Abstract
Circadian clocks govern temporal programs in the green lineage (Chloroplastida) as they do in other photosynthetic pro- and eukaryotes, bacteria, fungi, animals, and humans. Their physiological properties, including entrainment, phase responses, and temperature compensation, are well conserved. The involvement of transcriptional/translational feedback loops in the oscillatory machinery and reversible phosphorylation events are also maintained. Circadian clocks control a large variety of output rhythms in green algae and terrestrial plants, adjusting their metabolism and behavior to the day-night cycle. The angiosperm Arabidopsis (Arabidopsis thaliana) represents a well-studied circadian clock model. Several molecular components of its oscillatory machinery are conserved in other Chloroplastida, but their functions may differ. Conserved clock components include at least one member of the CIRCADIAN CLOCK ASSOCIATED1/REVEILLE and one of the PSEUDO RESPONSE REGULATOR family. The Arabidopsis evening complex members EARLY FLOWERING3 (ELF3), ELF4, and LUX ARRHYTHMO are found in the moss Physcomitrium patens and in the liverwort Marchantia polymorpha. In the flagellate chlorophyte alga Chlamydomonas reinhardtii, only homologs of ELF4 and LUX (named RHYTHM OF CHLOROPLAST ROC75) are present. Temporal ROC75 expression in C. reinhardtii is opposite to that of the angiosperm LUX, suggesting different clock mechanisms. In the picoalga Ostreococcus tauri, both ELF genes are missing, suggesting that it has a progenitor circadian “green” clock. Clock-relevant photoreceptors and thermosensors vary within the green lineage, except for the CRYPTOCHROMEs, whose variety and functions may differ. More genetically tractable models of Chloroplastida are needed to draw final conclusions about the gradual evolution of circadian clocks within the green lineage.
Some homologs of the angiosperm clock are conserved along the green lineage but may exhibit different functions, as observed in a flagellate chlorophyte alga.
Introduction
The daily environment influences the fitness and growth of photosynthetic organisms. Fluctuating conditions such as day–night or temperature cycles orchestrate their physiological processes. Cyanobacteria, algae, and terrestrial plants evolved an endogenous clock that drives a large variety of output processes, called circadian rhythms. This clock delivers an advantage in their fitness and survival as shown with competition experiments in cyanobacteria. Cyanobacterial fitness enhanced substantially when the circadian period was close to the period of the environmental cycle (Johnson et al., 1998). Also in land plants, the circadian clock increases plant growth, survival, and competitive advantage (Dodd et al., 2005). As cyanobacteria and photosynthetic eukaryotes produce the oxygen on Earth, are responsible for carbon dioxide fixation and form the base of food-webs, the influence of the circadian clock on their temporal programs is of key importance for life on Earth.
Circadian rhythms have key physiological properties that are maintained in all organisms, including prokaryotes (Kondo et al., 1993; Eelderink-Chen et al., 2021). They continue with a period of about 24 h when the plant is removed from the natural fluctuating conditions and put under free-running conditions of constant light or darkness and temperature (Hsu and Harmer, 2014). One of the first demonstrations of such an endogenous rhythm was done by the French scientist de Mairan (de Mairan, 1729) observing the leaf movement of a mimosa (Mimosa pudica) plant. Moreover, circadian rhythms can be entrained to Zeitgebers (Aschoff, 1954). Not only the daily light-dark (LD) cycles and temperature cycles, but also cycles of nutrients like endogenous sugars entrain the circadian clock (Haydon et al., 2013). The phases of circadian rhythms can be shifted either forward or backward depending on the time of the day and the organism. This relationship is summarized in phase-response curves. Finally, circadian rhythms are temperature compensated, meaning that their period is similar when the organism is released to different temperatures under free-running conditions (Q10 is ∼0.8–1.3) (Pittendrigh, 1993).
Circadian rhythms have been broadly studied in photosynthetic organisms, fungi, and animals. Despite the conservation of physiological properties and the discovery of transcriptional/translational feedback loops in the oscillatory system of these organisms, the molecular components of circadian clocks in cyanobacteria, plants, fungi, and animals are different. Exceptions are select kinases and the involvement of CRYPTOCHROMES (CRYs) in eukaryotes (Mittag et al., 2005; Doherty and Kay, 2010; Dunlap and Loros, 2017; Rosbash, 2021). Interestingly, peroxiredoxins are conserved markers of circadian rhythms (Edgar et al., 2012). Thus, metabolic switches of oxidation–reduction cycles of peroxiredoxin proteins have been found across the domains of life, including select animals, fungi, plants, algae, and cyanobacteria (Edgar et al., 2012). Currently, it is assumed that circadian rhythms emerged multiple times in evolution, underlining their importance (Rosbash, 2021).
In this review, we will focus on circadian clocks of the green lineage (Bowman et al., 2007; Bowman, 2013; Bowman et al., 2017). The green lineage (Figure 1) covers a broad range of photosynthetic eukaryotes with high relevance for life on Earth. It comprises chlorophyte algae ranging from unicells to the advanced Charophyceaen streptophyte macroalgae as well as all terrestrial plants. These include bryophytes, ferns, gymnosperms, and angiosperms. All these organisms are summarized as Chloroplastida (Adl et al., 2012). They represent primary endosymbionts whose plastids are derived by the engulfment of a cyanobacterium into a eukaryotic cell. Secondary endosymbionts (Keeling et al., 2005) are not part of this review. Here, we wish to highlight a recent review of the “brown” clock in secondary red photosynthetic organisms (Farré, 2020). Crop plants will also not be part of this review, as they will be covered in a separate article of this special issue. We also wish to highlight a recent review on clock evolution and climate change regarding bacterial rhythms (Jabbur and Johnson, 2022).
Figure 1.
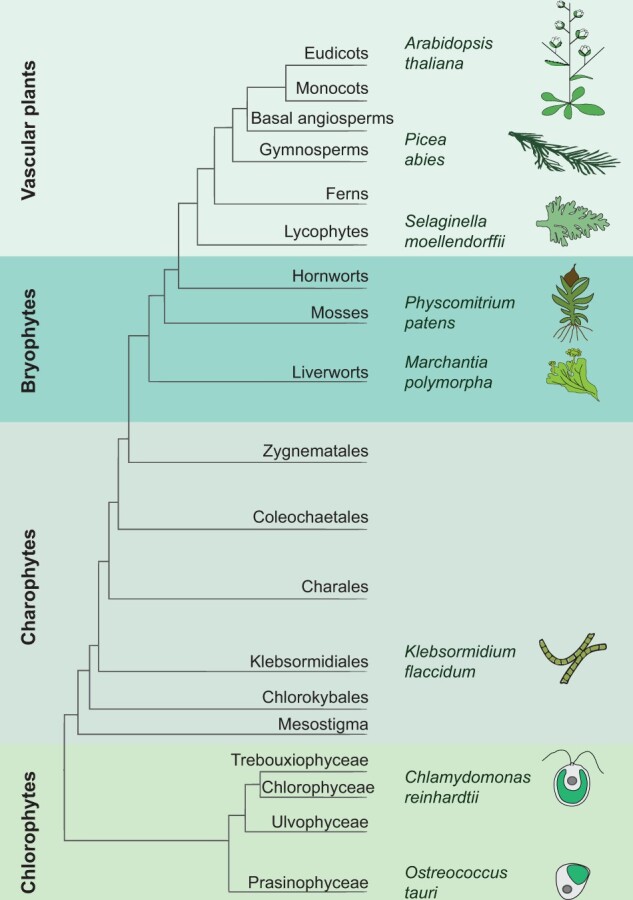
Schematic phylogenetic relationship in the green lineage. Only species discussed in this review have been highlighted. The phylogenetic tree was modified after Bowman et al. (2007) and Chang et al. (2016). It covers all green algae and terrestrial plants described in the review. Later phylogenetic trees have a focus on Charophytes (Bowman, 2013) or Bryophytes (Bowman et al., 2017).
Currently, there is a solid number of genome sequences from the green lineage and first functional data from select model organisms to study the clock traits of evolution in this lineage at the molecular basis. These studies will enable us to understand how photosynthetic eukaryotes that govern our planet adapted their lifestyles to temporal programs using clock systems. The focus of this review is thus on select model species of the green lineage (Figure 1) whose genomes are available and that are genetically tractable. Models include two green microalgal species, Ostreococcus tauri, a marine picoalga (Keeling, 2007), and Chlamydomonas reinhardtii, a freshwater alga, that lives primarily in wet soil (reviewed in Sasso et al., 2018). Under focus are also gametophyte-dominated plants, namely the liverwort Marchantia polymorpha and the bryophyte Physcomitrium patens (Linde et al., 2017; Rensing et al. 2020), formerly known as Physcomitrella patens. The sporophyte-dominated angiosperm Arabidopsis (Arabidopsis thaliana) is also part of this review, as it serves as the advanced model of the plant circadian system (Hsu and Harmer, 2014; Webb et al., 2019; Paajanen et al., 2021).
The variety of output processes and easily tractable circadian rhythms in the green lineage
In land plants, circadian rhythms of leaf, flower, and stomatal movement or circumnutation of inflorescence stems have been observed (de Mairan, 1729; Engelmann et al., 1992; Webb, 2003; Niinuma et al., 2005; Dornbusch et al., 2014; Yon et al., 2016; Creux et al., 2021). In the liverwort M. polymorpha, nyctinastic thallus movement was reported (Lagercrantz et al., 2020). As described above, sessile land plants profit from a clock-controlled program that initiates specific physiological processes within a given time frame. For example, it is advantageous for C3 and C4 plants to open their stomata during the day to gain carbon dioxide for the Calvin cycle of photosynthesis. Yet, environmental stresses may overrule the clock-driven processes. On a hot sunny day, the plant will close its stomata to avoid the loss of water. For the crosstalk of plants between circadian regulation and responses to the abiotic environment, we refer to a recent review (Paajanen et al., 2021).
