Abstract
The circadian clock facilitates the survival and reproduction of crop plants under harsh environmental conditions such as drought and osmotic and salinity stresses, mainly by reprogramming the endogenous transcriptional landscape. Nevertheless, the genome-wide roles of core clock components in rice (Oryza sativa L.) abiotic stress tolerance are largely uncharacterized. Here, we report that CIRCADIAN CLOCK ASSOCIATED1 (OsCCA1), a vital clock component in rice, is required for tolerance to salinity, osmotic, and drought stresses. DNA affinity purification sequencing coupled with transcriptome analysis identified 692 direct transcriptional target genes of OsCCA1. Among them, the genes involved in abscisic acid (ABA) signaling, including group A protein phosphatase 2C genes and basic region and leucine zipper 46 (OsbZIP46), were substantially enriched. Moreover, OsCCA1 could directly bind the promoters of OsPP108 and OsbZIP46 to activate their expression. Consistently, oscca1 null mutants generated via genome editing displayed enhanced sensitivities to ABA signaling. Together, our findings illustrate that OsCCA1 confers multiple abiotic stress tolerance likely by orchestrating ABA signaling, which links the circadian clock with ABA signaling in rice.
A rice core circadian clock component confers broad-spectrum tolerance to abiotic stress by transcriptional orchestration of multiple key genes in abscisic acid signaling.
Introduction
Land plants have evolved intricate circadian clock systems to synchronize their developmental and physiological behaviors to support growth and fitness by sensing and predicting the daily rhythmic environmental oscillations (Greenham and McClung, 2015; Sanchez and Kay, 2016). In the past decades, the roles of crop circadian clock components have mainly been characterized as the regulator of flowering time and adaptation to latitude cline (Wei et al., 2020a). Circadian clock components are also involved in biotic resistance and abiotic stress tolerance in both model plant Arabidopsis thaliana and staple crops. For instance, Arabidopsis GIGANTEA (GI), a key regulator of flowering time and circadian clock, can affect the adaptability to salt stress, through Salt Overly Sensitive 2 (SOS2)-mediated activation of an SOS-signaling pathway (Kim et al., 2013). In rice (Oryza sativa L.), the clock component O.sativa Pseudo Response Regulator 73 (OsPRR73) confers salt tolerance by repressing High-affinity K+ transporters 2;1 (OsHKT2;1) transcription, thereby reducing the accumulation of Na+ and reactive oxygen species (ROS), besides its role in controlling heading date (Liang et al., 2020; Wei et al., 2020b). In addition, our recent report indicated that a ternary repressive complex OsEC1 (O. sativa Evening Complex 1), comprised OsELF4a (O. sativa EARLY FLOWERING 4a), OsELF3-1, and OsLUX (O. sativa LUX ARRHYTHMO), is required for salt tolerance and heading date determination mainly through repressing OsGI transcription (Wang et al., 2021b). Together these findings suggest that rice circadian clock plays a vital role in coordinating plant development and stress response. Oryza sativa CIRCADIAN CLOCK ASSOCIATED 1 (hereafter as OsCCA1, a.k.a. OsLHY), a putative ortholog of Arabidopsis CCA1, was recently characterized as a vital circadian rhythm component in rice, with a transcriptional peak at morning (Sun et al., 2021). OsCCA1 acts as a master developmental regulator by integrating sugar response and strigolactone signaling to control tiller and panicle development (Wang et al., 2020), photoperiodic heading date, and nitrogen use efficiency (Zhang et al., 2020; Sun et al., 2021). Nevertheless, the roles of OsCCA1 and other rice core clock components in abiotic stress responses are largely unknown.
Abscisic acid (ABA) is a well-known plant hormone in regulating abiotic stress response, especially for salt and drought stress (Zhu, 2002; Miao et al., 2020). A canonical mechanism underlying ABA signal transduction has been well established. Briefly, ABA is perceived by the pyrabactin resistance 1 (PYR1)/PYR1-like (PYL) protein family which interacts with clade A type 2C protein phosphates (PP2Cs) to release sucrose nonfermenting 1-related protein kinase 2 (SnRK2). Once SnRK2s are activated through autophosphorylation, they further phosphorylate multiple downstream effectors, which are mainly transcription factors (TFs) such as NAC (NAM, CUC, and ATAF), basic leucine zipper (bZIP), ethylene-responsive factor/APETALA 2, and MYB (Antoni et al., 2013; Zhu, 2016). The core components of rice ABA signaling pathway have been identified as well, including 13 PYLs (Miao et al., 2018), 10 PP2Cs (Singh et al., 2015), and 10 SnRK2s (also known as osmotic stress/ABA-activated protein kinases, SAPKs; Kobayashi et al., 2004). Rice ABA receptor OsPYL /regulatory components of the ABA receptor interacts with a group A PP2C, OsPP2C30, which also follows a similar process to release kinase OsSAPK2 to reconstitute a core ABA signaling cascade (Kim et al., 2012). Most of rice PP2Cs family members play a considerable role in ABA signaling and stress tolerance (Singh et al., 2015; Zong et al., 2016; Bhatnagar et al., 2017; Miao et al., 2020). For instance, the transgenic plants of overexpression OsPP108 reduced the sensitive to ABA treatment and increased the tolerant to high salt and mannitol stresses during seed germination, root growth, and overall seedling growth (Singh et al., 2015). In addition, the bZIP family of transcription factors including OsbZIP46 were found to act in the ABA-dependent pathway to mediate downstream transcription cascades in response to stress condition (Xiang et al., 2008; Zou et al., 2008; Tang et al., 2012).
The close interplay between ABA and circadian clock has been implicated in Arabidopsis, as evidenced by the endogenous level of ABA and genes involved in ABA biosynthesis and response are under a tight control of circadian clock (Lee et al., 2006; Covington et al., 2008; Legnaioli et al., 2009; Liu et al., 2013). Timing of CAB expressing 1 (TOC1), a well-known Arabidopsis evening-phased key clock component, directly binds the promoter of ABA-BINDING PROTEIN/ GENOMES UNCOUPLED 5 (ABA-related gene) to modulate its circadian expression. Consequently, plants mis-expressing of TOC1 displayed altered survival rates to dehydration stress (Legnaioli et al., 2009). Moreover, a recent study has revealed that LHY controls ABA biosynthesis and downstream response, which enhances stress tolerance and alleviates the inhibitory effects of ABA on seed germination and plant growth (Adams et al., 2018). Similarly, soybean (Glycine max) GmLHY proteins negatively controls drought tolerance by regulating the expression of genes involved in ABA signaling pathway (Wang et al., 2021a). In rice, circadian clock-regulated Receptor for Activated C Kinase 1 enhances salt tolerance and substantially affects ABA accumulation (Zhang et al., 2018). However, the molecular mechanisms underlying the connection between circadian clock and ABA-signaling cascades have not been unveiled in rice.
In this report, we show that null mutants of OsCCA1 are more sensitive to salt, drought and osmotic stresses. By genome-wide identification of the direct targets of OsCCA1, we unexpectedly found that OsCCA1 functions as a key integrator in modulating ABA signaling pathway in rice. OsCCA1 directly binds the promoters of OsPP2C members and OsbZIP46 to activate their transcription, thereby controlling multiple stress-responsive genes responsible for tolerance to salt, osmotic and drought stresses. Our results support a role of OsCCA1 as a crucial molecular hub in linking rice circadian clock and transcriptional cascade of ABA signaling, thus to coordinate growth and abiotic stress responses.
Results
OsCCA1 positively regulates multiple stresses tolerance
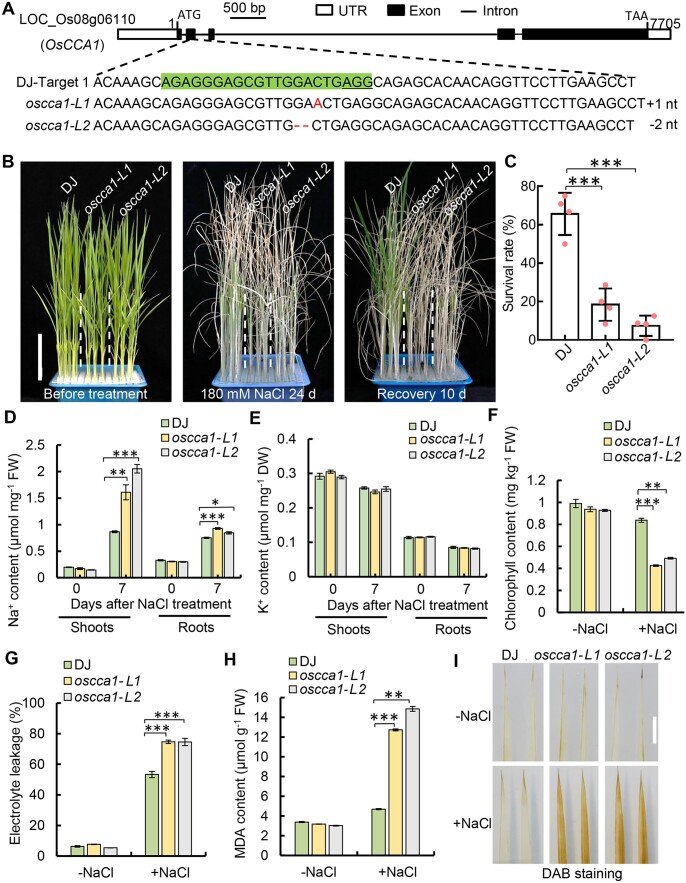
To unravel the role of OsCCA1 in rice abiotic stress tolerance, we first determined its transcriptome profiles under salt and drought stress conditions. The transcript level of OsCCA1 was drastically repressed by either NaCl or PEG-6000 which simulated the drought stress (Supplemental Figure S1). To further examine the function of OsCCA1 in abiotic stress response, two null mutant alleles of OsCCA1 were generated by CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/associated Protein 9) genome editing approach using rice plants in Dongjin (DJ) background (Figure 1A). One of the mutated alleles contained 1-bp insertion within the second exon of OsCCA1 open reading frame, which was named as oscca1-L1, while the other allele had a 2-bp deletion (oscca1-L2). Both alleles created a premature stop codon caused by the frame shift with a 27-aa truncated peptide (Supplemental Figure S2).
Figure 1.
