Abstract
Oral and craniofacial tissues are uniquely adapted for continuous and intricate functioning, including breathing, feeding, and communication. To achieve these vital processes, this complex is supported by incredible tissue diversity, variously composed of epithelia, vessels, cartilage, bone, teeth, ligaments, and muscles, as well as mesenchymal, adipose, and peripheral nervous tissue. Recent single cell and spatial multiomics assays—specifically, genomics, epigenomics, transcriptomics, proteomics, and metabolomics—have annotated known and new cell types and cell states in human tissues and animal models, but these concepts remain limitedly explored in the human postnatal oral and craniofacial complex. Here, we highlight the collaborative and coordinated efforts of the newly established Oral and Craniofacial Bionetwork as part of the Human Cell Atlas, which aims to leverage single cell and spatial multiomics approaches to first understand the cellular and molecular makeup of human oral and craniofacial tissues in health and to then address common and rare diseases. These powerful assays have already revealed the cell types that support oral tissues, and they will unravel cell types and molecular networks utilized across development, maintenance, and aging as well as those affected in diseases of the craniofacial complex. This level of integration and cell annotation with partner laboratories across the globe will be critical for understanding how multiple variables, such as age, sex, race, and ancestry, influence these oral and craniofacial niches. Here, we 1) highlight these recent collaborative efforts to employ new single cell and spatial approaches to resolve our collective biology at a higher resolution in health and disease, 2) discuss the vision behind the Oral and Craniofacial Bionetwork, 3) outline the stakeholders who contribute to and will benefit from this network, and 4) outline directions for creating the first Human Oral and Craniofacial Cell Atlas.
Keywords: craniofacial, oral mucosa, palate, saliva, tongue, periodontium, bone, musculoskeletal, single cell genomics, multiomics, spatial biology, Human Cell Atlas
Introduction: The Human Cell Atlas Oral and Craniofacial Bionetwork
Oral and craniofacial tissues support the human face and thus the concept of our individual and collective identity; biologically, they coordinate essential functions to sustain life, including breathing, feeding, and communication. Though often preventable, oral diseases are increasingly burdensome worldwide, affecting over one-third of the globe with a disproportionate effect on socially disadvantaged populations. In addition, chronic oral inflammatory diseases, such as periodontal disease, have been increasingly associated with >60 systemic diseases, such as cardiovascular diseases, diabetes, cancer, pneumonia, inflammatory bowel diseases, obesity, and premature birth (Schifter et al. 2010; Beck et al. 2019; Byrd and Gulati 2021). To achieve the goal of improved and precise whole-body health will require the creative and dedicated application of new toolkits to understand the human body in health and disease states in combination with extensive in vivo animal studies and in vitro systems.
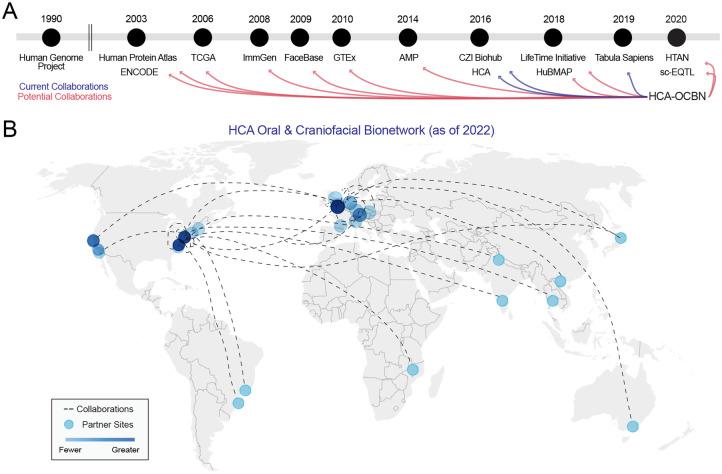
Recent advances in methods and the resolution of high-throughput single cell and spatial molecular profiling now appear to be the quantum leap that precision medicine has required, revolutionizing our ability to study tissue heterogeneity at a remarkable resolution and driving the scientific community to characterize all cells of the human body (Aldridge and Teichmann 2020). While the origins of the modern single cell revolution can be traced to quantitative polymerase chain reaction assays with individual neurons 3 decades ago (Eberwine et al. 1992), it was not until 2016 that the Human Cell Atlas (HCA; https://www.humancellatlas.org) initiative was officially launched (Fig. 1A). Since then, the HCA has established itself as an international and interdisciplinary collaborative effort to create reference cellular maps of the body across the life span (Regev et al. 2017; Regev et al. 2018).
Figure 1.
Collaboration opportunity across initiatives, networks, groups, and consortia. The Human Cell Atlas Oral and Craniofacial Bionetwork (HCA-OCBN; established 2020) is a collection of aligned investigators with the goal of mapping healthy human tissues of the oral and craniofacial complex. This network is using single cell and spatial multiomics to achieve these goals. (A) Since the Human Genome Project was completed in 2003, there have been several initiatives established that can benefit human oral and craniofacial research. There is enormous potential for fruitful collaboration between OCBN and initiatives such as FaceBase (established 2009), CZI Biohub (2016; Chan Zuckerberg Initiative), and HubMAP (2020; Human Biomolecular Atlas Program). Some of these groups are already collaborating with the OCBN (blue lines). (B) Collaboration within the OCBN can be as focused as sharing tissues and fluid samples or supporting computational analysis. As of May 2022, projects are growing monthly across new OCBN teams. Map generated from https://app.datawrapper.de/. AMP, Accelerating Medicines Partnership Program; ENCODE, Encyclopedia of DNA Elements; GTEx, Genotype-Tissue Expression project; HTAN, Human Tumor Atlas Network; ImmGen, Immunological Genome Project; sc-EQT, single-cell eQTLGen Consortium; TCGA, The Cancer Genome Atlas.
The HCA consortium sits uniquely among a lineage of initiatives, networks, groups, and other multiomic-focused consortia that came after the Human Genome Project was completed in 2003 (Fig. 1A) as it does not direct its associated investigators’ studies. Despite this leadership structure, various HCA bionetworks and single cell and spatial biology consortia have made landmark scientific advances, including the discovery of new and rare human cell type annotations, referred to as “cell subtypes” (Montoro et al. 2018; Park et al. 2018; Aizarani et al. 2019; Litvinukova et al. 2020; McDonald et al. 2021), as well as disease-associated cell “states” (Kinchen et al. 2018; Ledergor et al. 2018; Sade-Feldman et al. 2018; Martin et al. 2019; Vieira Braga et al. 2019; Reynolds et al. 2021).
To support the investigation and inclusion of healthy oral and craniofacial tissues in the first draft of the HCA, the Oral and Craniofacial Bionetwork (OCBN) was founded in 2020 as a research network focused on postnatal adult oral and craniofacial tissues within the HCA (Fig. 1). Additionally, other craniofacial organs and hard tissues, such as the brain, eye, nasal cavity, face skin, adipose, and musculoskeletal system (Baldwin et al. 2021), are described in allied HCA bionetworks. Here, we 1) highlight recent collaborative efforts to employ new single cell and spatial approaches to resolve our collective biology at a higher resolution in health and disease, 2) discuss the vision behind the OCBN, 3) outline the stakeholders who contribute to and will benefit from this network, and 4) outline directions for creating the first Human Oral and Craniofacial Cell Atlas.
A Roadmap for the Human Oral and Craniofacial Cell Atlas
Defining Oral and Craniofacial Tissue Heterogeneity
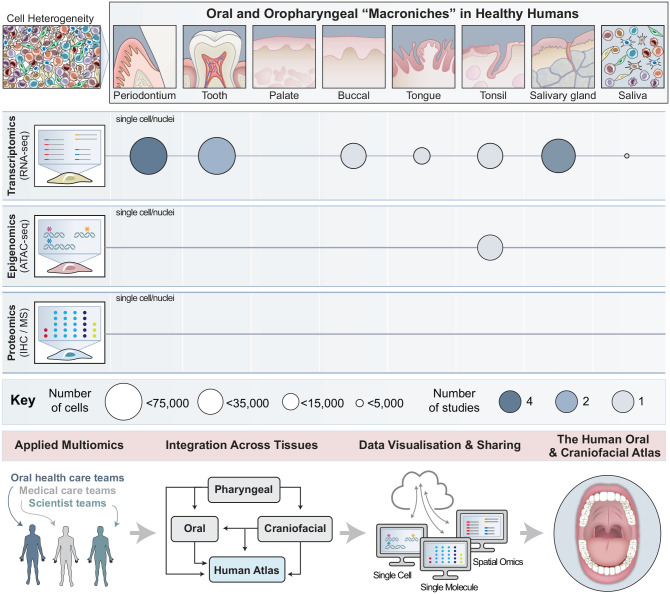
The oral and craniofacial complex is supported by highly diverse tissue niches, composed of epithelia, blood and lymphatic vessels, cartilage, bone, ligaments, and muscles, as well as adipose and peripheral nervous tissue (Nanci 2018). This anatomy has been well described for decades (Nanci 2018); however, recent genetic and genomics approaches based on mouse models have found intra- and interspecific niche heterogeneity among the periodontium (Nagata et al. 2021; Zhao and Sharpe 2021), tooth (Sharir et al. 2019; Takahashi et al. 2019; Chiba et al. 2020; Krivanek et al. 2020), salivary glands (Song et al. 2018; Hauser et al. 2020; Sekiguchi et al. 2020), palate (Byrd et al. 2019; Li et al. 2019; Han et al. 2021), buccal mucosa (Jones et al. 2019), and tongue (Tabula Muris et al. 2018; Almanzar et al. 2020). Some of these data have recently been integrated (Huang et al. 2020; Williams et al. 2021), though focused on healthy barrier epithelia, and there appears to be common and unique cell types among these niches.