In contrast to land plants, unicellular algae can move within the water column or in wet soil. Microalgal movement can be passive, driven by the current of the ocean, or active driven by cilia. In the biflagellate alga C. reinhardtii, tactic movements are controlled by the circadian clock. Green macroalgae of the species Aegagropila linnaei perform a circadian rhythm of buoyancy when formed into spherical aggregates (Cano-Ramirez and Dodd, 2018). Recordings of the rhythm of photo orientation in C. reinhardtii, also called phototaxis can be automatically collected and were even done on a spacecraft in space (Bruce, 1970; Mergenhagen and Mergenhagen, 1987). While this rhythm peaks during subjective day, the rhythm of chemotaxis to a nitrogen source (ammonium) is highest at subjective night (Byrne et al., 1992; reviewed in Mittag et al., 2005). Yet, the alga waits until the beginning of day to take up ammonium. Also, nitrite uptake occurs at dawn where the activity of nitrite reductase rises (Pajuelo et al., 1995). Thus, the unicellular algae show simple but coordinated clock-controlled behavioral responses. Moreover, rhythms of cell division, UV light sensitivity, stickiness to glass, and starch accumulation were reported (reviewed in Mittag et al., 2005; Matsuo and Ishiura, 2010).
The automated recording of circadian rhythms facilitates studies on their circadian clocks. In land plants, the measurement of the rhythm of leaf movement was automated. Leaf movements such as in A. thaliana were analyzed bioinformatically using video cameras or time lapse photography and subsequent bioinformatics analyses of leaf movements (Engelmann et al., 1992; Wagner et al., 2017). Simultaneously the invention of easily tractable reporters supported research in A. thaliana on chronobiology as well as the development of new model organisms in this field. Circadian controlled promoters were often fused to the reporter and this chimera served as a basis for genetic screens in the search for clock components. The most used reporter system involves bioluminescence and the enzyme LUCIFERASE (LUC). Based on the automated measurement of the natural rhythm of bioluminescence in a dinoflagellate alga (Broda et al., 1986), artificial-based bioluminescence imaging was deployed rapidly by the chronobiological community as a tool to track for example gene expression. LUC reporters from marine bacteria, from a soft coral or the firefly (Figure 2) were used to study circadian systems in O. tauri, C. reinhardtii, P. patens, M. polymorpha, and A. thaliana (Millar et al., 1995; Matsuo et al., 2006; Voytsekh et al., 2008; Thommen et al., 2012; Aoki et al., 2014; Linde et al., 2017).
Figure 2.
Measurement of circadian-controlled bioluminescence under a light–dark cycle and free-running conditions. A chimeric construct consisting of the clock-controlled cab2 promoter and the firefly LUC reporter is shown (Millar et al., 1995). The rhythm of bioluminescence is measured in relative luminescence units. Period, amplitude, and phase, peaking during the day, are indicated with arrows. Plants were grown in a LD cycle and then released under constant light (LL) and temperature. LD0 indicates the start of the day and LD12 the beginning of the night. Under constant light (LL), subjective days (white bars) and nights (gray bars) are highlighted. Hours indicate how long the plant has been kept under LL.
Components of the core oscillators in the green lineage
The core oscillatory system in the angiosperm model A. thaliana
The angiosperm clock model of A. thaliana (Figure 3) is the best studied in the green lineage. Its endogenous oscillator is composed of interlocking transcription–translation feedback loops (Hsu and Harmer, 2014). There are more than 20 clock and clock-related components known in A. thaliana. They are grouped into morning-, day-, afternoon-, and evening-phased components. The MYB-like transcription factors CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) are expressed at dawn (Schaffer et al., 1998; Wang and Tobin, 1998). They interact with each other and act as transcriptional repressors on genes containing the so-called evening element (EE) promoter motif (Harmer et al., 2000; Harmer and Kay, 2005). Knockout plants of cca1 or lhy show a shortened period in the expression of target genes and the double mutant shows an even shorter period (Green and Tobin, 1999; Mizoguchi et al., 2002). Initially, the PSEUDO RESPONSE REGULATOR 1 (PRR1), also known as TIMING OF CAB EXPRESSION 1 (TOC1), with peak expression in the early evening, was found to be a target of this repression and to directly regulate CCA1 and LHY expression. Based on these findings, the first single transcriptional–translational feedback loop was proposed (Alabadí et al., 2001). At first, it was suggested that TOC1 functions as an activator of morning-expressed oscillator genes (Pruneda-Paz et al., 2009). Later it was found that TOC1 acts as a repressor by its rhythmic association to promoters of oscillator genes, thus connecting the morning and evening feedback loops through the repressing activity of TOC1 (Huang et al., 2012; Nohales and Kay, 2016). Other members of the PRR family, PRR3, PRR5, PRR7, and PRR9, are also part of the plant oscillator whose transcripts increase over the course of the day. The PRR9 transcript peaks after dawn directly following CCA1/LHY expression, with PRR7, PRR5, and PRR3 following over the course of the day and TOC1 peaking in the evening (Matsushika et al., 2000; Farré and Liu, 2013). PRR9, PRR7, and PRR3 are partially redundant in repressing the morning genes CCA1 and LHY, forming a second loop (Nakamichi et al., 2010).
Figure 3.
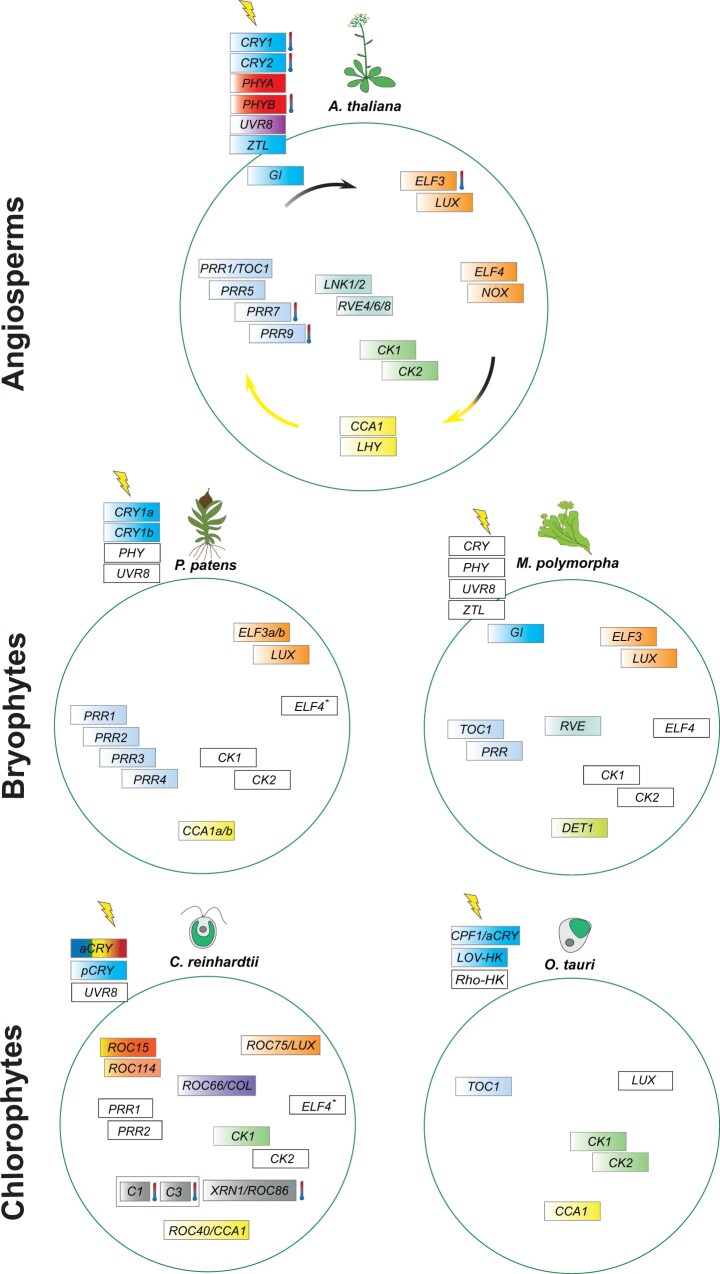
Comparison of molecular components of the circadian clock of select angiosperms, mosses, and chlorophyte algae. The molecular components of the circadian clock of select angiosperm, moss, and chlorophyte algae are shown. A simplified scheme of the angiosperm A. thaliana clock (Webb et al., 2019) is taken as a base model. The temporal series of the main oscillatory components that are shown in between the arrows is indicated by the flow of the arrows. It starts with CCA1/LHY expression at dawn (yellow color) and ends with the EC (black color). Functional details are explained in the main text. For clarity, arrows for other models than A. thaliana have been omitted as differences in expression can occur or expression times are not always known. Genes that are conserved in the central oscillator in mosses and chlorophyte algae, but not functionally characterized, are indicated by white boxes. Further clock components in a select liverwort (M. polymorpha) and a select flagellate alga (C. reinhardtii) are shown. The asterisk in ELF4 refers to complementation assays. Photoreceptors that have been characterized and found to participate in the circadian clock machinery are shown outside the green circles for each organism. Colors represent their absorption range. Representative photoreceptors that may relate to the clock are indicated by white boxes. Please note that PRR1 has no sequence similarity to TOC1 in some of the mentioned organisms. Not all potential clock-relevant photoreceptors have been added for clarity. For example, the genome of C. reinhardtii encodes 18 photoreceptors (Greiner et al., 2017). Several may be involved in the circadian clock, but only few have been studied so far. Clock components that can integrate temperature information and thus may be involved in temperature entrainment are labeled with a thermometer. aCRY, animal-like CRYPTOCHROME; C1, subunit 1 of the RNA-binding protein CHLAMY1; C3, subunit 3 of the RNA-binding protein CHLAMY1; CCA1, CIRCADIAN CLOCK-ASSOCIATED1; CK, CASEIN KINASE; COL, CONSTANS-like; CPF, CRYPTOCHROME PHOTOLYASE FAMILY; CRY1, Arabidopsis CRYPTOCHROME 1; CRY2, Arabidopsis CRYPTOCHROME 2; DET1, DEETIOLATED1, ELF, EARLY FLOWERING; GI, GIGANTEA; LHY, LATE ELONGATED HYPOCOTYL; LNK, NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED; LOV-HK, LOV-HISTIDINE KINASE, LUX, LUX ARRHYTHMO; NOX, BROTHER OF LUX ARRHYTHMO; pCRY, plant CRYPTOCHROME; PHYA, PHYTOCHROME A; PHYB, PHYTOCHROME B; PRR, PSEUDO RESPONSE REGULATOR; Rho-HK, RHODOPSIN-HISTIDINE KINASE; ROC, RHYTHM OF CHLOROPLAST; RVE, REVEILLE; TOC1, TIMING OF CAB 1 EXPRESSION; UVR-8, UV RESISTANCE LOCUS 8; XRN1, 5'-3' EXORIBONUCLEASE 1; ZTL, ZEITLUPE.
