GPX1, GPX4, CYTC, and CASP 3) selected from the identified pathways (Supplemental Fig. 2).
Fig. 2.
The GO analysis (A) with GO database and the pathway analysis (B) with KEGG database were applied for functional annotation. The significant GO terms and pathways were defined as P < 0.05 and FDR <0.05. (C) Heatmap showing the significant top 38 mRNAs affected by dietary Se and Vit. E deficiency in the pancreas of chicks on day 18. Yellow: increased; blue: decreased. P < 0.05 and FDR <0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Abbreviations: Se, selenium deficient; +Se, selenium adequate; –Vit. E, vitamin E deficient; +Vit. E, vitamin E adequate; Apaf-1, apoptotic peptidase activating factor 1; AIF, apoptosis inducing factor; BAX, BCL2 associated X; CASP 3, caspase 3; CASP 9, caspase 9; Cav1,caveolin 1; Cd44, CD44 molecule; CYTC, cytochrome c; Col 5, collage 5; GO, gene ontology; GPX1, glutathione peroxidase 1; GPX2, glutathione peroxidase 2; GPX3, glutathione peroxidase 3; GPX4, glutathione peroxidase 4; GLRX2, glutaredoxin 2; GR, glutathione reductase; GSS, glutathione synthetase; GSTM1, glutathione S-transferase mu 1; GSTP1, glutathione S-transferase pi 1; LAMB1, laminin subunit beta 1; KEGG, kyoto encyclopedia of genes and genomes; MsrB1, methionine sulfoxide reductase B1; MTHFD1, methylenetetrahydrofolate dehydrogenase, cyclohydrolase and for myltetrahydrofolate synthetase 1; p38, p38 kinase; PDK1, pyruvate dehydrogenase kinase 1; PI3K, phosphatidylinositol 3-kinase; Pxna, paxillin a; SELENOF, selenoprotein 15; SELENOK, selenoprotein K; SELENOH, selenoprotein H; SELENOM, selenoprotein M; SELENON, selenoprotein N; SELENOP, selenoprotein P; SELENOS, selenoprotein S; SELENOU, selenoprotein U; SELENOW, selenoprotein W; SPS2, selenophosphate synthetase 2; THBS, thrombospondin 1; TYMS, thymidylate synthetase.
3.3. GPX4 protein was depleted in the pancreas of chicks with NPA
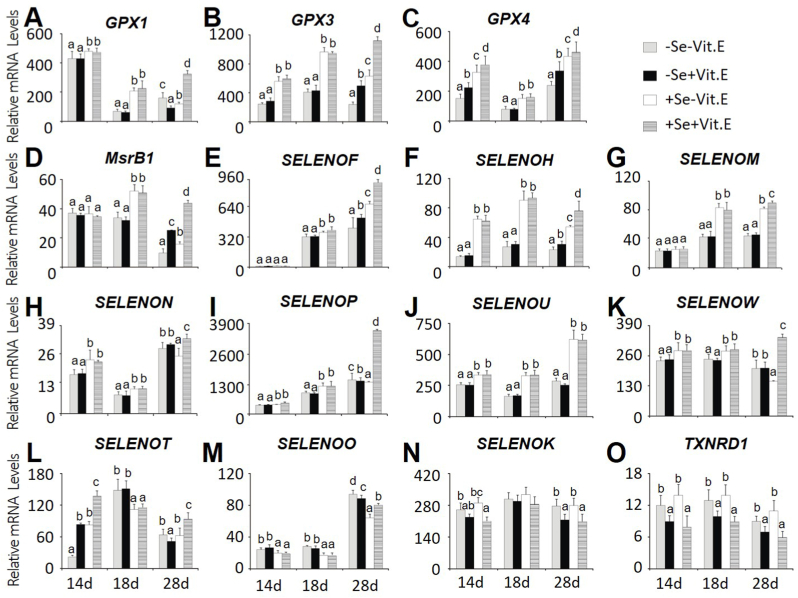
Expression of 11 of the 24 selenoproteingenesin pancreas (Fig. 3A–K) was down-regulated (P < 0.05) by dietary Se deficiency at most point times. Meanwhile, mRNA levels of SELENOT (Fig. 3L) in pancreas were up-regulated on days 14 and 28 and down-regulated (P < 0.05) on day 18 by dietary Se deficiency. However, mRNA levels of SELENOO (Fig. 3M) in pancreas were up-regulated (P < 0.05) by dietary Se deficiency at the three point times. Chicks fed the + Vit. E diets had 24% and 34-75% lower pancreas mRNA levels of SELENOK and TXNRD1 (Fig. 3N, O) than those fed the –Vit. E diets at the two levels of dietary Se concentrations, respectively. Meanwhile, pancreas mRNA levels of GPX2, SELENOI, SELENOPB, SELENOS, TXNRD2, TXNRD3, DIO1, DIO2 or DIO3 were not affected by dietary Se or Vit. E (Supplemental Table 4).
Fig. 3.
Effects of dietary Se and vitamin E concentrations on relative mRNA abundance of (A) GPX1, (B) GPX3, (C) GPX4, (D) MsrB1, (E) SELENOF, (F) SELENOH, (G) SELENOM, (H) SELENON, (I) SELENOP, (J) SELENOU, (K) SELENOW, (L) SELENOT, (M) SELENOO, (N) SELENOK, and (O) TXNRD1 in the chick pancreas on days 14, 18, and 28. Data are means ± SE, n = 5. Values within a given time differ (P < 0.05) without sharing a common superscript letter. Abbreviations: Se, selenium deficient; +Se, selenium adequate; –Vit. E, vitamin E deficient; +Vit. E, vitamin E adequate; GPX1, glutathione peroxidase 1; GPX3, glutathione peroxidase 3; GPX4, glutathione peroxidase 4; MsrB1, selenoprotein R; SELENOF, selenoprotein F; SELENOH, selenoprotein H; SELENOM, selenoprotein M; SELENON, selenoprotein N; SELENOP, selenoprotein P; SELENOU, selenoprotein U; SELENOW, selenoprotein W; SELENOT, selenoprotein T; SELENOO, selenoprotein O; SELENOK, selenoprotein K, and TXNRD1, Thioredoxin reductase 1.
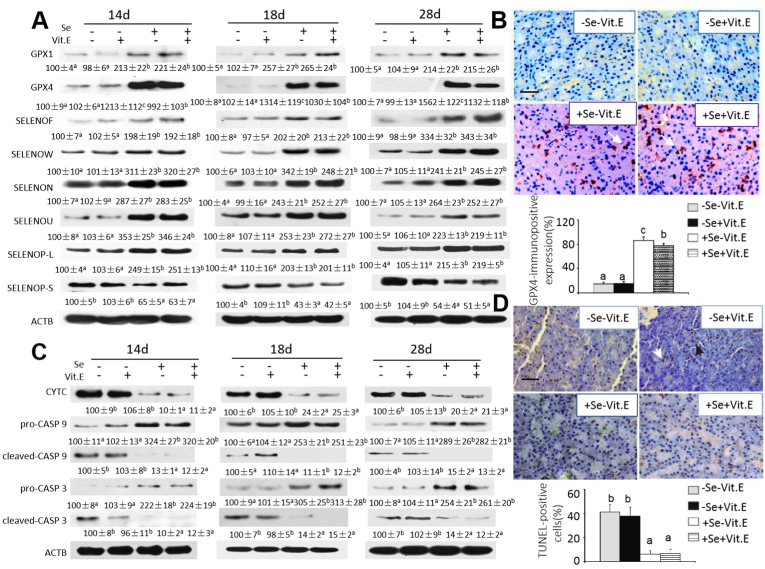
Compared with chicks fed the + Se diets, chicks fed the –Se diets had greater abundance (P < 0.05) of the short-form SELENOP, but lower abundance (P < 0.05) of the long-form SELENOP, GPX1, GPX4, SELENOF, SELENON, SELENOU, or SELENOW in the pancreas on days 14, 18, and 28 (Fig. 4A). Notably, pancreas GPX4 protein levels were decreased by 90–98% in chicks fed the –Se diets than those fed the +Se diets. While a strong positive immunohistochemical staining of GPX4 was seen in the cytoplasm and nuclei of pancreas of the +Se chicks, negligible staining occurred in that of –Se chicks on day 21(Fig. 4B). The relative staining density was 83% to 94% in chicks fed the +Se diets and only 10% in those fed the –Se diet (P < 0.05). Vit. E supplementation decreased (P < 0.05) GPX4 staining in the pancreas of +Se chicks.