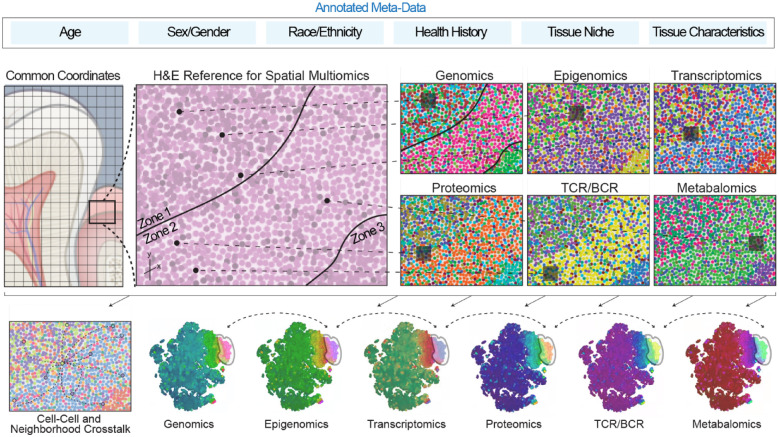
Importantly, oral and craniofacial tissue heterogeneity is not defined at just the level of biology (i.e., molecular, cellular, tissue, organ) as it is well established that human sex, age, and ancestry influence tissue physiology. There is an urgent need to increase representation in scientific research, as ~85% of today’s genomic data are derived from European ancestry; therefore, it is crucial to account not only for tissue heterogeneity but also for expanding into human geographic, sex/gender, age, ethnic, and ancestral diversity in the donor data sets (Figs. 2, 3). The OCBN takes this mission seriously among its investigators and study participants (Fig. 1B) with one funded study focused on generating healthy oral and craniofacial multiomic reference data from at least 8 global ancestries.
Figure 2.
A blueprint for the Human Oral and Craniofacial Cell Atlas. Oral and craniofacial tissue niches are incredibly diverse, including periodontium, tooth, palate, buccal mucosa, tongue, tonsils, salivary glands, and fluid from the saliva—these do not account for microniches that are unaccounted for among these tissue spaces. Each niche is supported by some combination of epithelia, cartilage, bone, ligaments, muscles, adipose tissue, blood and lymphatic vessels, and nerves, and these tissues are harmoniously integrated into the vital functions of communication, feeding, breathing, defense, sensing, and early digestion. The Human Oral and Craniofacial Cell Atlas, supported by the Oral and Craniofacial Bionetwork (OCBN), aims to create comprehensive and integrated cell atlases to understand the common and unique cell types that support these niches in health and to uncover which cell types and networks are affected in disease. We will do this using single cell and spatial multiomic approaches (transcriptome, epigenomic, proteomic), and we will incorporate additional omics technologies as assays are refined and available. Thus far, most work from the OCBN and others has focused on single cell transcriptomic (scRNAseq) and epigenomic (ATACseq) approaches from healthy adults, including nearly all major tissue niches and saliva. Work is currently being done with the OCBN for collecting, integrating, visualizing, sharing, and applying the knowledge gained from these early studies. The number of studies developed so far and the total number of cells profiled are depicted under each tissue. Tissue illustrations inspired by Huang et al. (2021). Credit: Heather McDonald, BioSerendipity, LLC.
Figure 3.
Applied spatial multiomics for coordinated and integrated analyses. Coordinated efforts across the Human Cell Atlas are working toward building a consensus of the necessary metadata (clinical and biological) to generate a common coordinate framework for the human body. Future work including these additional layers of data will highlight the diversity of cell types and states across humans considering age, sex, race, ethnicity, ancestry, and oral and systemic health history, as well as the specific niche, tissue orientation, and health status of that niche (healthy, inflamed) at the time of sample collection. There is an immense need to interrogate a higher number of molecular dimensions or human tissues—genome, transcriptome, epigenetic modifications, proteome, T cell receptor and B cell receptor repertoires (TCR/BCR), and metabolome of the collected sample itself—as no single “omics” technology can fully define the complexity of molecular mechanisms, but taken together, these integrated data have the potential to provide a more comprehensive landscape of basic biological processes and human disease. Multimodal sequencing has the capacity to move the field from descriptive “snapshots” toward a mechanistic understanding of gene regulatory networks and, importantly, to refine sources of cellular heterogeneity as already applied to the immune system. The use of multimodal single cell and spatial multiomics is therefore revolutionizing our understanding of cellular biology; however, relying on the dissociation of cells from their natural tissue environment limits our ability to understand the role of intrinsic and extrinsic factors that underpin cellular communication and organ function. Spatial multiomic approaches, which include information on the location of cells, will still need to be integrated with these single cell multiomic maps.
Collecting Oral and Craniofacial Atlases as Reference Data Sets
Significant progress has been made by the HCA community with the current total of profiled cells to date numbering about 40 million cells from 15 major organs (Lindeboom et al. 2021) (https://data.humancellatlas.org/). While there are an estimated 40 trillion cells in the human body, we estimate that there are about 2 trillion in the oral and craniofacial tissues. Although progress in the single cell profiling of human oral tissues has been slow in comparison, researchers have made significant progress as of 2022, publishing atlases of the oral mucosa, including the buccal mucosa, tongue, and gingiva (Caetano et al. 2021; Huang et al. 2021; Williams et al. 2021; Tabula Sapiens et al. 2022); major and minor salivary glands (Huang et al. 2021; Chen et al. 2022; Costa-da-Silva et al. 2022; Tabula Sapiens et al. 2022); dental pulp (Krivanek et al. 2020; Pagella et al. 2021; Opasawatchai et al. 2022); periodontal ligament (Pagella et al. 2021); tonsil (King et al. 2021); and even saliva itself (Choudhury et al. 2020; Fig. 2). Additional craniofacial niches, such as the temporomandibular joint and skeletal muscle, are planned and being executed as well.
In sum, these and other active studies have already identified regional differences in cellular types, proportions, states, phenotypes, and niche-dependent interactions and highlighted the importance of the tissue extracellular microenvironment in shaping the identity of resident epithelial, stromal, and immune cells. Within 18 mo (between September 2020 and March 2022), OCBN made significant strides forward to the modern age of single cell biology (Aldridge and Teichmann 2020), including nearly >250,000 cells representing the first cell types from 8 distinct niches from healthy adults (Fig. 2, Table). Within the OCBN, multidisciplinary teams are already conducting experiments incorporating single cell and spatial multiomics in pediatric and adult human subjects. Importantly, the rapid development of spatial technologies, such as sequencing- and FISH-based technologies, is now permitting the use of fresh-frozen and fixed paraffin-embedded samples, opening the possibility for analyzing human tissues from archived biobanks (Fig. 3).
Table.
