Abstract
Bacillus subtilis grown in media containing amino acids or glucose secretes acetate, pyruvate, and large quantities of acetoin into the growth medium. Acetoin can be reused by the bacteria during stationary phase when other carbon sources have been depleted. The acoABCL operon encodes the E1α, E1β, E2, and E3 subunits of the acetoin dehydrogenase complex in B. subtilis. Expression of this operon is induced by acetoin and repressed by glucose in the growth medium. The acoR gene is located downstream from the acoABCL operon and encodes a positive regulator which stimulates the transcription of the operon. The product of acoR has similarities to transcriptional activators of sigma 54-dependent promoters. The four genes of the operon are transcribed from a −12, −24 promoter, and transcription is abolished in acoR and sigL mutants. Deletion analysis showed that DNA sequences more than 85 bp upstream from the transcriptional start site are necessary for full induction of the operon. These upstream activating sequences are probably the targets of AcoR. Analysis of an acoR′-′lacZ strain of B. subtilis showed that the expression of acoR is not induced by acetoin and is repressed by the presence of glucose in the growth medium. Transcription of acoR is also negatively controlled by CcpA, a global regulator of carbon catabolite repression. A specific interaction of CcpA in the upstream region of acoR was demonstrated by DNase I footprinting experiments, suggesting that repression of transcription of acoR is mediated by the binding of CcpA to the promoter region of acoR.
During the growth of cultures of Bacillus subtilis, several products have been identified in the growth medium, such as lactate, acetate, succinate, acetoin, butanediol, and ethanol (5).
Acetoin (3-hydroxy 2-butanone) is a major catabolic product of B. subtilis grown aerobically in glucose media. Since it is neutral, this metabolite allows the bacteria to degrade large amounts of glucose without substantial acidification of the growth medium. Acetoin also serves as a carbon storage compound which is secreted into the growth medium and later reimported. In B. subtilis, the products of two genes, ilvBN (acetohydroxy acid synthase) and alsS (α-acetolactate synthase), are involved in the production of acetolactate from pyruvate. Acetolactate is converted to acetoin by spontaneous decarboxylation at low pH or via the action of alsD, encoding an acetolactate decarboxylase (37). Acetoin is reutilized during stationary phase when other carbon sources have been depleted. Many bacterial species are able to degrade acetoin: Micrococcus urea (20), Alcaligenes eutrophus (12), Enterococcus faecalis (10), Pelobacter carbinolicus (34), Klebsiella pneumoniae (9), and Clostridium magnum (22). Three genes forming an operon, acuABC, have been described in B. subtilis. The roles of the corresponding gene products are still unknown (16). Inactivation of the first gene of this operon resulted in diminished growth on acetoin and butanediol. Since utilization of acetoin is reduced but not abolished in an acuABC mutant, this result suggested that there is more than one pathway for acetoin utilization in B. subtilis. Recently, another gene cluster, the aco operon encoding the multicomponent acetoin dehydrogenase enzyme complex, was sequenced (18, 23). A plasmid encoding part of the α subunit of the acetoin dehydrogenase E1 was used to disrupt acoA, the first gene of this operon. This mutant was impaired in the expression of acetoin dehydrogenase E1 activity and for depletion of acetoin from the growth medium, indicating that this operon is the main system involved in the catabolism of acetoin (18). However, very little was known about the regulation of transcription of the aco operon in B. subtilis. The product of the acoR gene, which is located downstream from the aco operon, has similarities to transcriptional activators of sigma 54-dependent promoters. A putative −12, −24 promoter is located upstream from the acoA gene, strongly suggesting that the SigL sigma factor is necessary for its transcription. We studied the regulation of the expression of the aco operon in B. subtilis and found that transcription was strongly induced in the presence of acetoin in the growth medium and depended upon the presence of both AcoR and SigL.
MATERIALS AND METHODS
Bacterial strains and culture media.
The B. subtilis strains used in this work are listed in Table 1. Escherichia coli TGI [K-12 Δ(lac pro) supE thi hsd5/F′ traD36 proA+B+ laclq lacZΔM15] was used for cloning experiments. E. coli was grown in Luria-Bertani broth (38), and B. subtilis was grown in SP medium (8 g/liter of nutrient broth [Difco], 1 mM MgSO4, 10 mM KCl, 0.5 mM CaCl2, 10 μM MnCl2, 2 μM FeSO4) or in CSK medium. CSK medium is C medium (28) supplemented with potassium succinate (6 g/liter) and potassium glutamate (8 g/liter).
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype or description | Source or reference |
|---|---|---|
| 168 | trpC2 | 1 |
| QB5407 | trpC2 ccpA::spc | 36 |
| QB5505 | trpC2 sigL::aphA3 | 6 |
| QB7700 | trpC2 amyE acoA′-′lacZ | This work |
| QB7701 | trpC2 amyE acoA′-′lacZ ccpA::spc | This work |
| QB7702 | trpC2 amyE acoA′-′lacZ sigL::aphA3 | This work |
| QB7704 | trpC2 amyE acoA′-′lacZ acoR::aphA3 | This work |
| QB7713 | trpC2 acoR::pDIA5307 | This work |
| QB7714 | trpC2 acoR::pDIA5307 pACOR1 | This work |
| QB7719 | trpC2 amyE acoA′-′lacZ(ΔB) | This work |
| QB7720 | trpC2 amyE acoA′-′lacZ(ΔC) | This work |
| QB7721 | trpC2 amyE acoA′-′lacZ(ΔD) | This work |
| QB7724 | trpC2 acoA::pMutin4 | This work |
| QB7725 | trpC2 acoB::pMutin4 | This work |
| QB7726 | trpC2 acoC::pMutin4 | This work |
| QB7727 | trpC2 acoL::pMutin4 | This work |
| QB7728 | trpC2 amyE acoA′-′lacZ acuABC::aphA3 | This work |
| QB7733 | trpC2 acoR::pDIA5307 ccpA::spc pACOR1 | This work |
Transformation and phenotype characterization.
