Abstract
Catalase negatively regulates plant immunity and is targeted and degraded by ubiquitin E3 ligase.
Dear Editor,
The ubiquitin proteasome system (UPS) is an essential post-translational regulatory mechanism that controls plant growth, development, and immune responses (Moon et al., 2004; Xu and Xue, 2019). E3 ubiquitin ligases play vital roles in plant–pathogen interactions (Duplan and Rivas, 2014; Ning et al., 2016). The Magnaporthe oryzae effector AvrPiz-t interacts with the rice (Oryza sativa L.) E3 ubiquitin ligase AvrPiz-t Interacting Protein 6 (APIP6) to promote the degradation of both proteins (Park et al., 2012). Both ectopic expression of AvrPiz-t and silencing of APIP6 in rice plants reduce the flg22-triggered production of reactive oxygen species (ROS) (Park et al., 2012). However, how AvrPiz-t and APIP6 regulate ROS generation to modulate rice immunity remains unclear. A recent study demonstrated that AvrPiz-t structurally mimics the susceptibility protein RESISTANCE OF RICE TO DISEASES1 (ROD1) and both proteins can raise the activity of Catalase B (CatB) to eliminate ROS (Gao et al., 2021). Both AvrPiz-t and ROD1 are ubiquitinated by the same E3 ubiquitin ligase APIP6 (Park et al., 2012; Gao et al., 2021), indicating that the UPS may play a critical role in catalase-mediated ROS homeostasis. However, whether catalase is modified by ubiquitination is unknown (Wang et al., 2022).
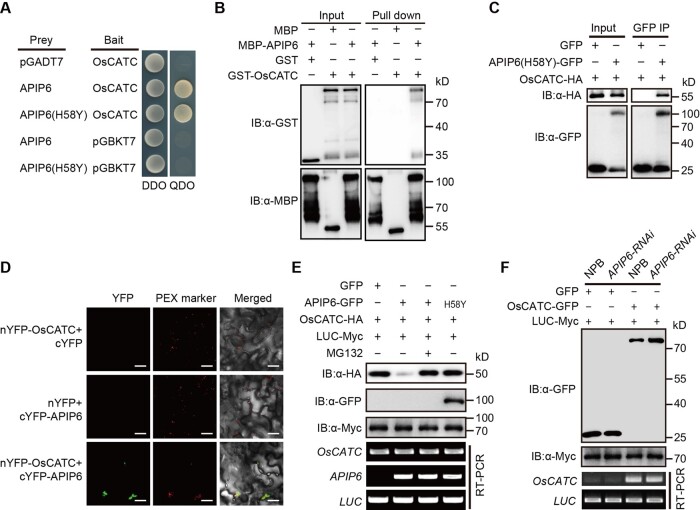
In this study, we found that OsCATC interacts with APIP6 in yeast (Saccharomyces cerevisiae) (Figure 1A) and confirmed this interaction by in vitro pull-down assays with GST and maltose binding protein (MBP) as the negative controls (Figure 1B). And then we performed a co-immunoprecipitation (Co-IP) assay in rice protoplasts using the more stable APIP6 mutant APIP6(H58Y) (Park et al., 2012), which also interacted with OsCATC in yeast (Figure 1A). Immunoblot analysis showed that OsCATC-HA specifically associated with APIP6(H58Y)-GFP (Figure 1C), suggesting that APIP6 interacts with OsCATC in vivo. OsCATC localized to peroxisomes in rice protoplasts (Supplemental Figure S1) and the bimolecular fluorescence complementation (BiFC) assays showed that the interaction occurred at the region where peroxisomes gathered (Figure 1D). These results indicated that APIP6 interacts with OsCATC in vitro and in vivo.
Figure 1.
The relationship between the catalase OsCATC and APIP6. A, OsCATC interacts with APIP6 and APIP6(H58Y) in yeast two-hybrid assay. Yeast cells were plated on SD/-Leu-Trp (DDO) and SD/-Leu-Trp-His-Ade (QDO) medium. B, Pull-down assay to confirm the interaction between OsCATC and APIP6. MBP and GST were used as the negative controls. C, Co-IP assay to detect the interaction between OsCATC and APIP6(H58Y) in rice protoplasts. The expressed proteins were immunoprecipitated with anti-GFP antibody. GFP was used as a negative control. IB, immunoblotting. D, BiFC assay showing the interaction of APIP6 and OsCATC in Nicotiana benthamiana leaves. OsPEX14-mCherry was used as a peroxisome marker. Bars: 20 μm. E, APIP6 promotes the degradation of OsCATC via the 26S proteasome pathway in rice protoplasts. OsCATC-HA was co-transfected with APIP6-GFP or APIP6(H58Y)-GFP in rice protoplasts. Samples were collected 24 h after transfection. The 26S proteasome inhibitor MG132 (50 μM) was added 12 h before sampling, with an equal volume of dimethyl sulfoxide (DMSO) as a control. LUC-Myc was used as an internal control. F, The expression level of OsCATC in the protoplasts of APIP6-RNAi plants. The OsCATC-GFP plasmids were transiently expressed in the protoplasts of NPB and APIP6-RNAi plants. Samples were collected 24 h after transfection for protein and RNA extraction. The extracted protein and RNA were used for western blot and RT-PCR analysis, respectively. LUC-Myc was used as an internal control. PEX, peroxisomes.