During the evening, the genes EARLY FLOWERING3 (ELF3), ELF4, and LUX ARRYTHMO (LUX) are expressed and interact to form the evening complex (EC; Nusinow et al., 2011; Herrero et al., 2012). The EC represses the expression of PRR9, PRR7, and TOC1, releasing the lock they form on CCA1 and LHY expression, giving way to a transcript increase of the two morning genes (Nohales and Kay, 2016). BROTHER OF LUX ARRHYTHMO (BOA/NOX) is another evening phased component that forms a complex with ELF3 and ELF4 and indirectly promotes expression of CCA1 (Dai et al., 2011; Nusinow et al., 2011). LUX also forms an autoinhibitory loop by repressing its own and the transcription of PRR9 (Helfer et al., 2011). In addition, CCA1 and LHY repress the expression of the EC genes LUX, ELF3 and ELF4 by binding to their EE motif, closing the loop (Adams et al., 2015; Nagel et al., 2015; Kamioka et al., 2016; Adams et al., 2018).
Furthermore, the activating MYB-like transcription factor REVEILLE 8 (RVE8) forms a complex with LNK1 (NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED 1) and LNK2 to induce TOC1 and PRR5 expression (Xie et al., 2014; Pérez-García et al., 2015). It was also shown that RVE4, RVE6, and RVE8 promote expression of PRRs and the EC (Farinas and Mas, 2011; Rawat et al., 2011; Hsu et al., 2013; Xie et al., 2014). Repression of these positive regulators is facilitated by the PRRs on RVE8 (Nakamichi, 2020) and on the LNKs (Nakamichi et al., 2012; Rugnone et al., 2013; Liu et al., 2016).
Light-dependent regulation by protein degradation is facilitated by the protein GIGANTEA (GI) together with the photoreceptor ZEITLUPE (ZTL). GI binds to the LOV domain of ZTL in a light-dependent manner, stabilizing both proteins (Kim et al., 2007). In darkness, this complex dissociates. GI and ZTL induce degradation of TOC1 and PRR5 via the proteasome pathway (Más et al., 2003; Kim et al., 2007). In turn, GI promotes transcription of the morning genes CCA1 and TOC1 (Martin-Tryon et al., 2007). GI is regulated by CCA1 binding to its CCA1 binding site, reducing its expression (Lu et al., 2012).
Recently, the circadian clock of the wild tobacco Nicotiana attenuata has been studied not only under laboratory conditions, but also in its native habitat (Yon et al., 2016; Cortés Llorca et al., 2020). Also, transcriptome dynamics of Arabidopsis halleri were investigated in natural environments under seasonal conditions (Nagano et al., 2019).
Clock components in the moss P. patens
A set of conserved circadian core components that share similarity in gene expression with vascular plants was found in the bryophyte P. patens. These genes include the Myb-like transcription factors PpCCA1a and PpCCA1b, homologs of CCA1/LHYI and the PRRs PpPRR1 to PpPRR4. One LUX putative ortholog (Phypa-47310) and two ELF3-like genes (Phypa-66647 and Phypa-165364) are described as putative circadian clock-related genes (Holm et al., 2010; Satbhai et al., 2011; Figure 3). However, homologous genes coding for proteins like TOC1, GI, or ZTL in A. thaliana are missing in P. patens (Linde et al., 2017). PpCCA1a and PpCCA1b both showed expression peaks like CCA1/LHY of A. thaliana at dawn in LD cycles and under continuous darkness, but not under continuous light. Like in A. thaliana, a knockout of the two genes resulted in a reduced amplitude and shorter period; a negative feedback regulation was suggested (Okada et al., 2009). The PRRs and ELF3-like genes have expression profiles similar to their A. thaliana counterparts (Holm et al., 2010). The function of the putative ELF4 ortholog in P. patens is poorly understood. However, constitutive expression of PpELF4 in the A. thaliana elf4 mutant completely rescued its early flowering phenotype. The same study suggests that PpELF4 can regulate the expression of CCA1, LHY, and PRRs, which implies an overlapping function with the circadian clock of A. thaliana (Zhao et al., 2019). Thus, the molecular components of the central moss clock seem to be closely related to the select angiosperm clock.
Time keeping in the liverwort M. polymorpha
There have been recent functional studies of clock gene homologs of the liverwort M. polymorpha. These include the PRR, RVE, and TOC1 genes (Figure 3) and suggest their involvement in the M. polymorpha circadian clock as well as experimental studies of ELF3, LUX, and GI (Linde et al., 2017). It was suggested that the core clock genes PRR, RVE, and TOC1 form a core transcriptional feedback loop (Lagercrantz et al., 2020). In addition, a DE-ETIOLATED1 homolog was studied in M. polymorpha as AtDET1 is associated with AtCCA1/LHY, which are missing in liverworts (Linde et al., 2017; Lagercrantz et al., 2021). The circadian rhythm of MpDET1 expression was disrupted in mutants of key clock and putative EC genes (Lagercrantz et al., 2021). MpDet1 knockdown disturbed the circadian rhythm of nyctinastic thallus movement and rhythmic gene expression patterns (Lagercrantz et al., 2021).
Comparative clock gene analysis in green chlorophyte algae compared to Streptophytes
Streptophytes comprise the freshwater Charophyceaen algal clade (Figure 1) as the most advanced green algae with similarities to land plants as well as all embryophyte plants including the mosses, ferns, gymnosperms, and angiosperms (Bowman, 2013). A recent study compared the angiosperm clock genes of A. thaliana with genes of other organisms of the green lineage. These include the gymnosperm Picea abies, the moss P. patens, the liverwort M. polymorpha, the fern Selaginella moellendorfii, a representative from Charophyceae Klebsormidium flaccidum, and the chlorophyte model algae C. reinhardtii and O. tauri (Linde et al., 2017).
The search for clock gene homologs revealed that the large REVEILLE (RVE) family consisting of the members CCA1 and LHY and other RVE genes is represented with at least one member in all investigated photosynthetic organisms of the green lineage (Linde et al., 2017; de los Reyes et al., 2017). This is also the case for the PRR family; at least one member is present in all investigated species of the green lineage. However, the sequence similarity may be relatively low. Thus, two TOC1 homologs were found in C. reinhardtii, named CrPRR1 and CrPRR2, but are rather distinct from the angiosperm TOC1 (Satbhai et al., 2011). In the case of M. polymorpha, the CCA1/LHY homolog is missing, as mentioned above, but a RVE member is present. Functional analysis is needed in all cases and has been executed in some members as described in other subchapters.
Genes of the angiosperm EC, ELF3, ELF4, and LUX are found in all members of the green lineage within the streptophytes with one exception. ELF3 seems to be absent in P. abies (Linde et al., 2017). This is also the case for the chlorophyte alga C. reinhardtii; EC members are found except for ELF3 (Mittag et al., 2005; Linde et al., 2017). In contrast, all above mentioned EC members seem to be missing in the picoalga O. tauri (Linde et al., 2017). However, a LUX(-like) was reported elsewhere (Serrano-Bueno et al., 2017; Matsuo et al., 2020) and awaits further functional characterization. These data indicate that O. tauri has the least conserved circadian key clock members.
The ZTL and GI duo that is closely associated with light input is present in all streptophytes, except for the moss P. patens and for the Charophyceae K. flaccidum regarding GI. Both ZTL and GI are missing in the select chlorophyte algae (Linde et al., 2017).