Standard procedures were used to transform E. coli (38), and transformants were selected on Luria-Bertani broth plates containing ampicillin (100 μg/ml). B. subtilis was transformed with plasmid or chromosomal DNA as previously described (1, 28), and transformants were selected on SP medium plates containing chloramphenicol (5 μg/ml), kanamycin (5 μg/ml), erythromycin (1 μg/ml) plus lincomycin (25 μg/ml), or spectinomycin (60 μg/ml). Amylase activity in B. subtilis was detected after growth on tryptose blood agar base (Difco) containing 10 g of hydrolyzed starch per liter (Connaught). Starch degradation was detected by sublimating iodine onto the plates.
DNA manipulations.
Standard procedures were used to extract plasmids from E. coli (38). Restriction enzymes, phage T4 DNA polymerase, phage T4 DNA ligase, and phage T4 polynucleotide kinase were used as recommended by the manufacturers. DNA fragments were purified from agarose gels with a Prep-A-Gene kit (Bio-Rad Laboratories). The PCR technique with Thermus aquaticus DNA polymerase was used for amplification. The oligonucleotide primers used included mismatches to create restriction sites.
Plasmid constructions.
pAC5 (29), a derivative of pAF1 (11), carries the pC194 chloramphenicol resistance gene cat and a lacZ gene between two fragments of the B. subtilis amyE gene. PCR was used to introduce EcoRI restriction sites at various positions upstream from acoA. PCR was performed with one oligonucleotide (5′-GGAGGATCCTCAGTTAATGACAAGCCTTC-3′) corresponding to the coding sequence of the acoA gene (codons 7 to 13) and one oligonucleotide corresponding to various positions in the acoA promoter region. The EcoRI-BamHI restriction fragments generated were inserted between the EcoRI and BamHI restriction sites of pAC5, creating translational fusions between codon 13 of acoA and codon 8 of lacZ. The DNA sequences of the different PCR fragments were verified by direct sequencing of the various corresponding plasmids. The resulting plasmids were linearized at the single PstI restriction site and integrated into the chromosome of strain 168 by homologous recombination at the amyE locus using chloramphenicol selection. The integrants carrying the translational fusions were named QB7700, QB7719, QB7720, and QB7721.
Gene disruptions and transcriptional fusions with pMutin4 (40) were constructed by PCR amplification of an internal segment of the target gene, ligation of the amplified DNA fragment into pMutin4, transformation of E. coli, and then insertion of the plasmid into the B. subtilis chromosome. The following oligonucleotides were used for PCR amplifications: 5′-GCCGAAGCTTGCCCAGGGAGTGCTTCCC-3′ and 5′-CGCGGATCCAGCAGCCGCCCGATCGGC-3′ for the acoA gene, 5′-GCCGAAGCTTGTCGCCGGGGGAGCGGCG-3′ and 5′-CGCGGATCCGACCTGCTTGCCGACTGC-3′ for the acoB gene, 5′-GCCGAAGCTTGGCGGTAAAAGTAGTGATG-3′ and 5′-CGCGGATCCGATCGTCAGCTGCGCGC-3′ for the acoC gene, and 5′-GCCGAAGCTTGACCGGCTGATCCCGGCT-3′ and 5′-CGCGGATCCGCCGATCTTCACATCCCC-3′ for the acoL gene. The target genes were interrupted by Campbell-type crossover integration. The integration of the recombinant plasmids fuses the target genes to the lacZ gene, leading to a transcriptional fusion.
The wild-type acoR gene was disrupted as follows. Two DNA fragments encoding the amino-terminal and the carboxy-terminal parts of acoR were synthesized by PCR with the following pairs of oligonucleotides, respectively: (i) 5′-GAAGAATTCGAAGGGGCATGCTGGACAGAAAC-3′ and 5′-GGAGGATCCCTGCCAATCCGGCATTGAGGTTC-3′ and (ii) 5′-CTGCTGCAGCGCGATCGCACCGAGGATATCCC-3′ and 5′-AAGAAGCTTTCCAGCGGCTTCCAGTAAAGCGG-3′ . The two DNA fragments were ligated into pHT181 (25) on each side of a DNA fragment containing aphA3 (39), leading to pACOR2. The interrupted gene was introduced into the chromosome of strain QB7700 to give strain QB7704. The wild-type acoR gene was cloned as follows. Strain QB7704 was transformed with a library of B. subtilis DNA established in E. coli by using the shuttle vector pHT315 (24). Transformants were isolated on CSK-erythromycin plates containing 10 mM acetoin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Several blue colonies were isolated. Plasmid DNA was extracted from cultures of one of these transformants, and the insert was characterized by DNA sequencing. The corresponding plasmid, which contained the entire wild-type acoR gene, was called pACOR1.
An acoR′-′lacZ transcriptional fusion was constructed in pDIA5307 (4) by PCR amplification of an internal fragment of the acoR gene, ligation of the amplified DNA fragment into pDIA5307, transformation of E. coli, and insertion of the recombinant plasmid into the chromosome of the 168 strain of B. subtilis, leading to strain QB7713. PCR amplification was done with two oligonucleotides: 5′-GAAGAATTCGAAGGGGCATGCTGGACAGAAAC-3′ and 5′-GGAGGATCCCTGCCAATCCGGCATTGAGGTTC-3′ .
Reverse transcriptase mapping of the mRNA start point in the acoA gene.
