Abstract
Basic helix–loop–helix (bHLH) transcription factors constitute a superfamily in eukaryotes, but their roles in plant immunity remain largely uncharacterized. We found that the transcript abundance in tomato (Solanum lycopersicum) leaves of one bHLH transcription factor-encoding gene, negative regulator of resistance to DC3000 1 (Nrd1), increased significantly after treatment with the immunity-inducing flgII-28 peptide. Plants carrying a loss-of-function mutation in Nrd1 (Δnrd1) showed enhanced resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 although early pattern-triggered immunity responses, such as generation of reactive oxygen species and activation of mitogen-activated protein kinases after treatment with flagellin-derived flg22 and flgII-28 peptides, were unaltered compared to wild-type plants. RNA-sequencing (RNA-seq) analysis identified a gene, Arabinogalactan protein 1 (Agp1), whose expression is strongly suppressed in an Nrd1-dependent manner. Agp1 encodes an arabinogalactan protein, and overexpression of the Agp1 gene in Nicotiana benthamiana led to ∼10-fold less Pst growth compared to the control. These results suggest that the Nrd1 protein promotes tomato susceptibility to Pst by suppressing the defense gene Agp1. RNA-seq also revealed that the loss of Nrd1 function has no effect on the transcript abundance of immunity-associated genes, including AvrPtoB tomato-interacting 9 (Bti9), Cold-shock protein receptor (Core), Flagellin sensing 2 (Fls2), Flagellin sensing (Fls3), and Wall-associated kinase 1 (Wak1) upon Pst inoculation, suggesting that the enhanced immunity observed in the Δnrd1 mutants is due to the activation of key PRR signaling components as well as the loss of Nrd1-regulated suppression of Agp1.
The tomato bHLH transcription factor Nrd1 negatively regulates tomato immunity to bacterial speck disease by suppressing a putative defense-related gene encoding an arabinogalactan protein.
Introduction
Plants have evolved sophisticated surveillance mechanisms to rapidly recognize and respond to pathogen attacks (Lolle et al., 2020; Zhou and Zhang, 2020). The first layer of plant immunity, referred as pattern-triggered immunity (PTI), is activated when plant cells detect microbe-associated molecular patterns (MAMPs) through transmembrane pattern recognition receptors (PRRs; DeFalco and Zipfel, 2021). Successful pathogens deploy effectors into plant cells that interfere with PTI, leading to effector-triggered susceptibility (ETS; Abramovitch et al., 2006). To defeat ETS, plants activate a more robust immune response, effector-triggered immunity (ETI), where nucleotide-binding leucine-rich repeat (NB-LRR or NLR) proteins directly or indirectly recognize a given effector, resulting in a hypersensitive cell death response (HR) and disease resistance (Jones and Dangl, 2006; Lolle et al., 2020). Although PRR-mediated PTI and NLR-mediated ETI involve different activation mechanisms and different early signaling components, recent evidence suggests that the two layers share some downstream components and both are needed to ensure robust immunity (Ngou et al., 2021; Yuan et al., 2021a, 2021b).
The interaction of tomato (Solanum lycopersicum) with the bacterial pathogen P.syringae pv. tomato (Pst) is a well-developed model system for understanding the molecular basis of plant immunity and bacterial pathogenesis (Martin, 2012; Xin et al., 2018; Roberts et al., 2019; Wu and Kamoun, 2021). When Pst enters the apoplastic space of tomato leaves, two flagellin-derived MAMPs, flg22 and flgII-28, are recognized by the tomato PRRs Flagellin sensing 2 (Fls2) and Flagellin sensing 3 (Fls3), respectively (Hind et al., 2016; Roberts et al., 2020; Zhang et al., 2020). MAMP detection activates early PTI responses such as production of reactive oxygen species (ROS), activation of the mitogen-activated protein kinase (MAPK) cascades, and transcriptional reprogramming of a subset of defense genes (Jia and Martin, 1999; Nguyen et al., 2010; Zipfel, 2014; Li et al., 2016). Two Pst effector proteins, AvrPto and AvrPtoB, bind and interfere with the intracellular protein kinase domain of Fls2, Fls3, and the co-receptor Bak1 thus disrupting the host response to these MAMPs (Xiang et al., 2008; Cheng et al., 2011; Hind et al., 2016). The two effectors are also recognized by the host kinase Pto and activate ETI through the NLR protein Prf (Kim et al., 2002; Pedley and Martin, 2003; Oh and Martin, 2011).
RNA-seq analyses have been used to identify immunity-associated genes in the tomato–Pst system by inoculating plants with Pst strains eliciting only the PTI or ETI response (Rosli et al., 2013; Pombo et al., 2014). A subset of FIRE (flagellin-induced, repressed by effectors) genes was identified and the cell wall-associated kinase, SlWak1, was demonstrated to play a critical role in the PTI signaling pathway (Rosli et al., 2013; Zhang et al., 2020). Similarly, a subset of ETI-specific genes whose expression was induced specifically during ETI was identified and one kinase, Epk1, was shown to play a role in the host response to three effector proteins (Pombo et al., 2014). These RNA-seq data provide a powerful resource for identifying additional immunity-associated genes involved in the tomato–Pst interaction.
We recently reported the generation of hundreds of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas)-mediated tomato lines that carry mutations in putative immunity-associated genes (Jacobs et al., 2017; Zheng et al., 2019; Zhang et al., 2020). The availability of these tomato mutant lines provides a robust resource for the research community to test the function of specific genes in plant immunity and other biological processes (Zheng et al., 2019; Roberts et al., 2020; Zhang et al., 2020). We initially screened homozygous mutant plants by inoculating them with various Pst strains, including DC3000, to determine if they play a demonstrable role in PTI or ETI. Additional experimental methods including a ROS assay, MAPK activation assay, reporter gene assay, and HR assay were also applied to the mutant collection to identify components of response pathways during the tomato–Pst interaction.
The basic helix–loop–helix (bHLH) proteins are a superfamily of transcription factors (TFs) that play an essential role in diverse biological processes in animals and plants (Heim et al., 2003; Toledo-Ortiz et al., 2003; Li et al., 2006; Kay et al., 2007; Sun et al., 2015; Wang et al., 2015a, 2015b). The bHLH family is defined by the bHLH signature domain, which consists of an N-terminal basic region functioning as a DNA-binding motif recognizing the E-box cis-element (CANNTG), and a C-terminal HLH region acting as a dimerization domain to form a homodimer or heterodimer required for TF functions (Toledo-Ortiz et al., 2003). The bHLH TFs can transcriptionally activate or suppress target genes by specifically binding to their promoters (Xu et al., 2014; Hu et al., 2020; Hussain et al., 2021). In tomato, approximately 160 bHLH protein-encoding genes were identified (Sun et al., 2015; Wang et al., 2015b), but only a few have been functionally characterized (Ling et al., 2002; Du et al., 2015; Schwartz et al., 2017; Kim and Mudgett, 2019) and even fewer have been reported to play a critical role in plant immunity (Schwartz et al., 2017; Kim and Mudgett, 2019).
The transcript abundance of one gene encoding a bHLH TF, referred to now as SlNrd1 (S. lycopersicumnegative regulator of resistance to DC3000 1, hereafter Nrd1), was previously found to be increased in tomato leaves specifically upon treatment with flgII-28. Here, through loss-of-function analyses we found that, unexpectedly, Nrd1 appears to act as a negative regulator in tomato immunity to P.syringae pv. tomato DC3000. Using CRISPR-generated Δnrd1 mutant plants and RNA-seq we identified a gene encoding an arabinogalactan protein (Agp1), whose expression was strongly suppressed in an Nrd1-dependent manner. Overexpression of Agp1 in Nicotiana benthamiana led to statistically significant less Pst growth, indicating Agp1 is a Nrd1-regulated defense gene against P.syringae.
Results
Identification of Nrd1 and generation of stable loss-of-function tomato mutants
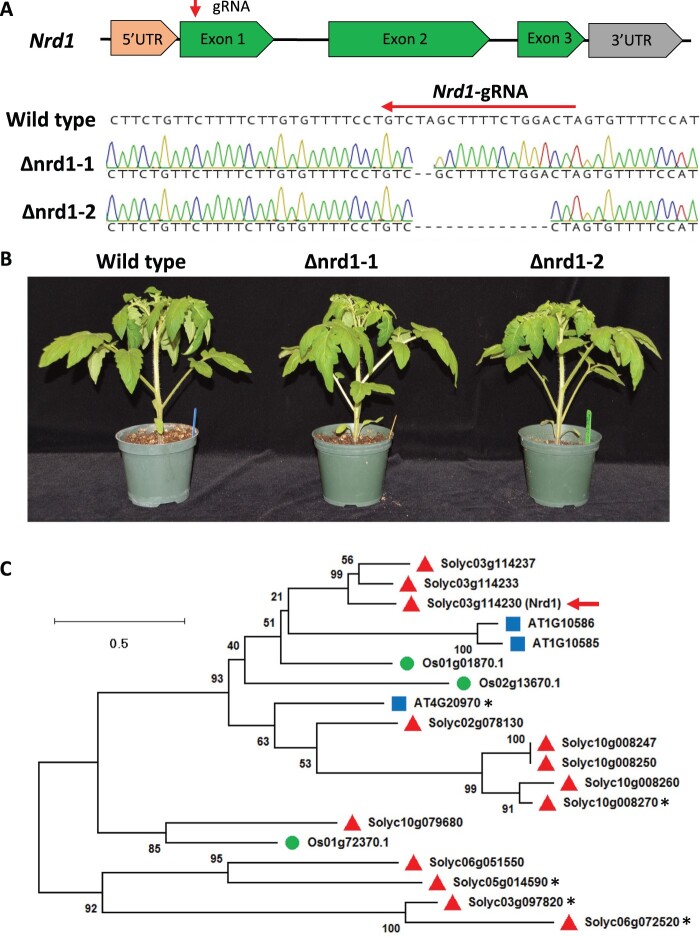
Previous analyses revealed that the transcript abundance of the tomato Nrd1 gene (Solyc03g114230) was significantly increased in leaves after treatment with 1-µM flgII-28 (Rosli et al., 2013; Roberts et al., 2020), suggesting it might play an important role in the tomato–Pst PTI response. To study the possible role of Nrd1 in tomato immunity, we generated T0 knockout mutant lines in tomato cultivar Rio Grande (RG)-PtoR using CRISPR/Cas9 with a guide RNA (5′-GTAGTCCAGAAAAGCTAGAC-3′; Figure 1A), which targets the first exon of the Nrd1 gene. Two independent Nrd1 homozygous mutants (Δnrd1-1 and Δnrd1-2) were derived and used in this study. The Δnrd1-1 mutant has a 2-bp deletion, whereas Δnrd1-2 contains a 13-bp deletion at the very 5′ end of the first exon of the Nrd1 gene. The deletions in the Δnrd1-1 and Δnrd1-2 lines introduce multiple amino acid substitutions around the cut site and eventually a premature stop codon at the 27th and 18th amino acid (aa) of the Nrd1 protein, respectively (Supplemental Figure S1). In addition, mutations in the Nrd1 gene did not allow retention of downstream open reading frames, further indicating they result in a loss-of-function of Nrd1 (Supplemental Figure S1). No morphological defects were observed in either of the two Nrd1 mutant plants when grown under greenhouse conditions (Figure 1B).
