Abstract
Karrikins (KARs) are chemicals in smoke that can enhance germination of many plants. Lettuce (Lactuca sativa) cv. Grand Rapids germinates in response to nanomolar karrikinolide (KAR1). Lettuce is much less responsive to KAR2 or a mixture of synthetic strigolactone analogs, rac-GR24. We investigated the molecular basis of selective and sensitive KAR1 perception in lettuce. The lettuce genome contains two copies of KARRIKIN INSENSITIVE2 (KAI2), which in Arabidopsis (Arabidopsis thaliana) encodes a receptor that is required for KAR responses. LsKAI2b is more highly expressed than LsKAI2a in dry achenes and during early stages of imbibition. Through cross-species complementation assays in Arabidopsis, we found that an LsKAI2b transgene confers robust responses to KAR1, but LsKAI2a does not. Therefore, LsKAI2b likely mediates KAR1 responses in lettuce. We compared homology models of KAI2 proteins from lettuce and a fire-follower, whispering bells (Emmenanthe penduliflora). This identified pocket residues 96, 124, 139, and 161 as candidates that influence the ligand specificity of KAI2. Further support for the importance of these residues was found through a broader comparison of pocket residues among 281 KAI2 proteins from 184 asterid species. Almost all KAI2 proteins had either Tyr or Phe identity at position 124. Genes encoding Y124-type KAI2 are more broadly distributed in asterids than in F124-type KAI2. Substitutions at residues 96, 124, 139, and 161 in Arabidopsis KAI2 produced a broad array of responses to KAR1, KAR2, and rac-GR24. This suggests that the diverse ligand preferences observed among KAI2 proteins in plants could have evolved through relatively few mutations.
Plants selectively detect germination stimulants in smoke and other butenolide chemicals as a result of evolutionary changes to the ligand-binding pocket of an enzyme-receptor protein.
Introduction
Plants use several strategies for regeneration in the postfire environment, including regrowth from surviving tissue (e.g. epicormic buds), physical release of seeds (e.g. serotiny), and germination from the soil seed bank. The ecological importance of postfire germination is perhaps best illustrated by fire ephemerals (or pyroendemics), short-lived plants that in some cases emerge only in the first one or two years after a fire. However, more than 1,200 plant species broadly distributed throughout the angiosperms show positive germination responses to aerosol smoke or smoke-water solutions. Searches for germination regulators among the thousands of compounds present in smoke have yielded a number of stimulants such as karrikins (KARs), glyceronitrile, and NO2, as well as inhibitors such as trimethylbutenolide and related furanones (Keeley and Fotheringham, 1997; Flematti et al., 2004; Light et al., 2010; Flematti et al., 2011; Nelson et al., 2012; Burger et al., 2018; Keeley and Pausas, 2018).
KARs are a class of butenolide molecules found in smoke and biochar that are produced by pyrolysis of carbohydrates (Flematti et al., 2004; Kochanek et al., 2016). KARs were discovered through bioassay-guided fractionation of smoke-water. This approach used several species that show sensitive responses to smoke-water, including lettuce (Lactuca sativa, Asterales) cv. Grand Rapids and the Australian fire-followers prickly conostylis (Conostylis aculeata, Commelinales) and queen triggerplant (Stylidium affine, Asterales), as biological readouts for the presence of germination stimulants. Karrikinolide (KAR1), the first karrikin to be identified, enhanced germination of these species at concentrations below 1 nM. (Flematti et al., 2004; van Staden et al., 2004) At least six KARs are found in smoke (Flematti et al., 2009). KAR1 is presumed to be the most potent KAR for many plants (Flematti et al., 2007; Sun et al., 2020). However, Arabidopsis (Arabidopsis thaliana) is more sensitive to KAR2 than KAR1 (Nelson et al., 2009, 2010).
KAR responses in plants are mediated by an ɑ/β-hydrolase protein, KARRIKIN INSENSITIVE2 (KAI2)/HYPOSENSITIVE TO LIGHT (HTL; Waters et al., 2012). Upon activation, KAI2/HTL associates with the F-box protein MORE AXILLARY GROWTH2 (MAX2)/DWARF3 (D3) and a subset of proteins in the SUPPRESSOR OF MAX2 1 (SMAX1)-LIKE (SMXL) family, SMAX1 and SMXL2. MAX2 acts within a Skp1-Cullin-F-box (SCF)-type E3 ubiquitin ligase complex to target SMAX1 and SMXL2 for proteasomal degradation (Stanga et al., 2013, 2016; Khosla et al., 2020; Zheng et al., 2020; Wang et al., 2020b). SMAX1, and presumably SMXL2, associates with TOPLESS (TPL) and TPL-related (TPR) transcriptional co-repressor proteins through an ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motif (Soundappan et al., 2015). Therefore, degradation of SMAX1 and SMXL2 is thought to relieve transcriptional repression and initiate downstream growth responses. At least some responses are mediated by the transcriptional regulators ELONGATED HYPOCOTYL5 (HY5), B-BOX DOMAIN PROTEIN 20 (BBX20), and BBX21 (Bursch et al., 2021). In addition to promoting seed germination, the KAR signaling pathway enhances seedling photomorphogenesis, anthocyanin accumulation, root hair density, root hair elongation, and abiotic stress tolerance; suppresses mesocotyl elongation in the dark; and enables symbiotic interactions with arbuscular mycorrhizal fungi (Nelson et al., 2010; Waters et al., 2012; Gutjahr et al., 2015; Li et al., 2017; Wang et al., 2018; Villaécija-Aguilar et al., 2019; Choi et al., 2020; Li et al., 2020; Shah et al., 2020; Zheng et al., 2020; Carbonnel et al., 2020a; 2020b; Bursch et al., 2021; Meng et al., 2022).
KAI2 is an ancient paralog of the strigolactone (SL) receptor DWARF14 (D14)/DECREASED APICAL DOMINANCE2 (DAD2)/RAMOSUS3 (RMS3) (Hamiaux et al., 2012; de Saint Germain et al., 2016; Yao et al., 2016). SLs are butenolide molecules like KARs, but have altogether different molecular structures, sources, and functions in plants. Canonical SLs consist of a tricyclic ABC ring connected by an enol–ether bond to a butenolide D-ring in the 2′R stereochemical configuration. Noncanonical SLs are similar, but lack cyclized ABC rings (Al-Babili and Bouwmeester, 2015; Yoneyama et al., 2018). SLs are carotenoid-derived plant hormones that regulate axillary bud outgrowth (i.e. shoot branching or tillering), leaf senescence, cambium development, and drought tolerance, among other developmental processes (Gomez-Roldan et al., 2008; Umehara et al., 2008; Agusti et al., 2011; Ueda and Kusaba, 2015; Li et al., 2020). SLs are also exuded into the soil, particularly under low N or P conditions, which promotes beneficial symbiotic interactions with arbuscular mycorrhizal fungi (Akiyama et al., 2005; Al-Babili and Bouwmeester, 2015; Yoneyama et al., 2018). Obligate parasitic plants in the Orobanchaceae, such as witchweeds (Striga spp.) and broomrapes (Orobanche, Phelipanche spp.) have evolved the ability to use very low levels of SLs in the rhizosphere as germination cues that indicate the presence of a potential host (Bouwmeester et al., 2021; Nelson, 2021).
SL signaling is highly similar to KAR signaling. Upon activation by SL, D14 works with SCFMAX2 to target a different subset of SMXL proteins (SMXL6, SMXL7, and SMXL8 in Arabidopsis, or the orthologous DWARF53 protein (D53) in rice (Oryza sativa)) for proteasomal degradation (Jiang et al., 2013; Zhou et al., 2013; Soundappan et al., 2015; Wang et al., 2015; Liang et al., 2016; Yao et al., 2016). In some conditions, D14-SCFMAX2 can also target SMAX1 and SMXL2 (Wang et al., 2020b; Li et al., 2022). Like SMAX1 and SMXL2, DWARF53 and its orthologs have one or more conserved EAR motifs and interact with TPL/TPR proteins (Jiang et al., 2013; Soundappan et al., 2015; Wang et al., 2015). D53 and its orthologs in dicots regulate downstream gene expression, at least in part, through interaction with SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors (Liu et al., 2017; Song et al., 2017; Xie et al., 2020). SMXL6 can also regulate transcription by binding DNA directly (Wang et al., 2020a).
KAI2 proteins in plants collectively perceive a diverse range of signals. In Arabidopsis, KAI2 mediates responses to GR24ent-5DS, a synthetic SL analog that has a D-ring in an unnatural 2′S configuration, as well as KARs (Waters et al., 2012; Scaffidi et al., 2014). There is substantial biochemical evidence that KAI2 can bind KAR1, which in combination with genetic evidence has led to the reasonable conclusion that KAI2 is a KAR receptor (Guo et al., 2013; Kagiyama et al., 2013; Toh et al., 2014; Xu et al., 2016, 2018; Lee et al., 2018; Bürger et al., 2019). However, it now seems more likely that KAI2 perceives an unknown KAR metabolite(s) (Waters et al., 2015; Xu et al., 2018; Khosla et al., 2020; Wang et al., 2020b; Sepulveda et al., 2022). Differential scanning fluorimetry (DSF) assays show that KAI2 undergoes thermal destabilization in vitro in the presence of GR24ent-5DS, but not KAR1 or KAR2 (Waters et al., 2015). Similarly, rac-GR24 or GR24ent-5DS, but not KAR1, promotes protein–protein interactions between KAI2 and MAX2, SMAX1, or SMXL2 (Xu et al., 2018; Khosla et al., 2020; Wang et al., 2020b). Therefore, unmodified KARs probably cannot activate KAI2 directly, but GR24ent-5DS can. Further supporting this idea, a KAI2 protein in pea (Pisum sativum) hydrolyzes GR24ent-5DS through nucleophilic attack by a Ser residue and is covalently modified at its catalytic His residue by the cleaved D-ring (Guercio et al., 2022). This is similar to the mechanism of SL perception by D14 (de Saint Germain et al., 2016; Yao et al., 2016; Chen and Shukla, 2022). The butenolide ring of KARs, however, does not have a cleavable enol–ether or ether linkage to the rest of the molecule. This raises doubt that KARs could activate KAI2 through the same mechanism. Putatively, KAI2 also perceives an undiscovered, endogenous signal in plants known as KAI2 ligand (KL) (Conn and Nelson, 2015). Evidence for KL includes kai2 and max2 mutant phenotypes that are opposite to the effects of KAR treatment, which are not observed in SL biosynthesis or signaling mutants (Nelson et al., 2011; Waters et al., 2012). In addition, highly conserved KAI2 paralogs (KAI2c) in root parasitic plants can rescue an Arabidopsis kai2 mutant but do not respond to KARs, suggesting they sense another signal in plants (Conn et al., 2015; Conn and Nelson, 2015). KAI2 has undergone an atypical degree of gene duplication in the Orobanchaceae (Lamiales), resulting in a parasite-specific clade of fast-evolving, divergent KAI2 paralogs (KAI2d) that can perceive different SLs and, in one case, isothiocyanates (Conn et al., 2015; Toh et al., 2015; Tsuchiya et al., 2015; de Saint Germain et al., 2021). D14 itself is another example of an SL receptor that was derived from KAI2 (Bythell-Douglas et al., 2017). Finally, a KAI2 representative from a grade that has undergone an intermediate level of purifying selection, Striga hermonthica KAI2i (ShKAI2i), confers KAR-specific responses and only weakly rescues Arabidopsis kai2 (Conn et al., 2015; Conn and Nelson, 2015).
