To sense potential mating partners’ positions, yeast uses a default polarity site to assemble a gradient tracking machine (GTM), which redistributes up the pheromone gradient. Here, Wang et al. show that GTM redistribution requires default-site inactivation and actin-independent vesicle delivery.
Abstract
The mating of budding yeast depends on chemotropism, a fundamental cellular process. Haploid yeast cells of opposite mating type signal their positions to one another through mating pheromones. We have proposed a deterministic gradient sensing model that explains how these cells orient toward their mating partners. Using the cell-cycle determined default polarity site (DS), cells assemble a gradient tracking machine (GTM) composed of signaling, polarity, and trafficking proteins. After assembly, the GTM redistributes up the gradient, aligns with the pheromone source, and triggers polarized growth toward the partner. Since positive feedback mechanisms drive polarized growth at the DS, it is unclear how the GTM is released for tracking. What prevents the GTM from triggering polarized growth at the DS? Here, we describe two mechanisms that are essential for tracking: inactivation of the Ras GTPase Bud1 and positioning of actin-independent vesicle delivery upgradient.
Introduction
Cellular responses to chemical gradients are important for a wide range of biological phenomena. The best-known gradient-stimulated outputs are chemotaxis (directed movement) and chemotropism (directed growth). For example, chemotaxis plays a vital role in development, immunity, wound healing, inflammation, and metastasis (Iijima et al., 2002) and chemotropism is integral to axon guidance (Hong and Nishiyama, 2010; Tojima et al., 2011), angiogenesis (Basile et al., 2004; English et al., 2001), pollen tube guidance (Kim et al., 2004; Palanivelu and Preuss, 2000), root hair growth (Palanivelu and Preuss, 2000), and fungal life cycles (Daniels et al., 2006; Snetselaar et al., 1996). Although they exhibit different behaviors, chemotactic and chemotropic cells face similar challenges. The responding cell must determine the direction of the gradient source by sensing small chemical concentration differences across its surface and then correctly polarize its cytoskeleton.
The unicellular eukaryote Saccharomyces cerevisiae is among the best-studied models of both cell-cycle control and chemotropism (Arkowitz, 2009). During vegetative growth, haploid yeast cells form new buds adjacent to their last division site, resulting in a characteristic axial budding pattern (Chant and Pringle, 1995). Late in the G1 phase of the cell cycle, the Axl2 cortical marker protein recruits the Bud5 guanine nucleotide exchange factor (GEF) to the axial bud site, where it activates its target, the Ras GTPase Bud1 (Marston et al., 2001; Roemer et al., 1996). Activated Bud1 interacts with the Rho GTPase Cdc42 and its GEF Cdc24, initiating local activation of Cdc42 (Zheng et al., 1995). This positional signal is greatly amplified by two positive feedback loops, and the resulting concentrated patch of active Cdc42 triggers the nucleation of actin cables and polarized delivery of secretory vesicles (Chant and Herskowitz, 1991; Drubin and Nelson, 1996; Irazoqui et al., 2003; Kang et al., 2010). During sexual reproduction, haploid yeast cells differentiate into gametes and fuse to form diploid zygotes. Each of the two haploid mating types, MATa and MATα, secretes a peptide pheromone that activates a G-protein-coupled receptor (GPCR) on cells of the opposite type. The pheromone-bound receptor activates its cognate heterotrimeric G protein, causing Gα-GTP to dissociate from Gβγ. Free Gβγ then signals the nucleus through the Fus3 MAPK cascade, inducing changes in gene expression and cell-cycle arrest in late G1. Gβγ also positions the eventual chemotropic growth site (CS) by linking the receptor to the machinery that nucleates actin cables via the Far1 scaffold protein (Butty et al., 1998; Nern and Arkowitz, 1998; Nern and Arkowitz, 1999). Actin-directed delivery of secretory vesicles to the CS results in the formation of mating projections, commonly known as shmoos. In mating mixtures, cells find and contact a partner by determining the direction of the most potent pheromone source and polarizing their growth toward it (hereafter referred to as shmooing; Jackson and Hartwell, 1990). However, when cells are treated with isotropic pheromone or are unable to sense a gradient, they shmoo adjacent to their last bud site, i.e., at the axial site where they would have budded next if not arrested in G1 (Dorer et al., 1995; Madden and Snyder, 1992). Hence, the axial bud site is also referred to as the default polarity site (DS; Dorer et al., 1995; Nern and Arkowitz, 1999).
Like all other chemosensing cells, budding yeast exhibit a remarkable ability to interpret chemoattractant gradients. It has been estimated that a 1% difference in receptor activation across the 5-μm diameter of a yeast cell is sufficient to elicit robust orientation (Segall, 1993). In mating mixtures, yeast cells almost invariably select a single partner, even when surrounded by multiple potential mates. Two types of models have been proposed to explain how mating yeast cells accurately position the CS in response to shallow and dynamic physiological gradients. Based on localization studies of polarity proteins involved in the nucleation of actin cables and recruitment of the vesicle fusion machinery in cells stimulated by isotropic pheromone or genetic activation of the pheromone pathway, Lew and colleagues proposed that the polarity site is established at random positions with respect to the gradient source and then moves by a biased random walk to the CS, where it is constrained by Gβγ (Dyer et al., 2013; McClure et al., 2015). They present evidence that the mobility of the polarity complex depends on F-actin and postulate that the moving complex leaves newly nucleated actin cables in its wake. These cables direct the nascent receptor and G protein to the plasma membrane (PM), where they sense the local pheromone concentration and determine the final position of the polarity site. Peter and colleagues also concluded that yeast gradient sensing is mediated by the biased wandering of the polarity complex based on localization studies of polarity, trafficking, and signaling proteins in cells subjected to artificial pheromone gradients and cells in mating mixtures (Hegemann et al., 2015). However, their model differs from that of Lew et al. in two significant ways: (1) polarity is established at the axial bud site (the DS), not randomly; (2) mobility of the polarity complex is independent of F-actin and requires low Cdc42 activity.
In contrast to the biased wandering hypothesis, we have proposed a deterministic model of gradient sensing based on localization studies of the receptor, its G protein, regulators and effectors of G-protein activity, and trafficking proteins in mating cells (Abdul-Ganiyu et al., 2021; Wang et al., 2019). In this model, yeast cells gain their gradient-sensing ability and orient toward their mating partners in four phases. During global internalization, the uniformly distributed receptor and G protein are removed from the PM. During assembly, mating yeast cells take advantage of the Bud1-positioned DS to assemble the signaling, polarity, and trafficking proteins into a gradient tracking machine (GTM). Assembly of the GTM starts with Far1-Cdc24-Bem1 localization to the DS and ends with the concentration of exocytic and endocytic activities upgradient and downgradient, respectively. During tracking, segregation of exocytosis and endocytosis incrementally redistributes the GTM up the gradient along the PM to the CS. Stabilization of the GTM occurs at the CS when vesicle delivery aligns with the pheromone gradient and the endocytic machinery surrounds the secretion site.
Although our model explains how yeast cells actively track pheromone gradients, it leaves a key question unanswered: how does tracking start? That is, how is the newly assembled GTM released from the DS? The challenge is to understand how the subtle directional information embedded in the extracellular pheromone gradient overrides the strong, feedback-amplified polarity of this intrinsic site. Here, we provide evidence for two mechanisms that explain how the GTM is freed from the DS to begin tracking. First, we show that the Bud1 GEF disappears from the PM in cells preparing to mate, whereas the Bud1 GTPase-activating protein (GAP), Bud2, polarizes to the DS and tracks with the receptor. Our data also suggest that tracking requires Bud1 inactivation. Second, we show that the mode of vesicle delivery changes as the GTM transitions from assembly to tracking and from tracking to stabilization. Although actin-dependent vesicle delivery (AD-VD) is active when the GTM is immobile, both before and after tracking, actin-independent vesicle delivery (AI-VD) is necessary and sufficient during tracking. Our findings suggest that tight regulation of both DS function and the modes of vesicle delivery are essential for yeast gradient sensing.
Results
Bud5-RFP fails to localize to the DS and disappears from the PM in mating cells, whereas RFP-Bud2 polarizes to the DS and tracks with the receptor
During the G1 phase of vegetative growth in haploid cells, the Bud1 GTPase is activated adjacent to the cytokinesis site, where it marks the DS as the bud site in the next cell cycle. Active Bud1 also positions shmoo formation at the DS in cells treated with isotropic pheromone. A longstanding question in the study of yeast mating is how shallow pheromone gradients compete with the polarization machinery at a cell’s DS to establish a CS aligned with that of its partner. We have shown that G1-arrested yeast cells preparing to mate assemble a GTM composed of signaling, polarity, and trafficking proteins at the DS, which enables them to find the closest mating partner (Wang et al., 2019). Moreover, the positioning of the GTM at the DS during mating, like the positioning of the daughter cell during vegetative growth, depends on Bud1. In the absence of Bud1, mating cells assemble multiple GTMs at random positions on the PM. These observations led us to hypothesize that the release of the GTM from the DS to start tracking requires the inactivation of Bud1.
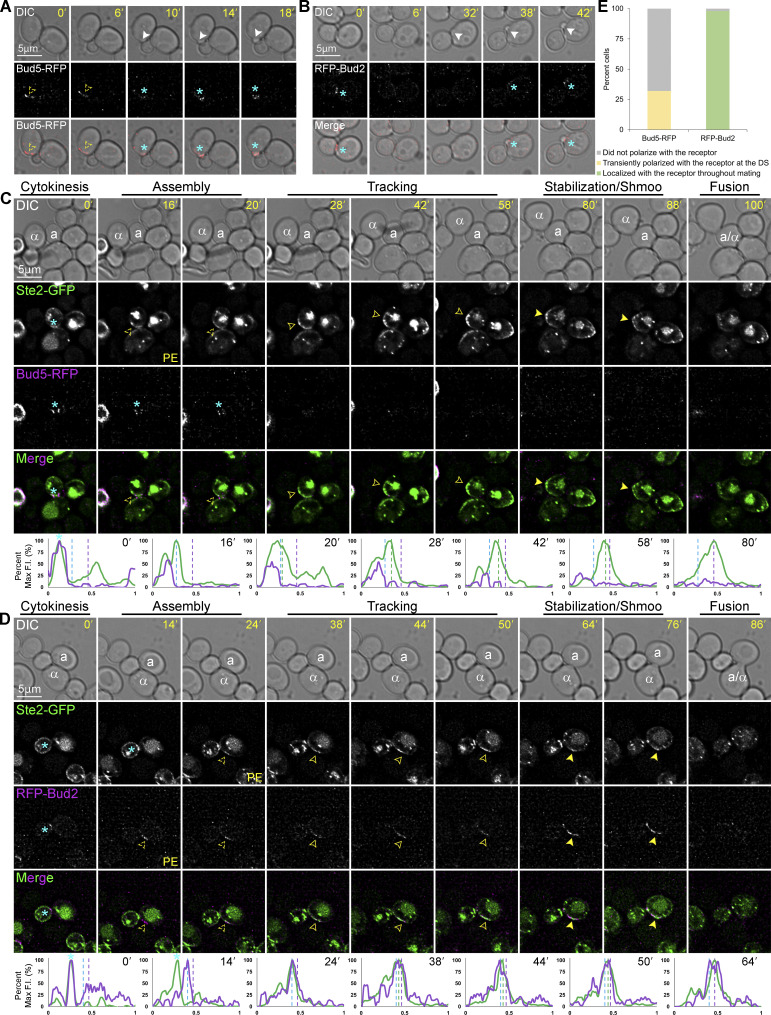
To test this idea, we engineered MATa cells coexpressing the receptor reporter (Ste2-GFP) as a proxy for the GTM, and either RFP-tagged Bud5, the Bud1 GEF (Marston et al., 2001), or RFP-tagged Bud2, the Bud1 GAP (Park et al., 1999). We took time-lapse images of these MATa cells during vegetative growth and in mating mixtures. In vegetative cells, Bud5-RFP localized to the DS after cytokinesis and before bud emergence (Fig. 1 A), whereas RFP-Bud2 concentrated at the bud neck but was not detectable at the PM between cytokinesis and bud emergence (Fig. 1 B). These observations are consistent with those reported by Park et al. (1999) and Marston et al. (2001). Conversely, in mating yeast, Bud5-RFP gradually disappeared from the mother–daughter neck and was never detectable at the DS or elsewhere on the PM (Fig. 1, C and E), whereas RFP-Bud2 polarized to the DS after cytokinesis and before the receptor (Fig. 1, D and E). The polarized RFP-Bud2 and receptor tracked together along the PM to the CS before shmoo formation and fusion. Coexpression of the receptor and Bud5 or Bud2 reporters had no effect on cell orientation (Fig. S1). These data suggest that Bud1 is inactivated in mating cells before tracking begins.