Figure 1.
Generation of tomato Δnrd1 mutants by CRISPR/Cas9. A, Schematics showing the guide-RNA (gRNA) target site and the missense mutations present in two independent Δnrd1 lines. The Δnrd1-1 line has a 2-bp deletion and the Δnrd1-2 line has a 13-bp deletion. Wild-type is Rio Grande (RG)-PtoR. UTR, untranslated region. B, Photographs of 4-week-old wild-type RG-PtoR and the two Δnrd1 mutant lines grown in the greenhouse. C, Phylogenetic tree of Nrd1 and related proteins. Amino acid sequences of Nrd1 and related proteins in Arabidopsis (blue squares), rice (green circles), and tomato (red triangles) were used to generate a maximum likelihood tree. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Numbers on branches indicate bootstrap support of the nodes (%). The red arrow indicates the Nrd1 protein. The asterisks indicate genes that have been reported to be implicated in immunity (Huibers et al., 2009; Wang et al., 2015b; Schwartz et al., 2017; Kim and Mudgett, 2019).
Nrd1 encodes a bHLH TF containing a domain that is known to bind the E-box motif (CANNTG) in the promoter sequence of target genes (Sun et al., 2015). To determine if Nrd1 has closely related proteins in tomato, Arabidopsis (Arabidopsis thaliana), or rice (Oryza sativa), we performed multiple Basic Local Alignment Search Tool (BLAST) searches of the NCBI databases using the Nrd1 protein sequence as the query sequence and obtained a limited number of protein hits. Phylogenetic analysis revealed that the Nrd1 protein has two closely related proteins in tomato, Solyc03g114233 and Solyc03g114237 (Figure 1C; Supplemental Figure S2A), with 60.3% and 65.0% similarity to the Nrd1 protein sequence, respectively. Nothing appears to be known about the biological functions of these two proteins, and they are newly annotated genes in the latest version of tomato reference genome (SL4.0; https://solgenomics.net). However, our RNA-seq data revealed very low transcript levels of Solyc03g114233 and Solyc03g114237 in leaves of both wild-type RG-PtoR plants and Δnrd1 mutants, whereas Nrd1 showed a higher transcript abundance after Pst inoculation (Supplemental Figure S2B). These results suggested that Nrd1, but not the two closely related genes, might play a role in the plant response to Pst. No putative orthologs of Nrd1 occur in Arabidopsis or rice, with the most closely related proteins (AT1G10585, AT1G10586, AT4G20970, and Os01g01870) having a very low sequence similarity (28.3%, 29.3%, 35.4%, and 38.3%, respectively) to Nrd1. Of these, only AT4G20970 has been previously associated with a biotic interaction due to its being induced by the downy mildew pathogen Hyaloperonospora arabidopsidis (Huibers et al., 2009).
Mutations in Nrd1 cause enhanced resistance to Pst in tomato
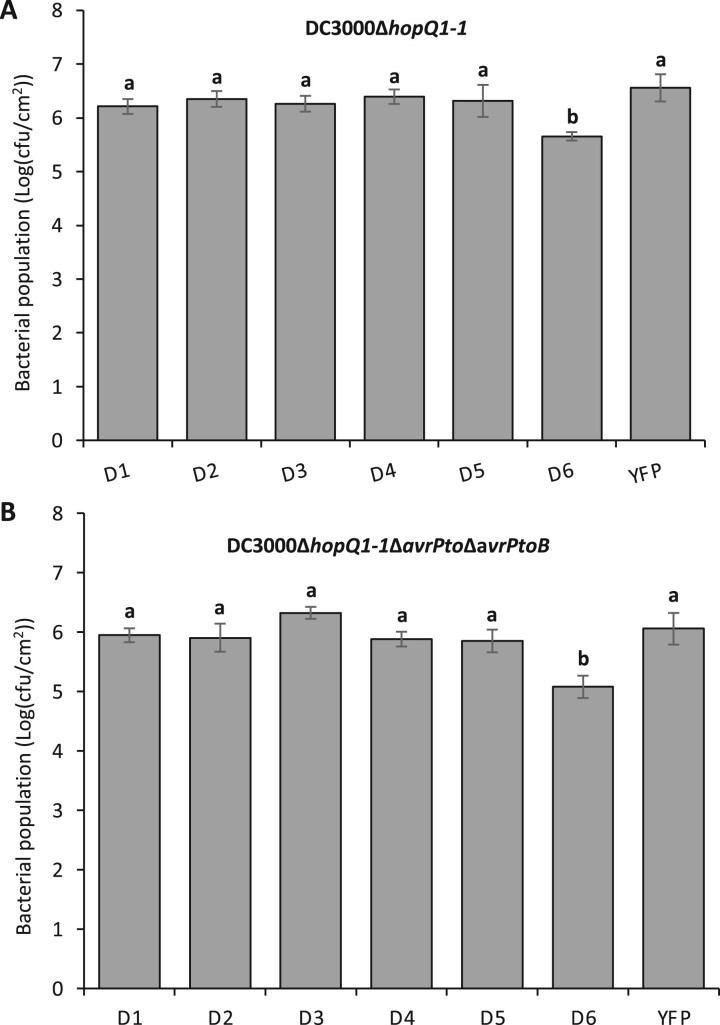
To test whether loss-of-function mutations in Nrd1 affect the ETI response to Pst, we vacuum-infiltrated Pst DC3000 into the two Δnrd1 mutants, wild-type RG-PtoR (which expresses the Pto and Prf genes allowing recognition of effectors AvrPto/AvrPtoB; Martin, 2012) and RG-prf3 (which has a mutation in Prf that makes the Pto pathway nonfunctional) plants (Figure 2A). We observed no significant difference in bacterial populations between the Δnrd1 mutants and wild-type RG-PtoR 2 days after inoculation, whereas bacterial populations were 10-fold more in RG-prf3 compared to Δnrd1 and RG-PtoR plants. Similarly, the Δnrd1 mutants and RG-PtoR plants had no disease symptoms whereas RG-prf3 showed severe disease symptoms 6 days after inoculation. These data indicate that Nrd1 does not have a major role in the ETI pathway acting against Pst DC3000.
Figure 2.
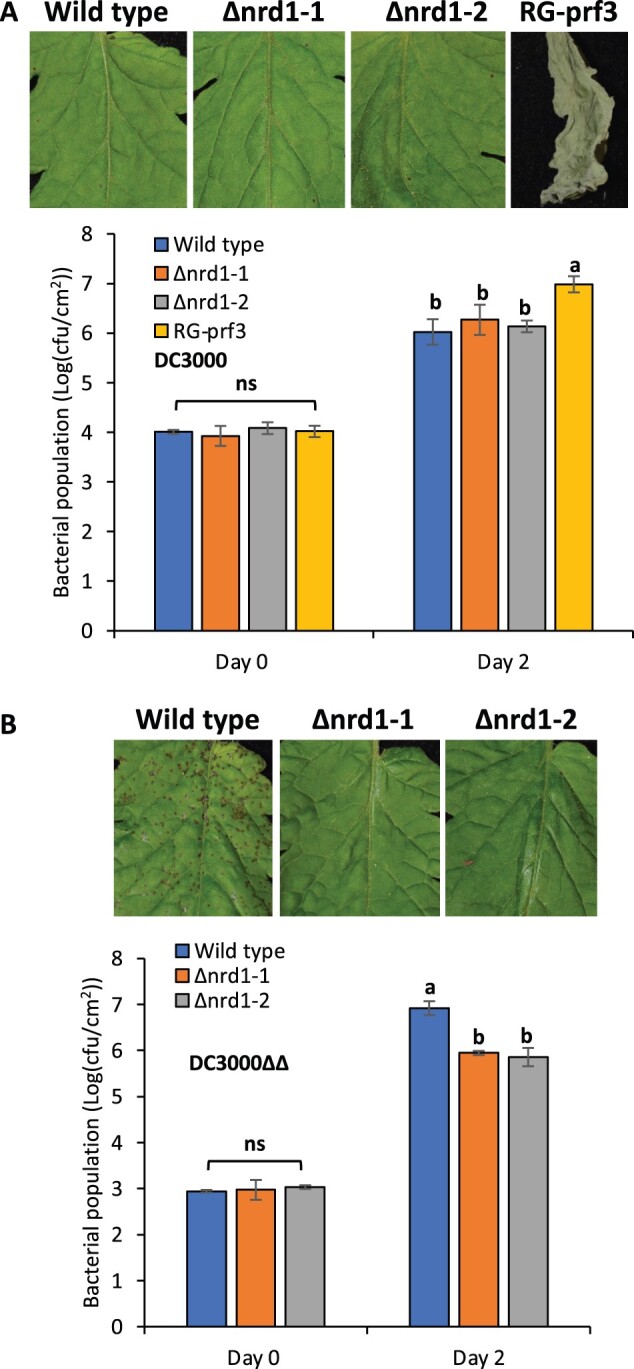
Investigation of ETI- and PTI-mediated immunity in the Δnrd1 mutants. Four-week-old Δnrd1 plants, Rio Grande (RG)-PtoR (wild-type), and RG-prf3 (a Prf mutant) plants were vacuum-infiltrated with: A, 1 × 106 cfu · mL−1 DC3000 or B, 5 × 104 cfu · mL−1 DC3000ΔavrPtoΔavrPtoB (DC3000ΔΔ). Photographs of disease symptoms were taken at 6 days (A) or 5 days (B) after inoculation. Bacterial populations were measured at 3 h (Day 0) and 2 days (Day 2) after infiltration. Cfu, colony-forming unit. Bars show means ± standard deviation (sd). Different letters indicate significant differences based on a one-way analysis of variance (ANOVA) followed by Student’s t test (P < 0.05). ns, no significant difference. Three plants for each genotype were tested per experiment. The experiment was performed three times with similar results.