Fig. 4.
Effects of dietary Se and vitamin E concentrations on pancreatic production of selenoproteins, immunohistochemistry of GPX4, production of apoptotic signal proteins, and apoptosis of chicks at various time points.
(A) Effects of dietary Se and Vit. E concentrations on pancreatic protein levels of selected selenoproteins in chick pancreas on days 14, 18, and 28. Data below the protein bands are means ± SE, n = 3. Values within a given protein differ (P < 0.05) without sharing a common superscript letter.
(B) Effects of dietary Se and Vit. E concentrations on immunohistochemistry of GPX4 in pancreas of chicks on day 21. The bar below figure shows means ± SE, n = 5. Means without sharing a common superscript letter are different (P < 0.05). Paraffin slices were treated according to the SABC immunohistochemical kit, and results were analyzed using a double-blind method. Five high-power fields ( × 400) were selected at random, and two pathologists evaluated scores independently. PBS, instead of the primary antibody, was used as negative control, and specimens were scored according to the intensity of the dye color and the number of positive cells. The intensity of the dye color was graded as 0 (no color), 1 (light yellow), 2 (light brown), or 3 (brown), and the number of positive cells was graded as 0 (<5%), 1 (5 - 25%), 2 (25 - 50%), 3 (51-75%), or 4 (>75%). The two grades were added together and specimens were assigned to one of 4 levels: 0–1 score (−), 2 scores (+), 3–4 scores (++), more than 5 scores (+++). The positive expression rate was expressed as the percent of the addition of (++) and (+++) to the total number.
(C) Effects of dietary Se and Vit. E concentrations on pancreatic protein levels of apoptotic signal proteins in chicks on days 14, 18, and 28. Data below the protein bands are means ± SE, n = 3.Values within a given protein differ (P < 0.05) without sharing a common superscript letter.
(D) Effects of dietary Se and Vit. E concentrations on apoptosis in pancreas of chicks on day 21 by TUNEL assay. TdT-mediated dUTP nick end labeling (TUNEL) was carried out for DNA fragmentation using a cell apoptosis detection kit (Roche, Basel, Switzerland) according to the manufacturer's protocol. Apoptotic cells of each slide were analyzed in at least 5 different fields by using the Image-proplus software (version 6.0 for windows, Media Cybernetics, Silver Spring, MD) with the aid of a microscope (Carl Zeiss, New York, NY) at 400× magnification. The number of positive cells was averaged for statistical analysis. TUNEL-positive myocyte nuclei (brown): white arrow; TUNEL-negative myocyte nuclei (blue): black arrow. The mean percentages of apoptosis were 41.4% (A) and 38.8% (B) in +Se–Vit. E, and +Se + Vit. E chicks, respectively. Mean percentages of apoptosis were 5.6% (C) and 6.1% (D) in pancreases in –Se–Vit. E and –Se + Vit. E chicks, respectively. Values are means ± SE, n = 5. Means without sharing a common superscript letter are different (P < 0.05).
Abbreviations: Se, selenium deficient; +Se, selenium adequate; –Vit. E, vitamin E deficient; +Vit. E, vitamin E adequate; ACTB, beta actin; cleaved-CASP 3, cleaved caspase 3; cleaved-CASP 9, cleaved caspase 9; CYTC, Cytochrome c; GPX1, glutathione peroxidase 1; GPX4, glutathione peroxidase 4; pro-CASP 3, pro caspase 3; pro-CASP 9, pro caspase 9; SELENOF, selenoprotein F; SELENOW, selenoprotein W; SELENON, selenoprotein N; SELENOU, selenoprotein U; SELENOP-L, long form of selenoprotein P; SELENOP-S, short form of selenoprotein P; TUNEL, TdT-mediated dUTP nick end labeling .
Whereas chicks fed the +Se diets had only 4%–5% TUNEL-positive or apoptotic cells in the pancreas, that rate reached 47% (P < 0.05) in those fed the –Se diets (Fig. 4D). Meanwhile, the –Se chicks had elevated amounts of (P < 0.05) CYTC (3.1 to 8.3-fold), cleaved-CASP 9 (6.3 to 7.8 fold), and cleaved-CASP 3 (5.6 to 7.9-fold, but lower amounts of (P < 0.05) pro-CASP 9 (60-69%) and pro-CASP 3 (54-67%) than the +Se groups in the pancreas on days 14, 18, and 28 (Fig. 4C).
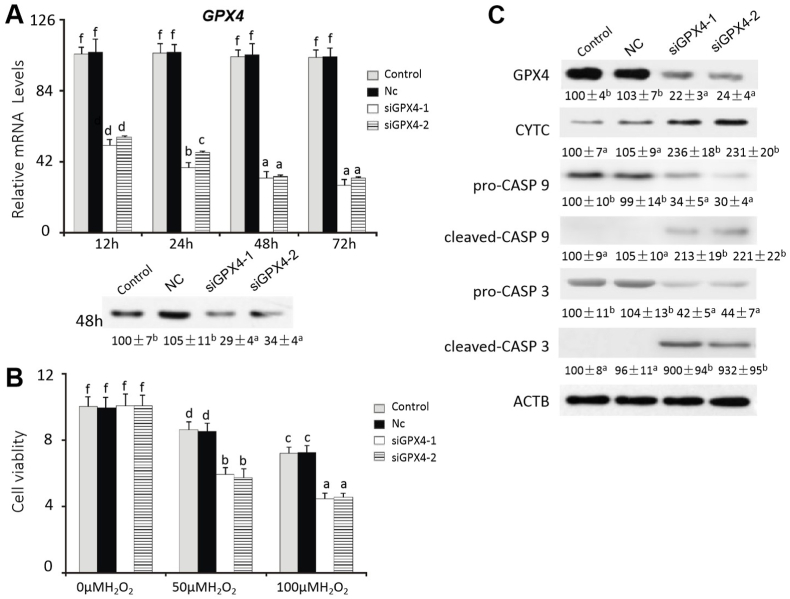
3.4. Expression of GPX4 in pancreatic acinar cells affected their susceptibility to H2O2-induced cell death
Compared with the control, GPX4 mRNA levels in the siGPX4-1 and siGPX4-2 transfected acinar cells were decreased (P < 0.05) by > 50% at 24 h, 48 h, and 72 h after the transfection. GPX4 proteins levels in the transfected cells were decreased by 66% to 71% (P < 0.05) at 48 h after the transfection (Fig. 5A). However, there was no significant difference in the mRNA and protein levels of GPX4 between the control (only reagents) and negative (siRNA) control groups. The siRNA transfections did not affect cell viability at 0 μM H2O2, but decreased it (P < 0.05) by 38-39% at 50 μM H2O2 and 41% to 42% at 100 μM H2O2, respectively, compared with the respective controls (Fig. 5B). The siRNA transfections increased (P < 0.05) CYTC (∼1.3-fold), cleaved-CASP 9 (∼1.2-fold), and cleaved-CASP 3 (∼8.2-fold (Fig. 5C), but decreased (P < 0.05) pro-CASP 9 (∼68%) and pro-CASP 3 (∼57%) in the acinar cells after the treatment of 50 μM H2O2 for 6 h (Fig. 5C).
Fig. 5.
Effects of GPX4 siRNA on H2O2-induced apoptosis in the pancreatic acinar cells of Se-adequate chicks.
(A) Effect of GPX4siRNA on relative mRNA abundances and protein levels of GPX4 in the acinar cells. Data are means ± SE, n = 5. Means without sharing a common superscript letter are different (P < 0.05).
(B) Effect of GPX4siRNA on cell viability of the pancreatic acinar cells treated with 0 μM, 50 μM, or 100 μM H2O2 for 6 h. Data are means ± SE, n = 6. Means without sharing a common superscript letter are different (P < 0.05).