These observations raise the question of how different ligand specificities have evolved in KAI2 proteins, enabling some plants to gain beneficial traits such as postfire germination or host-induced germination. We set out to determine the basis of highly sensitive germination responses to KAR in lettuce, which we reasoned might give clues to how some species have adapted to fire-prone ecosystems.
Results
Lettuce achenes are more sensitive to KAR1 than KAR2
We first tested whether lettuce achenes have different sensitivity to KAR1, KAR2, and rac-GR24, a racemic mixture of GR24ent-5DS and its 2′R-configured enantiomer GR245DS (Figure 1). Prior work had suggested that KAR1 is a more effective stimulant of lettuce germination than KAR2, but the compounds were tested in separate experiments, limiting a direct comparison (Flematti et al., 2007). We found that 1-µM KAR1 and KAR2 treatments induced ∼100% germination of lettuce in the dark, compared to ∼40% germination for mock-treated achenes (Figure 1B). 1-µM rac-GR24 also stimulated lettuce germination, but was less effective than either KAR. To determine whether KAR1 or KAR2 is more potent, we evaluated the effects of a range of KAR1 and KAR2 concentrations on lettuce germination. KAR1 induced nearly complete germination at 1 nM and higher concentrations (Figure 1C). In contrast, 1- and 10-nM KAR2 did not enhance germination, and 100-nM KAR2 had an intermediate effect compared to 1-µM KAR2.
Figure 1.
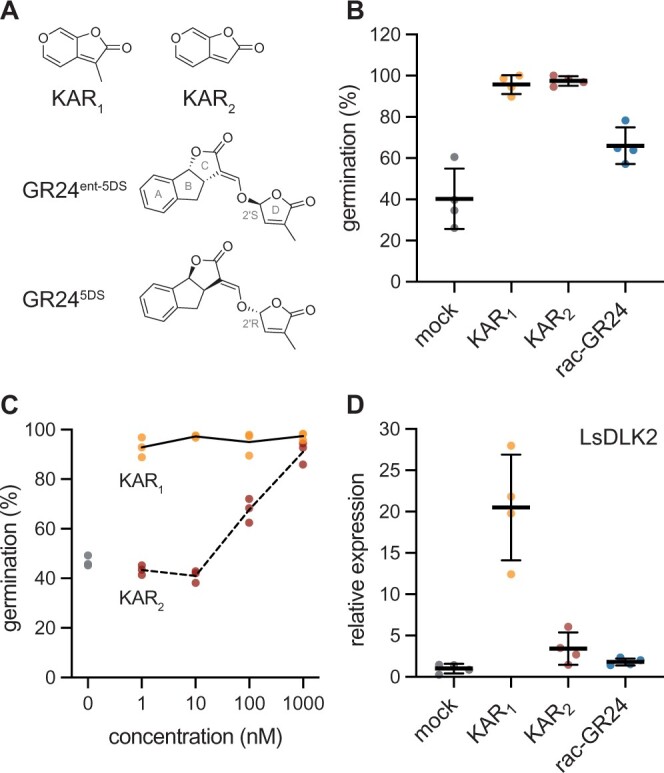
Lettuce achenes are highly sensitive to KAR1. A, Structures of KAR1, KAR2, GR24ent-5DS, and GR245DS. GR245DS is a strigolactone analog that mimics the stereochemical configuration of the SL 5-deoxystrigol. GR24ent-5DS is an enantiomer of GR245DS that has a methylbutenolide D-ring in 2′S configuration, which is not found in natural SLs. B, Lettuce germination in the presence of 0.1% (v/v) acetone or 1-µM KAR1, KAR2, or rac-GR24. Achenes were incubated 1 h in darkness, followed by a pulse of far-red light for 10 min, and the remaining 48 h in darkness at 20°C. n = 4 replicates of ∼50–70 achenes each; mean ± sd. C, Lettuce germination in the presence of a range of KAR1 and KAR2 concentrations after 1 h in darkness, followed by a pulse of far-red light for 10 min, and the remaining 48 h in darkness at 20°C. n = 3 replicates of ∼50–60 achenes each. D) RT-qPCR analysis of LsDLK2 expression relative to LsACT (actin) in lettuce achenes imbibed with 0.1% (v/v) acetone or 1-µM KAR1, KAR2, or rac-GR24 for 24 h in darkness at 20°C. n = 4 pools of achenes; mean ± sd. Values re-scaled to relative LsDLK2 expression in mock-treated achenes.
As an additional test of lettuce achene responses to KAR1, KAR2, and rac-GR24, we examined how each treatment affected the expression of D14-LIKE2 (DLK2). DLK2 is an ancient paralog of KAI2 and D14 that serves as a transcriptional marker of KAR/KL signaling in diverse angiosperms such as A. thaliana, Brassica tournefortii, O. sativa (rice), and Lotus japonicus (Waters et al., 2012; Sun et al., 2016, 2020; Carbonnel et al., 2020b). In achenes imbibed 24 h in the dark with KAR or rac-GR24 treatments, LsDLK2 (Lsat_1_v5_gn_8_94781) transcripts were induced approximately 20-fold by 1-µM KAR1 compared to mock treatment (Figure 1D). LsDLK2 transcripts were increased approximately four-fold by 1-µM KAR2 and approximately two-fold by 1-µM rac-GR24 compared to mock treatment, but these changes were not statistically significant (P > 0.05, Dunnett’s T3 multiple comparisons test). Altogether, these data indicate that lettuce achenes are much more sensitive to KAR1 than KAR2 or rac-GR24.
Lettuce has two KAI2 paralogs that have differential expression in seed
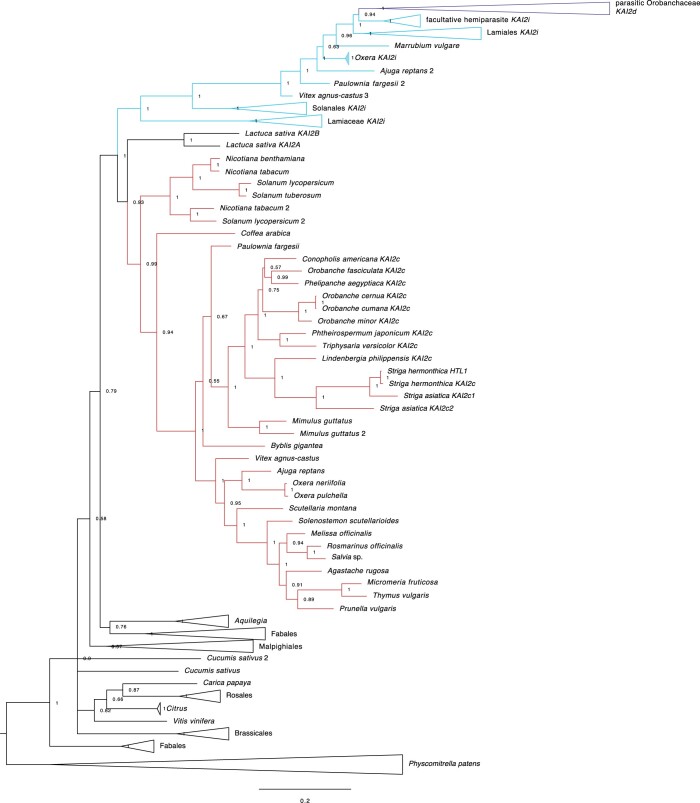
We hypothesized that altered ligand-specificity or -affinity of a KAI2 protein(s) may underlie the high sensitivity to KAR1 observed in lettuce. A BLAST search of the lettuce draft genome revealed two putative KAI2 orthologs that we designated LsKAI2a and LsKAI2b (Supplemental Table S1). We performed a phylogenetic analysis to determine whether either of these genes are related to ShKAI2i, which had previously been associated with selective responses to KAR1 (Conn et al., 2015; Conn and Nelson, 2015). We found that neither LsKAI2 paralog grouped with the lamiid KAI2i grade (Figure 2). Instead, LsKAI2a and LsKAI2b form a monophyletic sister group to the lamiid KAI2c clade, suggesting that they emerged from a gene duplication that occurred after the divergence of the lamiids and the campanulids.
Figure 2.
Lettuce KAI2 genes group with the conserved KAI2c clade. Bayesian phylogeny of KAI2 genes in dicots. Sequences from lamiids fall into the conserved clade (KAI2c), intermediate grade (KAI2i), and divergent clade (KAI2d; parasite-specific) that were previously described (Conn et al., 2015). Scale bar indicates substitutions per site.
We examined whether LsKAI2a and LsKAI2b are expressed at different levels in achenes, as it might highlight one gene as a more likely candidate for mediating KAR1 responses during germination. In dry, unimbibed achenes, LsKAI2b transcripts were approximately five-fold more abundant than LsKAI2a transcripts. LsKAI2b transcript abundance progressively declined after 6 and 24 h of imbibition, whereas LsKAI2a rose slightly after 24 h of imbibition (Figure 3). This suggested that LsKAI2b protein may be more abundant than LsKAI2a during the early stages of imbibition.
Figure 3.
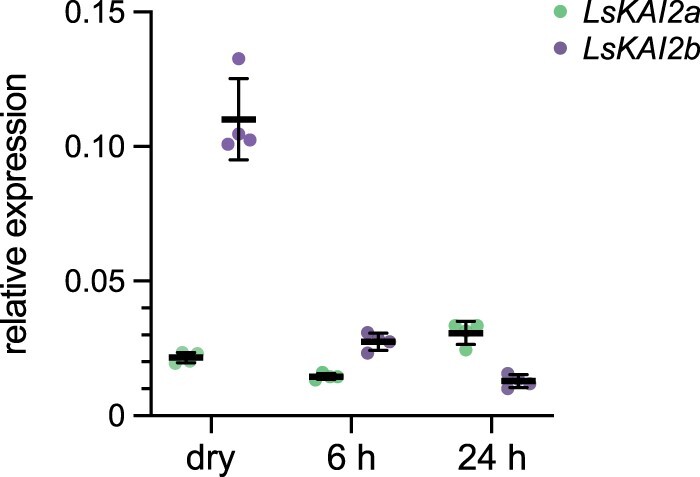
LsKAI2b transcripts are more abundant than LsKAI2a during early imbibition. RT–qPCR analysis of LsKAI2a and LsKAI2b expression relative to LsACT in lettuce achenes that were un-imbibed (dry), or imbibed in water for 1 h in darkness, followed by 10 min in far-red light, and the remaining time to 6 or 24 h in darkness at 20°C. n = 4 pools of achenes; mean ± sd.
LsKAI2b confers KAR1 responses to Arabidopsis seedlings
We then set out to determine whether LsKAI2a and LsKAI2b proteins have different ligand preferences. As discussed above, there is substantial evidence that KAR1 and KAR2 do not activate KAI2 directly. Until KAR metabolites are discovered, in vitro binding studies of KAI2 affinity for KARs may be misleading. Therefore, we used cross-species complementation assays to investigate the in vivo responses of LsKAI2a and LsKAI2b to KARs and rac-GR24. We cloned LsKAI2a and LsKAI2b genes (coding sequence including intron) into a plant transformation vector that drives transgene expression from an A. thaliana KAI2 (AtKAI2) promoter. We generated homozygous transgenic lines for AtKAI2p:LsKAI2A and AtKAI2p:LsKAI2B in an Arabidopsis d14 kai2 background, which does not respond to KARs or rac-GR24. As a control, we tested d14 kai2 lines that are transgenic for an Arabidopsis KAI2 coding sequence.
