Abstract
Understanding the evolution of antibody immunity following heterologous SAR-CoV-2 breakthrough infection will inform the development of next-generation vaccines. Here, we tracked SARS-CoV-2 receptor binding domain (RBD)-specific antibody responses up to six months following Omicron BA.1 breakthrough infection in mRNA-vaccinated individuals. Cross-reactive serum neutralizing antibody and memory B cell (MBC) responses declined by two- to four-fold through the study period. Breakthrough infection elicited minimal de novo Omicron-specific B cell responses but drove affinity maturation of pre-existing cross-reactive MBCs toward BA.1. Public clones dominated the neutralizing antibody response at both early and late time points, and their escape mutation profiles predicted newly emergent Omicron sublineages. The results demonstrate that heterologous SARS-CoV-2 variant exposure drives the evolution of B cell memory and suggest that convergent neutralizing antibody responses continue to shape viral evolution.
The emergence and global spread of the SARS-CoV-2 Omicron BA.1 variant in late 2021 resulted in the largest surge in COVID-19 caseloads to date (1). While currently available COVID-19 vaccines induced high levels of protection against pre-Omicron variants, the extensive immune evasiveness of Omicron resulted in significantly reduced vaccine efficacy and durability following both primary and booster immunization (2–5). Moreover, antigenically drifted sub-lineages of Omicron (e.g. BA.2, BA.2.12.1, BA.4/5, BA.2.75, BA.2.75.2, and BA.4.6) continue to emerge and supplant prior sub-variants (4, 6). The high prevalence of Omicron breakthrough infections led to the development and emergency use authorization of Omicron variant-based booster mRNA vaccines, despite limited immunogenicity and efficacy data in humans (2, 7). Thus, there is an urgent need to understand if and how secondary exposure to antigenically divergent variants, such as Omicron, shape SARS-CoV-2-specific B cell memory.
We and others have previously reported that the acute antibody response following Omicron BA.1 breakthrough infection is dominated by re-activated memory B cells induced by mRNA vaccination (8–11). In support of these findings, preliminary data from clinical trials evaluating the immunogenicity of variant-based booster vaccines demonstrated that BA.1-containing mRNA vaccines induce a modest improvement in peak serum neutralizing responses compared with ancestral Wuhan-1 immunization (12). Although these studies provide evidence for “original antigenic sin” in the early B cell response following Omicron breakthrough infection, if and how this response evolves over time remains unclear. To address these questions, we longitudinally profiled SARS-CoV-2-specific serological and memory B responses in mRNA-vaccinated donors up to six months following BA.1 breakthrough infection.
We initially characterized the antibody response to SARS-CoV-2 in a cohort of seven mRNA-1273 vaccinated donors 14 to 27 days (median = 23 days) after BA.1 breakthrough infection (8). To study the evolution of this response, we obtained blood samples from six of the seven participants at a follow-up appointment four to six months (median = 153 days) post-infection (Fig. 1A, Table S1). Three of the six donors experienced infection after two-dose mRNA-1273 vaccination while the remaining three donors were infected after a third booster dose. None of the donors reported a second breakthrough infection between the two sample collection time points.
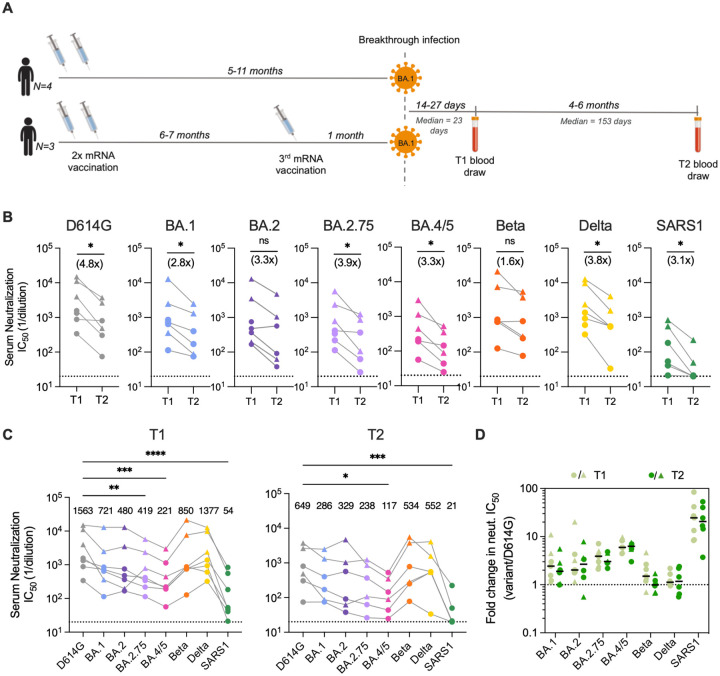
Figure 1. Serum neutralizing antibody responses induced following BA.1 breakthrough infection.