We next explored whether OsCATC is a substrate of APIP6. Immunoblot analysis revealed that OsCATC-HA protein abundance was lower in protoplasts co-transfected with APIP6-GFP compared with GFP alone or the APIP6(H58Y) mutant construct (Figure 1E). Furthermore, the 26S proteasome inhibitor MG132 blocked the degradation of OsCATC (Figure 1E), while the internal control LUC-Myc protein had a similar accumulation in those combinations. These results suggest that APIP6 promotes the degradation of OsCATC in vivo through the 26S proteasome-dependent pathway. To further confirm the degradation of OsCATC mediated by APIP6 in rice, we transiently expressed OsCATC-GFP plasmid in the protoplasts of APIP6-RNAi plants. The results showed that the protein levels, but not the transcriptional levels of OsCATC, were increased in APIP6-RNAi plants compared with the wild-type Nipponbare (NPB) (Figure 1F). To explore whether OsCATC has feedback regulation on APIP6, we transiently expressed APIP6(H58Y)-GFP plasmid in the protoplasts of oscatc mutants and found that both the protein and transcriptional levels of APIP6(H58Y)-GFP were similar in NPB and oscatc mutants (Supplemental Figure S2). These results suggested that APIP6 promotes the degradation of OsCATC in vivo and OsCATC has no obvious feedback regulation on APIP6.
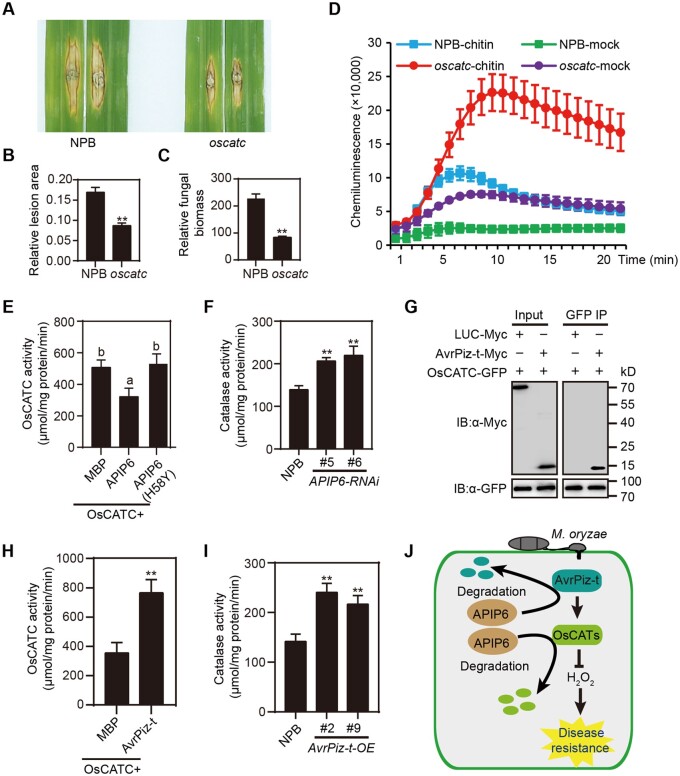
In Arabidopsis (Arabidopsis thaliana), the knock-out mutant of CATALASE 2 (CAT2) shows increased resistance to biotrophic pathogens (Yuan et al., 2017). A knockout mutant in rice CatB confers enhanced resistance to Xanthomonas oryzae pv. oryzae (Gao et al., 2021). These results suggested that catalase may play an important role in plant immunity. In rice, NOE1 encodes the catalase OsCATC, whose loss-of-function mutation noe1 accumulates hydrogen peroxide (H2O2) and exhibits programmed cell death phenotypes (Lin et al., 2012; Liang et al., 2015). To determine the biological roles of OsCATC in rice immunity, we performed punch inoculations and found that both lesion area and fungal biomass in the lesions were lower in the oscatc (noe1) mutant compared with NPB (Figure 2A–C). Consistent with previous results (Lin et al., 2012), the oscatc mutant accumulated higher levels of H2O2 than did NPB (Supplemental Figure S3). We also observed a much higher ROS production rate in the oscatc mutant than in NPB in response to chitin (Figure 2D). Taken together, these results demonstrated that the catalase OsCATC negatively regulates rice immunity by modulating the accumulation of H2O2.