Human Oral and Craniofacial Single Cell and Spatial Multiomic Data Sets (2020–2022).
| Study | Sample Type | Anatomic Information | Method | Clinical Annotations | Cell No. | Donors | Cell Ontology Class | Public Repository |
|---|---|---|---|---|---|---|---|---|
| Huang et al. (2021) | Gingival mucosa | Upper left maxillary lingual interproximal papilla | 10x scRNA-seq | Gingivitis | 6,683 | 4 | Basal keratinocyte 1 / 2 / 3 / 4Basal cycling keratinocyteSuprabasal keratinocyteMelanocyte | https://www.covid19cellatlas.org/byrd20/ |
| Merkel cell | ||||||||
| Langerhans cell | ||||||||
| Arterial endothelium | ||||||||
| Capillary endothelium | ||||||||
| Fibroblast | ||||||||
| Lymphatic endothelium | ||||||||
| Smooth muscle cell | ||||||||
| Macrophage | ||||||||
| Dendritic cell 1 / 2 | ||||||||
| Activated dendritic cell | ||||||||
| Plasmacytoid dendritic cell | ||||||||
| Mast cell | ||||||||
| Mucosal-associated invariant T cell | ||||||||
| Natural killer cell | ||||||||
| Cytotoxic T cell 1 | ||||||||
| Helper T cell | ||||||||
| Regulatory T cell | ||||||||
| γδ T cell | ||||||||
| T/NK cycling cell | ||||||||
| B cell | ||||||||
| Caetano et al. (2021) | Gingival mucosa | Upper right maxillary buccal margin | 10x scRNA-seq | Healthy, periodontitis | 12,411 | 4 | Epithelial cell 1 / 2Basal cycling keratinocyte | GSE152042 |
| Fibroblast 1 / stromal (S0) | ||||||||
| Fibroblast 2 / stromal (S1) | ||||||||
| Fibroblast 3 / stromal (S2) | ||||||||
| Fibroblast 4 / stromal (S4) | ||||||||
| Fibroblast 5 / stromal (S6) | ||||||||
| Myofibroblasts | ||||||||
| Endothelial cell 1 / 2 | ||||||||
| Perivascular cell | ||||||||
| Pericytes | ||||||||
| Macrophage 1 / 2 | ||||||||
| Dendritic cell | ||||||||
| Plasmacytoid dendritic cell | ||||||||
| Mast cells | ||||||||
| T cells | ||||||||
| IgG B cells | ||||||||
| Follicular B cell | ||||||||
| Memory B cell | ||||||||
| Williams et al. (2021) | Gingival mucosa | Upper right/left buccal margin, lower right buccal margin | 10x scRNA-seq | Healthy, periodontitis | 88,140 | 21 | Epithelial cell 1 / 2 / 3MelanocyteFibroblast 1 / 2 / 3 / 4 / 5 | https://oral.cellatlas.ioGSE164241 |
| Smooth muscle cell | ||||||||
| Vasculature endothelium cell 1 / 2 / 3 / 4 | ||||||||
| Lymphatic endothelium | ||||||||
| αβ CD4+ T cell | ||||||||
| Helper T cell 17 | ||||||||
| Mucosal-associated invariant T cell | ||||||||
| αβ CD8+ T cell | ||||||||
| γδ T cell | ||||||||
| Regulatory T cell | ||||||||
| Natural killer cell | ||||||||
| Neutrophil | ||||||||
| Macrophage | ||||||||
| Migratory dendritic cell | ||||||||
| Mast cell | ||||||||
| Plasmacytoid dendritic cell | ||||||||
| B cell | ||||||||
| Plasma cell | ||||||||
| Williams et al. (2021) | Buccal mucosa | Left buccal mucosa | 10x scRNA-seq | Healthy | 34,999 | 8 | Epithelial cell 1 / 2 / 3Melanocyte | https://oral.cellatlas.ioGSE164241 |
| Fibroblast 1 / 2 / 3 / 4 | ||||||||
| Smooth muscle cell | ||||||||
| αβ CD4+ T cell | ||||||||
| Helper T cell 17 | ||||||||
| Mucosal-associated invariant T cell | ||||||||
| αβ CD8+ T cell | ||||||||
| γδ T cell | ||||||||
| Regulatory T cell | ||||||||
| Natural killer cell | ||||||||
| Neutrophil | ||||||||
| Macrophage | ||||||||
| Migratory dendritic cell | ||||||||
| Mast cell | ||||||||
| Plasmacytoid dendritic cell | ||||||||
| B cell | ||||||||
| Plasma cell | ||||||||
| Tabula Sapiens et al. 2022 | Major salivary glands | Parotid; submandibular | 10x scRNA-seq; Smartseq2 | Healthy | 27,199 | 2 | Basal keratinocyte cellAcinar cell | https://tabula-sapiens-portal.ds.czbiohub.org |
| Salivary gland cell | ||||||||
| Duct epithelial cell | ||||||||
| Ionocyte | ||||||||
| Myoepithelial cell | ||||||||
| Fibroblast | ||||||||
| Endothelial cell | ||||||||
| Lymphatic endothelial cell | ||||||||
| Pericyte | ||||||||
| Adventitial cell | ||||||||
| Macrophage | ||||||||
| Monocyte | ||||||||
| Neutrophil | ||||||||
| NK cell | ||||||||
| B cell | ||||||||
| Naive B cell | ||||||||
| Plasma cell | ||||||||
| Memory B cell | ||||||||
| T cell | ||||||||
| CD4+ helper T cell | ||||||||
| αβ CD4+ T cell | ||||||||
| αβ CD8+ T cell | ||||||||
| Huang et al. (2021) | Minor salivary glands | Labial minor | 10x scRNA-seq | Healthy individuals | 7,107 | 5 | Basal keratinocyteMucous acinar cell | https://www.covid19cellatlas.org/byrd20/ |
| Serous acinar cell | ||||||||
| Duct epithelial cell | ||||||||
| Ionocyte | ||||||||
| Myoepithelial cell | ||||||||
| Fibroblast | ||||||||
| Arterial endothelium | ||||||||
| Capillary endothelium | ||||||||
| Venule endothelium | ||||||||
| Pericyte | ||||||||
| Smooth muscle cell | ||||||||
| Glial cell | ||||||||
| Macrophage 1 / 2 | ||||||||
| Mast cell | ||||||||
| Cytotoxic T cell 1 / 2 | ||||||||
| Helper T cell 1 | ||||||||
| Dendritic cell 2 | ||||||||
| B cell | ||||||||
| Plasma cell | ||||||||
| Erythrocyte | ||||||||
| Costa-da-Silva et al. (2022) | Minor salivary glands | Labial minor | 10x scRNA-seq | Healthy individuals | 21,402 | 4 | Serous acinar cell | GSE180544 |
| Seromucous acinar cell | ||||||||
| Mucous acinar cell | ||||||||
| Ductal epithelial cell | ||||||||
| Fibroblast | ||||||||
| Myoepithelial cell | ||||||||
| Pericyte/myofibroblast | ||||||||
| Vasculature endothelium | ||||||||
| Lymphatic endothelium | ||||||||
| Smooth muscle cell | ||||||||
| Myeloid immune cell | ||||||||
| Plasma cell | ||||||||
| T cell | ||||||||
| Pagella et al. (2021) | Periodontium | Periodontal ligament third molars | 10x scRNA-seq | Healthy individuals | 2,883 | 5 | Epithelial cell 1 / 2 / 3 / 4 / 5Mesenchymal stem cell | https://github.com/TheMoorLab/Tooth |
| Fibroblast | GSE161267 | |||||||
| Endothelial cell 1 / 2 | ||||||||
| Schwann cell | ||||||||
| Immune cell 1 / 2 / 3 | ||||||||
| Erythrocyte | ||||||||
| King et al. (2021) | Tonsil | Pediatric tonsils | scRNA-seq; scVDJ | Recurrent tonsilitis; obstructive sleep apnea | 32,607 | 7 | CD4+ NCM T cellCD4+ T cellTfH, T cell | https://www.tonsilimmune.orgE-MTAB-8999E-MTAB-9003 |
| TfR T cell | E-MTAB-9005 | |||||||
| Regulatory T cell | ||||||||
| CD8+ NCM T cell | ||||||||
| CD8+ cytotoxic T cell | ||||||||
| TIM3+ DN T cell | ||||||||
| Innate lymphoid cell | ||||||||
| NK Cell | ||||||||
| T cell cycling | ||||||||
| Macrophage precursor cell | ||||||||
| Macrophage 1 / 2 / 3 | ||||||||
| Dendritic cell 1 | ||||||||
| Plasmacytoid dendritic cell | ||||||||
| Follicular dendritic cell | ||||||||
| FCRL4+ marginal B cell | ||||||||
| Activated B cell | ||||||||
| Marginal B cell | ||||||||
| Naïve B cell | ||||||||
| PreGC B cell | ||||||||
| FCRL3-high B cell | ||||||||
| GC B cell | ||||||||
| DZ GC B cell | ||||||||
| B cell cycling | ||||||||
| Pre-plasmablast | ||||||||
| LZ GC B cell | ||||||||
| Plasmablast | ||||||||
| Choudhury et al. (2020) | Saliva | Adult saliva | Smart-seq2 | Healthy individuals | ~2,000 | 3 | Neutrophil | https://data.humancellatlas.org/explore/projects/60ea42e1-af49-42f5-8164-d641fdb696bc |
| Tabula Sapiens et al. 2022 | Tongue | Anterior, posterior | 10x scRNA-seq; Smartseq2 | Healthy individuals | 15,020 | 3 | Basal keratinocyte cellEpithelial cell | https://tabula-sapiens-portal.ds.czbiohub.org |
| Keratinocyte | ||||||||
| Fibroblast | ||||||||
| Arterial endothelial cell | ||||||||
| Capillary endothelial cell | ||||||||
| Venule endothelial cell | ||||||||
| Lymphatic endothelial cell | ||||||||
| Pericyte | ||||||||
| Skeletal muscle cell | ||||||||
| Schwann cell | ||||||||
| Immune cell | ||||||||
| Krivanek et al. 2020 | Dental pulp, apical papilla | Third molars | 10x scRNA-seq; Smartseq2 | Healthy individuals | 41,673 | 7 | Odontoblast | http://pklab.med.harvard.edu/ruslan/dental.atlas.html |
| Peri-odontoblastic cell | GSE146123 | |||||||
| Periodontal ligament cell | ||||||||
| Endothelial cell | ||||||||
| Perivascular cell | ||||||||
| Pulpal cell | ||||||||
| Glial cell | ||||||||
| Macrophage | ||||||||
| Neutrophil | ||||||||
| NK cell | ||||||||
| Lymphocyte | ||||||||
| Cycling cell | ||||||||
| Pagella et al. (2021) | Dental pulp | Third molars | 10x scRNA-seq | Healthy individuals | 32,378 | 5 | Epithelial cell | https://github.com/TheMoorLab/Tooth |
| Mesenchymal stem cell 1 / 2 / 3 | GSE161267 | |||||||
| Fibroblast 1 / 2 / 3 / 4 / 5 / 6 | ||||||||
| Endothelial cell 1 / 2 / 3 / 4 / 5 | ||||||||
| Odontoblast cell | ||||||||
| Non-myelinating Schwann cell | ||||||||
| Myelinating Schwann cell | ||||||||
| Immune cell 1 / 2 / 3 / 4 / 5 | ||||||||
| Erythrocyte | ||||||||
| Opasawatchai et al. (2022) | Dental pulp | Third molars | 10x scRNA-seq | Healthy and carious | 6,810 | 4 | Naive CD4/CD8 T cellEarly odontoblastMacrophage | GSE185222https://github.com/vclabsysbio/scRNAseq_Dentalpulp |