To test whether Nrd1 contributes to PTI acting against Pst, we vacuum-infiltrated the two Δnrd1 mutants and RG-PtoR with DC3000ΔavrPtoΔavrPtoB (DC3000ΔΔ; Figure 2B), which lacks the AvrPto and AvrPtoB effectors and therefore cannot activate ETI. Both mutant lines, Δnrd1-1 and Δnrd1-2, showed ∼10-fold smaller populations of Pst compared to wild-type RG-PtoR 2 days after bacterial inoculation. In addition, the Δnrd1 mutants developed much less symptoms of bacterial speck disease on leaves compared to RG-PtoR 5 days after inoculation. Thus, Nrd1 appears to act as a negative regulator of PTI against Pst DC3000, which was unexpected given that Nrd1 transcripts increase in abundance upon treatment with flgII-28, a MAMP, and we suspected it might make a positive contribution to PTI. The enhanced resistance in the Δnrd1 mutants to DC3000ΔavrPtoΔavrPtoB was not observed in experiments with four other Pst strains or with Xanthomonas campestris pv. vesicatoria (also known as X. euvesicatoria; Supplemental Table S1).
Mutations of Nrd1 do not affect MAMP-induced ROS production or MAPK activation
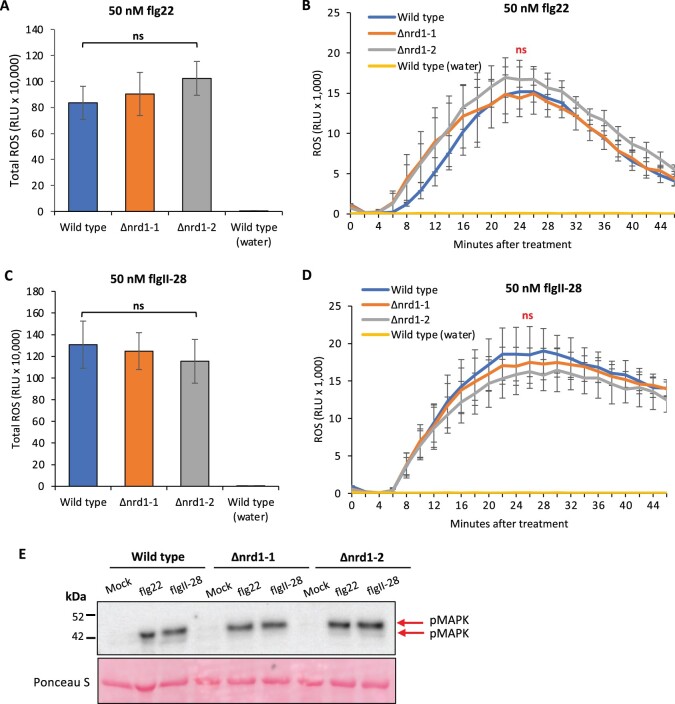
ROS production and MAPK activation are two early PTI-associated responses in bacterial-inoculated plants. To investigate whether Nrd1 contributes to these PTI responses, we performed ROS and MAPK activation assays using the two flagellin-derived peptides, flg22 and flgII-28. Leaves of wild-type RG-PtoR plants and the Δnrd1-1 and Δnrd1-2 mutant lines all produced a similar amount of ROS when treated with flg22 or flgII-28 (Figure 3, A–D). No evidence of constitutive generation of ROS in the Δnrd1-1 and Δnrd1-2 mutant lines was observed in experiments where leaves were mock treated with water only (Supplemental Figure S3). Similarly, we observed no difference in the ability of the Δnrd1-1 and Δnrd1-2 mutant lines and wild-type plants to activate MAPK phosphorylation in response to these two peptides or to mock treatment with water only (Figure 3E). Thus, Nrd1 appears to act downstream or independent of ROS and MAPK signaling pathways in the PTI response.
Figure 3.
Investigation of MAMP-induced ROS production and MAPK activation in the Δnrd1 mutants. A–D, Leaf discs from Δnrd1 or RG-PtoR wild-type plants were treated with 50-nM flg22 (A and B), 50-nM flgII-28 (C and D), or water only. Relative light units (RLU) were measured over 45 min. One-way ANOVA followed by Student’s t test (P < 0.05) was performed for total ROS (A and C) or at 24 min (peak readout) and 45 min after treatment with flg22 or flgII-28 (B and D). Bars represent means ± sd in (A, B, C, and D). No significant difference was observed between Δnrd1 and wild-type plants with either treatment. E, Leaf discs from wild-type RG-PtoR plants and Δnrd1 mutants were treated with 10-nM flg22, 25-nM flgII-28, or water (mock) for 10 min. Proteins were extracted from a pool of discs from plants of the three genotypes and subjected to immunoblotting using an anti-pMAPK antibody that detects phosphorylated MAPKs (red arrows). Ponceau staining shows equal loading of proteins.
RNA sequencing identifies Nrd1-regulated putative defense and susceptibility genes
Based on the enhanced resistance to Pst in the Δnrd1 mutants, we hypothesized that the increased abundance of the Nrd1 transcripts after flgII-28 treatment leads to increased Nrd1 protein that acts to suppress a subset of defense-related (D) genes and/or induces a subset of susceptibility (S) genes, thus promoting the growth of Pst. If this were the case, then in the Δnrd1 mutants, the Nrd1-regulated putative defense genes would be induced or no longer suppressed while the putative S genes would be suppressed, resulting in enhanced resistance to Pst infection. To identify Nrd1-regulated genes, we performed an RNA-seq analysis using the two Δnrd1 mutants and wild-type RG-PtoR plants inoculated with DC3000ΔavrPtoΔavrPtoB (Tables 1 and 2). Transcript levels were quantified as fragments per kilobase of transcript per million mapped fragments (FPKM) and ranged from 0 to approximately 10,000 for the genes predicted in the tomato genome.
Table 1.
Summary of genes with increased or decreased transcript abundance in the Δnrd1 lines compared to wild-type plants
| Comparison | Total no. of differentially expressed genes | No. of upregulated genes | No. of downregulated genes |
|---|---|---|---|
| Δnrd1-1/wild-type | 463 | 211 | 252 |
| Δnrd1-2/wild-type | 144 | 93 | 51 |
| Common | 51 | 43 | 8 |
The Δnrd1 and the wild-type Rio Grande (RG)-PtoR plant were inoculated with 5 × 106 cfu · mL−1 DC3000ΔavrPtoΔavrPtoB (DC3000ΔΔ) 6 h later. A ≥2-fold difference and adjusted P < 0.05 were used as cutoffs.
Table 2.
Nrd1-regulated putative defense-related genes and susceptibility genes identified by RNA-seq
| Gene class | Gene name | Gene ID | Description | Δnrd1-1/wild-type* | Adjusted P | Δnrd1-2/wild-type* | Adjusted P |
|---|---|---|---|---|---|---|---|
| Putative defense-related gene (upregulated in nrd1 mutants) | D1 | Solyc05g024190 | Chlorophyll synthase, chloroplastic | 2.857 | 0.001313 | 3.846 | 0.00792 |
| D2 | Solyc07g061790 | Heme-binding protein 2-like | 4.348 | 2.45E−05 | 6.667 | 2.66E−08 | |
| D3 | Solyc02g077330 | GDSL esterase/lipase | 2.778 | 0.014041 | 3.571 | 0.01104 | |
| D4 | Solyc12g009650 | Sl proline-rich protein | 2.222 | 6.79E−13 | 2.326 | 2.97E−05 | |
| D5 | Solyc11g019910 | Plant invertase/pectin methylesterase inhibitor superfamily protein | 2.128 | 0.00512 | 2.632 | 5.81E−05 | |
| D6 | Solyc08g078020 | Arabinogalactan (Agp1) | 3.125 | 3.81E−11 | 3.704 | 3.40E−09 | |
| Putative susceptibility genes (downregulated in nrd1 mutants) | S1 | Solyc03g112030 | Cytochrome P450 | 0.312 | 0.023826 | 0.415 | 0.00779 |
| S2 | Solyc02g088210 | SPX domain-containing protein 4 | 0.457 | 0.000645 | 0.355 | 5.14E−09 | |
| S3 | Solyc05g007440 | ARM repeat superfamily protein | 0.289 | 3.05E−30 | 0.348 | 4.42E−19 |
A total of 51 genes were differentially expressed in both Δnrd1-1 and Δnrd1-2 mutants compared to wild-type plants (Supplemental Table S2). From these, we selected six putative defense-related genes (fold-change ≥ 2 and adjusted P < 0.05) and three putative susceptibility genes (fold-change <0.5 and adjusted P < 0.05), based on two criteria: (1) the transcript abundance was ≥2 FPKM in either Δnrd1 mutants or wild-type plants and (2) the expression of putative Nrd1-regulated defense genes (upregulated in Δnrd1 mutants) was suppressed after flgII-28 treatment in wild-type plants, while the putative susceptibility (S) genes (downregulated in Δnrd1 mutants) were induced by flgII-28 in wild-type plants, based on previous RNA-seq data (Rosli et al., 2013; Supplemental Table S2). Using the motif-searching database PlantPan2.0 (Chow et al., 2016), we found one to five copies of the E-box element (CANNTG) in the promoters of these nine candidate genes suggesting that Nrd1 might directly bind to their promoters (Supplemental Figure S4).