We tested the inhibitory effects of 1-µM KAR1, KAR2, and rac-GR24 on hypocotyl elongation of seedlings grown under continuous red light (Figure 4). This assay provides a useful alternative to Arabidopsis germination tests, which are often challenging to perform consistently due to variable and labile seed dormancy. As expected, AtKAI2p:AtKAI2 rescued the elongated hypocotyl phenotype of d14 kai2 and restored responses to KAR1, KAR2, and rac-GR24. KAR2 caused a stronger reduction in hypocotyl elongation than KAR1, as observed in wild-type (wt) Col-0 Arabidopsis seedlings. Responses to rac-GR24 were partially reduced compared to wt due to the lack of D14-mediated signaling. The LsKAI2a transgene had mixed effects in different transgenic lines. All lines had reduced hypocotyl length under mock-treated conditions compared to d14 kai2, suggesting rescue of KL response. Only the line with the strongest LsKAI2a expression showed a response to KAR1 or KAR2, and this was weak compared to KAR responses in AtKAI2 transgenic lines (Figure 4;Supplemental Figure S1A). Inhibition of hypocotyl elongation by rac-GR24 was also weak, and was only observed in two transgenic lines. In contrast, LsKAI2b conferred a strong and specific response to KAR1. The degree of hypocotyl growth inhibition by KAR1 in LsKAI2b lines exceeded that observed in wt and AtKAI2 transgenic lines, despite lower levels of LsKAI2b transcripts compared to AtKAI2 (Figure 4;Supplemental Figure S1A). KAR2 did not affect hypocotyl elongation of AtKAI2p:LsKAI2b lines, and rac-GR24 had inconsistent and comparatively weak effects. Hypocotyl elongation under mock-treated conditions was only reduced in one transgenic line, which had the highest LsKAI2b expression. In terms of rescue of the reduced expression of DLK2 in d14 kai2 seedlings, neither LsKAI2a nor LsKAI2b were as effective as AtKAI2 (Supplemental Figure S1B).
Figure 4.
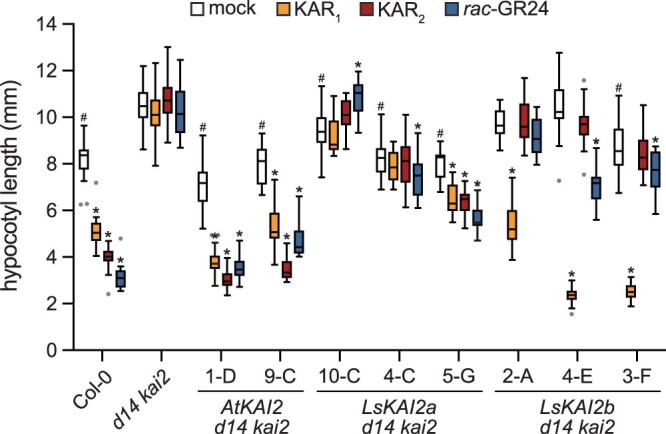
An LsKAI2b transgene confers strong KAR1 responses to Arabidopsis seedlings. Hypocotyl length of 5-day-old A. thaliana seedlings grown under red light on 0.5× MS media supplemented with 0.1% (v/v) acetone or 1-µM KAR1, KAR2, or rac-GR24. n = 20 seedlings. Box plots indicate median and quartiles with Tukey’s whiskers. Gray dots indicate outlier data beyond Tukey’s whiskers. *P < 0.01, Dunnett’s multiple comparisons test, treatment versus mock comparison within each line. #P < 0.01, Dunnett’s multiple comparisons test, comparison to d14 kai2, mock-treated samples only.
We compared the responses of wt and AtKAI2p:LsKAI2b d14 kai2 seedlings to a range of KAR1 concentrations (Supplemental Figure S1C). We found that 100 nM and higher concentrations of KAR1 caused a reduction in hypocotyl elongation of wt seedlings. In an LsKAI2b transgenic line, however, 1-nM KAR1 was sufficient to cause a similar response (Supplemental Figure S1C). In the presence of 1-µM KAR1, hypocotyl elongation was inhibited 78% in the LsKAI2b transgenic line compared to 38% inhibition in wt. As the transcript abundance of LsKAI2b was at least two-fold lower than endogenous KAI2 in wt Arabidopsis, this suggests that LsKAI2b is highly effective at transducing KAR1 responses (Supplemental Figure S1A).
Structural differences in the LsKAI2b pocket may influence KAR1 sensitivity
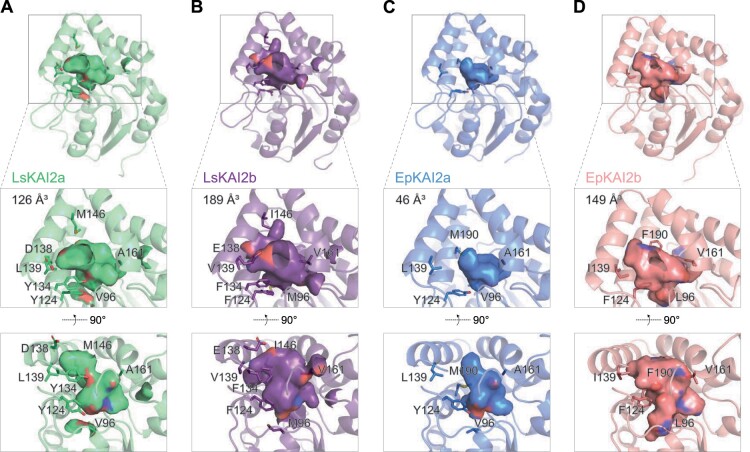
Although LsKAI2a and LsKAI2b proteins share 84% amino acid identity and 94% similarity, LsKAI2b is uniquely able to confer highly sensitive KAR1 responses to A. thaliana. We investigated which amino acid differences might alter the ligand specificity and/or affinity of LsKAI2b. To predict the overall structures and ligand-binding pocket morphologies of LsKAI2a and LsKAI2b, we generated protein structure homology models using Phyre2 with ShKAI2iB (PDB structure 5DNW) as a template (Kelley et al., 2015; Xu et al., 2016). Of the 43 amino acid differences between LsKAI2a and LsKAI2b, 7 are in pocket-defining residues: V/M96, Y/F124, Y/F134, D/E138E, L/V139, M/I146, and A/V161 (Figure 5; for consistency, equivalent AtKAI2 position numbers are used here and below). The differences at these positions are predicted to substantially enlarge the volume of the pocket in LsKAI2b relative to LsKAI2a (Figure 5, A and B). LsKAI2b has a pocket volume of 189 Å3, compared to 126 Å3 for LsKAI2a and 126 Å3 for AtKAI2 (PDB structure 5Z9G) (Lee et al., 2018). Particularly notable differences that affect pocket shape in lettuce KAI2 proteins are found among the residues that surround ɑ-helix 4; these residues are bulkier and more of them are positively charged in LsKAI2a than in LsKAI2b (Figure 5, A and B).
Figure 5.
KAI2b proteins in lettuce and E. penduliflora have enlarged ligand-binding pockets. Homology models of (A) LsKAI2a, (B) LsKAI2b, (C) EpKAI2a, and (D) EpKAI2b. Hydrophobic cavities and their volumes are shown. Pocket residues that differ between KAI2a and KAI2b in each species are indicated.
We hypothesized that similar changes to the volume or chemical properties of the ligand-binding pocket of KAI2 as those found in lettuce might have occurred in other species that have evolved sensitive responses to KAR1. The Californian chaparral species whispering bells (Emmenanthe penduliflora, Boraginales) is a smoke-responsive annual that primarily emerges in postfire sites. Its germination can be triggered by nitric oxides (e.g. NO2) from smoke as well as by <10-nM KAR1 (Keeley and Fotheringham, 1997; Flematti et al., 2004, 2007). To investigate KAI2 evolution in this fire-follower, we generated a de novo transcriptome assembly from RNA extracted from seedlings. We identified two KAI2 coding sequences (Supplemental Table S1). The predicted EpKAI2a and EpKAI2b protein sequences are 74% identical. Among the 70 amino acid differences, 5 are in pocket-defining residues (V/L96, Y/F124, L/I139, A/V161, and M/F190). As with lettuce KAI2 proteins, the differences at these five positions are predicted to substantially enlarge the pocket volume of EpKAI2b compared to EpKAI2a (Figure 5, C and D). The root mean square deviation (RMSD) for LsKAI2a and EpKAI2a models is 0.093 Å (Supplemental Figure S2). In contrast, comparisons of LsKAI2b and LsKAI2a, EpKAI2b and EpKAI2a, and LsKAI2b and EpKAI2b indicate larger RMSD values ranging from 0.42 to 0.63 Å (Supplemental Figure S2). These models suggest that EpKAI2b is a more likely candidate for a KAR1-specific receptor than EpKAI2a, which seems likely to have ligand preferences that are similar to LsKAI2a. Notably, four of the pocket-defining positions that distinguish EpKAI2a and EpKAI2b overlapped with those that distinguish LsKAI2a and LsKAI2b; specifically, positions 96, 124, 139, and 161.
Conserved pocket residue changes among two major groups of asterid KAI2 paralogs
We also compared the amino acid sequences of the parasitic plant proteins Phelipanche aegyptiaca KAI2c (PaKAI2c) and S. hermonthica KAI2c (ShKAI2c) to ShKAI2i, which confers sensitive responses to KAR1 to Arabidopsis (Conn and Nelson, 2015; Conn et al., 2015; Toh et al., 2015). Among the eight total positions that distinguish KAI2a and KAI2b pockets in either lettuce or E. penduliflora, seven substitutions (V96L, Y124F, E138D, L139V, M146I, C161V, and A/L190F) were observed in ShKAI2i relative to PaKAI2c and ShKAI2c (Supplemental Figure S3). This revealed several pocket residue substitutions in KAI2b/KAI2i proteins that were consistent in lettuce, E. penduliflora, and S. hermonthica: V96L/M, Y124F, L139V/I, and A/C161V. If EpKAI2b is selectively responsive to KAR1 like LsKAI2b and ShKAI2i, we hypothesized that these shared changes may cause their KAR1 specificity.
To investigate whether similar differences occur at these positions among KAI2 paralogs in other asterids, we performed an in-depth examination of predicted KAI2 protein sequences in de novo transcriptome assemblies that had been generated by the One Thousand Plants (1 KP) consortium (One Thousand Plant Transcriptomes Initiative, 2019). Using reciprocal BLAST searches of the 1 KP database, we identified 281 KAI2 predicted protein sequences from 184 asterid species (Supplemental Table S2). As suggested by the Y124F substitution that was shared by LsKAI2b, EpKAI2b, and ShKAI2i, we found that asterid KAI2 proteins could be split into two major groups based on Tyr or Phe identity at position 124. Only 4 of the 281 KAI2 predicted proteins (1.4%) were atypical, having neither Y124 nor F124 residues.
Almost all species (169 of 184, 91.8%) surveyed had at least one Y124-type KAI2 paralog. In contrast, F124-type KAI2 paralogs were found in less than half of the species (84 of 184, 45.7%; Figure 6; Supplemental Table S3). For 101 species, only one copy of KAI2 was identified; for 86 of these species (85.1%), the encoded protein was Y124 type and for 14 species (13.9%) it was F124-type. Although some KAI2 genes are likely to be missing in the de novo transcriptome assemblies (e.g. due to inadequate sequencing depth, noncomprehensive sampling of tissues for RNA, or assembly errors), the disparity in these distributions suggests that plants require Y124-type KAI2 proteins, while F124-type KAI2 proteins may have more auxiliary functions. Notably, an F124-type KAI2 was not observed in any of the 36 Asterales transcriptomes surveyed (Figure 6A). This suggested that the emergence of an F124-type KAI2 (i.e. LsKAI2b) in lettuce may have occurred independently within the Asterales lineage. In contrast, the presence of an F124-type KAI2 (i.e. EpKAI2b) in E. penduliflora is not unusual for the Boraginales.
