Abstract
Calmodulin-like proteins (CMLs) are calcium (Ca2+) sensors involved in plant growth and development as well as adaptation to environmental stresses; however, their roles in plant responses to cold are not well understood. To reveal the role of MsCML10 from alfalfa (Medicago sativa) in regulating cold tolerance, we examined transgenic alfalfa and Medicago truncatula overexpressing MsCML10, MsCML10-RNAi alfalfa, and a M. truncatula cml10-1 mutant and identified MsCML10-interacting proteins. MsCML10 and MtCML10 transcripts were induced by cold treatment. Upregulation or downregulation of MsCML10 resulted in increased or decreased cold tolerance, respectively, while cml10-1 showed decreased cold tolerance that was complemented by expressing MsCML10, suggesting that MsCML10 regulates cold tolerance. MsCML10 interacted with glutathione S-transferase (MsGSTU8) and fructose 1,6-biphosphate aldolase (MsFBA6), and the interaction depended on the presence of Ca2+. The altered activities of Glutathione S-transferase and FBA and levels of ROS and sugars were associated with MsCML10 transcript levels. We propose that MsCML10 decodes the cold-induced Ca2+ signal and regulates cold tolerance through activating MsGSTU8 and MsFBA6, leading to improved maintenance of ROS homeostasis and increased accumulation of sugars for osmoregulation, respectively.
A calmodulin-like protein activates glutathione S-transferase and fructose 1,6-biphosphate aldolase to improve cold tolerance in alfalfa.
Introduction
Cold stress adversely affects crop growth, productivity, and quality. Temperate plants increase their survival during freezing temperatures through a cold acclimation (CA) mechanism. Thousands of genes are reprogrammed in expression during CA, some of which encode key enzymes involved in antioxidant defense and biosynthesis of osmolytes such as soluble sugars and proline (Zhu, 2016). The antioxidant defense system functions to scavenge reactive oxygen species (ROS) which is usually accumulated under low-temperature conditions as a result of inhibition of Calvin–Benson cycle enzymes that limit the use of absorbed light energy by CO2 assimilation to maintain cell redox homeostasis (Suzuki et al., 2012). The antioxidant system consists of antioxidant enzymes, such as superoxide dismutase, catalase, ascorbate peroxidase, and glutathione (GSH) reductase, and nonenzyme antioxidants such as ascorbate, reduced GSH. Glutathione S-transferase (GST) is also an antioxidant enzyme that regulates multiple abiotic stress tolerance using GSH as a substrate to scavenge ROS (Xu et al., 2016).
Free calcium (Ca2+) is a universal signal involved in multiple cellular responses in plants (Reddy et al., 2011). Low temperature triggers Ca2+ channel activation, leading to a transient rise of free Ca2+ in the cytosol (Ding et al., 2019). The Ca2+ signal is perceived by Ca2+-binding proteins or Ca2+ sensors containing one to four helix–loop–helix structures called EF-hand motif(s) (Reddy et al., 2011). There are four types of Ca2+ sensors in plants, including calmodulin (CaM), CaM-like protein (CML), calcineurin B-like protein (CBL), and Ca2+-dependent protein kinase (CPK/CDPK) to decode Ca2+ signal by activating downstream proteins. CMLs in plants are specifically involved in the regulation of plant growth and development as well as responses to abiotic and biotic stresses (Reddy et al., 2011). Fifty CMLs have been identified in the genomes of Arabidopsis (Arabidopsis thaliana) and Medicago truncatula (Sun et al., 2020). In Arabidopsis, CML8, CML9, CML11, CML24, CML41, and CML43 are involved in plant innate immunity against bacteria (Chiasson et al., 2005; Ma et al., 2008; Leba et al., 2012; Cheng et al., 2016; Xu et al., 2017; Zhu et al., 2017), while CML9, CML20, CML37, and CML42 are responsive to abiotic stresses and regulate drought and salt tolerance positively or negatively (Magnan et al., 2008; Vadassery et al., 2012; Scholz et al., 2015; Wu et al., 2017; Zhu et al., 2022). CML24, CML25, CML36, and CML39 are involved in the regulation of flowering, pollen germination, and pollen tube elongation, or seed development and germination (Yang et al., 2014; Wang et al., 2015; Astegno et al., 2017; Midhat et al., 2018). In addition, CML10 interacts with phosphomannomutase and modulates stress responses by regulating ascorbate biosynthesis (Cho et al., 2016). CML24 regulates actin cytoskeleton and circadian oscillations through activating the circadian oscillator gene TIMING OF CAB2 EXPRESSION1 (Ruiz et al., 2018), and it regulates ALUMINUM-ACTIVATED MALATE TRANSPORTER1-dependent Al tolerance through interacting with CALMODULIN BINDING TRANSCRIPTION ACTIVATOR2, and WRKY46 (Zhu et al., 2022). CML38 confers inhibition of root growth through interacting with rapid alkalinization factor 1 (Campos et al., 2018). Overexpression of ShCML44 from wild tomato (Solanum habrochaites) results in enhanced multiple tolerance to cold, drought, and salinity stresses in transgenic tomato (Solanum lycopersicum; Munir et al., 2016). CsCML16, CsCML18-2, CsCML38, and CsCML42 are responsive to low temperature, salinity, and drought in tea plants (Camellia sinensis; Ma et al., 2019).
Alfalfa (Medicago sativa) is the most important perennial forage legume due to its excellent nutritional quality and high productivity. The difference in cold tolerance among alfalfa cultivars is associated with the accumulated sugars or total nonstructure carbohydrate levels in roots during CA (Cunningham et al., 2001; Seppanen et al., 2018). Higher levels of cryoprotective compounds such as raffinose family oligosaccharides (RFOs), sucrose, and proline are accumulated in the recurrently selected populations with superior freezing tolerance compared with the initial backgrounds during CA (Castonguay et al., 2011). Similarly, higher levels of soluble sugars such as sucrose, myo-inositol, galactinol, and raffinose are observed in M.sativa subsp. Medicagofalcata (L.), which is closely related to alfalfa, with greater cold tolerance than in alfalfa or in M. truncatula, a leguminous model plant with low cold tolerance, during CA (Zhang et al., 2011; Tan et al., 2013; Zhuo et al., 2013). In addition, the regulation of several cold-responsive genes from M. falcata, such as S-ADENOSYLMETHIONINE SYNTHETASE, ETHYLENE RESPONSE FACTOR (MfERF1), and AUXIN INDUCED IN ROOT CULTURE, on cold tolerance have been documented (Guo et al., 2014; Zhuo et al., 2018; Wang et al., 2021). Two CMLs associated with abiotic stress responses have been investigated. MtCML40 negatively regulates salt tolerance in M. truncatula although it is responsive to cold, salt, osmotic stress, and ABA (Zhang et al., 2019). MtCML42 regulates cold tolerance and early flowering through upregulating the CBF pathway and downregulating MtABI5, respectively (Sun et al., 2021). However, the function of CMLs in alfalfa and their regulation on cold tolerance are still unknown.
At least one low-temperature-responsive element is observed in the promoter region of six MtCML genes, including MtCML10 (Sun et al., 2020), implying that these MtCMLs are possibly involved in cold responses in Medicago plants. Given the important role of Ca2+ signaling in cold tolerance, the objective of this study was to study the regulation of MsCML10 and its homolog MtCML10 on cold tolerance. We documented that MsCML10 and MtCML10 could regulate cold tolerance through interactions with GST and fructose 1,6-biphosphate aldolase.
Results
Molecular characterization and spatial expression of MsCML10
The coding sequence of MsCML10 and its homologous gene MtCML10 were cloned from alfalfa and M. truncatula, respectively. MsCML10 had an ORF (open reading frame) of 453 bp, encoding a peptide consisting of 150 amino acids (GenBank accession No. OM049783) with a molecular weight of 17.16 kDa. MsCML10 was 100% identical to MtCML10 in amino acid sequence and had 78.67% identity with AtCML11 in Arabidopsis. Phylogenetic tree analysis showed that MsCML10 was mostly close to AtCML11 in Arabidopsis (Supplemental Figure S1). Structural domain analysis using SMART (http://smart.embl-heidelberg.de/) showed that MsCML10 had four EF-hand motifs similar to MtCML10 and AtCML11 (Supplemental Figure S2).