(C) Effect of GPX4siRNA on protein levels of GPX4 and apoptotic signal proteins in the pancreatic acinar cells treated with 50 μM H2O2 for 6 h. Data are means ± SE, n = 3. Values below the protein bands within a given protein differ (P < 0.05) without sharing a common superscript letter.
Abbreviations: ACTB, beta actin; cleaved-CASP 3, cleaved caspase 3; cleaved-CASP 9, cleaved caspase 9; Control, acinar cells without transfection; CYTC, cytochrome c; GPX4, glutathione peroxidase 4; Nc, negative siRNA transfected cells; pro-CASP 3, pro caspase 3; pro-CASP 9, pro caspase 9; siGPX4-1, GPX4 small interfering RNA 1; siGPX4-2, GPX4 small interfering RNA 2.
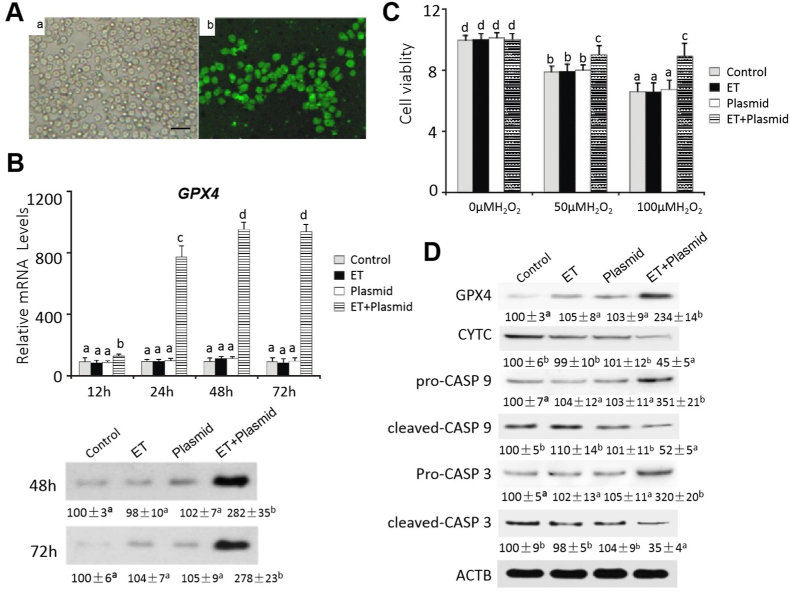
The transfection efficiency of pcDNA3.1(+)/GPX4-EGFP into the pancreatic acinar cells (48 h) were verified by immunofluorescence microscopy (Fig. 6A). Compared with the control, the pcDNA3.1(+)/GPX4-EGFP transfected cells had GPX4 mRNA levels elevated (P < 0.05) by 46%, 6.6-fold, 8.8-fold, and 8.5-fold at 12 h, 24 h, 48 h, and 72 h after the transfection, respectively (Fig. 6B). Their GPX4 protein levels were increased by 11.8-fold at 48 h or 72 h after the transfection. Their cell viability (Fi. 6C) was 13% and 28% greater (P < 0.05) than the controls after the treatments of 50 μM and 100 μM H2O2 for 6 h, respectively. Meanwhile, these cells had lower levels (P < 0.05) of CYTC (55%), cleaved-CASP 9 (48%), and cleaved-CASP 3 (65%), but greater levels (P < 0.05) of pro-CASP 9 (2.5-fold), and pro-CASP 3 (2.2-fold) than the controls after the treatment of 50 μM H2O2 for 6 h (Fig. 6D).
Fig. 6.
Effects of GPX4 overexpression on H2O2-induced apoptosis in the pancreatic acinar cells of Se-adequate chicks.
(A) Illustration of the control and GPX4 (OE) transfection into pancreatic acinar cells. a, bright filed of acinar cells (control without any transfection); b, 48 h after transfection of GPX4 (OE), fluorescence signal of GPX4 (green) in acinar cells. Images were taken with 800 × magnification. The image was a representative of five independent examinations.
(B) Effects of GPX4 overexpression on relative mRNA abundances (upper panel) and protein levels (lower panel) of GPX4 in the pancreatic acinar cells at different time points. Data are means ± SE, n = 5.
(C) Effect of GPX4 overexpression (48 h after transfection) on viability of the pancreatic acinar cells treated with 0 μM, 50 μM, or 100 μM H2O2 for 6 h. Data are means ± SE, n = 6.
(D) Effect of GPX4 overexpression (48 h after transfection) on protein levels of GPX4 and apoptotis signal proteins in the pancreatic acinar cells treated with 50 μM H2O2 for 6 h. Data are means ± SE, n = 3.
In B, C, and D, values within a given time, dose of H2O2, or protein differ (P < 0.05) without sharing a common superscript letter.
Abbreviations: ACTB, beta actin; cleaved-CASP 3, cleaved caspase 3; cleaved-CASP 9, cleaved caspase 9; Control, lysates of pancreatic acinar cells without transfection; ET, lysates of pancreatic acinar cells transfected with Entranster-H DNA transfection reagent only; CYTC, Cytochrome c; GPX4, glutathione peroxidase 4; ET + Plasmid, lysates of pancreatic acinar cells both transfected with pcDNA3.1/GPX4-EGFP and Entranster-H DNA transfection reagent; Plasmid, lysates of pancreatic acinar cells transfected with pcDNA3.1/GPX4-EGFP only; pro-CASP 3, pro caspase 3; pro-CASP 9, pro caspase 9.
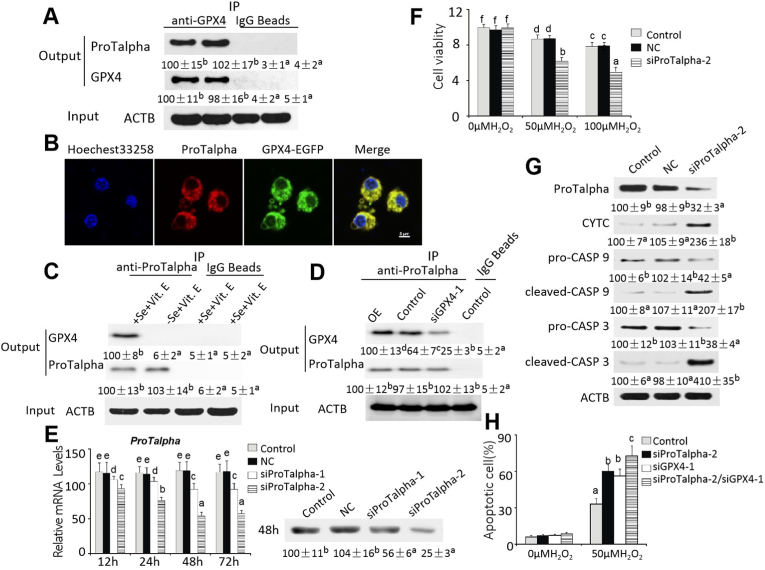
3.5. ProTalpha could interact with GPX4 and mediate its function
The GST-GPX4 pull-down and MS screening revealed 26 candidate proteins from the pancreatic acinar cell homogenates of chicks (2-wk old, +Se + Vit. E) that could potentially bind GPX4. While 6 of them had more than 2 matched peptides, ProTalpha had the highest number of matched peptides (Supplemental Table 5). When lysates of the pancreatic acinar cells of chicks (2-wk old, +Se + Vit. E) were incubated with the GPX4 antibody, the co-immuno-precipitated complex contained both GPX4 and ProTalpha proteins (Fig. 7A). The co-localization of these two proteins in the pancreatic acinar cells was verified by the co-immunofluorescence microscope assay (Fig. 7B) When the pancreas homogenates of chicks (18-day old) were incubated with the ProTalpha antibody, ProTalpha was pulled down from the homogenates of the +Se + Vit. E, but not the –Se + Vit. E chicks (Fig. 7C). Compared with the control, the ProTalpha antibody co-immuno-precipitated 56% more (P < 0.05) and 61% less (P < 0.05) of GPX4 protein from the lysates of pancreatic acinar cells transfected with the GPX4 (OE) and the siGPX4 constructs, respectively (Fig. 7D). The amounts of ProTalpha in the precipitated complex were not different among the three groups. To test if the ProTalpha deficiency induced apoptosis, we selected two regions of siRNA targeting to knock down expression of ProTalpha in the pancreatic acinar cells of chicks (+Se + Vit. E) (Fig. 7E). Compared with the control, the ProTalpha mRNA levels were decreased (P < 0.05) by 7% to 19% in the siProTalpha-1 and by > 50% in the siProTalpha-2 transfected cells, respectively. Meanwhile, the ProTalpha proteins levels in the siProTalpha-1 and siProTalpha-2 transfected cells were decreased by 44% and 75% (P < 0.05) at 48 h, respectively, compared with the control. Because of these differences between the two siRNAs, siProTalpha-2 was selected for the subsequent experiments. Compared with the control, the siProTalpha-2 transfected cells displayed 25% and 35% lower (P < 0.05) viability after the exposure to 50 μM and 100 μM H2O2, respectively (Fig. 7F). To determine the ProTalpha deficiency effect on apoptotic signaling in pancreatic acinar cells, we transfected them with siProTalpha-2. These cells had greater levels (P < 0.05) of CYTC (11.4-fold), cleaved-CASP 9 (1.1-fold), and cleaved-CASP 3 (3.1-fold), but lower levels (P < 0.05) of pro-CASP 9 (58%) and pro-CASP 3 (62%) after being treated with 50 μM H2O2 for 6 h than the controls (Fig. 7G). The H2O2-induced apoptosis rate was increased (P < 0.05) by 88% and 81% in the siGPX4 and siProTalpha-2 transfected acinar cells, respectively, and by 1.3-fold in those transfected with both siRNAs, compared with the control (Fig. 7H).