Figure 6.
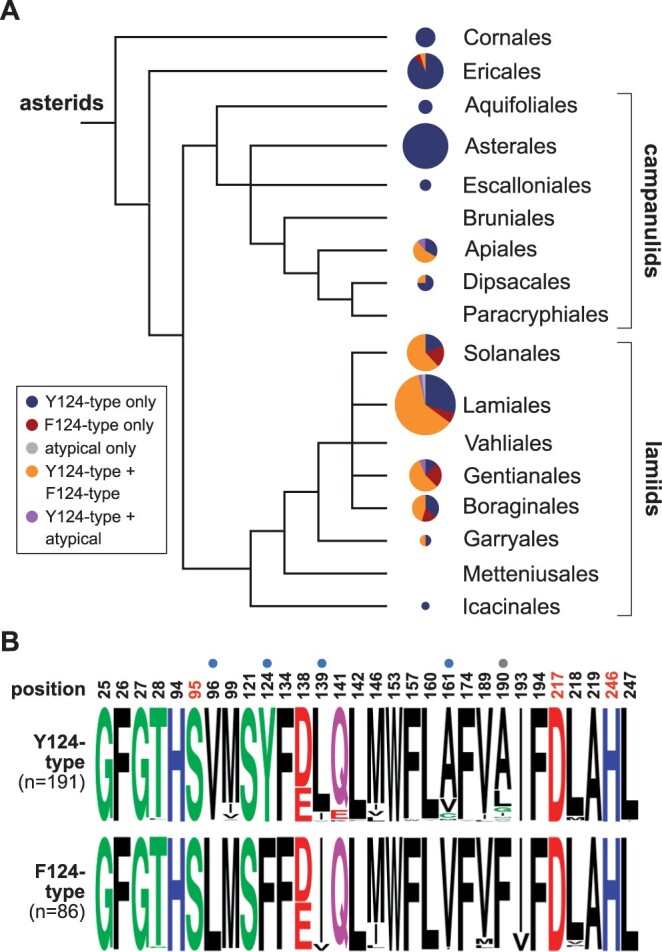
Two groups of asterid KAI2 proteins have conserved differences at five pocket positions. A, Distribution of KAI2 types in asterids. Phylogeny adapted from angiosperm phylogeny group IV system (Chase et al., 2016). Pie charts indicate the proportion of species for which only Y124-type KAI2, only F124-type KAI2, only atypical (neither Y124-type nor F124-type) KAI2, both Y124-type and F124-type KAI2, or both Y124-type and atypical KAI2 coding sequences were observed in de novo transcriptome assemblies from 1KP. The area of each pie chart is proportional to the number of species that were sampled from each order, from n = 1 for Icacinales to n = 57 for Lamiales. B, Frequency plots of amino acid composition in 277 asterid KAI2 proteins at 30 positions that form the ligand-binding pocket. Asterid KAI2 proteins were split into two groups based upon Tyr or Phe amino acid identity at position 124. Dots above residues indicate candidates for ligand specificity-determining residues based upon amino acid conservation within and across the two groups. Position 190 was de-prioritized because LsKAI2b does not have a Phe190 residue but is sensitive to KAR1 nonetheless.
We identified 30 positions that define the ligand-binding pocket of KAI2. We then examined amino acid conservation at these positions among Y124-type and F124-type KAI2 proteins in asterids. A high degree of conservation was observed within and across the two groups at 25 positions (Figure 6B). However, four positions in addition to 124 were well-conserved within each group and different between the groups. Positions 96, 124, 139, 161, and 190 stood out as candidates that might cause differences in ligand specificity between the KAI2 groups. This broader analysis gave us reason to exclude positions 134, 138, and 146, which had been identified in LsKAI2a and LsKAI2b comparisons, from further consideration, as similar amino acid compositions were found among the two KAI2 groups at these positions. Position 190 was also de-prioritized, as LsKAI2b does not share the F190 identity of other F124-type KAI2, but nonetheless confers sensitive KAR1 responses (Supplemental Figure S3).
The impact of four pocket residues on ligand specificity of KAI2
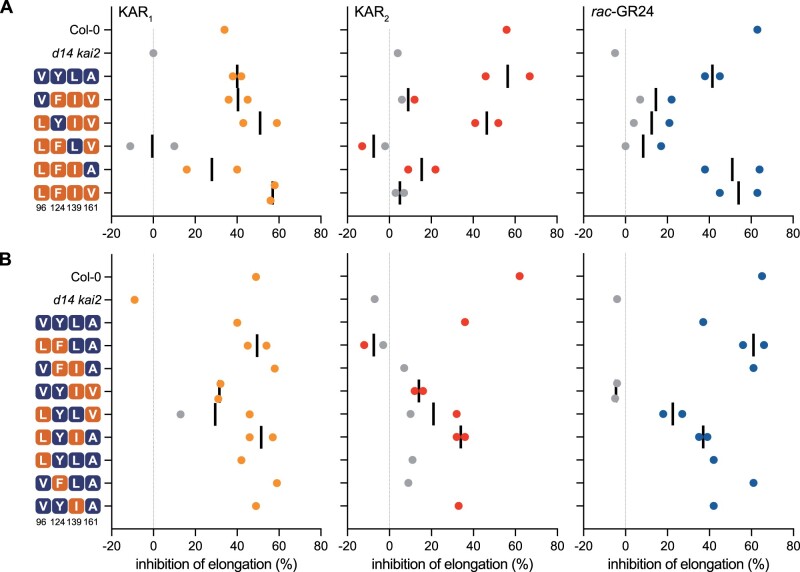
To investigate whether positions 96, 124, 139, and 161 influence ligand specificity, we generated a series of substitutions in AtKAI2 proteins. AtKAI2 shares V96, Y124, L139, and A161 identities with LsKAI2a, EpKAI2a, and most asterid Y124-type KAI2 proteins. We mimicked asterid F124-type KAI2 proteins at these positions by creating quadruple and triple mutant combinations of V96L, Y124F, L139I, and A161V substitutions in AtKAI2. (The variants are annotated here in superscripts by amino acid identities at positions 96, 124, 139, and 161, respectively, with non-AtKAI2 identities underlined.) The AtKAI2 variants were introduced into the Arabidopsis d14 kai2 background and homozygous transgenic lines were tested for responses to KAR1, KAR2, and rac-GR24. We observed a range of ligand specificities among the variants (Figure 7). AtKAI2VFIV was KAR1 specific, AtKAI2LYIV was KAR1- and KAR2 specific, AtKAI2LFIA showed reduced responses to KAR1 and KAR2, and the quadruple mutant AtKAI2LFIV was KAR1- and rac-GR24 specific (Figure 7A;Supplemental Figure S4). Interestingly, AtKAI2LFIV conferred a stronger response to KAR1 than triple-substituted variants or wt AtKAI2. AtKAI2LFLV may be KL specific, as it did not confer consistent responses to any of the treatments but did rescue hypocotyl elongation under mock-treated conditions.
Figure 7.
Pocket residues at positions 96, 124, 139, and 161 affect ligand specificity of AtKAI2. Inhibition of hypocotyl elongation by KAR1, KAR2, or rac-GR24 in 5-day-old seedlings grown in red light for transgenic lines of AtKAI2 variants with (A) quadruple- and triple-, or (B) double- and single substitutions. All transgenic lines are in the d14 kai2 double mutant background. Data are derived from hypocotyl length measurements shown in Supplemental Figure S4. Each data point represents growth inhibition for a unique genetic line. Gray points indicate data that were not significantly different from mock-treated controls for each transgenic line.
We attempted to establish a relationship between specific amino acid changes in KAI2 and ligand specificity. We noted that KAR2 response was highly reduced or absent in AtKAI2VFIV, AtKAI2LFIA, and AtKAI2LFIV lines (Figure 7A;Supplemental Figure S4A). This suggested that Y124F, L139I, or a combination of both substitutions abolishes KAR2 perception. Responses to rac-GR24 were highly reduced in AtKAI2VFIV and AtKAI2LYIV, which share L139I and A161V substitutions. This suggested that these positions may be relevant to rac-GR24 perception. However, it must be noted that AtKAI2LFIV also has L139I and A161V substitutions but remained responsive to rac-GR24 (Figure 7A;Supplemental Figure S4A). The shortest hypocotyls under mock-treated conditions, which may indicate KL responsiveness, were observed in AtKAI2LFIV lines (Supplemental Figure S4A).
To investigate these hypotheses further, we examined the effects of three of the four single substitutions and five of the six possible double substitution combinations at positions 96, 124, 139, and 161 (Figure 7B;Supplemental Figure S4B). Among the double-substituted mutants, AtKAI2LFLA and AtKAI2VFIA had similar effects; KAR2 response was lost, while KAR1 and rac-GR24 response remained. AtKAI2VYIV lost responses to rac-GR24 and had reduced responses to KAR2 and, to a lesser extent, KAR1. AtKAI2LYLV showed similar responses to KARs as AtKAI2VYIV, but was not as strongly affected in its rac-GR24 response. AtKAI2LYIA conferred similar responses to wt AtKAI2. Among the single-substituted mutants, AtKAI2LYLA had highly reduced responses to KAR2 and wt responses to KAR1 and rac-GR24. AtKAI2VFLA also showed highly reduced responses to KAR2, but had stronger responses to KAR1 and rac-GR24 than wt AtKAI2. AtKAI2VYIA had wt responses to KAR1, KAR2, and rac-GR24 (Figure 7B;Supplemental Figure S4B). AtKAI2LFLA seedlings showed the weakest rescue of hypocotyl elongation under mock-treated conditions, suggesting that KL perception may have been reduced more than in other variants (Supplemental Figure S4B).
From these data, we conclude that Y124F is sufficient to reduce KAR2 response. Indeed, every AtKAI2 transgene encoding a Y124F substitution conferred little or no response to KAR2. V96L also reduced KAR2 response as a single substitution, but did not have a consistent effect when combined with other substitutions. The effects of L139I and A161V substitutions were harder to decipher, but it is notable that five of the six AtKAI2 variants with A161V had the weakest responses to rac-GR24.
Discussion
We identified two receptors, LsKAI2a and LsKAI2b, encoded in the lettuce genome that might activate germination in the presence of nanomolar KAR1. We propose that LsKAI2b is responsible for germination responses to KAR1 in lettuce because (1) it is expressed more abundantly in achenes during early stages of imbibition than LsKAI2a and (2) it confers sensitive and specific responses to KAR1 when expressed in A. thaliana, while LsKAI2a does not. In the future, isolating loss-of-function mutations of these genes would be an ideal approach to conclusively determine their roles in KAR1-induced germination of lettuce. Interestingly, we found that a KAR1-responsive fire-follower, E. penduliflora, also encodes two KAI2 proteins in its genome. EpKAI2a is similar to LsKAI2a in terms of the predicted volume and morphology of its ligand-binding pocket. By comparison, LsKAI2b and EpKAI2b are predicted to have substantially enlarged pockets and share similar amino acid substitutions at a small set of pocket positions. We hypothesize that EpKAI2b enables KAR1 perception during seed germination of E. penduliflora, but this will require further investigation.