Altogether, this analysis suggests that some key clock genes of the Chloroplastida seem conserved from chlorophyte unicellular algae to advanced Charophyceae algae and land plants. However, the functional data on the involvement of these genes in the clock machinery of the respective organism are still under investigation. Complementation assays were done with ELF4 from C. reinhardtii and P. patens as mentioned above. Notably, both CrELF4 and PpELF4 rescued the A. thaliana elf4 mutant (Zhao et al., 2019) and are thus labeled with a star in Figure 3. The rescues involved the early flowering phenotype and affect the expression of circadian clock-controlled genes in A. thaliana.
The picoalga O. tauri
O. tauri, belonging to the Prasinophyceae (Figure 1) is a chlorophyte algal model with minimal cellular organization (Corellou et al., 2009). It contains two homologs of the A. thaliana core clock proteins. These are OtTOC1 and OtCCA1 with high conservation within the PRR, CCT, and MYB domains (Corellou et al., 2009; Figure 3). Members of the angiosperm EC, ELF3, and ELF4 have not been found in O. tauri (Linde et al., 2017). The O. tauri genome encodes genes containing GARP and B-box transcription factors, but it remains unknown if they are involved in its clock machinery (Corellou et al., 2009). A LUX-like protein was also reported (Serrano-Bueno et al., 2017; Matsuo et al., 2020). Expression profiles of OtTOC1 peak like the A. thaliana homolog at subjective dusk, whereas OtCCA1 expression has a broader and earlier peak compared to A. thaliana (Corellou et al., 2009; reviewed in Noordally and Millar, 2015). Notably, relevant clock-related kinases involved in the angiosperm clock, CASEIN KINASE1 (CK1) and CK2, are conserved in O. tauri (Box 1).
Box 1.
Posttranslational control and CKs
In all model species studied thus far including angiosperm plants, posttranslational control has been found to be of high importance (Yan et al., 2021). Among the diverse modifications of clock-related proteins ubiquitination, O-glycosylation, SUMOylation, methylation, and phosphorylation play important roles. We will not detail all these processes in this review because they have not been studied for all members in the green lineage. But we would like to highlight that reversible phosphorylation plays a major role in diverse organisms including not only algae and land plants (Kusakina and Dodd, 2012; Noordally and Millar, 2015; Krahmer et al., 2021), but also fungi (Liu et al., 2000), flies, and mammals (Narasimamurthy and Virshup, 2021). CKs (CK1 and CK2) are well conserved across species and are also found to be of importance in unicellular algae (Mittag et al., 2005; Schmidt et al., 2006; van Ooijen et al., 2013). The role of CKs has been demonstrated in the circadian systems of the chlorophyte algae O. tauri and C. reinhardtii, but also in the A. thaliana angiosperm clock. A CK1 knockdown shortens the period in C. reinhardtii (Schmidt et al., 2006) while inhibition of CK1 in O. tauri and A. thaliana lengthens the period (van Ooijen et al., 2013; Uehara et al., 2019). These effects underline that the clocks of O. tauri and A. thaliana may be closer in their mechanisms than the C. reinhardtii clock (Matsuo et al., 2020). It is also notable that CK1 is located in the flagella and the eyespot of C. reinhardtii (Schmidt et al., 2006). CK2 was also shown to be clock-relevant in O. tauri (Le Bihan et al., 2015). Data are still missing on CK2 and clock function in C. reinhardtii. In A. thaliana CK2 is involved in CCA1 and LHY phosphorylation (Sugano et al., 1998, 1999).
Another important feature in O. tauri is that 90% of its daily transcriptome is rhythmic (de los Reyes et al., 2017). Thus, this picoalga is heavily influenced by light switches. In contrast, there is a reduction in the size of diurnal transcriptomes in the flagellate alga C. reinhardtii (down to 70%) and in the land plant A. thaliana (only 40%, Covington et al., 2008; Zones et al., 2015; de los Reyes et al., 2017). Altogether, it seems that the clock of O. tauri is reduced in core clock components of the EC and may only have one transcription–translation feedback loop. However, this seems to be compensated by the high number of genes regulated by the daily change of light and darkness.
The biflagellated unicellular alga C. reinhardtii
The chlorophyte alga C. reinhardtii belongs to the Chlorophyceae (Figure 1). In contrast to O. tauri and all other presented plant models, C. reinhardtii is a motile organism that can use its flagella for clock-controlled tactic movements (Mittag et al., 2005).
Molecular characterization of a clock component was made by identification of the CHLAMY1 RNA-binding complex that binds to UG-repeat sequences in the 3′-UTR of mRNAs (reviewed in Mittag et al., 2005). CHLAMY1 consists of the subunits C1 and C3 (Zhao et al., 2004). An increase or decrease of the protein level of the C3 subunit, present in overexpression and knockdown lines, respectively, results in a shift in the acrophase, while changes in the level of C1 cause arrhythmicity (Iliev et al., 2006). However, it should be emphasized that the alterations on the C1 level automatically cause changes in the level of C3 due to a co-regulatory mechanism (Iliev et al., 2006). C3 has high sequence similarity to mammalian CUG-binding proteins that are involved in myotonic dystrophy (Zhao et al., 2004; Koshelev et al., 2010). Myotonic dystrophy is associated with variable circadian rhythms (Okumura et al., 2002).
Deeper insights into the clock of C. reinhardtii were obtained through forward genetic approaches screening a library of insertional mutants using a LUC reporter gene that is controlled by the psbD clock-controlled chloroplast promotor (Matsuo et al., 2006, 2008). The screen identified 105 genes that showed alterations in the amplitude, phase and/or period, termed “RHYTHM OF CHLOROPLAST” (ROC) mutants (Matsuo et al., 2008). Some of the proteins contain conserved domains that are also found in the core clock components of A. thaliana (reviewed in Matsuo and Ishiura, 2010; Ryo et al., 2016). ROC15 and ROC75 encode domains with sequence similarity to the GARP DNA-binding domain that can also be found in LUX. ROC15 is associated with the light signaling pathway (see section below). ROC40 encodes a Myb domain like CCA1/LHY. It has a UG-repeat in its 3′-UTR and thus may be connected to CHLAMY1 (Ryo et al., 2016). ROC15 and ROC40, mRNAs accumulate from subjective midnight until dawn.
ROC66 encodes a B-box and the CCT similar to CO proteins. Another CO family protein, which is different from ROC66, CrCO, affects circadian output regarding growth, diurnal starch content rhythms, and cell cycle-related gene expression (Serrano et al., 2009). It is notable that CrCO can complement the co mutation in A. thaliana regarding the timing of flowering phenotype. ROC114 encodes an F-box and is a potential ubiquitin ligase. As mentioned above, TOC1 homologs (CrPRR1 and CrPRR2) have been found in C. reinhardtii but are not functionally studied yet.
The LUX homolog ROC75 is a day-phase expressed nuclear-localized protein (Matsuo et al., 2020). ROC75 represses the key clock-related genes ROC15, ROC40, and ROC66, during subjective day. The acute downregulation of ROC15 and ROC40 mRNAs by light is, however, independent of ROC75.
Contrary to the evening expression of angiosperm LUX and the LUX-like transcript in O. tauri, the ROC75 transcript accumulates during daytime (Monnier et al., 2010; Matsuo et al., 2020). Thus, it was postulated that the C. reinhardtii clock may have a contrasting mechanism compared to A. thaliana and O. tauri (Matsuo et al., 2020). This hypothesis is consistent with a different effect on period caused by a knockdown mutant or inhibition of CK1 from O. tauri and A. thaliana compared to C. reinhardtii (Box 1; Schmidt et al., 2006; van Ooijen et al., 2013; Uehara et al., 2019). In general, CK1 and CK2 are involved in posttranslational regulation of the clock in several organisms and are recently even discussed with regard to temperature compensation (Kusakina and Dodd, 2012; Hu et al., 2021; Narasimamurthy and Virshup, 2021).
It may be that the circadian clock of flagellate algae has partially different properties compared to the clock of sessile plants and nonflagellate algae. The C. reinhardtii genome encodes conserved genes from the plant and animal kingdom, especially with regard to the flagella (Merchant et al., 2007). It should also be noted that six genes involved in flagellar function were included in the mutant clock screen suggesting the requirement of normal flagellar function to maintain the robustness of chloroplast rhythmicity (Matsuo et al., 2008). Also, CK1 is found in the flagellum and has many targets there (Schmidt et al., 2006; Boesger et al., 2012).
Sensors of light and temperature in the green lineage
Photoreceptors in land plants
In A. thaliana, four families of photoreceptors (Figure 3) have been described to be involved in the clock machinery: (1) PHYTOCHROMES (PHY); (2) CRYs; (3) the ZTL family, and UV RESISTANCE LOCUS 8 (UVR8; reviewed in Sanchez et al., 2020). These photoreceptors absorb light at specific wavelengths, ranging from the UV-B area to UV-A, blue and red/far red light.
It should be noted that the PHYs and CRYs seem to contribute only partially to clock entrainment in A. thaliana (reviewed in Sanchez et al., 2020). An Arabidopsis mutant lacking main photoreceptors such as phyA-201 phyB-5 cry1-201 cry2/fha-1 still shows clock entrainment (Balcerowicz et al., 2021). A recent study with RNA-sequencing time-series data identified light- and temperature-sensitive bursts of transcription factor activity. It suggests that PHY and CRY signaling is important for fine-tuning, but separate pathways can activate much of the program (Balcerowicz et al., 2021). Low-intensity UV-B radiation acts as an entraining signal for the clock. UVR8 and CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) are required for this process and their interactions were shown (Fehér et al., 2011; Podolec et al., 2021). We hypothesize that the coordination of several light sensors is needed for the final entrainment programs. Also, it should be kept in mind that the biological function of some light sensors such as for CRY3, a Drosophila, Arabidopsis, Synecococcus, and Human-CRY in A. thaliana, are not yet known.