OsCCA1 is required for salinity stress tolerance in rice. A, Loss-of-function mutants of OsCCA1 generated by CRISPR/Cas9-based genome editing which were named as oscca1-L1 and oscca1-L2, respectively. The target sequence is highlighted by the green color and the highlighted sequence that locates in the second exon of OsCCA1. B, The 4-week-old seedlings of DJ and oscca1-Ls before treatment (left), after treatment with 180-mM NaCl for 24 days (middle) and then recovered for 10 days (right). Scale bar = 5 cm. C, Statistical analysis of the survival rates of DJ and oscca1-Ls after recovery. Four biological replicates, and 24 plants were tested in each of biological replicates. Data are presented as mean ± sd. ***P ≤ 0.001, Student’s t test. D and E, Na+ and K+ contents in the shoots and roots of DJ and oscca1-Ls plants under non-NaCl (−NaCl) and 180-mM NaCl 7 days (+NaCl). Three biological replicates, six plants for each biological replicate. Data are presented as mean ± se. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, Student’s t test. F and G, Chlorophyll content (F) and the relative ion leakage (G) in leaves of 2-week-old plants treated with −NaCl or 180-mM NaCl for 7 days (+NaCl). Three biological replicates, three plants of each replicate. Data are presented as mean ± se. **P ≤ 0.01, ***P ≤ 0.001 were defined by Student’s t test. H, The concentrations of MDA in the leaves of 14-day-old plants after 180-mM NaCl treatment for 48 h. Three biological replicates, three plants of each replicate. Data are presented as mean ± se. **P ≤ 0.01 and ***P ≤ 0.001 were generated by Student’s t test. I, ROS content of the leaves of 14-day-old plants after 180-mM NaCl treatment for 3 days or not showed by DAB staining. Scale bar, 1 cm.
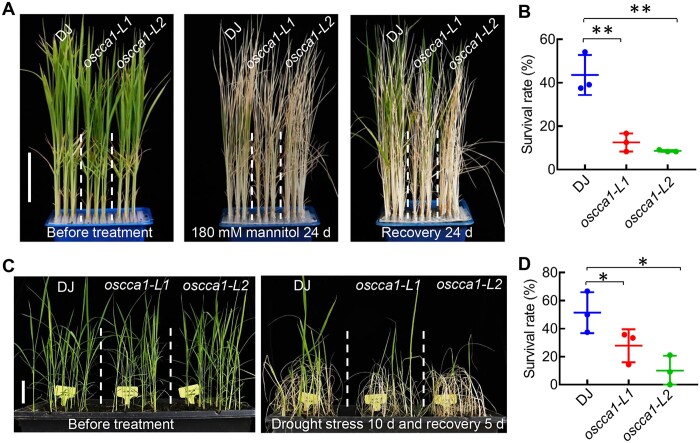
Next, the 4-week-old seedlings of T3 homozygous plants were used for determining the role of OsCCA1 in abiotic stress tolerance. In the normal growth conditions, the plant heights of oscca1-L1 and oscca1-L2 were slightly shorter than that of wild-type (WT) plants (Figures 1, B and 2, A, left), indicating OsCCA1 being a key regulator of plant growth. Remarkably, the oscca1-Ls plants were much more sensitive to 180-mM NaCl or mannitol (Figures 1, B and 2, A). The survival rates of oscca1-Ls plants treated by 180-mM NaCl or mannitol were only ∼10%–30%, whereas the survival rate of DJ plants was ranged from 50% to 70% (Figures 1, C and 2, B). Additionally, the grain yield and fertility of oscca1-Ls plants were substantially decreased when grown on soil irrigated with NaCl solutions (Supplemental Figure S3), indicating that OsCCA1 was required for sustaining the grain yield under salinity condition. Subsequently, we compared the Na+ and K+ contents in the shoots and roots in response to NaCl stress. Remarkably, a higher accumulation of Na+ was observed in both shoots and roots of oscca1-Ls mutant when treated with NaCl, although the K+ content was similar to that of DJ (Figure 1, D and E). Consistently, in the presence of NaCl stress, the chlorophyll content of oscca1-Ls mutants was markedly lower, while the ion leakage rate was much higher, in comparison with those of DJ plants (Figure 1, F and G). The content of malondialdehyde (MDA), an indicator of lipid peroxidation, was significantly increased in oscca1-Ls mutants (Figure 1H), suggesting higher ROS level in oscca1-Ls mutants, which was further confirmed by 3,3-diaminobenzidine (DAB) staining (Figure 1I). These data collectively indicated that OsCCA1 was required for salt stress tolerance. In addition, oscca1 mutants also displayed hypersensitivity to drought stress, with two to three-fold reduction of survival rate compared to WT plants (Figure 2, C and D). Together, these results support a notion that OsCCA1 plays an essential role in conferring rice tolerance to multiple abiotic stresses, including salt, osmotic, and drought.
Figure 2.
OsCCA1 confers osmotic and drought stresses tolerance in rice. A, The seedlings of DJ and oscca1-Ls mutants before treatment (left), after treatment with 180-mM mannitol for 24 days (middle) and recovered for 24 days (right). Scale bar, 5 cm. B, Statistical analysis of survival rates with mannitol treatment in (A). Data represent means ± sd. n = 3 biological replicates, 24 plants of each replicate. **P ≤ 0.01 indicates significant difference by Student’s t test. C, Phenotype of 3-week-old DJ and oscca1-Ls plants without stress treatment (left) and drought treatment for 10 days and recovery 5 days (right). Scale bar, 5 cm. D, Survival percentage of DJ and oscca1 lines after drought stress for 10 days. Data represent means ± sd. n = 3 biological replicates, 16 plants of each replicate. *P ≤ 0.05 indicates significant difference by Student’s t test.
Identification of the genome-wide direct targets of OsCCA1
As OsCCA1 is a Myb-type TF, its role in abiotic stress response is likely mediated through the modulation of target gene expression. In line with its transcriptional regulatory function, the OsCCA1 protein tagged with GFP, transiently expressed in the leaves of Nicotiana benthamiana, was localized in the nuclei of the epidermal cells, as indicated by the nuclear marker of H2B-mCherry (Supplemental Figure S4A). The temporal expression pattern of OsCCA1 in both short-day (SD) and long-day (LD) conditions was also determined by utilizing reverse transcription–quantitative polymerase chain reaction (RT–qPCR). Consistent with a recent report in which OsCCA1 was shown to be rhythmically expressed in many tissues (Wang et al., 2020), our results indicated that the transcript level of OsCCA1 displayed a robust oscillation pattern with a peak at dawn (ZT0, zeitgeber time) in both SD and LD conditions (Supplemental Figure S4B), indicating that OsCCA1 is a morning-phased clock gene and its phase is not affected by the external photoperiod.
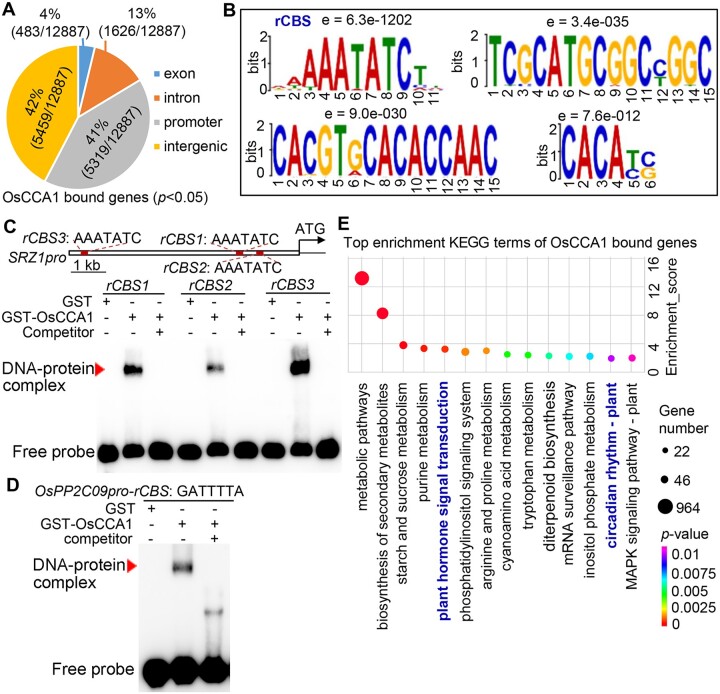
To further identify the direct transcriptional targets of OsCCA1, DNA affinity purification sequencing (DAP-seq) was employed to unravel its genome-wide binding sites (Bartlett et al., 2017). The in vitro-synthesized OsCCA1 protein was used to affinity purify the sheared genomic DNA of 21-day-old DJ plants, followed by deep sequencing (Wu et al., 2020). In total, 83% OsCCA1 binding peaks were distributed in the promoter or intergenic regions, while only 4% was in the exon and 13% was in the intron region (Figure 3A). Among the identified binding peaks, 5,319 were located within the 3-kb regions upstream of ATG (Figure 3A; Supplemental Data set 1). By using MEME suite, a motif with a core sequence of “AAAATATCT” was found as substantially enriched among the OsCCA1 binding regions (e value = 6.3e-1202, hereafter named as rice OsCCA1 binding site (rCBS; Figure 3B). It is noteworthy that the nucleotide sequence of rCBS is slightly deviated from Arabidopsis CCA1-binding site (CBS: AAA/CAATCT) but similar to Evening Element (EE: AAAATATCT; Harmer et al., 2000; Andronis et al., 2008), indicating that OsCCA1 might have a similar but slightly distinct binding property. As shown in Figure 3B, three additional potential OsCCA1 binding sites were also highly enriched. To validate the reliability of our DAP-seq result, electrophoretic mobility shift (EMSA) assays were performed using purified GST-OsCCA1 protein and labeled rCBS DNA probes. Evidently, OsCCA1 could bind the rCBS probes specifically (Figure 3, C and D), as supported by the reduction of binding signals in the presence of unlabeled competitors. Kyoto Encyclopedia of Genes and Genome (KEGG) cluster analysis of the potential OsCCA1-binding genes revealed that hormones signal transduction and circadian rhythm were enriched among the top biological processes (Figure 3E), further indicating that OsCCA1 controls multiple biological processes besides as a core clock component.
Figure 3.