We investigated how LsKAI2b mediates sensitive and specific responses to KAR1. By comparing the amino acid sequences of known (i.e. LsKAI2b and ShKAI2i) and putative (i.e. EpKAI2b) KAR1-responsive proteins to evolutionarily conserved paralogs from the same species that do not confer KAR1 responses (i.e. LsKAI2a, ShKAI2c, PaKAI2c, and putatively EpKAI2a), we surmised that the pocket residues at positions 96, 124, 139, and 161 may influence ligand specificity. We performed a broader comparison of KAI2 sequences from 184 asterids, including 158 eu-asterid (lamiids and campanulids) species and 26 outgroup species from the Ericales and Cornales. We found that the vast majority of sequences (98.6%) fall into two groups that are distinguished by the residue identity at position 124. The 158 eu-asterid species comprise 108 lamiids; of these, 94 (87.0%) have a Y124-type KAI2, while 76 (70.4%) have an F124-type KAI2. Of 50 campanulid species, all have a Y124-type KAI2, but only 6 (12.0%) have an F124-type KAI2. Similarly, 25 of 26 (96.2%) outgroup species are represented in the Y124 group, but only two outgroup species (7.7%) are represented in the F124 group. Amino acid identities at positions 96, 139, 161, and 190 were also well-conserved within each sequence type, but different across the sequence types. Thus, consensus combinations of V96, Y124, L139, A161, and A190 or L96, F124, I139, V161, and F190 are observed among asterid KAI2 proteins (Figure 6). The residues at these positions may have co-evolved. We do not yet know whether these combinations of amino acids evolved convergently among KAI2 paralogs in distinct lineages, or whether F124-type KAI2 paralogs were lost on many occasions within the asterids. However, the unequal representation of lamiid, campanulid, and outgroup species in the F124 group is suggestive of one KAI2 duplication shared among lamiids, with independent, smaller-scale duplications in the campanulids and Ericales.
We must be clear that the emergence of the two major types of KAI2 has not necessarily been driven by selection to respond to different KARs. KAR responses seem unlikely to be relevant to the ecology of many species that currently grow in nonfire-prone environments or do not follow fire, although environmental differences that were present during prior periods of plant evolution should not be discounted as potential selective forces. Regardless, we consider that KARs are useful as chemical probes that reveal differences in ligand specificity among KAI2 proteins, much in the same way that artificial ligands such as GR24 and other synthetic butenolide compounds have been used. Why then do so many species have multiple KAI2 paralogs with putatively different ligand preferences? One attractive possibility is that there is more than one endogenous KAI2 ligand (Carbonnel et al., 2020b). This could lead to the evolution of specialized perception of different KL molecules among paralogous KAI2 proteins. Relatedly, it is possible that some KAI2 proteins recognize external signals from the rhizosphere, perhaps produced by arbuscular mycorrhizal fungi or other soil microbes that are different from endogenous KL. Selection to ignore harmful allelochemicals could also drive diversification of KAI2 (Carbonnel et al., 2020b).
Pocket residues that influence ligand specificity of KAI2
Many studies have used structural comparisons, molecular dynamics modeling, and in vitro biochemical assays to identify residues that may influence the ligand specificities or affinities of KAI2 proteins, particularly with regard to SL perception (Nelson, 2021). Four recent studies, however, have examined how specific residues affect ligand preferences of KAI2 in vivo. Unlike its relative in the Brassicaceae, A. thaliana, the invasive, smoke-responsive species B. tournefortii is more sensitive to KAR1 than KAR2. Three KAI2 genes are present in the B. tournefortii genome, only two of which appear to encode functional proteins (Sun et al., 2020). BtKAI2b is the most highly expressed KAI2 paralog in seeds and seedlings. A BtKAI2b transgene confers stronger responses to KAR1 than KAR2 when expressed in Arabidopsis kai2, whereas BtKAI2a confers stronger or similar responses to KAR2 than KAR1. Swapping amino acid identities of BtKAI2a and BtKAI2b at positions 96 and 189 (per numbering in this study) switches their KAR preference in vivo and ability to respond to GR24ent-5DS in vitro. Of the two residues, position 96 is primarily responsible for determining KAR1 versus KAR2 preference. BtKAI2c has an unusual R96 residue that makes the protein unstable (Sun et al., 2020). Its orthologs in other Brassica spp., which have a combination of L96, F124, L139, and V161 residues, might mediate KL-specific responses similar to AtKAI2LFLV (Supplemental Figure S4). If so, perhaps the loss of BtKAI2c activity made B. tournefortii germination more dependent on external cues.
The legume L. japonicus has two KAI2 paralogs that show different ligand specificities. LjKAI2a responds similarly to KAR1 and KAR2, and responds better to GR24ent-5DS than GR245DS (Carbonnel et al., 2020b). In contrast, LjKAI2b responds to KAR1, has very little response to KAR2, and does not respond to either GR245DS or GR24ent-5DS. Pocket residues at positions 157, 160, 190, and 218 differ between LjKAI2a and LjKAI2b. An unusual Trp substitution for Phe at position 157 is primarily responsible for the reduced responses to GR24ent-5DS in LjKAI2b, although positions 160 and 190 may also contribute to a minor degree. The basis for different KAR responses has not yet been explored, or the role of position 218 (Carbonnel et al., 2020b). It is notable that LjKAI2a and LjKAI2b share V96, Y124, L139, A161 identities, and, therefore, would not have been anticipated to have different ligand specificities based upon our analysis.
Another legume, P. sativum (pea), also has two KAI2 paralogs. PsKAI2A protein appears to be KL responsive, while PsKAI2B is not (Guercio et al., 2022). Conversely, PsKAI2A does not respond to GR24ent-5DS, but PsKAI2B responds well in in vitro and in vivo assays. Several residue differences between these proteins alter the ligand-binding pocket morphology. Positions 160, 190, and 218 were particularly strong candidates for affecting the volume and solvent accessibility of the pocket. Changing PsKAI2B at positions 160 and 190 to PsKAI2A amino acid identities reduces its response to GR24ent-5DS in DSF assays. Reciprocal swaps at these positions in PsKAI2A did not increase GR24ent-5DS response, however, implying other amino acids also influence GR24ent-5DS perception (Guercio et al., 2022). Although we focused on different residues as potential determinants of KAI2 specificity, it is notable that in our experiments mutations in the adjacent position 161 of AtKAI2 were associated with reduced responses to rac-GR24 in vivo (Figure 7).
A fourth study examined the basis of SL perception by KAI2d proteins from parasitic plants. A KAI2d paralog from S. hermonthica, ShHTL7, confers exceptionally sensitive germination responses to SL when expressed in Arabidopsis (Toh et al., 2015). Ninety-two variants of AtKAI2 were analyzed that had single, double, or triple substitutions for ShHTL7 residues at pocket positions 26, 124, 142, 153, 157, 174, 190, and 194 (Arellano-Saab et al., 2021). The variant proteins that showed the strongest yeast two-hybrid interactions with a MAX2 fragment in the presence of rac-GR24 tended to have substitutions at positions 124, 157, or 190. In transgenic Arabidopsis lines, however, the strongest germination response to rac-GR24 was conferred by a variant with W153L, F157T, and G190T substitutions. This variant gained responsiveness to GR245DS, which has the 2′R stereochemical configuration of natural SLs, while retaining responses to KAR2 and putatively KL (Arellano-Saab et al., 2021).
It seems likely that multiple combinations of pocket residues can produce similar ligand specificities in KAI2 proteins. For example, the KAR1-responsive proteins ShKAI2i and LsKAI2b differ from the consensus for F124-type KAI2 at position 139 (both are Val), and LsKAI2b also has M96 and A190 residues. We found that position 124 is an important determinant of KAR2 responsiveness, but others showed that position 96 influences this (Sun et al., 2020). This poses challenges for forming a predictive model of ligand specificity based upon KAI2 amino acid sequences alone. It also implies that there are multiple evolutionary paths to produce convergent outcomes for KAI2 signaling. Several positions in KAI2 may be hotspots for the diversification of ligand preferences. In particular, residues at positions 96, 124, 157, 160/161, and 189/190 have been implicated in ligand selectivity by multiple studies (Sun et al., 2020; Carbonnel et al., 2020b; Arellano-Saab et al., 2021; Guercio et al., 2022).
Regardless of how KAI2 evolved to sense different compounds, potential benefits to agriculture may be achieved by understanding how to engineer the ligand preferences of KAI2. KAI2 variants could be introduced to crops as transgenes, or endogenous KAI2 genes could be edited in situ through CRISPR (clustered regularly interspaced short palindromic repeats)–Cas9-based technologies. This could then allow selective activation of KAI2 signaling, which controls diverse traits, through application of a synthetic KAI2 agonist. We were not able to fully recreate the ligand selectivity of LsKAI2b through modification of AtKAI2. Although we succeeded in removing KAR2 response while maintaining or enhancing KAR1 response, rac-GR24 response remained intact. One possible cause is that we performed substitutions at positions 96, 124, 139, and 161 with the consensus residues for F124-type KAI2 proteins in asterids, which differ slightly from LsKAI2b identities.
Evolution of smoke-induced germination
Several changes can be imagined to lead to the KAR1-dependent germination response observed in some fire-following species. First, a KAR1 receptor may evolve more robust signaling activity. This could occur through an increase in affinity for KAR1 (or rather, a presumed KAR1 metabolite). Alternatively, enhanced affinity of the receptor for its signaling partners upon activation may increase signal transduction. This appears to be the case for ShHTL7 (Wang et al., 2021). Although ShHTL7 transgenic lines respond to picomolar SL, the micromolar affinity of ShHTL7 for SL in vitro is comparable to other KAI2/HTL proteins in S. hermonthica (Toh et al., 2015; Tsuchiya et al., 2015; Wang et al., 2021). In contrast to the other KAI2/HTL proteins, however, ShHTL7 shows unusually high affinity for MAX2, which can be attributed to differences at five or fewer amino acids (Wang et al., 2021). It is unknown if ShHTL7 also has higher affinity for SMAX1 than other KAI2/HTL proteins. Second, increased expression of a KAR1-responsive receptor in seed may enable better germination responses to KAR1. As an example, we observed that LsKAI2b is more highly expressed than LsKAI2a in lettuce achenes (Figure 3). Similarly, the KAR1-preferring receptor BtKAI2b is more highly expressed in B. tournefourtii seed than other KAI2 paralogs (Sun et al., 2020). Third, if KARs are metabolized in vivo as hypothesized, increased expression or activity of an enzyme(s) involved in that process could increase the availability of bioactive signals that activate KAI2. The F-box protein KARRIKIN UPREGULATED F-BOX1 (KUF1) putatively inhibits conversion of KAR1 into an active ligand for KAI2 as well as biosynthesis of KL (Sepulveda et al., 2022). It may be that in some plants altered expression of KUF1 or its downstream target(s) enhances sensitivity to KAR1. Fourth, an increase in physiological dormancy may be required to make seed germination more strictly dependent upon KAI2 signaling. One way this might occur is through downregulation of gibberellin biosynthesis or signaling. Arabidopsis germination typically requires gibberellins, which counteract the dormancy-promoting effects of abscisic acid. However, loss of SMAX1 protein through mutation or KAI2-SCFMAX2 activity can bypass this requirement (Bunsick et al., 2020).