Overexpression of Agp1 in N. benthamiana significantly inhibits bacterial growth
The functional role of the nine candidate genes in tomato will need to be tested in the future by generation of stable tomato mutants. As an alternative, we chose to use the recently reported “agromonas” assay (Buscaill et al., 2021) to test the possible functions of the Nrd1-regulated genes in defense or susceptibility in N. benthamiana. In this assay, agroinfiltration is used first to overexpress the gene of interest in N. benthamiana leaves followed 2 days later by syringe-inoculation of the Pst strain DC3000ΔavrPtoΔavrPtoBΔhopQ1-1 or DC3000ΔhopQ1-1 at the same agroinfiltrated spots (Figure 4; Supplemental Figure S5). HopQ is recognized by NLR Roq in N. benthamiana and its deletion makes DC3000 virulent on this species (Wei et al., 2007; Schultink et al., 2017). We hypothesized that overexpression of an important defense gene would inhibit Pst growth, while overexpression of a key S gene would promote Pst growth. Among the nine candidate genes tested, overexpression in N. benthamiana leaves of the putative defense-related gene D6, Agp1 (Solyc08g078020), encoding an arabinogalactan protein, led to 8- to 10-fold less bacterial growth when inoculated with DC3000ΔavrPtoΔavrPtoBΔhopQ1-1 or DC3000ΔhopQ1-1, suggesting that Agp1 plays a role in tomato resistance to Pst. The expression of all proteins was confirmed by western blot (Supplemental Figure S5).
Figure 4.
Transient overexpression of candidate immunity-related proteins in N. benthamiana leaves followed by a bacterial pathogenicity assay. A and B, Leaves of 5-week-old N. benthamiana plants were syringe-infiltrated with Agrobacterium (1D1249) strains (Optical Density (OD) = 0.5) carrying a binary expression vector expressing each gene. Two days later, the same agroinfiltrated spots were syringe-infiltrated with 5 × 104 cfu · mL−1 DC3000ΔhopQ1-1 (A) or 5 × 104 cfu · mL−1 DC3000ΔhopQ1-1ΔavrPtoΔavrPtoB (B). Pst DC3000 populations were measured 2 days after the second infiltration. Cfu, colony-forming unit. Bars show means ± sd. Different letters indicate significant differences based on a one-way ANOVA followed by Student’s t test (P < 0.05). ns, no significant difference. Three (A) or four plants (B) were tested with each gene in each experiment. Each experiment was performed at least two times with similar results.
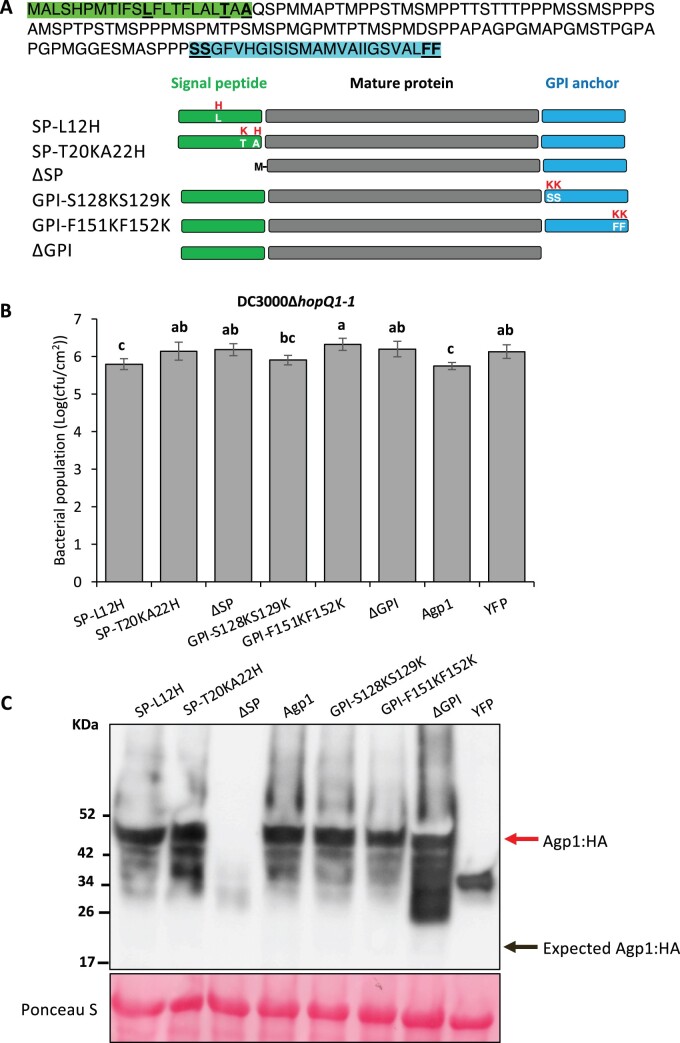
Agp1 has a predicted signal peptide (SP) and a glycosylphosphatidylinositol (GPI) lipid anchor and, like other arabinogalactan proteins, it is likely associated with the outer leaflet of the plasma membrane (Silva et al., 2020). To investigate the potential function of the Agp1 SP and putative GPI anchor in immunity, we introduced amino acid substitutions into the SP sequence (SP-L12H and SP-T20K/A22H) or GPI-anchor sequence (GPI-S128K/S129K and GPI-F151K/F152K), or deleted the entire SP (ΔSP) or GPI-anchor sequence (ΔGPI; Figure 5). We then performed the agromonas assay to test whether the effect of these substitutions on Agp1-mediated immunity to Pst. All the substitutions, except SP-L12H and GPI-S128K/S129K, impacted the ability of Agp1 to suppress Pst DC3000 growth compared to the wild-type Agp1 which, as expected, significantly inhibited bacterial growth in this assay (Figure 5). Each of the variant proteins was expressed similar to wild-type Agp1, except for the one lacking the entire SP protein, probably due to protein degradation (Figure 5). The mass of the Agp1 protein and its variants was more than twice that expected based solely on their amino acid sequences (22 kDa), which is possibly due to glycosylation, since Agp1 contains 28 predicted glycosylated sites (Steentoft et al., 2013; Supplemental Figure S6). Overall, these results showed the putative SP sequence and GPI-anchor sequence are essential for Agp1-mediated resistance to Pst.
Figure 5.
Analysis of the role in immunity to Pst of the Agp1 SP and putative GPI anchor. A, Top: amino acid sequence of the Agp1 protein. SP sequence is highlighted in green and the GPI-anchored sequence is highlighted in blue. Schematics show the substituted amino acids or deletions of the Agp1 protein, each fused to an HA epitope tag. B, Leaves of 5-week-old N. benthamiana plants were syringe-infiltrated with Agrobacterium (1D1249) strains (OD = 0.5) carrying a binary expression vector expressing each gene. Two days later, the same agroinfiltrated spots were syringe-infiltrated with 5 × 104 cfu · mL−1 DC3000ΔhopQ1-1. Pst DC3000 populations were measured 2 days after the second infiltration. Cfu, colony-forming unit. Bars show means ± sd. Different letters indicate significant differences based on a one-way ANOVA followed by Student’s t test (P < 0.05). Three plants were tested with each gene in each experiment. The experiments were performed twice with similar results. C, Proteins were extracted from N. benthamiana leaves expressing each Agp1:HA variant 2 days after agroinfiltration. Proteins were detected by immunoblotting with an α-HA antibody. The upper red arrow indicates the Agp1:HA fusion protein and the lower black arrow indicates the expected mass of the Agp1:HA protein.
Loss of Nrd1 function has no effect on the transcript abundance of multiple important PTI-associated genes
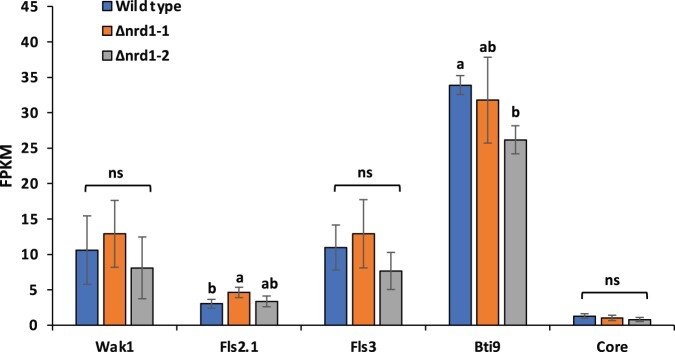
Multiple tomato immunity-associated genes including Bti9, Core, Fls2, Fls3, and Wak1 play important roles in PTI responses (Zeng et al., 2012; Rosli et al., 2013; Hind et al., 2016; Wang et al., 2016; Zheng et al., 2019; Roberts et al., 2020). These genes are greatly upregulated in wild-type RG-PtoR plants upon inoculation with the PTI-inducing strain DC3000ΔavrPtoΔavrPtoB (Rosli et al., 2013). We analyzed our RNA-Seq data to determine whether the loss of Nrd1 function affects transcript abundance of these immunity-associated genes upon inoculation with DC3000ΔavrPtoΔavrPtoB (Figure 6). The transcript abundance of each of the six genes was not significantly different in the Δnrd1 mutants compared to RG-PtoR except for Bti9 where there was an inexplicable difference in abundance between nrd1-2 and the wild-type and nrd1-1 plants. Therefore, the enhanced immunity observed in the Δnrd1 mutants is probably due to the activation of key components of PRR signaling (Fls2/Fls3/Wak1, etc.) as well as the loss of Nrd1-regulated suppression of the defense gene Agp1.
Figure 6.