Total RNA was isolated from B. subtilis 168 grown in CSK medium with or without 10 mM acetoin as the inducer. Exponentially grown cells were harvested at an optical density at 600 nm of 0.5, and RNA was extracted (15). One oligonucleotide (5′-CCTCAGTTAATGACAAGCCTTCTCG-3′) complementary to the acoA coding sequence was labeled with 10 U of polynucleotide kinase and 0.37 MBq of [γ−32P]ATP (15 TBq/mmol; Amersham). The DNA primer was elongated, and the product was analyzed as previously described (27).
DNase I footprinting.
DNA fragments used for DNase I footprinting were prepared by PCR using Pfu polymerase (Stratagene) and 20 pmol of each primer (5′-GGCGCTGTTAAGCACGATCGGCCTTGCG-3′ and 5′-CCGTTTTCTTAATCGGGCTTCGTCAACC-3′ ), one of which was previously labeled with T4 polynucleotide kinase (New England Biolabs) and [γ32P]ATP. The labeled PCR products were purified with the Qiagen PCR purification kit. Binding of CcpA, HPr, or HPr-Ser-P to these DNA probes was performed in a 20-μl volume containing 2 × 105 cpm of the 32P-labeled DNA fragments and 1 μg of poly(dI-dC) in 100 mM KCl, 10 mM HEPES (pH 7.6), 0.1 mM EDTA, 2 mM MgCl2, 1 mM dithiothreitol, and 10% glycerol. The DNA binding reaction was performed in the presence of 2 μM CcpA and 10 μM either HPr or HPr-Ser-P by incubating the assay mixture for 45 min at room temperature. The concentrations of MgCl2 and CaCl2 were adjusted to 1 and 0.5 mM, respectively, and 20 ng of DNase I (Worthington Biochemical, Freehold, N.J.) was added. The mixture was incubated at room temperature for 1 min, and the reaction was stopped by the addition of 7 volumes of stop buffer (0.4 M sodium acetate, 50 μg of calf thymus DNA/ml, 2.5 mM EDTA) followed by phenol extraction. The DNA fragments were ethanol precipitated, and an equivalent number of cpm (2 × 105) from each reaction was loaded on 7% polyacrylamide sequencing gels. A+G Maxam and Gilbert reactions (32) were carried out on the appropriate 32P-labeled DNA fragments, and the products were loaded alongside the DNase I footprinting reactions. Gels were dried and analyzed by autoradiography.
β-Galactosidase assays.
B. subtilis cells containing lacZ fusions were grown to an optical density at 600 nm of 1. β-Galactosidase specific activities were determined as previously described and are expressed as Miller units per milligram of protein (7). The values reported are averages of at least three independent assays.
Acetoin assays.
The acetoin assays were carried out as previously described (16) except that the reactions were done at room temperature for 15 min.
Glucose assays.
The glucose assays were performed by an enzymatic method using glucose oxidase and peroxidase as recommended by the manufacturer (Boehringer).
RESULTS
Induction of the aco operon in response to acetoin or glucose availability.
Four genes which probably form an operon encode the E1α, E1β, E2, and E3 subunits of the acetoin dehydrogenase multienzyme complex (18). It was previously shown that utilization of acetoin strongly depends upon the culture conditions. Addition of acetoin led to an increase in the specific activity of acetoin dehydrogenase, whereas addition of acetoin with glucose prevents this increase. An acoA′-′lacZ translational fusion was constructed and used to elucidate the regulation of the expression of the aco operon. The expression of the hybrid gene was studied in strain QB7700 grown in minimal CSK medium with and without 10 mM acetoin as the inducer. lacZ expression was strongly induced by acetoin in the growth medium, as shown in Table 2. The acuABC gene cluster plays a role in acetoin catabolism. The product of acuC shares similarities with eukaryotic histone deacetylase (16). In eukaryotes, histone deacetylation plays an important role in transcriptional regulation of cell cycle progression and developmental events. We constructed a strain in which the acuABC operon was deleted. The mutation was introduced by transformation into QB7700, leading to strain QB7728. We did not observe any modification of the induction of the acoA′-′lacZ fusion, and the repression effect observed in the presence of glucose was similar to that observed in the parental strain QB7700 (not shown). This indicates that the acu operon has no effect on the expression of the aco operon in B. subtilis. Addition of glucose repressed the expression of the lacZ fusion. CcpA, a member of the LacI-GalR family of repressors, is a regulator of catabolite repression in B. subtilis. It is a negative regulator of carbon utilization genes and is a positive effector of genes involved in the biosynthesis and secretion of acetate and acetoin (17, 19). The role of CcpA in glucose control was investigated by testing the effect of a ΔccpA mutation on the regulation of the aco operon. Chromosomal DNA from strain QB5407 ccpA::spc was introduced by transformation into strain QB7700, leading to strain QB7701. The effect of glucose was tested by comparing the β-galactosidase activity after the growth of strain QB7701 in CSK acetoin medium and in CSK acetoin glucose medium (Table 2). The expression of the acoA′-′lacZ fusion remained inducible by acetoin but was not repressed by glucose in the absence of CcpA in the cell.
TABLE 2.
Expression of the acoABCL operon and acoR
| Strain | Relevant genotype | β-Galactosidase specific activity (Miller units/mg of protein)
|
||
|---|---|---|---|---|
| CSK | CSK + 10 mM acetoin | CSK + 10 mM acetoin + 0.5% glucose | ||
| QB7700 | acoA′-′lacZ | 145 | 48,975 | 655 |
| QB7701 | acoA′-′lacZ ccpA::spc | 535 | 55,210 | 50,035 |
| QB7704 | acoA′-′lacZ acoR::aphA3 | 10 | 10 | 3 |
| QB7702 | acoA′-′lacZ sigL::aphA3 | 5 | 2 | 3 |
| QB7713 | acoR′-′lacZ | 155 | 140 | 10 |
| QB7714 | acoR′-′lacZ pACOR1 | 125 | 145 | 5 |
| QB7724 | acoA′-′lacZ pMutin4 | 355 | 3,875 | NDa |
| QB7725 | acoB′-′lacZ pMutin4 | 470 | 4,230 | ND |
| QB7726 | acoC′-′lacZ pMutin4 | 95 | 4,740 | ND |
| QB7727 | acoL′-′lacZ pMutin4 | 45 | 3,765 | ND |
| QB7733 | acoR′-′lacZ pACOR1 ccpA::spc | 215 | 230 | 220 |
ND, not determined.