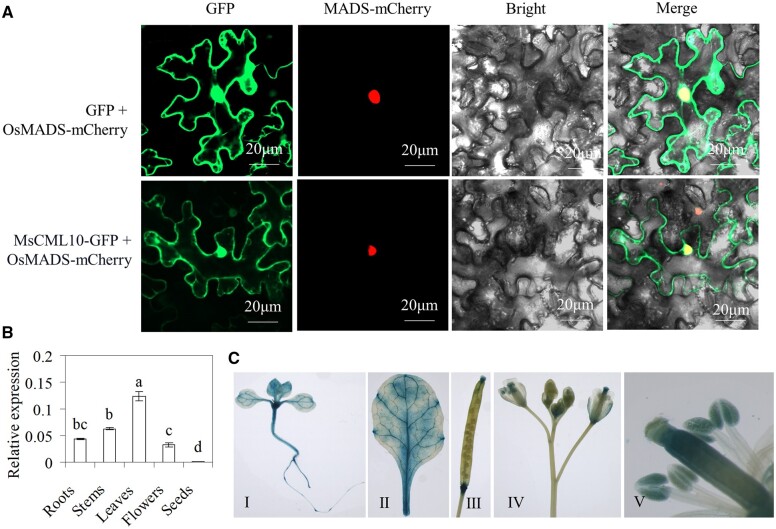
Analysis of subcellular localization of MsCML10 showed that the MsCML10-GFP fluorescence was observed in cytoplasm and nucleus, while the fluorescence of GFP or nuclear localization marker control was observed throughout cell or in the nucleus, respectively (Figure 1A). The results indicated that MsCML10 was located in the cytoplasm and nucleus. The MsCML10 transcript was detected in the roots, stems, leaves, flowers, and seeds of alfalfa, with the highest level in leaves (Figure 1B). Transgenic Arabidopsis expressing the β-glucuronidase (GUS) reporter gene driven by a 1,500-bp promoter of MsCML10 was generated. GUS staining results showed that GUS activity was observed in leaves, roots, anthers, stigma, and the ends of silique (Figure 1C), which is consistent with the MsCML10 transcript detected in alfalfa (Figure 1B).
Figure 1.
Subcellular localization of MsCML10 and the expression pattern. The vectors MsCML10::GFP or GFP in combination with mCherry-tagged MADS were co-transformed into N. benthamiana leaves for analysis of subcellular localization (A). Spatial expression of MsCML10 in alfalfa was analyzed using RT–qPCR (B). Seedlings (I), leaflet (II), silique (III), flower (IV), and stamen (V) in PMsCML10::GUS transgenic Arabidopsis was used for GUS staining (C). Means of three replicates and standard errors are presented; the same letter above the column indicates no significant difference at P < 0.05 using Duncan’s test.
MsCML10 and its homolog MtCML10 positively regulate cold tolerance in alfalfa and M. truncatula
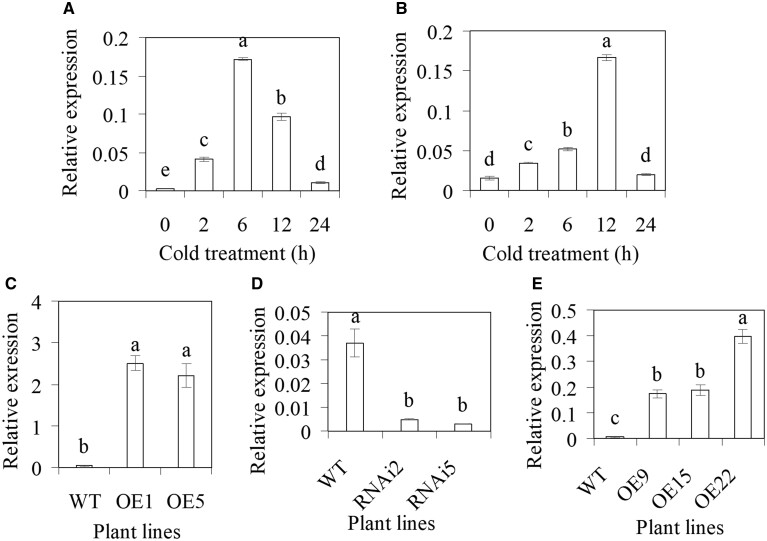
MsCML10 and MtCML10 were induced by cold treatment. The MsCML10 transcript was greatly induced after 2–12 h of cold treatment, with the highest level (40-fold) at 6 h (Figure 2A), while the MtCML10 transcript was induced after 2 to 12 h of cold treatment, with the highest level (10-fold) at 12 h (Figure 2B). In order to explore the role of MsCML10 in regulating cold tolerance, transgenic alfalfa plants overexpressing MsCML10 or downregulating MsCML10 by RNAi were generated. Two overexpressing lines (OE1 and OE5) and two RNAi lines (RNAi2 and RNAi5) showing greatly increased or decreased MsCML10 transcript levels were selected for further study (Figure 2, C and D).
Figure 2.
Analysis of transcript levels of MsCML10 and MtCML10 in response to cold and transgenic alfalfa and M. truncatula plants. Relative expression of MsCML10 (A) and MtCML10 (B) in response to cold treatment at 5°C was analyzed using RT–qPCR. Transcript levels of MsCML10 were analyzed in transgenic alfalfa overexpressing MsCML10 (C), RNAi (D), and in transgenic M. truncatula overexpressing MsCML10 (E). Means of three replicates and standard errors are presented; the same letter above the column indicates no significant difference at P < 0.05 using Duncan’s test.
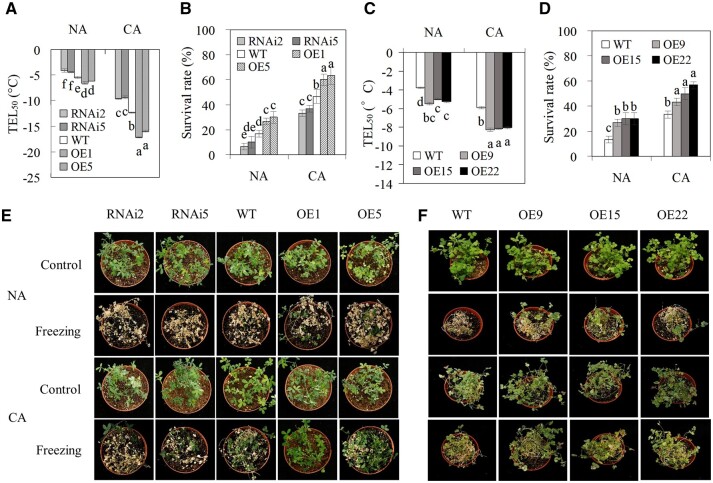
CA treatment resulted in a decrease in the temperature resulted in 50% electrolyte leakage (TEL50) and an increase in survival rate in all plant genotypes (Figure 3, A, B, and E), indicating that CA increased cold tolerance in alfalfa. Compared to the wild-type (WT), overexpression lines in alfalfa had lower levels of TEL50 and higher levels of survival rate, while RNAi lines had higher levels of TEL50 and lower levels of survival rate under both nonacclimated (NA) and CA conditions (Figure 3, A, B, and E). The results indicated that MsCML10 positively regulates cold tolerance in alfalfa.
Figure 3.
Analysis of cold tolerance in MsCML10-overexpressing and RNAi plants in comparison with the WT. The temperature resulting in TEL50 and survival rate in transgenic alfalfa and M. truncatula plants. Plants were placed in a growth chamber at 5°C for 7 days for CA treatment, while those at room temperature received the NA treatment. TEL50 was measured in MsCML10-RNAi and MsCML10-overexpressing alfalfa (A) as well as in MsCML10-overexpressing M. truncatula (C). Survival rates were measured after plants were treated by freezing at −5°C (NA plants) or −7°C (CA plants) for 6 h (B and D). After 2 days of recovery at room temperature, the freeze-treated and control alfalfa (E) and M. truncatula (F) plants were photographed. Means of three replicates and standard errors are presented; the same letter above the column indicates no significant difference at P < 0.05 using Duncan’s test.