Transcript abundance of selected immunity-associated genes in Rio Grande (RG)-PtoR (wild-type) and Δnrd1 mutant plants when inoculated with DC3000ΔavrPtoΔavrPtoB. RNA-seq analysis was performed using the two Δnrd1 mutants and wild-type RG-PtoR plants 6 h after inoculation with 5 × 106 cfu · mL−1 DC3000ΔavrPtoΔavrPtoB (DC3000ΔΔ). Four plants for each genotype were used. Bars show means ± sd. Different letters indicate significant differences based on a one-way ANOVA followed by Tukey’s HSD test (P < 0.05). ns, no significant difference.
Discussion
The Nrd1 gene was originally identified from a small subset of 44 genes whose transcript abundance in tomato leaves increased in response to flgII-28 but not in response to flg22 or csp22 (Rosli et al., 2013). This specificity was subsequently confirmed by reverse transcription quantitative real-time PCR (RT-qPCR) and Nrd1 is therefore useful as a reporter gene for the Fls3 pathway (Roberts et al., 2020). Because the gene is induced by flgII-28, we anticipated that a loss-of-function mutation in Nrd1 might lead to the loss of certain aspects of PTI. However, unexpectedly, two independent Δnrd1 mutants showed enhanced resistance specifically to Pst DC3000, indicating the Nrd1 protein acts as a negative regulator of resistance to this Pst strain. An RNA-seq analysis of the Δnrd1 mutants identified genes whose transcript abundance is either increased or decreased in an Nrd1-dependent manner and we hypothesized these genes might play a role in defense or susceptibility, respectively. The overexpression of one of the putative defense genes, Agp1, encoding an arabinogalactan protein, did in fact enhance resistance to DC3000, suggesting that it plays a role in the enhanced resistance of the Δnrd1 mutants. Here, we place Nrd1 in the context of previous reports of negative regulators of immunity, propose a model for Fls3-specific transcriptional reprogramming, discuss the possible role of Agp1 in defense and its regulation by Nrd1, and consider the prospect that Nrd1/Agp1 might be used to identify a unique component of Pst DC3000 that is involved in the enhanced resistance observed in the Δnrd1 mutants.
Negative regulators of plant immunity can be viewed as susceptibility (S) genes since their expression allows enhanced growth of the pathogen and accordingly enhanced disease (van Schie and Takken, 2014; Koseoglou et al., 2022). S genes have been classified into those that play a role in host recognition, suppression of host defenses, or in pathogen sustenance and they encode diverse proteins including transporters, protein kinases, membrane-associated proteins (e.g. Mlo), and enzymes (e.g. Dmr6; Zheng et al., 2013; van Schie and Takken, 2014; Santillan Martinez et al., 2020; Thomazella et al., 2021). Of particular relevance here, several S genes encode TFs in the bHLH, bZIP, ERF, and WRKY families (Jin et al., 2011; Fan et al., 2014; Wang et al., 2015a; Schwartz et al., 2017; Lu et al., 2020; Fang et al., 2021; Prior et al., 2021; Campos et al., 2022). Similar to Nrd1, a few bHLH TFs have been found previously to act as negative regulators of disease resistance in plants. For instance, two tomato bHLH genes, SlbHLH3 and SlbHLH6, are upregulated by the transcription activator-like effector AvrHah1 in Xanthomonas gardneri and promote susceptibility of tomato to bacterial spot disease (Schwartz et al., 2017). bHLH TFs in other plant species, including the well-characterized HBI1 in Arabidopsis thaliana, negatively regulate a subset of genes involved in plant immunity and mediate a tradeoff between growth and immunity in plants (Fan et al., 2014). In contrast to these bHLH negative regulators which are either induced by bacterial effectors (Schwartz et al., 2017) or suppressed by MAMPs or other bacterial components (Fan et al., 2014), the Nrd1 gene is induced specifically by a flagellin-derived MAMP flgII-28 but acts in a way that promotes bacterial pathogenesis.
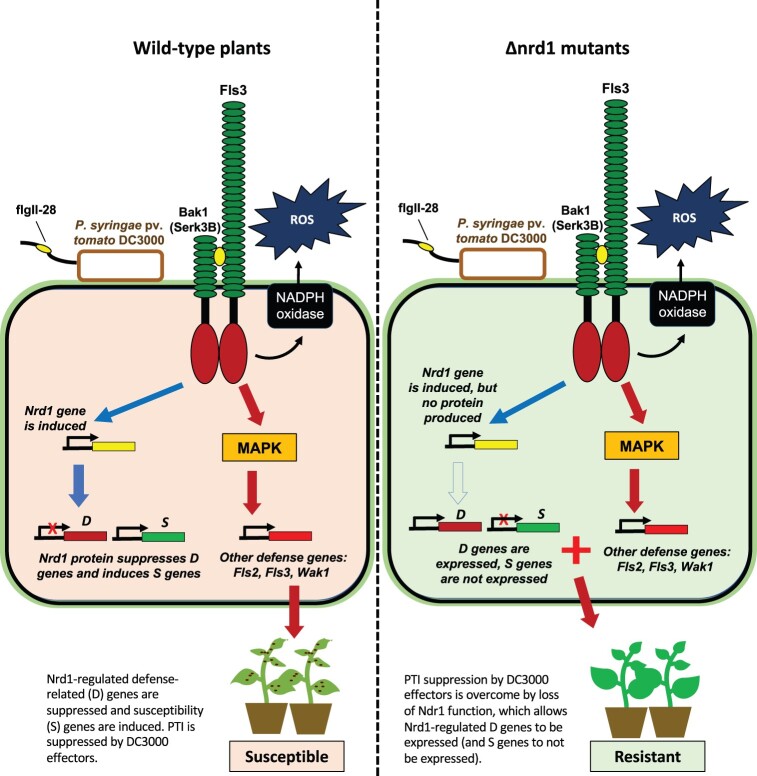
The tomato receptor Fls3 binds flgII-28 and works in concert with the co-receptor BAK1 (in tomato, Serk3A, and/or Serk3B) to activate intracellular signaling (Hind et al., 2016). Our present and previous RNA-Seq analysis and the phenotype of the Δnrd1 mutants together are consistent with a model in which Fls3 activates both resistance-enhancing and susceptibility-enhancing responses (Figure 7). To resist Pst infection, Fls3 and other PRRs activate PTI responses leading to the rapid generation of ROS, activation of MAPKs and extensive changes in transcriptional programming that inhibit Pst growth. The Fls3-activated pathway also results in the induction of Nrd1 gene expression and likely the increase of Nrd1 protein abundance which, we propose, suppresses a subset of defense genes and induces a subset of susceptibility genes promoting tomato susceptibility to Pst infection. In a loss-of-function mutation in Nrd1 the subset of defense genes, including Agp1, are no longer suppressed (or are induced) and S genes are not expressed, leading to enhanced Pst resistance (Figure 7). Additionally, in the Δnrd1 mutants, multiple well-characterized defense genes including Bti9, Core, Fls2, Fls3, and Wak1 are still induced upon Pst inoculation, and ROS production and MAPK activation are not compromised, suggesting that the observed increased resistance in the Δnrd1 mutants is due to the activation of key PRR signaling components as well as the loss of Nrd1-regulated suppression of some defense genes such as Agp1 and/or the loss of Nrd1-regulated induction of certain S genes.
Figure 7.
Proposed model for the enhanced resistance seen in Δnrd1 mutants. Fls3 appears to regulate two opposing host responses: (1) To resist Pst infection, Fls3 and other PRRs induce ROS, MAPK, and other defense responses which inhibit Pst growth. (2) Fls3 also induces Nrd1 gene expression, and increases Nrd1 protein abundance, which suppresses a subset of defense genes and also induces a subset of susceptibility genes further promoting susceptibility to Pst infection. When Nrd1 is mutated, the subset of defense genes, including Agp1, are no longer suppressed (or are induced) and S genes are not expressed leading to enhanced resistance to Pst.
The discovery that overexpression of the tomato Agp1 gene significantly reduced DC3000 populations in leaves further reinforces the importance of the plant cell wall as the location for key immunity-associated activities (Bacete et al., 2018; Molina et al., 2021). Arabinogalactan-proteins (AGPs) belong to a large family of cell wall hydroxyproline-rich glycoproteins that are involved in diverse biological processes including plant growth and development and plant–microbe interactions (Gaspar et al., 2004; Seifert and Roberts, 2007). Classical AGPs contain an N-terminal hydrophobic secretion signal, a central “PAST” domain (i.e. rich in Pro, Ala, Ser, and Thr) residues, and a hydrophobic C-terminal sequence that directs the attachment of GPI anchor (Silva et al., 2020), whose presence or absence has been demonstrated to play a major impact on the host immune response to pathogen infection (Butikofer et al., 2001). GPI modification also allows the defense-associated protein NDR1 to attach on the outer surface of the plasma membrane, thus positively regulating disease resistance to multiple bacterial and fungal pathogens (Century et al., 1997, 1995; Coppinger et al., 2004). In yeast, lesions in GPI-anchor production prevent certain proteins reaching the cell surface leading to cell wall defects and even death (Kinoshita et al., 1997). Consistent with this, we found removal of the GPI anchor from Agp1 caused a loss of N. benthamiana resistance to Pst DC3000, indicating the essential role of GPI anchor on Agp1 function in the immune response, likely by disrupting the association of the Agp1 protein with the extracellular face of the plasma membrane. Additionally, Agp1 is probably heavily glycosylated, a common post-translational modification in AGPs that might regulate protein conformation, activity and stability in host–pathogen interactions (Lin et al., 2020).