Light can also induce the expression of clock genes (Nohales and Kay, 2016). PHYB regulates the phase of several clock genes (Palágyi et al., 2010). It was also shown that a constitutively active allele of PHYB maintains circadian robustness in the absence of light (Jones et al., 2015). All these data implicate a complex interplay. The CRYs are involved in different physiological processes. Both CRY1 and CRY2 play an important role for the functioning of the endogenous oscillator (reviewed in Sanchez et al., 2020). A cry double mutant exhibits a longer period compared to wild-type (Devlin and Kay, 2000). Recently, it was shown that CRYs mediate blue light regulation of N6-methyladenosins modifications of mRNAs, especially those regulated by the circadian clock (Wang et al., 2021). This aspect of blue light signaling by CRYs allows an influence on posttranscriptional regulation.
The function of ZTL and GI has been detailed above. ZTL is found in select bryophytes, ferns, monocots, and dicots (Linde et al., 2017; Gil and Park, 2019), but is missing in O. tauri, C. reinhardtii, and P. patens. In P. patens two CRYs seem to be involved in blue light signaling in connection with the circadian clock (Ichikawa et al., 2004). It is too early to predict if the function of the CRYs fulfills the ZTL functions in plants where ZTL is missing. UVR8 is present in bryophytes (Soriano et al., 2018), but a clock function has not been described so far.
Photoperception in unicellular alga
Photoreceptors and their role for circadian clocks have been investigated in the model algae C. reinhardtii and O. tauri. Like A. thaliana, both algae have CRYs. However, the complexity of the CRY system in these algae is broader than in land plants (Petersen et al., 2021). The plant CRY in C. reinhardtii (called pCRY) has been shown to be involved in phase resetting of the circadian clock. A knock-down mutant results in period lengthening and after a few days under constant conditions in arrhythmicity (Müller et al., 2017). A functional connection to the clock-component ROC 75 was recently described (Kamrani et al., 2018). It is worth noting that the unicellular algae also has an animal-type CRY (aCRY) that is grouped closely to the CRYPTOCHROME PHOTOLYASE FAMILY (CPF1; Beel et al., 2012; Petersen et al., 2021). It absorbs not only in the blue range of the visible spectrum, but also in the yellow and red (Beel et al., 2012). Despite the dogma that CRYs have lost photolyase activity the algal animal-like CRYs that group to the CPF have maintained it (Coesel et al., 2009; Heijde et al., 2010; Franz et al., 2018). Moreover, the CPF1 from O. tauri exhibits clock function in a heterologous system (Heijde et al., 2010). aCRY from C. reinhardtii alters light-induction/reduction of some clock genes, like for the C3 subunit of the circadian RNA-binding protein CHLAMY 1 (Beel et al., 2012).
UV-B perception via UVR8 is conserved in C. reinhardtii (Tilbrook et al., 2016; Serrano-Bueno et al., 2017; Han et al., 2019). Like A. thaliana, UVR8 interacts with COP1. However, a UVR8 receptor seems to be missing in O. tauri (Han et al., 2019) as well as PHYs that are also absent in C. reinhardtii (Mittag et al., 2005; Han et al., 2019).
Furthermore, ROC15 of C. reinhardtii is involved in triggering the reset of the algal circadian clock along with the F-box protein ROC114 (Niwa et al., 2013). In a mutant called LRR that affects a RAS signaling-related leucine-rich-repeat protein, ROC15 degradation and resetting of the clock by red and violet light was impaired (Kinoshita et al., 2017).
In O. tauri, a LOV-HISTIDINE KINASE (LOV-HK) and a RHODOPSIN-HISTIDIN KINASE (Rho-HK) have been reported (Djouani-Tahri et al., 2011) and discussed (Thommen et al., 2015; Luck et al., 2019), to be involved in the circadian clock machinery.
These data highlight that conserved but also different types of known photoreceptors are involved in green algae and land plants (Figure 3). The genome of C. reinhardtii encodes for 18 different photoreceptors (Greiner et al., 2017) whose influence on the circadian clock is little understood.
Temperature sensing
Information on temperature sensing and signaling in the physiological range is still rare. In C. reinhardtii the two subunits of CHLAMY1 integrate changes in temperature in the physiological range with regard to protein expression (C3 subunit) or to reversible phosphorylation (C1 subunit) (Voytsekh et al., 2008). Two interaction partners of C3, the 5ʹ–3ʹ EXORIBONUCLEASE XRN1 (also known as ROC86; Dathe et al., 2012) and splicing variant 12 of a further RNA recognition motif-containing RNA-binding protein, MUSASHI, react to changes in ambient temperature with regard to their protein abundance (Li et al., 2018). It is assumed that these proteins are part of a thermal signaling network in the green alga.
In A. thaliana, the EC is temperature responsive. It was recently shown that ELF3 of A. thaliana Col-0 contains a polyglutamine (polyQ) repeat whose length corresponds with thermal responses. The ELF3 polyQ repeat is missing in stiff brome (Brachypodium distachyon), whereas potato (Solanum tuberosum) contains an intermediate in comparison to A. thaliana, suggesting that it is not universal for land plants (Jung et al., 2020). The temperature sensitivity of ELF3 is altered by the levels of ELF4. Plants lacking functional PRR7 and PRR9 are unable to entrain the clock to cold pulses after entrainment to thermocycles, which indicates a role in the clock temperature signaling (Salomé and McClung, 2005).
Another thermosensor in A. thaliana is the red-light photoreceptor PHYB (reviewed in Gil and Park, 2019). It senses temperature through the temperature-dependent reversion of the active Pfr to inactive Pr state (Legris et al., 2016). It was shown that PHYB associates in a temperature-dependent manner with promotors of target genes like the PHYTOCHROME-INTERACTING FACTORS (Jung et al., 2016; Legris et al., 2016). Also, for the photoreceptors CRY1 and CRY2 it was recently shown that their protein levels are substantially altered by temperature and that the photocycle is affected (Pooam et al., 2021).
A detailed overview of abiotic factors besides light and temperature influencing the period of the circadian clock is shown in (Webb et al., 2019) and will not be detailed here.
Conclusions
Genome-based data on circadian clock members of the green lineage and first functional studies suggest that several of its clock members are conserved and evolved from chlorophyte Prasinophyceaen picoalgae (Linde et al., 2017; Serrano-Bueno et al., 2017). The green lineage includes a large number of clock gene homologs that can be found in all/most of its members (Figure 3) and that are missing in fungi and animals except for CKs (Box 1) and CRYs (Figure 3). In several cases, functional data are still missing to draw final conclusions about a conserved mechanism of clocks in the green lineage. It seems that the clock complexity increases from the picoalga O. tauri to the flagellate C. reinhardtii and from the mosses to the angiosperm A. thaliana. The higher complexity may be since land plants are sessile and thus need more clock components to react to all possible environmental conditions. There is also some evidence that the clock-controlled tactic movements of the ciliated C. reinhardtii may result in an at least partially different clock mechanism. To confirm or disregard this hypothesis, studies on another flagellate green alga would be needed (see “Outstanding Questions Box”). More genetically tractable model organisms of the Chloroplastida, especially within the Charophytes (Figure 1), the transition zone of advanced streptophyte algae to terrestrial plants, would help to get a more comprehensive picture of circadian clocks in the green lineage.
Regarding the influence of light on the angiosperm clock, it is important to mention that the photoreceptor ZTL and GI have been found in all investigated streptophyte species (Linde et al., 2017) expect for P. patens and K. flaccidum where ZTL is present, but GI is missing. Both are absent in the select chlorophyte algae. These findings suggest that the negatively light regulated protein degradation mediated by ZTL is a component that arose during the transition to terrestrial habitats. Photoreceptor classes that can be found in most and all the presented species of this review, respectively, are the UVR8s that are only missing in O. tauri (Han et al., 2019) and the CRYs. CRY functions within the clock were already shown for C. reinhardtii, O. tauri, P. patens, and A. thaliana (Ichikawa et al., 2004; Beel et al., 2012; Müller et al., 2017; Sanchez et al., 2020). For this reason, it seems that CRYs are mainly conserved as mediators of the light input and clock control within the green lineage. The broad variety in their properties and functions has been discussed above and is especially visible in different algal species (Petersen et al., 2021). Apart from this, CRYs are involved in the clock-light input pathway in fungi and insects, whereas in mammals they have a role as key clock components within the endogenous oscillator (Foley and Emery, 2020).
ADVANCES .
Key components of the endogenous oscillator are conserved within the green lineage; their genomes encode at least one member of the PRR and at least one member of the RVE family.
The angiosperm EC members ELF3 and ELF4 are conserved to a large extent in streptophytes, but at least one of them is missing in select chlorophyte algae. Expression patterns of the EC LUX differ in A. thaliana and C. reinhardtii.
ZTL and GI are found in most streptophytes but are absent in the select chlorophyte algae.
The clock-related effect of CK1 differs in A. thaliana and O. tauri compared to that in the flagellate C. reinhardtii.
The variety of CRYs in algae is higher than in land plants, and their properties, as well as functions may differ.
OUTSTANDING QUESTIONS.
How conserved are circadian clock components and their mechanisms within the different phyla of Chloroplastida?
Which conserved clock genes along the green lineage have similar functions?
Have ZTL and GI been lost in the moss P. patens or have they evolved independently in liverworts and mosses?