Materials and methods
Materials and plant propagation
KAR1, KAR2, and rac-GR24 were synthesized and provided by Dr. Gavin Flematti and Dr. Adrian Scaffidi (University of Western Australia). Oligonucleotide primer sequences are described in Supplemental Table S4. Lettuce (L. sativa) cv. Grand Rapids achenes were sourced from a commercial supplier (185C, Stokes Seeds, Holland, MI, USA). The Arabidopsis (A. thaliana) double mutant line d14 htl-3 (here referred to as d14 kai2) was kindly provided by Dr. Peter McCourt (University of Toronto) and is previously described (Toh et al., 2014). Arabidopsis and whispering bells (E. penduliflora) plants were propagated in Sungro Professional Growing Mix under white light (∼110 µmol m−2 s−1; MaxLite LED T8 16.5W 4,000 k light-emitting diode bulbs) with 16-h light/8-h dark photoperiod at ∼21–24°C. Soil was supplemented with Gnatrol WDG (Valent Biosciences, Libertyville, IL, USA), Marathon (imidacloprid), and Osmocote 14–14–14 fertilizer.
Functional analysis of KAI2 genes from lettuce
The two KAI2 sequences from lettuce were obtained from the Lettuce Genome Resource (Reyes-Chin-Wo et al., 2017) through a TBLASTN search of the L. sativa genome (version 4) using the A. thaliana KAI2 (AtKAI2) protein sequence as a query. Each predicted protein sequence from lettuce was then used as a query in a reciprocal TBLASTN search of A. thaliana (Araport 11) transcripts (Berardini et al., 2015) to identify likely AtKAI2 orthologs. Lsat_1_v4_lg_4_361560640.361561729 was designated LsKAI2a and Lsat_1_v4_lg_4_361607540.361608739 was designated LsKAI2b. Each KAI2 paralog was amplified from lettuce genomic DNA using primers with Gateway attB adapters, and Gateway cloning (Thermo Fisher Scientific, Waltham, MA, USA) was used to shuttle each paralog via an entry vector into a plant binary destination vector (pKAI2pro-GW) that expresses genes under the control of the Arabidopsis KAI2 promoter (Waters et al., 2015). Constructs were transformed into Agrobacterium tumefaciens (GV3101 pMP90), and floral dip transformation of A. thaliana was performed in 5% sucrose (w/v) with 0.025% (v/v) Silwet-77. Germination and hypocotyl assays were performed in a HiPoint DCI-700 LED Z4 growth chamber (Taiwan HiPoint Corporation, Kaohsiung City, Taiwan).
Lettuce germination assays
Two layers of Whatman #1 filter paper (7 cm) were soaked with 2.5 mL of a treatment solution in a Petri dish. All aqueous solutions of KAR1, KAR2, and rac-GR24 were freshly prepared from 1,000X stocks in acetone stored at −20°C. Approximately 50 lettuce achenes were plated onto each dish in the dark. Petri dishes were sealed with Parafilm and immediately placed into a growth chamber. Plates were incubated at 20°C for 60 min in dark, 10 min in far-red light (730 nm, 26 µmol m−2 s−1), and 47 h in dark. Germination was indicated by the emergence of a radicle.
Gene expression analysis
Lettuce achenes were plated and light-treated as described for germination assays. Achenes were collected after 6 or 24 h of imbibition and flash-frozen in liquid nitrogen before storage at −80°C. RNA extraction was performed with Spectrum Plant Total RNA Kit (Sigma-Aldrich, St. Louis, MO, USA). DNase I (New England Biolabs, Ipswich, MA, USA) digestion to remove contaminating genomic DNA was performed after RNA extraction. RNA concentrations were measured using a Qubit RNA Broad-Range Assay Kit (Invitrogen, Carlsbad, CA, USA) and fluorometer. First-strand cDNA synthesis was performed with the Verso cDNA Synthesis Kit (Thermo Fisher Scientific) with random hexamer and anchored oligo dT primers. Reverse transcription–quantitative PCR (RT–qPCR) was performed on cDNA with Luna Universal qPCR Mastermix (New England Biolabs) in a CFX384 thermal cycler (Bio-Rad, Hercules, CA, USA). Amplification conditions were 95°C for 3 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min, followed by a melt-curve analysis. Arabidopsis thaliana seedlings were grown as described for hypocotyl elongation assays and harvested at 5 days old. Gene expression analysis was performed as described for lettuce, except an On-Column DNase I Digestion kit (Sigma-Aldrich) was used during RNA extraction.
Phylogenetic analysis of KAI2
The two KAI2 sequences from lettuce were obtained as described above. Additional KAI2 sequences from plant species representing the diversity of dicots, with Physcomitrium (Physcomitrella) patens as an outgroup, were collected from prior publications (Conn et al., 2015; Tsuchiya et al., 2015; Lopez-Obando et al., 2016; Yoshida et al., 2019). In total, 176 sequences from 56 species were combined, aligned, and manually adjusted with respect to predicted amino acid sequence. The nucleotide alignment was trimmed at the 5′- and 3′-ends to minimize gaps and regions of ambiguous alignment. Relative to the AtKAI2 coding sequence, the final alignment retained codons 7–266 and consisted of 819 characters. The alignment was converted to Nexus format using FaBox (Villesen 2007) and used to generate a Bayesian phylogeny in MrBayes version 3.2.5 (Ronquist and Huelsenbeck 2003) (Ronquist and Huelsenbeck, 2003) as previously described (Conn et al., 2015).
Hypocotyl assays
Seeds were surface-sterilized (5 min in 70% (v/v) EtOH with 0.05% (v/v) Triton X-100, followed by 70% (v/v) and 95% (v/v) EtOH washes, and air drying) and plated on 0.5× Murashige–Skoog Medium with MES Buffer and Vitamins (Research Products International), pH 5.7, solidified with 0.8% (w/v) Bacto agar supplemented with 0.1% (v/v) acetone or 1 µM KAR1, KAR2, or rac-GR24. Plates were stratified 3 days in dark at 4°C, then incubated in a growth chamber at 21°C for 3 h in white light (∼150 µmol m−2 s−1), 21-h darkness, and 4-day red light (660 nm, 30 µmol m−2 s−1). Seedlings were photographed and then hypocotyl lengths were measured using ImageJ (NIH).
Structural modeling and analyses
Homology models of LsKAI2A, LsKAI2b, EpKAI2a, and EpKAI2b structures were generated with Phyre2 using ShKAI2iB (PDB structure 5DNW) as a template (Kelley et al., 2015; Xu et al., 2016). Structural illustrations were generated using PyMOL. Pocket volume and solvent accessible surface area were determined via CASTp (Dundas et al., 2006). Residues defining the pocket were broadly identified via CASTp and then probed for position and conservation to identify a final pocket-defining residue list to examine for all species.
Emmenanthe penduliflora transcriptome assembly
RNA was extracted from 7-day-old E. penduliflora seedlings grown in 16:8 photoperiod on moistened filter paper using Spectrum Plant Total RNA kit (Sigma-Aldrich) after removal of seed coats. Library preparation was performed with 1,000 ng of RNA input using NEB Ultra II Directional RNA kit with mRNA isolation. Sequencing was performed on a Nextseq 500 instrument with NextSeq mid-output 2 × 75 kit (paired-end 75 bp reads), producing 38.4 M reads (∼5.8 Gbp). The raw RNA-seq reads are available in NCBI Sequence Read Archive SRR16264938. A de novo transcriptome was assembled from paired-end reads with Trinity 2.6.6 (Grabherr et al., 2011) ran with the “–no_bowtie” parameter. Putative homologs of KAI2 in E. penduliflora were identified by querying AtKAI2 against the transcriptome assembly in a custom BLAST search and validated by reciprocal BLAST. EpKAI2a and EpKAI2b coding sequences are provided in Supplemental Table S1.
Analysis of KAI2 evolution in asterids
A reciprocal BLAST strategy was used to conservatively identify KAI2 orthologs. An AtKAI2 query was used in a BLASTP search of asterids in The 1,000 Plants Project Database (version 5) (https://db.cngb.org/onekp/) (Carpenter et al., 2019; One Thousand Plant Transcriptomes Initiative, 2019). The 1,000 best BLASTP hits were used in reciprocal BLASTP comparisons to Arabidopsis KAI2, D14, and DLK2. About 164 BLAST matches with a match length less than 230 aa were filtered out as incompletely assembled genes or pseudogenes. A total of 21 proteins that had potentially ambiguous orthology based on a difference of less than 50 in BLASTP bit scores between the first and second best hits to Arabidopsis proteins were also removed. Two KAI2 from a “Mydocarpus sp.”, presumably mislabeled, were removed. The remaining 813 asterid proteins were composed of 352 KAI2, 257 D14, and 204 DLK2. Multiple transcriptome assemblies were present for some species. A total of 18 duplicates of KAI2 sequences from the same species were removed, leaving 334 KAI2 from 199 species. The nucleotide sequences corresponding to these 334 protein sequences were obtained through a TBLASTN search of asterids in 1KP. In two cases (Lindenbergia philippensis KAI2i and Oxera neriifolia_3), a sequence was included from Conn et al. (2015) that was not detected through the reciprocal BLAST search reported here. Only four Orobanchaceae were surveyed by 1KP (L. philippensis, Orobanche fasciculata, Epifagus virginiana, and Conopholis americana), only one of which has a KAI2d paralog; this prevented confusion about sequence conservation from the rapidly evolving parasite-specific KAI2d clade (Conn et al., 2015). Thirty-one sequences containing a frameshift mutation or premature stop codon were removed from the dataset. Eleven sequences were removed because a key site was missing or mutated (e.g. start codon was absent when sequence covered the 5′-end of the gene, or a Ser–His–Asp catalytic triad site was not conserved). These 42 sequences may be pseudogenes or have sequencing/assembly errors. In some cases, intron positions were corrected manually based on the presence of GT/AG splice site positions and homology in the amino acid sequence of this region to other sequences. Sequences from the same species were considered likely alleles rather than paralogs if their nucleotide identities were ≥95.0% in regions of alignment, as determined by Expasy SIM (Huang and Miller 1991); in this case, only one sequence was retained. Eleven putative allelic sequences were removed from the dataset. This left 281 KAI2 sequences from 184 species. Because some KAI2 proteins first obtained from BLASTP were later discarded due to problems with their corresponding nucleotide sequences, analyses on site conservation were conducted by translating nucleotide sequences into predicted protein sequences. We analyzed amino acid conservation among 277 predicted KAI2 proteins from 183 species that were classified as Y124- or F124-type based on the presence of Tyr or Phe at site 124. Only 4 proteins in the 281-sequence dataset did not meet this criterion; one is a KAI2d gene, and the other cases may be pseudogenes or incorrectly assembled. Asterid KAI2 protein sequences are provided in Supplemental Table S2. Multiple sequence alignments of KAI2 predicted proteins were performed in MEGA X and frequency plots of consensus sequences at pocket positions were visualized with WebLogo (Crooks et al., 2004; Kumar et al., 2018).