| Odontoblast | ||||||||
| CD4 T cell | ||||||||
| CD8 T cell | ||||||||
| Naive B cell | ||||||||
| Erythrocyte | ||||||||
| NK cell | ||||||||
| Granulocyte | ||||||||
| HSCs | ||||||||
| Vascular endothelium | ||||||||
| CD16 mono | ||||||||
| Immature erythrocyte | ||||||||
| Plasmacytoid dendritic cell | ||||||||
| Plasma cell | ||||||||
| Chen et al. (2022) | Major salivary glands | Parotid | 10x scRNA-seq | Healthy | 16,052 | 1 | B cellT cellFibroblast | GSE188478https://github.com/miao-OvO/PG-scRNA-seq |
| NK cell | ||||||||
| Serous acinar cell | ||||||||
| Myeloid cell | ||||||||
| Plasma cell | ||||||||
| Ductal epithelial cell | ||||||||
| Vascular endothelium | ||||||||
| Myoepithelial cell |
Currently available human data sets contributed to public repositories, including the Human Cell Atlas Data Coordination Portal (Choudhury et al. 2020; Krivanek et al. 2020; Caetano et al. 2021; Huang et al. 2021; King et al. 2021; Pagella et al. 2021; Williams et al. 2021; Chen et al. 2022; Costa-da-Silva et al. 2022; Opasawatchai et al. 2022; Tabula Sapiens et al. 2022).
While early efforts have primarily focused on the adult oral soft tissues, craniofacial structures such as the cranial ganglia, cranial vault, cranial base, nasopharynx, nasal bones and cartilages, neck, pinna, and middle ear bones will be essential to allow a more detailed investigation into the molecular mechanisms controlling tendogenesis, chondrogenesis, and osteogenesis, which support craniofacial function and enhance cranial skeletal repair, thereby leading to a better understanding of craniofacial anomalies. Achieving these goals will require more collaborative efforts among established and new OCBN investigators, other HCA networks, as well as other consortia.
Illuminating Intercellular Communication Networks between Defined Cell Types
With this reference data set, we aspire to accelerate discoveries in the domains of basic and clinically applied research with further downstream analyses. Expression profiling of different cell types in adult human tissues has shown how intercellular communication contributes to tissue function by coordinating cell functions in development and homeostasis; thus, when there are signaling defects, disease will follow. The study of intercellular communication has significantly accelerated with advances in the single cell field with several studies discovering novel signaling, mediating cellular differentiation and immune responses. Intercellular crosstalk has been investigated in oral tissues, with OCBN studies demonstrating how in periodontal disease there is a shift in the transcriptional signatures of stromal and epithelial oral mucosa cells to an inflammatory profile (Caetano et al. 2021; Williams et al. 2021). In disease, endothelial cells also showed upregulation of pathways related to lymphocyte adhesion and chemokine signaling (Williams et al. 2021).
Given that most cellular crosstalk is spatially restricted with signals working from 0 to 200 µm, spatial transcriptomics data are essential to understand intercellular communication in healthy and diseased tissues. To investigate how surrounding cells may regulate signaling, several computational methods have been recently developed to integrate spatial information with ligand-receptor analyses (Efremova et al. 2020; Dries et al. 2021). However, translating this to the clinic will require harmonized and consistent cell type annotation among different human atlases to allow for accurate intercellular communication network modeling, as disease networks consistently are sequenced and added to the HCA data sets. Harmonized nomenclature and annotation of cell types/states when integrating different source organ atlases is one of the current efforts of the HCA to achieve a unified reference cell ontology across tissues (Osumi-Sutherland et al. 2021).
Annotated Clinical and Biological Metadata for a Common Coordinate Framework
While these initial OCBN projects have started to construct high-resolution maps of organs and tissues, there is an unmet need to integrate these atlases to interrogate analogous cellular components across tissue niches and to develop a common coordinate framework for the healthy oral and craniofacial human tissues (Fig. 3). So far, most annotations of OCBN single cell genomics data sets have distinct cell type annotations, even within the same tissue (Table). This discordance makes it difficult to relate findings among studies, highlighting the need for a common “language” for cell annotation. This framework will aim to address clinical and spatial variability within studies and allow for more accurate and precise comparisons among data sets across the human body. Relevant clinical metadata will include but not be limited to donor gender, sex, ethnicity, age, tissue type, relevant clinical data, and technology used; spatial data will define the position, tissue plane, site, and size of anatomic structures (Table).
Furthermore, future work incorporating these clinical and biological data will highlight the niche-specific cellular diversity that may help to explain why some oral diseases manifest in some oral and craniofacial niches while sparing others. For example, understanding niche-specific cellular heterogeneity in pediatric tissues from neonatal to infancy, juvenile, and adolescence periods will allow the identification of cell states and cell lineages involved in tissue maturation and a better understanding of early disease onset in childhood. However, given the vast literature on immune training in early development, these efforts should also reveal the mechanisms of healthy and pathologic aging that may lead to early and accurate prognostic tools allowing for early intervention, similar to what is proposed by the LifeTime Initiative (Rajewsky et al. 2020, 2021).
This additional level of annotation is already relevant. For example, the concept of structural immunity (i.e., niche specific) has recently been described across the body, suggesting that each tissue’s cell-specific composition can instruct its niche-distinct immune response (Krausgruber et al. 2020). This lens will provide a new framework for interrogating human disease by mapping disease risk genes and for predicting cell type–specific and coregulated gene modules, as recently described in periodontitis (Williams et al. 2021). For the long-term success of the OCBN, high-quality and widely available single cell and spatial multiomic data sets will need to be published with detailed metadata for each experiment and study.
Integrated Analyses within and across Multimodal Data
A fundamental challenge when constructing biological systems is the correct definition of cell state, which is achieved by applying complementary approaches, such as molecular characterization (transcripts, distribution of chromatin marks and proteins) and functional testing (Fig. 3). Understanding cell-specific activity further requires proteomics, metabolomics, and functional assays to provide a direct readout of cellular activity. Furthermore, there is a need to interrogate a higher number of molecular dimensions. No single “omics” technology can fully define the complexity of molecular mechanisms, but taken together, these integrated data have the potential to provide a more comprehensive landscape of basic biological processes and human disease. Multimodal measurements, where distinct molecular parameters can be interrogated in the same cell, have been recently developed and are now allowing us to characterize cells, cell states, and transitions between cell states across multiple levels of regulation.
Multimodal sequencing has the capacity to move the field from descriptive “snapshots” toward a mechanistic understanding of gene regulatory networks and, importantly, to refine sources of cellular heterogeneity as already applied to the immune system (Hao et al. 2021). The use of multimodal single cell omics is therefore revolutionizing our understanding of cellular biology; however, relying on the dissociation of cells from their natural tissue environment limits our ability to understand the role of intrinsic and extrinsic factors that underpin cellular communication and organ function. Indeed, today in clinical settings, histopathology is a standard diagnostic tool as many diseases are defined by abnormal cellular organization. Additionally, many scientific discoveries rise from the understanding that cellular organization in tissues is highly connected to biological function. Thus, combining single cell molecular measurements with histology and microscopy assays will be required to ultimately generate biological insights into human health and disease.