Figure 1.
Localization of Bud5-RFP and RFP-Bud2 in vegetative and mating cells. (A–D) Representative time-lapse images. MATa cells coexpressing in situ-tagged Ste2-GFP and Bud5-RFP or RFP-Bud2 were imaged during vegetative growth or mixed with an equal number of MATα cells and imaged from cytokinesis to fusion. The mating partners are labeled a and α in the DIC images. The blue asterisk indicates reporter localization at the bud neck; PE indicates the first time point that reporter polarity is detectable at the DS; dashed arrowheads mark the signal peaks during the pause from PE to the initiation of tracking, which correlates with GTM assembly; closed arrowheads indicate redistribution (tracking) and mark the leading peak; filled arrowheads indicate stabilization at the CS and mark the signal peak of Ste2-GFP (the receptor). The plots show the distribution of each reporter on the PM at the indicated time points (10-point rolling average). The x-axes represent distances along the PM; the y-axes indicate the percent fluorescence intensity (F.I.) normalized to the maximum peak intensity for each reporter. The green plots show the signal distribution of Ste2-GFP at the indicated time points; the purple plots show the signal distribution of RFP-tagged reporters at the indicated time points; the dashed blue, green, and purple lines mark the DS peak, the leading peak, and the CS peak, respectively. Localization of (A) Bud5-RFP in a vegetative cell; (B) RFP-Bud2 in a vegetative cell; (C) Ste2-GFP and Bud5-RFP in a mating cell; (D) Ste2-GFP and RFP-Bud2 in a mating cell. (E) Bar graphs showing the proportion of mating cells in which the indicated RFP-labeled reporters colocalized with Ste2-GFP. Cells coexpressing in situ-tagged Ste2-GFP and Bud5-RFP (C) or RFP-Bud2 (D) that completed cytokinesis and formed zygotes were categorized as follows: the RFP-tagged reporter colocalized with Ste2-GFP from GTM assembly at the DS to cell fusion; the RFP-tagged reporter transiently colocalized with Ste2-GFP at the DS for up to two time points (10 min); the RFP-tagged reporter did not colocalize with Ste2-GFP. n = 66 for Ste2-GFP Bud5-RFP cells and 47 for Ste2-GFP RFP-Bud2 cells from two independent experiments.
Figure S1.
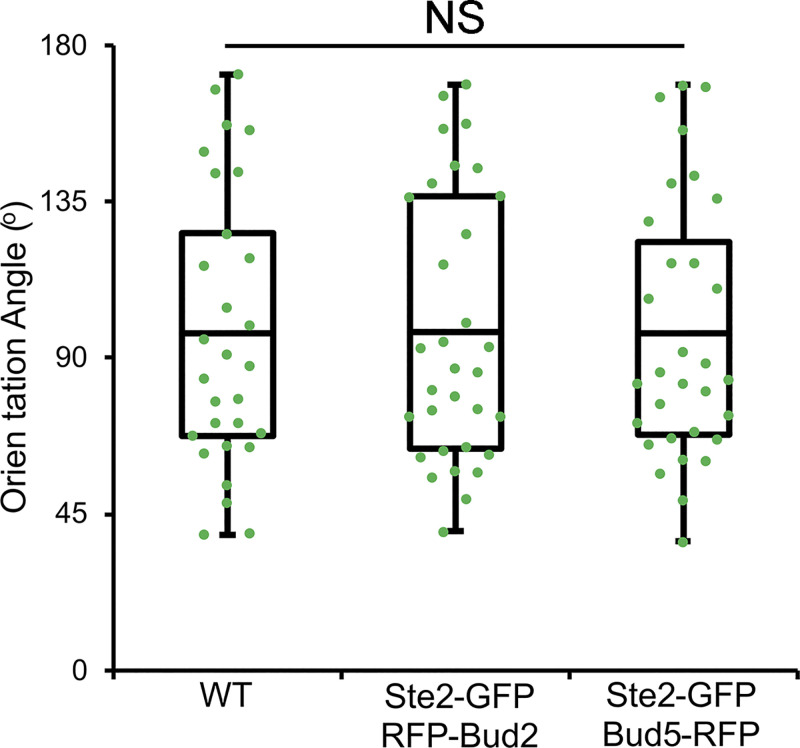
Coexpression of Ste2-GFP with RFP-Bud2 or Bud5-RFP has no effect on gradient tracking. MATa cells coexpressing in situ-tagged Ste2-GFP and Bud5-RFP or RFP-Bud2 were mixed with an equal number of MATα cells and imaged from cytokinesis to fusion. Orientation angle is defined by lines drawn from the center of the cell to the centers of the cytokinesis and fusion sites (Abdul-Ganiyu et al., 2021; Ismael et al., 2016). The scatter plots show the orientation angles measured in the indicated strains. The boxes enclose the middle two quartiles with the horizontal lines indicating the means; the whiskers show the top and bottom quartiles. Mean orientation angle ± SEM in degrees: WT = 97.2 ± 7.4 (n = 29); Ste2-GFP RFP-Bud2 = 97.4 ± 6.8 (n = 32); Ste2-GFP Bud5-RFP = 97.0 ± 6.8 (n = 31); P ≥ 0.97.
Tracking is defective in BUD1G12V/BUD1 cells and more rapidly initiated in bud1∆ cells
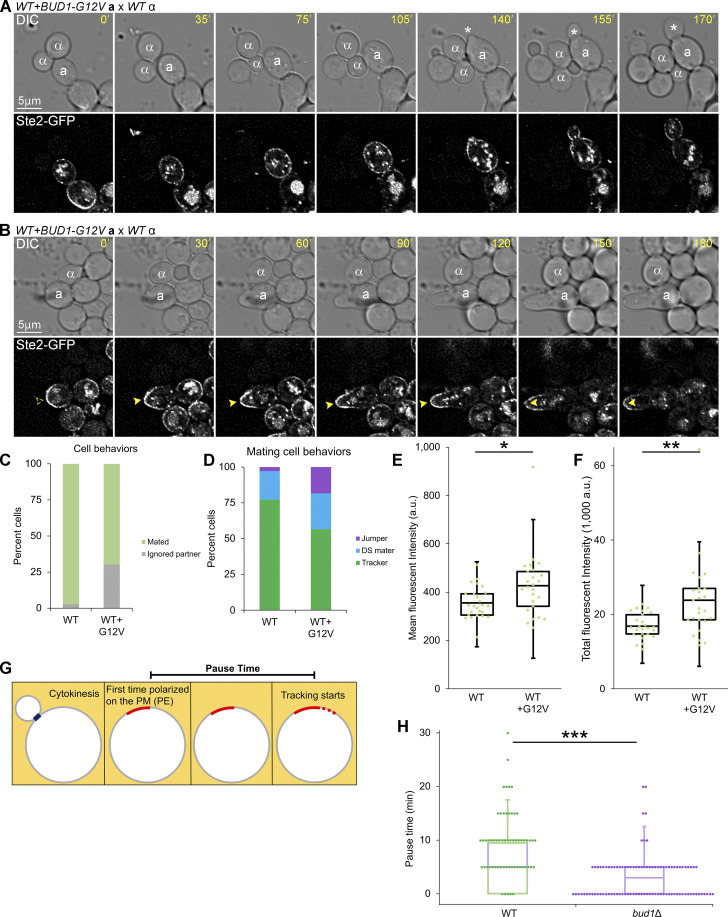
The absence of the Bud1 GEF (Bud5) from the DS along with the localization of the Bud1 GAP (Bud2) to the GTM during both assembly at the DS and redistribution suggested to us that Bud1 inactivation is required for gradient tracking. To test this, we imaged the receptor reporter in mating MATa BUD1 cells expressing a constitutively active form of Bud1, Bud1G12V, from a centromeric plasmid (hereafter, BUD1G12V/BUD1 cells; Fig. 2, A–D; Michelitch and Chant, 1996). The G12V amino acid substitution blocks the GTPase activity of Bud1, thereby locking it in the active state. Cells forced to express Bud1G12V in the absence of Bud1 are not viable. Consistent with our hypothesis, about 30% of the BUD1G12V/BUD1 cells ignored MATα cells with which they were in direct contact (Fig. 2 C), a behavior we see one-tenth as often in wild-type (WT) mating mixtures. These BUD1G12V/BUD1 cells either showed a prolonged G1 arrest before ultimately budding (Fig. 2 A), or they polarized the receptor and shmooed at the DS without regard to the positions of their potential partners (Fig. 2 B). Of the BUD1G12V/BUD1 cells that successfully formed zygotes, significantly fewer exhibited gradient tracking behavior as compared to control cells. They either fused with partners positioned near their DS or at the presumptive distal bud site (Fig. 2 D; Cullen and Sprague, 2012; Vasen et al., 2020). In the distal-mating class of cells, the receptor polarized directly to the fusion site rather than tracking from the DS toward the mating partner—a phenotype we call jumping (Abdul-Ganiyu et al., 2021). Considering both the cells that ignored potential partners and those that formed zygotes, about 40% of the BUD1G12V/BUD1 cells exhibit Ste2-GFP tracking as compared to 75% of the WT cells (Fig. 2, C and D). Measurement of Ste2-GFP PM signals demonstrated that the failure of BUD1G12V/BUD1 cells to track is not attributable to a defect in their ability to accumulate the receptor at the DS. In fact, the mean and total Ste2-GFP intensities were significantly higher in BUD1G12V/BUD1 cells one time point before shmooing at the DS than in WT cells one time point before tracking from the DS (Fig. 2, E and F). Given that WT Bud1 partially rescues DS function in cells coexpressing Bud1G12V, as evidenced by the viability of BUD1G12V/BUD1 cells, these data support our hypothesis that Bud1 must be inactivated to allow GTM tracking. To further test this conclusion, we evaluated the effect of bud1∆ on the pause time, which is defined as the delay between the initial polarization of the receptor at a GTM assembly site and the initiation of tracking (Fig. 2 G). We have previously shown that cells lacking Bud1 assemble functional GTMs, although they do so at multiple, random positions rather than at the DS (Wang et al., 2019). Notably, the mean pause time was significantly shorter in bud1∆ cells as compared with WT control cells (Fig. 2 H), consistent with the idea that Bud1 must be inactivated to enable tracking.
Figure 2.
Effects of GTP-locked and deletion alleles of BUD1 on gradient tracking. (A and B) MATa BUD1 cells coexpressing in situ-tagged Ste2-GFP and Bud1G12V from a centromeric plasmid were mixed with an equal number of MATα cells and imaged at 5-min intervals. The potential mating partners are labeled as a and α in the DIC images. The dashed arrowheads indicate polarity establishment; filled arrowheads indicate shmoo formation. Representative images showing MATa BUD1G12V/BUD1 cells that ignored potential mating partners with which they were in direct contact. (A) A BUD1G12V/BUD1 cell that arrested and elongated but ultimately resumed budding (white asterisks). (B) A BUD1G12V/BUD1 cell that polarized its receptor and shmooed but failed to orient toward and mate with its potential partner. (C) Bar graphs showing the proportion of WT cells and cells expressing Bud1G12V that ignored a potential mating partner. n = 171 for WT and 109 for WT + Bud1G12V from two independent experiments; P < 0.0001. Of the BUD1G12V/BUD1 cells that ignored partners, two-thirds behaved as shown in A and one-third behaved as shown in B. (D) MATa BUD1 cells expressing Bud1G12V are less likely than WT control cells to exhibit gradient-tracking behavior. MATa cells from B that completed cytokinesis and formed zygotes were scored as having mated at the default polarity site (DS maters), tracked to a chemotropic site (trackers), or localized directly to the fusion site (jumpers) based on the spatiotemporal dynamics of the Ste2-GFP reporter in time-lapse fluorescent images. n = 100 for WT cells and 76 for WT + Bud1G12V cells. P < 0.0001 for percent tracking and percent jumping; P = 0.21 (not significant) for percent default mating. (E and F) Box-scatter plots showing Ste2-GFP fluorescence intensity in arbitrary units (a.u.) at the end of GTM assembly. (E) The mean intensity of 30 pixels surrounding the peak signal ± SEM: WT = 355 ± 13 and WT + Bud1G12V = 426 ± 26. (F) Total intensity within the GTM ± SEM: 16,902 ± 667 and WT + Bud1G12V = 23,843 ± 2,114; n = 25 for both. *P < 0.02, **P = 0.003. (G) Cartoon illustrating the measurement of pause time. As described in Wang et al. (2019), pause time is the interval between PE and the initiation of tracking. (H) PE is the first time point that Ste2-GFP polarity is detected at the DS in WT cells and at random PM sites in bud1∆ cells. Box-scatter plots showing the distribution of receptor pause times in the indicated strains. Mean pause time ± SEM in minutes: WT = 9.4 ± 0.8; bud1Δ = 3.0 ± 0.4; n = 60 for WT and 100 for bud1Δ; ***P < 0.0001.