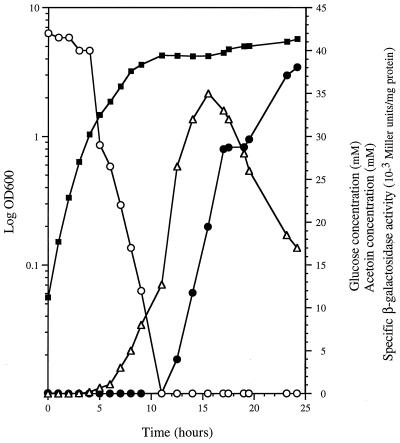
It was previously shown that different carbon sources influence the ability of B. subtilis to degrade acetoin (26). To assess the relationships among acetoin production, glucose degradation, and induction of the aco operon, strain QB7700 was grown in CSK medium containing 42 mM glucose. Glucose, acetoin, and lacZ were assayed at various times for 24 h (Fig. 1). During the exponential phase of growth, the glucose concentration decreased rapidly while acetoin production increased to 35 mM, indicating that most of the glucose was converted to acetoin in the culture medium. As expected, the acetoin concentration began to decline when glucose was completely metabolized. In parallel, the acoA′-′lacZ fusion present in the QB7700 strain was induced, indicating that acetoin produced during aerobic growth in the presence of glucose induced the aco operon. Strikingly, the complete disappearance of glucose from the medium marked the start of transcription of the aco operon.
FIG. 1.
Induction of the acoA′-′lacZ fusion after growth in glucose medium. B. subtilis strain QB7700 was cultured in CSK medium containing 42 mM glucose at 37°C. The optical density of the culture, at 600 nm (OD 600) was followed for 24 h, and the glucose and acetoin concentrations and LacZ activity were assayed. ■, optical density at 600 nm; ○, glucose concentration; Δ, acetoin concentration; ●, β-galactosidase specific activity.
Induction of expression of acoABCL genes.
The plasmid pMutin4 was used to construct transcriptional fusions with the lacZ gene. Integration of pMutin4 fuses the target genes to the lacZ gene and allows the expression of downstream genes from an IPTG-inducible promoter, pspac. These fusions were integrated by homologous recombination into the acoABCL genes in the chromosome, leading to strains QB7724, QB7725, QB7726, and QB7727, respectively. The recombination events were mediated by DNA fragments generated by PCR and corresponding to internal parts of each gene. These events inactivated the corresponding genes and fused them to the lacZ gene. The β-galactosidase activities of strains QB7724, QB7725, QB7726, and QB7727 were assayed after they were cultured in CSK medium containing or not containing acetoin (Table 2). The presence of 10 mM acetoin led to 10- to 100-fold-higher lacZ expression than in its absence. The induction by acetoin indicates that these four genes are coexpressed and probably belong to the same transcriptional unit. The level of β-galactosidase expression in strains containing transcriptional fusions constructed with pMutin4 was lower than that of the acoA′-′lacZ translational fusion in the vector pAC5. This is probably due to better translation of the acoA′-′lacZ mRNA. The acoR gene is located immediately downstream from the aco operon. Addition of 1 mM IPTG to the growth media of strains QB7724 and QB7727 did not change the lacZ expression (not shown). This result indicates that the expression of the acoR gene is not affected by the pspac promoter inserted into the acoA or acoL gene. This could be due to premature termination of transcription initiated at the pspac promoter, suggesting that the aco operon and the acoR genes are distinct transcriptional units.
The acoR and sigL gene products are involved in aco expression.
The four genes, acoABCL, of the operon are followed by acoR, a gene which encodes a peptide of 67 kDa with similarities to activators of sigma 54-dependent promoters. This family of activators contains a region of 220 to 240 residues called the central domain, which is specifically required for the formation of open complexes between RNA polymerase and the −12, −24 promoters, probably by interacting with the RNA polymerase or with sigma 54. An aphA3 cassette was inserted into the acoR gene by a double-crossover event. This mutation, which inactivates the acoR gene, was introduced into the chromosome of QB7700, leading to strain QB7704. β-Galactosidase activity was assayed in QB7704 cultures in CSK medium with or without acetoin and glucose (Table 2). The product of acoR was found to be necessary for the expression of the operon. This strongly suggests that the promoter of the aco operon is recognized by an RNA polymerase associated with the SigL sigma factor. Therefore, a sigL::aphA3 null mutation was introduced by transformation into QB7700, leading to strain QB7702. The β-galactosidase activity was assayed in cultures of this strain grown in CSK medium with or without acetoin as the inducer (Table 2). There was no LacZ activity, indicating that a SigL-dependent promoter is involved in the transcription of the aco operon.
Promoter and control regions located upstream from the acoABCL operon.