(A) Timeline of vaccination, BA.1 breakthrough infection, and sample collections. (B) Paired analysis of serum neutralizing activity against SARS-CoV-2 D614G and BA.1, BA.2, BA.2.75, BA.4/5, Beta, and Delta variants, and SARS-CoV (SARS1) at 1-month (T1) and 5–6-month (T2) time points, as determined via a MLV-based pseudovirus neutralization assay. Connected data points represent paired samples for each donor, and the median fold change in serum titer between the two time points is shown in parentheses. Dotted lines represent the lower limit of detection of the assay. (C) Serum neutralizing titers against SARS-CoV-2 variants and SARS-CoV in samples collected at (left) 1-month and (right) 5–6-month post-breakthrough infection for each donor. Median titers are shown above the data points. Dotted lines represent the lower limit of detection of the assay. (D) Fold change in serum neutralizing titers for the indicated SARS-CoV-2 variants and SARS-CoV relative to SARS-CoV-2 D614G at early (T1) and late (T2) time points. Black bars represent median fold changes. Dotted line indicates no change in IC50. Breakthrough infection donors infected after two-dose mRNA vaccination (n = 4) are shown as circles and those infected after a third mRNA dose (n = 3) are shown as triangles. One two-dose vaccinated breakthrough donor was lost to follow-up at the second time point. Statistical comparisons were determined by (B) Wilcoxon matched-pairs signed rank test, (C) Friedman’s one-way ANOVA with Dunn’s multiple comparisons, or (D) mixed model ANOVA. *P < 0.05; **P < 0.01; ****P < 0.0001; ns, not significant.
To evaluate serum neutralization breadth and potency, we tested the plasma samples for neutralizing activity against SARS-CoV-2 D614G, emergent variants (BA.1, BA.2, BA.4/5, BA.2.75, Beta, and Delta), and the more evolutionarily divergent sarbecovirus SARS-CoV, in a murine-leukemia virus (MLV)-based pseudovirus assay. Paired comparisons within each participant revealed that serum neutralizing titers against D614G declined by a median of 4.8-fold at 5- to 6-months post-infection relative to those observed within one-month post-infection (Fig. 1B). Correspondingly, we observed lower serum neutralizing titers against Omicron subvariants (2.8 to 3.9-fold, respectively), Beta (1.6-fold), Delta (3.8-fold), and SARS-CoV (3.1-fold) at the 5- to 6-month time point relative to the early time point (Fig. 1B). Despite this waning of neutralizing antibody titers over time, all of the donor sera displayed detectable neutralizing activity against all of the SARS-CoV-2 variants tested at the 5–6 month time point (median titers ranging from 117 to 552) (Fig. 1C). Notably, titers remained within 3-fold of that observed for D614G for all variants except BA.4/5, which showed the greatest degree of escape from serum neutralizing antibodies (5.5-fold reduction from D614G), consistent with published serological studies (4, 5). Furthermore, the fold reduction in serum neutralizing titer for SARS-CoV-2 VOCs relative to D614G remained similar at both time points, suggesting maintained serum neutralization breadth over time (Fig. 1D). We observed minimal cross-neutralizing activity against SARS-CoV (median titer = 21) in all donors, suggesting that serum neutralization breadth remained limited to SARS-CoV-2 variants (Fig. 1C). We conclude that serum neutralizing titers wane over the course 6-months following Omicron BA.1 breakthrough infection but nevertheless remain at detectable levels across a diverse range of SARS-CoV-2 variants through 6 months.