Transgenic M. truncatula plants overexpressing MsCML10 were generated. Three transgenic lines (OE9, OE15, and OE22) showing greatly increased MsCML10 transcript were selected (Figure 2E). Similar to alfalfa plants, CA treatment resulted in a decrease in TEL50 and an increase in survival rate in both transgenic lines and the WT plants (Figure 3, C, D, and F), while transgenic lines had lower levels of TEL50 and higher levels of survival rate than the WT under NA and CA conditions (Figure 3, C, D, and F). In addition, a homozygous mutation line with Tnt1 retrotransposon insertion in the fourth exon of MtCML10, cml10-1, was identified (Supplemental Figure S3a). Compared to the WT, no MtCML10 transcript was detected in cml10-1 (Supplemental Figure S3b). Higher TEL50 and lower survival rate were observed in cml10-1 than in the WT under both NA and CA conditions (Supplemental Figure S3, c–e), indicating that MtCML10 was essential for cold tolerance in M. truncatula. The complementary plants (MsCML10::cml10-1) with a similar or slightly higher level of MsCML10 transcript as that in the WT were generated (Supplemental Figure S4a). The complementary plant had the similar level of TEL50 to that in the WT, but lower level was observed in complementing plant than in cml10-1 (Supplemental Figure S4b). The results indicated that the defect in cold tolerance in cml10-1 could be complemented by expressing MsCML10.
Ca2+-dependent interaction of MsCML10 with MsGSTU8 and MsFBA6
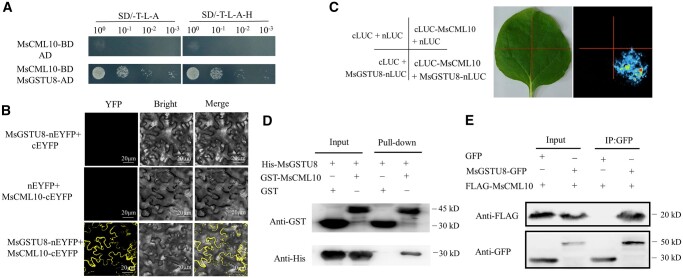
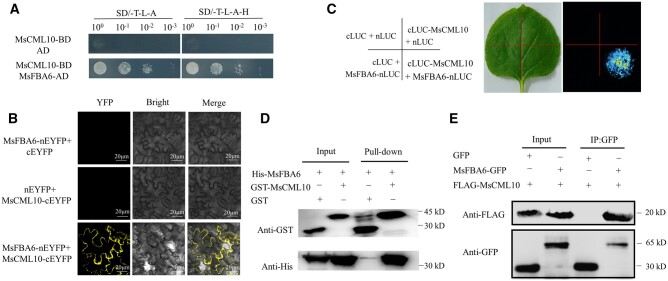
For understanding how MsCML10 regulates cold tolerance, a yeast two-hybrid (Y2H) screen was performed to identify proteins that interact with MsCML10. Several interacting clones were isolated, among them are MsGST8 and MsFBA6. To verify the interaction, the full length of CDS of MsGST8 and MsFBA6 was used for Y2H analysis. The results showed that MsGST8 and MsFBA6 possibly interact with MsCML10 (Figures 4, A and 5, A). The interactions were further confirmed by using bimolecular fluorescence complementation (BiFC), firefly luciferase complementation imaging (LCI), GST pull-down and co-immunoprecipitation (Co-IP) assays. Co-expression of MsCML10-cYFP and YFPN-MsGSTU8 or YFPN-MsFBA6 in Nicotianabenthamiana leaves resulted in a strong YFP fluorescence in the cytosol in BiFC assay, while no fluorescence was shown in the negative controls (Figures 4b and 5b). The results indicated that GSTU8 and FBA6 were cytoplasmic isozymes. MtGSTU8 protein is predicted to be localized in the cytoplasm (Hasan et al., 2021). Phylogenetic tree analysis showed that MsGSTU8 and MsFBA6 were mostly similar to AtGSTU19 (Supplemental Figure S5) and cytoplasmic FBAs in Arabidopsis, respectively (Supplemental Figure S6). The interactions were also observed in the LCI assay (Figures 4, C and 5, C). The pull-down assay showed that MsGSTU8 or MsFBA6 interacts with GST-MsCML10, but not with GST alone (Figures 4, D and 5, D). The interactions were further verified using Co-IP assay when MsGSTU8-GFP or MsFBA6-GFP was co-expressed with Flag-MsCML10 in leaf cells of N. benthamiana. Immunoblot analysis using an anti-Flag antibody showed that both MsGSTU8 and MsFBA6 interacted with MsCML10 (Figures 4, E and 5, E). The above results indicated that MsCML10 interacted with MsGST8 and MsFBA6.
Figure 4.
Analysis of interaction of MsCML10 with MsGSTU8. Interaction of MsCML10 with MsGSTU8 was analyzed using the methods of Y2H (A), BiFC (B), the LCI (C), pull-down (D), and Co-IP (E). GST-MsCML10 fusion protein was used as bait and His-MsGSTU8 fusion protein as prey in the pull-down assay. Anti-His and anti-GST antibodies were used to detect bait and prey proteins, and the His and GST proteins were used as negative controls (D). Flag-MsCML10 and MsGSTU8-GFP were co-expressed in leaves of N. benthamiana. Total proteins were subjected to immunoprecipitation with anti-GFP beads. The presence of Flag-MsCML10 was detected by anti-Flag immunoblotting (E).
Figure 5.
Analysis of interaction of MsCML10 with MsFBA6. Interaction of MsCML10 with MsFBA6 was analyzed using the methods of Y2H (A), BiFC (B), the LCI (C), pull-down (D), and Co-IP (E). GST-MsCML10 fusion protein was used as bait and His-MsFBA6 fusion protein as prey in the pull-down assay. Anti-His and anti-GST antibodies were used to detect bait and prey proteins, and the His and GST proteins were used as negative controls (D). Flag-MsCML10 and MsFBA6-GFP were co-expressed in leaves of N. benthamiana. Total proteins were subjected to immunoprecipitation with anti-GFP beads. The presence of Flag-MsCML10 was detected by anti-Flag immunoblotting (E).
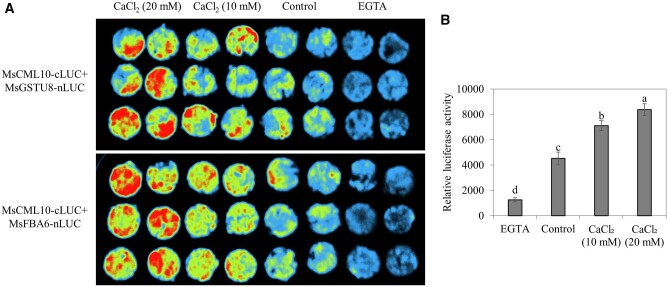
To investigate whether of the interaction between MsCML10 and MsGSTU8 or MsFBA6 was dependent upon Ca2+, a transient FLC assay was performed in the presence of CaCl2 or EGTA. The results showed that CaCl2-treated leaves had stronger interaction signal, while the EGTA-treated leaves had weaker signal as compared with the control (Figure 6, A and B), indicating that the interaction between MsCML10 and MsGSTU8 or MsFBA6 was dependent upon Ca2+.
Figure 6.
Analysis of Ca2+-dependent interaction of MsCML10 with MsGSTU8 or MsFBA6. Nicotiana benthamiana leaves co-transformed using the mixed Agrobacterium harboring 35S::cLUC-MsCML10 and 35S::MsGSTU8-nLUC or 35S::MsFBA6-nLUC were placed on 4% agar containing 10- or 20-mM CaCl2, 20-mM EGTA or without any supplement as control for 48 h, followed by capturing the luciferase image (A) or measuring the fluorescence intensity as the relative LUC activity (B). Means of the data from eight-leaf discs and standard errors are presented; the same letter above the column indicates no significant difference at P < 0.05 using Duncan’s test.