Identification of genome-wide direct targets of OsCCA1. A, The number of bound genes by OsCCA1 obtained by DAP-seq with P < 0.05 and their distribution in the whole genome. The promoter regions were determined as the binding peaks within 3,000-bp upstream of ATG. B, The identified binding motifs of OsCCA1 protein by MEME-ChIP. The core sequence of “AAAATATCT” was substantially enriched among the OsCCA1 binding regions (e-value = 6.3e-1202) and was named as rCBS. C and D, Results of EMSAs confirming OsCCA1 bound to the AAATATC motif of SRZ1pro and OsPP2C09pro. The unlabeled probe was taken as competitor. GST alone was used as negative control of the binding. Red arrowhead indicates the DNA–protein complex. E, The top KEGG-enriched terms of OsCCA1-bound genes by DAP-seq.
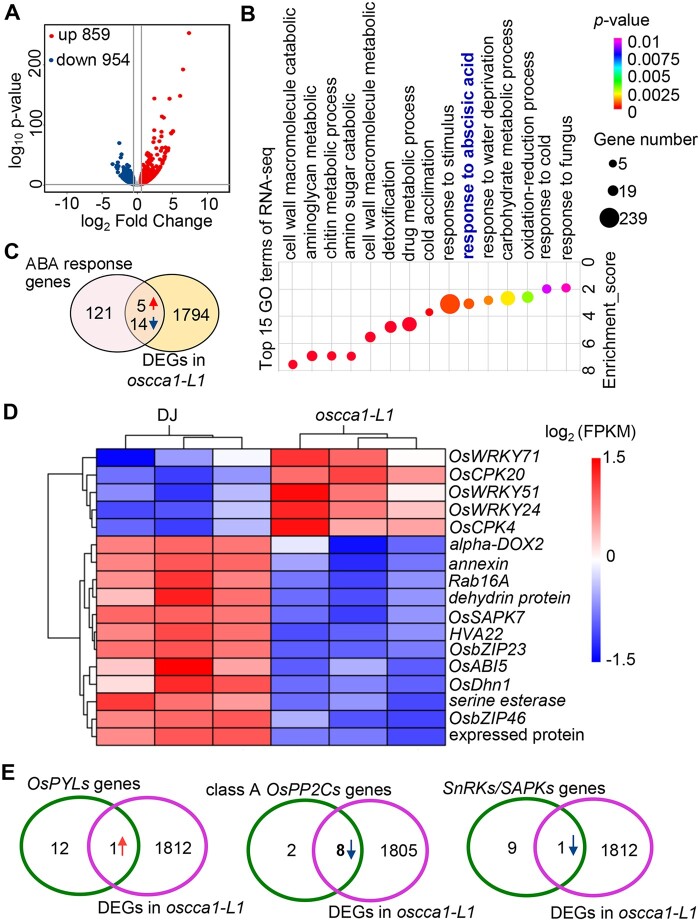
To help define the potential direct transcriptional targets of OsCCA1 in response to abiotic stress such as salinity condition, we conducted RNA-sequencing (RNA-seq) using 3-week-old oscca1-L1 seedlings treated with 180-mM NaCl at ZT0 for 24 h. Using the threshold of P < 0.05 and the value of log2 (fold change) over 1.5 relative to DJ, we obtained 1,813 differentially expressed genes (DEGs) including 859 upregulated and 954 downregulated genes in oscca1-L1 mutant (Figure 4A; Supplemental Data set 2). Gene ontology (GO) analysis revealed that the response to abscisic acid (GO:0009737) cluster was among the top 15 enriched GO terms (Figure 4B). As ABA-signaling plays a key role in salt- and drought-stress response in plants (Zhu, 2002), we compared the 1,813 DEGs in oscca1-L1 mutant and 140 known genes involved in ABA signaling (http://amigo.geneontology.org/amigo/term/GO:0009737). As shown in the Venn diagram in Figure 4C, 19(13.57%) overlapping genes were found, including 5 upregulated genes and 14 downregulated genes, implicating that OsCCA1 is likely involved in ABA signaling. For example, OsbZIP46, OsbZIP23, and ABA INSENSITIVE 5 (OsABI5) were downregulated (Figure 4D), while OsWRKY71, -51 and -24, and calcium-dependent protein kinase 4 (OsCPK4) and -20 were upregulated in oscca1-L1 mutants (Figure 4D). To further reveal the role of OsCCA1 in ABA signaling, we compared the expression of known players including class A PP2C members, OsPYLs and SAPKs in the identified DEGs. Strikingly, 8 out of 10 class-A PP2Cs were downregulated in oscca1 mutant plants (Figure 4E). Moreover, the transcriptional abundance of OsPYL6, the member of ABA receptor subfamily I, was significantly higher in oscca1 mutants, while OsSAPK3, one of the SnRKs family member, was lower (Figure 4E), implying that OsCCA1 orchestrates ABA signaling through multiple targets to confer salinity stress tolerance.
Figure 4.
OsCCA1 modulates ABA signaling pathway in response to high salinity stress condition. A, The number of the upregulated and downregulated genes of oscca1-L1 mutant compared with DJ. The samples were harvested at ZT0 from 3-week-old seedlings after treatment with 180-mM NaCl for 24 h. B, The top 15 enriched GO terms of DEGs in oscca1 under salt stress condition. P-value was determined by Fisher’s test. C, Venn diagram showing the overlapped DEGs in oscca1 mutant with ABA response genes. The red and blue arrows showed the upregulated and downregulated genes, respectively. D, The heatmap showing the transcriptional abundances of 19 ABA response genes in (C). E, Venn diagram showing the OsPYLs genes (left), class A OsPP2Cs genes (middle), SnRKs/SAPKs genes (right) that overlap with DEGs in oscca1 under NaCl stress condition, respectively. The red and blue arrows represent the upregulated and downregulated genes, respectively.
OsCCA1 is involved in the regulation of ABA signaling
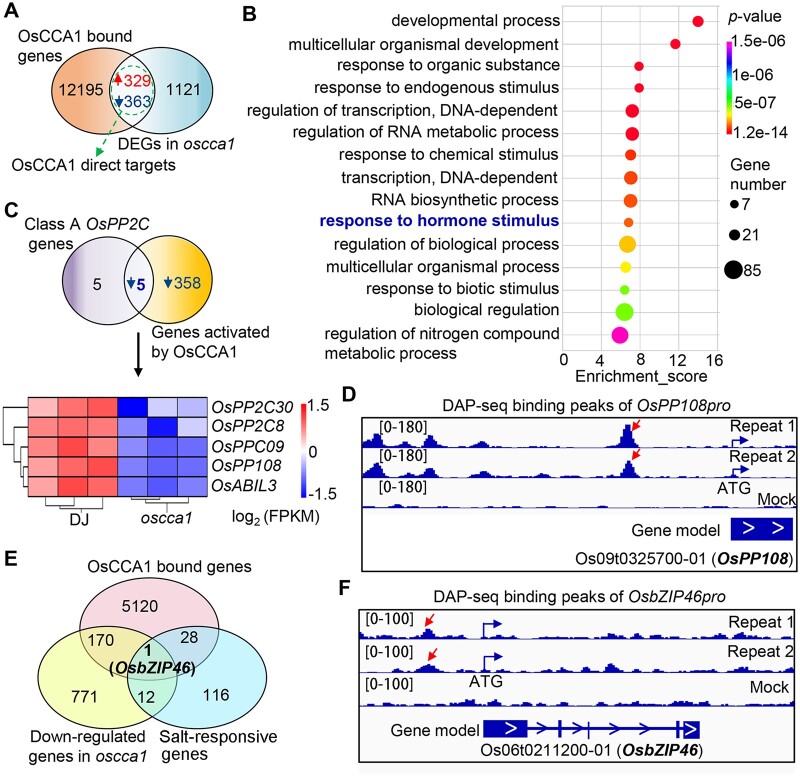
To further identify the direct transcriptional targets of OsCCA1, by comparing DEGs in oscca1 with OsCCA1 potential target genes, we identified 692 potential OsCCA1 direct target genes (Figure 5A). GO analysis of these genes indicated that “response to hormone stimulus” was among the top 15 enriched terms (Figure 5B). Additionally, the biological process of “regulation of nitrogen compound metabolic process” was also enriched, in line with the function of OsCCA1 in mediating nitrogen use efficiency (Zhang et al., 2020). Strikingly, we found that, among 10 class A OsPP2C members, 5 of them were identified as potential direct targets of OsCCA1 (Figure 5C). Heatmap analysis clearly demonstrated that the 5 class-A PP2Cs members (OsPP108/OsPP2C68, OsPP2C09, OsABIL3, OsPP2C30, and OsPP2C8) were downregulated in oscca1 mutant (Figure 5C). Visual checking of the binding peaks of OsCCA1 confirmed that at least one conspicuous peak was located in their promoters (Figure 5D; Supplemental Figure S5), indicating that OsCCA1 orchestrates ABA-signaling pathway by predominantly regulating multiple components of class A PP2Cs members at the transcriptional level. To further unveil the candidate genes of OsCCA1 in response to salt stress, we also performed Venn diagram analysis using the downregulated genes in oscca1, the OsCCA1-bound genes in which the binding peaks locate upstream of ATG, and known salt-responsive genes (Wei et al., 2020b). Interestingly, we found only one candidate gene, OsbZIP46 (Figure 5E). OsbZIP46 is induced by ABA and acts as a positive regulator of drought tolerance in an ABA-dependent pathway (Tang et al., 2012). Overexpression of a constitutive activation form of OsbZIP46 (OsbZIP46CA1) substantially increases drought and osmotic stress tolerance (Tang et al., 2012, 2016). Our DAP-seq results indicated an obvious OsCCA1-binding peak located in the OsbZIP46 promoter 1,263-bp upstream of start codon (Figure 5F). Remarkably, when compared the downregulated DEGs in oscca1 with the upregulated genes in OsbZIP46CA1 overexpression plants, 20.5% (78/379) OsbZIP46-controlled genes were also modulated by OsCCA1 (Supplemental Figure S6A). In particular, the transcript abundance of Responsive to ABA 21 (RAB21), RAB16B, Late embryogenesis abundant 3 (OsLEA3) and OsLEA14, four known direct target genes of OsbZIP46, were all lower in oscca1 plants (Supplemental Figure S6B), suggesting that OsbZIP46 partially mediates the response of OsCCA1 in abiotic stress such as drought. Given the function of OsbZIP46 in abiotic stress adaption is tightly linked to ABA signaling, we proposed that OsCCA1 is likely to orchestrate ABA signaling via a multifaceted mechanism by modulating OsPYL6, class A PP2Cs, OsSAPK3, and OsbZIP46.