Do flagellate algae have an individual additional set of clock components?
Are there main differences in the circadian clock between sessile plants and motile flagellate algae?
How conserved is photo perception in the green lineage?
How conserved is temperature sensing in the green lineage?
Acknowledgments
We thank Volker Wagner and Ellen McPherson for helpful comments on the review and Andreas Holzinger for his helpful comments on streptophyte algae.
Funding
Our work was supported by the Deutsche Forschungsgemeinschaft (DFG) by grant Mi373/16-1 to M.M.
Conflict of interest statement. None declared.
Contributor Information
Jan Petersen, Matthias Schleiden Institute of Genetics, Bioinformatics and Molecular Botany, Friedrich Schiller University Jena, Jena 07743, Germany.
Anxhela Rredhi, Matthias Schleiden Institute of Genetics, Bioinformatics and Molecular Botany, Friedrich Schiller University Jena, Jena 07743, Germany.
Julie Szyttenholm, Matthias Schleiden Institute of Genetics, Bioinformatics and Molecular Botany, Friedrich Schiller University Jena, Jena 07743, Germany.
Maria Mittag, Matthias Schleiden Institute of Genetics, Bioinformatics and Molecular Botany, Friedrich Schiller University Jena, Jena 07743, Germany.
J.P., A.R., J.S., and M.M. designed Figures 1 and 3. J.P., A.R., and M.M. designed Figure 2. J.P., A.R., J.S., and M.M. wrote the paper.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Maria Mittag (M.Mittag@uni-jena.de).
References
- Adams S, Grundy J, Veflingstad SR, Dyer NP, Hannah MA, Ott S, Carré IA (2018) Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol 220: 893–907 [DOI] [PubMed] [Google Scholar]
- Adams S, Manfield I, Stockley P, Carré IA (2015) Revised morning loops of the Arabidopsis circadian clock based on analyses of direct regulatory interactions. PLoS One 10: e0143943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adl SM, Simpson AGB, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al. (2012) The revised classification of eukaryotes. J Eukaryot Microbiol 59: 429–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Aoki S, Okada R, Satbhai SB (2014) Transformation and measurement of bioluminescence rhythms in the moss Physcomitrella patens. Methods Mol Biol 1158: 325–336 [DOI] [PubMed] [Google Scholar]
- Aschoff J (1954) Zeitgeber der tierischen Tagesperiodik. Naturwissenschaften 41: 49–56 [Google Scholar]
- Balcerowicz M, Mahjoub M, Nguyen D, Lan H, Stoeckle D, Conde S, Jaeger KE, Wigge PA, Ezer D (2021) An early-morning gene network controlled by phytochromes and cryptochromes regulates photomorphogenesis pathways in Arabidopsis. Mol Plant 14: 983–996 [DOI] [PubMed] [Google Scholar]
- Beel B, Prager K, Spexard M, Sasso S, Weiss D, Müller N, Heinnickel M, Dewez D, Ikoma D, Grossman AR, et al. (2012) A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell 24: 2992–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan T, Hindle M, Martin SF, Barrios-Llerena ME, Krahmer J, Kis K, Millar AJ, van Ooijen G (2015) Label-free quantitative analysis of the casein kinase 2-responsive phosphoproteome of the marine minimal model species Ostreococcus tauri. Proteomics 15: 4135–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesger J, Wagner V, Weisheit W, Mittag M (2012) Application of phosphoproteomics to find targets of casein kinase 1 in the flagellum of Chlamydomonas. Int J Plant Genomics 2012: 581460 doi: 10.1155/2012/581460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL (2013) Walkabout on the long branches of plant evolution. Curr Opin Plant Biol 16: 70–77 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK, Sakakibara K (2007) Green genes-comparative genomics of the green branch of life. Cell 129: 229–234 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F, et al. (2017) Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171: 287–304.e15 [DOI] [PubMed] [Google Scholar]
- Broda H, Van Gooch VD, Taylor W, Aiuto N, Hastings JW (1986) Acquisition of circadian bioluminescence data in Gonyaulax and an effect of the measurement procedure on the period of the rhythm. J Biol Rhythms 1: 251–263 [DOI] [PubMed] [Google Scholar]
- Bruce VG (1970) The biological clock in Chlamydomonas reinhardi. J Protozool 17: 328–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne TE, Wells MR, Johnson CH (1992) Circadian rhythms of chemotaxis to ammonium and of methylammonium uptake in Chlamydomonas. Plant Physiol 98: 879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Ramirez DL, Dodd AN (2018) New connections between circadian rhythms, photosynthesis, and environmental adaptation. Plant Cell Environ 41: 2515–2517 [DOI] [PubMed] [Google Scholar]
- Chang C, Bowman JL, Meyerowitz EM (2016) Field Guide to Plant Model Systems. Cell 167: 325–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coesel S, Mangogna M, Ishikawa T, Heijde M, Rogato A, Finazzi G, Todo T, Bowler C, Falciatore A (2009) Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Rep 10: 655–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corellou F, Schwartz C, Motta JP, Djouani-Tahri EB, Sanchez F, Bougeta FY (2009) Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote Ostreococcus. Plant Cell 21: 3436–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés Llorca L, Li R, Yon F, Schäfer M, Halitschke R, Robert CAM, Kim SG, Baldwin IT (2020) ZEITLUPE facilitates the rhythmic movements of Nicotiana attenuata flowers. Plant J 103: 308–322 [DOI] [PubMed] [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL (2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creux NM, Brown EA, Garner AG, Saeed S, Scher CL, Holalu S V., Yang D, Maloof JN, Blackman BK, Harmer SL (2021) Flower orientation influences floral temperature, pollinator visits and plant fitness. New Phytol 232: 868–879 [DOI] [PubMed] [Google Scholar]
- Dai S, Wei X, Pei L, Thompson RL, Liu Y, Heard JE, Ruff TG, Beachy RN (2011) BROTHER OF LUX ARRHYTHMO is a component of the Arabidopsis circadian clock. Plant Cell 23: 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe H, Prager K, Mittag M (2012) Novel interaction of two clock-relevant RNA-binding proteins C3 and XRN1 in Chlamydomonas reinhardtii. FEBS Lett 586: 3969–3973 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Kay SA (2000) Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12: 2499–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djouani-Tahri EB, Christie JM, Sanchez-Ferandin S, Sanchez F, Bouget FY, Corellou F (2011) A eukaryotic LOV-histidine kinase with circadian clock function in the picoalga Ostreococcus. Plant J 65: 578–588 [DOI] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR (2005) Cell biology: Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Doherty CJ, Kay SA (2010) Circadian control of global gene expression patterns. Annu Rev Genet 44: 419–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornbusch T, Michaud O, Xenarios I, Fankhauser C (2014) Differentially phased leaf growth and movements in Arabidopsis depend on coordinated circadian and light regulation. Plant Cell 26: 3911–3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ (2017) Making time: conservation of biological clocks from fungi to animals. Microbiol Spectr 5, doi: 10.1128/microbiolspec.funk-0039-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RS, Green EW, Zhao Y, Van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, et al. (2012) Peroxiredoxins are conserved markers of circadian rhythms. Nature 485: 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eelderink-Chen Z, Bosman J, Sartor F, Dodd AN, Kovács ÁT, Merrow M (2021) A circadian clock in a nonphotosynthetic prokaryote. Sci Adv 7: eabe2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann W, Simon K, Jyh Phen C (1992) Leaf movement rhythm in Arabidopsis thaliana. Zeitschr Naturforsch - Sect C J Biosci 47: 925–928 [Google Scholar]
- Farinas B, Mas P (2011) Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J 66: 318–329 [DOI] [PubMed] [Google Scholar]
- Farré EM (2020) The brown clock: circadian rhythms in stramenopiles. Physiol Plant 169: 430–441 [DOI] [PubMed] [Google Scholar]
- Farré EM, Liu T (2013) The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr Opin Plant Biol 16: 621–629 [DOI] [PubMed] [Google Scholar]
- Fehér B, Kozma-Bognár L, Kevei É, Hajdu A, Binkert M, Davis SJ, Schäfer E, Ulm R, Nagy F (2011) Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana. Plant J 67: 37–48 [DOI] [PubMed] [Google Scholar]
- Foley LE, Emery P (2020) Drosophila cryptochrome: variations in blue. J Biol Rhythms 35: 16–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz S, Ignatz E, Wenzel S, Zielosko H, Ngurah Putu EPG, Maestre-Reyna M, Tsai MD, Yamamoto J, Mittag M, Essen LO (2018) Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii. Nucleic Acids Res 46: 8010–8022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil KE, Park CM (2019) Thermal adaptation and plasticity of the plant circadian clock. New Phytol 221: 1215–1229 [DOI] [PubMed] [Google Scholar]
- Green RM, Tobin EM (1999) Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA 96: 4176–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner A, Kelterborn S, Evers H, Kreimer G, Sizova I, Hegemann P (2017) Targeting of photoreceptor genes in Chlamydomonas reinhardtii via zinc-finger nucleases and CRISPR/Cas9. Plant Cell 29: 2498–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Chang X, Zhang Z, Chen H, He H, Zhong B, Deng XW (2019) Origin and evolution of core components responsible for monitoring light environment changes during plant terrestrialization. Mol Plant 12: 847–862 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Kay SA (2005) Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17: 1926–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon MJ, Mielczarek O, Robertson FC, Hubbard KE, Webb AAR (2013) Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502: 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M, Zabulon G, Corellou F, Ishikawa T, Brazard J, Usman A, Sanchez F, Plaza P, Martin M, Falciatore A, et al. (2010) Characterization of two members of the cryptochrome/photolyase family from Ostreococcus tauri provides insights into the origin and evolution of cryptochromes. Plant Cell Environ 33: 1614–1626 [DOI] [PubMed] [Google Scholar]
- Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA (2011) LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol 21: 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, et al. (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm K, Källman T, Gyllenstrand N, Hedman H, Lagercrantz U (2010) Does the core circadian clock in the moss Physcomitrella patens (Bryophyta) comprise a single loop? BMC Plant Biol 10: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Devisetty UK, Harmer SL (2013) Accurate timekeeping is controlled by a cycling activator in Arabidopsis. Elife 2: e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Harmer SL (2014) Wheels within wheels: the plant circadian system. Trends Plant Sci 19: 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Liu X, Lu Q, Yang Y, He Q, Liu Y, Liu X (2021) FRQ-CK1 interaction underlies temperature compensation of the Neurospora circadian clock. MBio 12: e0142521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 335: 75–79 [DOI] [PubMed] [Google Scholar]
- Ichikawa K, Sugita M, Imaizumi T, Wada M, Aoki S (2004) Differential expression on a daily basis of plastid sigma factor genes from the moss Physcomitrella patens. Regulatory interactions among PpSig5, the circadian clock, and blue light signaling mediated by cryptochromes. Plant Physiol 136: 4285–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev D, Voytsekh O, Schmidt EM, Fiedler M, Nykytenko A, Mittag M (2006) A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol 142: 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbur ML, Johnson CH (2022) Spectres of clock evolution: past, present, and yet to come. Front Physiol 12: 815847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH, Golden SS, Kondo T (1998) Adaptive significance of circadian programs in cyanobacteria. Trends Microbiol 6: 407–410 [DOI] [PubMed] [Google Scholar]
- Jones MA, Hu W, Litthauer S, Lagarias JC, Harmer SL (2015) A constitutively active allele of phytochrome B maintains circadian robustness in the absence of light. Plant Physiol 169: 814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Barbosa AD, Hutin S, Kumita JR, Gao M, Derwort D, Silva CS, Lai X, Pierre E, Geng F, et al. (2020) A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585: 256–260 [DOI] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S, et al. (2016) Phytochromes function as thermosensors in Arabidopsis. Science 354: 886–889 [DOI] [PubMed] [Google Scholar]
- Kamioka M, Takao S, Suzuki T, Taki K, Higashiyam T, Kinoshita T, Nakamichi N (2016) Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28: 696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamrani YY, Matsuo T, Mittag M, Minagawa J (2018) ROC75 is an attenuator for the circadian clock that controls LHCSR3 expression. Plant Cell Physiol 59: 2602–2607 [DOI] [PubMed] [Google Scholar]
- Keeling PJ (2007) Ostreococcus tauri: seeing through the genes to the genome. Trends Genet 23: 151–154 [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW (2005) The tree of eukaryotes. Trends Ecol Evol 20: 670–676 [DOI] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Niwa Y, Onai K, Yamano T, Fukuzawa H, Ishiura M, Matsuo T (2017) CSL encodes a leucine-rich-repeat protein implicated in red/violet light signaling to the circadian clock in Chlamydomonas. PLoS Genet 13: e1006645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Strayer CA, Kulkarni RD, Taylor W, Ishiura M, Golden SS, Johnson CH (1993) Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA 90: 5672–5676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshelev M, Sarma S, Price RE, Wehrens XHT, Cooper TA (2010) Heart-specific overexpression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum Mol Genet 19: 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer J, Hindle M, Perby LK, Mogensen HK, Nielsen TH, Halliday KJ, VanOoijen G, LeBihan T, Millar AJ (2021) The circadian clock gene circuit controls protein and phosphoprotein rhythms in Arabidopsis thaliana. Mol Cell Proteomics 21: 100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakina J, Dodd AN (2012) Phosphorylation in the plant circadian system. Trends Plant Sci 17: 575–583 [DOI] [PubMed] [Google Scholar]
- Lagercrantz U, Billhardt A, Rousku SN, Leso M, Reza SH, Eklund DM (2021) DE-ETIOLATED1 has a role in the circadian clock of the liverwort Marchantia polymorpha. New Phytol 232: 595–609 [DOI] [PubMed] [Google Scholar]
- Lagercrantz U, Billhardt A, Rousku SN, Ljung K, Eklund DM (2020) Nyctinastic thallus movement in the liverwort Marchantia polymorpha is regulated by a circadian clock. Sci Rep 10: 8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie ES, Rojas CC, Neme M, Hiltbrunner A, Wigge PA, Schäfer E, Vierstra RD, Casal JJ (2016) Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354: 897–900 [DOI] [PubMed] [Google Scholar]
- Li W, Flores DC, Füßel J, Euteneuer J, Dathe H, Zou Y, Weisheit W, Wagner V, Petersen J, Mittag M (2018) A musashi splice variant and its interaction partners influence temperature acclimation in Chlamydomonas. Plant Physiol 178: 1489–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde AM, Eklund DM, Kubota A, Pederson ERA, Holm K, Gyllenstrand N, Nishihama R, Cronberg N, Muranaka T, Oyama T, et al. (2017) Early evolution of the land plant circadian clock. New Phytol 216: 576–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TL, Newton L, Liu MJ, Shiu SH, Farré EM (2016) A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol 170: 528–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Loros J, Dunlap JC (2000) Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc Natl Acad Sci USA 97: 234–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Reyes P, Romero-Campero FJ, Teresa Ruiz M, Romero JM, Valverde F (2017) Evolution of daily gene co-expression patterns from algae to plants. Front Plant Sci 8: 1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Webb CJ, Knowles SM, Kim SHJ, Wang Z, Tobin EM (2012) CCA1 and ELF3 interact in the control of hypocotyl length and flowering time in arabidopsis. Plant Physiol 158: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck M, Velázquez Escobar F, Glass K, Sabotke MI, Hagedorn R, Corellou F, Siebert F, Hildebrandt P, Hegemann P (2019) Photoreactions of the histidine kinase rhodopsin Ot-HKR from the marine picoalga Ostreococcus tauri. Biochemistry 58: 1878–1891 [DOI] [PubMed] [Google Scholar]
- de Mairan J (1729) Observation botanique. Histoire de l’Academie Royale) des Sciences. Paris, l'Imprimerie Royale, pp 35–36
- Martin-Tryon EL, Kreps JA, Harmer SL (2007) GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol 143: 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P, Kim WY, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Matsuo T, Iida T, Ohmura A, Gururaj M, Kato D, Mutoh R, Ihara K, Ishiura M (2020) The role of ROC75 as a daytime component of the circadian oscillator in Chlamydomonas reinhardtii. PLOS Genet 16: e1008814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Ishiura M (2010) New insights into the circadian clock in Chlamydomonas. Int Rev Cell Mol Biol 280: 281–314 [DOI] [PubMed] [Google Scholar]
- Matsuo T, Okamoto K, Onai K, Niwa Y, Shimogawara K, Ishiura M (2008) A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev 22: 918–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Onai K, Okamoto K, Minagawa J, Ishiura M (2006) Real-time monitoring of chloroplast gene expression by a luciferase reporter: evidence for nuclear regulation of chloroplast circadian period. Mol Cell Biol 26: 863–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T (2000) Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol 41: 1002–1012 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergenhagen D, Mergenhagen E (1987) The biological clock of Chlamydomonas reinhardii in space. Eur J Cell Biol 43: 203–207 [PubMed] [Google Scholar]
- Millar AJ, Straume M, Chory J, Chua NH, Kay SA (1995) The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267: 1163–1166 [DOI] [PubMed] [Google Scholar]
- Mittag M, Kiaulehn S, Johnson CH (2005) The circadian clock in Chlamydomonas reinhardtii. What is it for? What is it similar to? Plant Physiol 137: 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carré IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Monnier A, Liverani S, Bouvet R, Jesson B, Smith JQ, Mosser J, Corellou F, Bouget FY (2010) Orchestrated transcription of biological processes in the marine picoeukaryote Ostreococcus exposed to light/dark cycles. BMC Genomics 11: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller N, Wenzel S, Zou Y, Künzel S, Sasso S, Weiß D, Prager K, Grossman A, Kottke T, Mittag M (2017) A plant cryptochrome controls key features of the Chlamydomonas circadian clock and its life cycle. Plant Physiol 174: 185–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano AJ, Kawagoe T, Sugisaka J, Honjo MN, Iwayama K, Kudoh H (2019) Annual transcriptome dynamics in natural environments reveals plant seasonal adaptation. Nat Plants 5: 74–83 [DOI] [PubMed] [Google Scholar]
- Nagel DH, Doherty CJ, Pruneda-Paz JL, Schmitz RJ, Ecker JR, Kay SA (2015) Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc Natl Acad Sci USA 112: E4802–E4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N (2020) The transcriptional network in the Arabidopsis circadian clock system. Genes (Basel) 11: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Kamioka M, Suzukie T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T (2012) Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci USA 109: 17123–17128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimamurthy R, Virshup DM (2021) The phosphorylation switch that regulates ticking of the circadian clock. Mol Cell 81: 1133–1146 [DOI] [PubMed] [Google Scholar]