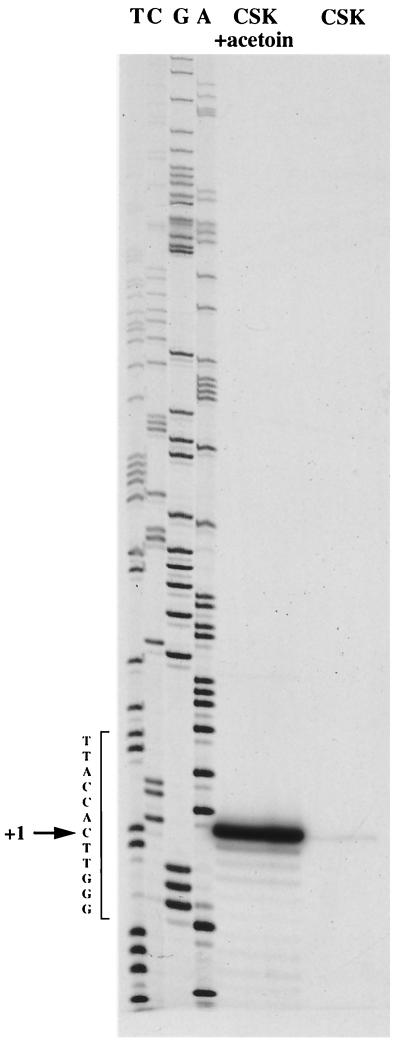
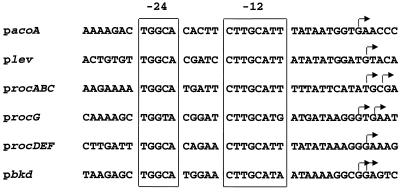
In gram-negative and gram-positive bacteria, all promoters recognized by holoenzyme containing ς54 possess at least one conserved GG doublet around position −26 followed by a GC doublet at a distance of 10 bp (2, 33). The transcription start site of acoABCL was mapped by primer extension using reverse transcriptase (Fig. 2). RNA was extracted from uninduced cells grown in CSK medium or induced cells grown in CSK medium containing 10 mM acetoin. A unique band was observed when RNA extracted under induced conditions was used, allowing the definition of the promoter. An alignment of the deduced promoter region with five other −12, −24 SigL-dependent promoters from B. subtilis is shown in Fig. 3. The promoter contains −12 and −24 regions identical to those observed in sigma 54-dependent promoters, for instance, the levanase and rocABC promoters (4, 6). A second DNA sequence with similarities to the −12, −24 promoter is present 135 bp upstream from the transcription start site. A DNA fragment lacking this similar sequence was synthesized by PCR and fused upstream from the lacZ gene. This construction was reintroduced at the amyE locus of B. subtilis by using the plasmid pDH32 (35). The resulting strain did not express LacZ activity in CSK medium containing acetoin as the inducer (not shown). This result indicates that only one −12, −24 promoter identified by primer extension (shown in Fig. 2) is involved in the transcription of the acoABCL operon.
FIG. 2.
Reverse transcriptase mapping of the transcriptional start site of the acoA gene. RNA was extracted from B. subtilis 168 grown in the presence (+ acetoin) or absence of 10 mM acetoin. The position of the cDNA-extended fragment was compared to that of fragments obtained by sequencing an M13 recombinant phage containing the promoter region, with the same oligonucleotide used as a primer. The transcriptional start site is indicated by an arrow.
FIG. 3.
Nucleotide sequences of promoter regions of the acoABCL operon (pacoA), the levanase operon (plev) (6), the rocABC operon (procABC) (4), the rocG gene (procG) (3), the rocDEF operon (procDEF) (14), and the bkd operon (pbkd) (8). The transcriptional start sites are indicated by arrows. The boxes indicate conserved DNA sequences around positions −12 and −24 with respect to the transcriptional start sites.
The aco operon requires upstream sequences for its expression.
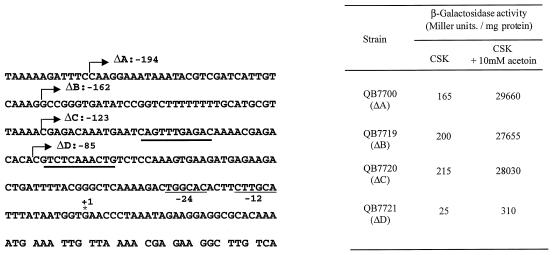
Central-domain activators contact ς54-holoenzyme through DNA looping after binding to the enhancer, also called upstream activating sequences (UAS), typically 80 to 60 bp upstream from the transcription start site. A notable exception is rocG in B. subtilis, which is transcribed by a SigL-containing RNA polymerase and requires RocR, a member of the NtrC/NifA family of proteins. Unlike other ς54-dependent genes, rocG has no UAS; instead, its expression is dependent on a sequence called DAS (downstream activating sequence) located downstream from the rocG coding region. This DAS also serves as a UAS for rocABC (3). To identify any such sequences associated with acoA expression, deletions ending upstream from the transcriptional start site were introduced. A set of DNA fragments from which part of the upstream region was missing was obtained by PCR synthesis (see Materials and Methods). These fragments were then inserted upstream from the lacZ gene in pAC5. The deletion end points are indicated in Fig. 4. The fusions were reintroduced as single copies at the amyE locus of B. subtilis. The levels of lacZ expression in this strain were analyzed and are shown in Fig. 4. In the ΔB and ΔC deletions, lacZ expression was identical to that obtained in QB7700. In the ΔD deletion, lacZ expression was strongly reduced, indicating that full induction of the aco operon requires DNA sequences located between positions −123 and −85. Interestingly, this DNA region includes three copies of a hexanucleotide sequence, 5′-GAGACA-3′ , and also a palindromic sequence of 11 bp centered on position −91. Possibly these direct repeats or the palindromic sequence are involved in the binding of AcoR.
FIG. 4.
Nucleotide sequence of the acoA upstream region. The deletion end points are indicated by bent arrows and numbered with respect to the transcriptional start site, labeled with an asterisk. −12, −24 sequences are indicated. The boldly underlined regions indicate putative palindromic UAS. The effects of upstream deletions on expression of the acoA′-′lacZ translational fusion are indicated on the right. β-Galactosidase specific activity was determined in extracts prepared from exponentially growing cells in CSK medium containing 10 mM acetoin as the inducer.
Analysis of acoR gene expression.