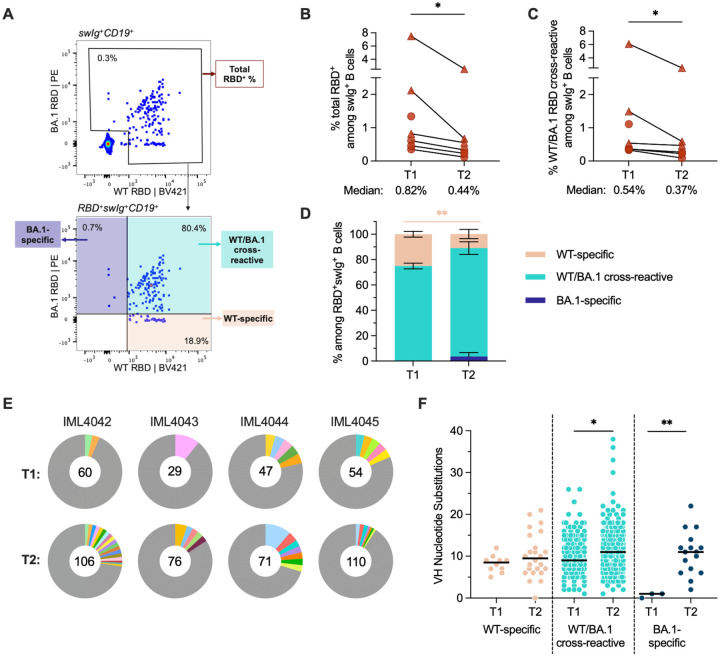
Next, we assessed the magnitude and cross-reactivity of the antigen-specific B cell response via flow cytometric enumeration of B cells stained with differentially labeled wildtype (Wuhan-1; WT) and BA.1 RBD tetramers (Fig. 2A, Fig. S1A). At the 5–6-month time point, total RBD-reactive B cells (WT and/or BA.1-reactive) and WT/BA.1 cross-reactive B cells comprised a median of 0.44% (ranging 0.12–2.53%) and 0.37% (ranging 0.12–2.53%) of class-switched (IgG+ or IgA+) B cells, respectively (Fig. 2B, 2C, Fig. S1A). Thus, 86% (ranging 69–100%) of all RBD+ class-switched B cells at 5–6 months post-infection displayed BA.1/WT cross-reactivity, compared with 75% at 1-month post-infection (ranging 65–81%) (Fig. 2D, Fig. S2). Correspondingly, WT-specific B cells decreased from 25% of all RBD+ class-switched B cells at 1 month to 11% at 5–6 months (Fig. 2D, Fig. S2). Consistent with the waning of serum neutralizing titers over time, we also observed a modest but significant decline (1.1 to 3.7-fold) in the frequencies of WT/BA.1 cross-reactive B cells at 5–6 months relative to the 1-month time point (Fig. 2C). At the late time point, we also detected the emergence of a BA.1-specific B cell population (average = 3% of class-switched B cells) in 3 of the 6 individuals, although the magnitude of this response varied widely among individuals (ranging from 1–18%) (Fig. 2D, Fig. S2). In summary, Omicron BA.1 breakthrough infection induces a WT/BA.1 cross-reactive B cell response at early time points post-infection and this response only modestly declines over the course of 6 months.
Figure 2. SARS-CoV-2 RBD-specific memory B cell responses following BA.1 breakthrough infection.