Fig. 7.
The protein protein interaction between GPX4 and ProTalpha and the impact on apoptosis in the pancreatic acinar cells.
(A) A representative image (n = 3) of immunoprecipitation of ProTalpha from the lysates of pancreatic acinar cells of chicks (2-wk old, +Se + Vit. E) by anti-GPX4 antibody.
(B) Co-immunofluorescence staining of ProTalpha (the anti-ProTalpha antibody was labeled with a red fluorescent dye DyLight™ 594) and GPX4 (green) in pancreatic acinar cells after transfected with GPX4 (OE) 48 h. The merged image of yellow fluorescence represents the co-localization of ProTalpha and GPX4 in the acinar cells. Nuclei were labeled using Hoechest33258 staining. Scale = 5 μm.
(C) A representative image (n = 3) of immunoprecipitation of GPX4 from lysates of pancreases of chicks (2-wk old, +Se + Vit. E verse –Se + Vit. E) by anti-ProTalpha antibody.
(D) A representative image (n = 3) of immunoprecipitation of GPX4 from the lysates of pancreatic acinar cells transfected with the GPX4 (OE) or siGPX4 construct by anti-ProTalpha antibody. Control, lysates of acinar cells without transfection; OE, lysates of acinar cells transfected with pcDNA3.1/GPX4-EGFP and Entranster-H DNA transfection reagent; siGPX4-1, lysates of acinar cells transfected with GPX4-siRNA-1 and Lipofect AMINE 2000 transfection reagent.
(E) Effect of ProTalpha siRNA on relative mRNA abundance and protein levels of ProTalpha in pancreatic acinar cells at different time points. Data are means ± SE, n = 6.
(F) Effect of ProTalpha siRNA (48 h after transfection) on viability of pancreatic acinar cells treated with 0 μM, 50 μM, or 100 μM H2O2 for 6 h. Data are means ± SE, n = 6.
(G) Effect of ProTalpha siRNA (48 h after transfection) on protein levels of ProTalpha, pro-CASP 9, cleaved-CASP 9, pro-CASP 3, and cleaved-CASP 3 of pancreatic acinar cells treated with 50 μM H2O2 for 6 h. Data are means ± SE, n = 3.
(H) Effect of ProTalpha and/or GPX4 siRNA (48 h after transfection) on apoptosis of pancreatic acinar cells treated with 0 μM or 50 μM H2O2 for 6 h. Data are means ± SE, n = 6.
In A, C, D, E, F, G, and H, values within a given gene, protein, time, or dose of H2O2 differ (P < 0.05) without sharing a common superscript letter.
Abbreviations: ACTB, beta actin; cleaved-CASP 3, cleaved caspase 3; cleaved-CASP 9, cleaved caspase 9; Control, acinar cells without transfection; CYTC, cytochrome c; GPX4, glutathione peroxidase 4; Nc, acinar cells both transfected with negative control siRNA and Lipofect AMINE 2000 transfection reagent; ProTalpha, prothymosin alpha; pro-CASP 3, pro caspase 3; pro-CASP 9, pro caspase 9; siGPX4-1, lysates of pancreatic acinar cells both transfected with GPX4 small interfering RNA 1 and Lipofect AMINE 2000 transfection reagent; siProTalpha-1, lysates of pancreatic acinar cells both transfected with ProTalpha small interfering RNA 1 and Lipofect AMINE 2000 transfection reagent; siProTalpha-2, lysates of pancreatic acinar cells both transfected with ProTalpha small interfering RNA 2 and Lipofect AMINE 2000 transfection reagent; siProTalpha-2/siGPX4-1, lysates of pancreatic acinar cells transfected with ProTalpha small interfering RNA 2, GPX4 small interfering RNA 1 and Lipofect AMINE 2000 transfection reagent.
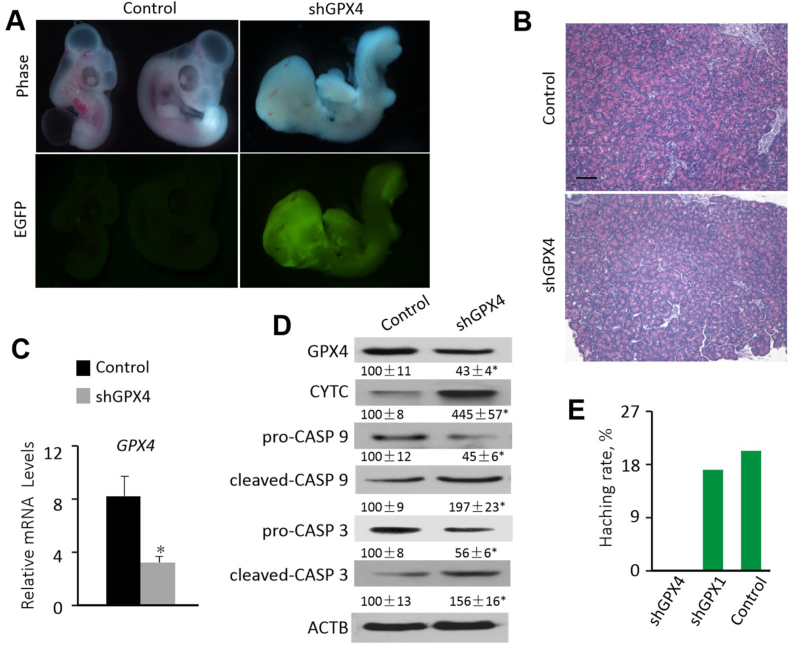
3.6. Suppression of GPX4 impaired embryonic survival and pancreatic integrity of chicks
Because pancreatic GPX4 was depleted in chicks with NPA and altering GPX4 expression in pancreatic acinar cells affected apoptotic signaling and cell death, we next sought to silence GPX4 expression through subgerminal cavity injection of the shRNA-expressing lentivirus in chick embryo to determine its physiological role and relevance. The recombinant lentiviral vector sequences were confirmed (Supplemental Fig. 2B). The presences of the EGFP (465 bp, Supplemental Fig. 2C) and the U6-CMV (684 bp, Supplemental Fig. 2D) in the blood samples of the G0 of GPX4-shRNA transgenic chicken embryos (day 18) was verified. The expression of the EGFP protein was seen in the transgenic embryo on day 8 (Fig. 8A). Compared with control, the shGPX4 chicken embryonic pancreas (day 18) showed acinar cell swelling, granular degeneration, and vacuolar degeneration (Fig. 8B). However, there was no significant difference in pancreatic length or weight (Supplemental Figs. 2E and F). In the day 18 embryonic pancreas, shGPX4 decreased (P < 0.05) GPX4 mRNA levels (34%), GPX4 protein (57%), pro-CASP 9 (55%), and pro-CASP 3 (44%), but increased (P < 0.05) CYTC (3.5-fold), cleaved-CASP 9 (97%), and cleaved-CASP 3 (56%) compared with the controls (Fig. 8 C, D). Hatching rate was 20.3% and 17.3% for control eggs and eggs injected with shGPX1, respectively. However, the rate was dropped to 0% for eggs injected with shGPX4 (Fig. 8E).