The intergenic region located between the end of acoL and the beginning of acoR is 118 bp long, suggesting that acoR may be part of the acoABCL operon. This possibility was tested by constructing a B. subtilis strain in which the lacZ gene was integrated and fused into the acoR gene. A DNA fragment encoding an internal part of acoR was obtained by PCR synthesis and inserted into pDIA5307 upstream from the lacZ gene. The recombinant plasmid was integrated by a single-crossover event into the chromosome of strain 168, leading to strain QB7713. As the integration of the lacZ gene inactivates the acoR gene, the plasmid pACOR1, which contains the wild-type acoR, was introduced by transformation into strain QB7713, leading to strain QB7714. The level of lacZ expression was assayed in extracts of cultures of strains QB7714 and QB7713 grown in CSK medium containing 10 mM acetoin (Table 2). The results indicate that the transcription of acoR was not induced by acetoin and did not require the wild-type acoR gene for its expression. Therefore, the acoR gene and the acoABCL operon are organized as two distinct transcriptional units and acoR is not autoregulated. In addition, the transcription of acoR is strongly repressed by the presence of glucose in the growth medium. A ΔccpA::spc null mutation was introduced by transformation into strain QB7714, leading to strain QB7733 (Table 1). The properties of this strain show that the repression by glucose of the acoR transcription depended upon CcpA (Table 2).
Catabolite repression of aco operon expression.
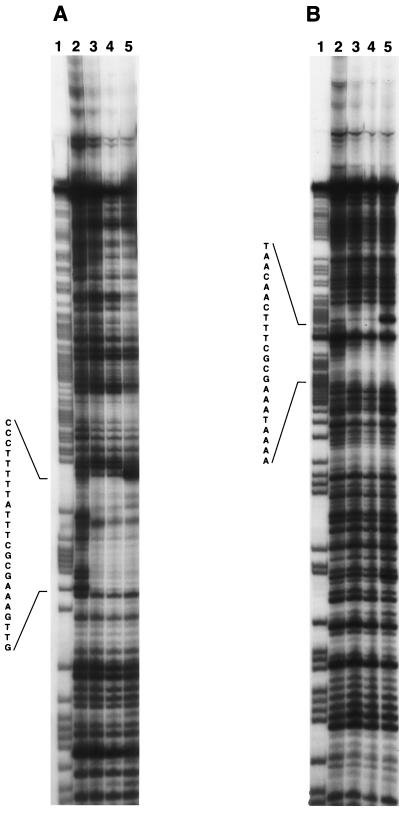
The acoABCL operon of B. subtilis is subject to carbon catabolite repression by glucose via CcpA (Table 2). Several CRE-like sequences could be involved in carbon catabolite repression by glucose. A DNA sequence, 5′-TGATTTTACGGGCTCA-3′, is located between positions −34 and −49 from the transcription start site of the acoABCL operon. Another DNA sequence, 5′-TGAATTCGGTCCCA-3′, is located in the beginning of the coding sequence of acoR (codons 1 to 5). Mutants were constructed containing either point mutations or deletions of these CRE-like sequences. The glucose effect in the resulting strains was assayed and indicated that neither CRE-like sequence is involved in catabolite control by glucose (not shown). Two other CRE-like sequences are located in the intergenic region between acoL and acoR (Fig. 5). In order to test the binding of CcpA in the upstream region of acoR, DNase I footprinting experiments were performed. A 239-bp DNA fragment containing both CRE-like sequences located between the end of acoL and the beginning of acoR was used as a template. The results of the DNase I footprinting experiments are presented in Fig. 6. In the presence of CcpA, a clear protection pattern was observed in a single region extending from 45 to 71 bases upstream from the ATG start codon of acoR. A specific interaction between CcpA and HPr-Ser-P has been demonstrated by several in vitro techniques (19). Sites of hypersensitivity to DNase I digestion were observed when CcpA and HPr-Ser-P were present in the reaction mixture. Similar alterations of the pattern of DNase I digestion were previously observed outside the CRE sequence in other systems (31).
FIG. 5.
Organization of the acoR promoter region. The DNA sequence located between the end of acoL and the beginning of acoR is shown. DNA regions with similarities to the CRE consensus sequence are underlined. Putative −10 and −35 regions of the acoR promoter are indicated in boldface letters. Regions protected by CcpA are marked by brackets.
FIG. 6.
DNase I footprinting analysis of CcpA binding to the acoR promoter region. Lanes containing 2 × 105 cpm of the labeled nontemplate strand (A) and template strand (B) of acoR are shown. Fragments were incubated in the presence of 2 μM purified CcpA. A + G Maxam and Gilbert reaction products of the appropriate DNA fragments were loaded in lanes 1. Lanes 2 through 5 were as follows: lanes 2, no protein; lanes 3, 2 μM CcpA; lanes 4, 2 μM CcpA and 10 μM HPr; and lanes 5, 2 μM CcpA and 10 μM HPr-Ser-P. Regions protected by CcpA are indicated by DNA sequences.