Overcoming data set–specific batch effects through data integration of such population-level single cell data has remained a limitation. New computational tools recently developed outside the OCBN address this (Sikkema et al. 2022), allowing for the integration of multimodal health and disease data sets. However, computational tools to integrate high-resolution molecular and spatial information are still being established; there is no clear method that accounts for anatomic differences among individuals. The advantage of these newer integration assays is for the annotation of regional/cell molecular identities within the tissue architecture, unravelling cell-cell communication with a spatial context and clarifying tissue microniches, now referenced as “cell neighborhoods” within tissues (Nitzan et al. 2019).
Understanding the cellular context, including extracellular components and signaling molecules that contribute to organ homeostasis, will help to further identify the functions of specific cell types and interactions as well as provide mechanistic insights into fundamental biological processes in health and disease. Next, it will be necessary to integrate human multiomics data with common model organisms as well as patient-derived experimental disease models during the progression from health to disease. The interrogation of these in parallel will be essential for functional assays for data validation and the development of new testable hypotheses, ultimately accelerating targeted follow-up studies to enter the clinical research space.
Clinically Relevant Innovation, Discovery, and Collaboration
For the construction of future oral and craniofacial atlases in disease, it remains paramount to assess the functions, gene expression, and intercellular interactions of all resident cells in healthy tissue as a reference. This level of annotation will allow for a clearer understanding of how these processes are disrupted in disease states. For instance, small subsets of cells are important in the pathogenesis of a variety of complex diseases (Regev et al. 2017), and studying the breakdown of immune mechanisms and dysregulated proinflammatory pathways on a cell-by-cell basis presents an opportunity to understand how perturbed molecular pathways and processes can lead to disease. Understanding these processes may identify molecular mechanisms that lead to improved targetedtherapies—for instance, in human craniofacial birth defects such as orofacial clefting or craniosynostosis. This is foundational knowledge that we intend to be publicly available to advance oral health across the globe.
The clinical significance of single cell approaches has been successfully demonstrated in various human diseases by allowing the identification of disease-associated cell phenotypes—for example, malignant tumor cells within a tumor’s mass (Tirosh et al. 2016) or the identification of immune cells that can predict clinical outcomes and enhance treatment strategies (Sade-Feldman et al. 2018). In the oral and craniofacial region, there are now glimpses of what is possible. For example, stromal cells promoting neutrophil migration in health that expand in disease have been identified (Williams et al. 2021), and an IgG plasma B cell response was identified as a hallmark of periodontitis (Caetano et al. 2021; Williams et al. 2021). The impact of single cell approaches in understanding human oral disease was further demonstrated during the COVID-19 pandemic, when the oral cavity was proved to be an important site for infection with saliva as a potential route of transmission (Huang et al. 2021). This is a minute sampling of the ever-growing clinical advances made possible through single cell approaches.
Many oral conditions will benefit from the molecular characterization of cellular subpopulations. It will provide valuable insights into factors that affect disease progression of head and neck tumors, such as oral carcinoma; potentially malignant disorders of the oral cavity, such as proliferative verrucous leukoplakia (Thompson et al. 2021); oral mucosal diseases, such as lichen planus and vesiculobullous diseases; salivary gland disorders, such as Sjögren’s syndrome; odontogenic and bony pathologies; and trisomy 21. Deepening our understanding of molecular characterization of cellular subpopulations will uncover the processes involved in oral manifestations of systemic disease, especially gastroenterology diseases such as Crohn’s disease, rheumatologic conditions such as lupus and Sjögren’s disease, and systemic hematologic diseases including white and red cell dyscrasias, sickle cell anaemia, or leukemia. To succeed in achieving these aims, the network actively seeks to engage all stakeholders, including patients, oral physicians, researchers, cell biologists, and data scientists, all of whom share in the collective vision of developing our understanding of oral health and disease—all while adhering to our values of transparency and open science (Regev et al. 2018).
Discussion
A Phased Strategy for the OCBN
Organizing such a large-scale project is dependent on a multidisciplinary approach relying on the close collaboration between oral surgeons and oral health care providers (to devise quality metrics to obtain and collect clinical samples), with wet laboratory scientists and bioinformaticians (for sample processing, data processing, and analysis). From the clinician’s view, it is crucial to carefully consider 1) sample collection criteria checkpoints, such as medical history screening; 2) the recording of the precise anatomic location of each sample; and 3) review and collection of associated donor and sample metadata, including health and disease states. Furthermore, this sort of collaboration will accelerate a shift from description to knowledge and the solving of complex clinical questions.
While work from the OCBN has just begun to reveal the diverse tissues and fluids of the oral and craniofacial complex (phase 1), we are already planning to conduct studies that illuminate how these niches harmoniously integrate into the vital functions of communication, defense, breathing, and digestion (phase 2). The OCBN is committed to establishing an initial version of the oral and craniofacial atlas within 2 y with phase 1 of all other HCA bionetwork atlases. As such, the OCBN is currently integrating the existing OCBN data sets (Table) with new unpublished data using harmonized nomenclature and annotation of cell types.
By taking an agnostic approach to tissue and organ physical location (e.g., not oral but airway; not skin but stratified squamous epithelia) should allow for shared discoveries to accelerate human health clinical benefit. For example, while the oral cavity is an important ecosystem, it is also a crossroads that is affected by the condition of other, even distant, body sites. This is not a surprise; these tissues are intimately connected to the nervous, immune, cardiovascular, and endocrine systems (Tirosh et al. 2016). In a bidirectional manner, the condition of the oral cavity can affect distant sites as well (Beck et al. 2019). Phase 1 (oral and craniofacial atlas) and phase 2 (integration of oral and other tissue atlases) of the OCBN will complement other consortia and other model organism databases, such as FaceBase (Samuels et al. 2020). Furthermore, although the OCBN occupies a unique gap within current multiomic initiatives (Fig. 1A), partnership with these other groups outside the HCA, within the HCA, and within the OCBN (Fig. 1B) is essential to the successful integration of the healthy oral and craniofacial data and will facilitate clinical applications.
Progress toward comprehensive mapping of oral and craniofacial tissues requires not only careful experimental design to robustly capture variation within and across individuals but, importantly, a physiologic insight to interpret data and curate data sets. To this end, it is essential to increase dialogue among biologists, computational data scientists, clinicians, pathologists, and statisticians to achieve a consensus on data curation and cell annotation and to deliver data analysis platforms that are relevant and user-friendly (Fig. 4). Moreover, public engagement involving different communities and research participants will be essential to articulate the motivations of the project and to raise awareness of its ambitions and research priorities. In addition, public data portals and biorepositories that enable users to easily access and analyze HCA data are essential to the HCA goal of inclusivity, integrity, and data sharing (https://data.humancellatlas.org; https://oral.cellatlas.io; https://www.covid19cellatlas.org/byrd20/).
Figure 4.
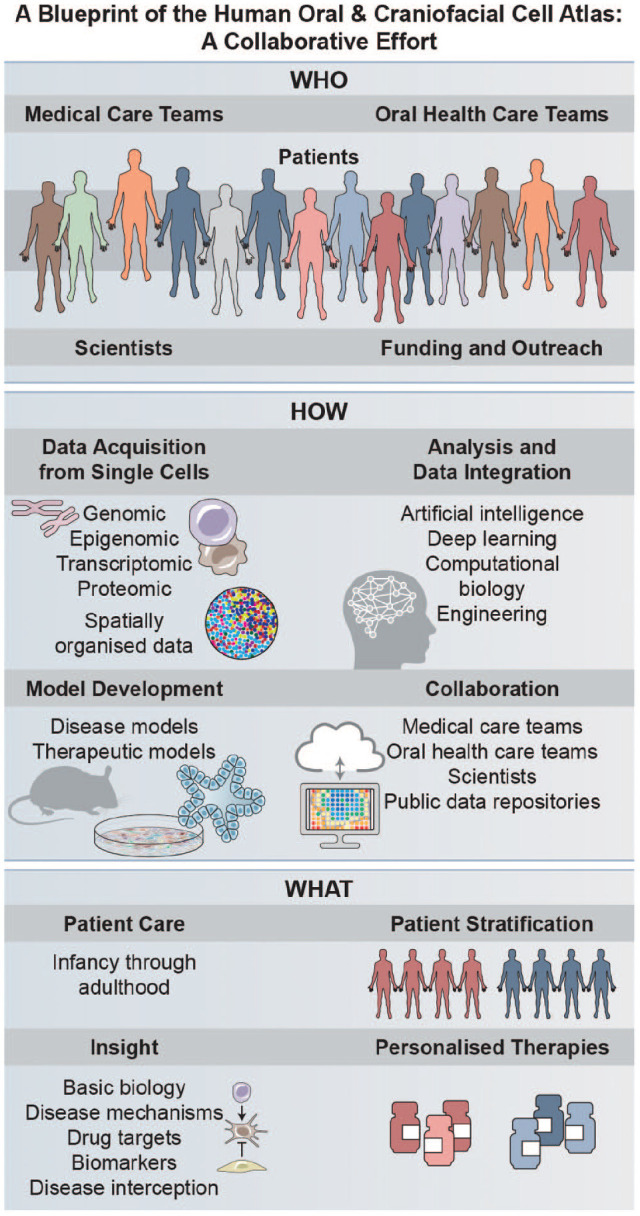
The vision for a globally representative Human Oral and Craniofacial Cell Atlas. To accomplish the goals of the OCBN, we will require the development of inter- and intrainstitute partnerships to connect oral health care teams (namely but not exclusively: oral pathology, oral surgeons and periodontists, endodontics, oral medicine, oral radiology, and general dentists) with medical care teams (namely but not exclusively: ear/nose/throat, dermatology, gastroenterology, pulmonology, allergists, endocrinologists, pediatricians, neurologists, pathologists, clinical genetics, oncologists, radiologists as well as internal and emergency medicine providers). This collaborative approach will allow us to lead the burgeoning field of integrated precision oral health through the application of innovative investigative approaches (single cell, spatially mapped genomics, epigenomics, transcriptomics, and proteomics as well as other “omics” approaches such as metabolomics and microbiomics). Furthermore, defining oral and craniofacial tissue mucosal niches across the life span and building on those reference data will provide unparalleled opportunities to interrogate the mechanisms of common and rare oral and craniofacial diseases across the life span. We will be working collaboratively with other research centers, investigator networks, and related consortia to make data and samples available to the broader research community to address common and rare health concerns globally; to develop scalable representative disease and therapeutic models; and to further integrate, analyze, and discover actionable targets through artificial intelligence/machine learning technologies. In total, our network should have a positive impact on precision medicine for all patients. Credit: Heather McDonald, BioSerendipity, LLC.
