Abstract
Disrupted development of the gut microbiota is a contributing cause of childhood malnutrition. Bifidobacterium longum subspecies infantis (B. infantis) is a prominent early colonizer of the infant gut that utilizes human milk oligosaccharides (HMO). We found that the absolute abundance of B. infantis is lower in 3-to-24-month-old Bangladeshi infants with severe acute malnutrition (SAM) compared to their healthy age-matched counterparts. A single-blind, placebo-controlled trial (SYNERGIE) was conducted in 2 to 6 month-old Bangladeshi infants with SAM. A commercial USA donor-derived B. infantis strain (EVC001) was administered daily with or without the HMO lacto-N-neotetraose for 28 days. This intervention increased fecal B. infantis abundance in infants with SAM, although the increase was 10-100-fold lower than the abundance in healthy controls. EVC001 treatment also promoted weight gain and reduced markers of intestinal inflammation in infants with SAM. We subsequently cultured fecal B. infantis strains from Bangladeshi infants and colonized gnotobiotic mice with these cultured strains. The gnotobiotic mice were fed a weaning diet representative of that consumed by 6-month-old Bangladeshi infants, with or without HMO supplementation. One B. infantis strain, Bg_2D9, that expressed two gene clusters involved in uptake and utilization of N-glycans and plant-derived polysaccharides exhibited superior fitness over EVC001. The fitness advantage of Bg_2D9 over EVC001 was confirmed in a gnotobiotic mouse model of mother-to-infant gut microbiota transmission where dams received a pre-treatment fecal community from a SAM infant in the SYNERGIE trial. Whether Bg_2D9 is superior to EVC001 for treating malnourished infants who consume diets with limited human breastmilk content requires further clinical testing.
Editor’s Summary
Bifidobacterium infantis, a gut bacterium uniquely adapted to metabolizing breastmilk carbohydrates, is deficient in Bangladeshi infants with severe acute malnutrition (SAM). A USA-derived B. infantis strain improved weight gain and reduced intestinal inflammation in infants with SAM. A strain cultured from a healthy Bangladeshi child with expanded capacity to metabolize both plant and milk carbohydrates achieved greater colonization and augmented weight gain in gnotobiotic mice colonized with a fecal microbial community from an infant with SAM. This probiotic strain may help to treat acutely malnourished infants, expecially those who have a breastmilk-poor diet.
One sentence summary
Restoring B. infantis to the gut of malnourished infants boosts weight gain and reduces inflammatory markers.
INTRODUCTION
The global burden of childhood undernutrition is great, causing 3.1 million deaths annually and accounting for 21% of life-years lost among children younger than 5 years (1). Over 18 million children in this age range are affected by severe acute malnutrition (SAM), the most extreme form of undernutrition. SAM is responsible for nearly half of all undernutrition-related mortality; it is postulated to be caused by a number of factors, including, for example, very low dietary energy intake and nutrient loss due to infection (2). SAM is manifest by markedly impaired ponderal growth (wasting) and is defined clinically by a mid-upper arm circumference (MUAC) of less than 11.5 cm and/or a weight-for-length (height) score that is greater than 3 standard deviations below the mean value for a multi-national World Health Organization cohort of infants and children (WLZ <−3). SAM is associated with an enteropathy manifest as disruption of epithelial tight junctions and increased gut permeability, leading to intestinal and systemic inflammation (3). Notably, four million children under 6-months-of-age are affected by SAM; they are particularly vulnerable, with increased immediate risk of infection and death, as well as long-term impairment of growth and cognitive development (4,5). Current management approaches include treatment with broad spectrum antibiotics and nutritional supplementation with energy-dense therapeutic foods (6,7). Whereas these approaches reduce mortality rates (8), many children emerge with persistent moderate acute malnutrition (MAM) and remain susceptible to relapse to SAM.
The current study explores the role of bifidobacteria, notably Bifidobacterium longum subspecies infantis (B. infantis), in the pathogenesis of SAM and as probiotic treatment. Recent work has shown that acute malnutrition in infants and children is associated with impaired development of the gut microbiota (9,10). Preclinical and clinical studies indicate that this disrupted co-development of the gut microbiota and host is a contributing cause, not simply an effect, of acute malnutrition (11,12). The gut microbiota of healthy breastfed infants in the first few months of life is dominated by members of the genus Bifidobacterium (13) including B. breve, B. bifidum, B. longum subspecies longum (B. longum), and B. infantis. Among these bifidobacteria, B. infantis is uniquely adapted to the gut of the breastfed infant as it possesses five distinct gene clusters (commonly referred to as H1-H5), including a 43-kb gene cluster (H1) that encodes a variety of glycoside hydrolases and oligosaccharide transporters, that enable this subspecies to metabolize the full repertoire of the several hundred known human milk oligosaccharides (HMOs) (14-16). Observational studies of Bangladeshi infants have documented a positive association between the abundance of bifidobacteria in feces in the first 4 months of life and responses to oral and parenteral vaccines (17). Clinical studies of exclusively breastfed U.S. infants who received a probiotic strain of B. infantis (EVC001) revealed that colonization was associated with reductions in gut inflammation markers (18). Similar beneficial effects of B. infantis have been observed in preterm infants who are at elevated risk for nosocomial infections (19) and in experimental models of necrotizing enterocolitis (20).
A meta-analysis of HMOs in mothers representing different geographies, socio-economic features and stages of lactation identified LNT (lacto-N-tetraose) and LNnT (lacto-N-neotetraose) as among the most abundant structures present, accounting for up to 20% of total HMOs (21). More complex HMOs based on the LNT and LNnT core can comprise up to 70% of total HMOs and over 90% in “non-secretor” mothers who lack a functional fucosyltransferase 2 (FUT2) gene (22,23). B. infantis strains characterized to date can grow in vitro with HMOs as a sole carbon source; this phenotype is believed to involve internalization of the HMO by a specific ATP-binding cassette (ABC) transporter and its subsequent catabolism by intracellular glycoside hydrolases to monosaccharides. With the exception of fucose, which is catabolized to pyruvate and 1,2-propanediol via the intermediate fuconolactone (24), monosaccharides from HMOs are substrates for the “bifid shunt” (fructose-6-phosphate phosphoketolase pathway), which produces ATP, acetate and lactate as end-products (25). Despite the broad conservation of the H1 gene cluster among B. infantis strains, strain-level variation exists for genes in the H5 cluster encoding the ABC transporter and associated solute binding protein (Blon_2175-2177) involved in the uptake of LNT (15,26). Therefore, a reduced capacity to utilize LNT, and its derivatives, poses a competitive disadvantage to such B. infantis strains (27). At the same time, in many low-income settings, animal milk, gruels and complementary foods are often introduced into the diet at a very early age, creating a nutrient landscape that may not be optimal for B. infantis strains.
A general challenge in the field of human microbiome research is to identify alterations in microbial community composition and expressed functions, establish causal relationships between these configurational changes and disease pathogenesis, and then develop strategies for effectively repairing community function to restore health. If restoration involves administration of microbes, then decisions have to be made about which strains to select. The fitness of strains in the gut ecosystems of individuals will reflect a given strain’s capacity to adapt to host habitat and environmental features; this may require searches for strains that have evolved in the microbial communities of the populations of individuals where treatment is being directed. The current study identifies B. infantis deficiency as a feature of the fecal microbiota of children with SAM compared to their age-matched healthy counterparts and offers an approach for identifying B. infantis strains that may be better suited for repairing the perturbed gut microbiota of infants presenting with SAM, at a time when their diets are rich in glycans derived from sources other than breastmilk.
RESULTS
B. infantis in the gut microbiota of Bangladeshi infants with SAM
B. infantis abundance was measured using a qPCR assay directed to the nanH2/ exo-α-sialidase gene (Blon_2348) in the H1 locus that is uniquely present in this subspecies (29) (Fig. 1). This qPCR assay was applied to DNA isolated from fecal samples collected from 3-24-month-old Bangladeshi children with SAM (n=102) and age-matched Bangladeshi children without a wasting disorder (WLZ ≥-2) (n=49) (see table S1A for clinical characteristics). All children lived in Dhaka, Bangladesh, with many residing in Mirpur, an urban slum district within the city. Primers used to measure total bifidobacterial load were targeted to the 16S rRNA gene (28). The specificity of targeting for both sets of primers was confirmed using a reference collection of cultured gut bacterial strains with sequenced genomes (table S1B, C).
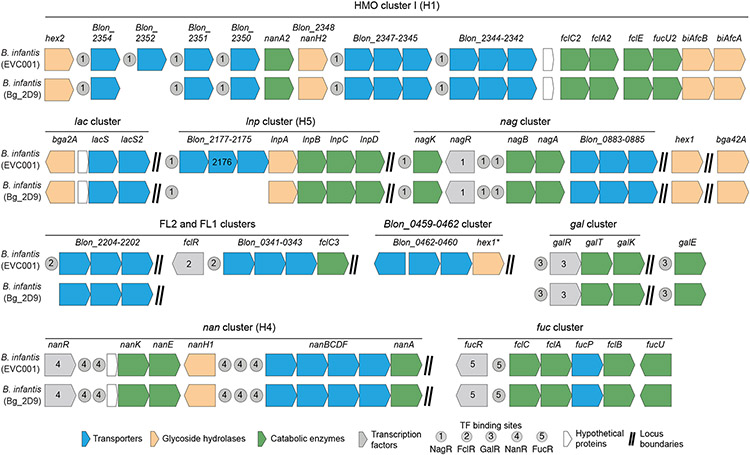
Fig. 1. HMO utilization genes in B. infantis EVC001 and Bg_2D9 strains.
The H1-H5 gene clusters encode proteins that transport and metabolize HMOs and are characteristic features of B. longum subspecies infantis (15). EVC001 is a USA donor-derived probiotic B. infantis strain. Bg_2D9 was isolated from a 12-month-old healthy Bangladeshi child. Shown is the reconstruction of the H1-H5 clusters in these two strains (also see table S3). Abbreviations: Blon, B. longum locus tag; FL1/FL2, fucosyllactose 1 and fucosyllactose 2; Nan, N-acetylneuraminic acid.
In a cross-sectional study of healthy breastfed Bangladeshi infants and children, maximal amounts of B. infantis were documented in feces at the end of the first postnatal month, with no statistically significant diminution in B. infantis absolute abundance through 12 months of age (blue points, Fig. 2A). During this period, ~75% of all bifidobacterial strains detected in fecal samples were B. infantis (Blon_2348-positive), and then its abundance declined progressively by >4 orders of magnitude between 12 and 24 months of age (Fig. 2A; table S1A). The overall abundance of members of the genus Bifidobacterium remained high throughout the 24-month period (Fig. 2B; table S1A), reflecting an increase in other bifidobacterial species (table S1D).
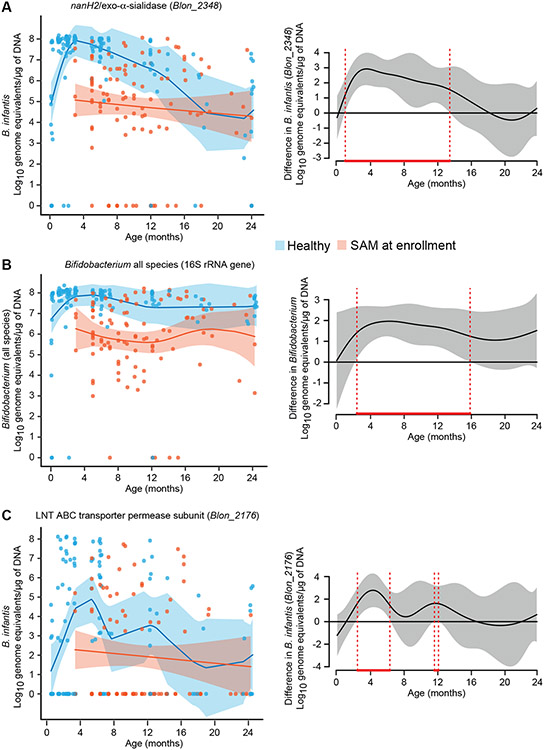
Fig. 2. B. infantis abundance in fecal samples from healthy Bangladeshi infants and those with SAM.