The GST activity and ROS level is affected by MsCML10 expression
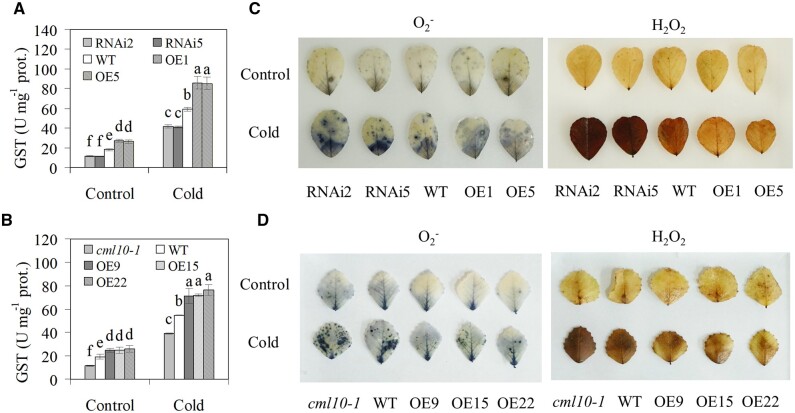
The GST activity was increased in all genotypes of alfalfa plants after cold treatment (Figure 7A). Compared to the WT, GST activity was higher in MsCML10 overexpressing lines but lower in RNAi lines under both NA and CA conditions (Figure 7A). Likely, higher GST activity was observed in overexpressing lines in M. truncatula, but lower level in cml10-1 was observed as compared with the WT under both NA and CA conditions (Figure 7B). ROS levels in the plants overexpressing and RNAi lines were examined. Leaves in overexpressing lines were less stained by 3,3-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) than WT plants, while the staining was substantially stronger in the RNAi lines (Figure 7C). Similar results were also shown in overexpressing lines in M. truncatula and cml10-1 (Figure 7D). The results indicated that lower levels of ROS (H2O2 and −) in overexpressing lines and higher level in RNAi and cml10-1 lines were accumulated as compared with the WT after CA treatment.
Figure 7.
Analysis of GST activity and ROS levels in response to cold. GST activity in MsCML10-RNAi and MsCML10-overexpressing alfalfa (A) as well as in MsCML10-overexpressing M. truncatula and cml10-1 (B) in comparison with the WT were measured after 7 days of cold treatment at 5°C. H2O2 and in MsCML10-RNAi and MsCML10-overexpressing alfalfa (C) as well as in MsCML10-overexpressing M. truncatula and cml10-1 (D) were measured after 7 days of cold treatment at 5°C. Means of three replicates and standard errors are presented; the same letter above the column indicates no significant difference at P < 0.05 using Duncan’s test.
The FBA activity and sugar accumulation is affected by MsCML10 expression
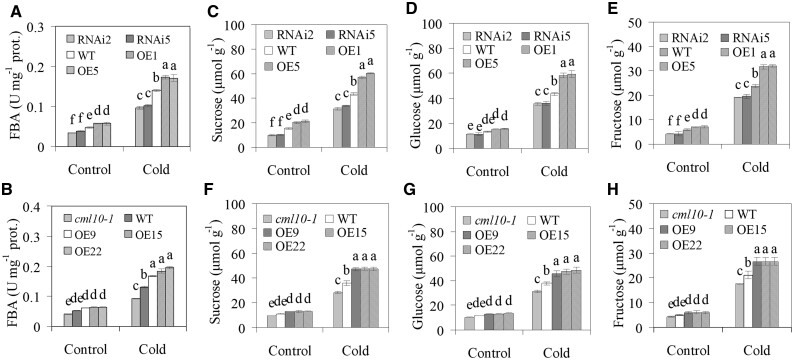
Fructose 1,6-bisphosphate aldolase activity was increased in all genotypes of alfalfa and M. truncatula plants after CA treatment (Figure 8, A and E). Compared to the WT, FBA activity was higher in overexpressing alfalfa and M. truncatula lines and lower in alfalfa RNAi lines and cml10-1 under both NA and CA conditions (Figure 8, A and E). Soluble sugars (sucrose, glucose, and fructose) levels showed the similar alterations with FBA activity in the different genotypes of plants (Figure 8, B–D and F–H). The results indicated that the alterations in sugars were associated with the altered FBA activity.
Figure 8.
Analysis of FBA activity and sugar concentrations in response to cold. FBA activity in MsCML10-RNAi and MsCML10-overexpressing alfalfa (A) as well as in MsCML10-overexpressing M. truncatula and cml10-1 (B) in comparison with the WT were measured after 7 days of cold treatment at 4°C. Sucrose (C and F), glucose (D and G), and fructose (E and H) in MsCML10-RNAi and MsCML10-overexpressing alfalfa (C, D, and E) as well as in MsCML10-overexpressing M. truncatula and cml10-1 (F, G, and H) were measured after 7 days of cold treatment at 4°C. Means of three replicates and standard errors are presented; the same letter above the column indicates no significant difference at P < 0.05 using Duncan’s test.
Discussion
A transient increase in cytosolic Ca2+ concentration is common in plants in response to abiotic stress. The Ca2+ signal is decoded by Ca2+ sensors including CaM, CML, CBL, and CDPK through activating interaction proteins and regulating downstream cellular responses (Lecourieux et al., 2006). MsCML10 from alfalfa was characterized in the present study. It is identical to MtCML10 in amino acid sequence and mostly similar to AtCML11 in Arabidopsis. EF-hand motifs are the sites binding with Ca2+, which is the conserved domain in Ca2+-binding proteins (Reddy et al., 2011). One to four EF-hand motifs are present in MtCMLs in M. truncatula (Sun et al., 2020). Four EF-hand motifs were observed in MsCML10. In addition, MsCML10 was located in the cytoplasm and nucleus, although no nuclear signal peptide was observed in the MsCML10 sequence. The fused MsCML10-GFP is still small (∼44.16 kDa); thus, it is presumed that MsCML10 was localized in the cytoplasm, but it may diffuse nonspecifically into nucleus through nuclear pores like MtCML42 (Sun et al., 2021).
Although some CML members are responsive to abiotic stresses in Arabidopsis (Magnan et al., 2008; Vadassery et al., 2012; Scholz et al., 2015; Wu et al., 2017), their role in regulation of cold tolerance has not been documented. Similar to MtCML42 in M. truncatula (Sun et al., 2021), MsCML10 and MtCML10 transcripts were greatly induced by cold, while the induction of MsCML10 was earlier with a higher level than that of MtCML10. Overexpression of MsCML10 resulted in enhanced cold tolerance in alfalfa and M. truncatula, while downregulating MsCML10 and mutation of MtCML10 resulted in decreased cold tolerance in alfalfa RNAi lines and cml10-1, respectively. In addition, the defect in cold tolerance in cml10-1 was complemented by expressing MsCML10. The results suggest that MsCML10 and MtCML10 positively regulate cold tolerance in alfalfa and M. truncatula. MtCML42 regulates cold tolerance through upregulating the CBF pathway and MtGolS expression for increased RFO accumulation in M. truncatula (Sun et al., 2021).
MsGSTU8 was documented to interact with MsCML10 and the interaction is dependent upon Ca2+. GSTU is a plant-specific GST that plays a key role in ROS scavenging (Xu et al., 2016). GSTU19 confers multiple tolerance to salt, drought and methyl viologen-induced oxidative stress in Arabidopsis (Xu et al., 2016). GSTU can be activated by phytohormone signaling. TaGST1 is activated by BRASSINAZOLE-RESISTANT 2 for regulation of drought tolerance in wheat (Triticum aestivum; Cui et al., 2019). PtrGSTU17 expression is regulated by ERF9 for regulation of cold tolerance by maintaining ROS homeostasis in Poncirus trifoliata (Zhang et al., 2022). GST activity was increased in all genotypes of plants and a higher level was obtained in MsCML10 overexpressing alfalfa and M. truncatula and lower level was in RNAi alfalfa and cml10-1 mutant as compared with the WT after CA treatment. In addition, lower levels of ROS (H2O2 and −) were observed in overexpressing lines of MsCML10, while higher levels were in the RNAi lines and cml10-1 mutant than in the WT, suggesting that the altered ROS levels were associated with GST activity and MsCML10 and MtCML10 transcript levels. ROS production is unavoidable under low temperature as a result of inhibition of the Calvin–Benson cycle enzyme activity that limits the use of absorbed light energy by CO2 assimilation (Suzuki et al., 2012). Thus, ROS homeostasis is important for plant CA. Our results suggest that MsCML10 regulates cold tolerance by activating MsGSTU8 to maintain ROS homeostasis.