Figure 2.
The catalase OsCATC negatively regulates rice innate immunity to the blast fungus M. oryzae. A, Punch inoculation of the NPB and the oscatc mutant. The leaves were photographed 14 days after inoculation. B and C, Relative lesion area and relative fungal biomass of NPB and the oscatc mutant in A. D, Chitin-induced ROS burst in NPB and oscatc mutant leaves. Leaf disks from NPB and oscatc plants were treated with 8 nM chitin and water. Data shown as means ± sd (n = 3). E, APIP6 suppresses the H2O2 degradation activity of OsCATC in the presence of rice extracts. Recombinant purified OsCATC was used for catalase assays and mixed with MBP, MBP-APIP6, or MBP-APIP6(H58Y) for 1 h at 30°C in the presence of rice extracts. Data shown as means ± sd. Significant differences (P < 0.01) were determined by Duncan’s new multiple range test and indicated with different letters (n = 3). F, Catalase activity in APIP6-RNAi plants. G, Co-IP assay to confirm the interaction between OsCATC and AvrPiz-t in rice protoplast. AvrPiz-t-Myc or LUC-Myc was co-expressed with OsCATC-GFP in NPB protoplasts. LUC-Myc was used as a negative control. IPs were performed using anti-GFP antibody and the associated proteins were detected by IB with anti-Myc antibody. H, AvrPiz-t enhances the H2O2 degradation activity of OsCATC. Purified OsCATC protein was mixed with MBP or MBP-AvrPiz-t for 1 h at 4°C in 50 mM KH2PO4 and 20 mM H2O2 for in vitro catalase assay. MBP was used as a negative control. I, Catalase activity in AvrPiz-t-OE plants. J, A proposed working model for OsCATC-mediated rice immune responses. Data shown as means ± sd (n = 3) in (B, C, F, H, and I). Asterisks represent statistically significant differences between samples (**P < 0.01 by Student’s t test).
We explored whether APIP6 affects the biochemical activity of OsCATC by performing catalase activity assays. After 1 h of incubation, APIP6 attenuated the efficiency of OsCATC-mediated H2O2 scavenging by promoting OsCATC degradation (Figure 2E and Supplemental Figure S4). Moreover, APIP6-RNAi lines had higher catalase activity (Figure 2F), suggesting that APIP6 negatively regulates catalase-mediated ROS scavenging by targeting OsCATC for degradation. As AvrPiz-t interacts with CatB and improves the efficiency of CatB-mediated H2O2 scavenging (Gao et al., 2021), we established that AvrPiz-t interacts with OsCATC by Co-IP and AvrPiz-t also enhances the efficiency of OsCATC-mediated H2O2 scavenging (Figure 2, G and H and Supplemental Figure S5). Furthermore, catalase activity in AvrPiz-t ectopic expression plants was higher than in NPB (Figure 2I).
Based on these findings and results from a previous study (Park et al., 2012), we proposed the following working model to illustrate the relationship between APIP6, AvrPiz-t, and OsCATC in rice (Figure 2J). When rice plants are infected by M. oryzae, the secreted effector AvrPiz-t stimulates OsCATC activity to degrade H2O2 to create more favorable conditions for M. oryzae infection. To defend against pathogen attack, the E3 ubiquitin ligase APIP6 targets both AvrPiz-t and OsCATC for degradation to increase ROS production for the activation of rice defense responses.
This study revealed the E3 ubiquitin ligase-mediated degradation of a catalase protein in plants. The rice genome encodes three CAT genes: OsCATA, CatB (OsCATB), and OsCATC (Ye et al., 2011; Gao et al., 2021). APIP6 may also target OsCATA and CatB for degradation. Gao et al. (2021) and this study showed that AvrPiz-t targets CatB and OsCATC to strengthen catalase activity for H2O2 scavenging. ROD1 is degraded by the ubiquitin ligases APIP6 and its homologous protein RIP1 (Gao et al., 2021). Based on these results, all three CAT proteins may participate in the AvrPiz-t-ROD1-CATs-APIP6/RIP1 hierarchical regulatory immunity network. The predominant source of catalase activity in rice leaves may be derived from OsCATC oligomers (Zhang et al., 2016), indicating that rice catalases may function as a homologous or heterologous complex. Whether catalases participate in rice immunity as a complex and how they fine-tune ROS levels during the arms race between rice and pathogens remain unclear. Although the mutants oscatc and CR-catb (genome-edited at CatB) showed enhanced disease resistance by increasing ROS levels, these plants suffered obvious growth penalties (Lin et al., 2012; Gao et al., 2021). How to harness catalase genes to generate plants with greater disease resistance warrants further investigation.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Subcellular localization of OsCATC-GFP in rice protoplasts.