(A-C) qPCR assay primers were directed against the NanH2 exo-α-sialidase (Blon_2348) gene of B. infantis (A), the 16S rRNA gene of all bifidobacterial species (B), and the LNT ABC transporter permease subunit (Blon_2176) gene of B. infantis (C). Fecal biospecimens were assayed from healthy children (n=130 samples) and children with SAM (n=102 samples). Scatterplots (left panels) display the absolute abundance of target genes as normalized to log10 transformed genome equivalents per μg of fecal DNA as a function of age at the time of specimen collection. Samples from healthy infants and children are indicated by blue dots/shading, whereas those from individuals with SAM are denoted in red. A generalized additive model-derived best fit line (±2 SEM) is shown. Plot difference curves (right panels) depict statistically significant differences in the best fit lines (areas bounded by red dashed vertical lines), between the two models (healthy vs SAM) (see also table S1).
The nanH2-based qPCR assay showed that fecal amounts of B. infantis were on average 2-3 orders of magnitude lower in 3 to 13-month-old Bangladeshi children with SAM (Fig. 2A red points) compared to their healthy age-matched counterparts (P<0.05, generalized additive model; Fig. 2A, red bounded region in right panel). No significant differences were evident between the ages of 14-24 months (P>0.05; generalized additive model). These findings were consistent with our observation that Bangladeshi infants with SAM on average received less than 10% of the recommended daily volume of breast milk (table S2C). Total bifidobacterial abundances were also significantly lower in fecal samples from 3 to 16 month-old children with SAM compared to their age-matched healthy counterparts (P<0.05, generalized additive model; Fig. 2B). Notably, compared to healthy infants, the fecal microbial communities of infants with SAM were dominated by pathobionts including Escherichia, Shigella, Klebsiella and Streptococcus species (fig. S1A, B, table S1D).
Blon_2176, part of the H5 gene cluster, encodes the permease component of the ABC transporter for a prominent lacto-N-biose type I tetrasaccharide in breast milk. A qPCR assay using primers directed against Blon_2176 revealed that the mean absolute abundance of this gene was >1000-fold lower in both healthy infants and infants with SAM compared to the abundance of the B. infantis Blon_2348 gene (Fig. 2A, C). Moreover, there was a significant difference in the percentage of fecal samples from children with SAM compared to healthy Bangladeshi children that had detectable amounts of Blon_2176 (38% versus 66%, respectively; P<0.05, two proportion Z test; table S1A).
Colonization of Bangladeshi infants with SAM by milk-adapted B. infantis EVC001
Given the deficiency of B. infantis in the fecal microbiota of infants with SAM, we performed a pilot single-blind, randomized clinical SYNERGIE trial (SYNbiotic for Emergency Relief of Gut Instability and Enteropathy) to assess the extent to which EVC001 (a commercially available, USA infant-derived B. infantis strain with intact H1-H5 gene clusters) could colonize the gut in this population (18, 27, 30) (Fig. 1). Infants with SAM between 2 and 6 months of age (mean 4.1±1.1 months; WLZ < -3) were eligible for enrollment in the SYNERGIE trial after they had completed a standardized protocol for initial management of SAM (see table S2A for CONSORT diagram). Sixty-two enrolled infants were subsequently randomly assigned to one of three treatment groups: once daily administration of 8x109 colony-forming units (CFU) of B. infantis EVC001 alone (probiotic arm; n=20); the same dose of B. infantis EVC001 plus 1.6 g of the purified HMO LNnT (synbiotic arm; n=21); 625 mg/day of lactose (placebo arm; n=21) (Fig. 3A; table S2B).
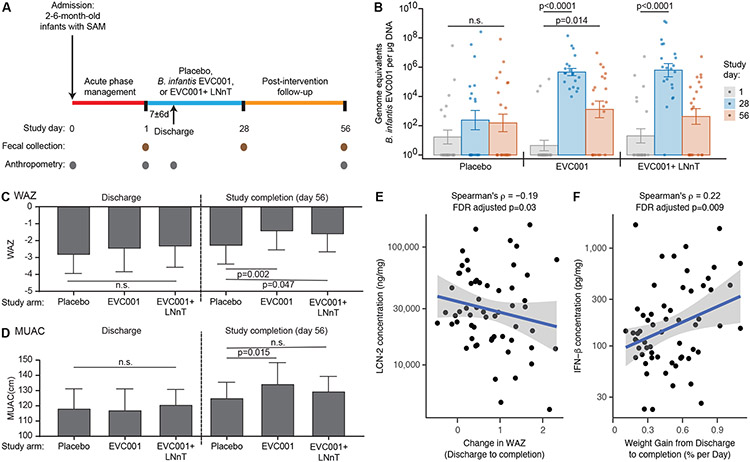
Fig. 3. SYNERGIE clinical trial.
(A) Shown is the design of the SYNERGIE clinical study. (B) The absolute abundance of B. infantis EVC001 in fecal samples collected from trial participants was measured using qPCR primers directed to a region of the epsJ gene. Fecal samples were collected on the indicated study days from participants in each arm of the study: placebo (lactose), probiotic (EVC001) and synbiotic (EVC001 + LNnT). Each dot represents a study participant. Barplots represent mean values ± SEM; n=20-21 participants per study arm. The P values indicated for within group comparisons were determined using Dunn’s test with Benjamini-Hochberg correction; n.s., not significant. (C, D) Effect of the interventions on weight-for-age z scores (WAZ; panel C) and Mid-Upper Arm Circumference (MUAC, panel D) at the end of the study (day 56) compared to the time of hospital discharge. Bar plots represent group means; error bars represent standard deviations. P values were calculated using the Mann-Whitney U test. (E) Spearman correlation between the concentration of fecal lipocalin-2 (LCN-2) and the change in WAZ from hospital discharge to study completion. (F) Spearman correlation between the concentration of fecal interferon-β (IFN-β) and the rate of weight gain in infants between discharge and study completion (Spearman’s ρ and FDR adjusted P values for each correlation are shown in panels E and F).
At enrollment, there were no statistically significant differences between the three groups with respect to socio-demographic or clinical characteristics (table S2C, D). Forty-two of the 62 infants had bilateral pedal edema, however, there were no statistically significant differences in the representation of this Kwashiorkor-like phenotype among the three arms (p=0.54; Kruskal-Wallis test; table S2D). Participants were treated with the probiotic, synbiotic or placebo daily for 28 days, starting in hospital and continuing at home after discharge. They were subsequently followed for an additional 28 days after cessation of their treatment intervention. Hospital discharge occurred when an infant had achieved a WLZ better than −3, or for those with Kwashiorkor, when their edema had resolved. There were no statistically significant differences between the three groups in the duration of their hospital stay after treatment was initiated (p=0.89; Kruskal-Wallis test; table S2D). Fecal samples were collected on day 1 (enrollment/baseline), day 28 and day 56 for analysis of the absolute abundance of B. infantis and for quantification of markers of intestinal inflammation. Anthropometry was performed on admission to the hospital of infants with SAM just prior to intervention with one of the two treatments or placebo (day 1), at the time of hospital discharge (day 28; end of treatment) and at study completion (day 56) (table S2C).
EVC001 contains a 123 bp sequence in epsJ (putative family 2 glycosyltransferase) that was not present in the genomes of six other B. infantis isolates that we had cultured from Bangladeshi children (table S3A). We designed qPCR primers targeting this region (table S1B) and confirmed their specificity for EVC001 using a panel of 31 sequenced human gut-derived strains, including 16 bifidobacteria (table S1C). Based on this qPCR assay, we observed a statistically significant increase in the abundance of EVC001 in the fecal microbiota at day 28 compared to baseline in infants with SAM who received EVC001 alone as well as in those who received EVC001 plus LNnT (P<0.0001 and P<0.0001 respectively; Dunn’s test with Benjamini-Hochberg correction) (Fig. 3B). At day 28, amounts of EVC001 in feces from infants in the EVC001 and EVC001+LNnT groups were significantly higher than in infants in the placebo group (P=0.004 and P=0.001 respectively; Dunn’s test with Benjamini-Hochberg correction). By day 56, the abundance of EVC001 had fallen in both the probiotic and synbiotic treatment groups compared to day 28, although it remained significantly higher than prior to intervention in the probiotic (EVC001 alone) arm (P=0.014; Dunn’s test with Benjamini-Hochberg correction (Fig. 3B).
Gut bacterial community composition was assessed by sequencing of PCR amplicons generated from variable region 4 of 16S rRNA genes present in the fecal samples collected before and after the interventions (table S2E). No significant differences were noted in the changes in the relative abundance of specific bacterial taxa between treatment groups from baseline to day 28 (P>0.05, Kruskal-Wallis test with Benjamini-Hochberg correction). However, in all groups, Enterobacteriaceae were significantly reduced from baseline to day 28 (P<0.001, Mann Whitney U test with Benjamini-Hochberg correction), falling in the EVC001 group from a relative abundance of 79±29% (mean±SD) at baseline to 42±26% at day 28. However, colonization by the probiotic EVC001 strain in this pilot study of infants with SAM was approximately 1-2 orders of magnitude lower than the average total abundance of B. infantis in age-matched healthy children living in the same community (Fig. 2A, table S1A).
Effects of colonization with EVC001 on host phenotypes
Due to the presence of bipedal edema in 42 of the 62 infants (68%) at enrollment, it was not possible to compare weights between groups until edema had resolved at the time of hospital discharge. At discharge, there were no statistically significant differences in weight-for-age z scores (WAZ) or mid-upper arm circumference (MUAC) between the intervention groups (P=0.361 and P=0.624 respectively, Kruskal-Wallis test; table S2F). However, at study completion (day 56), WAZ and MUAC in infants treated with EVC001 were significantly greater than for infants in the placebo arm, indicating an improvement in weight gain [P=0.002 WAZ; P=0.015, MUAC; Mann-Whitney U test; Fig. 3C, D, table S2F); WAZ was also significantly higher in children receiving EVC001+LNnT compared to children in the placebo arm at study completion (P=0.047; Mann-Whitney U test; Fig. 3C, table S2F). At the end of the 2-month study, ponderal growth (WLZ) had improved in all groups, although the differences between treatment groups were not statistically significant (P=0.22, Kruskal-Wallis test; table S2F). Finally, at study completion, linear growth [defined by length (height)-for-age z score (LAZ)] trended higher in children who had received EVC001 compared to those in the placebo arm (P=0.08, Kruskal-Wallis test; table S2F).
A previous study showed that stable colonization of healthy breastfed infants in the US with EVC001 resulted in reduced fecal markers associated with gut inflammation and intestinal permeability compared to a control group that did not receive EVC001 (18). Therefore, we measured lipocalin-2 (LCN-2), myeloperoxidase (MPO), calprotectin, and several cytokines (IFN-β, IL-17A, IL-1β and IL-6) in fecal samples collected from study participants on study day 1, at the time of hospital discharge, and on days 28 and 56 (table S2G). We did not observe statistically significant differences in these markers between the groups in this pilot study. However, the amount of LCN-2 was negatively correlated with the change in WAZ from hospital discharge to study completion (Spearman’s ρ= −0.19, FDR adjusted P=0.03; Fig. 3E). In addition, we observed a significant positive correlation between the amount of IFN-β at day 28 (31) and the rate of weight gain in infants with SAM between discharge and study completion (ρ=0.22, FDR adjusted P=0.009; Fig. 3F).