Interaction of MsCML10 with MsFBA6 was observed in cytosol and depended upon the presence of Ca2+. The interaction localization and similarity of MsFBA6 with Arabidopsis cytoplasmic FBAs suggest that MsFBA6 is a cytoplasmic isozyme. FBA activity was increased in all plants after CA treatment, and higher activity of FBA was observed in MsCML10 overexpressing alfalfa and M. truncatula plants, while lower activity was in RNAi and cml10-1 plants, suggesting that FBA activity was activated by MsCML10 or MtCML10 in alfalfa and M. truncatula. Cytoplasmic FBA catalyzes the reversible conversion of fructose-1,6-diphosphate (FBP) into dihydroxyacetone phosphate and glyceraldehyde-3-phosphate during glycolysis and gluconeogenesis and is involved in sugar metabolism (Ronimus and Morgan, 2003). Sugar accumulation plays an important role in winter hardening in alfalfa (Cunningham et al., 2001; Seppanen et al., 2018) and in cold tolerance in M. truncatula (Zhang et al., 2011). Sucrose, glucose, and fructose concentrations were increased after cold treatment in alfalfa and M. truncatula plants, with higher levels in MsCML10 overexpressing plants but lower levels in RNAi and cml10-1 plants. The altered sugar concentrations were associated with changes in FBA activity. Transgenic tomato plants overexpressing SiFBA5 from Saussurea involucrata show increased cold tolerance and biomass (Mu et al., 2021). Overexpression of FBA7 results in increased net photosynthetic rate and stem thickness in N. benthamian plants (Cai et al., 2016). Nevertheless, our results suggest that interaction of MsCML10 with MsFBA6 resulted in accumulated sugars that was associated with the increased cold tolerance in alfalfa and M. truncatula plants.
Conclusions
MsCML10 and MtCML10 transcripts were induced in response to cold. Upregulation or downregulation of MsCML10 resulted in increased or decreased cold tolerance, respectively, while cml10-1 showed decreased cold tolerance that could be complemented by expressing MsCML10. The Ca2+-dependent interaction of MsCML10 with MsGSTU8 and MsFBA6 and the altered activities of GST and FBA and levels of ROS and sugars were associated with the MsCML10 transcript level. It is suggested that MsCML10 decodes the cold-induced Ca2+ signal and regulates cold tolerance through activating MsGSTU8 and MsFBA6, which leads to maintaining ROS homeostasis and increased accumulation of sugars for osmoregulation, respectively.
Materials and methods
Plant growth and stress treatments
A Tnt1 retrotransposon inserted mutant (NF19519) was obtained from the Noble Research Institute (https://medicago-mutant.noble.org/mutant/) for selecting homozygous line cml10-1 using retrotransposon display PCR as described (Tadege et al., 2008). Alfalfa (M.sativa L. cv. Regen-SY4D), Mtruncatula cv. R108 and cml10-1, as well as the transgenic plants, were grown in plastic pots of 15 cm in diameter in a greenhouse at 25°C under natural light, as described previously (Sun et al., 2021). Alfalfa was placed in a growth chamber at 5°C under 14 h of light for cold treatment and used to isolate total RNA. Sterilized seeds of Arabidopsis (A.thaliana) and N.benthamiana were placed on 1/2 MS medium for germination at 24°C for 7 days, and the seedlings were transplanted to a mixture of peat and perlite (3:1, v/v) and grown in a growth chamber at 22°C under 14 h of light for 3–4 weeks.
Isolation of total RNA and analysis of reverse transcription quantitative PCR and RT–PCR
Total RNA was isolated using the RNAprep Pure Plant Kit (Tiangen, Beijing, China), and 1 μg RNA was used for synthesis of the first strand cDNA using the TaKaRa PrimeScript RT kit with gDNA Eraser according to the manufacturer’s protocol. Reverse transcription–quantitative PCR (RT–qPCR) was conducted using Thermal Cycler Dice Real Time System II (Takara, Shiga, Japan) with three biological replicates of independent samples. The RT–qPCR solution (10 μL) contained 0.2-μM forward primer and reverse primer, 200-ng cDNA, and 5-μL SYBR qPCR master mix. The RT–qPCR solution (50 μL) contained 0.2-μM forward primer and reverse primer, 200-ng cDNA, 1 μL Ex Taq DNA polymerase, and 5-μL deoxyribonucleotide triphosphate (Takara, Dalian, China). The MsActin and MtActin7 (Medtr3g095530) genes that showed 100% identity in nucleic acid sequence were used as the internal controls. Relative expression was calculated using the normalized (2−ΔΔCt) value. The primers used for RT–qPCR and RT–PCR analysis are listed in Supplemental Table S1. DNAMAN software (Lynnon Biosoft, Vaudreuil, Quebec, Canada) was used for analysis of amino acid sequence.
Analysis of subcellular localization of MsCML10
The coding sequence of MsCML10 without stop codon was cloned into the vector pCAMBIA-1305 and fused with GFP driven by CaMV 35S promoter. The construct (35S::MsCML10-GFP) or the control vector (35S::GFP) in combination with 35S:OsMADS-mCherry vector were used to co-transform leaves of 1-month-old N. benthamiana plants using the Agrobacterium tumefaciens-mediated transformation method (Kumar and Kirti, 2010). The plants were incubated at 25°C for 72 h in a growth chamber, and the transformed leaves were observed and photographed using a confocal laser scanning microscope (Zeiss LSM800, Germany), using excitation/emission wavelengths 488/498–540 nm for GFP and 552/600–640 nm for mCherry.
Generation of transgenic plants
The coding sequence of MsCML10 driven by the CaMV 35S promoter was cloned into the pCAMBIA3301 binary expression vector (35S::MsCML10). To construct a complementing vector (PMtCML10::MsCML10), a 1,500-bp promoter of MtCML10 upstream initiation codon was used to drive MsCML10 in the binary expression vector pCAMBIA3301. To construct a RNAi vector, the sense fragment of MsCML10 was constructed on the pFGC5941 vector using the XhoI and NcoI sites (Kerschen et al., 2004), and a 201-bp of antisense fragment was further cloned into the above vector using the SmaI and BamHI sites. Transgenic alfalfa or M. truncatula plants were generated using A. tumefaciens strain EHA105 containing the above vectors (Sun et al., 2021). For investigation of spatial expression, a 1,500-bp promoter of MsCML10 upstream initiation codon (https://medicagohapmap2.org/) was used to drive GUS in the binary expression vector pCAMBIA3301. The A. tumefaciens strain EHA105 containing the above vector (PMsCML10::GUS) was used to transform the WT Arabidopsis (Col0) by the method of floral dip.
GUS staining
The seedlings and a specific organ were immersed in GUS staining solution consisting of 50-mM sodium phosphate buffer (pH 7.2), 2-mM X-Gluc, 2-mM K4Fe(CN)6, 10-mM EDTA, 2-mM K3Fe(CN)6, and 0.1% (v/v) Triton X-100, with 30 min of vacuum. After incubating overnight at 37°C in the dark, the green tissues were decolorized in 70% (v/v) ethanol to remove chlorophyll for photography.
Evaluation of cold tolerance
Cold tolerance was evaluated based on survival rate and the temperature resulting in TEL50. Two-month-old alfalfa and M. truncatula plants were placed in a growth chamber for 7 days of CA at 5°C or at room temperature as NA control. TEL50 was measured based on ion leakage of leaflets after 1 h of treatment at freezing temperatures and calculated by a fitted mode plot (Geng et al., 2021). For measurement of survival rate, 1-month-old plants were placed in a growth chamber with temperatures decreasing from 25°C to −5°C for NA plants or to −7°C for CA plants linearly within 6 h and maintained for 6 h, followed by moving to a fridge at 4°C overnight and then at room temperature for 2 days of recovery. The surviving seedlings were counted to calculate survival rate. The experiments consisted of three pots as repeats, and each pot contained seven individual plants.