Supplemental Figure S2. The expression level of APIP6 in the protoplasts of oscatc mutants.
Supplemental Figure S3. H2O2 contents in 6-week-old NPB and oscatc mutant plants.
Supplemental Figure S4. OsCATC and APIP6 protein abundance in Figure 2E.
Supplemental Figure S5. OsCATC and AvrPiz-t protein abundance in Figure 2H.
Funding
This work was supported by the National Natural Science Foundation of China (31822041, 32161143009, and 31972225), the China Postdoctoral Science Foundation (2020M680131), and the Shenzhen Science and Technology Program (KQTD20180411143628272).
Conflict of interest statement. The authors declare that there is no conflict of interest.
Supplementary Material
Contributor Information
Xiaoman You, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, 440307 Shenzhen, China; State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China.
Fan Zhang, State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China.
Zheng Liu, State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China.
Min Wang, State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China.
Xiao Xu, State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China.
Feng He, State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China.
Debao Wang, State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China.
Ruyi Wang, State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China.
Yiqin Wang, State Key Laboratory of Plant Genomics, Institute of Genetics and Developmental Biology, The Innovative Academy for Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
Guirong Wang, Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, 440307 Shenzhen, China; State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China.
Chengcai Chu, Guangdong Laboratory for Lingnan Modern Agricultural Science and Technology, College of Agriculture, South China Agricultural University, Guangzhou 510000, China.
Guo-Liang Wang, Department of Plant Pathology, The Ohio State University, Columbus, Ohio 43210, USA.
Yuese Ning, State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China.
These authors contributed equally (X.Y. and F.Z.)
Y.N. supervised the project. Y.N., X.Y., and F.Z. conceived and designed the research plans. Z.L. and M.W. performed the reverse transcription PCR and subcellular localization assay. X.X., F.H., and D.W. performed immunoblots. R.W. and Y.W. preformed blast inoculation. X.Y., G.W., C.C., G.-L.W., and Y.N. analyzed the data. X.Y., G.-L.W., and Y.N. wrote the manuscript. All authors revised the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Yuese Ning (ningyuese@caas.cn).
References
- Duplan V, Rivas S (2014) E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front Plant Sci 5: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, He Y, Yin X, Zhong X, Yan B, Wu Y, Chen J, Li X, Zhai K, Huang Y, et al. (2021) Ca2+ sensor-mediated ROS scavenging suppresses rice immunity and is exploited by a fungal effector. Cell 184: 5391–5404 [DOI] [PubMed] [Google Scholar]
- Liang C, Zheng G, Li W, Wang Y, Hu B, Wang H, Wu H, Qian Y, Zhu XG, Tan DX, et al. (2015) Melatonin delays leaf senescence and enhances salt stress tolerance in rice. J Pineal Res 59: 91–101 [DOI] [PubMed] [Google Scholar]
- Lin A, Wang Y, Tang J, Xue P, Li C, Liu L, Hu B, Yang F, Loake GJ, Chu C (2012) Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol 158: 451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y, Wang R, Shi X, Zhou X, Wang GL (2016) A layered defense strategy mediated by rice E3 ubiquitin ligases against diverse pathogens. Mol Plant 9: 1096–1098 [DOI] [PubMed] [Google Scholar]
- Park CH, Chen S, Shirsekar G, Zhou B, Khang CH, Songkumarn P, Afzal AJ, Ning Y, Wang R, Bellizzi M, et al. (2012) The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24: 4748–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xu X, Wang G-L, Ning Y (2022) Ubiquitination of susceptibility proteins modulates rice broad-spectrum resistance. Trends Plant Sci 27: 322–324 [DOI] [PubMed] [Google Scholar]
- Xu FQ, Xue HW (2019) The ubiquitin-proteasome system in plant responses to environments. Plant Cell Environ 42: 2931–2944 [DOI] [PubMed] [Google Scholar]
- Ye N, Zhu G, Liu Y, Li Y, Zhang J (2011) ABA controls H2O2 accumulation through the induction of OsCATB in rice leaves under water stress. Plant Cell Physiol 52: 689–698 [DOI] [PubMed] [Google Scholar]
- Yuan HM, Liu WC, Lu YT (2017) CATALASE2 coordinates SA-mediated repression of both auxin accumulation and JA biosynthesis in plant defenses. Cell Host Microbe 21: 143–155 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Xu Y, Xie Z, Li X, He ZH, Peng XX (2016) Association-dissociation of glycolate oxidase with catalase in rice: a potential switch to modulate intracellular H2O2 levels. Mol Plant 9: 737–748 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.