More than half of the infants with SAM in the SYNERGIE study were not receiving any breastmilk at the time of hospital admission: 15/21 (71%) of the children who were subsequently randomized to the synbiotic arm, 8/20 (40%) in the probiotic arm, and 11/21 (52%) in the placebo arm. Even among those infants who were receiving breastmilk at the time of admission, consumption was only 18±13% of the recommended daily volume for aged-matched healthy infants (table S2C). This raised the question of whether the limited persistence of colonization with the EVC001 strain compared to that previously observed in exclusively breastfed healthy US infants reflected the reduced prevalence of breast feeding and lower quantities of breastmilk consumption by Bangladeshi infants with SAM. Therefore, we next examined gut microbial communities of Bangladeshi children to search for B. infantis strains with a competitive advantage over other endogenous B. infantis strains as well as EVC001 that rely upon breastmilk oligosaccharides as their primary source of carbon.
In silico and in vitro characterization of Bangladeshi B. infantis strains
We characterized the genomic features of 10 B. infantis strains; six we had cultured from fecal samples collected from three healthy and one undernourished infants/children, aged 6-24 months, living in Mirpur, two strains from Malawian infants (MC1, MC2), a USA donor-derived type strain (ATCC 15697), plus EVC001 (table S3A). To do so, we turned to microbial community SEED (mcSEED), which has been used for manual curation of >2600 bacterial taxa cultured from the human gut microbiota (32). mcSEED ‘subsystems’ encompass functions (enzymes, transporters, transcriptional regulators) that reflect current knowledge of specific metabolic pathways, or groups of pathways represented in this set of human gut genomes (33). mcSEED ‘metabolic modules’ are lists of genes that can be more granular than a subsystem (e.g., uptake of a nutrient separately from its metabolism) (32,33). mcSEED-based in silico metabolic reconstructions allowed us to predict the ability of these strains to synthesize amino acids and B vitamins and utilize various carbohydrates (table S3B-D).
Table S3E,F summarizes the results of our in silico analysis of the ability of each strain to import and utilize HMOs. All these B. infantis strains have transporters for prominent neutral/fucosylated HMOs, with the notable exception of LNT (table S3E). Unlike the USA donor-derived ATCC15697 type strain and EVC001, only one of the Bangladeshi strains (Bg_463) possessed an ortholog of the canonical, biochemically characterized LNT transporter, Blon_2175-2177 (26). In contrast, all strains possessed a transporter for LNnT, the prominent type II [Gal(β1-4)GlcNAc-containing] isomer of LNT (Blon_2342-2344 or Blon_2345-2347).
We subsequently performed a series of in vitro growth experiments using four Bangladesh B. infantis strains plus EVC001. Monocultures were grown for 30 h under anaerobic conditions at 37 °C in a low carbohydrate DeMan-Rogosa-Sharpe (lc-MRS) medium that was supplemented with purified HMOs prominently represented in breast milk [LNT, LNnT, 2’-fucosyllactose (2’-FL), 3’- and 6’-sialyllactose (3’-SL, 6’-SL)]; lactose served as a positive control (n=3 replicates for each strain under each condition). Each of the strains tested exhibited slower growth rates in the presence of sialylated HMOs (3’-SL and 6’-SL) compared to neutral or fucosylated carbohydrate structures (fig. S2). [To date, no SL-specific transporter has been characterized in bifidobacteria, though the exo-α-sialidase NanH2 (Blon_2348, GH33) that is ubiquitous among B. infantis strains cleaves the sialic acid residue from both 3'-SL and 6'-SL]. We found that strains predicted to have the full complement of downstream catabolic enzymes and transporters for either LNnT or 2’-FL [Blon_2345-2347 and Blon_2202-2204 respectively (26, 34)], were able to grow in the presence of the corresponding HMO (table S3E, F). An unanticipated result was the comparable growth of strains in the presence of LNT (fig. S2A), suggesting that the three Bangladeshi strains that lack the Blon_2175-2177 ABC transporter have an alternative mechanism for LNT uptake/utilization whose efficiency is comparable to the canonical LNT transporter, at least in vitro in the absence of competition. Based on these in vitro observations, we performed a follow-up in vivo study involving gnotobiotic mice colonized with a consortium of B. infantis strains where this canonical LNT transporter was either present (EVC001, Bg_463) or absent (Bg_2D9, Bg_1G8, PS064).
In vivo competition between Bangladeshi B. infantis strains and EVC001 in gnotobiotic mice fed HMO-supplemented Bangladeshi diets
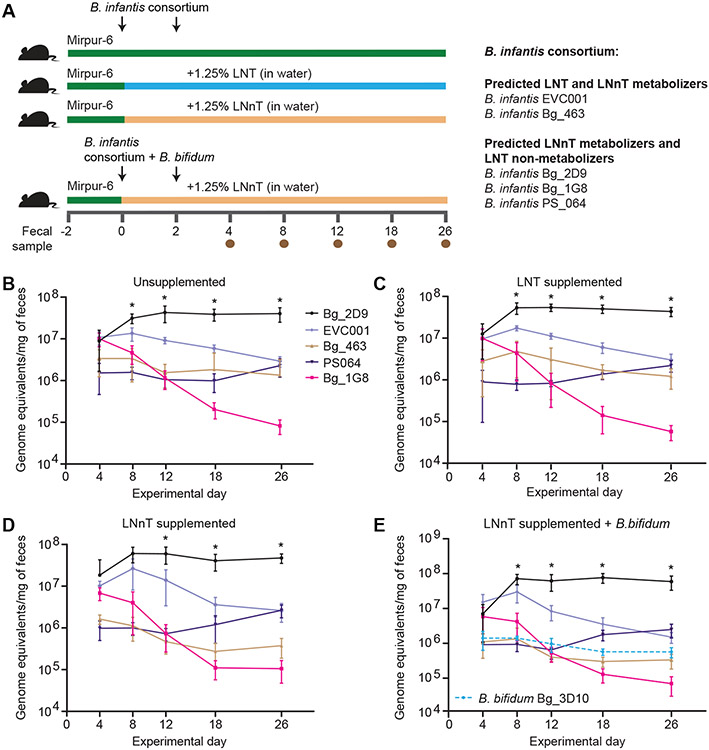
To compare the in vivo fitness of the Bangladeshi gut microbial B. infantis strains, 5-week-old germ-free C57BL/6J mice were started on a sterilized bovine milk powder/rice-lentil based chow similar to the diet consumed by 6-month-old children living in the Mirpur slum of Dhaka (table S4). After 2 days of consumption of this diet (Mirpur-6), the drinking water of one group of animals was supplemented with 12.5 g/L LNT, a second group received 12.5 g/L LNnT, and a third control group received unsupplemented water. All mice in these three arms (n=6-7/treatment group) were subsequently colonized with a consortium of five B. infantis strains followed 2 days later by a second gavage with the same consortium (Fig. 4A). In a fourth arm, mice were colonized with the 5-member B. infantis consortium plus a B. bifidum strain (Bg_3D10; table S3A) that we isolated from the fecal microbiota of a healthy 12-month-old Mirpur child; these mice were treated with LNnT-supplemented drinking water (Fig. 4A). Analysis of the genome of this B. bifidum strain indicated that it contained genes encoding membrane-bound extracellular lacto-N-biosidase I (LnbB) and extracellular exo-β-(1-3)-N-acetylglucosaminidase (BbhI) (table S3F). These enzymes endowed it with the capacity to degrade LNT and LNnT (35, 36) resulting in release of lacto-N-biose and lactose. The presence of B. bifidum could potentially limit the amount of LNnT available to other microbial consumers through a mechanism that is not dependent upon its ability to directly import LNnT. Mice in all four treatment groups were fed the Mirpur-6 diet ad libitum for 4 weeks. Every 2 days, fecal samples were collected and water, with or without HMO supplementation, was replenished daily.
Fig. 4. In vivo competition between B. infantis strains in gnotobiotic mice.
(A) Shown is the experimental design of the study examining the fitness of five B. infantis strains in gnotobiotic mice consuming the Mirpur-6 diet ± LNT or LNnT. (B-D) Shown are results of competition experiments involving the 5-member consortium of B. infantis strains introduced into gnotobiotic mice. (E) The same 5-member B. infantis consortium shown in panels B-D was introduced together with a B. bifidum strain isolated from a healthy 12-month-old Bangladeshi child into gnotobiotic mice. Absolute abundances of the different strains, as a function of time and HMO supplementation, were determined by short read shotgun sequencing of fecal DNA. Mean values ± SD are plotted; n=6-7 mice per group; n=1 experiment. Timepoints where the absolute abundance of the B. infantis strain Bg_2D9 was higher than other consortium members were identified using a mixed effects linear model followed by Tukey’s multiple comparison test. *, Padj<0.05.
There were no significant differences in body weights (measured every 2 days) between the four treatment groups (P>0.05, two-way repeated measures ANOVA; table S5A). The absolute abundance of each B. infantis strain in each of the treatment groups was determined on experimental days 4, 8, 12, 18 and 26 by short read shotgun sequencing of fecal DNA. The Bg_2D9 strain, which was isolated from a healthy 12-month-old child from Mirpur and lacked the LNT transporter (Blon_2175-2177), became the dominant member over time, achieving an absolute abundance on day 18 that was ≥10-fold higher than that for each of the other microbial community members in the three groups that had received the 5-strain consortium (FDR-adjusted P< 0.01, mixed effects model followed by Tukey’s multiple comparison test) (Fig. 4B-D, table S5B). During the first 18 days of colonization, EVC001 was the second most abundant strain, after which time it no longer exhibited a competitive advantage (Fig. 4B-D). In the 6-member community containing B. bifidum, Bg_2D9 dominated the community by day 8 and maintained its higher absolute abundance compared to each of the other strains for the duration of the experiment (Fig. 4E, table S5B).
Comparison of the Bg_2D9 genome to those of other consortium members
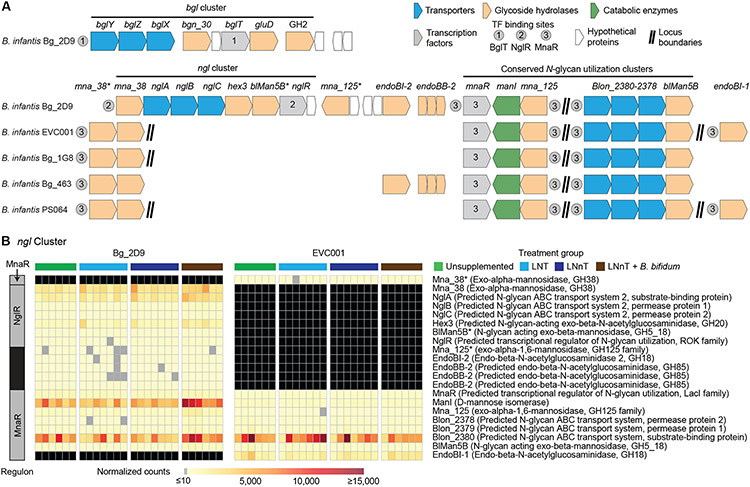
To identify features that might explain the competitive advantage of the Bg_2D9 strain, we compared its genome to the genomes of each of the other four B. infantis strains in the consortium. Using a minimum threshold of 95% sequence identity for orthologous genes, we identified 267 genes that were unique to the Bg_2D9 strain (Fig. 5A). Most of these 267 genes encoded short proteins or were mobile elements, but they included members of a predicted α-glucoside utilization gene cluster (bgl) and an N-glycan utilization gene cluster (ngl) (Fig. 5A, table S3G-I).
Fig. 5. Sugar utilization gene clusters in B. infantis strains.