Figure 5.
OsCCA1 directly regulates genes involved in ABA signaling. A, Venn diagram showing a comparison of DAP-seq binding genes with the identified DEGs in oscca1 mutant by RNA-seq. The overlapped genes were considered as OsCCA1 direct targets. B, The top 15 enriched GO terms of the OsCCA1 direct targets showed in (A). P-value was defined by Fisher’s test. C, Venn diagram depicting the overlapped gene number between OsCCA1 activated direct target genes and class A PP2C members. The heatmap showing the transcriptional abundances of five overlapped genes in oscca1 mutants and DJ plants. The scale bar showing the log2 (FPKM). D, The binding peaks (repeats 1 and 2) and negative control (mock) of OsCCA1 in OsPP108p (−2,625 bp) by DAP-seq. The [0–180] shows the scale bar of binding peak that refers to the height of the peak. E, Venn diagram showing OsbZIP46 is the only gene among OsCCA1-bound genes, downregulated genes in oscca1 mutant, and the known salt-responsive genes. F, The binding peaks (repeats 1 and 2) and negative control (mock) of OsCCA1 in the promoter (−1,263 bp) of OsbZIP46 by DAP-seq. The [0–100] shows the scale bar of binding peak that refers to the height of the peak.
OsCCA1 transcriptionally activates OsPP108 and OsbZIP46
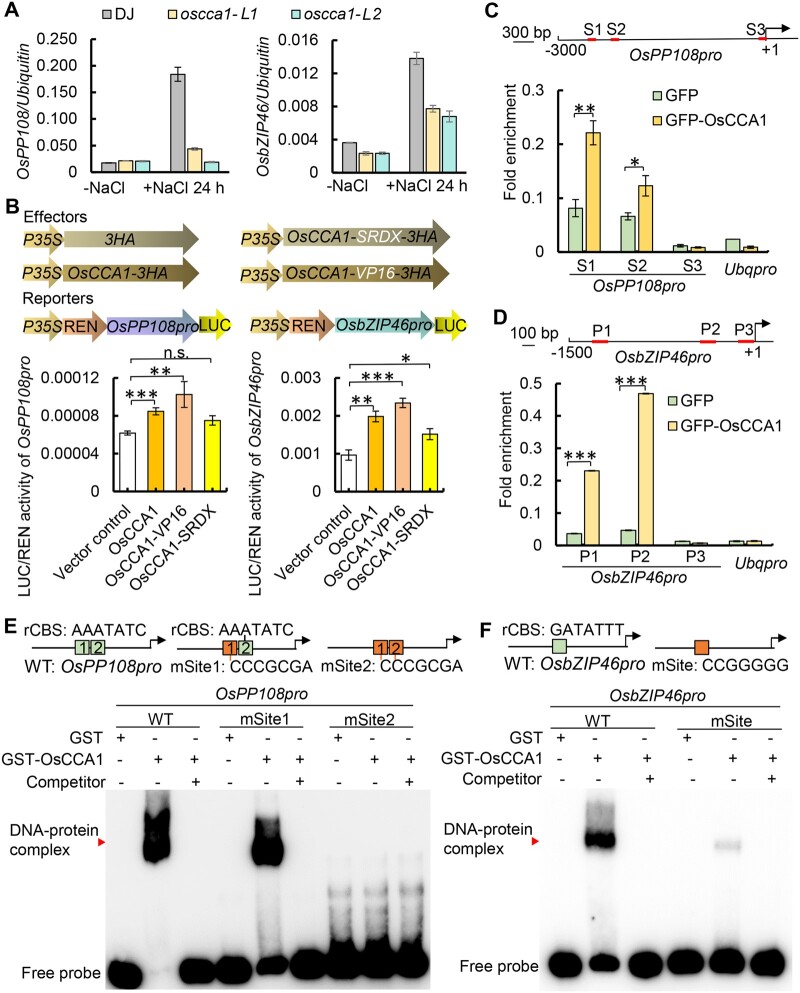
Since class-A OsPP2Cs and OsbZIP46 appeared to be major direct targets of OsCCA1, we were prompted to test the roles of OsCCA1–OsPP2Cs and OsCCA1–OsbZIP46 regulatory modules in salt stress response. Initially, we examined the transcript levels of OsPP2Cs and OsbZIP46 in both the normal growth condition and in the presence of salt stress. While there were no substantial significant differences in gene expression under normal growth condition, upon the treatment of NaCl for 24 h started at ZT0, the transcript abundance of OsPP108 and OsbZIP46 were substantially significantly lower in oscca1 mutants (Figure 6A), indicating that OsCCA1 is required for salt-induced expression of OsPP108 and OsbZIP46. Similarly, other class-A PP2C genes such as OsABIL3, OsPP2C30, and OsPP2C8 were also downregulated in oscca1-Ls plants upon NaCl treatment (Supplemental Figure S7, A, C, D). To determine if OsbZIP46 mediated-transcriptional cascade was affected by OsCCA1, the expression of OsbZIP46 downstream targets, including OsLEA3, OsLEA14, RAB21, and RAB16B, were determined by RT-qPCR. Clearly, the transcript levels of these OsbZIP46 target genes were much lower in oscca1-Ls than in WT plants under salt stress (Supplemental Figure S8, A–D). The role of OsCCA1 in OsPP108 and OsbZIP46 expression were directly examined in vivo by monitoring bioluminescence signals of OsPP108p:LUC and OsbZIP46p:LUC using a rice protoplast transient expression system. When OsCCA1-HA, OsCCA1-VP16-HA, and OsCCA1-SRDX-HA were well expressed in the transformed rice protoplast (Supplemental Figure S9), OsCCA1 and OsCCA1-VP16 could significantly promotes OsPP108 and OsbZIP46 transcription (Figure 6B), but the activation ability of OsCCA1-SRDX was weaker. These results further confirmed that OsCCA1 may function as a transcriptional activator of OsPP108 and OsbZIP46 in response to stress condition. To substantiate the idea in which OsCCA1 protein could physically associate with OsPP108 and OsbZIP46 promoters, chromatin immunoprecipitation (ChIP)-qPCR and EMSAs were conducted. We found that the amplicons of S1 (−2,406 bp) and S2 (−2,301 bp) of OsPP108 promoter, P1 and P2 regions of OsbZIP46, which contain the “GATATTT” element, were significantly accumulated in the immunoprecipitation of OsCCA1-GFP compared to the GFP-alone control (Figure 6, C and D), indicating that OsCCA1 is capable of binding to their promoters. Furthermore, EMSA assay showed that the purified GST-tagged OsCCA1 protein but not the GST alone could directly bind to the rCBS sites in vitro within the promoters of OsPP108 and OsbZIP46, but not the mutated rCBS sites (Figure 6, E and F). Furthermore, the unlabeled probes could effectively compete with the binding (Figure 6, E and F), indicating that OsCCA1 could directly bind to the promoters of OsPP108 and OsbZIP46 through the rCBS elements. Similarly, our EMSA assay confirmed that OsCCA1 could bind the promoter of OsABIL3 via rCBS, but not to the mutated rCBS sites (Supplemental Figure S7B), supporting that rCBS is a bona fide binding site of OsCCA1. Taken together, our results indicated that ABA signaling key components such as OsPP108 and OsbZIP46 are direct transcriptional targets of OsCCA1, which may form genetic cascades regulating multiple stress tolerance in rice.
Figure 6.
OsCCA1 directly binds the promoters of OsPP108 and OsbZIP46 to activate their transcription. A, The transcription levels of OsPP108 (left) and OsbZIP46 (right) in DJ and oscca1-Ls plants under normal condition (−NaCl) and NaCl-treated 24 h (+NaCl 24 h). At least three biological replicates were performed. The error bars represent se. B, Transient expression assays in rice protoplast showing the transcriptional activities of OsPP108 and OsbZIP46 were activated by OsCCA1. The upper parts show the effectors and reporters, respectively. OsPP108p:LUC and OsbZIP46p:LUC were co-transformed with empty vector control (empty) or OsCCA1 overexpression vectors (OsCCA1, OsCCA1-VP16, and OsCCA1-SRDX) as indicated. At least three biological replicates performed. The error bars represent se. P-values were generated by Student’s t test. The abbreviation of n.s. stands for non-significant with P > 0.05, *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001 indicate significance difference. C and D, ChIP-qPCR assay showing OsCCA1 binds the promoters of OsPP108 and OsbZIP46. Top scheme illustrates the locations of the amplicons for ChIP assay. Two biological replicates were performed with similar results. The error bars represent se. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001 indicate significant difference by Student’s t test. E and F, EMSA assay showing that OsCCA1 directly binds to the rCBSs of OsPP108 and OsbZIP46 promoter. The purified GST-OsCCA1 protein was incubated with the indicated the probes designed for DAP-seq binding region of OsPP108 or OsbZIP46 promoter (WT or mSite), and unlabeled probe (WT) or mSite (the orange box showing the mutated site) was used as competitor. GST alone was used as negative control of the binding. Red arrowhead indicates the DNA–protein complex. Two biological replicates were conducted with similar result.