Fig. 8.
Effects of the shGPX4 transfection on chicken embryo of the EGFP expression, pancreatic histology, GPX4 mRNA expression, production of pancreatic apoptotic signal proteins, and hatch rate.
(A) Expression of EGFP of transgenic chicken embryo (day 8). shGPX4, eggs injected with pLL3.7-GPX4-shRNA; control, eggs injected with empty vector.
(B) H&E staining of paraffin-embedded chicken embryonic pancreas (day 18) sections. Compared with the control, the shGPX4 chicken embryonic pancreas showed acinar cell swelling, granular degeneration and vacuolar degeneration. Images were taken with 400 × magnification. The image was a representative of five independent examinations.
(C) Effects of the shGPX4 transfection on relative mRNA abundances of GPX4 in the day18 embryonic pancreas. Data are means ± SE, n = 5, *P < 0.05 compared with the control.
(D) Effects of the shGPX4 transfection on protein levels of GPX4 and apoptotic signal proteins in the day18 embryonic pancreas. Data are means ± SE, n = 5, *P < 0.05 compared with the control.
(E) Effects of shGPX4 and shGPX1 transfections on egg hatching rates. GPX1, eggs injected with pLL3.7-GPX1-shRNA; shGPX4, eggs injected with pLL3.7-GPX4-shRNA; Control, eggs injected with empty vector.
Abbreviations: ACTB, beta actin; cleaved-CASP 3, cleaved caspase 3; cleaved-CASP 9, cleaved caspase 9; Control, eggs injected with the empty vector; CYTC, Cytochrome c; GPX1, glutathione peroxidase 1; GPX4, glutathione peroxidase 4; pro-CASP 3, pro caspase 3; pro-CASP 9, pro caspase 9; shGPX1, chicken eggs injected with pLL3.7-GPX1-shRNA; shGPX4, chicken eggs injected with pLL3.7-GPX4-shRNA.
4. Discussion
Our present study reveals a key role and novel mechanism of GPX4 in the development of NPA induced by dietary Se-deficiency in broiler chicks. We have taken stepwise approaches to make and support our findings. Firstly, we fed chicks a practical, low-Se diet and successfully replicated the typical symptoms of NPA. Secondly, we performed global gene expression and target pathway analyses in the pancreas of chicks and postulated a cascade event of depletion of GPX4, up-regulation of apoptosis signaling, and development of NPA. Thirdly, we specifically altered GPX4 expression by silencing or overexpressing the gene in the pancreatic acinar cells and demonstrated an inverse relationship between GPX4 expression and susceptibility of acinar cells to H2O2-induced apoptosis. Fourthly, we applied pull down, mass spectrometry, and cell biology approaches to unveil ProTalpha as a novel binding protein for GPX4 to exert its role in protecting against the H2O2-induced apoptosis of acinar cells. Finally, we silenced expression of GPX4 in the chick embryo and verified the physiological relevance of the GPX4 role and pathway demonstrated in the acinar cells.
Pancreas was recognized as one of the most susceptible organs to dietary Se/Vit. E deficiencies in poultry, and NPA was reported in 1950–60s as a classical Se/Vit. E deficiency disease [6,14,50]. It is quite remarkable that our practical, low-Se diets could duplicate the typical NPA in the currently-used commercial chicks in terms of occurrence time and rate (∼18 days old of age, > 80%), overt signs (pancreas weight, length, and index), and histological characterization (hyaline body formation, vacuolation and cytoplasmic shrinkage, and disrupted mitochondria), compared with the earlier observations [13,14,50]. Because the genetic background, metabolic capacity such as growth rate, and housing conditions have been substantially changed between chicks raised 60 years ago and now, this incredible reproducibility of NPA illustrates that the disease is induced by dietary Se/Vit. E deficiency per se, independent of genetic context of chicks or other environmental factors.
The DGE analysis demonstrated a stark contrast between down-regulation of selenoprotein genes and up-regulation of apoptosis signaling-related genes in the pancreas of –Se chicks suffered with a high incidence of NPA. The followed Q-PCR, Western blot, and IHC analyses clearly confirmed these changes. Although mRNA levels of at least 12 selenoproteins and protein levels of 7 selenoproteins were decreased by dietary Se deficiency and all those changes could have contributed to the development of NPA, we focused on the role and mechanism of GPX4 in the regard. This was because pancreatic GPX4 protein was nearly diminished in the -Se chicks, representing one of the most decreased selenoproteins by dietary Se deficiency. Such depletion of avian GPX4 gave rise to an extreme comparison with the strong resistance of mammalian GPX4 expression to Se deprivation [51]. Previous research has demonstrated that tissue-specific differences in selenoprotein synthesis and sensitivity to Se deprivation are associated with efficiency of Se retention, levels of Sec insertion sequence (SECIS) binding protein 2 (SBP2) [52] and selenocysteine-specific elongation factor [53], and effects on selenocysteyl-tRNA population [54]. It remains to be determined if the low ranking of GPX4 in avian selenoprotein hierarchy is due to a low SECIS-SBP2 affinity [55]. Because NPA is a poultry-specific, but not mammals-prone, disease, the distinct differences in GPX4 expression responses to dietary Se deficiency between the avian and mammalian species could implicate a special role of GPX4 in the development of NPA. In fact, the GPX4-immunopositive staining was localized in the acinar cell nucleus and cytoplasm of pancreas in +Se chicks. The decrease or loss of GPX4 staining in these subcellular locations of –Se chicks corresponded well to the intracellular vacuolation and hyaline body formation, followed by loss of acinar zonation [50]. Furthermore, GPX4 is the only known selenoperoxidase that reduces phospholipid hydroperoxides in membrane and interacts with Vit. E [5,18]. A global knockout of GPX4 causes early embryonic lethality [56], and GPX4 involves in different types of cell death including necrosis, apoptosis, and ferroptosis [[57], [58], [59]]. Our basis for postulating a functional link between GPX4 depletion and the development of NPA was derived from the disrupted mitochondria, elevated apoptotic proteins (cleaved-CASP 3, cleaved-CASP 9, and CYTC), and decreased pro-apoptotic protein (pro-CASP 3 and pro-CASP 9) in the pancreas of –Se chicks. While roles and responses of caspases 3 and 9 and CYTC in ROS-related apoptosis are well-documented [35,60], their cross-talking with GPX4 was clearly elucidated as a cause-effect event only in the present study. When GPX4 expression was suppressed in the freshly isolated chicken pancreatic acinar cells, the apoptotic proteins (CYTC, cleaved CASP 3, and cleaved CASP 9) and the proapoptotic proteins (pro-CASP 3 and pro-CASP 9) were up- and down-regulated, respectively. These changes were the exact same as observed in the pancreas of –Se chicks with NPA. In contrast, overexpression of GPX4 in the cells resulted in the exact opposite changes. Altogether, GPX4 could act as a major selenoprotein involved in the apoptotic signaling and cell death occurred in NPA of –Se chicks.