(A) Shown are a β-glucoside utilization (bgl) gene cluster in Bg_2D9 and the N-glycan utilization (ngl) clusters in B. infantis strains of the 5-member consortium introduced into gnotobiotic mice. Predicted transcription factor binding sites are denoted by grey circles. Gene locus tags and annotations are given in table S3. (B) Expression of ngl cluster genes in the B. infantis Bg_2D9 and EVC001 strains. Mice fed either Mirpur-6, Mirpur-6 + 1.25% LNT (in their drinking water) or Mirpur-6 + 1.25% LNnT (in their drinking water) were colonized with a 5-member consortium of B. infantis strains including Bg_2D9 and EVC001. In a fourth group, B. bifidum was included in the 5-member consortium and mice were given drinking water supplemented with 1.25% LNnT. The black bar in the leftmost column indicates that the transcription factor has not been characterized. Cells in the heatmap that are colored black denote the absence of an ortholog in a strain and cells colored gray depict low expression (≤10 DESeq2-normalized read counts).
The bgl cluster contained three glycoside hydrolases, an ABC transport system and a TetR family transcriptional regulator (bglT). Among 34 published B. infantis genomes (37), only three (Bg_2D9, Bg_2C3, and Malawi_MC1; all described here) possess this locus (table S3I). Given that β-1,3-linked glucosides are common constituents of plant cell wall polysaccharides and approximately 60% by weight of the Mirpur-6 diet is plant-based (table S4A), we reasoned that a unique α-glucoside utilization cluster in Bg_2D9 could provide one explanation for its fitness advantage over the other members of the B. infantis consortium introduced into gnotobiotic mice.
Asparagine-linked glycans (N-glycans) have structural similarities to HMOs and are abundant in human and bovine milk where they decorate numerous proteins including lactoferrin and immunoglobulins (38). Previous reports have indicated that B. infantis ATCC 15697 and EVC001 are capable of utilizing a wide array of N-glycans both in vitro and in vivo (39, 40). We identified a ngl cluster in the Bg_2D9 genome that contained two endo-β-N-acetylglucosaminidases, EndoBI-2 and EndoBB-2 (GH85) (Fig. 5A, table S3G). The ngl cluster also contained genes encoding an ABC transport system predicted to transport N-glycans, glycoside hydrolases involved in degradation of N-glycans, and a transcriptional regulator NglR (Fig. 5A). Extending our analysis to 336 bifidobacterial genomes including 34 B. infantis isolates (37) revealed that the Bg_2D9 strain was one of only six that contained the ngl cluster (table S3I). EVC001 was predicted to have N-glycan metabolizing capabilities through an alternative pathway that includes EndoBI-1, a β-mannosidase (BlMan5B) and another predicted N-glycan transporter (Blon_2378-2380) (41), plus a α-mannosidase, Mna_125 and mannose isomerase (ManI) (Fig. 5A).
Analysis of gene expression in consortium members
We subsequently performed microbial RNA-Seq of cecal contents harvested at the time of euthanasia of gnotobiotic mice colonized with the five B. infantis strains with or without B. bifidum. We examined expression of the β-glucoside utilization (bgl) and N-glycan utilization (ngl) loci, as well as other genes involved in LNT and LNnT utilization, as a function of the presence or absence of these HMOs in the drinking water. Transcript counts were mapped to the genomes of consortium members, then normalized and analyzed to identify differentially expressed genes.
All eight genes comprising the bgl cluster were expressed in the Bg_2D9 strain, but there was no difference in their expression based on the presence or absence of either LNT or LNnT (fig. S3A, tables S5C, S6). The bgl cluster comprises a regulon controlled by a TetR family regulator, BglT (Fig. 5A, table S7). We postulated that this regulator potentially responded to β-gluco-oligosaccharides originating from the grain components (rice, lentil) of the Mirpur-6 diet (42).
There were no statistically significant effects of HMO supplementation on expression of genes in the ngl cluster of Bg_2D9 that are regulated by NglR or MnaR (Fig. 5B, tables S5C, S6, S7). Compared to animals receiving the Mirpur-6 diet alone, supplementation with LNT produced a statistically significant increase in expression (log 2-fold difference >1.5 and FDR-adjusted P<0.05) of genes in the H1 cluster that encoded six type II HMO transporter proteins (Fig. 1, fig. S3B and tables S5C, S6). In LNnT-supplemented mice, with the exception of Blon_2345, expression of these genes was also significantly elevated in Bg_2D9 compared to their expression in animals receiving unsupplemented water (log 2-fold difference >1.5; FDR-adjusted P<0.05; fig. S3B, tables S5C, S6). The absence of statistically significant induction of expression of other glycan transporter genes in the presence of LNT raises the possibility that this H1 cluster encodes a transport system that might also import LNT and LNnT.
LNnT supplementation also significantly increased expression of several genes required for HMO metabolism in the Bg_2D9 strain, including nagA (N-acetylglucosamine-6-phosphate deacetylase) and nagB (glucosamine-6-phosphate deaminase) which are involved in GlcNAc catabolism; the lnp cluster (H5) involved in lacto-N-biose/galacto-N-biose catabolism; the predicted HMO transporters Blon_2350 and Blon_2351 in the H1 cluster (log2-fold difference >1.5 and FDR-adjusted P<0.05; fig. S3B; tables S5C, S6). In contrast, expression of genes involved in sialic acid utilization including components of the nan cluster (H4) sialic acid ABC transporter (nanB and nanD) and a N-acetylneuraminate lyase (nanA) was reduced significantly in Bg_2D9 in both LNT-supplemented and LNnT-supplemented animals (log 2-fold difference >1.2, FDR-adjusted P<0.05; fig S3B; tables S5C, S6).
Finally, when comparing the LNnT-supplemented groups of gnotobiotic mice with or without B. bifidum, expression of most genes involved in HMO and N-glycan metabolism did not differ significantly (fig. S3B, tables S5C, S6). However, we observed a significant increase in the expression of H4 cluster genes involved in sialic acid catabolism in the Bg_2D9 genome (Fig. 1) when B. bifidum was present (log 2-fold difference >1.8, FDR-adjusted P<0.5; fig S3B; tables S5C, S6). Consistent with these observations, B. bifidum has been reported to produce an extracellular sialidase (table S3F) that can liberate sialylated glycans present in bovine milk, which would then be available for cross-feeding of other members of the microbial community (43,44). Collectively, our analyses indicate that among the strains evaluated, Bg_2D9 had the biggest endowment of glycoside hydrolases and candidate transporters for N-glycan utilization.
Prevalence of B. infantis strains possessing the ngl transporter gene in Bangladeshi children
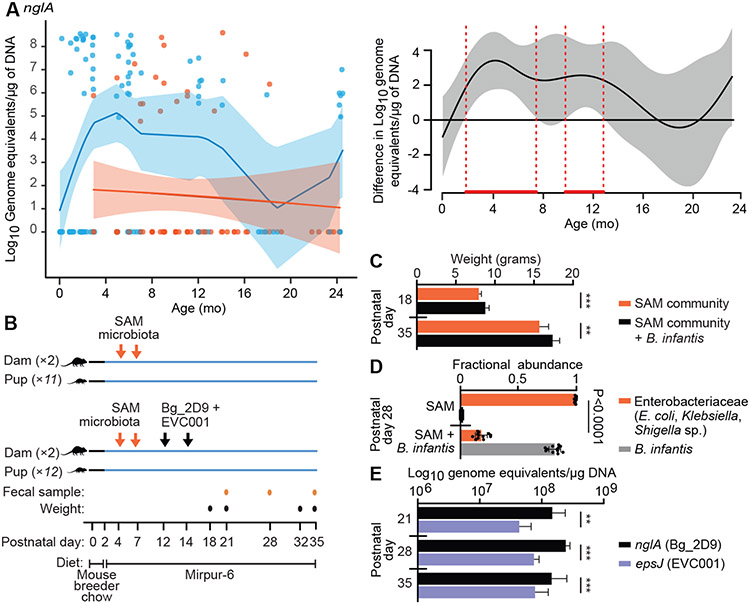
We returned to the collection of fecal DNA samples from the cross-sectional survey of Bangladeshi infants and children described in table S1A and applied a qPCR assay that used primers against the nglA component of the ABC transport system identified in the genome of the B. infantis Bg_2D9 strain (see table S1B,C for primer design and the results of specificity tests). This assay disclosed that fecal amounts of nglA were on average 2-3 orders of magnitude lower in 3 to 13-month-old children with SAM compared to their healthy age-matched counterparts (P<0.05, generalized additive model; red-bounded region in Fig. 6A), whereas no significant differences were evident between 14 to 24-month-old children (P>0.05; Generalized additive model). Notably, of the samples for which sufficient DNA was available to assay, 55 of 117 (47%) from healthy children and only 18 out of 83 (22%) from children with SAM were positive for nglA (P<0.05, two proportion Z-test; table S1A). Importantly, there were no fecal samples that were positive for nglA and negative for the characteristic B. infantis Blon_2348/nanH2 sialidase gene, suggesting that when detected, nglA was present within genomes belonging to strains of B. infantis.
Fig 6. Colonization by B. infantis Bg_2D9 of gnotobiotic mice harboring a SAM microbiota.
(A) The absolute abundance of the nglA subunit of the B. infantis N-glycan ABC transport system (NglABC) was measured in fecal samples from Bangladeshi children using qPCR. Fecal biospecimens from healthy children (n=117 samples) and children with SAM (n=83 samples) were assayed. The scatterplot (left panel) displays the absolute abundance of nglA as a function of age at the time of specimen collection. Samples from healthy infants and children are indicated by blue points/shading, whereas those from individuals with SAM are denoted in red. A generalized additive model-derived best fit line (±2 SEM) is shown. A plot difference curve (right panel) showing statistically significant differences in the best fit lines between the two models (healthy vs SAM) is indicated by areas bounded by red dashed vertical lines. (B) Shown is the experimental design of the gnotobiotic mouse study examining colonization of the gut microbiota of pups whose mothers received a fecal microbiota sample from a SAM donor with or without B. infantis strains Bg_2D9 and EVC001. (C) Shown is the weight of pups on postnatal days 18 and 35 (mean ± SD; n=11 pups in the SAM alone group and n=12 pups in the SAM plus B. infantis group, n=1 experiment). ***, P<0.001; **, P<0.01, two-way repeated measures ANOVA followed by Šidák’s multiple comparison test (Table S5D). (D) Shown are the results of a 16S rRNA-based analysis of the relative abundances of ASVs assigned to Enterobacteriaceae and bifidobacteria present in the fecal microbiota of P28 pups belonging to the two treatment groups. (E) Shown are the absolute abundances of Bg_2D9 and EVC001 in feces collected from pups at P21, P28 and P35 as defined by qPCR using strain-specific primers targeting the nglA and epsJ genes, respectively. ***, P<0.001; **, P<0.01, two-tailed Wilcoxon matched-pairs signed rank test.
Competition between B. infantis Bg_2D9 and EVC001 in gnotobiotic mice harboring a fecal microbiota from children with SAM
We subsequently used gnotobiotic mice to test the relative capacities of B. infantis Bg_2D9 and EVC001 to establish themselves in a fecal microbiota sample obtained from a 5-month-old infant with SAM in the SYNERGIE trial prior to the probiotic intervention. (The experimental design is summarized in Fig. 6B). Germ-free pregnant C57BL/6J dams were initially housed in the same isolator that contained 2 cages with 2 dams/cage. Animals were fed a standard breeder chow. On day 2 after parturition, both groups of dams were switched to the Mirpur-6 diet. On postpartum day 4, both dams in each group were gavaged with the fecal microbial community from the infant with SAM. The gavage was repeated three days later and on day 11, one of the two cages was moved to a separate isolator. On postpartum days 12 and 14, another type of gavage was performed, this one consisting of a mixture containing equivalent concentrations of Bg_2D9 and EVC001 that was administered to both dams in one of the two groups; both dams in the other group received a sham gavage. Pups (n=11-12/treatment group) were maintained with their dams until postnatal day 23 (time of weaning), after which they were provided the Mirpur-6 diet, exclusively.