Y2H assay
The Y2H assay was conducted using Matchmaker Gold Y2H System (Clontech, Mountain View, CA, USA) according to the manufacturer’s instructions. The coding sequence of MsCML10 was cloned into the pGBKT7 vector as a bait that was then transformed into the yeast strain Y2HGold. The transformed Y2HGold yeast strain was then transformed with the alfalfa cDNA library and placed on the synthetic dropout (SD) medium–Trp–Leu–Ade containing 10-mM 3-AT for selection at 30°C. Positive clones were selected for sequencing. The interactions between MsCML10 and MsGSTU8 or MsFBA6 were verified by Y2H assay as described above. The coding sequence of MsGSTU8 and MsFBA6 was inserted into the pGADT7 vector to produce the prey constructs. The prey vectors pGADT7-MsGSTU8 or pGADT7-MsFBA6 were transformed into the Y2HGold yeast strain expressing bait. The yeast cells were placed on SD medium–Trp–Leu–Ade–His containing 10 mM 3-AT for selection as above.
BiFC assay
The coding sequence of MsCML10 without stop codon was fused with EYFP on the vector P2YC, while MsGSTU8 or MsFBA6 were fused with EYFP at the N-terminal of MsGSTU8 or MsFBA6 on vector P2YN, respectively. Agrobacteriumtumefaciens containing MsCML10-cYFP and nYFP-MsGSTU8 or nYFP-MsFBA6 were used to co-transform the leaves of 1-month-old N. benthamiana, separately. Fluorescence of YFP was observed and photographed under a confocal laser scanning microscope (Zeiss LSM800, Germany), using excitation/emission wavelength 510 nm.
Firefly LCI analysis
The coding sequence of MsCML10 was fused with cLUC on the vector pCAMBIA-cLUC, while MsGSTU8 or MsFBA6 was fused with nLUC on the vector pCAMBIA-nLUC (Ding et al., 2021). Agrobacteriumtumefasciens GV3101 harboring 35S::cLUC-MsCML10 and 35S::MsGSTU8-nLUC or 35S::MsFBA6-nLUC were used to co-transform leaves of 1-month-old N. benthamiana plants (Pandey et al., 2015). For determination of Ca2+ dependence of the interaction, leaf discs of N. benthamiana were co-transformed using the above Agrobacterium strain and placed on 4% (w/v) agar containing 20-mM EGTA, 10-mM or 20-mM CaCl2, or without any supplement as control for 48 h. The leaf discs were used to capture the luciferase image (Lumazone Pylon 2048B, Princeton, USA; Ding et al., 2021), or placed in a 96-well microtiter plate containing 200 μL of 1-mM fluorescein for 10 min to measure the fluorescence intensity as the relative LUC activity.
Pull-down assay
The coding sequence of MsCML10 was cloned into pGEX-4T-1, while MsGSTU8 and MsFBA6 were cloned into pET-30a, separately. The GST-tagged MsCML10 and His-tagged MsGSTU8 or MsFBA6 protein was expressed and purified using columns packed with GST-agarose beads (Sangon Biotech, Shanghai, China) and a Ni-NTA Resin column (TransGen Biotech, Beijing, China), respectively, according to the manufacturer’s instructions. The pull-down assay was performed as described by Zhang et al. (2016). GST-tagged MsCML10 or GST was immobilized on GST beads which were then suspended in Lysing buffer (150-mM NaCl, 10% (v/v) glycerol, 100-mM Tris–HCl, pH 7.5), and the same amount of MsGSTU8-His or MsFBA6-His protein was added. The mixture was rotated and incubated for 2 h at 4°C, followed by centrifugation for 1 min at 5,000g at 4°C. The pellet was washed 5 times with Lysing buffer and resuspended in 50 µL of 2× SDS protein electrophoresis loading buffer. A 10-µL sample was loaded on 10% (w/v) SDS–PAGE gel for electrophoresis, followed by transferring onto PVDF membrane (Millipore, Bedford, MA, USA). Immunoblotting was conducted using anti-GST or anti-His antibody.
Co-IP assay
Co-IP assay was performed as described by Zhu et al. (2022). The full-length CDS fragment of CML10 and the coding sequences of GSTU8 and FBA6 without stop codon were cloned, respectively, into the vector pCAMBIA-1305 harboring 35S:3×FLAG or 35S:GFP to obtain the recombinant constructs of FLAG-CML10, GSTU8-GFP and FBA6-GFP. The A. tumefaciens strain GV3101 harboring the Flag-CML10 and GSTU8-GFP or FBA6-GFP constructs was used to co-transform leaves of 4-week-old N. benthamiana plants. After 48 h, total protein was extracted from the transformed leaves using NB1 buffer containing 50 mM Tris–MES buffer (pH 8.0), 0.5-M sucrose, 1-mM MgCl2, 10-mM EDTA, 5-mM DTT, and protease inhibitors, followed by incubating with anti-GFP antibody and magnetic beads for 2 h at 4°C with rotation. After three washes using wash buffer, the protein was eluted by boiling the beads with SDS buffer. Immunoprecipitated proteins were analyzed using SDS–PAGE and immunoblotted with anti-GFP or anti-Flag antibody.
Measurements of enzyme activities
Fresh leaves (0.1 g) were frozen in liquid nitrogen and grinded in 1 ml of 100-mM phosphate buffer (PBS) containing 1-mM GSH (pH 6.5), followed by centrifugation at 12,000g for 20 min at 4°C. The supernatants were collected for measurement of GST activity (Xu et al., 2016). The reaction solution (1 mL) contained 50-mM PBS (pH 7.4), 1-mM GSH, 0.5-mM 1-chloro-2, 4-Dinitrobenzene (CDNB), and 0.1-mL enzyme extract. The reaction was initiated by adding CDNB, and changes in absorbance at 340 nm within 1 min at 25°C were recorded to calculate GST activity based on the extinction coefficient of CDNB (9.6 mM−1·cm−1). For extraction of FBA, fresh leaves (0.25 g) were ground in a mortar with a pestle in 3-mL precooled extraction buffer containing 50-mM KH2PO4 buffer (pH 7.0), 10% (v/v) glycerol, 4-mM MgCl2, 1-mM EDTA, and 5-mM dithiothreitol. After centrifugation at 12,000g for 20 min at 4°C, the supernatants were used to determine FBA activity (Kamies et al., 2017). The reaction solution (1 mL) contained 50-mM HEPES–KOH buffer (pH 7.3), 1-mM EDTA, 0.1-mM NADH, 0.75 units of glycerol-3-phosphate dehydrogenase, 10 units of phosphotriose isomerase, and 100 µL of enzyme extract. The reaction was initiated by adding 0.1 mL of 4-mM FBP. The decrease in absorbance at 340 nm due to NADH oxidation within one minute at 25°C was recorded to calculate enzyme activity based on the extinction coefficient of NADH (6.22 mM−1·cm−1). One unit of enzyme activity was defined as the amount of enzyme required for catalyzing the conversion of 1 µmol substrate within one minute. Protein concentration was determined using Coomassie Brilliant Blue G-250 (Bradford, 1976).
DAB and NBT staining
The leaflets detached from 2-month-old plants were immersed in DAB solution (1 mg mL−1) for 1 h to detect H2O2 or in NBT solution (5 mg mL−1) for 12 h in the dark to detect − (Gou et al., 2020). The leaflets were decolorized in 95% (v/v) ethanol to remove chlorophyll for photography.
Measurements of soluble sugars
Fresh leaves (0.5 g) were frozen in liquid nitrogen and ground into a powder with addition of 5 mL of 80% (v/v) ethanol for extraction in a water bath at 80°C for 30 min. The mixture was cooled at room temperature and centrifuged at 12,000g for 20 min. The supernatant was quickly frozen in liquid nitrogen and vacuum-dried, followed by dissolving in 500 μL of deionized water and passing through a 0.22-μm Millipore membrane (Millipore, Bedford, MA, USA). The filtrate (30 μL) was injected into a Waters 2695 separation system, supplied with an amino bonded column (Zorbax NH2; 250 × 4.6 mm; Agilent Technologies Inc., Santa Clara, CA, USA) and Waters 2414 refractive index detector. The sugar concentration was calculated based on the standard curve for each sugar and calibrated according to the recovery throughout the analysis procedure (Sun et al., 2021).