DISCUSSION
B. subtilis cells growing in medium containing an excess of carbon source excrete a large number of organic compounds, including pyruvate, succinate, acetate, and acetoin. It was previously proposed that the conversion of pyruvate to acetoin or butanediol prevents overacidification of the growth medium during exponential growth (16). Acetoin is secreted during the exponential growth phase and also serves as a carbon storage compound which can be reused when other carbon sources have been exhausted. It is interesting to note that the CcpA protein is required for the expression of the als gene, involved in the biosynthesis of acetoin, and of the ack and pta genes, which are involved in the production of acetate. Conversely, CcpA is a repressor of the transcription of the acu and aco operons. B. subtilis therefore has a fine-tuning system controlling both the synthesis and the degradation of secondary metabolites, such as acetate and acetoin. We studied the regulation of transcription of the aco operon. It contains four genes, acoABCL, encoding the acetoin dehydrogenase complex. Transcription of this operon is strongly induced in the presence of acetoin in the growth medium. A regulatory gene, called acoR, is required for the positive regulation of this operon. This gene encodes a positive regulator containing a central domain which is probably involved in the activation of the −12, −24 promoter. In this family of transcriptional activators, there are several domains with different functions. Typically, the amino-terminal domain is the signal reception domain. The amino-terminal part of AcoR is a large domain of 300 residues, a characteristic which is shared with AcoR from A. eutrophus (21) and BkdR from B. subtilis (8). This amino-terminal domain is probably involved in the control of AcoR activity in response to the presence of the internal inducer. We also characterized a DNA sequence located between positions −85 and −123 with respect to the acoA transcription start site and demonstrated that it is involved in the transcription of the gene. This sequence contains a perfect palindromic structure of 2 × 11 bp. Since the upstream part of the palindromic structure is located between the deletion ΔC and the deletion ΔD, it is tempting to speculate that this sequence is the target of AcoR. Glucose represses the transcription of the aco operon, and this catabolic control depends upon the catabolite control protein A (CcpA). CcpA is a pleiotropic repressor and binds to cis-active operator sequences. An interaction of HPr-Ser-P with CcpA has been demonstrated in vitro, and the resulting complex binds specifically to the CRE sequence in several genes of B. subtilis. In addition to HPr, an HPr-like protein, Crh, exhibiting 45% identity with HPr, participates in catabolite repression (13). A DNA sequence located between positions −50 and −38 in the promoter of the levanase operon shows similarities to the CRE sequence. This sequence plays a role in catabolite repression of the lev operon (30, 31). Interestingly, there is a similar DNA sequence at approximately the same position in the promoter region of acoA. Point mutations introduced by site-directed mutagenesis did not affect repression by glucose. The transcription of acoR itself is repressed by glucose, and a DNA sequence located upstream from the start codon of acoR is protected by CcpA against DNase I digestion. This protected region contains the sequence 5′-TGAAAGCGCTTTAT-3′, which matches the CRE consensus sequence proposed by Weickert and Chambliss (41) except for the last 2 bases. Similar deviations from the CRE consensus sequence were previously observed in other systems (19). Sites of hypersensitivity to DNase I digestion were observed when HPr-Ser-P was present in the reaction mixture, suggesting that binding of CcpA and HPr-Ser-P might induce changes in the DNA structure. The protected region contains the sequence 5′-TGAAAGCGCTTTAT-3′, which matches the CRE consensus sequence. The protected sequence overlaps a 5′-TTGAAA-3′ sequence which is the presumed −35 sequence of the acoR promoter. Therefore, we conclude that repression of transcription of acoR is probably mediated by the binding of CcpA to the promoter region of acoR. It is likely that most if not all the catabolite repression of acetoin utilization by glucose in B. subtilis can be attributed to the control of transcription of acoR by CcpA.
ACKNOWLEDGMENTS
We thank Isabelle Martin-Verstraete for helpful discussion, Anne Galinier for the gift of CcpA, HPr, and HPr-Ser-P, Alex Edelman for correcting the manuscript, and Christine Dugast for expert secretarial assistance.
This work was supported by research funds from the Institut Pasteur, the Centre National de la Recherche Scientifique, and the Ministère de la Recherche. Naima Ould Ali is the recipient of a grant from the French Ministry of Cooperation and the Algerian Ministry of Higher Education and Scientific Research.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrios H, Valderrama B, Morett E. Compilation and analysis of ς54-dependent promoter sequences. Nucleic Acids Res. 1999;27:4305–4313. doi: 10.1093/nar/27.22.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belitsky B R, Sonenshein A L. An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proc Natl Acad Sci USA. 1999;96:10290–10295. doi: 10.1073/pnas.96.18.10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calogero S, Gardan R, Glaser P, Schweizer J, Rapoport G, Débarbouillé M. RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J Bacteriol. 1994;176:1234–1241. doi: 10.1128/jb.176.5.1234-1241.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruz-Ramos H, Hoffmann T, Marino M, Nedjari H, Presecan-Siedel E, Dressen O, Glaser P, Jahn D. Fermentative metabolism of Bacillus subtilis: physiology and regulation of gene expression. J Bacteriol. 2000;182:3072–3080. doi: 10.1128/jb.182.11.3072-3080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Débarbouillé M, Martin-Verstraete I, Kunst F, Rapoport G. The transcriptional regulator LevR of Bacillus subtilis has domains homologous to both ς54- and phosphotransferase system-dependent regulators. Proc Natl Acad Sci USA. 1991;88:2212–2216. doi: 10.1073/pnas.88.6.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Débarbouillé M, Martin-Verstraete I, Kunst F, Rapoport G. The Bacillus subtilis sigL gene encodes an equivalent of ς54 from Gram-negative bacteria. Proc Natl Acad Sci USA. 1991;88:9092–9096. doi: 10.1073/pnas.88.20.9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Débarbouillé M, Gardan R, Arnaud M, Rapoport G. Role of BkdR, a transcriptional activator of the SigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J Bacteriol. 