Pups were weighed on postnatal days 18 (P18), P21, P32 and P35 (day of euthanasia). Animals in the B. infantis-treated group had significantly greater weights at all time points compared to pups whose mothers had only received the SAM microbiota (P<0.01, 2-way repeated measures ANOVA with Šidák’s correction for multiple comparisons; table S5D, Fig. 6C).
Fecal samples were collected from the four dams on P11, P21 and P28 and from their pups on P21, P28 and P35. Sequencing amplicons generated from the 16S rRNA genes present in fecal samples from the four dams collected prior to the B. infantis gavage on P11 revealed that they were colonized almost exclusively (>99% relative abundance) by Enterobacteriaceae. Specifically, there were amplicon sequence variants (ASVs) belonging to Enterococcus (61±11%; mean±SD), Escherichia/Shigella (24±12%) and Klebsiella (14±7%). In the dams that received B. infantis gavages, Enterobacteriaceae were reduced to 64±9% relative abundance in post-partum day 28 fecal samples, with bifidobacteria accounting for 35±9% of their communities (table S5E). No bifidobacteria were detected in dams (or their pups) that did not receive the B. infantis gavage. Suppression of Enterobacteriaceae was even more pronounced in weaned (P28) pups of the B. infantis recipients: 18±5% relative abundance for Enterococcus, Escherichia/Shigella and Klebsiella ASVs combined compared to 99±1% aggregate relative abundance of these taxa in pups of mothers that received only the SAM microbiota (table S5E, Fig. 6D). These data are consistent with our observations in the SYNERGIE trial, although the magnitude of suppression of Enterobacteriaceae achieved was larger in the mouse study where B. infantis achieved approximately two orders of magnitude greater absolute abundance than in the probiotic-treated infants in the SYNERGIE trial.
Strain-specific qPCR revealed that dams that had received the B. infantis gavage were colonized with both strains at P35, with Bg_2D9 being significantly more abundant than EVC001 (8.17 vs 7.67 log10 genome equivalents/μg DNA; P=0.012, paired t-test). Pups of mothers that had received the Bg_2D9 plus EVC001 strain mixture were colonized with high amounts (>8 log10 genome equivalents/μg DNA) of B. infantis at the earliest time point sampled (P21) (Fig. 6E). The absolute abundance of Bg_2D9 attained was greater than that for EVC001 and the difference persisted until euthanasia (P35) (P< 0.01, two-tailed Wilcoxon matched-pairs signed rank test). Thus, this maternal-pup transmission model provided preclinical evidence of the superior competitiveness of the Bg_2D9 strain over the EVC001 strain in the context of a donor SAM microbiota and the Mirpur-6 diet.
DISCUSSION
Disrupted co-development of the gut microbiota and its host during early postnatal development is one manifestation of moderate to severe acute malnutrition. A recent randomized controlled trial of a microbiota-directed complementary food (MDCF) formulation designed to repair features of disrupted gut microbiota development evident in 12 to 18-month-old Bangladeshi children with moderate acute malnutrition living in an urban slum in Dhaka Bangladesh demonstrated improvements in weight gain that were superior to those obtained with a standard nutritional supplement (12). Gut microbiota repair was accompanied by increases in 70 plasma proteins and mediators of various aspects of musculoskeletal and brain development, as well as decreases in proteins involved in inflammatory responses. The SYNERGIE clinical study described here focused on the more severe form of wasting, SAM, manifest in younger (2 to 6-month-old) infants living in the same urban slum in Dhaka Bangladesh. Compared to chronologically age-matched healthy infants, those with SAM exhibited lower absolute abundances of a key early colonizer of the gut that is a highly efficient consumer of human milk oligosaccharides, B. longum subsp infantis. The SYNERGIE trial examined the extent to which treatment with EVC001, a commercially available B. infantis strain isolated from a healthy infant in the USA, with or without LNnT supplementation, could colonize the gut microbiota of the Bangladeshi infants with SAM. Daily treatment with the probiotic alone, synbiotic (EVC001 plus LNnT) or placebo (lactose) began after participants had completed an acute phase management protocol for SAM (45) and continued for 28 days, followed by an additional 28 days of follow-up. Despite the relatively short exposure period and small cohort size, engraftment of EVC001 was similar in the probiotic and synbiotic arms and was associated with statistically significant improvements in ponderal growth (WAZ and MUAC) and changes in fecal markers indicating reduced gut inflammation. In addition, we observed a positive correlation between fecal IFN-β and the rate of weight gain. A recent study of EVC001-supplemented U.S. infants also revealed suppression of intestinal inflammation markers, a response that was associated with a higher abundance of bifidobacteria in their feces. Moreover, in vitro assays of fecal water from these EVC001-treated U.S. infants disclosed a shift in T cell polarization away from a T helper 2 (Th2) state towards a Th1 state, with accompanying suppression of Th17 cytokines and an increase in IFN-β (46). These data raise the possibility that increased intestinal IFN-β may be one factor that contributed to weight gain in children treated with EVC001 in the SYNERGIE study.
The mean absolute abundance of B. infantis EVC001 achieved in the SYNERGIE study was ~10-100-fold lower than that found in age-matched, healthy breastfed children from the same locale and was not significantly improved by co-administration with LNnT. Breastmilk consumption by the infants with SAM in our pilot study was low suggesting that other bifidogenic factors found in breastmilk may have been limiting for robust colonization by EVC001. Thus, alternative approaches to restoring B. infantis in the gut microbiota of such infants to levels found in healthy infants, might be expected to provide even greater clinical responses than were obtained in our study. As described here, one strategy for achieving this goal involved searching for B. infantis strains adapted to the gut environments of infants living in impoverished communities. This search was guided by the reality that whereas exclusive breastfeeding of infants is recommended by the WHO for the first 6 months, in many low-income settings, gruels, animal milk and complementary foods are often introduced into the diet at an early age for economic or cultural reasons. Given that this dietary landscape may not be optimal for B. infantis strains adapted to thrive in the gut microbiota of infants who are exclusively/predominantly breastfed, we conducted an in vivo screen of a consortium of B. infantis strains cultured from the fecal microbiota of Bangladeshi infants living in the same urban slum community. This screen was executed in gnotobiotic mice fed a diet that reflected the early complementary feeding practices of these children. The staged in vivo competition experiments in gnotobiotic mice revealed a strain (Bg_2D9) isolated from a healthy 12-month-old child that expressed a distinctive repertoire of carbohydrate utilizing genes and had a marked fitness advantage over the other, predominantly milk-adapted, strains that lacked these genes. Two expressed gene clusters in particular distinguished this isolate from the others. One gene cluster encoded enzymes that utilize β-1,3-linked glucosides, which are common constituents of plant cell wall polysaccharides, and the other gene cluster encoded components needed for utilization of N-linked glycans represented in human and bovine milk proteins. These components included two endo-β-N-acetylglucosaminidases for releasing sugar moieties of N-glycans, an ABC transport system, and multiple intracellular glycoside hydrolases involved in utilizing HMOs that may also contribute to metabolism of complex N-glycans (fig. S4). We postulate that these endo-β-N-acetylglucosaminidases (EndoBI-2 and EndoBB-2) may be able to release sugar moieties from N-glycans, which are further transported into the cell via NglABC or Blon_2378-2380. Internalized sugar moieties would then be degraded from their non-reducing ends by the orchestrated action of exo-acting glycoside hydrolases; these glycoside hydrolases include Bga2A, Hex1, Hex2, NanH2, BiAfcA and BiAfcB, which act on linkages found in both HMOs and complex N-glycans containing GlcNAc, fucose, and NANa residues.
The fitness advantage of the B. infantis Bg_2D9 strain in gnotobiotic mice was maintained even when the diet was supplemented with the HMOs, LNT and LNnT, suggesting that the abundant non-human milk glycans (e.g., β-1,3-linked glucosides and N-linked glycans) present in the Mirpur-6 diet may have a dominant effect on shaping the structure of this defined gut microbial community. An alternative possibility is that the Bg_2D9 strain acquired an enhanced capacity to utilize various host mucin glycan degradation products, which play an important role in localizing beneficial mutualists to the intestinal mucosa (47).
One limitation of the clinical study is that we did not include an arm with LNnT alone due to our concern that in the absence of a ‘consumer’ such as B. infantis, a high dose of this prebiotic may cause GI intolerance /adverse events in an already vulnerable study population. A limitation of the preclinical studies is that although the ngl and bgl gene clusters in Bg_2D9 appear to be involved in the uptake and utilization of N-glycans and plant-derived polysaccharides, establishing that they are important for the fitness of Bg_2D9 in vivo requires testing their functions using targeted genetic deletion and complementation studies. Moreover, experimental tests of the carbohydrate structures that they are able to utilize are needed. Another limitation is the degree to which these or related genomic features are represented in other B. infantis isolates recovered from various populations where there is early postnatal consumption of substantial amounts of dietary plant glycans.
Our follow-on studies involved an intergenerational gnotobiotic mouse model of microbial transmission from dam to pups. These experiments revealed that when administered to dams that had previously been colonized with a SAM community, Bg_2D9 became the dominant member of the gut microbiota of their offspring, and this was associated with suppression of Enterobacteriaceae and superior weight gain in the pups. Translating these preclinical results into studies to assess the clinical benefits of Bg_2D9 on restoration of healthy growth in infants with SAM will require development of stable, affordable probiotic formulations that can be administered in low-income settings and tested in larger, sufficiently-powered trials. Our finding that two of the most prominent HMOs, LNT and LNnT, did not produce marked effects on the fitness of B. infantis Bg_2D9 in the staged competition experiments in gnotobiotic mice suggests that Bg_2D9 could be effective as a probiotic without the need for HMO supplements and the added cost that they bring. In addition, the potential for maternal-infant transmission of B. infantis raises the question of whether Bg_2D9 could be administered as a probiotic to women during pregnancy. Future clinical studies should ideally include a mechanistic component using multi-omics approaches to define the gut microbiota and host physiologic state prior to, during and after probiotic treatment (11, 12). The resulting data sets would enable the role of Bg_2D9 and its effects on gut microbiota repair and health to be more clearly and comprehensively characterized. Finally, the analysis of B. infantis EVC001 and Bg_2D9 strains illustrates how searching for different strains that have evolved to adapt to host habitats with different environmental exposures could yield single-strain or multi-strain probiotics or synbiotics with broader host ranges, higher engraftment efficiencies, and thus more generalizable and impactful health benefits.
MATERIALS AND METHODS
Study Design
The SYNbiotic for Emergency Relief of Gut Instability and Enteropathy (SYNERGIE) was a three-arm, single-blind, parallel assignment trial designed to assess the extent of colonization of the SAM infant microbiota B. infantis EVC001 strain administered daily for 28 days either alone or in combination with a human milk oligosaccharide (LNnT), compared to a placebo treatment (lactose). The primary outcome measure was the abundance of B. infantis in the feces of study participants as measured by qPCR after 28 days of supplementation (EVC001 versus placebo, and EVC001 + LNnT versus placebo). Secondary outcome measures included assessment of the bacterial composition of the gut microbiota of participants, changes in their anthropometric indices, and changes in markers of intestinal inflammation after 28 days of treatment. The goal of SYNGERIE was to generate data to justify whether a larger, appropriately powered trial to assess the impact of treatment on clinical outcomes.