The molecular mechanisms of AGPs in plant–microbe interactions remain largely unknown. The accumulation of AGPs was found to be one of the earliest observable changes near bacterial infection sites in Arabidopsis leaves, and the authors speculated they crosslinked with other polymers to entrap bacteria in conjunction with ROS and peroxidases (Mitchell et al., 2014). It also has been proposed that GPI-anchored proteins can be involved in signaling via phospholipase cleavage of the protein from the lipid anchor or via interactions with other plasma membrane or cell wall-associated proteins that are able to activate signaling pathways (Schultz et al., 1998; Schultz and Harrison, 2008; Yeats et al., 2018; Zhou, 2019). It is intriguing to speculate that GPI-anchored Agp1 might act in a complex with PRRs and modulate ligand recognition specificity (Yeats et al., 2018; Zhou, 2019) or that Agp1 interacts with the cell wall-associated kinase SlWak1 (Zhang et al., 2020) after the release of Agp1 from the plasma membrane by cleavage of the GPI anchor; AGP epitopes have been reported to co-localize with Waks in tobacco (Nicotiana tabacum) protoplasts (Gens et al., 2000). Degradation products of AGPs could also function as damage-associated molecular patterns (DAMPs) eliciting a defense response (Villa-Rivera et al., 2021). In this regard, Arabidopsis WAK1 has been demonstrated to be a receptor of oligogalacturonides (OGs), an important component of some DAMPs (Brutus et al., 2010). The observation that AGPs localize in lipid rafts where many receptor proteins are clustered further supports the hypothesis that Agp1 might associate with certain defense-associated receptors (Ellis et al., 2010). Although these various studies suggest possible molecular mechanisms of AGPs in plant–microbe interaction, more experiments are needed to understand how Agp1 enhances plant defense against Pst.
Our RNA-seq analysis identified a small number of genes whose transcript abundance was statistically significantly different in the Δnrd1 mutants compared to wild-type RG-PtoR. Using criteria based on transcript abundance and the effect of flgII-28 on gene expression we focused on nine genes which we hypothesized could contribute to either defense (D) or susceptibility (S). Each of these genes contains one or more E-box elements in their promoter which raised the possibility that their expression might be regulated, at least in part, by direct binding of the Nrd1 protein to these elements. However, we were unable to detect such binding using electrophoretic mobility shift assays with the two E-box elements present in the Agp1 promoter. The mechanism by which Nrd1 leads to changes in transcript abundance of the D and S genes therefore remains unknown and could involve Nrd1 binding to another cis-element, or an indirect mechanism such as Nrd1 interaction with other TFs, or a role of Nrd1 in inducing expression of another TF which then regulates the D and S genes.
Loss-of-function mutations in S genes offer a promising approach to enhancing broad-spectrum disease resistance, if the mutation does not have pleiotropic detrimental effects (Koseoglou et al., 2022). There are several examples of this strategy in the literature, although none yet involve a bHLH TF (Seifert and Roberts, 2007; Zheng et al., 2013; van Schie and Takken, 2014; Sun et al., 2016; Santillan Martinez et al., 2020; Hanika et al., 2021; Thomazella et al., 2021). In contrast to such broad-spectrum activity, the enhanced resistance in the Δnrd1 mutants appears specific to Pst DC3000 as the Δnrd1 mutants were susceptible to four other strains of Pst and to the bacterial pathogen Xanthomonas (Supplemental Table S1). In light of this, although we saw no detrimental morphological or growth defects in the Δnrd1 mutants they will likely not be generally useful for controlling bacterial speck disease. However, our results do raise the possibility that DC3000 expresses a unique component, lacking in other Pst strains, that is recognized by the Δnrd1 mutants. The future identification of such a Pst component might lead to the discovery of a novel host recognition mechanisms.
Materials and methods
Generation of Nrd1 tomato mutants using CRISPR/Cas9
To generate the Δnrd1 mutants in the tomato (Solanum lycopersicum) cultivar Rio Grande (RG)-PtoR, which has the Pto and Prf genes, we designed a guide RNA (5′-GTAGTCCAGAAAAGCTAGAC-3′) that targets the first exon of Nrd1 using the software Geneious R11 (Kearse et al., 2012). The gRNA cassette was cloned into the p201N:Cas9 binary vector as described previously (Jacobs et al., 2017). Tomato transformation was performed at the Biotechnology Center at the Boyce Thompson Institute as described previously (Zhang et al., 2020). Mutations were confirmed by Sanger sequencing at the Biotechnology Resource Center (BRC) at Cornell University.
Phylogenetic analyses
The Nrd1 protein sequence was used as a query sequence to search for related sequences in tomato, Arabidopsis, and rice using NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Amino acid alignments were performed by ClustalW (https://www.genome.jp/tools-bin/clustalw). In addition, other tomato bHLH genes were included since they have been characterized previously (Ling et al., 2002; Du et al., 2015; Schwartz et al., 2017; Wang et al., 2015b; Kim and Mudgett, 2019). Phylogenetic trees were constructed with MEGA-X (Kumar et al., 2018) using the maximum likelihood method and JTT matrix-based model (Jones et al., 1992). Bootstrap analysis with 1,000 replicates was performed. Positions containing gaps and missing data were eliminated.
Bacterial inoculation
Four-week-old Δnrd1 and wild-type plants were vacuum-infiltrated with the various Pst DC3000 strains at different titers, including DC3000ΔavrPtoΔavrPtoB (DC3000ΔΔ) or DC3000ΔavrPtoΔavrPtoBΔfliC (DC3000ΔΔΔ) at 5 × 104 cfu · mL−1 or DC3000 at 1 × 106 cfu−1. Bacterial populations were measured at 3 h (Day 0) and 2 days after inoculation (Day 2). Photographs of disease symptoms were taken 5 or 6 days after bacterial inoculation.
ROS assay
ROS production was measured as described previously (Clarke et al., 2013). In brief, leaf discs were collected and floated in water overnight. Water was then removed and replaced with a solution containing flg22 (QRLSTGSRINSAKDDAAGLQIA) or flgII-28 (ESTNILQRMRELAVQSRNDSNSSTDRDA) at the indicated concentrations, in combination with 34 µg · mL−1 luminol (Sigma-Aldrich) and 20 µg · mL−1 horseradish peroxidase. ROS production was measured using a Synergy 2 microplate reader (BioTek).
MAPK phosphorylation assay
MAPK phosphorylation assay was performed as described previously (Zhang et al., 2020). Six leaf discs of Δnrd1 mutant and wild-type plants were floated in water overnight. The leaf discs were then incubated with flg22 or flgII-28 at desired concentrations, or water only for 10 min, and immediately frozen in liquid nitrogen. Protein was extracted using buffer containing 50-mM Tris–HCl (pH 7.5), 10% glycerol (v/v), 2-mM ethylenediaminetetraacetic acid (EDTA), 1% Triton X-100 (v/v), 5-mM dithiothreitol (DTT), 1% protease inhibitor cocktail (Sigma-Aldrich; v/v), 0.5% Phosphatase inhibitor cocktail 2 (Sigma-Aldrich; v/v). MAPK phosphorylation was determined using an anti-phospho-p44/42 MAPK(Erk1/2) antibody (anti-pMAPK; Cell Signaling).
Construct generation
The coding region of each putative defense or susceptibility gene was amplified from tomato cDNA using Phusion Hot Start II DNA polymerase (ThermoFisher Scientific) and gene-specific primers (Supplemental Table S3) and cloned into pJLSmart (Mathieu et al., 2014) by Gibson assembly. The gene expression cassette in pJLSmart was then cloned into the destiny vector pGWB414 via recombination reactions using LR Clonase II (ThermoFisher Scientific). Vectors were confirmed by Sanger sequencing and transformed into Agrobacterium strain 1D1249 for transient expression and agromonas assays in N. benthamiana.
Amino acid substitutions in the SP and putative GPI-anchor sequences of the Agp1 protein were determined using SignalP-5.0 (Almagro Armenteros et al., 2019) and NetGPI-1.1 (Gíslason et al., 2021). Amino acid substitutions were generated with the Q5 site-directed mutagenesis kit (NEB) with specific primers (Supplemental Table S3). The SP sequence (retaining the ATG) and the GPI sequence were deleted by PCR with specific primers using Phusion Hot Start II DNA polymerase (Supplemental Table S3). All mutated fragments were first cloned into pJLSmart by Gibson assembly and then pGWB414 by LR reaction.
Agromonas assay
The agromonas assays were performed as described (Buscaill et al., 2021). Briefly, Agrobacterium strains 1D1249 carrying a binary vector (pGWB414) expressing the gene of interest were syringe infiltrated into leaves of 4-week-old N. benthamiana plants. Two days later, the same agroinfiltrated spots were syringe infiltrated with either DC3000ΔhopQ1-1 or DC3000ΔhopQ1-1ΔavrPtoΔavrPtoB at 5 × 104 cfu · mL−1. Bacterial populations were measured by serial dilutions on LB medium supplemented with 10-μg · mL−1 cetrimide, 10-μg · mL−1 fucidin, and 50-μg · mL−1 cephaloridine (CFC; Oxoid C-F-C Supplement) 2 days after Pst inoculation.
Immunoblotting
Total protein was extracted from N. benthamiana leaves using 250-μL extraction buffer consisting of 62.5-mM Tris–HCl (pH 6.8), 2% sodium dodecyl sulfate (v/v), 10% glycerol (v/v), and 5% β-mercaptoethanol (v/v). A 12-μL soluble protein solution mixed with 4× Laemmli sample buffer were boiled at 95°C for 5 min before loaded for gel electrophoresis. Protein was loaded on 4%–20% precast sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel (Bio-Rad), blotted on polyvinylidene fluoride or polyvinylidene difluoride membrane (Merck Millipore), inoculated with α-HA primary antibody (1:7,000; v/v) and α-rat-HRP secondary antibody (1:10,000; v/v), and developed with Piece ECL plus substrate (Thermo Scientific) for 5 min.