The receptor polarizes to the DS but fails to track in bem1ΔCPX cells
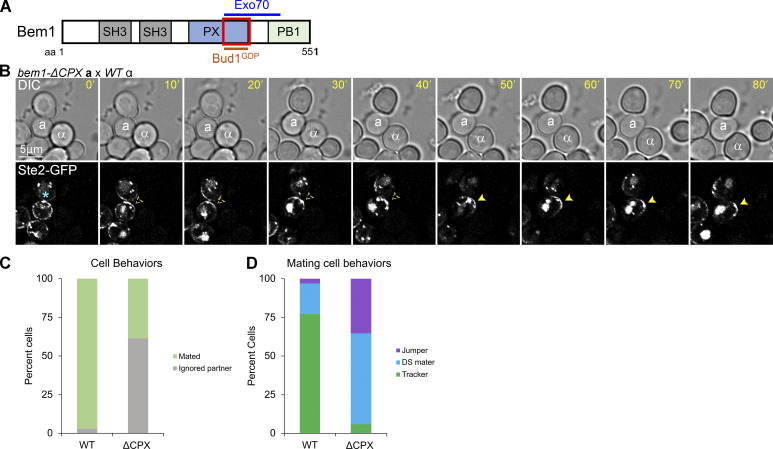
The polarization of RFP-Bud2 to the DS before the receptor in mating cells suggested that Bud1 is inactivated early during GTM assembly. Miller et al. (2019) demonstrated that inactive Bud1 (hereafter, Bud1GDP) directly interacts with the C-terminal half of the Bem1 Phox homology domain (hereafter the CPX domain; Fig. 3 A). In mating cells, the earliest event in GTM assembly detected thus far is the localization of Far1 to the DS, presumably in a complex with Cdc24 (Wang et al., 2019). We postulated that Far1–Cdc24 recruitment to the assembly site depends on the reported interaction of Cdc24 with Bem1 (Butty et al., 1998; Nern and Arkowitz, 1998; Nern and Arkowitz, 1999), and further, that Bud1GDP initiates GTM assembly by recruiting Bem1 to the DS. To test this conjecture, we took time-lapse images of MATa cells expressing Bem1ΔCPX and the receptor reporter in mating mixtures. In the absence of Bud1GDP–Bem1 interaction, we expected a failure to recruit Far1–Cdc24, and therefore, no receptor polarization to the DS. Surprisingly, the receptor polarized to the DS in bem1ΔCPX cells just as well as in the WT cells (Fig. 3 B). However, bem1ΔCPX cells showed almost no evidence of gradient-sensing. Their polarized receptor crescents rarely tracked toward potential mating partners, and they nearly always shmooed and mated at the DS. Considering both the cells that ignored potential partners and those that formed zygotes, about 2% of the bem1ΔCPX mutants exhibited Ste2-GFP tracking (Fig. 3, C and D). These observations suggest that the interaction between inactive Bud1 and Bem1 is not required for GTM assembly at the DS, but that the Bem1 CPX domain is required for tracking from the DS.
Figure 3.
Localization of the receptor in mating Bem1ΔCPX cells. (A) Diagram showing the Bem1 domains that interact with Exo70 and inactive Bud1. The red box indicates the region deleted in bem1ΔCPX. (B) MATa bem1ΔCPX cells expressing in situ-tagged Ste2-GFP were mixed with an equal number of MATα cells and imaged at 5-min intervals. Representative images showing a MATa bem1ΔCPX cell that ignored a potential partner with which it was in direct contact. The potential mating partners are labeled as a and α in the DIC images. The blue asterisk indicates receptor localization at the bud neck; dashed arrowheads indicate polarity establishment of the receptor at the DS; filled arrowheads indicate stabilization of the receptor and shmoo formation. (C) Bar graph showing the proportion of WT and bem1ΔCPX cells that ignored a potential mating partner with which they were in direct contact. n = 171 for WT and 160 for bem1ΔCPX from two independent experiments; P < 0.0001. (D) MATa bem1ΔCPX cells are less likely than WT control cells to exhibit gradient-tracking behavior. MATa cells from B that completed cytokinesis and formed zygotes were scored as having mated at the default polarity site (DS maters), tracked to a chemotropic site (trackers), or localized directly to the fusion site (jumpers) based on the spatiotemporal dynamics of the Ste2-GFP reporter in time-lapse fluorescent images. n = 100 for WT and 51 for bem1ΔCPX; P < 0.0001 for percent tracking.
Receptor tracking is defective in exo70 and sec3 mutants compromised in actin-independent exocyst localization
We have shown that the receptor tracks normally from randomly positioned GTM assembly sites in bud1Δ cells (Wang et al., 2019). Therefore, the inability of the receptor to track in bem1ΔCPX cells must be because the CPX domain of Bem1 provides a critical tracking function independent of Bud1, and not because tracking depends on the interaction of Bud1GDP with Bem1. We have also shown that the initiation of tracking correlates with the concentration of Sec3, a component of the vesicle-tethering exocyst complex, on the upgradient side of the GTM (Wang et al., 2019). Notably, Bem1 has been reported to direct vesicle delivery independent of F-actin through the interaction of its Phox homology domain with another component of the exocyst, Exo70 (Liu and Novick, 2014; Fig. 3 A). Exo70 and Sec3 are unique among the eight exocyst components in their direct recruitment to the PM independent of F-actin (Boyd et al., 2004; Guo et al., 2001; Hutagalung et al., 2009; Liu et al., 2018) and are partially redundant for positioning the exocyst complex at discrete locations on the PM (Boyd et al., 2004; Liu et al., 2018). Because truncating the C-terminal half of the Bem1 Phox homology domain likely abolishes the Bem1-Exo70 interaction (Liu and Novick, 2014), we hypothesized that the inability of bem1ΔCPX cells to track is due to the loss of AI-VD.
The C-domain of Exo70 is essential for Bem1–Exo70 interaction (Fig. 4 A), and its deletion (denoted ∆dC) blocks AI-VD of Exo70 without affecting its AD-VD (Hutagalung et al., 2009; Liu and Novick, 2014). To determine whether the failure of the GTM to track in bem1ΔCPX cells might be due to the loss of Bem1–Exo70 directed AI-VD, we took time-lapse images of MATa exo70ΔdC cells expressing Ste2-GFP in mating mixtures (Fig. 4 B). Consistent with our hypothesis, about 40% of the exo70ΔdC cells ignored MATα cells with which they were in direct contact, a behavior we see ten times less often in WT mating mixtures (Fig. 4, B and F). In most of these cells, the receptor polarized to the DS but did not track toward proximal partners; consequently, such cells shmooed at the DS (Fig. 4 B). Of the exo70ΔdC cells that successfully formed zygotes, a significantly larger fraction mated at their DS or at the presumptive distal bud site as compared with the control cells, while a significantly smaller fraction exhibited gradient tracking and chemotropic mating (Fig. 4 G). Considering both the cells that ignored potential partners and those that formed zygotes, about 11% of the exo70ΔdC mutants exhibit Ste2-GFP tracking as compared with 75% of the WT cells (Fig. 4, F and G). These data support the idea that AI-VD is required for tracking. However, given the deletion of the entire 169-residue C-domain, it is possible that the severe tracking defect seen in exo70ΔdC cells reflects diminished interaction between Exo70ΔdC and Rho3 (Hutagalung et al., 2009), Cdc42 (Liu and Novick, 2014), Sec6 (Dong et al., 2005), PIP2 (Zhang et al., 2008), or unknown Exo70 binding partners.
Figure 4.
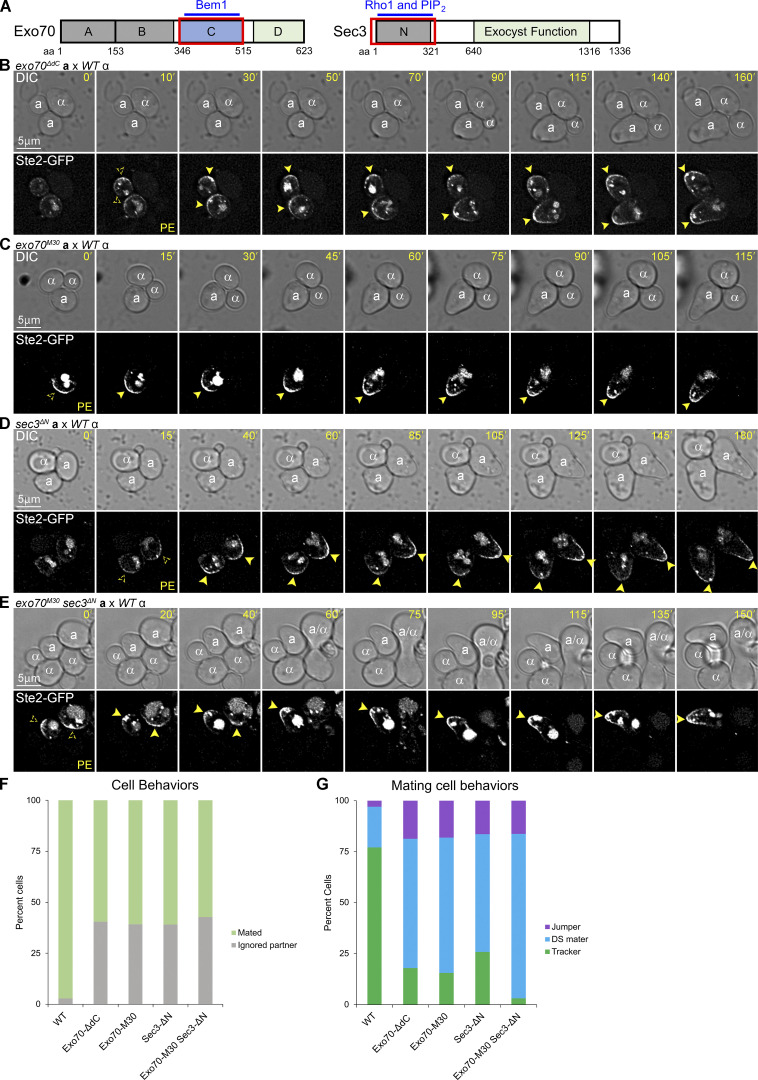
Localization of Ste2-GFP in exo70ΔdC, exo70M30, sec3ΔN, and exo70M30sec3ΔN cells in mating mixtures. (A) Diagrams showing the domains of Exo70 and Sec3 that interact with the indicated proteins and lipids. The red boxes indicate the deleted regions. (B–E) Time-lapse images of mating mixtures. MATa exo70ΔdC, MATa exo70M30, MATa sec3ΔN, and MATa exo70M30sec3ΔN cells expressing in situ-tagged Ste2-GFP were mixed with an equal number of MATα cells and imaged at 5-min intervals. The potential mating partners are labeled as a and α in the DIC images; the dashed arrowheads indicate receptor polarity establishment (PE); filled arrowheads indicate stabilization of the receptor and shmoo formation. Representative images show MATa cells of the indicated genotypes in which the receptor polarized but failed to track to the adjacent MATα cell (B–E), or which polarized the receptor and mated at the DS (E). (F) Bar graph showing the proportion of WT, exo70ΔdC, exo70M30, sec3ΔN, and exo70M30sec3ΔN cells that ignored a potential mating partner with which they were in direct contact. n = 171 for WT, 242 for exo70ΔdC, 156 for sec3ΔN, and 264 for exo70M30sec3ΔN from two independent experiments; P < 0.0001 for each mutant compared to WT. (G) MATa exo70ΔdC, exo70M30, sec3ΔN, and exo70M30sec3ΔN cells are less likely than WT control cells to exhibit gradient-tracking behavior. MATa cells from F that completed cytokinesis and formed zygotes were scored as having mated at the default polarity site (DS maters), tracked to a chemotropic site (trackers), or localized directly to the fusion site (jumpers) based on the spatiotemporal dynamics of the Ste2-GFP reporter in time-lapse fluorescent images. n = 100 for WT, 123 for exo70ΔdC, 110 for exo70M30, 85 for sec3ΔN, and 98 for exo70M30sec3ΔN; P < 0.0001 for percent tracking when comparing WT with each of the mutant strains and when comparing the exo70M30sec3ΔN double mutant with each of the single mutant strains.