Gnotobiotic mouse studies were designed to compare the fitness of EVC001 with several B. infantis strains that had been cultured from the fecal microbiota of Bangladeshi infants. Animals were fed a diet representative of that consumed by 6 month-old Bangladeshi children living in an urban slum and given drinking with or without supplementation with two prominent HMOs. B. infantis strains were introduced into germ-free mice in two different formats. The first format involved recently weaned animals who received a consortium of five B. infantis strains with or without B. bifidum. The second format was designed to mimic mother-to-infant transmission; it entailed initial colonization of female germ-free mice (dams) with an intact uncultured microbiota obtained from an untreated infant participant in the SYNERGIE trial followed by simultaneous gavage of B. infantis EVC001 and Bg2D9. Transfer of the B. infantis strains and their absolute abundance was defined by quantitative PCR of fecal DNA. Dams were randomly assigned to each treatment group. All data were generated from all samples without knowledge of treatment group. No data were excluded and there were no outliers.
Human Study
The SYNERGIE study was approved by the Institutional Review Board of the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b) (Protocol # PR-17112) and registered at ClinicalTrials.gov (“Pilot of a Prebiotic and Probiotic Trial in Young Infants With Severe Acute Malnutrition” NCT03666572). The study was conducted between September 2018 and March 2020 as a single-blind randomized trial involving 2-6 month-old infants of either sex presenting with a WLZ score <−3, or bilateral pedal edema, who had completed an acute phase management protocol for SAM (45) in the in-patient ward of Dhaka Hospital at icddr,b. Other inclusion criteria were that the child’s caregiver (i) was willing to provide consent for enrollment; (ii) lived locally (<15 km) and (iii) was willing to stay in the Nutritional Rehabilitation Unit of the Dhaka Hospital for the duration of the hospital stay (~15 days). Infants were excluded if they had septic shock or pneumonia requiring assisted ventilation, acute kidney injury, congenital defects interfering with feeding such as cleft palate, chromosomal anomalies, jaundice, tuberculosis, or ongoing maternal antibiotic usage for breastfeeding infants.
Each participant was assigned to receive the standard of care diet (F-75 formula during the acute phase treatment of SAM, F-100 formula during hospital stay for nutritional rehabilitation, and standardized infant formula upon discharge). After acute phase management, infants were transferred to the Nutritional Rehabilitation Unit (NRU), enrolled and randomized to receive either 8x109 CFU B. infantis EVC001, 8x109 CFU B. infantis EVC001 plus LNnT, or placebo for 4 weeks, after which time they were followed for 4 additional weeks (Fig. 3A, table S2A,B). Randomization was performed in a 1:1 ratio using stratification by age (2-3.9 months vs. 4-6 months) to ensure equal allocation of infants by age group. Treatments were assigned using random permuted blocks of 3. The enrollment target was 54 participants, with 18 infants (9+9 from each age bin) allocated to each arm.
B. infantis EVC001 was administered as a single daily dose mixed with 5 mL of milk (breastmilk or F-100). Each sachet containing 1.6 g of LNnT was mixed with 200 mL of F-100 (WHO, 2002) and administered daily after the completion of the antibiotic component of their in-patient acute phase management protocol. The protocol for discharge from the NRU required each participant to achieve a WLZ ≥−3 (ideally, >−2), and in the case of infants with edematous malnutrition, resolution of their edema. While at home, 1.6 g LNnT was given twice daily by the caregiver, each time mixed with 120 mL of breastmilk or infant formula without prebiotics or HMOs (Mead Johnson). Refrigerated storage of the probiotic, consumption of LNnT and morbidity were all monitored twice a week by field research assistants. Parents/caregivers in the study were blinded to treatment assignment; icddr,b study staff were not blinded since they needed to prepare the LNnT in the infant formula while the infants were hospitalized.
The amount of breastmilk consumed was measured by the test weighing method (i.e., weighing the infant before and after feeding) at the time of enrollment. Breastfeeding was encouraged between feeds throughout the study. Infants who were not breastfed received infant formula at home in addition to their allotted LNnT supplements. Notably, a group of non-malnourished infants (WLZ >−1) who were hospitalized with infections and treated with antibiotics were also given EVC001 in the SYNERGIE study; the results of this analysis will be reported separately.
Fecal samples and anthropometric data were obtained prior to the start of supplementation (day 1), at the end of supplementation (day 28) and 4-weeks following cessation of treatment (day 56). To preserve bacterial community composition at the time of fecal collection, fecal swabs for DNA-based assays were placed in pre-labeled, buffered lysis tubes (Zymo Research) that were flash frozen in liquid nitrogen within 20 minutes of defecation (48). Fecal specimens used for measurement of intestinal inflammation markers were flash frozen without added buffer. Samples were stored at −80°C prior to being shipped to Evolve BioSystems, Inc. (Davis, CA) where assays of EVC001 colonization and markers of intestinal inflammation were performed. Data were analyzed from 21 participants who completed the synbiotic arm, 20 from the probiotic arm and 21 from the placebo arm.
qPCR assays of the absolute abundance of B. infantis in human fecal samples
PCR primers -
A multiplex quantitative polymerase chain reaction (qPCR) assay was developed to quantify B. infantis using primers directed at Blon_2348 (29), Blon_2176 and Bifidobacterium spp 16S rRNA (28). A fourth assay (primers/probe combination) was designed to quantify the absolute abundance of EVC001. The EVC001 genome was first fragmented in silico into 20bp k-mers; all tiled k-mers were aligned with the genomes of sequenced B. infantis strains isolated from Bangladeshi children, [plus the B. breve type strain (ATCC15700) as an additional control for specificity] to identify sequences (k-mers) containing ≥2 base mismatches. Contiguous k-mers were stitched together, leading to the identification of a 123 bp sequence in epsJ (locus tag N_00627) that was represented in the EVC001 genome but rarely present in Bangladeshi samples. We also developed a qPCR assay directed against a 95bp target in the nglA gene, a component of an ABC transporter that is predicted to import N-glycans, that was present in B. infantis Bg_2D9 but not EVC001 (table S3I).
Custom Taqman™ probes (Applied Biosystems™) with FAM (Blon_2348), VIC (Blon_2176) and JUN (Bifidobacterium spp) dyes and non-fluorescent QSY quencher were obtained from Thermo Fisher Scientific. Primers were purchased from Integrated DNA Technologies (IDT, Coralville, IA). Forward and reverse primer sets were tested in silico using the NCBI Primer-Blast program with default settings against the NCBI non-redundant nucleotide database to predict specificity and cross-reactivity. The Oligoanalyzer tool (IDT) was used to assess the melting temperature compatibility of primers for multiplexing. To determine sensitivity, a standard curve was generated for the Blon_2348 and Blon_2176 qPCR assays with seven 10-fold dilutions of DNA purified from B. infantis ATCC 15697.
A similar approach was used for the epsJ and nglA assays, except that DNA purified from B. infantis Bg_2D9 and EVC001, respectively, was used to generate the standard curves. To ascertain the specificity of the assays, the multiplex qPCR panel was run with purified nucleic acids from 16 Bifidobacterium strains and 15 non-Bifidobacterium human gut bacterial isolates selected based on their 16S rRNA gene sequence similarity to members of Bifidobacterium (table S1C).
Conditions for PCR -
PCR reaction mixtures contained (i) 900 nmol forward and reverse primers and 250 nmol Taqman probes for Blon_2348, Blon_2176 and nglA assays, and 150nM of both 16S rRNA gene primers for Bifidobacterium sp, (ii) 5μL Taqman™ Multiplex Master Mix, (iii) 2.5μL genomic DNA (≤7.5 ng total DNA mass; prepared from fecal samples as previously described; ref. 11), and (iv) nuclease-free water to make up a total reaction volume of 10. Assays were performed in duplicate in a 384-well plate format using an Applied Biosystems Quantstudio 6 Flex qPCR instrument. Temperature parameters were as follows: 50 °C for 2 minutes and 95 °C for 10 minutes followed by 40 cycles of 95 °C for 10 seconds and 60 °C for 30 seconds. With the exception of the nglA assay, standard curves were generated for every PCR plate run using seven serial 10-fold dilutions of purified DNA from B. infantis type strain (ATCC15697). For the nglA assay, standard curves were generated using seven serial 10-fold dilutions of purified DNA from B. infantis Bg_2D9.
A singleplex qPCR assay was run using the epsJ primers. PCR reactions consisted of (i) 900 nmol forward and reverse primers and 250 nmol FAM-labeled TaqMan probe, (ii) 10 μL of TaqMan Universal Master Mix II containing uracil N-glycosylase (UNG, Applied Biosystems), (iii) 5 μL of a given fecal genomic DNA preparation (diluted 1:10), and (iv) nuclease-free water to make up a total reaction volume of 20 μL. Assays were performed in duplicate in 96-well plate format using an QuantStudio 3 instrument (ThermoFischer). Cycling parameters were as follows: 50 °C for 2 minutes and 95 °C for 10 minutes, followed by 40 cycles of 95 °C for 10 seconds, and 60 °C for 30 seconds. Standard curves were generated for each PCR plate using six 10-fold dilutions of genomic DNA extracted from known CFU counts of B. infantis EVC001.
Analysis of data -
Based on the slope and intercept derived from linear regression for cycle thresholds against copy number for the reference, the abundance of each of the three targets was calculated for each sample on the plate. Amplification efficiencies were calculated from the slope [94.3±2.1% (mean±SD) for the Bifidobacterium genus 16S rRNA gene assay, 89.6±3.1% for the Blon_2348 assay, 87.8±2.3% for the Blon_2176 assay and 85.7±4.1% for the nglA assay]. R2 values for each standard curve exceeded 0.99. Raw data were normalized for input DNA concentration and expressed in genome equivalents per μg of fecal DNA. [Note that since in silico alignment of the 16S rRNA gene primers against the genome of B. infantis ATCC 15697 using the NCBI Primer-BLAST program (49) identified four identical target sequences, calculation of total Bifidobacterium abundance in fecal samples was based on the assumption that four copies of this gene are present in each B. infantis genome, whereas abundances of the other PCR targets (Blon_2348, Blon_2176, epsJ and nglA) were each based on a single copy/genome]. Due to non-normal distribution (Shapiro-Wilk Normality Test, P< 0.001) qPCR target abundance data were log10-transformed and the Mann-Whitney U test was used to determine statistical significance of differences between healthy and SAM children in age bins of 3-12 months, 12-18 months and 18-24 months. A pseudocount of 1 was added to all counts before log10 transformation to convert values below the limit of detection limit to 0. Log10-transformed individual qPCR target abundance data were fitted with a generalized additive model using the “gam” function from the “mgcv 1.8-31” package in R as a Gaussian family. A generalized cross validation (GCV) method was used for smoothing, and age at the time of sample collection was used as a predictor to compare the healthy versus SAM models. The “plot_diff” function from “itsadug 2.4” package in R was utilized to generate difference curves for the two groups.
Log10-transformed epsJ qPCR values generated from fecal samples collected during the SYNERGIE study were compared across each timepoint within treatment arms using a Dunn's Test of Multiple Comparisons with Benjamini-Hochberg adjustment in R using the “dunn_test” function from the “rstatix 3.6.2 “package.
GNOTOBIOTIC MOUSE STUDIES
All mouse experiments were carried out using protocols approved by the Washington University in St. Louis Institutional Animal Care and Use Committee (IACUC; Protocol # 20-0322). Mice were housed in plastic flexible film gnotobiotic isolators (Class Biologically Clean Ltd., Madison, WI) at 23 °C under a strict 12-hour light cycle (lights on at 0600h). Cages contained autoclaved paper ‘shepherd shacks’ for environmental enrichment.