Finally, the Human Oral and Craniofacial Cell Atlas encourages and supports the participation of scientists and clinicians from countries around the globe by recognizing the need to integrate different ethnicities, environments, and regional diseases, and we invite any interested stakeholder to join the network, participate in our meetings, and contribute data for the integrated atlas (see contact details in Conclusions). We are aware of potential challenges and limitations, including available resources and tissue sampling, but we are committed to shared protocols (Greenwell-Wild et al. 2021), open and immediate data release, and prioritizing international collaborative work. Collective development of research ideas and equitable partnerships guided under the HCA Ethics Working Group is our priority. To enable the advancement of such large-scale projects, we will aim to implement ethically responsible, socially robust, and legally compliant research from the beginning. Continuous monitoring in the form of yearly consortium meetings should be tasked with the analysis of emerging ethical and societal aspects from the use of these new technologies. These meetings will also attempt to identify weaknesses or areas of inadequate progress and take actions to flag areas of expertise that are missing. The task forces formed during the initial meetings should meet regularly to apply concrete changes for patient groups, researchers, or data collectors.
Conclusions
In sum, there is an enormous opportunity for integrated and precise oral health care initiatives that leverage the accessibility of this space to improve oral and systemic health. Profiling of these conditions from a patient-centered perspective will increase our understanding of disease cellular origin, mechanisms, etiology, and diagnostics—rendering them experimentally tractable to test new hypotheses for better diagnosis and drug discovery. To achieve this grand vision requires the collaboration of a multidisciplinary international team—spanning the basic and computational sciences to clinical practice (Fig. 4). This team science approach will be key to achieving inclusive, ancestrally diverse, open access, multiomic reference atlases of the human oral and craniofacial tissues and fluids across the life span. This open data resource will provide quantitative, multiscale information sufficient to build integrated prediction models of key oral and craniofacial cell and tissue states to develop breakthroughs in oral health for all. If you want to join the network, know more about our work, and collaborate with the network either by facilitating access to samples or by contributing with data sets for the OCBN atlas, please contact oral@humancellatlas.org.
Author Contributions
A.J. Caetano, contributed to data analysis, drafted and critically revised the manuscript; I. Sequeira, K.M. Byrd, contributed to conception, design and data analysis, drafted and critically revised the manuscript; P.T. Sharpe, A. Kimple, M.A. Shazib, A. Opasawatchai, B.M. Warner, J. Krivanek, K. Kretzschmar, L.K. McKay, M. Freire, O. Klein, P.R. Tata, S. Pringle, S. Teichmann, D.W. Williams, I. Miller Zmora, Q. Easter, W.J. Lu, P. Perez, T. Pranzatelli, S. Lwin, R. Boucher, M. Bush, C.D. Conde, M. Haniffa, J.S. Hagood, A.A. Volponi, V. Yianni, J. Macken, M. Efremova, E. Todres, B. Matuck, F. Fortune, A.O. Pisco, D. Boffelli, Y. Kapila, F. Momen-Heravi, A. Gulati, N. Moutsopoulos, P. Beachy, S. Wallet, T. Biddinger, D. Pereira, R. Kumar, contributed to data analysis, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Acknowledgments
We thank Nancy R. Gough, BioSerendipity, LLC, for assistance with Figures 2 and 4. We also acknowledge the Human Cell Atlas Oral and Craniofacial Bionetwork for comments on the manuscript. We are especially thankful for the generous support from the Human Cell Atlas, especially from Sarah Teichmann and her team. This publication is part of the Human Cell Atlas: http://www.humancellatlas.org/publications.
Due to space limitations, we apologize for being unable to cite all the relevant primary literature that has contributed to this roadmap
Footnotes
Declaration of Conflicting Interests: The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Although the authors view each of these as noncompeting financial interests, K.M. Byrd, A.J. Caetano, and I. Sequeira are active members of the Human Cell Atlas; furthermore, in the last year, K.M. Byrd has been a scientific advisor at Arcato Laboratories, and I. Sequeira is a consultant for L’Oréal Research and Innovation.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: For this work, A.J. Caetano was funded by the National Institute for Health Research’s Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the National Institute for Health Research, or the Department of Health. Work in the laboratories of N. Moutsopoulos and B.M. Warner is supported by the Intramural Research Program of the National Institute of Dental and Craniofacial Research (National Institutes of Health). K.K. is funded by the German Cancer Aid (via MSNZ Würzburg), the European Union (ERC Starting Grant agreement 101042738/OralNiche), the Interdisciplinary Centre for Clinical Research at the Medical Faculty of the University of Würzburg (IZKF project B-435), the Single Cell Center Würzburg, and the Foundation Tumour Research Head-Neck. I. Sequeira was funded by a Barts Charity Lectureship (MGU045), the Royal Society (RGS\R2\202291), and the CZI Pediatric Networks for Human Cell Atlas. This study is supported by the Medical College of Saint Bartholomew’s Hospital Trust, an independent registered charity that promotes and advances medical and dental education and research at Barts and The London School of Medicine and Dentistry (I. Sequeira and J. Macken) and by Fundação para a Ciência e a Tecnologia (2020.08715.BD, I. Sequeira and D. Pereira). K.M. Byrd was funded by Volpe Researcher Scholar start-up funds (ADASRI) and the CZI Pediatric Networks for Human Cell Atlas.