- Niinuma K, Someya N, Kimura M, Yamaguchi I, Hamamoto H (2005) Circadian rhythm of circumnutation in inflorescence stems of Arabidopsis. Plant Cell Physiol 46: 1423–1427 [DOI] [PubMed] [Google Scholar]
- Niwa Y, Matsuo T, Onai K, Kato D, Tachikawa M, Ishiura M (2013) Phase-resetting mechanism of the circadian clock in Chlamydomonas reinhardtii. Proc Natl Acad Sci 110: 13666–13671doi: 10.1073/pnas.1220004110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohales MA, Kay SA (2016) Molecular mechanisms at the core of the plant circadian oscillator. Nat Struct Mol Biol 23: 1061–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordally ZB, Millar AJ (2015) Clocks in algae. Biochemistry 54: 171–183 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4-ELF3-"LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada R, Kondo S, Satbhai SB, Yamaguchi N, Tsukuda M, Aoki S (2009) Functional characterization of CCA1/LHY homolog genes, PpCCA1a and PpCCA1b, in the moss Physcomitrella patens. Plant J 60: 551–563 [DOI] [PubMed] [Google Scholar]
- Okumura KI, Aso Y, Tayama K, Yoshida N, Takiguchi Y, Takemura Y, Inukai T (2002) Myotonic dystrophy associated with variable circadian rhythms of serum cortisol and isolated thyrotropin deficiency. Am J Med Sci 324: 158–160 [DOI] [PubMed] [Google Scholar]
- van Ooijen G, Hindle M, Martin SF, Barrios-Llerena M, Sanchez F, Bouget FY, O’Neill JS, Le Bihan T, Millar AJ (2013) Functional analysis of casein kinase 1 in a minimal circadian system. PLoS One 8: e70021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paajanen P, Lane de Barros Dantas L, Dodd AN (2021) Layers of crosstalk between circadian regulation and environmental signalling in plants. Curr Biol 31: R399–R413 [DOI] [PubMed] [Google Scholar]
- Pajuelo E, Pajuelo P, Clemente MT, Márquez AJ (1995) Regulation of the expression of ferredoxin-nitrite reductase in synchronous cultures of Chlamydomonas reinhardtii. Biochim Biophys Acta (BBA)/Protein Struct Mol 1249: 72–78 [DOI] [PubMed] [Google Scholar]
- Palágyi A, Terecskei K, Ádám É, Kevei É, Kircher S, Mérai Z, Schäfer E, Nagy F, Kozma-Bognár L (2010) Functional analysis of amino-terminal domains of the photoreceptor phytochrome B. Plant Physiol 153: 1834–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-García P, Ma Y, Yanovsky MJ, Mas P (2015) Time-dependent sequestration of RVE8 by LNK proteins shapes the diurnal oscillation of anthocyanin biosynthesis. Proc Natl Acad Sci USA 112: 5249–5253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Rredhi A, Szyttenholm J, Oldemeyer S, Kottke T, Mittag M (2021) The world of algae reveals a broad variety of cryptochrome properties and functions. Front Plant Sci 12: 766509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS (1993) Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol 55: 17–54 [DOI] [PubMed] [Google Scholar]
- Podolec R, Demarsy E, Ulm R (2021) Perception and signaling of ultraviolet-B radiation in plants. Annu Rev Plant Biol 72: 793–822 [DOI] [PubMed] [Google Scholar]
- Pooam M, Dixon N, Hilvert M, Misko P, Waters K, Jourdan N, Drahy S, Mills S, Engle D, Link J, et al. (2021) Effect of temperature on the Arabidopsis cryptochrome photocycle. Physiol Plant 172: 1653–1661 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA (2009) A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, Phinney BS, Harmer SL (2011) REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet 7: e1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Goffinet B, Meyberg R, Wu SZ, Bezanilla M (2020) The moss Physcomitrium (Physcomitrella) patens: a model organism for non-seed plants. Plant Cell 32: 1361–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M (2021) Circadian rhythms and the transcriptional feedback loop (Nobel Lecture). Angew Chem 60: 8650–8666 [DOI] [PubMed] [Google Scholar]
- Rugnone ML, Soverna AF, Sanchez SE, Schlaen RG, Hernando CE, Seymour DK, Mancini E, Chernomoretz A, Weigel D, Mas P, et al. (2013) LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc Natl Acad Sci USA 110: 12120–12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryo M, Matsuo T, Yamashino T, Ichinose M, Sugita M, Aokia S (2016) Diversity of plant circadian clocks: insights from studies of Chlamydomonas reinhardtii and Physcomitrella patens. Plant Signal Behav 11: e1116661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, McClung CR (2005) Pseudo-response Regulator 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez SE, Rugnone ML, Kay SA (2020) Light perception: a matter of time. Mol Plant 13: 363–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasso S, Stibor H, Mittag M, Grossman AR (2018) The natural history of model organisms from molecular manipulation of domesticated Chlamydomonas reinhardtii to survival in nature. Elife 7: e39233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satbhai SB, Yamashino T, Okada R, Nomoto Y, Mizuno T, Tezuka Y, Itoh T, Tomita M, Otsuki S, Aoki S (2011) Pseudo-response regulator (PRR) homologues of the moss Physcomitrella patens: insights into the evolution of the prr family in land plants. DNA Res 18: 39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Schmidt M, Geßner G, Luff M, Heiland I, Wagner V, Kaminski M, Geimer S, Eitzinger N, Reißenweber T, Voytsekh O, et al. (2006) Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell 18: 1908–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Bueno G, Romero-Campero FJ, Lucas-Reina E, Romero JM, Valverde F (2017) Evolution of photoperiod sensing in plants and algae. Curr Opin Plant Biol 37: 10–17 [DOI] [PubMed] [Google Scholar]
- Serrano G, Herrera-Palau R, Romero JM, Serrano A, Coupland G, Valverde F (2009) Chlamydomonas CONSTANS and the evolution of plant photoperiodic signaling. Curr Biol 19: 359–368 [DOI] [PubMed] [Google Scholar]
- Soriano G, Cloix C, Heilmann M, Núñez-Olivera E, Martínez-Abaigar J, Jenkins GI (2018) Evolutionary conservation of structure and function of the UVR8 photoreceptor from the liverwort Marchantia polymorpha and the moss Physcomitrella patens. New Phytol 217: 151–162 [DOI] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Green RM, Wang ZY, Tobin EM (1998) Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc Natl Acad Sci USA 95: 11020–11025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Ong MS, Green RM, Tobin EM (1999) The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proc Natl Acad Sci USA 96: 12362–12366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thommen Q, Pfeuty B, Corellou F, Bouget FY, Lefranc M (2012) Robust and flexible response of the Ostreococcus tauri circadian clock to light/dark cycles of varying photoperiod. FEBS J 279: 3432–3448 [DOI] [PubMed] [Google Scholar]
- Thommen Q, Pfeuty B, Schatt P, Bijoux A, Bouget FY, Lefranc M (2015) Probing entrainment of Ostreococcus tauri circadian clock by blue and green light through a mathematical modeling approach. Front Genet 6: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilbrook K, Dubois M, Crocco CD, Yin R, Chappuis R, Allorent G, Schmid-Siegert E, Goldschmidt-Clermont M, Ulma R (2016) UV-B perception and acclimation in Chlamydomonas reinhardtii. Plant Cell 28: 966–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara TN, Mizutani Y, Kuwata K, Hirota T, Sato A, Mizoi J, Takao S, Matsuo H, Suzuki T, Ito S, et al. (2019) Casein kinase 1 family regulates PRR5 and TOC1 in the Arabidopsis circadian clock. Proc Natl Acad Sci USA 166: 11528–11536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytsekh O, Seitz SB, Iliev D, Mittag M (2008) Both subunits of the circadian RNA-binding protein CHLAMY1 can integrate temperature information. Plant Physiol 147: 2179–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner L, Schmal C, Staiger D, Danisman S (2017) The plant leaf movement analyzer (PALMA): A simple tool for the analysis of periodic cotyledon and leaf movement in Arabidopsis thaliana. Plant Methods 13: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jiang B, Gu L, Chen Y, Mora M, Zhu M, Noory E, Wang Q, Lin C (2021) A photoregulatory mechanism of the circadian clock in Arabidopsis. Nat Plants 7: 1397–1408 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Webb AAR (2003) The physiology of circadian rhythms in plants. New Phytol 160: 281–303 [DOI] [PubMed] [Google Scholar]
- Webb AAR, Seki M, Satake A, Caldana C (2019) Continuous dynamic adjustment of the plant circadian oscillator. Nat Commun 10: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Wang P, Liu X, Yuan L, Wang L, Zhang C, Li Y, Xing H, Zhi L, Yue Z, et al. (2014) LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 26: 2843–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Kim YJ, Somers DE (2021) Post‐translational mechanisms of plant circadian regulation. Genes (Basel) 12: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yon F, Joo Y, Cortés Llorca L, Rothe E, Baldwin IT, Kim SG (2016) Silencing Nicotiana attenuata LHY and ZTL alters circadian rhythms in flowers. New Phytol 209: 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Schneid C, Iliev D, Schmidt EM, Wagner V, Wollnik F, Mittag M (2004) The circadian RNA-binding protein CHLAMY 1 represents a novel type heteromer of RNA recognition motif and lysine homology domain-containing subunits. Eukaryot Cell 3: 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Lin K, Gan S, Li G (2019) Heterologous expression of ELF4 from Chlamydomonas reinhardtii and Physcomitrella patens delays flowering in Arabidopsis thaliana. Plant Syst Evol 305: 777–785 [Google Scholar]
- Zones JM, Blaby IK, Merchant SS, Umen JG (2015) High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 27: 2743–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]