Quantifying absolute abundances of B. infantis and B. bifidum strains in mice colonized with the bifidobacterial consortium
The absolute abundances of B. infantis strains in fecal samples collected from colonized mice in the experiment described in Fig. 4A were defined by short read shotgun sequencing of community DNA (COPRO-Seq; 50). To determine absolute abundance, 6.7 x 106 cells of Alicyclobacillus acidiphilus DSM 14558 and 29.8 x 106 cells of Agrobacterium radiobacter DSM 30147 were added to each weighed frozen fecal pellet collected from each animal on study days 4, 8, 12, 18, 26 (51). Fecal pellets were then subjected to bead beading for 4 minutes (Mini-BeadBeater-8, BioSpec) in a mixture containing 500 μL of extraction buffer [200 mM NaCl, 200 mM Tris (pH 8), 20 mM EDTA], 210 μL of 20% SDS, 500 μL of phenol/ chloroform/isoamyl alcohol (pH 7.9) (25:24:1; Ambion), and 250 μL of 0.1 mm-diameter zirconia beads (BioSpec Products). Samples were centrifuged at 4 °C for 4 minutes at 3,220 x g. The aqueous phase was collected; nucleic acids were purified using QIAquick columns (Qiagen) and eluted from the columns into 10 mM Tris-Cl, pH 8.5. DNA concentration was quantified (Quant-iT dsDNA assay kit, broad sensitivity; ThermoFisher), and adjusted to 0.75 ng/μL with UltraPure water (Milli-Q). COPRO-Seq libraries were prepared using the Nextera DNA Library Prep kit protocol (Illumina) and custom barcoded primers (52). Barcoded libraries were sequenced on an Illumina NextSeq instrument [75 nt uni-directional reads; 2.71 ± 1.38 x 106 reads/sample (mean ± S.D.)].
Reads were de-multiplexed and mapped to the sequenced whole genomes of the five Bifidobacterium strains, plus five “distractor” genomes (Lactobacillus ruminis ATCC 27782, Olsenella uli DSM 7084, Pasteurella multocida USDA-ARS-USMARC 60385, Prevotella dentalis DSM 3688 and Staphylococcus saprophyticus ATCC 15305). The proportion of total reads mapping to the five distractor genomes for each sample was used to set a conservative threshold (mean + 2SD) for colonization of an organism in an animal. For each member of the community, absolute abundance was calculated by multiplying the normalized counts of strains by the abundance of Alicyclobacillus acidiphilus (cell number per normalized count) divided by the sample weight (53). Mixed-effects linear models followed by Tukey’s post-hoc test (GraphPad Prism, Version 8) were applied to test for significant interaction of time and abundance of the strains.
Microbial RNA-Seq
Cecal contents harvested at the time of euthanasia from gnotobiotic mice in the experiment described in Fig. 4A were flash frozen in liquid nitrogen and stored at −80 °C. For RNA extraction, cecal samples were kept on ice and the following reagents were added in the following order: (i) 250 μL of acid-washed glass beads (212-300 μm diameter; Millipore Sigma; catalog number G1277), (ii) 500 μL of 2X Buffer B (200 mM NaCl, 20 mM EDTA), (iii) 210 μL of 20% SDS, and (iv) 500 μL phenol:chloroform:isoamyl alcohol (25:24:1, pH 4.5; ThermoFisher). The mixture was homogenized using a bead beater (Mini-BeadBeater-8, BioSpec) at room temperature for a total of 5 minutes, with a pause for 2 minutes on ice after the first three minutes. The mixture was centrifuged (7000 x g for 10 minutes at 4°C) and RNA was isolated from 500μL of the aqueous phase using a previously described protocol (54). RNA integrity and fragment size were assessed [4200 TapeStation System (Agilent)] followed by elimination of genomic DNA by using two sequential DNAase treatments [Baseline-ZERO DNase (Lucigen) and Turbo DNAse (Invitrogen)]. The absence of genomic DNA was verified by qPCR using primers against Bifidobacterium spp 16S rRNA gene (28). Total RNA was purified (MEGAclear Transcription Clean-Up Kit; ThermoFisher) and quantified (Qubit RNA BR Assay Kit;Invitrogen); a 1 μg aliquot was subsequently depleted of ribosomal RNA using the Ribo-Zero (Epidemiology/Bacteria) kit (Illumina) followed by ethanol precipitation. The SMARTer Stranded RNASeq kit (Takara Bio USA) was used to prepare double-stranded cDNA and indexed libraries. Libraries were sequenced using an Illumina NextSeq platform [70 nt unidirectional reads; 5.3x107 ± 2.8x106 reads/sample (mean ± SD); n=26 samples]. The first five cycles of sequencing were omitted as this library preparation strategy introduces three non-templated deoxyguanines. Reads were demultiplexed, checked for quality using FastQC and were mapped to the genomes of the members of the consortia. Transcript counts were normalized and analyzed using the DESeq2 package in R (version 4.0.2; 55) at the level of individual strains. For each strain, raw count data were fitted to a negative binomial generalized linear model using the DESeq2 workflow to identify statistically significant differences in gene expression (log2 fold difference and Wald test P value) in the following groups of mice: (i) LNT-supplemented versus unsupplemented, (ii) LNnT-supplemented versus unsupplemented and (iii) LNnT-supplemented and colonized with or without B. bifidum.
Statistical Analysis
Clinical data from the SYNERGIE study were entered into pre-tested Case Report Forms (CRFs) and analyzed using SPSS (20.0 version, Armonk, NY). Demographic, clinical and socioeconomic data were expressed as median and interquartile range (IQR) for asymmetric quantitative data. For categorical data, frequency with proportional estimates was used. A Kruskal-Wallis H test was used to assess the statistical significance of differences between the three treatment arms. Mann-Whitney U tests were used to determine statistically significant differences in anthropometric measures between pairs of treatment groups at the indicated time points. The statistical significance of differences in EVC001 engraftment over time within treatment groups was determined using a Dunn's Test of Multiple Comparisons with Benjamini-Hochberg adjustment in R (“dunn_test” function from the rstatix 3.6.2 package). A Kruskal-Wallis test with FDR correction was used to assess the statistical significance of the effects of the different interventions on markers of gut inflammation at each timepoint. Correlations between fecal markers and anthropometric parameters were calculated as Spearman’s ρ, with FDR adjusted P values.
Supplementary Material
Acknowledgements:
We thank the families of the Bangladesh study cohorts. We are indebted to the staff and health care workers at the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b) for their contributions to recruitment and enrollment of mothers as well as collection of biospecimens and data from their offspring. The authors are also grateful to core donors that provide unrestricted support to icddr,b, including the Government of the People’s Republic of Bangladesh, Global Affairs Canada (GAC), Swedish International Development Cooperation Agency (Sida) and the Department for International Development (UK-Aid). We thank Su Deng, Justin Serugo, Marty Meier, J. Hoisington-López, Sabrina Wagoner, Maria Karlsson and David O’Donnell for their superb technical assistance; Nicholas Griffin for his help with generalized additive models, Hao-Wei Chang and Chandni Desai for their assistance in archiving and depositing data, and Laura Kyro for her help with graphical design. We are grateful to Heather Brown (Evolve Biosystems Inc.) for her assistance with the analysis of fecal marker data, and for the insights provided by Daniela Barile, Carlito Lebrilla and David Mills (UC Davis) throughout this work. We thank with Glycosyn, GeneChem and Glycom for their generous provision of HMOs used in this study, and Mead Johnson Nutrition for the infant formula used in the SYNERGIE trial.
Funding:
This work was funded by the Bill and Melinda Gates Foundation [OPP1134649 to JIG; OPP1179599 to icddr,b and Evolve BioSystems Inc.] and by the National Institutes of Health (DK30292 to JIG).
Footnotes
Competing interests: S.F., B.H., G.C., R.F., R.M., R.M.D. and D.K. are employees of Evolve BioSystems Inc., a company that is developing and marketing next-generation probiotics designed to promote healthy gut microbiota development in infants. B. infantis EVC001 is covered by US patent 10,716,816 and International Patent Application PCT/US2015/057226 entitled “Activated bifidobacteria and methods of use thereof” with D.K. as co-inventor. A patent application has been filed for use of the B. infantis Bg_2D9 strain for the treatment of malnutrition with M.J.B., K.A., S.N., T.A. and J.I.G. as co-inventors. The other authors declare no competing interests.
Data and materials availability: V4-16S rRNA gene amplicon sequences in raw format prior to post-processing and data analysis, COPRO-Seq, microbial RNA-Seq datasets generated from the cecal contents of gnotobiotic mice colonized with bifidobacterial consortia, plus genome assemblies from the cultured bifidobacterial strains have been deposited at the European Nucleotide Archive under study accession number PRJEB45396. Human biospecimens and bacterial strains cultured from fecal samples collected from Bangladeshi children are the property of icddr,b and are available under a Materials Transfer Agreement (MTA) upon request to J.I.G. MTAs exist between icddr,b, and Washington University and between icddr,b and Evolve BioSystems Inc. for the use of these samples.