To minimize the possibility of pleiotropic effects, we took advantage of the well-characterized M30 allele of exo70, which encodes six charge-change amino acid substitutions (K/R to A) in the C-domain (Liu and Novick, 2014). The Exo70M30 mutant protein shows only residual binding to Bem1 in vitro, whereas its interactions with Rho3, Cdc42, PIP2, and other subunits of the exocyst are unaffected (Liu and Novick, 2014); like Exo70ΔdC, Exo70M30 cannot localize to the PM independent of F-actin (Liu and Novick, 2014). In mating mixtures, the effects of exo70M30 and exo70ΔdC were indistinguishable. Both alleles conferred a severe defect in gradient tracking without affecting mating at the default sites (Fig. 4, B, C, F, and G). Considering both the cells that ignored potential partners and those that formed zygotes, about 11% of the exo70M30 mutants exhibit Ste2-GFP tracking (Fig. 4, F and G).
Although disruption of Bem1–Exo70 binding dramatically reduced tracking, it did not eliminate it. A priori, the residual tracking ability of exo70ΔdC and exo70M30 cells could require AD-VD or it could depend on the partial maintenance of AI-VD by the functional redundancy of Sec3 with the C-domain of Exo70 (Guo et al., 2001; Hutagalung et al., 2009). To distinguish between these possibilities, we took advantage of the well-characterized sec3∆N allele. Without its N-terminus (residues 1–320), Sec3 does not bind Cdc42 (Guo et al., 2001; Zhang et al., 2001) or PIP2 (Zhang et al., 2008); it localizes to the PM by AD-VD but not by AI-VD (Zhang et al., 2008). In mating mixtures, sec3ΔN cells showed a tracking defect similar to that conferred by exo70ΔdC and exo70M30. About 16% of the sec3ΔN mutants exhibit Ste2-GFP tracking (Fig. 4, D, F and G). In contrast, Ste2-GFP tracking is observed in only about 2% of exo70M30 sec3ΔN double mutant cells (Fig. 4, E–G)—sixfold and ninefold less than in the exo70M30 and sec3ΔN single mutants, respectively. Since neither Exo70M30 nor Sec3ΔN can localize to the PM independent of F-actin, and because AD-VD is unaffected in exo70M30 sec3ΔN cells (which bud and shmoo normally), these data strongly suggest that AI-VD is necessary and sufficient for gradient tracking.
AD-VD markers do not track with the receptor but polarize strongly at the eventual chemotropic site in mating cells
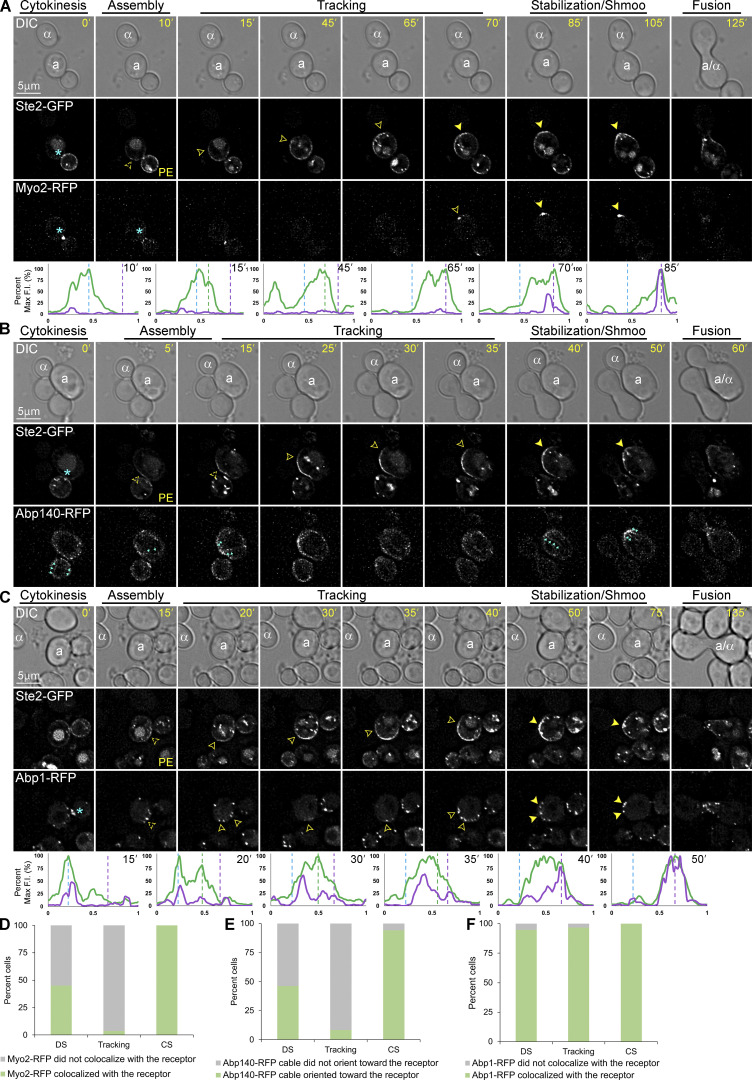
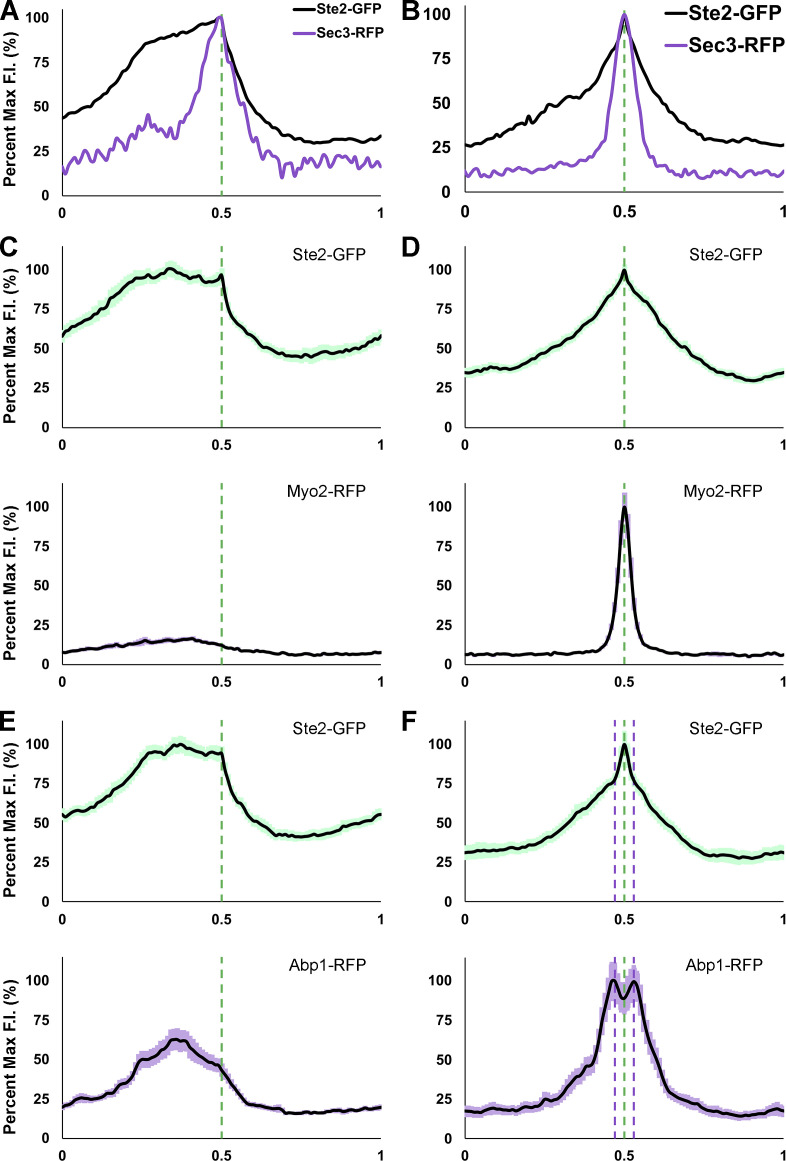
It has been proposed that the movement of the polarity complex along the cell cortex during yeast gradient sensing is driven by vesicles delivered to the PM on actin filaments (Dyer et al., 2013; McClure et al., 2015). Although AD-VD appears unable to compensate for the lack of AI-VD during gradient tracking, vesicle delivery via actin cables could contribute to the process. To determine whether AD-VD is associated with gradient tracking in mating cells, we engineered MATa cells coexpressing the receptor reporter and RFP-tagged Myo2, a marker for vesicles delivered to the PM on actin cables (Johnston et al., 1991); RFP-tagged Abp140, a marker for actin cables (Asakura et al., 1998); or RFP-tagged Abp1, a marker for actin patches (Drubin et al., 1988). We took time-lapse images of these MATa cells from cytokinesis to fusion as they mated with MATα cells (Fig. 5). In Wang et al. (2019), we demonstrated that Sec3-RFP robustly polarizes to the DS with the receptor, tracks on the leading side of the receptor crescent (Fig. 6 A), and becomes highly concentrated at the eventual CS (Fig. 6 B). Unlike Sec3-RFP, Myo2-RFP polarized to the DS in only 45% of the cells examined (n = 51; Fig. 5 A) and tracked with the leading receptor peak in only 4% of the cells examined (n = 56; Figs. 5, A and D; and 6 C). Like Sec3-RFP, Myo2-RFP invariably polarized sharply at the CS (n = 70; Figs. 5, A and D; and 6 D). Similarly, we detected actin cables marked by Abp140-RFP oriented toward the receptor at the DS during assembly (46%; n = 52) and at the CS after stabilization (94%; n = 87), but seldom during tracking (8%; n = 37; Fig. 5, B and E). On the contrary, Abp1-RFP polarized to the DS (95%; n = 58), tracked behind the receptor (96%; n = 59), and polarized as peaks surrounding the eventual CS (100%; n = 84, Figs. 5, C and F; and 6, E and F). This is consistent with our conclusion, based on time-lapse imaging of Sla1-RFP, that receptor-driven endocytosis is maximal behind the peak of the tracking receptor and surrounds the eventual CS (Wang et al., 2019). Together, these observations suggest that AD-VD does not contribute to GTM tracking, but is operative during GTM assembly at the DS and stabilization at the eventual CS.
Figure 5.
Localization of Myo2-RFP, Abp140-RFP, and Abp1-RFP in mating cells. (A–C) Representative time-lapse images. MATa cells coexpressing in situ-tagged Ste2-GFP and Myo2-RFP, Abp140-RFP, or Abp1-RFP were mixed with an equal number of MATα cells and imaged from cytokinesis to fusion. The mating partners are labeled a and α in the DIC images. The blue asterisk indicates reporter localization at the bud neck; PE indicates the first time point that reporter polarity is detectable at the DS; dashed arrowheads mark the signal peaks during the pause from PE to the initiation of tracking, which correlates with GTM assembly; closed arrowheads indicate redistribution and mark the leading peak; filled arrowheads indicate stabilization at the CS and mark the signal peak of Ste2-GFP; the cyan arrow heads mark the actin cables. The plots show the distribution of each reporter on the PM at the indicated time points (10-point rolling average). The x-axes represent distances along the PM; the y-axes indicate the percent fluorescence intensity (F.I.) normalized as described in the Materials and methods. The green plots show the signal distribution of Ste2-GFP at the indicated time points; the purple plots show the signal distribution of RFP-tagged reporters at the indicated time points; the dashed blue, green, and purple lines mark the DS peak, the leading peak, and the CS peak, respectively. Localization of (A) Ste2-GFP and Myo2-RFP in a mating cell; (B) Ste2-GFP and Abp140-RFP in a mating cell; (C) Ste2-GFP and Abp1-RFP in a mating cell. (D–F) Bar graphs showing the proportion of mating cells in which the indicated AD-VD reporters colocalized with (Myo2-RFP and Abp1-RFP) or oriented towards (Abp140-RFP) the Ste2-GFP crescent at the DS, while tracking, and at the CS. (D) Ste2-GFP Myo2-RFP cells. n = 51 for DS, 56 for tracking, and 70 for CS from two independent experiments. (E) Ste2-GFP Abp140-RFP cells. n = 52 for DS, 36 for tracking, and 87 for CS from two independent experiments. (F) Ste2-GFP Abp1-RFP, n = 58 for DS, 59 for tracking, and 84 for CS from two independent experiments.
Figure 6.
Average PM distribution of Myo2-RFP and Abp1-RFP relative to Ste2-GFP in mating cells during tracking and in shmoos. (A and B) Illustration of Sec3-RFP distribution relative to receptor distribution during tracking (A) and in shmoos (B; Wang et al., 2019). (C–F) The PM signals of 30 cells two time points before stabilization (tracking) and two time points before fusion (shmooing) were quantified with ImageJ, normalized for cell size, and averaged as described in Materials and methods. The plots show the mean signal distribution ± SEM (light shadow) of Ste2-GFP (green) and Myo2-RFP (purple) or Abp1-RFP (purple). The dashed green lines mark the leading receptor peak and the shmoo tip during tracking and in shmoos; the dashed purple lines mark the surrounding peaks of Abp1-RFP in shmoos. Average distributions of (C) Ste2-GFP and Myo2-RFP during tracking; (D) Ste2-GFP and Myo2-RFP in shmoos; (E) Ste2-GFP and Abp1-RFP during tracking; (F) Ste2-GFP and Abp1-RFP in shmoos.