AtKAI2 variant generation and analysis
A binary plant transformation plasmid expressing an AtKAI2 coding sequence under control of an AtKAI2 promoter (pKAI2pro-AtKAI2) was modified (Waters et al., 2015). A set of five oligonucleotides ranging from 41 to 56 nt in length (Supplemental Table S4) were designed to span a central portion of the AtKAI2 sequence encoding amino acids 96, 124, 139, and 161 when ligated end-to-end. Wt and mutant versions of the oligonucleotides were synthesized and phosphorylated with T4 polynucleotide kinase. Different combinations of wt and mutant phosphorylated oligonucleotides were annealed to four bridging oligonucleotides that were each complementary to the ends of two phosphorylated oligonucleotides. T4 DNA ligase was used to join the adjacent phosphorylated oligonucleotides into a continuous single strand of DNA. The new strand was amplified with Phusion high-fidelity DNA polymerase and inserted through Gibson assembly (New England Biolabs) into pKAI2pro-AtKAI2 that had first been linearized by PCR to drop out the central region being swapped and digested with DpnI to remove un-linearized, methylated plasmid from the reaction. Sanger-sequence-verified pKAI2pro-AtKAI2 variant plasmids were introduced into the d14 kai2 mutant background via floral dip transformation. Transformants were selected at the seedling stage with hygromycin. Lines with single T-DNA insertion events were brought to homozygosity and characterized.
Statistical analysis
Statistical analysis was performed in Prism version 9 (GraphPad, La Jolla, CA, USA). Post hoc statistical comparisons were performed after ANOVA or two-way ANOVA. Box plots show the median, 25th percentile, and 75th percentile. Tukey whiskers on box plots extend 1.5 times the interquartile range beyond the 25th/75th percentile up to the minimum/maximum value in the data set. Outlier data beyond Tukey whiskers are shown as individual points.
Accession numbers
Sequence data from this article can be found in NCBI GenBank data libraries under accession numbers: EpKAI2a, ON707005; EpKAI2b, ON707006. Other KAI2 sequences from lettuce and 1KP databases are provided in Supplemental Tables S1 and S2. Emmenanthe penduliflora RNA-seq, NCBI Sequence Read Archive SRR16264938.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transgene expression levels and KAR1 sensitivity in lettuce KAI2 transgenic lines.
Supplemental Figure S2. Comparisons of lettuce and E. penduliflora KAI2 structures within and across species.
Supplemental Figure S3. Sequence comparison of several characterized KAI2 proteins.
Supplemental Figure S4. Hypocotyl elongation responses to KARs and rac-GR24 conferred by AtKAI2 variants.
Supplemental Table S1. KAI2 coding sequences from L. sativa and E. penduliflora.
Supplemental Table S2. Asterid KAI2 protein sequences.
Supplemental Table S3. Summary of KAI2 type distribution in asterids.
Supplemental Table S4. Oligonucleotide primer sequences.
Supplementary Material
Acknowledgments
We thank Dr. Gavin Flematti and Dr. Adrian Scaffidi (University of Western of Australia) for providing KAR1, KAR2, and rac-GR24; Dr. Winslow Briggs (Carnegie Institution for Science) for providing Emmenanthe penduliflora seed; and Elise Landsbergen for extraction of RNA from E. penduliflora. RNA-seq library preparation and sequencing were performed by the Genomics Core at University of California, Riverside (UCR) and de novo transcriptome assembly was performed on the High Performance Computer Cluster at UCR.
Funding
Funding was provided by the National Science Foundation (NSF-IOS 1856741 to D.C.N.; NSF-CAREER 2047396 and NSF-EAGER 2028283 to N.S.; NSF-CAREER 2046256 to D.K.; and NSF Research Traineeship (NRT) Program Grant DGE-1922642 “Plants3D” to S.E.M.) and the United States Department of Agriculture (NIFA Award 2017-38422-27135 to S.E.M.; Hatch project CA-R-BPS-5209-H to D.C.N.; and Hatch project CA-R-BPS-5154-H to D.K.).
Conflict of interest statement. N.S. has an equity interest in OerthBio-LLC and serves on the company’s Scientific Advisory Board.
Responsibilities of the author for contact
It is the responsibility of the author for contact to ensure that all scientists who have contributed substantially to the conception, design or execution of the work described in the manuscript are included as authors, in accordance with the guidelines from the Committee on Publication Ethics (COPE) (http://publicationethics.org/resources/guidelines). It is the responsibility of the author for contact also to ensure that all authors agree to the list of authors and the identified contributions of those authors.
Contributor Information
Stephanie E Martinez, Department of Botany and Plant Sciences, University of California, Riverside, California 92521, USA.
Caitlin E Conn, Department of Biology, Berry College, Mount Berry, Georgia 30149, USA.
Angelica M Guercio, Department of Plant Biology, University of California, Davis, California 95616, USA.
Claudia Sepulveda, Department of Botany and Plant Sciences, University of California, Riverside, California 92521, USA.
Christopher J Fiscus, Department of Botany and Plant Sciences, University of California, Riverside, California 92521, USA.
Daniel Koenig, Department of Botany and Plant Sciences, University of California, Riverside, California 92521, USA.
Nitzan Shabek, Department of Plant Biology, University of California, Davis, California 95616, USA.
David C Nelson, Department of Botany and Plant Sciences, University of California, Riverside, California 92521, USA.
S.E.M., C.E.C., and C.S. cloned lettuce and Arabidopsis variant KAI2 genes, and generated Arabidopsis transgenic lines. S.E.M. analyzed germination and gene expression of lettuce, and phenotyped Arabidopsis transgenic lines. C.E.C. performed phylogenetic analysis of KAI2; A.M.G. generated homology models of KAI2 proteins and performed structural analyses; C.J.F. generated a de novo transcriptome assembly of Emmenanthe penduliflora and identified EpKAI2 genes; D.C.N. conceived the project and wrote the article with contributions from all the authors; D.K., N.S., and D.C.N. secured funding to support the research, provided technical training, and assisted with data interpretation. D.C.N. agrees to serve as the author responsible for contact and distribution of data and resources.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is David C. Nelson (david.nelson@ucr.edu).
References
- Agusti J, Herold S, Schwarz M, Sanchez P, Ljung K, Dun EA, Brewer PB, Beveridge CA, Sieberer T, Sehr EM, et al. (2011) Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc Natl Acad Sci USA 108:20242–20247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki KI, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Al-Babili S, Bouwmeester HJ (2015) Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol 66:161–186 [DOI] [PubMed] [Google Scholar]
- Arellano-Saab A, Bunsick M, Al Galib H, Zhao W, Schuetz S, Bradley JM, Xu Z, Adityani C, Subha A, McKay H, et al. (2021) Three mutations repurpose a plant karrikin receptor to a strigolactone receptor. Proc Natl Acad Sci USA 118: e2103175118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E (2015) The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53:474–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester H, Li C, Thiombiano B, Rahimi M, Dong L (2021) Adaptation of the parasitic plant lifecycle: germination is controlled by essential host signaling molecules. Plant Physiol 185:1292–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunsick M, Toh S, Wong C, Xu Z, Ly G, McErlean CSP, Pescetto G, Nemrish KE, Sung P, Li JD, et al. (2020) SMAX1-dependent seed germination bypasses GA signalling in Arabidopsis and Striga. Nat Plants 6:646–652 [DOI] [PubMed] [Google Scholar]
- Burger BV, Pošta M, Light ME, Kulkarni MG, Viviers MZ, Van Staden J (2018) More butenolides from plant-derived smoke with germination inhibitory activity against karrikinolide. S Afr J Bot 115:256–263 [Google Scholar]
- Bürger M, Mashiguchi K, Lee HJ, Nakano M, Takemoto K, Seto Y, Yamaguchi S, Chory J. ( 2019) Structural basis of karrikin and non-natural strigolactone perception in Physcomitrella patens. Cell Rep 26:855–865.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursch K, Niemann ET, Nelson DC, Johansson H (2021) Karrikins control seedling photomorphogenesis and anthocyanin biosynthesis through a HY5-BBX transcriptional module. Plant J 107:1346–1362 [DOI] [PubMed] [Google Scholar]
- Bythell-Douglas R, Rothfels CJ, Stevenson DWD, Graham SW, Wong GKS, Nelson DC, Bennett T (2017) Evolution of strigolactone receptors by gradual neo-functionalization of KAI2 paralogues. BMC Biol 15:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnel S, Das D, Varshney K, Kolodziej MC, Villaécija-Aguilar JA, Gutjahr C (2020a) The karrikin signaling regulator SMAX1 controls Lotus japonicus root and root hair development by suppressing ethylene biosynthesis. Proc Natl Acad Sci USA 117: 21757–21765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnel S, Torabi S, Griesmann M, Bleek E, Tang Y, Buchka S, Basso V, Shindo M, Boyer F-D, Wang TL, et al. (2020b) Lotus japonicus karrikin receptors display divergent ligand-binding specificities and organ-dependent redundancy. PLoS Genet 16:e1009249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter EJ, Matasci N, Ayyampalayam S, Wu S, Sun J, Yu J, Vieira FR, Bowler C, Dorrell RG, Gitzendanner MA, et al. (2019) Access to RNA-sequencing data from 1,173 plant species: the 1000 Plant transcriptomes initiative (1KP). Gigascience 8: giz126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase MW, Christenhusz MJM, Fay MF (2016) An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J 181:1– 20 [Google Scholar]
- Chen J, Shukla D (2022) Multiple modes of substrate hydrolysis-induced covalent modification of strigolactone receptors. bioRxiv 2022.04.22.489200 [Google Scholar]
- Choi J, Lee T, Cho J, Servante EK, Pucker B, Summers W, Bowden S, Rahimi M, An K, An G, et al. (2020) The negative regulator SMAX1 controls mycorrhizal symbiosis and strigolactone biosynthesis in rice. Nat Commun 11:2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn CE, Bythell-Douglas R, Neumann D, Yoshida S, Whittington B, Westwood JH, Shirasu K, Bond CS, Dyer KA, Nelson DC (2015) PLANT EVOLUTION. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349:540–543 [DOI] [PubMed] [Google Scholar]
- Conn CE, Nelson DC (2015) Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front Plant Sci 6:1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res 34:W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flematti GR, Ghisalberti EL, Dixon KW, Trengove RD (2004) A compound from smoke that promotes seed germination. Science 305:977. [DOI] [PubMed] [Google Scholar]
- Flematti GR, Ghisalberti EL, Dixon KW, Trengove RD (2009) Identification of alkyl substituted 2H-furo[2,3-c]pyran-2-ones as germination stimulants present in smoke. J Agric Food Chem 57:9475–9480 [DOI] [PubMed] [Google Scholar]
- Flematti GR, Goddard-Borger ED, Merritt DJ, Ghisalberti EL, Dixon KW, Trengove RD (2007) Preparation of 2H-furo[2,3-c]pyran-2-one derivatives and evaluation of their germination-promoting activity. J Agric Food Chem 55:2189–2194 [DOI] [PubMed] [Google Scholar]
- Flematti GR, Merritt DJ, Piggott MJ, Trengove RD, Smith SM, Dixon KW, Ghisalberti EL (2011) Burning vegetation produces cyanohydrins that liberate cyanide and stimulate seed germination. Nat Commun 2:360. [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455:189–194 [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guercio AM, Torabi S, Cornu D, Dalmais M, Bendahmane A, Le Signor C, Pillot J-P, Le Bris P, Boyer FD, Rameau C, et al. (2022) Structural and functional analyses explain Pea KAI2 receptor diversity and reveal stereoselective catalysis during signal perception. Commun Biol 5:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Zheng Z, La Clair JJ, Chory J, Noel JP (2013) Smoke-derived karrikin perception by the α/β-hydrolase KAI2 from Arabidopsis. Proc Natl Acad Sci USA 110: 8284–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Gobbato E, Choi J, Riemann M, Johnston MG, Summers W, Carbonnel S, Mansfield C, Yang SY, Nadal M, et al. (2015) Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350:1521–1524 [DOI] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RSM, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC (2012) DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol 22:2032–2036 [DOI] [PubMed] [Google Scholar]