Statistical analysis
The physiological measurements were repeated 3 times from different plant samples. All data were subjected to analysis of variances according to the model for completely randomized design using an SPSS program (SPSS Inc., Chicago, IL, USA). Differences among means of treatments and plant lines were evaluated using Duncan’s test at 0.05 probability level.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers (OM049783).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree of MsCML10 and 50 AtCMLs.
Supplemental Figure S2. Multiple alignment of MsCML10 with MtCML10 and AtCML11.
Supplemental Figure S3. Analysis of cold tolerance in cml10-1 plants in comparison with the WT.
Supplemental Figure S4. Analysis of the complementary plants as compared with the cml42 mutant and the WT.
Supplemental Figure S5. Phylogenetic tree of MsGSTU8 and AtGSTs.
Supplemental Figure S6. Phylogenetic tree of MsFBA6 and AtFBAs.
Supplemental Table S1. Primer sequences used for RT–qPCR and RT–PCR and the accession numbers of the analyzed genes.
Supplementary Material
Contributor Information
Shuhan Yu, College of Grassland Science, Nanjing Agricultural University, Nanjing 210095, China.
Jiaxuan Wu, College of Grassland Science, Nanjing Agricultural University, Nanjing 210095, China.
Yanmei Sun, College of Grassland Science, Nanjing Agricultural University, Nanjing 210095, China.
Haifeng Zhu, College of Grassland Science, Nanjing Agricultural University, Nanjing 210095, China.
Qiguo Sun, College of Grassland Science, Nanjing Agricultural University, Nanjing 210095, China.
Pengcheng Zhao, College of Grassland Science, Nanjing Agricultural University, Nanjing 210095, China.
Risheng Huang, College of Grassland Science, Nanjing Agricultural University, Nanjing 210095, China.
Zhenfei Guo, College of Grassland Science, Nanjing Agricultural University, Nanjing 210095, China.
S.Y., J.W., and Y.S. performed the experiments. S.Y., H.Z., Q.S., R.H., and P.Z. analyzed the experimental results. S.Y. and Z.G. wrote the manuscript. Z.G. conceived the research and designed the experiments.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Zhenfei Guo (zfguo@njau.edu.cn).
Funding
This work was supported by the National Natural Science Foundation of China (Grant number: 32030074).
Conflict of interest statement. None declared.
References
- Astegno A, Bonza MC, Vallone R, La Verde V, D'Onofrio M, Luoni L, Molesini B, Dominici P (2017) Arabidopsis calmodulin-like protein CML36 is a calcium (Ca2+) sensor that interacts with the plasma membrane Ca2+-ATPase isoform ACA8 and stimulates its activity. J Biol Chem 292: 15049–15061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai BB, Li Q, Xu YC, Yang L, Bi HG, Ai XZ (2016) Genome-wide analysis of the fructose 1,6-bisphosphate aldolase (FBA) gene family and functional characterization of FBA7 in tomato. Plant Physiol Biochem 108: 251–265 [DOI] [PubMed] [Google Scholar]
- Campos WF, Dressano K, Ceciliato PHO, Guerrero-Abad JC, Silva AL, Fiori CS, do Canto AM, Bergonci T, Claus LAN, Silva-Filho MC, et al. (2018) Arabidopsis thaliana rapid alkalinization factor 1-mediated root growth inhibition is dependent on calmodulin-like protein 38. J Biol Chem 293: 2159–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castonguay Y, Bertrand A, Michaud R, Laberge S (2011) Cold-induced biochemical and molecular changes in alfalfa populations selectively improved for freezing tolerance. Crop Sci 51: 2132–2144 [Google Scholar]
- Cheng HQ, Han LB, Yang CL, Wu XM, Zhong NQ, Wu JH, Wang FX, Wang HY, Xia GX (2016) The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection. J Exp Bot 67: 1935–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiasson D, Ekengren SK, Martin GB, Dobney SL, Snedden WA (2005) Calmodulin-like proteins from Arabidopsis and tomato are involved in host defense against Pseudomonas syringae pv. tomato. Plant Mol Biol 58: 887–897 [DOI] [PubMed] [Google Scholar]
- Cho KM, Nguyen HTK, Kim SY, Shin JS, Cho DH, Hong SB, Shin JS, Ok SH (2016) CML10, a variant of calmodulin, modulates ascorbic acid synthesis. New Phytol 209: 664–678 [DOI] [PubMed] [Google Scholar]
- Cui XY, Gao Y, Guo J, Yu TF, Zheng WJ, Liu YW, Chen J, Xu ZS, Ma YZ (2019) BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol 180: 605–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham SM, Gana JA, Volenec JJ, Teuber LR (2001) Winter hardiness, root physiology, and gene expression in successive fall dormancy selections from ‘Mesilla’ and ‘CUF 101’ alfalfa. Crop Sci 41: 1091–1098 [Google Scholar]
- Ding L, Wu Z, Teng R, Xu S, Cao X, Yuan G, Zhang D, Teng N (2021) LlWRKY39 is involved in thermotolerance by activating LlMBF1c and interacting with LlCaM3 in lily (Lilium longiflorum). Hortic Res 8: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding YL, Shi YT, Yang SH (2019) Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol 222: 1690–1704 [DOI] [PubMed] [Google Scholar]
- Geng B, Wang Q, Huang R, Liu Y, Guo Z, Lu S (2021) A novel LRR-RLK (CTLK) confers cold tolerance through regulation on the C-repeat-binding factor pathway, antioxidants, and proline accumulation. Plant J 108: 1679–1689 [DOI] [PubMed] [Google Scholar]
- Gou L, Zhuo C, Lu S, Guo Z (2020) A universal stress protein from Medicago falcata (MfUSP1) confers multiple stress tolerance by regulating antioxidant defense and proline accumulation. Environ Exp Bot 178: 104168 [Google Scholar]
- Guo Z, Tan J, Zhuo C, Wang C, Xiang X, Wang Z (2014) Abscisic acid, H2O2 and nitric oxide interactions mediated cold-induced S-adenosylmethionine synthetase in Medicago sativa subsp. falcata that confers cold tolerance through up-regulating polyamine oxidation. Plant Biotechn J 12: 601–612 [DOI] [PubMed] [Google Scholar]
- Hasan MS, Singh V, Islam S, Islam MS, Ahsan R, Kaundal A, Islam T, Ghosh A (2021) Genome-wide identification and expression profiling of glutathione S-transferase family under multiple abiotic and biotic stresses in Medicago truncatula L. PLoS One 16: e0247170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamies R, Farrant JM, Tadele Z, Cannarozzi G, Rafudeen MS (2017) A proteomic approach to investigate the drought response in the orphan crop Eragrostis tef. Proteomes 5: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschen A, Napoli CA, Jorgensen RA, Muller AE (2004) Effectiveness of RNA interference in transgenic plants. FEBS Lett 566: 223–228 [DOI] [PubMed] [Google Scholar]
- Kumar KRR, Kirti PB (2010) A mitogen-activated protein kinase, AhMPK6 from peanut localizes to the nucleus and also induces defense responses upon transient expression in tobacco. Plant Physiol Biochem 48: 481–486 [DOI] [PubMed] [Google Scholar]
- Leba LJ, Cheval C, Ortiz-Martin I, Ranty B, Beuzon CR, Galaud JP, Aldon D (2012) CML9, an Arabidopsis calmodulin-like protein, contributes to plant innate immunity through a flagellin-dependent signalling pathway. Plant J 71: 976–989 [DOI] [PubMed] [Google Scholar]
- Lecourieux D, Raneva R, Pugin A (2006) Calcium in plant defence-signalling pathways. New Phytol 171: 249–269 [DOI] [PubMed] [Google Scholar]
- Ma Q, Zhou Q, Chen C, Cui Q, Zhao Y, Wang K, Arkorful E, Chen X, Sun K, Li X (2019) Isolation and expression analysis of CsCML genes in response to abiotic stresses in the tea plant (Camellia sinensis). Sci Rep 9: 8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Smigel A, Tsai YC, Braam J, Berkowitz GA (2008) Innate immunity signaling: cytosolic Ca2+elevation as an early signal is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol 148: 818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnan F, Ranty B, Charpenteau M, Sotta B, Galaud JP, Aldon D (2008) Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J 56: 575–589 [DOI] [PubMed] [Google Scholar]