It was exciting for us to reveal the protein protein interaction between GPX4 and ProTalpha and to confer their roles in regulating intracellular caspase activity, CYTC release, and apoptosis in the pancreatic acinar cells. This discovery offers a novel mechanism to explain how GPX4 deficiency could lead to apoptosis and the development of NPA in –Se chicks. Among the GPX4-binding protein candidates identified from the GST-GPX4 pull down and MS screening, ProTalpha showed the highest fidelity. Subsequent co-immuno precipitation and co-fluorescence localization assays clearly confirmed their interactions in the acinar cells. The anti-ProTalpha antibody pulled down GPX4 protein from the pancreatic homogenates of +Se chicks, but not those of –Se chicks. More convincingly, using siRNA to silence the expression of these two proteins led to similar increases in apoptotic signaling proteins (cleaved-CASP 3 and 9 and CYTC), susceptibility to H2O2-mediated loss of viability, and apoptosis. These changes imply equal importance of the two proteins in controlling apoptotic process. The indispensability of each protein or more accurately the interdependence of each other in forming a complex to exert their anti-apoptotic function in acinar cells was further supported by only a moderate increase of the H2O2-induced apoptosis resulted from the double silences of both genes simultaneously compared with the single silence [61,62]. Illustrating the interaction and mutual dependence of these two protein not only offers a novel role of GPX4 but also a new control mechanism of apoptosis. A previous study indicated that activation of ferroptosis led to increases of the 70 kDa heat shock cognate protein (HSC70)-mediated lysosomal delivery and degradation of GPX4. Immunoprecipitation assays suggested interactions between legumain, heat shock protein 90 (HSP90), HSC70, lysossomal associated protein 2a (lamp-2a) and GPX4 [63]. In this study, we have added ProTalpha as a new binding protein of GPX4 for its role in protecting against ROS-induced apoptosis. Meanwhile, Markova et al. reported that CYTC was transformed from anti-to pro-oxidant when interacting with truncated oncoprotein ProTalpha [35]. Because ProTalpha is an abundant nuclear protein involved in cellular processes intricately linked to development such as cell proliferation, revealing its interaction with GPX4 may give us a new avenue to study other unrecognized functions of GPX4. It remains a fascinating question if the co-expression and interaction of GPX4 and ProTalpha exist in other tissues, in particular those with preferential Se supply like the endocrine glands.
The outcomes of target silencing GPX4 expression through subgerminal cavity injection of the shRNA-expressing lentivirus in the chick embryo supported the relevance of the association between down-regulation of GPX4 and development of NPA in –Se chicks and the direct role of altering GPX4 expression in regulating apoptotic signaling and death in the pancreatic acinar cells. Lentiviral vector-mediated gene knockdown has been successfully used to transfect different types of cells in vivo with several technical advantages [64], and is a very useful tool for sensory regeneration research of embryonic pancreas, particularly for in vivo use [65]. In this study, the shRNA sequences were designed according to the siGPX4 sequence that could inhibit the expression of GPX4 gene in chicken embryos. When the GPX1 shRNA injection did not decrease the hatching rate compared with the vector controls, the GPX4 shRNA produced zero hatching rate (100% mortality rate) of embryos. Although the control hatching rate, probably due to the subgerminal cavity injection procedure, was rather low, the difference between the GPX1 and GPX4 shRNA injections was evident. While the shGPX4-induced embryonic lethality confirms the findings from previous mouse research [66], the most relevant finding for the present study was the activation of mitochondrial apoptotic pathway by the GPX4 shRNA in the day-18 embryonic pancreas.
Although we applied multiple robust methods to determine overall metabolic changes in the pancreas of –Se verse + Se chicks and specific responses to alterations of GPX4 and ProTalpha in the pancreatic acinar cells, our findings on the novel role and mechanism of GPX4 in the development of NPA still have limitations. Ideally, a pancreas-specific knockout of GPX4 in chicks should have been the best model for our objective. However, generating such a sophisticated model in poultry presents many technical challenges although our ongoing research has succeeded in producing serval lines of global knockouts of avian selenoprotein genes. This renders our current approaches as the most practical or feasible alternative. Another finding requiring substantialization is the protein protein interaction between GPX4 and ProTalpha. Five other proteins were also identified from the GST-GPX4 pull down screening. It will be interesting to explore if and how these proteins bind GPX4 and affect the interaction between GPX4 and ProTalpha. In addition, it is necessary to develop effective antibodies specifically against many of the avian selenoproteins to assess their contributions, in relation to GPX4, to the development of NPA.
5. Conclusion
In the present study, we have successfully replicated the classical Se/Vit. E deficiency disease NPA in broiler chicks fed a practical, low-Se diet. A combined global gene expression profiling with target pathway characterization in the pancreas of -Se chicks depicted a key role of GPX4 deficiency in the apoptotic signaling involved in the onset of NPA. The functional link between GPX4 depletion and the elevated apoptotic signaling and cell death induced by H2O2 was elucidated by silencing and overexpressing GPX4 gene in the pancreatic acinar cells. An array of pull down, mass spectrometry, and cell biology methods were used to unveil ProTalpha as a novel binding protein for GPX4 and a mutual dependence of these two proteins to form a complex in exerting their roles in regulating H2O2-induced apoptosis of acinar cells. The GPX4 shRNA was used to silence expression of GPX4 in the chick embryos and confirmed the in vivo relevance of the GPX4 role and pathway shown ex vivo and in the acinar cells. While our findings provide a novel molecular mechanism for the pathogenesis of NPA as a classical Se/Vit. E deficiency disease in chicks reported 60 years ago, unveiling the protein protein interaction between GPX4 and ProTalpha in regulating apoptosis adds a new concept to both Se biochemistry and cell biology.
Funding
This research was supported in part by National Natural Science Foundation of China (32002216 and 32172772).
Contributions of authors
XGL: conceived and designed the research; FZR: supervised the research and obtained funding; JQH and YYJ: conducted the research and analyzed the data; JQH and XGL: wrote and edited the paper; XGL and JQH: had primary responsibility for final content; and all authors: read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102482.
Contributor Information
Jia-Qiang Huang, Email: jqhuang@cau.edu.cn.
Xin Gen Lei, Email: XL20@cornell.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Thompson J.N., Scott M.L. Impaired lipid and vitamin E absorption related to atrophy of the pancreas in selenium-deficient chicks. J. Nutr. 1970;100(7):797–809. doi: 10.1093/jn/100.7.797. [DOI] [PubMed] [Google Scholar]
- 2.Cantor A.H., Moorhead P.D., Musser M.A. Comparative effects of sodium selenite and selenomethionine upon nutritional muscular dystrophy, selenium-dependent glutathione peroxidase, and tissue selenium concentrations of Turkey poults. Poultry Sci. 1982;61(3):478–484. doi: 10.3382/ps.0610478. [DOI] [PubMed] [Google Scholar]