The receptor tracks in myo2-16 cells mated with WT MATα cells at the restrictive temperature
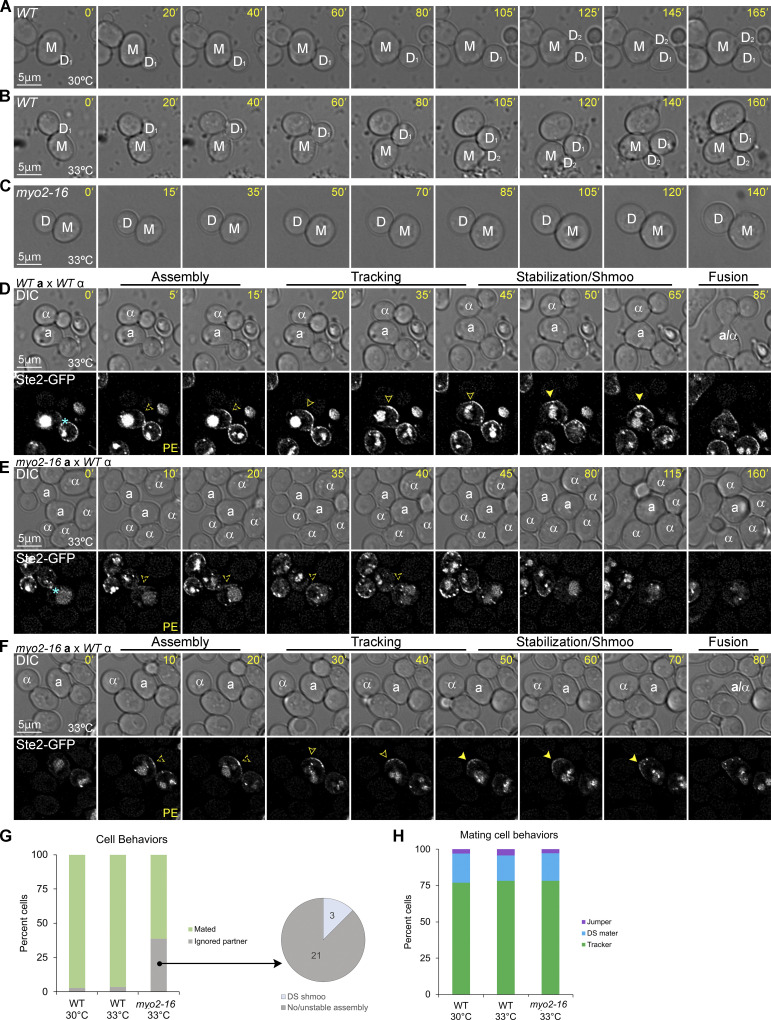
As a second way of asking whether AD-VD is required for gradient tracking, we engineered MATa cells expressing Ste2-GFP and the temperature-sensitive allele myo2-16 (Schott et al., 1999). Myo2, the type V myosin motor protein in yeast, docks post-Golgi vesicles to actin cables and carries them to sites of actin-dependent polarized secretion on the PM (Johnston et al., 1991). At the restrictive temperature of 33°C, the myo2-16 mutant protein cannot dock post-Golgi vesicles to actin cables but has no effect on actin-cable assembly (Schott et al., 1999). As a result, AD-VD is blocked in the mutant cells. As previously reported for other myo2-16 strains (Schott et al., 1999), the mutant cells were unable to initiate the growth of a new daughter cell (bud) at the restrictive temperature (Fig. 7, A–C). We took time-lapse images of WT MATa cells and myo2-16 MATa cells in mating mixtures at 33°C (Fig. 7, D–F). Although the restrictive temperature had no effect on tracking and mating in WT cells (Fig. 7, D, G, and H), about 40% of the MATa myo2-16 cells ignored MATα cells with which they were in direct contact (Fig. 7, E and G). Unlike the BUD1G12V/BUD1 and the exo70ΔdC mutant cells that ignored potential mating partners due to defects in tracking, most of the partner-ignoring myo2-16 cells exhibited a defect in GTM assembly. They were either unable to polarize the receptor to the DS or the polarized receptor signal became undetectable over time. This phenotype is consistent with our observation that Myo2-RFP polarized to the DS in about 45% of the WT cells, suggesting that AD-VD contributes to GTM assembly. In the cohort of myo2-16 cells that successfully mated (about 60%), the receptor polarized to the DS, tracked along the PM, and stabilized at the CS shortly before cell fusion, phenocopying receptor behavior in WT cells (Fig. 7, F–H). This result demonstrates that blocking AD-VD does not affect tracking.
Figure 7.
Localization of Ste2-GFP in myo2-16 mutant cells mated at the restrictive temperature. (A–C) Representative time-lapse images of vegetative cells at the myo2-16 permissive and restrictive temperatures. Mother and daughter cells are labeled M and D. DIC images of (A) a WT cell at 30°C, (B) a WT cell at 33°C, and (C) a myo2-16 cell at 33°C. (D–F) Representative time-lapse images of cells in mating mixtures at 33°C. MATa MYO2 (WT) and MATa myo2-16 mutant cells expressing Ste2-GFP were mated at the restrictive temperature of 33°C. Potential mating partners are labeled a and α in the DIC images. The blue asterisk indicates receptor localization at the bud neck; PE indicates the first time point that reporter polarity is detectable at the DS; dashed arrowheads mark the signal peaks during the pause from PE to the initiation of tracking, which correlates with GTM assembly; closed arrowheads indicate redistribution and mark the leading peak; filled arrowheads indicate stabilization at the CS and mark the signal peak of Ste2-GFP. Representative images of (D) a mating WT cell, (E) a myo2-16 cell ignoring potential partners in contact with it, and (F) a mating myo2-16 cell. (G) Bar graph showing the proportion of WT and myo2-16 cells that mated with an adjacent MATα cell. n = 171 for WT at 30°C, 56 for WT at 33°C, and 62 for myo2-16 at 33°C from two independent experiments; no significant difference when comparing WT at 30°C with WT at 33°C; P < 0.0001 when comparing myo2-16 to WT at 30 or 33°C. The pie graph shows the behaviors of the myo2-16 cells that ignored potential mating partners (the number of cells in each category is indicated). (H) MATa myo2-16 cells and WT control cells exhibit indistinguishable gradient-tracking behavior. MATa cells from B that completed cytokinesis and formed zygotes were scored as having mated at the default polarity site (DS maters), tracked to a chemotropic site (trackers), or localized directly to the fusion site (jumpers) based on the spatiotemporal dynamics of the Ste2-GFP reporter in time-lapse fluorescent images. n = 100 for WT at 30°C, 46 for WT at 33°C, and 37 for myo2-16 at 33°C; no significant difference when comparing any two of these mating mixtures for percent tracking, percent default mating, or percent jumping.
Discussion
Significant progress has recently been made in understanding how mating yeast cells interpret pheromone gradients and accurately position their chemotropic growth sites. Emerging evidence suggests that yeast are not global gradient sensors able to orient and polarize directly toward their mating partners. Rather, yeast cells become competent to sense pheromone gradients by colocalizing the pheromone-signaling and vesicle-trafficking systems at an intrinsically determined polarity site, the DS (Hegemann et al., 2015; Park et al., 1999; Wang et al., 2019)—assembling what we call the gradient tracking machine. Within the GTM, the pheromone gradient determines the pattern of dimeric receptor species, which segregates anterograde and retrograde vesicle trafficking, resulting in the redistribution of the GTM toward the pheromone source and stabilization at the CS (Abdul-Ganiyu et al., 2021; Wang et al., 2019). At the level of the receptor, G protein, RGS protein Sst2, scaffold protein Far1, and exocyst component Sec3, GTM redistribution appears to be deterministic. The primary pheromone sensing components and pioneer exocytic marker move together—directly and steadily upgradient from the DS to the CS. This model not only provides a framework for understanding how yeast cells decode shallow pheromone gradients but also explains how a weak spatial signal can position the CS despite the strong intrinsic polarity of the DS. Instead of a global gradient-sensing mechanism that competes with the DS, a mobile GTM is assembled at the DS, which then incrementally redistributes toward the mating partner. However, concentrating the key polarity and secretory proteins at the DS presents its own problems as follows: what prevents polarized growth at that site? And how is the GTM released for tracking after its assembly is complete? Here we describe two mechanisms that are essential for tracking. First, the Ras GTPase Bud1 must be inactivated to allow GTM release. Second, actin-independent—but not actin-dependent—vesicle delivery must be targeted upgradient to drive GTM redistribution (Fig. 8).
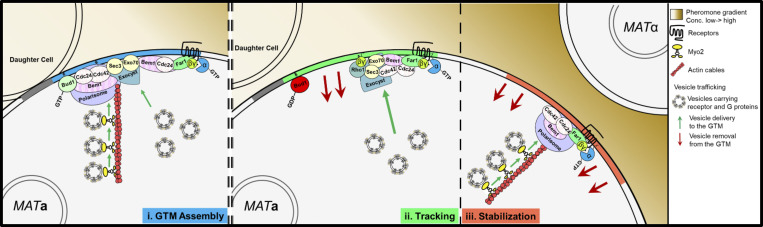
Figure 8.
The mode of vesicle delivery controls GTM mobility. (i) During assembly, vesicles carrying the receptor and G proteins are delivered to the active Bud1-marked DS along and independent of actin cables. (ii) Inactivation of Bud1 and cessation of AD-VD is essential for tracking to start. During tracking, AI-VD is biased upgradient within the GTM, where the active receptor and G protein are enriched. Free Gβγ directs exocyst assembly, and hence vesicle docking, up the gradient independent of actin cables by recruiting Exo70 and Sec3 via its interactions with Far1–(Cdc24–Bem1)–Cdc42 and Rho1. (iii) When the GTM stabilizes at the CS, actin cables are nucleated, and robust vesicle delivery drives polarized growth and cell fusion.
The bud-positioning function of the DS must be inactivated to release the GTM
During the GTM assembly process in cells preparing to mate, we found that the Bud1 GEF became undetectable, whereas the Bud1 GAP polarized to the DS. Subsequently, the Bud1 GAP tracked with the receptor to the CS. These results suggest that Bud1 is inactivated during GTM assembly and that it remains inactive throughout tracking. Is Bud1 inactivation required for tracking to begin? Even when coexpressed with WT Bud1, which is necessary for viability, GTP-locked Bud1 conferred a severe defect in tracking, where Ste2-GFP tracking was seen in about half as many BUD1G12V/BUD1 cells as in WT cells (P < 0.0001). Taken together with our previous finding that bud1Δ cells assemble multiple GTMs at random positions on the PM—a maladaptive phenotype that occasionally results in bud1Δ cells fusing with multiple partners and forming heterokaryons rather than zygotes (Wang et al., 2019)—we conclude that Bud1 plays an important albeit transitory role in yeast mating. It is initially required to promote the assembly of a single GTM at a specific point in the cell cycle (late G1) and at a specific cortical position (the DS). It must then be inactivated and remain inactive to permit tracking. In this view, the Bud1 GAP travels with the GTM to ensure that Bud1 stays off. Without this protection, stochastic activation of Bud1 could trigger positive feedback amplification of the Cdc42–Cdc24–Bem1 loop, leading to local nucleation of actin cables (Chant and Herskowitz, 1991; Drubin and Nelson, 1996; Irazoqui et al., 2003; Kang et al., 2010; Zheng et al., 1995). This would likely cause the GTM to stall before aligning with the gradient source, as we observed in BUD1G12V/BUD1 cells. Although it is formally possible that Bud1-GDP localizes with the GTM and contributes to its function, we observed no tracking defect in bud1∆ cells20.