Contributing Authors: Human Cell Atlas Oral and Craniofacial Bionetwork: A. Caetano, P. Sharpe, A.A. Volponi, V. Yianni (Faculty of Dentistry, Oral and Craniofacial Sciences, Centre for Craniofacial and Regenerative Biology, King’s College London, UK); M. Bush, L.K. McKay, S. Wallet (Division of Oral and Craniofacial Health Sciences, Adams School of Dentistry, University of North Carolina, Chapel Hill, NC, USA); M.A. Shazib (High Point University School of Dental Medicine and Oral Health, High Point, NC, USA); A. Kimple (Department of Otolaryngology–Head and Neck Surgery, School of Medicine, University of North Carolina, Chapel Hill, NC, USA); K.M. Byrd, Q. Easter, T. Weaver (Lab of Oral and Craniofacial Innovation, ADA Science and Research Institute, Gaithersburg, MD, USA); I. Sequeira, R. Kumar, D. Pereira, J. Macken, F. Fortune (Institute of Dentistry, Barts Centre for Squamous Cancer, Barts and The London School of Medicine and Dentistry, Queen Mary University London, London, UK); M. Efremova (Barts Cancer Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University London, London, UK); S. Teichmann (Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, UK; Cavendish Laboratory/Department of Physics, University of Cambridge, Cambridge, UK); K. Kretzschmar (Mildred Scheel Early Career Centre for Cancer Research Würzburg, University Hospital Würzburg, Würzburg, Germany); E. Todres (Broad Institute of MIT and Harvard, Cambridge, MA, USA); D. Boffelli (Department of Pediatrics, University of California, San Francisco, CA, USA; Institute for Human Genetics, University of California, San Francisco, CA, USA); James S. Hagood (Program for Rare and Interstitial Lung Disease, University of North Carolina, Chapel Hill, NC, USA); O. Klein (Department of Orofacial Sciences and Program in Craniofacial Biology, Department of Pediatrics and Institute for Human Genetics, University of California, San Francisco, CA, USA; Cedars-Sinai Guerin Children’s, Los Angeles, CA, USA); R. Boucher (Marsico Lung Institute/UNC Cystic Fibrosis Center, University of North Carolina, Chapel Hill, NC, USA); S. Lwin (St John’s Institute of Dermatology, Guy’s Hospital, King’s College London, London, UK); T. Pranzatelli (AAV Biology Lab, National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD, USA; Biological Sciences Program, University of Maryland, College Park, MD, USA); P. Beachy, W.J. Lu (Institute for Stem Cell Biology and Regenerative Medicine, School of Medicine, Stanford University, Stanford, CA, USA); I. Miller Zmora (Program in Craniofacial Biology and Department of Orofacial Sciences, University of California, San Francisco, CA, USA); N. Moutsopoulos, D.W. Williams (Oral Immunity and Inflammation Section, National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD, USA); S. Pringle (Department of Rheumatology and Clinical Immunology, University Medical Center Groningen, Groningen, the Netherlands); J. Krivanek (Department of Histology and Embryology, Faculty of Medicine, Masaryk University, Brno, Czech Republic); B.M. Warner, P. Perez (Salivary Disorders Unit, Sjögren’s Syndrome Clinic, National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD, USA); A. Opasawatchai (Department of Oral Microbiology, Faculty of Dentistry, Mahidol University, Bangkok, Thailand; Integrative Computational Bioscience Center, Mahidol University, Nakorn Pathom, Thailand); B. Matuck (Department of Pathology, School of Medicine, University of Sao Paulo, Sao Paulo, Brazil); M. Freire (Department of Genomic Medicine and Infectious Disease and Department of Infectious Disease, J. Craig Venter Institute, La Jolla, CA, USA); P.R. Tata (Department of Cell Biology, Duke Cancer Institute, School of Medicine, Duke University, Durham, NC, USA; Regeneration Next, Duke University, Durham, NC, USA); C.D. Conde (Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, UK); M. Haniffa (Biosciences Institute, Newcastle University, Newcastle upon Tyne, UK; Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, UK; Department of Dermatology and NIHR Newcastle Biomedical Research Centre, Newcastle Hospitals NHS Foundation Trust, Newcastle upon Tyne, UK); A.O. Pisco (Chan Zuckerberg Biohub, San Francisco, CA, USA); F. Momen-Heravi (College of Dental Medicine, Columbia University, New York, New York, USA); Y. Kapila (School of Dentistry, University of California, Los Angeles, CA, USA); A. Gulati (Division of Gastroenterology, Department of Pediatrics, University of North Carolina, Chapel Hill, NC, USA).
ORCID iD: A.J. Caetano  https://orcid.org/0000-0003-4588-3241
https://orcid.org/0000-0003-4588-3241
Contributor Information
Human Cell Atlas Oral and Craniofacial Bionetwork:
A. Caetano, P. Sharpe, A.A. Volponi, V. Yianni, M. Bush, L.K. McKay, S. Wallet, M.A. Shazib, A. Kimple, K.M. Byrd, Q. Easter, T. Weaver, I. Sequeira, R. Kumar, D. Pereira, J. Macken, F. Fortune, M. Efremova, S. Teichmann, K. Kretzschmar, E. Todres, D. Boffelli, James S. Hagood, O. Klein, R. Boucher, S. Lwin, T. Pranzatelli, P. Beachy, W.J. Lu, I. Miller Zmora, N. Moutsopoulos, D.W. Williams, S. Pringle, J. Krivanek, B.M. Warner, P. Perez, A. Opasawatchai, B. Matuck, M. Freire, P.R. Tata, C.D. Conde, M. Haniffa, A.O. Pisco, F. Momen-Heravi, Y. Kapila, and A. Gulati
References
- Aizarani N, Saviano A, Sagar, Mailly L, Durand S, Herman JS, Pessaux P, Baumert TF, Grün D. 2019. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 572(7768):199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge S, Teichmann SA. 2020. Single cell transcriptomics comes of age. Nat Commun. 11(1):4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almanzar N, Antony J, Baghel AS, Bakerman I, Bansal I, Barres BA, Beachy PA, Berdnik D, Bilen B, Brownfield D, et al. 2020. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature. 583(7817):590–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin MJ, Cribbs AP, Guilak F, Snelling SJB. 2021. Mapping the musculoskeletal system one cell at a time. Nat Rev Rheumatol. 17(5):247–248. [DOI] [PubMed] [Google Scholar]
- Beck JD, Papapanou PN, Philips KH, Offenbacher S. 2019. Periodontal medicine: 100 years of progress. J Dent Res. 98(10):1053–1062. [DOI] [PubMed] [Google Scholar]
- Byrd KM, Gulati AS. 2021. The “gum-gut” axis in inflammatory bowel diseases: a hypothesis-driven review of associations and advances. Front Immunol. 12:620124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd KM, Piehl NC, Patel JH, Huh WJ, Sequeira I, Lough KJ, Wagner BL, Marangoni P, Watt FM, Klein OD, et al. 2019. Heterogeneity within stratified epithelial stem cell populations maintains the oral mucosa in response to physiological stress. Cell Stem Cell. 25(6):814–829.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano AJ, Yianni V, Volponi A, Booth V, D’Agostino EM, Sharpe P. 2021. Defining human mesenchymal and epithelial heterogeneity in response to oral inflammatory disease. Elife. 10:e62810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Lin W, Gan J, Lu W, Wang M, Wang X, Yi J, Zhao Z. 2022. Transcriptomic mapping of human parotid gland at single-cell resolution. J Dent Res [epub ahead of print 26 Feb 2022] in press. doi: 10.1177/00220345221076069 [DOI] [PubMed] [Google Scholar]
- Chiba Y, Saito K, Martin D, Boger ET, Rhodes C, Yoshizaki K, Nakamura T, Yamada A, Morell RJ, Yamada Y, et al. 2020. Single-cell RNA-sequencing from mouse incisor reveals dental epithelial cell-type specific genes. Front Cell Dev Biol. 8:841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury SN, Novotny M, Aevermann BD, Lee S, Mandava A, Qian Y, Scheuermann R, Freire M. 2020. A protocol for revealing oral neutrophil heterogeneity by single-cell immune profiling in human saliva. Research Square. doi: 10.21203/rs.3.pex-953/v1 [DOI] [Google Scholar]
- Costa-da-Silva AC, Aure MH, Dodge J, Martin D, Dhamala S, Cho M, Rose JJ, Bassim CW, Ambatipudi K, Hakim FT, et al. 2022. Salivary ZG16B expression loss follows exocrine gland dysfunction related to oral chronic graft-versus-host disease. iScience. 25(1):103592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dries R, Zhu Q, Dong R, Eng CL, Li H, Liu K, Fu Y, Zhao T, Sarkar A, Bao F, et al. 2021. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 22(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M, Coleman P. 1992. Analysis of gene expression in single live neurons. Proc Natl Acad Sci U S A. 89(7):3010–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. 2020. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 15(4):1484–1506. [DOI] [PubMed] [Google Scholar]
- Greenwell-Wild T, Williams DW, Moutsopoulos NM. 2021. Dissociation of human oral mucosal tissue for single-cell applications. STAR Protoc. 2(4):100908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Feng J, Guo T, Loh YE, Yuan Y, Ho TV, Cho CK, Li J, Jing J, Janeckova E, et al. 2021. Runx2-Twist1 interaction coordinates cranial neural crest guidance of soft palate myogenesis. Elife. 10:e62387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Hao S, Andersen-Nissen E, Mauck WM, 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. 2021. Integrated analysis of multimodal single-cell data. Cell. 184(13):3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser BR, Aure MH, Kelly MC. Genomics and Computational Biology Core, Hoffman MP, Chibly AM. 2020. Generation of a single-cell RNAseq atlas of murine salivary gland development. iScience. 23(12):101838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Perez P, Kato T, Mikami Y, Okuda K, Gilmore RC, Conde CD, Gasmi B, Stein S, Beach M, et al. 2021. SARS-CoV-2 infection of the oral cavity and saliva. Nat Med. 27(5):892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Perez P, Kato T, Mikami Y, Okuda K, Gilmore RC, Domínguez Conde C, Gasmi B, Stein S, Beach M, et al. 2020. Integrated single-cell atlases reveal an oral SARS-CoV-2 infection and transmission axis. medRxiv. Preprint. doi: 10.1101/2020.10.26.20219089 [DOI] [Google Scholar]