OsCCA1 negatively modulates ABA signaling
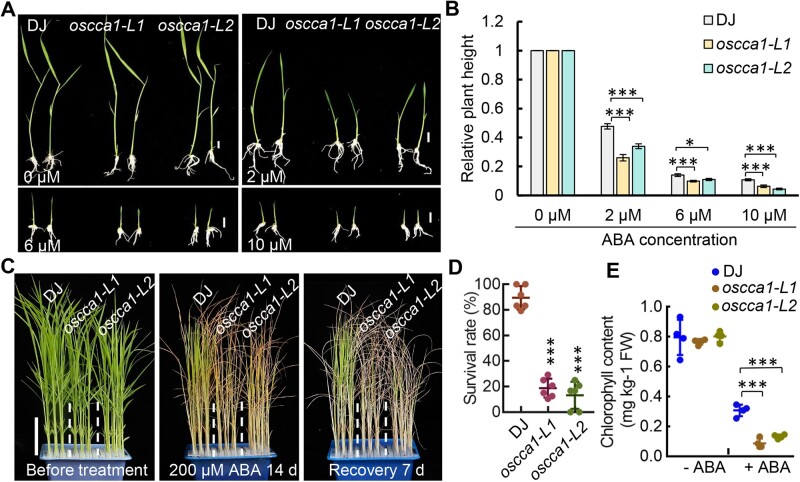
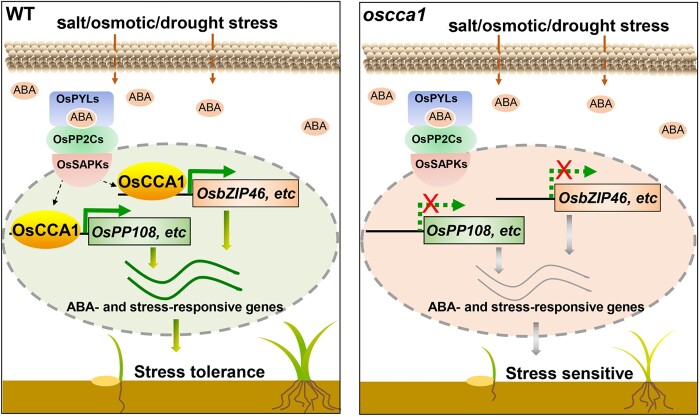
To investigate if OsCCA1 is directly involved in ABA signaling, ABA-mediated plant development inhibition assays were conducted. We found all tested ABA concentrations could effectively inhibit plant growth of both DJ and oscca1 mutants in a dose-dependent manner (Figure 7, A and B). Importantly, the relative plant height of oscca1 was reduced more pronouncedly by ABA, compared with DJ control (Figure 7, A and B), suggesting that oscca1 was hypersensitive to ABA treatment. We also treated the 3-week-old seedlings of DJ and oscca1 mutants, the more tolerant stage to ABA treatment, with 200-μM ABA to calculate the survival rate. After growing for 2 weeks in ABA containing solution followed by 7 days recovery, the survival rates of oscca1 mutants were significantly lower than that of DJ plants (Figure 7, C and D), further confirmed that oscca1 mutant was more sensitive to ABA. Consistently, the chlorophyll contents were lower in oscca1-Ls mutants under ABA treatment with no difference in normal culture (Figure 7E). In addition, we found the plant height of oscca1-Ls was slightly reduced relative to WT which could not be rescued by spraying gibberellic acid (GA) (Supplemental Figure S10), indicating a diminished GA signaling but not GA biosynthesis, existed in oscca1-Ls. Thus, we proposed that in the presence of abiotic stress such as salt, osmotic and drought, OsCCA1 could activate the transcription of multiple class-A OsPP2C members and OsbZIP46, together with other components of ABA signaling pathway such as OsPYL6, OsSAPK3, and OsABI5, etc., which subsequently control the expression of a wide spectrum of stress-responsive genes to confer multiple-stress tolerance (Figure 8, left). In oscca1 mutant plants, the expressions of stress-inducible OsPP108 and OsbZIP46 were substantially attenuated, hence resulted in hypersensitivity to abiotic stresses (Figure 8, right).
Figure 7.
The oscca1 mutant plants are more sensitive to ABA-inhibited seedling growth. A, Seedling growth inhibition of oscca1 mutants by ABA treatment with the indicated concentration for 7 days. Scale bar, 1 cm. B, Statistic analysis of relative plant height of DJ and oscca1 mutants in (A). Values are means ±se (n ≥ 17), two biological replicates with similar result. *P ≤ 0.05 and ***P ≤ 0.001 indicate significant difference by Student’s t test. C, 3-week-old seedlings of DJ and oscca1 mutants before treatment (left), after treatment with 200 μM ABA for 14 days (middle) and recovered for additional 7 days (right). Scale bar, 5 cm. D, The survival rates of DJ and oscca1 mutant plants after ABA treatment in (C). Data represent means ± sd. n = 6 biological replicates, 24 plants for each biological replicate. ***P ≤ 0.001 indicates significant difference by Student’s t test. E, Chlorophyll content in leaves of the plants treated with or without 200-μM ABA for 14 days, respectively. Four biological replicates were conducted, six plants of each replicate. Data are presented as mean ± sd. ***P ≤ 0.001 indicates significant difference by Student’s t test.
Figure 8.
A proposed molecular model of OsCCA1 orchestrating ABA signaling to confer multiple abiotic stress tolerance. Under stress condition in the WT (left), endogenous ABA levels increase rapidly, and OsCCA1 orchestrates ABA signaling by binding to the rCBS motifs within the promoters of multiple components of ABA signaling, such as OsPP108, regulating their expression and thereby negatively modulating the ABA signaling pathway. OsCCA1 also directly activates the transcription of OsbZIP46, a well-established key positive regulator of abiotic stress tolerance, which allows OsCCA1 to modulate a wide spectrum of stress-responsive genes by forming a transcriptional cascade, in both ABA dependent or independent manner. In the absence of OsCCA1 (right), the dual regulations disappeared, resulting in the enhanced sensitivity to ABA signaling and correspondingly the abiotic stresses.
Discussion
The allelic variants of plant core clock genes have been selected for agricultural breeding, particularly for modification of flowering time (Wei et al., 2020a). The crop circadian clocks also regulate a variety of plant physiological, metabolic, and stress responses, which are amenable for improving latitude adaption and crop yield (Steed et al., 2021). Here we unmasked the role of a rice core clock component OsCCA1 in multiple abiotic stress tolerance through direct binding and regulation of OsPP2Cs and OsbZIP46 genes to orchestrate ABA signaling. Our findings hence provide insights into the molecular mechanism by which a clock component reinforces adaptation and fitness under harsh growth conditions, hence providing a promising candidate in developing stress-tolerant cultivars.
A large number of the genes involved in abiotic stress response was intimately controlled by the circadian clock. In Arabidopsis, ∼50% of drought-responsive and 40% of cold-responsive genes display circadian rhythmicity (Bieniawska et al., 2008; Covington et al., 2008). Daily rhythmic expression of the abiotic stress-responsive genes was also described in major crop plants, such as barley (Hordeum vulgar) and soybean (Habte et al., 2014; Marcolino-Gomes et al., 2014). In contrast, how stress signaling interplays with circadian clock to balance growth and defense is largely uncharacterized. In this study, we employed DAP-seq to identify the direct regulons of OsCCA1 involved in stress response (Cao et al., 2020; Wu et al., 2020). We uncovered a prominent OsCCA1 binding element with the core sequence of “AAAATATCT,” and named it as rCBS (Figure 3, B–E), which is similar with EE bound by Arabidopsis CCA1 (Harmer et al., 2000), but there are three other binding motifs that are different from EE. The difference is unlikely due to the technical method used, as the DNA libraries of DAP-seq are constructed using native genomic DNA, which can preserve the chromatin modifications (such as DNA methylation) that are known to impact TF binding. Hence the DAP-seq results are expected to provide higher fidelity than the ChIP-seq results (Bartlett et al., 2017). Previous studies have shown that MYB DNA-binding domain of OsCCA1 protein is more closely related to those of maize (Zea mays) ZmCCA1s and sorghum (Sorghum bicolor) SbLHY than that of Arabidopsis CCA1 (Wang et al., 2020). The binding specificity to distinct cis-element is likely due to the evolutionary divergence of the OsCCA1 and AtCCA1 protein.