REFERENCES AND NOTES
- 1.UNICEF. Malnutrition rates remain alarming: stunting is declining too slowly while wasting still impacts the lives of far too many young children. http://data.unicef.org/topic/nutrition/malnutrition/#. (2018). [Google Scholar]
- 2.Uauy R, Desjeux JF, Ahmed T, Hossain M, Brewster D, Forbes D, Caton H, Kleinman RE, Global efforts to address severe acute malnutrition. J. Pediatr. Gastroenterol. Nutr 55, 476–481 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Amadi B, Besa E, Zyambo K, Kaonga P, Louis-Auguste J, Chandwe K, Tarr PI, M Denno D, Nataro JP, Faubion W, Sailer A, Yeruva S, Brantner T, Murray J, Prendergast AJ, Turner JR, Kelly P, Impaired Barrier Function and Autoantibody Generation in Malnutrition Enteropathy in Zambia. EBioMedicine 22, 191–199. (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerac M, McGrath M, Grijalva-Eternod C, Bizouerne C, Saxton J, Bailey H, et al. Management of Acute Malnutrition in Infants (MAMI) Project. Technical review: current evidence, policies, practices and program outcomes. (2011). [Google Scholar]
- 5.Walker S, Wachs TD, Gardner JM, Lozoff B, Wassermann GA, Pollitt E, Carter JA, International Child Development Steering Group, Child development: risk factors for adverse outcomes in developing countries. Lancet 369, 145–57 (2007). [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization, World Food Programme. United Nations System Standing Committee on Nutrition, United Nations Children’s Fund. Community-based management of severe acute malnutrition. (2007). www.who.int/nutrition/topics/Statement_community_based_man_sev_acute_mal_eng.pdf. [Google Scholar]
- 7.Ashworth A, Khanum S, Jackson A, Schofield C. Guidelines for the inpatient treatment of severely malnourished children (2003). www.who.int/nutrition/publications/guide_inpatient_text.pdf. [Google Scholar]
- 8.Hossain M, Chisti MJ, Hossain MI, Mahfuz M, Islam MM, Ahmed T, Efficacy of World Health Organization guideline in facility-based reduction of mortality in severely malnourished children from low and middle income countries: A systematic review and meta-analysis. J. Paediatr. Child Health 53, 474–479 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, Destefano J, Meier MF, Muegge BD, Barratt MJ, VanArendonk LG, Zhang Q, Province MA, Petri WA, Ahmed T, Gordon JI, Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510, 417–421 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raman AS, Gehrig JL, Venkatesh S, Chang HW, Hibberd MC, Subramanian S, Kang G, Bessong PO, Lima AAM, Kosek MN, Petri WA, Rodionov DA, Arzamasov AA, Leyn SA, Osterman AL, Huq S, Mostafa I, Islam M, Mahfuz M, Haque R, Ahmed T, Barratt MJ, and Gordon JI, A sparse covarying unit that describes healthy and impaired human gut microbiota development. Science 365, eaau4735 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gehrig JL, Venkatesh S, Chang HW, Hibberd MC, Kung VL, Cheng J, Chen RY, Subramanian S, Cowardin CA, Meier M, O’Donnell D, Talcott M, Spears LD, Semenkovich CC, Henrissat B, Giannone RJ, Hettich RL, Ilkayeva O, Muehlbauer M, Newgard CB, Sawyer C, Head RD, Rodionov DA, Arzamasov AA, Leyn SA, Osterman AL, Hossain Md I., Islam M, Choudhury N, Sarker SA, Huq S, Mahnud I, Mostafa I, Mahfuz M, Barratt MJ, Ahmed T, Gordon JI. Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science 365, eaau4732 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen RY, Mostafa I, Hibberd MC, Das S, Mahfuz M, Naila NN, Islam Md. M., Huq S, Alam Md. A, Zaman MU, Raman AS, Webber D, Zhou C, Sundaresan V, Ahsan K, Meier MF, Barratt MJ, Ahmed T, Gordon JI. A microbiota-directed food intervention for undernourished children. N Engl J Med. 384, 1517–1528 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrido D, Barile D, Mills DA. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv. Nutr 3, 415S–421S (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U S A 105, 18964–18969 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl. Environ. Microbiol 76, 7373–81 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Underwood MA, German JB, Lebrilla CB, Mills DA, Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut. Pediatr. Res 77, 229–235 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, Qadri F, Underwood MS, Mills DA, Stephensen CB. Stool microbiota and vaccine responses of infants. Pediatrics 134, e362–72. (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henrick B, Chew S, Mitchell R R, Contreras L, Casaburi G, Frese S, Smilowitz J, Underwood M. Restoring Bifidobacterium infantis EVC001 to the infant gut microbiome significantly reduces intestinal inflammation. Curr. Dev. Nutr 3, nzz049.OR12-01-19 (2019). [Google Scholar]
- 19.Nguyen M, Holdbrooks H, Mishra P, Abrantes MA, Eskew S, Garma M, Oca CG, McGuckin C, Hein CB, Mitchell RD, Kazi S, Chew S, Casaburi G, Brown HK, Frese SA, Henrick BM. Impact of Probiotic B. infantis EVC001 Feeding in premature infants on the gut microbiome, nosocomially acquired antibiotic resistance, and enteric inflammation. Front. Pediatr 16, 9:618009. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Underwood MA, Arriola J, Gerber CW, Kaveti A, Kalanetra KM, Kananurak A, Bevins CL, Mills DA, Dvorak B, Bifidobacterium longum subsp. infantis in experimental necrotizing enterocolitis: alterations in inflammation, innate immune response, and the microbiota. Pediatr Res. 76, 326–333 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thurl S, Munzert M, Boehm G, Matthews C, Stahl B, Systematic review of the concentrations of oligosaccharides in human milk. Nutr. Rev 75, 920–933 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thurl S, Munzert M, Henker J, Boehm G, Müller-Werner B, Jelinek J, Stahl B. Variation of human milk oligosaccharides in relation to milk groups and lactational periods. Br. J. Nutr 104, 1261–1271 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Totten SM, Zivkovic AM, Wu S, Ngyuen U, Freeman SL, Ruhaak LR, Darboe MK, German JB, Prentice AM, Lebrilla CB, Comprehensive profiles of human milk oligosaccharides yield highly sensitive and specific markers for determining secretor status in lactating mothers. J. Proteome Res 11, 6124–6133 (2012). [DOI] [PubMed] [Google Scholar]
- 24.James K, Bottacini F, Contreras J, Vigoureux M, Egan M, Motherway MO, Holmes E, van Sinderen D. Metabolism of the predominant human milk oligosaccharide fucosyllactose by an infant gut commensal. Sci Reports 9, 15427 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomson P, Medina DA, Garrido D. Human milk oligosaccharides and infant gut bifidobacteria: Molecular strategies for their utilization. Food Microbiol. 75, 37–46 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Garrido D, Kim JH, German JB, Raybould HE, Mills DA. Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS One 6, e17315 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duar RM, Casaburi G, Mitchell RD, Scofield LNC, Ortega Ramirez CA, Barile D, Henrick BM, Frese SA. Comparative genome analysis of Bifidobacterium longum subsp. infantis strains reveals variation in human milk oligosaccharide utilization genes among commercial probiotics. Nutrients 12, 3247 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penders J, Vink C, Driessen C, London N, Thijs C, Stobberingh EE, Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR, FEMS Microbiol Letters 243, 141–147 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Lawley B, Munro K, Hughes A Hodgkinson A, Prosser CG, Lowry D, Zhou SJ, Makrides M, Gibson RA, Lay C, Chew C, Lee PS, Wong KH, Tannock GW Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets. Peer J. 5, e3375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casaburi G, Karav S, Frese S, Henrick B. Gut barrier function is improved in infants colonized by Bifidobacterium longum subsp. infantis EVC001. Curr. Dev. Nutr 3, Suppl 1, nzz048.FS04-05-19 (2019). [Google Scholar]
- 31.Kumaran Satyanarayanan S, El Kebir D, Soboh S, Butenko S, Sekheri M, Saadi J, Peled N, Assi S, Othman A, Schif-Zuck S, Feuermann Y, Barkan D, Sher N, Filep JG, Ariel A, IFN-β is a macrophage-derived effector cytokine facilitating the resolution of bacterial inflammation. Nat Commun. 10, 3471 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodionov DA, Arzamasov AA, Khoroshkin MS, Iablokov SN, Leyn SA, Peterson SN, Novichkov PS, Osterman AL, Micronutrient requirements and sharing capabilities of the human gut microbiome. Front. Microbiol 10, 1316 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang H-Y, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–702 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakanaka M, Hansen ME, Gotoh A, Katoh T, Yoshida K, Odamaki T, Yachi H, Sugiyama Y, Kurihara S, Hirose J, Urashima T, Xiao J-Z, Kitaoka M, Fukiya S, Yokota A, Lo Leggio L, Hachem MA, Katayama T, Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Sci Adv 5, eaaw7696 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wada J, Ando T, Kiyohara M, Ashida H, Kitaoka M, Yamaguchi M, Kumagai H, Katayama T, Yamamoto K, Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Applied Environ. Microbiol 74, 3996–4004 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miwa M, Horimoto T, Kiyohara M, Katayama T, Kitaoka M, Ashida H, Yamamoto K. Cooperation of β-galactosidase and β-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology 20, 1402–1409 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, Chlenski P, Conrad N, Dickerman A, Dietrich EM, Gabbard JL, Gerdes S, Guard A, Kenyon RW, Machi D, Mao C, Murphy-Olson D, Nguyen M, Nordberg EK, Olsen GJ, Olson RD, Overbeek JC, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomas C, VanOeffelen M, Vonstein V, Warren AS, Xia F, Xie D, Yoo H, Stevens R. The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res. 48, D606–D612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nwosu CC, Aldredge DL, Lee H, Lerno LA, Zivkovic AM, German JB, Lebrilla CB, Comparison of the human and bovine milk N-glycome via high-performance microfluidic chip liquid chromatography and tandem mass spectrometry. J. Proteome Res 11, 2912–2924 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garrido D, Nwosu C, Ruiz-Moyano S, Aldredge D, German JB, Lebrilla CB, Mills DA. Endo-β-N-acetylglucosaminidases from infant gut-associated bifidobacteria release complex N-glycans from human milk glycoproteins. Mol. Cell Proteomics 11, 775–785 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karav S, Le Parc A, de Moura Bell J. M. Leite Nobrega, Frese SA, Kirmiz N, Block DE, Barile D, Mills DA. Oligosaccharides released from milk glycoproteins are selective growth substrates for infant-associated Bifidobacteria. Appl. Environ. Microbiol 82, 3622–3630 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cordeiro RL, Pirolla R, Persinoti GF, Gozzo FC, de Giuseppe PO, Murakami MT. N-glycan utilization by Bifidobacterium gut symbionts involves a specialist β-Mannosidase. J. Mol. Biol 431, 732–747 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Demirbas A. β-Glucan and mineral nutrient contents of cereals grown in Turkey. Food Chemistry 90, 773–777 (2005). [Google Scholar]
- 43.Kiyohara M, Tanigawa K, Chaiwangsri T, Katayama T, Ashida H, Yamamoto K, An exo-alpha-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology 21, 437–447 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Egan M, O’Connell Motherway M, van Sinderen D. A GntR-type transcriptional repressor controls sialic acid utilization in Bifidobacterium breve UCC2003. FEMS Microbiol. Lett 362, 1–9 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Ahmed T, Ali M, Ullah MM, Choudhury IA, Haque ME, Salam MA, Rabbani GH, Suskind RM, Fuchs GJ. Mortality in severely malnourished children with diarrhoea and use of a standardised management protocol. Lancet 5, 1919–1922 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Henrick B, Rodriguez L, Lakshmikanth T, Pou C, Henckel E, Arzoomand A, Olin A, Bifidobacteria-mediated immune system imprinting early in life. Cell 184, 1–15 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. A refined palate: bacterial consumption of host glycans in the gut. Glycobiology 23, 1038–1046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahfuz M, Das S, Mazumder RN, Rahman MM, Haque R, Bhuiyan MMR, Akhter H, Sarker Md S.A., Mondal D, Muaz SSA, Karmim ASMB, Borowitz SM, Moskaluk CA, Barratt MJ, Petri WA, Gordon JI, Ahmed T, Bangladesh environmental enteric dysfunction (BEED) study: protocol for a community-based intervention study to validate non-invasive biomarkers of environmental enteric dysfunction. BMJ Open 7, e017768 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, (1), 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL, Gordon JI, Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 11, e1001637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stämmler F, Gläsner J, Hiergeist A, Holler E, Weber D, Oefner PJ, Gessner A, Sprang R. Adjusting microbiome profiles for differences in microbial load by spike-in bacteria. Microbiome 4, 28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adey A Morrison HG, Asan HG, Xun X, Kitzman JO, Turner EH, Stackhouse B MacKenzie AP Caruccio NC, Zhang X, Shendure J, Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 11, R119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolf AR, Wesener DA, Cheng J, Houston-Ludlam AN, Beller ZW, Hibberd MC, Giannone RJ, Peters SL, Hettich RL, Leyn SA, Rodionov DA, Osterman AL, Gordon JI, Bioremediation of a common product of food processing by a human gut bacterium. Cell Host & Microbe 26, 463–477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hibberd MC, Wu M, Rodionov DA, Li X, Cheng J, Griffin NW, Barratt MJ, Giannone RJ, Hettich RL, Osterman AL, Gordon JI, The effects of micronutrient deficiencies on bacterial species from the human gut microbiota. Sci. Transl. Med 9, eaal4069 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods. 13, 581–583. (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mallick H, McIver LJ, Rahnavard A, Ma S, Zhang Y, Nguyen LH, Tickle TL, Weingart G, Ren B, Schwager E, Subramanian A, Lu Y, Waldron L, Paulson JN, Franzosa EA, Corrada Bravo H, Huttenhower C, Multivariable Association in Population-scale Meta-omics Studies. bioRxiv 01.20.427420 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turner S S, Pryer KM KM, Miao VP, Palmer JD, Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol 1999 46, 327–38 (1999). [DOI] [PubMed] [Google Scholar]
- 59.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Price MN, Arkin AP, PaperBLAST: Text mining papers for information about homologs. mSystems. 2, e00039–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B, The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saier MH, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res. 44, D372–379 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Novichkov PS, Rodionov DA, Stavrovskaya ED, Novichkova ES, Kazakov AE, Gelfand MS, Arkin AP, Mironov AA, Dubchak I. RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res. 38, W299–W307 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arzamasov AA, van Sinderen D and Rodionov DA. Comparative Genomics Reveals the Regulatory Complexity of Bifidobacterial Arabinose and Arabino-Oligosaccharide Utilization. Front Microbiol. 9, 776 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J, Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altschul SF and Lipman DJ. Protein database searches for multiple alignments. Proc. Natl. Acad. Sci. U. S. A 87, 5509–5513 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.