- 3.Tadashi N., Langevin M.L., Combs G.F., Scott M.L. Biochemical and histochemical studies of the selenium-deficient pancreas in chicks. J. Nutr. 1973;103(3):444–453. doi: 10.1093/jn/103.3.444. [DOI] [PubMed] [Google Scholar]
- 4.Avanzo J.L., Junior C., Cesar M. Role of antioxidant systems in induced nutritional pancreatic atrophy in chicken, Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2002;131(4):815–823. doi: 10.1016/s1096-4959(02)00040-4. [DOI] [PubMed] [Google Scholar]
- 5.Carlson B.A., Tobe R., Yefremova E., Tsuji P.A., Hoffmann V.J., Schweizer U., Gladyshev V.N., Hatfield D.L., Conrad M. Glutathione peroxidase 4 and vitamin E cooperatively prevent hepatocellular degeneration. Redox Biol. 2016;9:22–31. doi: 10.1016/j.redox.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitacre M.E., Combs G.F., Combs S.B., Parker R.S. Influence of dietary vitamin E on nutritional pancreatic atrophy in selenium-deficient chicks. J. Nutr. 1987;117(3):460–467. doi: 10.1093/jn/117.3.460. [DOI] [PubMed] [Google Scholar]
- 7.Li S., Gao F., Huang J., Wu Y., Wu S., Lei X.G. Regulation and function of avian selenogenome. BBA-Gen. Subj. 2018;1862(11):2473–2479. doi: 10.1016/j.bbagen.2018.03.029. [DOI] [PubMed] [Google Scholar]
- 8.Sun L.H., Huang J.Q., Deng J., Lei X.G. Avian selenogenome: response to dietary Se and vitamin E deficiency and supplementation. Poultry Sci. 2018;98:4247–4254. doi: 10.3382/ps/pey408. [DOI] [PubMed] [Google Scholar]
- 9.Zhu S.Y., Liu L.L., Huang Y.Q., Li X.W., Milton T., Dai X.Y., Li Y.H., Li J.L. In silico analysis of selenoprotein N (Gallus gallus): absence of EF-hand motif and the role of CUGS-helix domain in antioxidant protection. Metallomics. 2021;13(3) doi: 10.1093/mtomcs/mfab004. [DOI] [PubMed] [Google Scholar]
- 10.Kryukov G., Castellano S., Novoselov S.V., Lobanov A.V., Gladvshev V.N. Characterization of mammalian selenoproteomes. Science. 2003;300(5624):1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 11.Wang S., Zhao X., Liu Q., Wang Y., Li S., Xu S. Selenoprotein K protects skeletal muscle from damage and is required for satellite cells-mediated myogenic differentiation. Redox Biol. 2022;50 doi: 10.1016/j.redox.2022.102255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long W., Zou X., Zhang X. Transcriptome analysis of canola (Brassica napus) under salt stress at the germination stage. PLoS One. 2015;10(2) doi: 10.1371/journal.pone.0116217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bunk M.J., Combs G.F. Relationship of selenium-dependent glutathione peroxidase activity and nutritional pancreatic atrophy in selenium-deficient chicks. J. Nutr. 1981;111(9):1611–1620. doi: 10.1093/jn/111.9.1611. [DOI] [PubMed] [Google Scholar]
- 14.Whitacre M.E., Combs G.F. Selenium and mitochondrial integrity in the pancreas of the chick. J. Nutr. 1983;113(10):1972–1983. doi: 10.1093/jn/113.10.1972. [DOI] [PubMed] [Google Scholar]
- 15.Gregolin M.C. The selenoenzyme phospholipid hydroperoxide glutathione peroxidase. BBA - Gen Subjects. 1985;839(1):62–70. doi: 10.1016/0304-4165(85)90182-5. [DOI] [PubMed] [Google Scholar]
- 16.Root E.J., Combs G.F. Disruption of endoplasmic reticulum is the primary ultrastructural lesion of the pancreas in the selenium-deficient chick. Proc. Soc. Exp. Biol. Med. 1988;187(4):513–521. doi: 10.3181/00379727-187-42697. [DOI] [PubMed] [Google Scholar]
- 17.Ingold I., Berndt C., Schmitt S., Doll S., Poschmann G., Buday K., Roveri A., Peng X., Freitas F.P., Seibt T. Selenium utilization by GPX4 is required to prevent hydroperoxide-induced ferroptosis. Cell. 2017:423–434. doi: 10.1016/j.cell.2017.11.048. [DOI] [PubMed] [Google Scholar]
- 18.Hu Q., Zhang Y., Lou H., Ou Z., Ju Z. GPX4 and vitamin E cooperatively protect hematopoietic stem and progenitor cells from lipid peroxidation and ferroptosis. Cell Death Dis. 2021;12(7):706. doi: 10.1038/s41419-021-04008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiorino, Matilde Conrad, Marcus Ursini, Fulvio, GPX4 Lipid peroxidation, and cell death: discoveries, rediscoveries, and open issues. Antioxid. redox signal. 2018;29(1):61–74. doi: 10.1089/ars.2017.7115. [DOI] [PubMed] [Google Scholar]
- 20.Fradejas-Villar N., Zhao W., Reuter U., Doengi M., Ingold I., Bohleber S., Conrad M., Schweizer U. Missense mutation in selenocysteine synthase causes cardio-respiratory failure and perinatal death in mice which can be compensated by selenium-independent GPX4. Redox Biol. 2021;48 doi: 10.1016/j.redox.2021.102188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao Y., Chen Z., Zhang H., Chen C., Zeng M., Yunis J. Selenium-GPX4 axis protects follicular helper T cells from ferroptosis. Nat. Immunol. 2021;22(9):1127–1139. doi: 10.1038/s41590-021-00996-0. [DOI] [PubMed] [Google Scholar]
- 22.Fan R., Sui J., Dong X., Jing B., Gao Z. Wedelolactone alleviates acute pancreatitis and associated lung injury via GPX4 mediated suppression of pyroptosis and ferroptosis. Free Radic. Biol. Med. 2021;173:29–40. doi: 10.1016/j.freeradbiomed.2021.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Xin G.L., Evenson J.K., Thompson K.M., Sunde R.A. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J. Nutr. 1995;125(6):1438–1446. doi: 10.1093/jn/125.6.1438. [DOI] [PubMed] [Google Scholar]
- 24.Huang J.Q., Ren F.Z., Jiang Y.Y., Chen X., Lei X.G. Selenoproteins protect against avian nutritional muscular dystrophy by metabolizing peroxides and regulating redox/apoptotic signaling. Free Radic. Biol. Med. 2015;83:129–138. doi: 10.1016/j.freeradbiomed.2015.01.033. [DOI] [PubMed] [Google Scholar]
- 25.Huang J.Q., Zhou J.C., Wu Y.Y., Ren F.Z., Lei X.G. Role of glutathione peroxidase 1 in glucose and lipid metabolism-related diseases. Free Radic. Biol. Med. 2018;127:108–115. doi: 10.1016/j.freeradbiomed.2018.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li T., Zhang J., Wang P.J., Zhang Z.W., Huang J.Q. Selenoproteins protect against avian liver necrosis by metabolizing peroxides and regulating receptor interacting serine threonine kinase 1/receptor interacting serine threonine kinase 3/mixed lineage kinase domain-like and mitogen-activated protein kinase signaling. Front. Physiol. 2021;12(1216) doi: 10.3389/fphys.2021.696256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canli Ö., Alankuş Y.B., Grootjans S., Vegi N., Hültner L., Hoppe P.S., Schroeder T., Vandenabeele P., Bornkamm G.W., Greten F.R. Glutathione peroxidase 4 prevents necroptosis in mouse erythroid precursors. Blood. 2016;127(1):139–148. doi: 10.1182/blood-2015-06-654194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu P., Zhu J., Yuan G., Li D., Wen Y., Huang S., Lv Z., Guo Y., Cheng J. The effects of selenium on GPX4-mediated lipid peroxidation and apoptosis in germ cells. J. Appl. Toxicol. 2021:1–13. doi: 10.1002/jat.4273. [DOI] [PubMed] [Google Scholar]
- 29.Ursini F. GPX4 is the controller of a specific form of programmed cell death executed by lipid peroxidation products. Free Radic. Biol. Med. 2018;124:559–560. [Google Scholar]
- 30.Ran Q., Liang H., Gu M., Qi W., Walter C.A., Roberts L.J., 2nd, Herman B., Richardson A., Van Remmen H. Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. J. Biol. Chem. 2004;279(53):55137–55146. doi: 10.1074/jbc.M410387200. [DOI] [PubMed] [Google Scholar]
- 31.Moreira D., Díaz-Jullien C., Concepción G.C., Sarandeses S., Barbeito Pablo, Freire Manuel. The influence of phosphorylation of prothymosin α on its nuclear import and antiapoptotic activity. Biochem. Cell. Biol. 2013;91(4):265–269. doi: 10.1139/bcb-2012-0090. [DOI] [PubMed] [Google Scholar]