Transcript abundance of multiple genes encoding ABA biosynthetic enzymes oscillates in Arabidopsis, tomato (Solanum lycopersicum), maize, and sugarcane (Saccharum officinarum), indicating a rhythmic control of ABA level by the circadian clocks (Thompson et al., 2000; Covington et al., 2008; Khan et al., 2010; Hotta et al., 2013). In turn, ABA feedback modulates the Arabidopsis core oscillator activity (Hanano et al., 2006), in which TOC1 may play a paramount role. Very recently, PRR5 and PRR7, the two members of PRR gene family, were found to physically interact with ABI5 and promote its transcriptional activity, thereby positively modulating ABA signaling in a redundant manner (Yang et al., 2021). OsABI5, a homolog of Arabidopsis ABI5, was also an OsCCA1’s potential direct target and was downregulated in oscca1 mutants (Figures 4, E and 5, C). In contrast, the rice WRKY genes such as OsWRKY71, OsWRKY51, and OsWRKY24, together with OsCPK20, OsCPK4 were upregulated in oscca1 mutants (Figure 4E). The promoter of OsCPK4 could be bound by OsCCA1 protein according to the DAP-seq data (Supplemental Data set 1). OsWRKY71, OsWRKY51, and OsCPK20, OsCPK4 were formerly shown to enhance rice resistance to virulent bacterial pathogens (Liu et al., 2007; Fu et al., 2014; Bundó and Coca, 2016; Hwang et al., 2016), suggesting that OsCCA1 might also be required for biotic stress resistance as its Arabidopsis putative ortholog (Wang et al., 2011). In addition, OsWRKYs were previously shown to function as positive or negative regulators of ABA signaling (Xie et al., 2005), and OsCPK4 regulates the salt and drought stress response via reducing oxidative damage (Campo et al., 2014), implicating that they mediate the OsCCA1-ABA signaling module to confer abiotic stress tolerance directly or indirectly. Unlike other processes, the interplay between ABA signaling and CCA1 or its homologs was less well characterized. For example, Arabidopsis CCA1 was shown to be essential for temporal control of the expression of defense genes which permits plants to anticipate pathogen infection (Wang et al., 2011), and to control resistance to aphids by timing basal indole glucosinolate levels (Lei et al., 2019). Moreover, CCA1 was implicated in the regulation thermal response, ROS homeostasis, and abiotic stress responses in Arabidopsis (Lai et al., 2012; Seo et al., 2012; Nagel et al., 2015; Nitschke et al., 2016; Zha et al., 2017). In this study, by genome-wide identification of direct transcriptional targets of OsCCA1 using DAP-seq and RNA-seq approaches, we unexpectedly manifested that OsCCA1 plays a prominent role in modulating multiple ABA signaling core components including clade-A OsPP2C members and OsbZIP46 (Figure 5). In addition, we found that OsABI5, a homolog of Arabidopsis ABI5, was also a potential direct target of OsCCA1 and was downregulated in oscca1 mutants (Figures 4, E and 5, C). Therefore, it is imperative to determine if OsCCA1 can directly bind and modulate the expression of OsABI5 in the near future. Moreover, the rice WRKY genes, such as OsWRKY71, OsWRKY51, and OsWRKY24, were upregulated in oscca1 mutants as well. OsWRKY24 was previously shown as a negative regulator of ABA signaling, further implicating that OsCCA1 orchestrates ABA signaling to confer abiotic stress tolerance through multiple targets (Zhang et al., 2009). Interestingly, in Arabidopsis and soybean, LHY and its homolog were recently shown to play an important role in the modulation of ABA biosynthesis and ABA responses (Adams et al., 2018; Wang et al., 2021a). Hence, it is conceivable that ABA signaling, one of the most important hormone signal transduction mechanisms for abiotic stress, is under an intimate control of the circadian clock. On the other hand, there is a divergence in linking the functions of the key clock components with transcriptional targets encompassing distinct cis-regulatory elements in mono and dicotyledonous plants. Taken together, our findings abrogate the notion that the environmental factor-circadian clock interplay in rice could have been simply extrapolated from what has been learned in Arabidopsis, similar as shown in soybean (Li et al., 2019). Meanwhile, whether ABA signaling component-SnRK2/SAPKs are involved in feedback regulation of OsCCA1, and the possibility that OsCCA1 can associate with other proteins in modulating ABA signaling and stress response awaits a full elucidation in the future. Finally, the clade-A PP2Cs are differentially regulated by subgroup-A bZIP TFs in feedback regulatory loops. For example, OsbZIP23 directly binds to the promoters of nine rice clade-A PP2Cs under stress conditions (Zong et al., 2016), and OsbZIP12 and OsbZIP46 directly bind to OsPP2C09 promoter to mediate the ABA response and drought stress tolerance (Miao et al., 2020). Hence, the complex feedback regulation between bZIP TFs such as OsbZIP46 and OsPP2Cs remains to be further investigated. Additionally, ABA has a dose-dependent effect in regulating plant response to adversity stress condition, and over-sensitive ABA signaling may trigger senescence. For example, Zou et al., have reported that over-expression of OsABI5 in rice is hypersensitivity to ABA, but also is sensitive to salt stress. Similarly, overexpression of OsNAC2 causes sensitivity to ABA and early senescence (Mao et al., 2017). Interestingly, we found the ROS level was dramatically increased in oscca1 mutant in the presence of NaCl (Figure 1, H and I). It is widely known that ROS acts in ABA mediated stress responses to sustain plant survival under adverse growth conditions, however, once the ROS level accumulated over the capacity of the detoxifying machinery, it will cause oxidative damage to cellular constituents (Lee and Park, 2012). Furthermore, besides of the reduced transcript level of OsPP108 and OsZIP46, we found the expression level of OsPYL6 was also upregulated in oscca1 mutant (Supplemental Data set 2), which may collectively render oscca1 mutants more sensitive to ABA and accumulate high ROS level to ultimately trigger senescence. Nevertheless, the possibility that other major downstream genes of OsCCA1 contribute to its effect on abiotic stress and ABA signaling, directly or indirectly, cannot exclusively rule out, which awaits to be fully revealed.
In summary, we have identified genome-wide direct targets of OsCCA1 and its binding core elements, which will be a valuable resource to systematically uncover OsCCA1 mediated circadian outputs. OsCCA1 appears to confer multiple abiotic stress responses by orchestrating ABA signaling pathways. OsCCA1 is previously considered as a core clock component in regulating heading date and nitrogen use efficiency (Zhang et al., 2020). Our findings support a notion that OsCCA1 serves as a key circadian hub in fine-tuning plant growth and abiotic stress tolerance, consistent with the idea that the clock-regulated transcriptional networks are crucial not only for plant adaption to the latitudinal cline, but also to the harsh growth conditions. Elucidation of the underlying mechanisms by which circadian clock coordinates growth and environmental response will surely facilitate the development of strategies in improving rice and other cereal crops. The supreme haplotypes/elite alleles of circadian core components are likely valuable resources for future chronoculture guided crop breeding.
Materials and methods
Plant materials and growth condition
All of the rice (O.sativa L.) seedlings used in this study were generated in the background of japonica cultivar “Dongjin.” For oscca1-L1 and oscca1-L2 null mutant plants, the single-guide RNAs (sgRNAs) constructs for the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9-mediated knock out of OsCCA1 were generated according to previously described method (Ma et al., 2015). The sgRNAs were designed to target the second exon of OsCCA1, which were then cloned into the pYLCRISPR/Cas9Pubi-MH vector (Ma et al., 2015). These transgenic plants were obtained by using Agrobacteria tumefaciens EHA105-mediated transformation (Hiei et al., 1994). Homozygous insertion and deletion mutants at the target sites were characterized according to PCR scoring followed by DNA sequencing. The primers were listed in Supplemental Table S1. Seeds were sown and cultured in the greenhouse on May 1, then transplanted to paddy field a month later. These plants were grown in the Beijing experimental field under natural LD conditions till October. Seedlings were also grown in culture rooms under SD (10-h light, 30°C/14-h dark, 25°C) conditions with 50%–70% relative humidity.
Stress treatment
For NaCl or mannitol treatment, seedlings grown in bottomless 96-well plate were cultured in liquid Yoshida’s solution under SD conditions. Four-week-old plants were transferred to nutrient solution containing 180-mM NaCl, 180-mM mannitol for 24 additional days or 200-mM ABA, followed for 10, 24, or 7 days recovery, then the survival rate was calculated. For drought stress testing at seedling stage, oscca1-Ls and DJ plants were grown on the soil. Then the water supply was stopped at 21 days. After 10 days when all of the leaves wilted, plants were rewatered and the survival rate were recorded. For ABA and GA treatment at the germination stage, the seeds of DJ and oscca1 mutants were sterilized and placed on half-strength MS medium with 0, 2, 6, and 10 µM ABA (ABA treatment) or containing 0, 1, and 10 µM GA (GA treatment), then were grown in light incubator with 28°C for 7 days and measured the plant height using ImageJ.
Chlorophyll and ion leakage measurements
Total chlorophyll content and relative membrane ion leakage were calculated according to the previously reported method (Wei et al., 2020b). Absorbance was measured at optical density 645 (OD645) and OD663 nm with microplate reader (Infinite M200 Pro; Tecan, Männedorf, Switzerland). The ion leakage C1 and C2 were determined by conductivity meter (alalis, CD400).
Determination of the MDA and DAB staining
According to the methods described previously, the content of MDA and DAB staining were measured in the leaves under normal and NaCl stress conditions (Wei et al., 2020b). About 0.1 g of ground sample powder were extracted with 3 mL of 10% (w/v) trichloroacetic acid (TCA), then 2 mL supernatant liquid were mixture with 2 mL of 0.6% (w/v) thiobarbituric acid (made in 10% TCA) and reacted 15 min with boiling bath. The absorbance of these supernatants was read at OD450, OD532, and OD600 nm with microplate reader (Infinite M200 Pro, Tecan). The rice leaves were immersed in 10-mL staining solution with 1-mg mL−1 DAB containing 10-mM MES (pH 6.5) in the dark overnight and shaking at 100 rpm. Then used 90% ethanol for boiling water bath to remove the chlorophyll.
Measurement of Na+ and K+ concentration
To determine the relative ion accumulation of Na+ and K+ in shoots and roots, the materials collected and determination methods were according to the protocol described previously (Wei et al., 2020b). The shoots and roots of 2-week-old seedlings under NaCl stress for 7 days or not were collected respectively. Then all materials were washed with ddH2O, dried and weighed. After that, all the samples were digested 12 h with nitric acid under darkness condition and heated at 200°C for 8 h. The contents of Na+ and K+ were detected by inductively coupled plasma optical emission spectrometer (ICAP6300).
Subcellular localization of OsCCA1
The coding sequences of OsCCA1 was amplified and inserted into EcoRI and XhoI sites of the pENTR2B vector, then fused in the pMDC45 through LR recombination resulting in the 35S:GFP-OsCCA1 plasmid. The Agrobacteria with GFP fusion plasmid and 35S:H2B-mCherry were co-infiltrated into leaves of N.benthamiana. After 2 days of the infection, fluorescence signals were observed by confocal laser scanning microscope (Olympus, Tokyo, Japan; FV 1000). The main parameters were recorded as follows: average setting was 2, zoom was ×1.0, the values of three gains of negative control GFP were 350 V (EGFP), 566 V (RFP), and 120 V (TD1), and the gains of GFP-OsCCA1 were 358 V (EGFP), 513 V (RFP), and 144 V (TD1), respectively.
RNA extraction and qPCR assays
Total RNA was extracted from the 3-week-old seedling leaves under normal or treatment with 180 mM NaCl for 24 h conditions using the TRIzol reagent (Invitrogen, Waltham, MA, USA), and according to PrimeScript RT Reagent Kit (Takara, Shiga, Japan) to obtain cDNA. The templates of cDNA diluted 10-fold were tested the relative transcription levels of OsCCA1, OsPP108, OsbZIP46, and other salt responsive genes by RT-qPCR using gene-specific primers, and Ubiquitin was used for internal control. SYBR Green Real-Time Master Mix (Toyobo, Osaka, Japan) were used in a 15-μL reaction volume and ran on QuantStudio 3 instrument (Applied Biosystems, Waltham, MA, USA) with this program: 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 15 s. These primers are listed in Supplemental Table S1.
RNA-seq analysis
Seedlings were grown under 12-h light/12-h dark conditions 28°C for 21 days and the penultimate leaf blades were harvested at ZT0 treated with NaCl for 24 h. Total RNAs were extracted using TRIzol reagent. RNA-seq were performed at BerryGenomics (Beijing, China). Qualified RNA samples using NEB Next Ultra RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) generated sequencing libraries, which were then sequenced on an Illumina NovaSeq6000 platform and paired-end reads were created. TopHat (version 2.1.1) was used to analyze the RNA-seq clean reads reference the rice genome (the rice annotation project, RAP-DB). Genes with q < 0.05 and |log2_ratio| > 1.5 were identified as DEGs using DEseq (version 1.10.1). The analysis pipelines of RNA-seq have been upload in the Github (Tian et al., 2021). GO analysis of these DEGs were conducted by agriGO version 2.0 (http://systemsbiology.cau.edu.cn/agriGOv2/) or AmiGO version 2 (http://amigo.geneontology.org/amigo). Venn diagrams were created by VENNY version 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/index.html).