- Midhat U, Ting MKY, Teresinski HJ, Snedden WA (2018) The calmodulin-like protein, CML39, is involved in regulating seed development, germination, and fruit development in Arabidopsis. Plant Mol Biol 96: 375–392 [DOI] [PubMed] [Google Scholar]
- Mu J, Fu Y, Liu B, Zhang Y, Wang A, Li Y, Zhu J (2021) SiFBA5, a cold-responsive factor from Saussurea involucrata promotes cold resilience and biomass increase in transgenic tomato plants under cold stress. BMC Plant Biol 21: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir S, Liu H, Xing Y, Hussain S, Ouyang B, Zhang Y, Li H, Ye Z (2016) Overexpression of calmodulin-like (ShCML44) stress-responsive gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses. Sci Rep 6: 31772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Kanwar P, Singh A, Steinhorst L, Pandey A, Yadav AK, Tokas I, Sanyal SK, Kim BG, et al. (2015) Calcineurin B-Like protein-interacting protein kinase CIPK21 regulates osmotic and salt stress responses in Arabidopsis. Plant Physiol 169: 780–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy ASN, Ali GS, Celesnik H, Day IS (2011) Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23: 2010–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronimus RS, Morgan HW (2003) Distribution and phylogenies of enzymes of the Embden-Meyerhof-Parnas pathway from archaea and hyperthermophilic bacteria support a gluconeogenic origin of metabolism. Archaea 1: 199–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz MCM, Hubbard KE, Gardner MJ, Jung HJ, Aubry S, Hotta CT, Mohd-Noh NI, Robertson FC, Hearn TJ, et al. (2018) Circadian oscillations of cytosolic free calcium regulate the Arabidopsis circadian clock. Nat Plants 4: 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz SS, Reichelt M, Vadassery J, Mithofer A (2015) Calmodulin-like protein CML37 is a positive regulator of ABA during drought stress in Arabidopsis. Plant Signal Behav 10: e1011951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppanen MM, Alitalo V, Backstrom HK, Makiniemi K, Jokela V, Falghera-Winseman L, Khazaei H (2018) Growth, freezing tolerance, and yield performance of alfalfa (Medicago sativa L.) cultivars grown under controlled and field conditions in northern latitudes. Can J Plant Sci 98: 1109–1118 [Google Scholar]
- Sun Q, Huang R, Zhu H, Sun Y, Guo Z (2021) A novel Medicago truncatula calmodulin-like protein (MtCML42) regulates cold tolerance and flowering time. Plant J 108: 1069–1082 [DOI] [PubMed] [Google Scholar]
- Sun Q, Yu S, Guo Z (2020) Calmodulin-Like (CML) gene family in Medicago truncatula: genome-wide identification, characterization and expression analysis. Inter J Mol Sci 21: 7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Koussevitzky S, Mittler R, Miller G (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35: 259–270 [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen JQ, He J, Tu HD, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. (2008) Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Tan J, Wang C, Xiang B, Han R, Guo Z (2013) Hydrogen peroxide and nitric oxide mediated cold- and dehydration-induced myo-inositol phosphate synthase that confers multiple resistances to abiotic stresses. Plant Cell Environ 36: 288–299 [DOI] [PubMed] [Google Scholar]
- Vadassery J, Reichelt M, Hause B, Gershenzon J, Boland W, Mithofer A (2012) CML42-mediated calcium signaling coordinates responses to spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol 159: 1159–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Shi H, Huang R, Ye R, Luo Y, Guo Z, Lu S (2021) AIR12 confers cold tolerance through regulation of the CBF cold response pathway and ascorbate homeostasis. Plant Cell Environ 44: 1522–1533 [DOI] [PubMed] [Google Scholar]
- Wang SS, Diao WZ, Yang X, Qiao Z, Wang M, Acharya BR, Zhang W (2015) Arabidopsis thaliana CML25 mediates the Ca2+ regulation of K+ transmembrane trafficking during pollen germination and tube elongation. Plant Cell Environ 38: 2372–2386 [DOI] [PubMed] [Google Scholar]
- Wu X, Qiao Z, Liu H, Acharya BR, Li C, Zhang W (2017) CML20, an Arabidopsis calmodulin-like protein, negatively regulates guard cell ABA signaling and drought stress tolerance. Front Plant Sci 8: 824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Cheval C, Laohavisit A, Hocking B, Chiasson D, Olsson TSG, Shirasu K, Faulkner C, Gilliham M (2017) A calmodulin-like protein regulates plasmodesmal closure during bacterial immune responses. New Phytol 215: 77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Tian YS, Xing XJ, Peng RH, Zhu B, Gao JJ, Yao QH (2016) Over-expression of AtGSTU19 provides tolerance to salt, drought and methyl viologen stresses in Arabidopsis. Physiol Plant 156: 164–175 [DOI] [PubMed] [Google Scholar]
- Yang X, Wang SS, Wang M, Qiao Z, Bao CC, Zhang W (2014) Arabidopsis thaliana calmodulin-like protein CML24 regulates pollen tube growth by modulating the actin cytoskeleton and controlling the cytosolic Ca2+ concentration. Plant Mol Biol 86: 225–236 [DOI] [PubMed] [Google Scholar]
- Zhang LL, Zhao MG, Tian QY, Zhang WH (2011) Comparative studies on tolerance of Medicago truncatula and Medicago falcata to freezing. Planta 234: 445–457 [DOI] [PubMed] [Google Scholar]
- Zhang XX, Wang TZ, Liu M, Sun W, Zhang WH (2019) Calmodulin-like gene MtCML40 is involved in salt tolerance by regulating MtHKTs transporters in Medicago truncatula. Environ Exp Bot 157: 79–90 [Google Scholar]
- Zhang Y, Ming R, Khan M, Wang Y, Dahro B, Xiao W, Li C, Liu J-H (2022) ERF9 of Poncirus trifoliata (L.) Raf. undergoes feedback regulation by ethylene and modulates cold tolerance via regulating a glutathione S-transferase U17 gene. Plant Biotechn J 20: 183–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZQ, Hu XN, Zhang YQ, Miao ZY, Xie C, Meng XZ, Deng J, Wen JQ, Mysore KS, Frugie F, et al. (2016) Opposing control by transcription factors MYB61 and MYB3 increases freezing tolerance by relieving C-repeat binding factor suppression. Plant Physiol 172: 1306–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XY, Robe E, Jomat L, Aldon D, Mazars C, Galaud JP (2017) CML8, an Arabidopsis calmodulin-like protein, plays a role in Pseudomonas syringae plant immunity. Plant Cell Physiol 58: 307–319 [DOI] [PubMed] [Google Scholar]
- Zhu X, Wang P, Bai Z, Herde M, Ma Y, Li N, Liu S, Huang CF, Cui R, Ma H, et al. (2022) Calmodulin-like protein CML24 interacts with CAMTA2 and WRKY46 to regulate ALMT1-dependent Al resistance in Arabidopsis thaliana. New Phytol 233: 2471–2487 [DOI] [PubMed] [Google Scholar]
- Zhuo C, Liang L, Zhao Y, Guo Z, Lu S (2018) A cold responsive ethylene responsive factor from Medicago falcata confers cold tolerance by up-regulation of polyamine turnover, antioxidant protection, and proline accumulation. Plant Cell Environ 41: 2021–2032 [DOI] [PubMed] [Google Scholar]
- Zhuo C, Wang T, Lu S, Zhao Y, Li X, Guo Z (2013) A cold responsive galactinol synthase gene from Medicago falcata (MfGolS1) is induced by myo-inositol and confers multiple tolerances to abiotic stresses. Physiol Plant 149: 67–78 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers (OM049783).