RNA-seq
Five-week-old wild-type RG-PtoR plants and the two lines of Δnrd1 mutants were vacuum infiltrated with a suspension of DC3000ΔavrPtoΔavrPtoB at 5 × 106 cfu · mL−1. Four biological replicates were performed for each treatment. Tissue samples were collected at 6 h after infiltration. Total RNA was isolated with the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. RNA was treated with DNase by column-based purification (RNase-Free DNase Kit, Qiagen). RNA libraries were prepared and sequenced on an Illumina HiSeq 4000 system. Raw RNA-seq reads were processed to remove adaptors and low-quality sequences using Trimmomatic (version 0.36) with default parameters (Bolger et al., 2014). The remaining cleaned reads were aligned to the ribosomal RNA database (Quast et al., 2013) using bowtie (version 1.1.2; Langmead, 2010) allowing up to three mismatches, and those aligned were discarded. The remaining cleaned reads were mapped to the tomato reference genome (SL4.0 and ITAG4.1) using HISAT2 (version 2.1.0; Kim et al., 2019) with default parameters. Based on the alignments, raw read counts for each gene were calculated using HTSeq-count (Anders et al., 2015) and normalized to FPKM. Raw read counts were then fed to DESeq2 (Love et al., 2014) to identify differentially expressed genes, with a cutoff of adjusted P < 0.05 and fold change >2.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers XM_010320613 (Nrd1, Solyc03g114230) and NM_001247216.2 (Agp1, Solyc08g078020).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. The wild-type and mutated Nrd1 protein sequences.
Supplemental Figure S2. Analysis of the two most closely related genes to Nrd1 in tomato.
Supplemental Figure S3. The Δnrd1 mutants do not constitutively produce ROS.
Supplemental Figure S4. Predicted E-box elements (CANNTG) in Nrd1-regulated putative defense and susceptibility genes.
Supplemental Figure S5. Transient overexpression of putative susceptibility genes proteins in N. benthamiana leaves followed by a bacterial pathogenicity assay and confirming expression of the D and S proteins.
Supplemental Figure S6. Prediction of glycosylation sites in the Agp1 protein.
Supplemental Table S1. Summary of disease assays with the Δnrd1 mutant plants.
Supplemental Table S2. The 51 Nrd1-regulated putative defense and susceptibility genes identified by RNA-Seq.
Supplemental Table S3. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Liam Cleary, Brian Bell, Jay Miller, and Joe Ettenberger for plant care, and Joyce Van Eck for tomato transformation.
Funding
The funding was provided by National Science Foundation grant IOS-1546625 (G.B.M. and Z.F.).
Conflict of interest statement. The authors declare that they have no conflict of interest.
Contributor Information
Ning Zhang, Boyce Thompson Institute for Plant Research, Ithaca, New York 14853, USA; Plant Pathology and Plant-Microbe Biology Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853, USA.
Chloe Hecht, Boyce Thompson Institute for Plant Research, Ithaca, New York 14853, USA.
Xuepeng Sun, Boyce Thompson Institute for Plant Research, Ithaca, New York 14853, USA.
Zhangjun Fei, Boyce Thompson Institute for Plant Research, Ithaca, New York 14853, USA; Plant Pathology and Plant-Microbe Biology Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853, USA; USDA-ARS Robert W. Holley Center for Agriculture and Health, Ithaca, New York 14853, USA.
Gregory B Martin, Boyce Thompson Institute for Plant Research, Ithaca, New York 14853, USA; Plant Pathology and Plant-Microbe Biology Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853, USA.
G.B.M. and N.Z. conceived and designed the experiments. N.Z. designed gRNAs, constructed vectors, performed genotyping and phenotyping experiments, and analyzed the data. C.H. performed ROS assays. Z.F. and X.S. analyzed RNA-seq data. N.Z. and G.B.M. interpreted the data and wrote the manuscript. All the authors read and approved the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Gregory B. Martin (gbm7@cornell.edu).
References
- Abramovitch RB, Anderson JC, Martin GB (2006) Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol 7: 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37: 420–423 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacete L, Melida H, Miedes E, Molina A (2018) Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. Plant J 93: 614–636 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaill P, Sanguankiattichai N, Lee YJ, Kourelis J, Preston G, van der Hoorn RAL (2021) Agromonas: a rapid disease assay for Pseudomonas syringae growth in agroinfiltrated leaves. Plant J 105: 831–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butikofer P, Malherbe T, Boschung M, Roditi I (2001) GPI-anchored proteins: now you see’em, now you don’t. FASEB J 15: 545–548 [DOI] [PubMed] [Google Scholar]
- Campos MD, Félix M, Patanita M, Materatski P, Albuquerque A, Ribeiro JA, Varanda C (2022) Defense strategies: the role of transcription factors in tomato-pathogen interaction. Biology 11: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ (1997) NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278: 1963–1965 [DOI] [PubMed] [Google Scholar]
- Century KS, Holub EB, Staskawicz BJ (1995) NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc Natl Acad Sci USA 92: 6597–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Munkvold KR, Gao H, Mathieu J, Schwizer S, Wang S, Yan YB, Wang J, Martin GB, Chai J (2011) Structural analysis of Pseudomonas syringae AvrPtoB bound to host BAK1 reveals two similar kinase-interacting domains in a type III effector. Cell Host Microbe 10: 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CN, Zheng HQ, Wu NY, Chien CH, Huang HD, Lee TY, Chiang-Hsieh YF, Hou PF, Yang TY, Chang WC (2016) PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res 44: D1154–D1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke CR, Chinchilla D, Hind SR, Taguchi F, Miki R, Ichinose Y, Martin GB, Leman S, Felix G, Vinatzer BA (2013) Allelic variation in two distinct Pseudomonas syringae flagellin epitopes modulates the strength of plant immune responses but not bacterial motility. New Phytol 200: 847–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppinger P, Repetti PP, Day B, Dahlbeck D, Mehlert A, Staskawicz BJ (2004) Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J 40: 225–237 [DOI] [PubMed] [Google Scholar]
- DeFalco TA, Zipfel C (2021) Molecular mechanisms of early plant pattern-triggered immune signaling. Mol Cell 81: 3449–3467 [DOI] [PubMed] [Google Scholar]
- Du J, Huang Z, Wang B, Sun H, Chen C, Ling HQ, Wu H (2015) SlbHLH068 interacts with FER to regulate the iron-deficiency response in tomato. Ann Bot 116: 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M, Egelund J, Schultz CJ, Bacic A (2010) Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol 153: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Bai MY, Kim JG, Wang T, Oh E, Chen L, Park CH, Son SH, Kim SK, Mudgett MB, et al. (2014) The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell 26: 828–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Meng X, Zhang J, Xia M, Cao S, Tang X, Fan T (2021) AtWRKY1 negatively regulates the response of Arabidopsis thaliana to Pst. DC3000. Plant Physiol Biochem 166: 799–806 [DOI] [PubMed] [Google Scholar]
- Gaspar YM, Nam J, Schultz CJ, Lee LY, Gilson PR, Gelvin SB, Bacic A (2004) Characterization of the Arabidopsis lysine-rich arabinogalactan-protein AtAGP17 mutant (rat1) that results in a decreased efficiency of agrobacterium transformation. Plant Physiol 135: 2162–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gens JS, Fujiki M, Pickard BG (2000) Arabinogalactan protein and wall-associated kinase in a plasmalemmal reticulum with specialized vertices. Protoplasma 212: 115–134 [DOI] [PubMed] [Google Scholar]
- Gíslason MH, Nielsen H, Almagro Armenteros JJ, Johansen AR (2021) Prediction of GPI-anchored proteins with pointer neural networks. Curr Res Biotechnol 3: 6–13 [Google Scholar]
- Hanika K, Schipper D, Chinnappa S, Oortwijn M, Schouten HJ, Thomma B, Bai Y (2021) Impairment of tomato WAT1 enhances resistance to vascular wilt fungi despite severe growth defects. Front Plant Sci 12: 721674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Hind SR, Strickler SR, Boyle PC, Dunham DM, Bao Z, O'Doherty IM, Baccile JA, Hoki JS, Viox EG, Clarke CR, et al. (2016) Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat Plants 2: 16128. [DOI] [PubMed] [Google Scholar]
- Hu DG, Wang N, Wang DH, Cheng L, Wang YX, Zhao YW, Ding JY, Gu KD, Xiao X, Hao YJ (2020) A basic/helix-loop-helix transcription factor controls leaf shape by regulating auxin signaling in apple. New Phytol 228: 1897–1913 [DOI] [PubMed] [Google Scholar]
- Huibers RP, de Jong M, Dekter RW, Van den Ackerveken G (2009) Disease-specific expression of host genes during downy mildew infection of Arabidopsis. Mol Plant Microbe Interact 22: 1104–1115 [DOI] [PubMed] [Google Scholar]
- Hussain A, Noman A, Arif M, Farooq S, Khan MI, Cheng P, Qari SH, Anwar M, Hashem M, Ashraf MF, et al. (2021) A basic helix-loop-helix transcription factor CabHLH113 positively regulate pepper immunity against Ralstonia solanacearum. Microb Pathog 156: 104909. [DOI] [PubMed] [Google Scholar]
- Jacobs TB, Zhang N, Patel D, Martin GB (2017) Generation of a collection of mutant tomato lines using pooled CRISPR libraries. Plant Physiol 174: 2023–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Martin GB (1999) Rapid transcript accumulation of pathogenesis-related genes during an incompatible interaction in bacterial speck disease-resistant tomato plants. Plant Mol Biol 40: 455–465 [DOI] [PubMed] [Google Scholar]
- Jin J, Hewezi T, Baum TJ (2011) The Arabidopsis bHLH25 and bHLH27 transcription factors contribute to susceptibility to the cyst nematode Heterodera schachtii. Plant J 65: 319–328 [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kay S, Hahn S, Marois E, Hause G, Bonas U (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318: 648–651 [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Mudgett MB (2019) Tomato bHLH132 transcription factor controls growth and defense and is activated by Xanthomonas euvesicatoria effector XopD during pathogenesis. Mol Plant Microbe Interact 32: 1614–1622 [DOI] [PubMed] [Google Scholar]
- Kim YJ, Lin NC, Martin GB (2002) Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109: 589–598 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Ohishi K, Takeda J (1997) GPI-anchor synthesis in mammalian cells: genes, their products, and a deficiency. J Biochem 122: 251–257 [DOI] [PubMed] [Google Scholar]