Actin-independent vesicle delivery is necessary and sufficient for GTM tracking
The exocyst pioneer proteins, Sec3 and Exo70, localize to PM secretion sites by both vesicular transport on actin cables (AD-VD) and by direct interaction with protein and lipid partners independent of actin cables (AI-VD). Based on the localization of Sec3, we previously concluded that vesicle delivery is involved in all three phases of gradient sensing after global internalization (Wang et al., 2019). We showed that Sec3-RFP polarizes to the DS during GTM assembly, concentrates to the upgradient side of the GTM during tracking, and sharply peaks in the center of the GTM after stabilization. The observations reported here suggest a relationship between the mode of vesicle delivery and the phase of gradient sensing. Vesicles are delivered to the PM by both actin-cable-dependent and actin-cable-independent mechanisms during GTM assembly and stabilization, but only by actin-cable-independent mechanisms when the GTM is tracking. Our conclusion that AI-VD is both necessary and sufficient to drive tracking is based on the following observations. From other studies, we know that actin-independent recruitment of Exo70 and Sec3 to the PM (and hence, assembly of the exocyst) requires direct interaction with Bem1 and Cdc42, respectively (Boyd et al., 2004; Guo et al., 2001; Hutagalung et al., 2009; Liu et al., 2018; Liu and Novick, 2014; Zhang et al., 2001; Zhang et al., 2008). We found that specifically disabling the actin-independent PM recruitment domains of either Exo70 (exo70ΔdC, exo70M30) or Sec3 (sec3∆N) confers a dramatic defect in gradient tracking, whereas preventing the actin-independent localization of both Exo70 and Sec3 (exo70M30 sec3∆N) or preventing Bem1 from binding to Exo70 (bem1ΔCPX) nearly abrogates GTM function. As exo70M30 sec3∆N cells bud and shmoo normally—processes that require AD-VD—their failure to track must be due to the loss of AI-VD. The findings that the Myo2 motor protein, which transports vesicles to the PM along actin cables, is neither required nor polarized to the secretion site when the GTM is moving upgradient supports our conclusion that AD-VD is not active during the tracking phase of chemotropic mating.
The apparent contradiction between AI-VD-driven GTM movement and AD-VD-driven polarity-complex wandering (Dyer et al., 2013; McClure et al., 2015) can be attributed to differences in the experiments on which these claims are based. Although we studied the redistribution of the pheromone sensing, signaling, and protein trafficking components of the GTM under physiological conditions—i.e., in mating cells—Lew and associates studied the movement of downstream polarity proteins in the absence of gradient stimulation. In one study, they found that when cdc24-m1 mutant cells, in which Gβγ-Far1 cannot interact with Cdc24 (Nern and Arkowitz, 1999), were treated with a low dose of isotropic pheromone, the addition of the actin-depolymerizing drug latrunculin A (LatA) decreased the mobility of Bem1 and the formin regulator, Spa2, which are both markers for the polarity complex (Dyer et al., 2013). They also showed that conditional inactivation of the myo2-16 mutant protein in cdc24-m1 cells treated with a high dose of isotropic pheromone reduced the mobility of Spa2 by about half (Dyer et al., 2013). In a second study, the Lew group reported that LatA decreased Spa2 wandering in cells genetically engineered to induce the pheromone response pathway in the absence of pheromone (McClure et al., 2015). Although it is clear from these data that AD-VD contributes to polarity-complex mobility under the conditions tested, actin filaments associated with wandering polarity markers have not been visualized. Moreover, Peter and colleagues found that latrunculin does not affect the chemotropic movement of a Cdc24 reporter in cells stimulated with an artificial gradient and that the mobility of the receptor and Exo70 is actin-independent in cells treated with a low dose of isotropic pheromone (Hegemann et al., 2015). A point of agreement is that vesicle delivery is the engine that drives the redistribution of the receptor and G protein as well as the wandering of the polarity complex.
What directs AI-VD upgradient to the CS in mating cells?
According to the model we proposed in Wang et al. (2019) and Abdul-Ganiyu et al. (2021), a greater proportion of receptors are activated and protected from endocytosis on the upgradient side of the tracking GTM; consequently, this region is enriched in activated Gα and free Gβγ. As free Gβγ interacts directly with Far1–(Cdc24–Bem1)–Cdc42 (Butty et al., 1998; Nern and Arkowitz, 1998; Nern and Arkowitz, 1999) and associates with active Rho1 (Bar et al., 2003), it is a likely candidate to direct the actin-independent localization of Exo70 (via Bem1) and Sec3 (via Cdc42 and possibly Rho1). Moreover, we have recently shown that phosphorylated Gβ concentrates on the leading side of the GTM and acts as a directional cue (Abdul-Ganiyu et al., 2021). Therefore, we propose that Gβγ recruits Exo70 and Sec3 upgradient within the GTM through their mutually interacting partners, and thereby biases AI-VD toward the gradient source (Fig. 8). This adds an important feature to our understanding of deterministic gradient tracking. Free Gβγ, which rapidly reports receptor activity, guides the GTM by moving the exocyst. It will be interesting to determine whether the pheromone-induced phosphorylation of Gβ increases the affinity of Gβγ for Far1 and/or Rho1, as this could underlie additional positive feedback loops.
Switching modes of vesicle delivery: Stability vs. mobility
Why does the mode of vesicle secretion change as mating cells progress from GTM assembly to tracking and from tracking to stabilization? Robust polarized growth such as bud and shmoo formation requires rapid, focused, and stable vesicle delivery along actin cables. In contrast, gradient tracking requires dynamic positioning of the secretory site in response to the pheromone gradient and vesicle delivery at a rate that does not result in polarized growth. AI-VD positioned by a heterotrimeric G protein whose local activity and concentration directly reflect that of the pheromone receptor is likely a faster and more flexible way to effect tracking than a mechanism that depends on the polymerization and depolymerization of actin cables. We propose that the high rate of vesicle delivery along actin cables (Boyd et al., 2004), which is needed to assemble the GTM and polarize growth at the CS, must be turned off to permit gradient tracking; there is a tradeoff between faster vesicle delivery and GTM mobility. Despite distinctions in the underlying mechanisms, the distantly related yeast, Schizosaccharomyces pombe, also modulates the position and mode of vesicle delivery when mating (Bendezu and Martin, 2013). While in the exploratory phase, fission yeast sequentially establish Cdc42 polarity sites at discrete positions around the PM but do not secrete cell wall synthetases or polarize their growth. Once a cell aligns a polarity site with that of a mating partner, it enters the committed phase. A single reinforced Cdc42 zone promotes secretion of the cell wall synthetases and shmooing.
What controls the transition from AD-VD at a fixed site during GTM assembly to AI-VD-powered GTM tracking, and back to AD-VD at a fixed site after GTM stabilization? Our results suggest that Bud1 must be inactivated to enable tracking. As Bud1 positions Far1–(Cdc24–Bem1)–Cdc42 to nucleate actin cables during bud emergence, it likely plays the same role during GTM assembly. As active Bud1 disappears, the Far1–(Cdc24–Bem1)–Cdc42 complex is freed to interact with Gβγ. In this view, Bud1 inactivation is a prerequisite to turning off AD-VD and allowing Gβγ-positioned AI-VD to predominate during the transition from assembly to tracking, although other factors are likely to contribute to this switch. For example, Hegemann et al. (2015) have proposed that Fus3 activity, through its effect on Far1 expression and nuclear export, decreases Cdc42 activity during actin-independent polarity-complex wandering and increases it when wandering stops. These observations are consistent with our findings that the F-actin and Myo2 markers of AD-VD become detectable again only after the GTM stabilizes at the CS, suggesting that actin cables cannot be nucleated if the polarity complex is mobile. In this view, the positional stability of the GTM, along with Fus3-regulated Cdc42 activity (Hegemann et al., 2015), influences where and when actin cables form, thus aligning AD-VD with the pheromone source. As Spa2 tracks from the DS to the CS in mating cells (Waszczak et al., 2019), and because the formin protein, Bni1, moves with the polarity complex in cells stimulated by artificial pheromone gradients (Hegemann et al., 2015), we speculate that the machinery for nucleating actin cables travels with the GTM in “stand by” mode, ready to initiate AD-VD when the CS is reached.
Intrinsic polarity may be integral to the differentiation of many cell types
Depending on environmental conditions, haploid yeast cells choose one of three distinct fates late in the G1 phase of the cell cycle. When well nourished, they commit to mitosis and begin to polarize the growth of a daughter cell or bud at the DS, concomitant with the initiation of S phase; in mating mixtures, they assemble the GTM at the DS preparatory to locating and fusing with a proximal partner; when starved, they form long, chained projections called pseudohyphae, likely at the DS, which are thought to be used to forage for nutrients (Gimeno et al., 1992). In addition to being determined at a unique point in the cell cycle (late G1), we infer that yeast cell fate is regulated at a unique cortical site (the DS). The importance of DS regulation during budding is well documented (Bi and Park, 2012). The work we have presented here, and previously (Wang et al., 2019), demonstrates the essential role of DS regulation during mating. Other studies have shown that yeast cells cannot initiate pseudohyphal growth in the absence of Bud1, Bud2, or Bud5, suggesting that DS regulation is essential for cellular differentiation in response to starvation (Roberts and Fink, 1994). Thus, in S. cerevisiae, the specific structure generated at the intrinsically determined polarity site depends on the environmental input.
Like S. cerevisiae, most cell types in higher eukaryotes begin to differentiate when their progenitor cells complete a division cycle, and like the DS in yeast, polarity sites on the PM are associated with cell division (Motegi et al., 2020). Moreover, the basic components and systems required to generate cell polarity and direct vesicle secretion are highly conserved across the eukaryota. Historically, cells were thought to polarize in response to environmental cues as they differentiated (Motegi et al., 2020). However, recent evidence in many models suggests that the establishment of intrinsic polarity—independent of environmental stimuli—precedes and is required for proper differentiation (Motegi et al., 2020; Naganathan et al., 2018; Taniguchi et al., 2017; Zenker et al., 2018; Zhu and Zernicka-Goetz, 2020). For example, actomyosin flows generate mechanical constraints that result in the establishment of polarity, which subsequently determines cell fate in M. musculus and C. elegans (Naganathan et al., 2018; Zhu and Zernicka-Goetz, 2020); during inner–outer lineage differentiation at the eight-cell stage of mice embryonic development, F-actin and polarity-related proteins, such as PKC, PARs, and Ezrin, are gradually translocated from the division plane to the apical cortex of the outer cells before fate determination (Zenker et al., 2018); and, human pluripotent stem cells autonomously develop polarity before differentiating (Taniguchi et al., 2017). It remains to be seen whether these and other differentiating cell types use division-marked polarity site(s) to assemble protein complexes that enable them to respond to environmental cues. If so, it will be interesting to determine whether these protein complexes relocate to environmentally determined positions, how the pre-existing polarity sites are regulated, and whether different modes of vesicle delivery are involved in these processes.
Materials and methods
Molecular and microbiological techniques
Standard methods were used for microbial culture and molecular manipulation, as described previously (Ausubel et al., 1994; Guthrie and Fink, 2002; Sherman et al, 1986).
Yeast strain construction
The yeast strains used in this study are listed in Table S1. They were derived by the transformation of strain 15Dau (MATa ade1 his2 leu2-3,-112 trp1 ura3Δ), which is congenic with strain BF264-15D (Reed et al., 1985). In situ-tagged strains XWY143, XWY145, XWY164, XWY169, XWY176, XWY180, XWY184, XWY188, XWY192, and CPY136 were generated by integrating Bsu36I-cut XWB121, XbaI-cut XWB123, PstI-cut XWB143, BlpI-cut XWB148, SalI-cut XWB156, SalI-cut XWB162, StuI-cut XWB163, BlpI-cut XWB182, SacI-cut XWB183, and BsmI-cut CPB032 into strain XWY065, respectively. Strain XWY185 was generated by transforming XWB173 into XWY065. Strain CPY137 was generated by integrating BsmI-cut CPB032 into XWY192. All genomic modifications were confirmed by sequencing (UIC Research Resource Center Sequencing Core).