- Jones KB, Furukawa S, Marangoni P, Ma H, Pinkard H, D’Urso R, Zilionis R, Klein AM, Klein OD. 2019. Quantitative clonal analysis and single-cell transcriptomics reveal division kinetics, hierarchy, and fate of oral epithelial progenitor cells. Cell Stem Cell. 24(1):183–192.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen J, Chen HH, Parikh K, Antanaviciute A, Jagielowicz M, Fawkner-Corbett D, Ashley N, Cubitt L, Mellado-Gomez E, Attar M, et al. 2018. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell. 175(2):372–386.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King HW, Orban N, Riches JC, Clear AJ, Warnes G, Teichmann SA, James LK. 2021. Single-cell analysis of human B cell maturation predicts how antibody class switching shapes selection dynamics. Sci Immunol. 6(56):eabe6291. [DOI] [PubMed] [Google Scholar]
- Krausgruber T, Fortelny N, Fife-Gernedl V, Senekowitsch M, Schuster LC, Lercher A, Nemc A, Schmidl C, Rendeiro AF, Bergthaler A, et al. 2020. Structural cells are key regulators of organ-specific immune responses. Nature. 583(7815):296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivanek J, Soldatov RA, Kastriti ME, Chontorotzea T, Herdina AN, Petersen J, Szarowska B, Landova M, Matejova VK, Holla LI, et al. 2020. Dental cell type atlas reveals stem and differentiated cell types in mouse and human teeth. Nat Commun. 11(1):4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledergor G, Weiner A, Zada M, Wang SY, Cohen YC, Gatt ME, Snir N, Magen H, Koren-Michowitz M, Herzog-Tzarfati K, et al. 2018. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat Med. 24(12):1867–1876. [DOI] [PubMed] [Google Scholar]
- Li H, Jones KL, Hooper JE, Williams T. 2019. The molecular anatomy of mammalian upper lip and primary palate fusion at single cell resolution. Development. 146(12):dev174888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom RGH, Regev A, Teichmann SA. 2021. Towards a human cell atlas: taking notes from the past. Trends Genet. 37(7):625–630. [DOI] [PubMed] [Google Scholar]
- Litvinukova M, Talavera-Lopez C, Maatz H, Reichart D, Worth CL, Lindberg EL, Kanda M, Polanski K, Heinig M, Lee M, et al. 2020. Cells of the adult human heart. Nature. 588(7838):466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JC, Chang C, Boschetti G, Ungaro R, Giri M, Grout JA, Gettler K, Chuang LS, Nayar S, Greenstein AJ, et al. 2019. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 178(6):1493–1508.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MM, Khoo WH, Ng PY, Xiao Y, Zamerli J, Thatcher P, Kyaw W, Pathmanandavel K, Grootveld AK, Moran I, et al. 2021. Osteoclasts recycle via osteomorphs during RANKL-stimulated bone resorption. Cell. 184(7):1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, et al. 2018. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 560(7718):319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata M, Chu AKY, Ono N, Welch JD, Ono W. 2021. Single-cell transcriptomic analysis reveals developmental relationships and specific markers of mouse periodontium cellular subsets. Front Dent Med. 2:679937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanci A. 2018. Ten Cate’s oral histology: development, structure, and function. 9th ed. St. Louis (MO): Elsevier. [Google Scholar]
- Nitzan M, Karaiskos N, Friedman N, Rajewsky N. 2019. Gene expression cartography. Nature. 576(7785):132–137. [DOI] [PubMed] [Google Scholar]
- Opasawatchai A, Nguantad S, Sriwilai B, Matangkasombut P, Matangkasombut O, Srisatjaluk R, Charoensawan V. 2022. Single-cell transcriptomic profiling of human dental pulp in sound and carious teeth: a pilot study. Front Dent Med [epub ahead of print 5 Jan 2022] in press. doi: 10.3389/fdmed.2021.806294 [DOI] [Google Scholar]
- Osumi-Sutherland D, Xu C, Keays M, Levine AP, Kharchenko PV, Regev A, Lein E, Teichmann SA. 2021. Cell type ontologies of the human cell atlas. Nat Cell Biol. 23(11):1129–1135. [DOI] [PubMed] [Google Scholar]
- Pagella P, de Vargas Roditi L, Stadlinger B, Moor AE, Mitsiadis TA. 2021. A single-cell atlas of human teeth. iScience. 24(5):102405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, Li M, Barasch J, Susztak K. 2018. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 360(6390):758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky N, Almouzni G, Gorski SA, Aerts S, Amit I, Bertero MG, Bock C, Bredenoord AL, Cavalli G, Chiocca S, et al. 2020. Lifetime and improving European healthcare through cell-based interceptive medicine. Nature. 587(7834):377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky N, Almouzni G, Gorski SA, Aerts S, Amit I, Bertero MG, Bock C, Bredenoord AL, Cavalli G, Chiocca S, et al. 2021. Publisher correction: Lifetime and improving European healthcare through cell-based interceptive medicine. Nature. 592(7852):E8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, et al. 2017. The Human Cell Atlas. Elife. 6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A, Teichmann SA, Rozenblatt-Rosen O, Stubbington MJT, Ardlie KG, Amit I, Arlotta P, Bader GD, Benoist C, Biton M, et al. 2018. The human cell atlas white paper. arXiv. Preprint. doi: 10.48550/arXiv.1810.05192 [DOI] [Google Scholar]
- Reynolds G, Vegh P, Fletcher J, Poyner EFM, Stephenson E, Goh I, Botting RA, Huang N, Olabi B, Dubois A, et al. 2021. Developmental cell programs are co-opted in inflammatory skin disease. Science. 371(6527): eaba6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, et al. 2018. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 175(4):998–1013.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels BD, Aho R, Brinkley JF, Bugacov A, Feingold E, Fisher S, Gonzalez-Reiche AS, Hacia JG, Hallgrimsson B, Hansen K, et al. 2020. FaceBase 3: analytical tools and FAIR resources for craniofacial and dental research. Development. 147(18):dev191213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schifter M, Yeoh SC, Coleman H, Georgiou A. 2010. Oral mucosal diseases: the inflammatory dermatoses. Aust Dent J. 55 Suppl 1:23–38. [DOI] [PubMed] [Google Scholar]
- Sekiguchi R, Martin D. Genomics and Computational Biology Core, Yamada KM. 2020. Single-cell RNA-seq identifies cell diversity in embryonic salivary glands. J Dent Res. 99(1):69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharir A, Marangoni P, Zilionis R, Wan M, Wald T, Hu JK, Kawaguchi K, Castillo-Azofeifa D, Epstein L, Harrington K, et al. 2019. A large pool of actively cycling progenitors orchestrates self-renewal and injury repair of an ectodermal appendage. Nat Cell Biol. 21(9):1102–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikkema L, Strobl D, Zappia L, Madissoon E, Markov N, Zaragosi L, Ansari M, Arguel M, Apperloo L, Bécavin C, et al. 2022. An integrated cell atlas of the human lung in health and disease. bioRxiv. Preprint. doi: 10.1101/2022.03.10.483747 [DOI] [Google Scholar]
- Song EC, Min S, Oyelakin A, Smalley K, Bard JE, Liao L, Xu J, Romano RA. 2018. Genetic and scRNA-seq analysis reveals distinct cell populations that contribute to salivary gland development and maintenance. Sci Rep. 8(1):14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabula Muris Consortium, Overall Coordination, Logistical Coordination, Organ Collection and Processing, Library Preparation and Sequencing, Computational Data Analysis, Cell Type Annotation, Writing Group, Supplemental Text Writing Group, Principal Investigators. 2018. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 562(7727):367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabula Sapiens Consortium, Jones RC, Karkanias J, Krasnow MA, Pisco AO, Quake SR, Salzman J, Yosef N, Bulthaup B, Brown P, et al. 2022. The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science. 376(6594):eabl4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Nagata M, Gupta A, Matsushita Y, Yamaguchi T, Mizuhashi K, Maki K, Ruellas AC, Cevidanes LS, Kronenberg HM, et al. 2019. Autocrine regulation of mesenchymal progenitor cell fates orchestrates tooth eruption. Proc Natl Acad Sci U S A. 116(2):575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LDR, Fitzpatrick SG, Müller S, Eisenberg E, Upadhyaya JD, Lingen MW, Vigneswaran N, Woo SB, Bhattacharyya I, Bilodeau EA, et al. 2021. Proliferative verrucous leukoplakia: an expert consensus guideline for standardized assessment and reporting. Head Neck Pathol. 15(2):572–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH, 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. 2016. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 352(6282):189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, Brouwer S, Gomes T, Hesse L, Jiang J, et al. 2019. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med. 25(7):1153–1163. [DOI] [PubMed] [Google Scholar]
- Williams DW, Greenwell-Wild T, Brenchley L, Dutzan N, Overmiller A, Sawaya AP, Webb S, Martin D, NIDCD/NIDCR Genomics and Computational Biology Core, Hajishengallis G, et al. 2021. Human oral mucosa cell atlas reveals a stromal-neutrophil axis regulating tissue immunity. Cell. 184(15):4090–4104.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sharpe PT. 2021. Telocytes regulate macrophages in periodontal disease. bioRxiv. Preprint. doi: 10.1101/2021.06.03.446871 [DOI] [PMC free article] [PubMed] [Google Scholar]