- Huang X, Miller W (1991) A time-efficient, linear-space local similarity algorithm. Adv Appl Mathe 12:337–357 [Google Scholar]
- Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, et al. (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504:401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiyama M, Hirano Y, Mori T, Kim S-Y, Kyozuka J, Seto Y, Yamaguchi S, Hakoshima T (2013) Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18:147–160 [DOI] [PubMed] [Google Scholar]
- Keeley JE, Fotheringham CJ (1997) Trace gas emissions and smoke-induced seed germination. Science 276: 1248–1250 [Google Scholar]
- Keeley JE, Pausas JG (2018) Evolution of “smoke” induced seed germination in pyroendemic plants. South Afr J Bot 115:251–255 [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla A, Morffy N, Li Q, Faure L, Chang SH, Yao J, Zheng J, Cai ML, Stanga JP, Flematti GR, et al. (2020) Structure-function analysis of SMAX1 reveals domains that mediate its karrikin-induced proteolysis and interaction with the receptor KAI2. Plant Cell 32: 2639–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanek J, Long RL, Lisle AT, Flematti GR (2016) Karrikins identified in biochars indicate post-fire chemical cues can influence community diversity and plant development. PLoS One 11: e0161234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Kim K, Lee S, Lee S, Hwang E (2018) Functional analysis of a missense allele of KARRIKIN-INSENSITIVE2 that impairs ligand-binding and downstream signaling in Arabidopsis thaliana. J Exp Bot 69: 3609–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ward S, Li P, Bennett T, Leyser O (2016) SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms. Plant Cell 28:1581–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light ME, Burger BV, Staerk D, Kohout L, Van Staden J (2010) Butenolides from plant-derived smoke: natural plant-growth regulators with antagonistic actions on seed germination. J Nat Prod 73:267–269 [DOI] [PubMed] [Google Scholar]
- Liu J, Cheng X, Liu P, Sun J (2017) miR156-targeted SBP-box transcription factors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiol 174:1931–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Nguyen KH, Chu HD, Van Ha C, Watanabe Y, Osakabe Y, Leyva-González MA, Sato M, Toyooka K, Voges L, et al. (2017) The karrikin receptor KAI2 promotes drought resistance in Arabidopsis thaliana. PLoS Genet 13:e1007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Nguyen KH, Chu HD, Watanabe Y, Osakabe Y, Sato M, Toyooka K, Seo M, Tian L, Tian C, et al. (2020) Comparative functional analyses of DWARF14 and KARRIKIN INSENSITIVE 2 in drought adaptation of Arabidopsis thaliana. Plant J 103:111–127 [DOI] [PubMed] [Google Scholar]
- Li Q, Martín-Fontecha ES, Khosla A, White ARF, Chang S, Cubas P, Nelson DC (2022) The strigolactone receptor D14 targets SMAX1 for degradation in response to GR24 treatment and osmotic stress. Plant Commun 3: 100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Obando M, Conn CE, Hoffmann B, Bythell-Douglas R, Nelson DC, Rameau C, Bonhomme S (2016) Structural modelling and transcriptional responses highlight a clade of PpKAI2-LIKE genes as candidate receptors for strigolactones in Physcomitrella patens. Planta 243:1441–1453 [DOI] [PubMed] [Google Scholar]
- Meng Y, Varshney K, Incze N, Badics E, Kamran M, Davies SF, Oppermann LMF, Magne K, Dalmais M, Bendahmane A, et al. (2022) KARRIKIN INSENSITIVE2 regulates leaf development, root system architecture and arbuscular-mycorrhizal symbiosis in Brachypodium distachyon. Plant J 109:1559–1574 [DOI] [PubMed] [Google Scholar]
- Nelson DC (2021) The mechanism of host-induced germination in root parasitic plants. Plant Physiol 185: 1353–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Flematti GR, Ghisalberti EL, Dixon KW, Smith SM (2012) Regulation of seed germination and seedling growth by chemical signals from burning vegetation. Annu Rev Plant Biol 63: 107–130 [DOI] [PubMed] [Google Scholar]
- Nelson DC, Flematti GR, Riseborough J-A, Ghisalberti EL, Dixon KW, Smith SM (2010) Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc Natl Acad Sci USA 107:7095–7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Riseborough JA, Flematti GR, Stevens J, Ghisalberti EL, Dixon KW, Smith SM (2009) Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol 149: 863–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 108:8897–8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- One Thousand Plant Transcriptomes Initiative (2019) One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574: 679–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Chin-Wo S, Wang Z, Yang X, Kozik A, Arikit S, Song C, Xia L, Froenicke L, Lavelle DO, Truco MJ, et al. (2017) Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat Commun 8:14953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP (2003) MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- de Saint Germain A, Clavé G, Badet-Denisot MA, Pillot JP, Cornu D, Le Caer JP, Burger M, Pelissier F, Retailleau P, Turnbull C, et al. (2016) An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat Chem Biol 12: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A, Jacobs A, Brun G, Pouvreau JB, Braem L, Cornu D, Clavé G, Baudu E, Steinmetz V, Servajean V, et al. (2021) A Phelipanche ramosa KAI2 protein perceives strigolactones and isothiocyanates enzymatically. Plant Commun 2:100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi A, Waters MT, Sun YK, Skelton BW, Dixon KW, Ghisalberti EL, Flematti GR, Smith SM (2014) Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol 165:1221–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda C, Guzmán MA, Li Q, Villaécija-Aguilar JA, Martinez SE, Kamran M, Khosla A, Liu W, Gendron JM, Gutjahr C, et al. (2022) KARRIKIN UP-REGULATED F-BOX 1 (KUF1) imposes negative feedback regulation of karrikin and KAI2 ligand metabolism in Arabidopsis thaliana. Proc Natl Acad Sci USA 119:e2112820119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah FA, Wei X, Wang Q, Liu W, Wang D, Yao Y, Hu H, Chen X, Huang S, Hou J, et al. (2020) Karrikin improves osmotic and salt stress tolerance via the regulation of the redox homeostasis in the oil Plant Sapium sebiferum. Front Plant Sci 11: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Lu Z, Yu H, Shao G, Xiong J, Meng X, Jing Y, Liu G, Xiong G, Duan J, et al. (2017) IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res 27:1128–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan I, Bennett T, Morffy N, Liang Y, Stanga JP, Abbas A, Leyser O, Nelson DC (2015) SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27: 3143–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Staden J, Jäger AK, Light ME, Burger BV, Brown NAC, Thomas TH (2004) Isolation of the major germination cue from plant-derived smoke. S Afr J Bot 70: 654–659 [Google Scholar]
- Stanga JP, Morffy N, Nelson DC (2016) Functional redundancy in the control of seedling growth by the karrikin signaling pathway. Planta 243: 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga JP, Smith SM, Briggs WR, Nelson DC (2013) SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol 163:318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YK, Flematti GR, Smith SM, Waters MT (2016) Reporter gene-facilitated detection of compounds in Arabidopsis leaf extracts that activate the Karrikin signaling pathway. Front Plant Sci 7: 1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YK, Yao J, Scaffidi A, Melville KT, Davies SF, Bond CS, Smith SM, Flematti GR, Waters MT (2020) Divergent receptor proteins confer responses to different karrikins in two ephemeral weeds. Nat Commun 11:1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S, Holbrook-Smith D, Stogios PJ, Onopriyenko O, Lumba S, Tsuchiya Y, Savchenko A, McCourt P (2015) Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350: 203–207 [DOI] [PubMed] [Google Scholar]
- Toh S, Holbrook-Smith D, Stokes ME, Tsuchiya Y, McCourt P (2014) Detection of parasitic plant suicide germination compounds using a high-throughput Arabidopsis HTL/KAI2 strigolactone perception system. Chem Biol 21:988–998 [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Yoshimura M, Sato Y, Kuwata K, Toh S, Holbrook-Smith D, Zhang H, McCourt P, Itami K, Kinoshita T, et al. (2015) PARASITIC PLANTS. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349:864–868 [DOI] [PubMed] [Google Scholar]
- Ueda H, Kusaba M (2015) Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiol 169:138–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455:195–200 [DOI] [PubMed] [Google Scholar]
- Villaécija-Aguilar JA, Hamon-Josse M, Carbonnel S, Kretschmar A, Schmid C, Dawid C, Bennett T, Gutjahr C (2019) SMAX1/SMXL2 regulate root and root hair development downstream of KAI2-mediated signalling in Arabidopsis. PLoS Genet 15:e1008327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villesen P (2007) FaBox: an online tooxbox for fasta sequences. Mol Ecol Notes 7: 965–968 [Google Scholar]
- Wang L, Wang B, Jiang L, Liu X, Li X, Lu Z, Meng X, Wang Y, Smith SM, Li J (2015) Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27:3128–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang B, Yu H, Guo H, Lin T, Kou L, Wang A, Shao N, Ma H, Xiong G, et al. (2020a) Transcriptional regulation of strigolactone signalling in Arabidopsis. Nature 583:277–281 [DOI] [PubMed] [Google Scholar]
- Wang L, Waters MT, Smith SM (2018) Karrikin-KAI2 signalling provides Arabidopsis seeds with tolerance to abiotic stress and inhibits germination under conditions unfavourable to seedling establishment. New Phytol 219:605–618 [DOI] [PubMed] [Google Scholar]
- Wang L, Xu Q, Yu H, Ma H, Li X, Yang J, Chu J, Xie Q, Wang Y, Smith SM, et al. (2020b) Strigolactone and karrikin signaling pathways elicit ubiquitination and proteolysis of SMXL2 to regulate hypocotyl elongation in Arabidopsis. Plant Cell 32:2251–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yao R, Du X, Guo L, Chen L, Xie D, Smith SM (2021) Molecular basis for high ligand sensitivity and selectivity of strigolactone receptors in Striga. Plant Physiol 185:1411–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139:1285–1295 [DOI] [PubMed] [Google Scholar]
- Waters MT, Scaffidi A, Moulin SLY, Sun YK, Flematti GR, Smith SM (2015) A selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27:1925–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Liu Y, Ma M, Zhou Q, Zhao Y, Zhao B, Wang B, Wei H, Wang H (2020) Arabidopsis FHY3 and FAR1 integrate light and strigolactone signaling to regulate branching. Nat Commun 11:1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Miyakawa T, Nakamura H, Nakamura A, Imamura Y, Asami T, Tanokura M (2016) Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica. Sci Rep 6:31386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Miyakawa T, Nosaki S, Nakamura A, Lyu Y, Nakamura H, Ohto U, Ishida H, Shimizu T, Asami T, et al. (2018) Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga. Nat Commun 9:3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Chen L, et al. (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536:469–473 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Yoneyama K, Kisugi T, Nomura T, Nakatani Y, Akiyama K, McErlean CSP (2018) Which are the major players, canonical or non-canonical strigolactones? J Exp Bot 69: 2231–2239 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Kim S, Wafula EK, Tanskanen J, Kim YM, Honaas L, Yang Z, Spallek T, Conn CE, Ichihashi Y, et al. (2019) Genome sequence of striga asiatica provides insight into the evolution of plant parasitism. Curr Biol 29: 3041–3052.e4 [DOI] [PubMed] [Google Scholar]
- Zheng J, Hong K, Zeng L, Wang L, Kang S, Qu M, Dai J, Zou L, Zhu L, Tang Z, et al. (2020) Karrikin signaling acts parallel to and additively with strigolactone signaling to regulate rice mesocotyl elongation in darkness. Plant Cell 32:2780–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, et al. (2013) D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504: 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.