DAP-seq sampling and data analysis
DAP-seq was conducted by following the method described in previous study (Bartlett et al., 2017), and performed at Bluescape Hebei Biotech to purify the gDNA from the leaves of 30-day-old seedlings. Fragmented gDNA were constructed into libraries using the NEXTFLEX Rapid DNA Seq Kit (PerkinElmer, Inc., Austin, TX, USA). The coding sequence of OsCCA1 was cloned into a pFN19K HaloTag T7 SP6 Flexi vector and was expressed using the TNT SP6 Coupled Wheat Germ Extract System (Promega, Madison, WI, USA). Magne Halo Tag Beads (Promega) were utilized to purify and capture the expressed protein. The OsCCA1-bound beads were incubated with adapter ligated gDNA libraries. Eluted DNA, were sequenced on Illumina NavoSeq6000 with two technical duplicates. Without the addition of protein to beads were taken as the input negative control DAP libraries. DAP-seq reads were aligned to the reference genome (RAP) using Bowtie2 (Langmead and Salzberg, 2012). MACS2 callpeak (Zhang et al., 2008) and IDR software were used to merge the peaks of the two biological duplicates with P < 0.05, and to score the reliability of these repeated peaks. MEME-CHIP software was adopted to analyze the conservative motifs in the peaks area (Machanick and Bailey, 2011). The bound peaks were annotated using Homer software (Heinz et al., 2010). We performed KEGG analysis using the KOBAS version 2.0 database (http://kobas.cbi.pku.edu.cn/). We defined target genes that contained DAP-seq peaks located within 3-kb upstream of ATG.
Transcriptional activity assay in protoplast
To evaluate the transcriptional activity of OsCCA1, the 2,000-bp OsbZIP46 and 2,500-bp OsPP108 promoters were inserted into Hind III and Bam HI sites of pGreenll-0800-LUC vector and fused with LUC to, respectively, obtain the reporter constructs OsPP108p:LUC and OsbZIP46p:LUC which also contains the 35S:REN (Renilla) cassette. The primers are listed in Supplemental Table S1. For the effecter constructs UC18-OsCCA1-3HA, UC18-OsCCA1-VP16-3HA, UC18-OsCCA1-SRDX-3HA, and UC18-3HA as a negative control, all reporters and effectors were simultaneously transformed into rice protoplast that were separated from 2-week-old etiolated seedlings and incubated in darkness about 16 h. Then, the protoplasts were lysed with diluted 1× passive lysis buffer (Promega; E1910). After cell lysis, 5 μL of protein extractions were, respectively, mixed with 15-μL LUC and REN Assay Substrate and measured with a luminometer (Promega GloMax Muti Jr). Finally, the 15 μL of protein extractions were mixed with 7.5-μL protein loading buffer, and were fractionated by 8% SDS–PAGE (acrylamide: bisacrylamide, 37.5:1) gels. HA tagged OsCCA1, OsCCA1-VP16, and OsCCA1-SRDX were detected by western blotting using 1:1,000 monoclonal primary anti-HA antibody (Roche, Basel, Switzerland; 11867423001).
ChIP-qPCR
ChIP assay was performed based on the previous report with slight modification (Wei et al., 2020b). First, we separated the protoplast form the 2- week-old etiolated seedlings, and transformed the plasmids 35S:GFP-999 or 35S:OsCCA1-GFP-999, respectively, which were incubated in darkness for 16 h and then cultured in light for 1 h. Then these protoplasts were collected and cross-linked with 1% (v/v) formaldehyde for 10 min. The nuclear proteins were extracted and purified, GFP-Trap magnetic agarose (Chromotek, Munich, Germany) were used for immunoprecipitation. The ChIPed DNA fragments were diluted 10-folds and used for qPCR, and the primers are listed in Supplemental Table S1. The fold enrichment was calculated according to the formula Log2(CTInput - CTIP).
EMSA
The GST–OsCCA1 fusion protein were expressed in Escherichia coli strain BL21 and purified according to the published protocol (Li et al., 2020). The EMSA was performed by the LightShfit Chemiluminescent EMSA kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. About 6-μL-purified GST-tagged OsCCA1 or GST protein and 0.5 μL of each biotin labeled probe were mixed into a reaction of 20 μL at 4°C for 1 h. The unlabeled probe (0.5 μL) was added in this reaction used as competitor. The probes sequence of the EMSA is listed in the Supplemental Table S1.
Data analysis
All data are presented herein as the means ± standard deviations or standard error, represented by error bars. Data analyses were completed by comparing the raw data of all the individuals using the Microsoft EXCEL program (Microsoft, Redmond, WA, USA). Diagrams were prepared using Adobe Photoshop (Adobe Systems, San Jose, CA, USA). The levels of significance were determined using Student’s t test: *P < 0.05; **P < 0.01; ***P < 0.001, ns, not significant.
Accession numbers
Sequences mentioned in this article can be download from Rice Genome Annotation Project Database (http://rice.plantbiology.msu.edu/) under the following accession numbers: OsCCA1, LOC_Os08g06110; OsPP108, LOC_Os09g15670; OsbZIP46, LOC_Os06g10880; OsLEA3, LOC_Os05g46480; OsLEA14, LOC_Os01g50910; RAB21, LOC_Os11g26790; RAB16B, LOC_11g26780; OsPP2C30, LOC_Os03g16170; OsPP2C8, and LOC_Os01g46760; OsABIL3, LOC_Os05g46040. The RNA-seq and DAP-seq source data are deposited in the National Center for Biotechnology Information under the Bioproject accession number PRJNA754696. All study data are included in the article and/or Supplementary appendix.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transcription profiles of OsCCA1 treated with NaCl or mimicked drought stress by addition of PEG6000.
Supplemental Figure S2. Characterization of oscca1-Ls generated by genome editing.
Supplemental Figure S3. Analysis of agronomic traits of oscca1 mutants.
Supplemental Figure S4. Subcellular localization and temporal expression pattern of OsCCA1.
Supplemental Figure S5. OsCCA1 may participate in the ABA signaling pathway.
Supplemental Figure S6. The RNA-seq data showed OsCCA1 regulates many OsbZIP46-mediated stress-responsive genes.
Supplemental Figure S7. OsCCA1 may directly regulate multiple class A PP2Cs members.
Supplemental Figure S8. OsCCA1 regulates many OsbZIP46-mediated stress-responsive genes.
Supplemental Figure S9. Protein expression of OsCCA1-HA, OsCCA1-VP16-HA, and OsCCA1-SRDX-HA in transformed rice protoplasts was detected by western blotting.
Supplemental Figure S10. Height of DJ and oscca1 plants after GA treatment.
Supplemental Table S1 . The primers used in this study.
Supplemental Data set 1. Identified OsCCA1-bound genes by DAP-seq.
Supplemental Data set 2. The DEGs of oscca1-L1 mutants by RNA-seq.
Supplementary Material
Acknowledgments
We thank Ms. Jingquan Li from Plant Science Facility of the Institute of Botany, Chinese Academy of Sciences for her excellent technical assistance on confocal microscopy. We thank Dr. JC Jang (Ohio State University) for his critical reading and comments.
Funding
This work was supported by National Natural Science Foundation of China (31770287), National Key Research and Development Program of China (2016YFD0100604) and Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27030206) to L.W.
Conflict of interest statement. The authors declare no conflict of interest.
Contributor Information
Hua Wei, Key Laboratory of Plant Molecular Physiology, CAS Center for Excellence in Molecular Plant Sciences, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Hang Xu, Key Laboratory of Plant Molecular Physiology, CAS Center for Excellence in Molecular Plant Sciences, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Chen Su, Key Laboratory of Plant Molecular Physiology, CAS Center for Excellence in Molecular Plant Sciences, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Xiling Wang, Key Laboratory of Plant Molecular Physiology, CAS Center for Excellence in Molecular Plant Sciences, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Lei Wang, Key Laboratory of Plant Molecular Physiology, CAS Center for Excellence in Molecular Plant Sciences, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Research design: H.W. and L.W.; Research: H.W., H.X., C.S., and X.W.; Data analysis and discussion: H.W., H.X., C.S., X.W., and L.W.; Manuscript writing: H.W. and L.W.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Lei Wang (wanglei@ibcas.ac.cn).