- 32.Qi X., Lai W., Du F. Novel small molecules relieve prothymosin α-mediated inhibition of apoptosome formation by blocking its interaction with Apaf-1. Biochemistry. 2010;49(9):1923–1930. doi: 10.1021/bi9022329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Acehan D., Jiang X., Morgan D.G., Heuser J.E., Wang X., Akey C.W. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol. Cell. 2002;9(2):423–432. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- 34.Jiang X., Kim H.E., Shu H., Zhao Y., Zhang H., Kofron J., Donnelly J., Burns D., Ng S.C., Rosenberg S. Distinctive roles of phap proteins and prothymosin- in a death regulatory pathway. Science. 2003;299(5604):223–226. doi: 10.1126/science.1076807. [DOI] [PubMed] [Google Scholar]
- 35.Markova O.V., Evstafieva A.G., Mansurova S.E., Moussine S.S., Skulachev V.P. Cytochrome c is transformed from anti- to pro-oxidant when interacting with truncated oncoprotein prothymosin alpha. BBA-Bioenergetics. 2003;1557(1–3):109–117. doi: 10.1016/s0005-2728(03)00003-3. [DOI] [PubMed] [Google Scholar]
- 36.Malicet C., Giroux V., Vasseur S., Dagorn J.C., Neira J.L., Iovanna J.L. Regulation of apoptosis by the p8/prothymosin complex. Proc. Natl. Acad. Sci. U.S.A. 2006;103(8):2671–2676. doi: 10.1073/pnas.0508955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Padmanabhan B., Nakamura Y., Yokoyama S. Structural analysis of the complex of Keap1 with a prothymosin α peptide. Acta Crystallogr. 2008;64(Pt 4):233–238. doi: 10.1107/S1744309108004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segade F., Gómez-Márquez J. Prothymosin α. Int. J. Biochem. Cell Biol. 1999;31(11):1243–1248. doi: 10.1016/s1357-2725(99)00094-1. [DOI] [PubMed] [Google Scholar]
- 39.Ueda H. Prothymosin α and cell death mode switch, a novel target for the prevention of cerebral ischemia-induced damage. Pharmacol. Therapeut. 2009;123(3):323–333. doi: 10.1016/j.pharmthera.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Huang J.Q., Li D.L., Zhao H., Sun L.H., Xia X.J. The selenium deficiency disease exudative diathesis in chicks is associated with downregulation of seven common selenoprotein genes in liver and muscle. J. Nutr. 2011;141(9):1605–1610. doi: 10.3945/jn.111.145722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 42.Moreno M.E., Rez-Conde C.P., Mara C.C. Speciation of inorganic selenium in environmental matrices by flow injection analysis-hydride generation-atomic fluorescence spectrometry. Comparison of off-line, pseudo on-line and on-line extraction and reduction methods. J. Anal. Atomic Spectrom. 2000;15(6):681–686. [Google Scholar]
- 43.Zhang X., Xiong W., Chen L.L., Huang J.Q., Lei X.G. Selenoprotein V protects against endoplasmic reticulum stress and oxidative injury induced by pro-oxidants. Free Radic. Biol. Med. 2020;160:670–679. doi: 10.1016/j.freeradbiomed.2020.08.011. [DOI] [PubMed] [Google Scholar]
- 44.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L., Smyth G.K., Wei S. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 46.Yan W.A., Klee E.W., Thompson E.A., Perez E.A., Middha S., Oberg A.L., Therneau T.M., Smith D.I., Poland G.A., Wieben E.D. 3' tag digital gene expression profiling of human brain and universal reference RNA using Illumina Genome Analyzer. BMC Genom. 2009;10(1):1–11. doi: 10.1186/1471-2164-10-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chi Q., Zhang Q., Lu Y., Zhang Y., Xu S., Li S. Roles of selenoprotein S in reactive oxygen species-dependent neutrophil extracellular trap formation induced by selenium-deficient arteritis. Redox Biol. 2021;44 doi: 10.1016/j.redox.2021.102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen L.L., Huang J.Q., Wu Y.Y., Chen L.B., Li S.P., Zhang X., Wu S., Ren F.Z., Lei X.G. Loss of Selenov predisposes mice to extra fat accumulation and attenuated energy expenditure. Redox Biol. 2021;45 doi: 10.1016/j.redox.2021.102048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen L.L., Huang J.Q., Xiao Y., Wu Y.Y., Ren F.Z., Lei X.G. Knockout of selenoprotein V affects regulation of selenoprotein expression by dietary selenium and fat intakes in mice. J. Nutr. 2020;150:483–491. doi: 10.1093/jn/nxz287. [DOI] [PubMed] [Google Scholar]
- 50.Gries C.L., Scott M.L. Pathology of selenium deficiency in the chick. J. Nutr. 1972;102(10):1287–1296. doi: 10.1093/jn/102.10.1287. [DOI] [PubMed] [Google Scholar]
- 51.Lei X.G., Evenson J.K., Thompson K.M., Sunde R.A. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J. Nutr. 1995;125(6):1438–1446. doi: 10.1093/jn/125.6.1438. [DOI] [PubMed] [Google Scholar]
- 52.Paul R., Copeland, Julia E., Fletcher, Bradley A., Carlson Dolph. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tujebajeva R.M., Copeland P.R., Xu X.i., Carlson B.A., Berry M.J. Decoding apparatus for eukaryot selenocysteine insertion. EMBO Rep. 2000;1(2):158–163. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jung J.E., Karoor V., Sandbaken M.G., Lee B.J., Wahba A.J. Utilization of selenocysteyl-tRNA[Ser]Sec and seryl-tRNA[Ser]Sec in protein synthesis. J. Biol. Chem. 1994;269(47):29739–29745. [PubMed] [Google Scholar]
- 55.Low C.S., Grundner-Culemann E., Harney W.J., Berry J.M. SECIS-SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J. 2000;19(24):6882–6890. doi: 10.1093/emboj/19.24.6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imai H., Hirao F., Sakamoto T., Sekine K., Mizukura Y., Saito M., Kitamoto T., Hayasaka M., Hanaoka K., Nakagawa Y. Early embryonic lethality caused by targeted disruption of the mouse PHGPx gene. Biochem. Bioph. Res. Co. 2003;305(2):278–286. doi: 10.1016/s0006-291x(03)00734-4. [DOI] [PubMed] [Google Scholar]
- 57.Bersuker K., Hendricks J.M., Li Z., Magtanong L., Olzmann J.A. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575(7784):688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doll S., Freitas F.P., Shah R., Aldrovandi M., Conrad M. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575(7784):693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 59.Gaschler M.M., Andia A.A., Liu H., Csuka J.M., Hurlocker B., Vaiana C.A., Heindel D.W., Zuckerman D.S., Bos P.H., Reznik E. FINO 2 initiates ferroptosis through GPX4 inactivation and iron oxidation. Nat. Chem. Biol. 2018;14(5):507–515. doi: 10.1038/s41589-018-0031-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bossy-Wetzel E., Green D.R. Caspases induce cytochrome c release from mitochondria by activating cytosolic Factors. J. Biol. Chem. 1999;274(25):17484–17490. doi: 10.1074/jbc.274.25.17484. [DOI] [PubMed] [Google Scholar]
- 61.Hu S., Sechi M., Singh P.K., Dai L., Neamati N. A novel redox modulator induces a GPX4-mediated cell death that is dependent on iron and reactive oxygen species. J. Med. Chem. 2020;63(17):9838–9855. doi: 10.1021/acs.jmedchem.0c01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Q., Zhan S., Liu Y., Han F., Huang Z.W. Low-Se diet can affect sperm quality and testicular glutathione peroxidase-4 activity in rats. Biol. Trace Elem. Res. 2021;199(10):3752–3758. doi: 10.1007/s12011-020-02515-y. [DOI] [PubMed] [Google Scholar]
- 63.Chen C., Wang D., Yu Y., Zhao T., Tan X. Legumain promotes tubular ferroptosis by facilitating chaperone-mediated autophagy of GPX4 in AKI. Cell Death Dis. 2021;12(1):65. doi: 10.1038/s41419-020-03362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jia Y., Xiang M., Wu H., Shen C.L. Intratympanic injection of shRNA-expressing lentivirus causes gene silencing in the inner ear in chicken. Neurosci. Lett. 2012;510(2):132–137. doi: 10.1016/j.neulet.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 65.Bukrinsky M.I., Haggerty S., Dempsey M.P., Sharova N., Adzhubei A., Spitz L., Lewis P., Goldfarb D., Emerman M., Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection ofnon-dividing cells. Nature. 1993;365(6447):666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brütsch S., Wang C.C., Li L., Stender H., Neziroglu N., Richter C., Kuhn H., Borchert A. Expression of inactive glutathione peroxidase 4 leads to embryonic lethality, and inactivation of the Alox15 gene does not rescue such knock-in mice. Antioxidants Redox Signal. 2014;22(4):281–293. doi: 10.1089/ars.2014.5967. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.