1999;181:2059–2066. doi: 10.1128/jb.181.7.2059-2066.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng W-L, Chang H-Y, Peng H-L. Acetoin catabolic system of Klebsiella pneumoniae CG43: sequence, expression, and organization of the aco operon. J Bacteriol. 1994;176:3527–3535. doi: 10.1128/jb.176.12.3527-3535.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolin M J. Diacetyl oxidation by Streptococcus faecalis, a lipoic acid-dependent reaction. J Bacteriol. 1955;69:51–58. doi: 10.1128/jb.69.1.51-58.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouet A, Sonenshein L. A target for carbon source-dependent negative regulation of the citB promoter of Bacillus subtilis. J Bacteriol. 1990;172:835–844. doi: 10.1128/jb.172.2.835-844.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frund C, Priefert H, Steinbüchel A, Schlegel H G. Biochemical and genetic analysis of acetoin catabolism in Alcaligenes eutrophus. J Bacteriol. 1989;171:6539–6548. doi: 10.1128/jb.171.12.6539-6548.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galinier A, Haiech J, Kilhoffer M C, Jaquinod M, Stülke J, Deutscher J, Martin-Verstraete I. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc Natl Acad Sci USA. 1997;94:8439–8444. doi: 10.1073/pnas.94.16.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardan R, Rapoport G, Débarbouillé M. Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. J Mol Biol. 1995;249:843–856. doi: 10.1006/jmbi.1995.0342. [DOI] [PubMed] [Google Scholar]
- 15.Glatron M-F, Rapoport G. Biosynthesis of the parasporal inclusion of Bacillus thuringiensis: half-life of its corresponding messenger RNA. Biochimie. 1972;54:1291–1301. doi: 10.1016/s0300-9084(72)80070-1. [DOI] [PubMed] [Google Scholar]
- 16.Grundy F J, Waters D A, Takova T Y, Henkin T M. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol Microbiol. 1993;10:259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- 17.Henkin T M. The role of the CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol Lett. 1996;135:9–15. doi: 10.1111/j.1574-6968.1996.tb07959.x. [DOI] [PubMed] [Google Scholar]
- 18.Huang M, Oppermann-Sanio F B, Steinbüchel A. Biochemical and molecular characterization of the Bacillus subtilis acetoin catabolic pathway. J Bacteriol. 1999;181:3837–3841. doi: 10.1128/jb.181.12.3837-3841.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global mechanism for the Gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 20.Juni E, Heyn G A. A cyclic pathway for the bacterial dissimilation of 2.3-butamediol, acetylmethylcarbinol, and diacetyl. II. The synthesis of diacetylmethylcarbinol from diacetyl, a new diphosphothiamin catalyzed reaction. J Bacteriol. 1956;72:746–753. doi: 10.1128/jb.72.6.746-753.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krüger N, Steinbüchel A. Identification of acoR, a regulatory gene for the expression of genes essential for acetoin catabolism in Alcaligenes eutrophus H16. J Bacteriol. 1992;174:746–753. doi: 10.1128/jb.174.13.4391-4400.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krüger N, Oppermann F B, Lorenzl H, Steinbüchel A. Biochemical and molecular characterization of the Clostridium magnum acetoin dehydrogenase enzyme system. J Bacteriol. 1994;176:3614–3630. doi: 10.1128/jb.176.12.3614-3630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 24.Lereclus D, Arantes O, Chaufaux J, Lecadet M-M. Transformation and expression of a cloned δ-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol Lett. 1989;60:211–218. doi: 10.1016/0378-1097(89)90511-9. [DOI] [PubMed] [Google Scholar]
- 25.Lereclus D, Arantes O. spbA locus ensures the segregational stability of pHT1030, a novel type of Gram-positive replicon. Mol Microbiol. 1992;6:35–46. doi: 10.1111/j.1365-2958.1992.tb00835.x. [DOI] [PubMed] [Google Scholar]
- 26.Lopez J, Fortnagel P. The regulation of the butanediol cycle in Bacillus subtilis. Biochim Biophys Acta. 1972;279:554–560. doi: 10.1016/0304-4165(72)90177-8. [DOI] [PubMed] [Google Scholar]
- 27.Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. Induction and metabolite regulation of levanase synthesis in Bacillus subtilis. J Bacteriol. 1989;171:1885–1892. doi: 10.1128/jb.171.4.1885-1892.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. Levanase operon of Bacillus subtilis includes a fructose-specific phosphotransferase system regulating the expression of the operon. J Mol Biol. 1990;214:657–671. doi: 10.1016/0022-2836(90)90284-S. [DOI] [PubMed] [Google Scholar]
- 29.Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. Mutagenesis of the Bacillus subtilis “−12, −24” promoter of the levanase operon and evidence for the existence of an Upstream Activating Sequence. J Mol Biol. 1992;226:85–99. doi: 10.1016/0022-2836(92)90126-5. [DOI] [PubMed] [Google Scholar]
- 30.Martin-Verstraete I, Stülke J, Klier A, Rapoport G. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J Bacteriol. 1995;177:6019–6027. doi: 10.1128/jb.177.23.6919-6927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Verstraete I, Deutscher J, Galinier A. Phosphorylation of HPr and Crh by HprK, early steps in the catabolite repression signalling pathway for the Bacillus subtilis levanase operon. J Bacteriol. 1999;181:2966–2969. doi: 10.1128/jb.181.9.2966-2969.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 33.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 34.Oppermann F B, Schmidt B, Steinbüchel A. Purification and characterization of acetoin 2,6-dichlorophenolindophenol oxidoreductase, dihydrolipoamide dehydrogenase, and dihydrolipoamide acetyltransferase of the Pelobacter carbinolicus acetoin dehydrogenase enzyme system. J Bacteriol. 1991;173:757–767. doi: 10.1128/jb.173.2.757-767.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perego M. Integrational vectors for genetic manipulation in Bacillus subtilis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 615–624. [Google Scholar]
- 36.Presecan-Siedel E, Galinier A, Longin R, Deutscher J, Danchin A, Glaser P, Martin-Verstraete I. Catabolite regulation of the pta gene as part of carbon flow pathway in Bacillus subtilis. J Bacteriol. 1999;181:6889–6897. doi: 10.1128/jb.181.22.6889-6897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renna M C, Najimudin N, Winik L R, Zahler S A. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J Bacteriol. 1993;175:3863–3875. doi: 10.1128/jb.175.12.3863-3875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Trieu-Cuot P, Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene. 1983;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- 40.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 41.Weickert M J, Chambliss G H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]