- Koseoglou E, van der Wolf JM, Visser RGF, Bai Y (2022) Susceptibility reversed: modified plant susceptibility genes for resistance to bacteria. Trends Plant Sci 27: 69–79 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35: 1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B (2010) Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics Chapter 11: Unit 11.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Meng X, Shan L, He P (2016) Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe 19: 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, et al. (2006) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol 141: 1167–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Qing X, Liao J, Zhuo K (2020) Role of protein glycosylation in host-pathogen interaction. Cells 9: 1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M (2002) The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA 99: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolle S, Stevens D, Coaker G (2020) Plant NLR-triggered immunity: from receptor activation to downstream signaling. Curr Opin Immunol 62: 99–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Deng F, Jia J, Chen X, Li J, Wen Q, Li T, Meng Y, Shan W (2020) The Arabidopsis thaliana gene AtERF019 negatively regulates plant resistance to Phytophthora parasitica by suppressing PAMP-triggered immunity. Mol Plant Pathol 21: 1179–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GB (2012) Suppression and activation of the plant immune system by Pseudomonas syringae effectors AvrPto and AvrPtoB. InMartin F, Kamoun S, eds, Effectors in Plant-Microbe Interactions. Wiley-Blackwell, Ames, IA, pp 123–154 [Google Scholar]
- Mathieu J, Schwizer S, Martin GB (2014) Pto kinase binds two domains of AvrPtoB and its proximity to the effector E3 ligase determines if it evades degradation and activates plant immunity. PLoS Pathog 10: e1004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell K, Brown I, Knox P, Mansfield J (2014) The role of cell wall-based defences in the early restriction of non-pathogenic hrp mutant bacteria in Arabidopsis. Phytochemistry 112: 139–150 [DOI] [PubMed] [Google Scholar]
- Molina A, Miedes E, Bacete L, Rodriguez T, Melida H, Denance N, Sanchez-Vallet A, Riviere MP, Lopez G, Freydier A, et al. (2021). Arabidopsis cell wall composition determines disease resistance specificity and fitness. Proc Natl Acad Sci USA 118: e2010243118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou BPM, Ahn HK, Ding P, Jones JDG (2021) Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592: 110–115 [DOI] [PubMed] [Google Scholar]
- Nguyen HP, Chakravarthy S, Velásquez AC, McLane HS, Zeng L, Park D-W, Collmer A, Martin GB (2010) Methods to study PAMP-triggered immunity using tomato and Nicotiana benthamiana. Mol Plant-Microbe Interact 23: 991–999 [DOI] [PubMed] [Google Scholar]
- Oh C-S, Martin GB (2011) Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci 16: 132–140 [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB (2003) Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Ann Rev Phytopathol 41: 215–243 [DOI] [PubMed] [Google Scholar]
- Pombo MA, Zheng Y, Fernandez-Pozo N, Dunham DM, Fei Z, Martin GB (2014) Transcriptomic analysis reveals tomato genes whose expression is induced specifically during effector-triggered immunity and identifies the Epk1 protein kinase which is required for the host response to three bacterial effector proteins. Genome Biol 15: 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior MJ, Selvanayagam J, Kim JG, Tomar M, Jonikas M, Mudgett MB, Smeekens S, Hanson J, Frommer WB (2021) Arabidopsis bZIP11 is a susceptibility factor during Pseudomonas syringae infection. Mol Plant Microbe Interact 34: 439–447 [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R, Liu AE, Wan L, Geiger AM, Hind SR, Rosli HG, Martin GB (2020) Molecular characterization of differences between the tomato immune receptors Flagellin Sensing 3 and Flagellin Sensing 2. Plant Physiol 183: 1825–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R, Mainiero S, Powell AF, Liu AE, Shi K, Hind SR, Strickler SR, Collmer A, Martin GB (2019) Natural variation for unusual host responses and flagellin-mediated immunity against Pseudomonas syringae in genetically diverse tomato accessions. New Phytol 223: 447–461 [DOI] [PubMed] [Google Scholar]
- Rosli HG, Zheng Y, Pombo MA, Zhong S, Bombarely A, Fei Z, Collmer A, Martin GB (2013) Transcriptomics-based screen for genes induced by flagellin and repressed by pathogen effectors identifies a cell wall-associated kinase involved in plant immunity. Genome Biol 14: R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santillan Martinez MI, Bracuto V, Koseoglou E, Appiano M, Jacobsen E, Visser RGF, Wolters AA, Bai Y (2020) CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol 20: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultink A, Qi T, Lee A, Steinbrenner AD, Staskawicz B (2017) Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J 92: 787–795 [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Gilson P, Oxley D, Youl JJ, Bacic A (1998) GPI-anchors on arabinogalactanproteins: implications for signalling in plants. Trends Plant Sci 3: 426–431 [Google Scholar]
- Schultz CJ, Harrison MJ (2008) Novel plant and fungal AGP-like proteins in the Medicago truncatula-Glomus intraradices arbuscular mycorrhizal symbiosis. Mycorrhiza 18: 403–412 [DOI] [PubMed] [Google Scholar]
- Schwartz AR, Morbitzer R, Lahaye T, Staskawicz BJ (2017) TALE-induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proc Natl Acad Sci USA 114: E897–E903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ, Roberts K (2007) The biology of arabinogalactan proteins. Annu Rev Plant Biol 58: 137–161 [DOI] [PubMed] [Google Scholar]
- Silva J, Ferraz R, Dupree P, Showalter AM, Coimbra S (2020) Three decades of advances in arabinogalactan-protein biosynthesis. Front Plant Sci 11: 610377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, et al. (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J 32: 1478–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Fan HJ, Ling HQ (2015) Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics 16: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Wolters AM, Vossen JH, Rouwet ME, Loonen AE, Jacobsen E, Visser RG, Bai Y (2016) Silencing of six susceptibility genes results in potato late blight resistance. Transgenic Res 25: 731–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomazella DPT, Seong K, Mackelprang R, Dahlbeck D, Geng Y, Gill US, Qi T, Pham J, Giuseppe P, Lee CY, et al. (2021) Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc Natl Acad Sci USA 118: e2026152118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie CC, Takken FL (2014) Susceptibility genes 101: how to be a good host. Annu Rev Phytopathol 52: 551–581 [DOI] [PubMed] [Google Scholar]
- Villa-Rivera MG, Cano-Camacho H, Lopez-Romero E, Zavala-Paramo MG (2021) The role of arabinogalactan type II degradation in plant-microbe interactions. Front Microbiol 12: 730543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Lin R, Feng J, Qiu D, Chen W, Xu S (2015a) Wheat bHLH transcription factor gene, TabHLH060, enhances susceptibility of transgenic Arabidopsis thaliana to Pseudomonas syringae. Physiol Mol Plant Pathol 90: 123–130 [Google Scholar]
- Wang J, Hu Z, Zhao T, Yang Y, Chen T, Yang M, Yu W, Zhang B (2015b) Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum). BMC Genomics 16: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Albert M, Einig E, Furst U, Krust D, Felix G (2016) The pattern-recognition receptor CORE of Solanaceae detects bacterial cold-shock protein. Nat Plants 2: 16185 [DOI] [PubMed] [Google Scholar]
- Wei CF, Kvitko BH, Shimizu R, Crabill E, Alfano JR, Lin NC, Martin GB, Huang HC, Collmer A (2007) A Pseudomonas syringae pv. Tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J 51: 32–46 [DOI] [PubMed] [Google Scholar]
- Wu C-H, Kamoun S (2021) Tomato Prf requires NLR helpers NRC2 and NRC3 to confer resistance against the bacterial speck pathogen Pseudomonas syringae pv. tomato. Acta Hortic 1316: 61–66 [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, et al. (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 18: 74–80 [DOI] [PubMed] [Google Scholar]
- Xin XF, Kvitko B, He SY (2018) Pseudomonas syringae: what it takes to be a pathogen. Nat Rev Microbiol 16: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Kapos P, Cheng YT, Li M, Zhang Y, Li X (2014) NLR-associating transcription factor bHLH84 and its paralogs function redundantly in plant immunity. PloS Pathog 10: e1004312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Bacic A, Johnson KL (2018) Plant glycosylphosphatidylinositol anchored proteins at the plasma membrane-cell wall nexus. J Integr Plant Biol 60: 649–669 [DOI] [PubMed] [Google Scholar]
- Yuan M, Jiang Z, Bi G, Nomura K, Liu M, Wang Y, Cai B, Zhou JM, He SY, Xin XF (2021a) Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592: 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Ngou BPM, Ding P, Xin XF (2021b) PTI-ETI crosstalk: an integrative view of plant immunity. Curr Opin Plant Biol 62: 102030. [DOI] [PubMed] [Google Scholar]
- Zeng L, Velasquez AC, Munkvold KR, Zhang J, Martin GB (2012) A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J 69: 92–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Pombo MA, Rosli HG, Martin GB (2020) Tomato wall-associated kinase SlWak1 acts in an Fls2- and Fls3-dependent manner to promote apoplastic immune responses to Pseudomonas syringae. Plant Physiol 183: 1869–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Roberts HM, Van Eck J, Martin GB (2020) Generation and molecular characterization of CRISPR/Cas9-induced mutations in 63 immunity-associated genes in tomato reveals specificity and a range of gene modifications. Front Plant Sci 11: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Zhang N, Martin GB, Fei Z (2019) Plant Genome Editing Database (PGED): a call for submission of information about genome-edited plant mutants. Mol Plant 12: 127–129 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Nonomura T, Appiano M, Pavan S, Matsuda Y, Toyoda H, Wolters AM, Visser RG, Bai Y (2013) Loss of function in Mlo orthologs reduces susceptibility of pepper and tomato to powdery mildew disease caused by Leveillula taurica. PLoS One 8: e70723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Zhang Y (2020) Plant immunity: danger perception and signaling. Cell 181: 978–989 [DOI] [PubMed] [Google Scholar]
- Zhou K (2019) Glycosylphosphatidylinositol-anchored proteins in Arabidopsis and one of their common roles in signaling transduction. Front Plant Sci 10: 1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C (2014) Plant pattern-recognition receptors. Trends Immunol 35: 345–351 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.