Plasmid construction
The plasmids used in this study are listed in Table S2. All genes were directly amplified from DSY129 genomic DNA except for the gene encoding the RFP protein, which was amplified from DSB405. XWB121, YIplac211-Pbud2-RFP-BUD21-405 was constructed by PCR-amplifying the BUD2 promoter with the oligos 5′-ATCAGAATTCTATCA-ATGCTAGCGTTGAGATGT-3' and 5′-ATCAGGTACCAATATGATACAATCAAAGATAA-ACACCACG-3′, the RFP fragment with the oligos 5′-ATCGGGTACCATGGTTTCAAAAGG-TGAAGAAGATAATATG-3' and 5′-ATCAGTCGACTTTATATAATTCATCCATACCACCA-GTTG-3′, and BUD2 bases 1–405 with the oligos 5′-ATCAGTCGACATGAGCTCCAACAAT-GAACC-3' and 5′-CAGTAAGCTTGATCAGACATGTTAGCA-ATTTCTTG-3'. The EcoRI- and KpnI-digested Pbud2 PCR product, the KpnI- and SalI-digested RFP PCR product, and the SalI- and HindIII-digested BUD21-405 PCR product were inserted into YIplac211. XWB123, YIplac211-BUD51141-1926-RFP was constructed by PCR-amplifying BUD5 bases 1141–1926 with the oligos 5′-ATCAGCATGCGATGAATTGTGCGGTGCAAC-3' and 5′-ATCAGGTACCGGT-AAGCCTTGGAACCTTAGC-3′, and RFP fragment with oligos 5′-ATCGGGTACCATGGTTT-CAAAAGGTGAAGAAGATAATATG-3' and 5′-ATCAGAATTCTTATTTATATAATTCATC-CATACCACCAGTTG-3'. The SphI- and KpnI-cut BUD51141-1926 PCR product and the KpnI- and EcoRI-digested RFP PCR product were inserted into YIplac211. XWB143, YIplac211-BEM1649-1032; 1225-1656, was constructed by PCR-amplifying BEM1 bases 649–1032 with oligos 5′-CAGTAAGCTTATTGCCACGGGGTACGCAACAGGTAAC-3' and 5′-TCCAC-CTGCATCCCTCAACTTACCGGCC-3′ and BEM1 bases 1225–1656 with oligos 5′-GAAAGA-GACGAAAATCAAAACAATATTAAAAC-3' and 5′-ATCAGAATTCTCAAATATCGTGAA-CGGAAATTTTCAG-3'. The HindIII-digested BEM1649-1032 PCR product and the EcoRI- digested BEM11225-1656 PCR product were inserted into YIplca211. XWB148, YIplac211-MYO24261-4722-RFP was constructed by PCR-amplifying MYO2 bases 4261–4772 with oligos 5′-CAGTAAGCTTGAGGAATGGTGCAAGACGCATGG-3' and 5′-ATCGGGATCCGTGGC-CGTCTTGAACGACTTG-3′, and RFP fragment with oligos 5′-ATCGGGATCCATGGTTTCA-AAAGGTGAAGAAGATAATATG-3' and 5′-ATCAGAATTCTTATTTATATAATTCATCCA-TACCACCAGTTG-3'. The HindIII- and BamHI-digested MYO24261-4772 PCR product and the BamHI- and EcoRI-digested RFP PCR product were inserted into YIplac211. XWB156, YIPlac211-ABP11414-1776-RFP was constructed by PCR-amplifying ABP1 bases 1411–1776 with oligos 5′-ATCGCCCCAAGCTTACCTTCTAG-3' and 5′-ATCAGGTACCGTTGCCCAAAGA-CACATAATTGC-3′, and RFP fragment as described in XWB123. The HindIII- and KpnI-digested Abp11141-1776 PCR product and the KpnI- and EcoRI-digested RFP PCR product were inserted into YIplac211. XWB162, YIplac211-ABP140610-1885-RFP was constructed by PCR-amplifying ABP140 bases 610–1885 with oligos 5′-CAGTAAGCTTGCTAATGATGGCTCTA-CAAGTACC-3' and 5′-ATCAGGTACCTTGAGGAACGTCAAACACAGC-3′, and RFP fragment as described in XWB123. The HindIII- and KpnI-digested Abp140610-1885 PCR product and the KpnI- and EcoRI-digested RFP PCR product were inserted into YIplac211. XWB163, YIPlac204-myo23388-4475 was constructed by PCR-amplifying MYO2 bases 3388–4475 with oligos 5′-CAGTGT-CGACTGATGCTTGAGAATTCCGACTTATCTCC-3′ and 5′-ATCGGGATCCTTAGTGGCC-GTCTTGAACGACTTG-3'. The underlined sequence indicates the additional stop codon introduced after base 4475 for generating the myo2-16 mutant. The SalI- and BamHI-digested PCR product was inserted into YIplac204. XWB171, YCplac33-Pbud1-BUD1 was generated by PCR amplifying the BUD1 promoter and BUD1 with oligos 5′-TTTCTCGAATTCTCAGGTAGTAC-TG-3' and 5′-CAGTAAGCTTCTATAGAATAGTGCAAGTGGAAGCG-3′. The EcoRI- and HindIII-digested PCR product was inserted into YCplac33. XWB173, YCplac33-Pbud1-BUD1G12V was generated by site-directed mutagenesis XWB171 using oligos 5′-GTAGTATTGGGTGCTGTTGGTGTCGGTAAATCCTGC-3′ and 5′-GCAGGATTTACCG-ACACCAACAGCACCCAATACTAC-3'. The underlined bases indicate the point mutation introduced by this mutagenesis. XWB182, YIPlac211-Exo70715-1035-linker-1546-1872 was generated by PCR amplifying the EXO70 bases 715–1035 using oligos 5′-ATCAGGTACCATGAACAGC-TACACGGAGGCC-3' and 5′-GAATAATGACTGTGTTACTTGACGTACTTCTTG-3′, and EXO70 bases 1546–1872 using oligos 5′-GTGGGTACCGGTTCGGGTGTATCAGATTGGAG-AGATTTAACTGC-3′ and 5'- ATCAGAATTCCTATCTCACTAATTGGTTAAGAACAGT-AG-3'. The underlined sequence indicates the peptidyl linker introduced for proper Exo70 function. The KpnI-digested EXO70715-1035 PCR product and the EcoRI-digested EXO701546-1872 PCR product were inserted into YIplca211. CPB032, YIplac211-EXO70M30 was generated by PCR amplifying the EXO70 bases 600–1872 containing the M30 mutations (K354A/K355A/E488A/K489A/E505A/R506A) using oligos 5′-ATCAGGTACCATACTCGTCGGTGAAAGGAG-3' and 5′-ATCAGAATTCCTATCTCAC-TAATTGGTTAAGAACAGTAG-3′ from NRB1572 as a template. The KpnI- and EcoRI-digested PCR product was inserted into YIplca204. XWB183, YIplac211-Psec3-SEC3961-1599 was generated by PCR amplifying the SEC3 promoter using oligos 5'-CAGTAAGCTTGGAATAGTACGCATTCCTGTTCAAG-3′ and 5′-AATATAGAATAAATTGCGTAGCCTTGTTACC-3′, and SEC3 bases 961–1599 with a start codon introduced using oligos 5′-ATGACATTGAATGAAGTGAATAAAAGATACGAG-3′ and 5′-ATCAGAATTCTTAGATCGAGTCATTCAAATCGCTTCC-3′. The HindIII-digested SEC3 promoter PCR product and the EcoRI-digested SEC3961-1599 PCR product were inserted into YIplac211. CPB032, YIplac211-EXO70M30, was generated by PCR amplifying the EXO70 bases 600–1872 containing the M30 mutations (K354A/K355A/E488A/K489A/E505A/R506A) using oligos 5′-ATCAGGTACCATACTCGTCGGTGAAAGGAG-3′ and 5′-ATCAGAATTCCTATCTCAC-TAATTGGTTAAGAACAGTAG-3′ from NRB1572 as a template. The KpnI- and EcoRI-digested PCR product was inserted into YIplac204.
Time-lapse imaging of mating mixtures
The time-lapse imaging experiments were performed as described in Wang et al. (2019). WT MATα cells and experimental MATa cells were grown to mid-log phase in synthetic 2% dextrose medium at 30°C, mixed 1:1, and spread at a density of 14,000 cells/mm2 on agarose pads made from synthetic dextrose medium. Mating mixtures were maintained at 30°C using a DeltaVision environment control chamber except as noted below. DIC and fluorescent images were acquired from 12 fields at 2- or 5-min intervals using a DeltaVision Elite Deconvolution Microscope (GE Healthcare Bio-Sciences) with a 60× oil immersion objective and a Front Illuminated sCMOS camera. To minimize phototoxicity, five z-sections 0.5 µm apart were acquired around the center slice of each cell at each time point. Identical light-emitting diode intensities and exposure times were used to image cells expressing the GFP-tagged reporters (10% maximum intensity at 461–489 nm for 200 ms) and the RFP-tagged reporters (10% maximum intensity at 529–556 nm for 200 ms). Representative fluorescent images were deconvolved using Huygens Essential software (Scientific Volume Imaging) in standard mode. To image the temperature-sensitive myo2-16 strain, the cells were grown to mid-log phase at 25°C and incubated at 33°C for 30 min before mixing and spreading onto the agarose pads. Mating mixtures were maintained at 33°C.
Image analysis
For the two-reporter plots (Fig. 6), PM fluorescence signal intensities were quantified by tracing the circumference of the center-slice DIC images using the segmented line tool of ImageJ; pixel values were determined from the corresponding raw fluorescent images after subtracting the background. Cell circumferences were normalized to 100 points (Wang et al., 2019) and the mean distribution of the receptor was generated by aligning the leading Ste2-GFP peaks with each other during tracking and with the center of the fusion site at the prezygote stage. To determine the proportion of cells that ignored a potential partner (Figs. 2 C and 7 B), all MATa cells directly in contact with one or more MATα cell(s) were scored for mating. Ste2-GFP signal intensities at the DS of WT and BUD1G12V/BUD1 cells were quantified by tracing the crescent of the center-slice GFP images using the segmented line tool of ImageJ. The mean fluorescence intensities of 30 pixels surrounding the peak signal (Fig. 2 E) and the total fluorescence intensities of the receptor crescent (Fig. 2 F) were determined one time point before tracking in WT cells and one time point before morphogenesis in BUD1G12V/BUD1 cells that ignored potential mating partners.
Online supplemental material
Table S1 lists the yeast strains used in this study. Table S2 lists the plasmids used in this study. Fig. S1 shows the effect of coexpressing Ste2-GFP and RFP-Bud2 or Ste2-GFP and Bud5-RFP on gradient tracking.
Supplementary Material
lists yeast strains used in this study.
lists plasmids used in this study.
Acknowledgments
We thank Rob Arkowitz, Edward Draper, Holly Stratton, and members of the Stone lab for helpful discussions and critical reading of the manuscript, and Dr. Peter Novick for generously sharing the exo70M30 construct.
This work was supported by National Science Foundation grant 1818067 (D.E. Stone).
The authors declare no competing financial interests.
Author contributions: X. Wang and D.E. Stone conceived the project, designed the experiments, analyzed the data, and wrote the manuscript. C.-Y. Pai analyzed the Ste2-GFP signals in the BUD1/BUD1G12V cells, constructed the exo70M30 and exo70M30 sec3ΔN strains, and imaged the exo70M30, sec3ΔN, and exo70M30 sec3ΔN cells in mating mixtures. X. Wang performed all the other experiments, developed the data analysis methods, and made all the figures.