References
- Adams S, Grundy J, Veflingstad SR, Dyer NP, Hannah MA, Ott S, Carre IA (2018) Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytologist 220: 893–907 [DOI] [PubMed] [Google Scholar]
- Andronis C, Barak S, Knowles SM, Sugano S, Tobin EM (2008) The clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis. Molar Plant 1: 58–67 [DOI] [PubMed] [Google Scholar]
- Antoni R, Gonzalez-Guzman M, Rodriguez L, Peirats-Llobet M, Pizzio GA, Fernandez MA, De Winne N, De Jaeger G, Dietrich D, Bennett MJ, et al. (2013) PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol 161: 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett A, O'Malley RC, Huang SC, Galli M, Nery JR, Gallavotti A, Ecker JR (2017) Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat Protoc 12: 1659–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar N, Min MK, Choi EH, Kim N, Moon SJ, Yoon I, Kwon T, Jung KH, Kim BG (2017) The protein phosphatase 2C clade A protein OsPP2C51 positively regulates seed germination by directly inactivating OsbZIP10. Plant Mol Biol 93: 389–401 [DOI] [PubMed] [Google Scholar]
- Bieniawska Z, Espinoza C, Schlereth A, Sulpice R, Hincha DK, Hannah MA (2008) Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol 147: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundó M, Coca M (2016) Enhancing blast disease resistance by overexpression of the calcium-dependent protein kinase OsCPK4 in rice. Plant Biotechnol J 14: 1357–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo S, Baldrich P, Messeguer J, Lalanne E, Coca M, San Segundo B (2014) Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol 165: 688–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Zeng H, Ku L, Ren Z, Han Y, Su H, Dou D, Liu H, Dong Y, Zhu F, et al. (2020) ZmIBH1-1 regulates plant architecture in maize. J Exp Bot 71: 2943–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL (2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genom Biol 9: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Yu X, An C (2014) OsCPK20 positively regulates Arabidopsis resistance against Pseudomonas syringae pv. tomato and rice resistance against Magnaporthe grisea. Acta Physiol Plant 36: 273–282 [DOI] [PubMed] [Google Scholar]
- Greenham K, McClung CR (2015) Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet 16: 598–610 [DOI] [PubMed] [Google Scholar]
- Habte E, Mueller LM, Shtaya M, Davis SJ, von Korff M (2014) Osmotic stress at the barley root affects expression of circadian clock genes in the shoot. Plant Cell Environ 37: 1321–1337 [DOI] [PubMed] [Google Scholar]
- Hanano S, Domagalska MA, Nagy F, Davis SJ (2006) Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells 11: 1381–1392 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch LB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Effient Transformation of rice (oryza sativa L.) mediated by agrobacterium and sequence-analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hotta CT, Nishiyama MY Jr, Souza GM (2013) Circadian rhythms of sense and antisense transcription in sugarcane, a highly polyploid crop. PLoS One 8: e71847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SH, Kwon SI, Jang JY, Fang IL, Lee H, Choi C, Park S, Ahn I, Bae SC, Hwang DJ (2016) OsWRKY51, a rice transcription factor, functions as a positive regulator in defense response against Xanthomonas oryzae pv. oryzae. Plant Cell Rep 35: 1975–1985 [DOI] [PubMed] [Google Scholar]
- Khan S, Rowe SC, Harmon FG (2010) Coordination of the maize transcriptome by a conserved circadian clock. BMC Plant Biol 10: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Hwang H, Hong JW, Lee YN, Ahn IP, Yoon IS, Yoo SD, Lee S, Lee SC, Kim BG (2012) A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J Exp Bot 63: 1013–1024 [DOI] [PubMed] [Google Scholar]
- Kim H, Lee K, Hwang H, Bhatnagar N, Kim DY, Yoon IS, Byun MO, Kim ST, Jung KH, Kim BG (2014) Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J Exp Bot 65: 453–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Ali Z, Park HJ, Park SJ, Cha JY, Perez-Hormaeche J, Quintero FJ, Shin G, Kim MR, Qiang Z, et al. (2013) Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat Commun 4: 1352. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T (2004) Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16: 1163–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JHM, Dijkwel PP (2012) CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc Natl Acad Sci USA 109: 17129–17134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Piao HL, Kim HY, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee IJ, Hwang I (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Lee S, Park CM (2012) Regulation of reactive oxygen species generation under drought conditions in Arabidopsis. Plant Signal Behav 7: 599–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnaioli T, Cuevas J, Mas P (2009) TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J 28: 3745–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei JX, Jayaprakasha GK, Singh J, Uckoo R, Borrego EJ, Finlayson S, Kolomiets M, Patil BS, Braam J, Zhu-Salzmana K (2019) CIRCADIAN CLOCK-ASSOCIATED1 controls resistance to aphids by altering indole glucosinolate production. Plant Physiol 181: 1344–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Cao L, Mwimba M, Zhou Y, Li L, Zhou M, Schnable PS, O'Rourke JA, Dong X, Wang W (2019) Comprehensive mapping of abiotic stress inputs into the soybean circadian clock. Proc Natl Acad Sci USA 116: 23840–23849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang Y, He Y, Wang Y, Wang L (2020) Pseudo response regulators regulate photoperiodic hypocotyl growth by repressing PIF4/5 transcription. Plant Physiol 183: 686–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Zhang Z, Cheng N, Liu H, Song S, Hu Y, Zhou X, Zhang J, Xing Y (2020) The transcriptional repressor OsPRR73 links circadian clock and photoperiod pathway to control heading date in rice. Plant Cell Environ 44: 842–855 [DOI] [PubMed] [Google Scholar]
- Liu T, Carlsson J, Takeuchi T, Newton L, Farre EM (2013) Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J 76: 101–114 [DOI] [PubMed] [Google Scholar]
- Liu X, Bai X, Wang X, Chu C (2007) OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol 164: 969–979 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. (2015) A Robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL (2011) MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Lu S, Lv B, Zhang B, Shen J, He J, Luo L, Xi D, Chen X, Ming F (2017) A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol 174: 1747–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcolino-Gomes J, Rodrigues FA, Fuganti-Pagliarini R, Bendix C, Nakayama TJ, Celaya B, Correa Molinari HB, Neves de Oliveira MC, Harmon FG, Nepomuceno A (2014) Diurnal oscillations of soybean circadian clock and drought responsive genes. PLoS One 9: e86402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao C, Xiao L, Huaa K, Zou C, Zhao Y, Bressan RA, Zhu JK (2018) Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc Natl Acad Sci USA 115: 6058–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Li X, Li X, Tan W, You A, Wu S, Tao Y, Chen C, Wang J, Zhang D, et al. (2020) OsPP2C09, a negative regulatory factor in abscisic acid signalling, plays an essential role in balancing plant growth and drought tolerance in rice. New Phytologist 227: 1417–1433 [DOI] [PubMed] [Google Scholar]
- Nagel DH, Doherty CJ, Pruneda-Paz JL, Schmitz RJ, Ecker JR, Kay SA (2015) Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc Natl Acad Sci USA 112: e4802–e4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke S, Cortleven A, Iven T, Feussner I, Havaux M, Riefler M, Schmulling T (2016) Circadian stress regimes affect the circadian clock and cause jasmonic acid-dependent cell death in cytokinin-deficient Arabidopsis plants. Plant Cell 28: 1616–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez SE, Kay SA (2016) The plant circadian clock: from a simple timekeeper to a complex developmental manager. Cold Spring Harbor Perspect Biol 8: a027748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Park MJ, Lim MH, Kim SG, Lee M, Baldwin IT, Park CM (2012) A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24: 2427–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Jha SK, Bagri J, Pandey GK (2015) ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS One 10: e0125168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed G, Ramirez DC, Hannah MA, Webb AAR (2021) Chronoculture, harnessing the circadian clock to improve crop yield and sustainability. Science 372: eabc9141. [DOI] [PubMed] [Google Scholar]
- Sun C, Zhang K, Zhou Y, Xiang L, He C, Zhong C, Li K, Wang Q, Yang C, Wang Q, et al. (2021) Dual function of clock component OsLHY sets critical day length for photoperiodic flowering in rice. Plant Biotechnol J 19: 1644–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Ma S, Zong W, Yang N, Lv Y, Yan C, Guo Z, Li J, Li X, Xiang Y, et al. (2016) MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice. Plant Cell 28: 2161–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Zhang H, Li X, Xiao J, Xiong L (2012) Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol 158: 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Parker RA, Morpeth DR, Burbidge A, Taylor IB (2000) Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol Biol 42: 833–845 [DOI] [PubMed] [Google Scholar]
- Tian W, Wang R, Bo C, Yu Y, Zhang Y, Shin GI, Kim WY, Wang L (2021) SDC mediates DNA methylation-controlled clock pace by interacting with ZTL in Arabidopsis. Nucleic Acids Res 49: 3764–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Han T, Song Q, Ye W, Song X, Chu J, Li J, Chen ZJ (2020) The rice circadian clock regulates tiller growth and panicle development through strigolactone signaling and sugar sensing. Plant Cell 32: 3124–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Bu T, Cheng Q, Dong L, Su T, Chen Z, Kong F, Gong Z, Liu B, Li M (2021a) Two homologous LHY pairs negatively control soybean drought tolerance by repressing the abscisic acid responses. New Phytologist 229: 2660–2675 [DOI] [PubMed] [Google Scholar]
- Wang W, Barnaby JY, Tada Y, Li H, Tor M, Caldelari D, Lee DU, Fu XD, Dong XN (2011) Timing of plant immune responses by a central circadian regulator. Nature 470: 110–U126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, He Y, Wei H, Wang L (2021b) A clock regulatory module is required for salt tolerance and control of heading date in rice. Plant Cell Environ 44: 3283–3301 [DOI] [PubMed] [Google Scholar]
- Wei H, Wang X, He Y, Xu H, Wang L (2020b) Clock component OsPRR73 positively regulates rice salt tolerance by modulating OsHKT2;1-mediated sodium homeostasis. EMBO J 40: e105086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Wang X, Xu H, Wang L (2020a) Molecular basis of heading date control in rice. aBIOTECH 1: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu VW, Thieme N, Huberman LB, Dietschmann A, Kowbel DJ, Lee J, Calhoun S, Singan VR, Lipzen A, Xiong Y, et al. (2020) The regulatory and transcriptional landscape associated with carbon utilization in a filamentous fungus. Proc Natl Acad Sci USA 117: 6003–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye H, Xiong L (2008) Characterization of OsbZIP23 as a key pPlayer of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148: 1938–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, Shen QJ (2005) Annotations and functional analyses of the rice WRKY Gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 137: 176–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Han X, Yang J, Jiang Y, Hu Y (2021) The Arabidopsis circadian clock protein PRR5 interacts with and stimulates ABI5 to modulate abscisic acid signaling during seed germination. Plant Cell 33: 3022–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha P, Jing YJ, Xu G, Lin RC (2017) PICKLE chromatin-remodeling factor controls thermosensory hypocotyl growth of Arabidopsis. Plant Cell Environ 40: 2426–2436 [DOI] [PubMed] [Google Scholar]
- Zhang D, Wang Y, Shen J, Yin J, Li D, Gao Y, Xu W, Liang J (2018) OsRACK1A, encodes a circadian clock-regulated WD40 protein, negatively affect salt tolerance in rice. Rice 11: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhang Y, Li K, Yan M, Zhang J, Yu M, Tang S, Wang L, Qu H, Luo L, et al. (2020) Nitrogen mediates flowering time and nitrogen use efficiency via floral regulators in rice. Curr Biol 31: 671–683 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genom Biolol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZL, Shin M, Zou X, Huang J, Ho TH, Shen QJ (2009) A negative regulator encoded by a rice WRKY gene represses both abscisic acid and gibberellins signaling in aleurone cells. Plant Mol Biol 70139–151 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Ann Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong W, Tang N, Yang J, Peng L, Ma SQ, Xu Y, Li GL, Xiong LZ (2016) Feedback Regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol 171: 2810–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M, Guan Y, Ren H, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66: 675–683 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.