References
- Abdul-Ganiyu, R., Venegas L.A., Wang X., Puerner C., Arkowitz R.A., Kay B.K., and Stone D.E.. 2021. Phosphorylated Gbeta is a directional cue during yeast gradient tracking. Sci. Signal. 14:eabf4710. 10.1126/scisignal.abf4710 [DOI] [PubMed] [Google Scholar]
- Arkowitz, R.A. 2009. Chemical gradients and chemotropism in yeast. Cold Spring Harbor Perspect. Biol. 1:a001958. 10.1101/cshperspect.a001958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura, T., Sasaki T., Nagano F., Satoh A., Obaishi H., Nishioka H., Imamura H., Hotta K., Tanaka K., Nakanishi H., and Takai Y.. 1998. Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene. 16:121–130. 10.1038/sj.onc.1201487 [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., and Struhl K.. 1994. Current Protocols in Molecular Biology. John Wiley and Sons, Inc [Google Scholar]
- Bar, E.E., Ellicott A.T., and Stone D.E.. 2003. Gbetagamma recruits Rho1 to the site of polarized growth during mating in budding yeast. J. Biol. Chem. 278:21798–21804. 10.1074/jbc.M212636200 [DOI] [PubMed] [Google Scholar]
- Basile, J.R., Barac A., Zhu T., Guan K.L., and Gutkind J.S.. 2004. Class IV semaphorins promote angiogenesis by stimulating Rho-initiated pathways through plexin-B. Cancer Res. 64:5212–5224. 10.1158/0008-5472.CAN-04-0126 [DOI] [PubMed] [Google Scholar]
- Bendezu, F.O., and Martin S.G.. 2013. Cdc42 explores the cell periphery for mate selection in fission yeast. Curr. Biol. 23:42–47. 10.1016/j.cub.2012.10.042 [DOI] [PubMed] [Google Scholar]
- Bi, E., and Park H.O.. 2012. Cell polarization and cytokinesis in budding yeast. Genetics. 191:347–387. 10.1534/genetics.111.132886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd, C., Hughes T., Pypaert M., and Novick P.. 2004. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J. Cell Biol. 167:889–901. 10.1083/jcb.200408124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butty, A.C., Pryciak P.M., Huang L.S., Herskowitz I., and Peter M.. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science. 282:1511–1516. 10.1126/science.282.5393.1511 [DOI] [PubMed] [Google Scholar]
- Chant, J., and Herskowitz I.. 1991. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 65:1203–1212. 10.1016/0092-8674(91)90015-q [DOI] [PubMed] [Google Scholar]
- Chant, J., and Pringle J.R.. 1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129:751–765. 10.1083/jcb.129.3.751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, P.J., and Sprague G.F. Jr. 2012. The regulation of filamentous growth in yeast. Genetics. 190:23–49. 10.1534/genetics.111.127456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, K.J., Srikantha T., Lockhart S.R., Pujol C., and Soll D.R.. 2006. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J. 25:2240–2252. 10.1038/sj.emboj.7601099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, G., Hutagalung A.H., Fu C., Novick P., and Reinisch K.M.. 2005. The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat. Struct. Mol. Biol. 12:1094–1100. 10.1038/nsmb1017 [DOI] [PubMed] [Google Scholar]
- Dorer, R., Pryciak P.M., and Hartwell L.H.. 1995. Saccharomyces cerevisiae cells execute a default pathway to select a mate in the absence of pheromone gradients. J. Cell Biol. 131:845–861. 10.1083/jcb.131.4.845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin, D.G., Miller K.G., and Botstein D.. 1988. Yeast actin-binding proteins: Evidence for a role in morphogenesis. J. Cell Biol. 107:2551–2561. 10.1083/jcb.107.6.2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin, D.G., and Nelson W.J.. 1996. Origins of cell polarity. Cell. 84:335–344. 10.1016/s0092-8674(00)81278-7 [DOI] [PubMed] [Google Scholar]
- Dyer, J.M., Savage N.S., Jin M., Zyla T.R., Elston T.C., and Lew D.J.. 2013. Tracking shallow chemical gradients by actin-driven wandering of the polarization site. Curr. Biol. 23:32–41. 10.1016/j.cub.2012.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, D., Garcia J.G., and Brindley D.N.. 2001. Platelet-released phospholipids link haemostasis and angiogenesis. Cardiovasc. Res. 49:588–599. 10.1016/s0008-6363(00)00230-3 [DOI] [PubMed] [Google Scholar]
- Gimeno, C.J., Ljungdahl P.O., Styles C.A., and Fink G.R.. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS. Cell. 68:1077–1090. 10.1016/0092-8674(92)90079-r [DOI] [PubMed] [Google Scholar]
- Guo, W., Tamanoi F., and Novick P.. 2001. Spatial regulation of the exocyst complex by Rho1 GTPase. Nat. Cell Biol. 3:353–360. 10.1038/35070029 [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and Fink G.R.. 2002. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego, CA. 325–456 [Google Scholar]
- Hegemann, B., Unger M., Lee S.S., Stoffel-Studer I., van den Heuvel J., Pelet S., Koeppl H., and Peter M.. 2015. A cellular system for spatial signal decoding in chemical gradients. Dev. Cell. 35:458–470. 10.1016/j.devcel.2015.10.013 [DOI] [PubMed] [Google Scholar]
- Hong, K., and Nishiyama M.. 2010. From guidance signals to movement: Signaling molecules governing growth cone turning. Neuroscientist. 16:65–78. 10.1177/1073858409340702 [DOI] [PubMed] [Google Scholar]
- Hutagalung, A.H., Coleman J., Pypaert M., and Novick P.J.. 2009. An internal domain of Exo70p is required for actin-independent localization and mediates assembly of specific exocyst components. Mol. Biol. Cell. 20:153–163. 10.1091/mbc.e08-02-0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima, M., Huang Y.E., and Devreotes P.. 2002. Temporal and spatial regulation of chemotaxis. Dev. Cell. 3:469–478. 10.1016/s1534-5807(02)00292-7 [DOI] [PubMed] [Google Scholar]
- Irazoqui, J.E., Gladfelter A.S., and Lew D.J.. 2003. Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 5:1062–1070. 10.1038/ncb1068 [DOI] [PubMed] [Google Scholar]
- Ismael, A., Tian W., Waszczak N., Wang X., Cao Y., Suchkov D., Bar E., Metodiev M.V., Liang J., Arkowitz R.A., and Stone D.E.. 2016. Gβ promotes pheromone receptor polarization and yeast chemotropism by inhibiting receptor phosphorylation. Sci. Signal. 9:ra38. 10.1126/scisignal.aad4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, C.L., and Hartwell L.H.. 1990. Courtship in S. cerevisiae: Both cell types choose mating partners by responding to the strongest pheromone signal. Cell. 63:1039–1051. 10.1016/0092-8674(90)90507-b [DOI] [PubMed] [Google Scholar]
- Johnston, G.C., Prendergast J.A., and Singer R.A.. 1991. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J. Cell Biol. 113:539–551. 10.1083/jcb.113.3.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, P.J., Beven L., Hariharan S., and Park H.O.. 2010. The Rsr1/Bud1 GTPase interacts with itself and the Cdc42 GTPase during bud-site selection and polarity establishment in budding yeast. Mol. Biol. Cell. 21:3007–3016. 10.1091/mbc.E10-03-0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Dong J., and Lord E.M.. 2004. Pollen tube guidance: The role of adhesion and chemotropic molecules. Curr. Top. Dev. Biol. 61:61–79. 10.1016/S0070-2153(04)61003-9 [DOI] [PubMed] [Google Scholar]
- Liu, D., Li X., Shen D., and Novick P.. 2018. Two subunits of the exocyst, Sec3p and Exo70p, can function exclusively on the plasma membrane. Mol. Biol. Cell. 29:736–750. 10.1091/mbc.E17-08-0518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D., and Novick P.. 2014. Bem1p contributes to secretory pathway polarization through a direct interaction with Exo70p. J. Cell Biol. 207:59–72. 10.1083/jcb.201404122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden, K., and Snyder M.. 1992. Specification of sites for polarized growth in Saccharomyces cerevisiae and the influence of external factors on site selection. Mol. Biol. Cell. 3:1025–1035. 10.1091/mbc.3.9.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston, A.L., Chen T., Yang M.C., Belhumeur P., and Chant J.. 2001. A localized GTPase exchange factor, Bud5, determines the orientation of division axes in yeast. Curr. Biol. 11:803–807. 10.1016/s0960-9822(01)00230-5 [DOI] [PubMed] [Google Scholar]
- McClure, A.W., Minakova M., Dyer J.M., Zyla T.R., Elston T.C., and Lew D.J.. 2015. Role of polarized G protein signaling in tracking pheromone gradients. Dev. Cell. 35:471–482. 10.1016/j.devcel.2015.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitch, M., and Chant J.. 1996. A mechanism of Bud1p GTPase action suggested by mutational analysis and immunolocalization. Curr. Biol. 6:446–454. 10.1016/s0960-9822(02)00512-2 [DOI] [PubMed] [Google Scholar]
- Miller, K.E., Lo W.C., Chou C.S., and Park H.O.. 2019. Temporal regulation of cell polarity via the interaction of the Ras GTPase Rsr1 and the scaffold protein Bem1. Mol. Biol. Cell. 30:2543–2557. 10.1091/mbc.E19-02-0106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi, F., Plachta N., and Viasnoff V.. 2020. Novel approaches to link apicobasal polarity to cell fate specification. Curr. Opin. Cell Biol. 62:78–85. 10.1016/j.ceb.2019.09.003 [DOI] [PubMed] [Google Scholar]
- Naganathan, S.R., Furthauer S., Rodriguez J., Fievet B.T., Julicher F., Ahringer J., Cannistraci C.V., and Grill S.W.. 2018. Morphogenetic degeneracies in the actomyosin cortex. Elife. 7:e37677. 10.7554/eLife.37677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern, A., and Arkowitz R.A.. 1998. A GTP-exchange factor required for cell orientation. Nature. 391:195–198. 10.1038/34458 [DOI] [PubMed] [Google Scholar]
- Nern, A., and Arkowitz R.A.. 1999. A Cdc24p-Far1p-Gbetagamma protein complex required for yeast orientation during mating. J. Cell Biol. 144:1187–1202. 10.1083/jcb.144.6.1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanivelu, R., and Preuss D.. 2000. Pollen tube targeting and axon guidance: Parallels in tip growth mechanisms. Trends Cell Biol. 10:517–524. 10.1016/s0962-8924(00)01849-3 [DOI] [PubMed] [Google Scholar]
- Park, H.O., Sanson A., and Herskowitz I.. 1999. Localization of Bud2p, a GTPase-activating protein necessary for programming cell polarity in yeast to the presumptive bud site. Genes Dev. 13:1912–1917. 10.1101/gad.13.15.1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, S.I., Hadwiger J.A., and Lorincz A.T.. 1985. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc. Natl. Acad. Sci. USA. 82:4055–4059. 10.1073/pnas.82.12.4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, R.L., and Fink G.R.. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: Mating and invasive growth. Genes Dev. 8:2974–2985. 10.1101/gad.8.24.2974 [DOI] [PubMed] [Google Scholar]
- Roemer, T., Madden K., Chang J., and Snyder M.. 1996. Selection of axial growth sites in yeast requires Axl2p, a novel plasma membrane glycoprotein. Genes Dev. 10:777–793. 10.1101/gad.10.7.777 [DOI] [PubMed] [Google Scholar]
- Schott, D., Ho J., Pruyne D., and Bretscher A.. 1999. The COOH-terminal domain of Myo2p, a yeast myosin V, has a direct role in secretory vesicle targeting. J. Cell Biol. 147:791–808. 10.1083/jcb.147.4.791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall, J.E. 1993. Polarization of yeast cells in spatial gradients of alpha mating factor. Proc. Natl. Acad. Sci. USA. 90:8332–8336. 10.1073/pnas.90.18.8332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman,F., Fink G.R., and Hicks J.B., editors. 1986. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- Snetselaar, K.M., Bolker M., and Kahmann R.. 1996. Ustilago maydis mating hyphae orient their growth toward pheromone sources. Fungal Genet. Biol. 20:299–312. 10.1006/fgbi.1996.0044 [DOI] [PubMed] [Google Scholar]
- Taniguchi, K., Shao Y., Townshend R.F., Cortez C.L., Harris C.E., Meshinchi S., Kalantry S., Fu J., O’Shea K.S., and Gumucio D.L.. 2017. An apicosome initiates self-organizing morphogenesis of human pluripotent stem cells. J. Cell Biol. 216:3981–3990. 10.1083/jcb.201704085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojima, T., Hines J.H., Henley J.R., and Kamiguchi H.. 2011. Second messengers and membrane trafficking direct and organize growth cone steering. Nat. Rev. Neurosci. 12:191–203. 10.1038/nrn2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasen, G., Dunayevich P., Constantinou A., and Colman-Lerner A.. 2020. GPCR receptor phosphorylation and endocytosis are not necessary to switch polarized growth between internal cues during pheromone response in S. cerevisiae. Commun. Integr. Biol. 13:128–139. 10.1080/19420889.2020.1806667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Tian W., Banh B.T., Statler B.M., Liang J., and Stone D.E.. 2019. Mating yeast cells use an intrinsic polarity site to assemble a pheromone-gradient tracking machine. J. Cell Biol. 218:3730–3752. 10.1083/jcb.201901155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszczak, N., DeFlorio R., Ismael A., Cheng N., Stone D.E., and Metodiev M.V.. 2019. Quantitative proteomics reveals a Gα/MAPK signaling hub that controls pheromone-induced cellular polarization in yeast. J. Proteomics. 207:103467. 10.1016/j.jprot.2019.103467 [DOI] [PubMed] [Google Scholar]
- Zenker, J., White M.D., Gasnier M., Alvarez Y.D., Lim H.Y.G., Bissiere S., Biro M., and Plachta N.. 2018. Expanding actin rings zipper the mouse embryo for blastocyst formation. Cell. 173:776–791.e17. 10.1016/j.cell.2018.02.035 [DOI] [PubMed] [Google Scholar]
- Zhang, X., Bi E., Novick P., Du L., Kozminski K.G., Lipschutz J.H., and Guo W.. 2001. Cdc42 interacts with the exocyst and regulates polarized secretion. J. Biol. Chem. 276:46745–46750. 10.1074/jbc.M107464200 [DOI] [PubMed] [Google Scholar]
- Zhang, X., Orlando K., He B., Xi F., Zhang J., Zajac A., and Guo W.. 2008. Membrane association and functional regulation of Sec3 by phospholipids and Cdc42. J. Cell Biol. 180:145–158. 10.1083/jcb.200704128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y., Bender A., and Cerione R.A.. 1995. Interactions among proteins involved in bud-site selection and bud-site assembly in Saccharomyces cerevisiae. J. Biol. Chem. 270:626–630. 10.1074/jbc.270.2.626 [DOI] [PubMed] [Google Scholar]
- Zhu, M., and Zernicka-Goetz M.. 2020. Building an apical domain in the early mouse embryo: Lessons, challenges and perspectives. Curr. Opin. Cell Biol. 62:144–149. 10.1016/j.ceb.2019.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists yeast strains used in this study.
lists plasmids used in this study.