(A) Representative fluorescence-activated cell sorting gating strategy used to enumerate frequencies of (top) total (WT+BA.1) RBD-reactive B cells among classswitched (IgG+ or IgA+) CD19+ B cells and (bottom) WT-specific, BA.1-specific, and WT/BA.1 cross-reactive B cells among total RBD-reactive, class-switched (IgG+ or IgA+) CD19+ B cells. (B-C) Frequencies of (B) total RBD-reactive or (C) WT/BA.1 RBD cross-reactive B cells among class-switched CD19+ B cells at 1-month (T1) and 5–6-month (T2) time points. Connected data points represent paired samples for each donor. Donors infected after two-dose mRNA vaccination (n = 4) are shown as circles and those infected after a third mRNA dose (n = 3) are shown as triangles. One two-dose vaccinated breakthrough donor was censored at the second time point. (D) Mean proportions of RBD-reactive, class-switched B cells that display WT-specific, BA.1-specific or WT/BA.1-cross-reactive binding at each time point. Error bars indicate standard error of mean. (E) Clonal lineage analysis of RBD-directed antibodies isolated from four donors at the early (T1) and late (T2) time points. Clonally expanded lineages (defined as antibodies with the same heavy and light chain germlines, same CDR3 lengths, and ≥ 80% CDRH3 sequence identity) are represented as colored slices. Each colored slice represents a clonal lineage with the size of the slice proportional to the lineage size. Unique clones are combined into a single gray segment. The number of antibodies is shown in the center of each pie. Three of the donors (IML4042, IML4043, and IML4044) experienced BA.1 breakthrough infection following two-dose mRNA vaccination and the remaining donor (IML4045) was infected after a booster immunization. (F) Levels of somatic hypermutation, as determined by the number of nucleotide substitutions in the variable heavy (VH) region, at the early and late time points among WT-specific, WT/BA.1 cross-reactive, and BA.1-specific antibodies. Medians are shown by black bars. Statistical significance was determined by (B and C) Wilcoxon matched-pairs signed rank test or (D and F) Mann-Whitney U test. swIg+, class-switched immunoglobulin. PE, phycoerythrin; *P < 0.05; **P < 0.01.
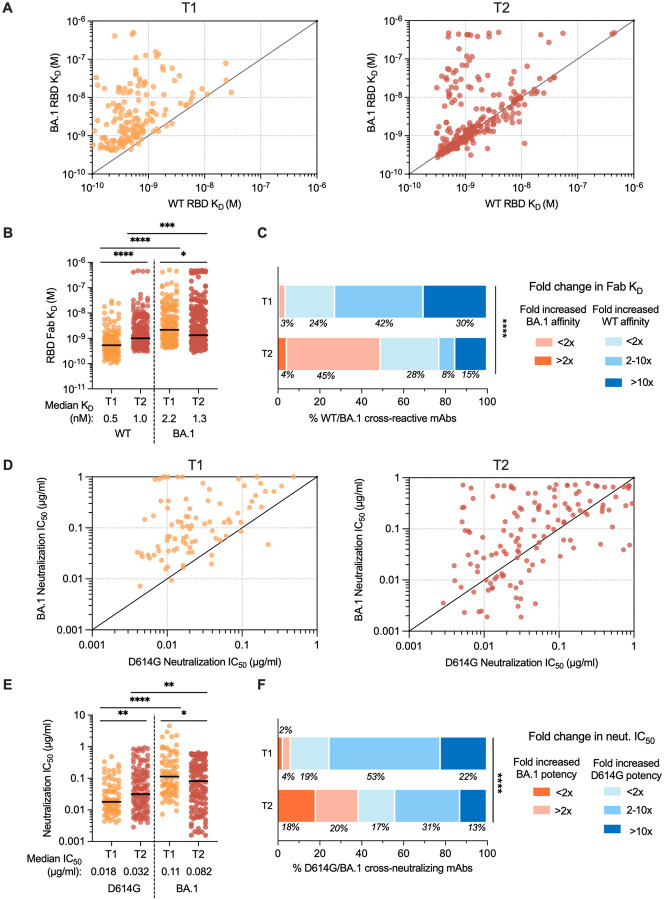
To compare the molecular characteristics of antibodies isolated at early and late time points following BA.1 breakthrough infection, we single-cell sorted 71 to 110 class-switched RBD-reactive B cells from four of the five previously studied donors (donors IML4042, IML4043, IML4044, IML4045) at 139 to 170 days after breakthrough infection and expressed a total of 363 natively paired antibodies as full-length IgGs (Fig. S1B) (8). Similar to the antibodies characterized from the acute time point, the newly isolated antibodies primarily recognized both WT and BA.1 RBD antigens (73–97%), exhibited a high degree of clonal diversity, and displayed preferential usage of certain VH germline genes (IGHV1–46, 1–69, 3–13, 3–53, 3–66, 3–9, and 4–31 germline genes at both time points) (Fig. 2E, Fig. S3 and S4). The level of SHM in the cross-reactive antibodies increased from a median of 9 VH nucleotide substitutions at 1-month to 11 VH nucleotide substitutions by 5–6 months, potentially suggesting affinity maturation in secondary germinal centers (Fig. 2F). Consistent with their higher levels of SHM, the antibodies isolated at 5–6 months displayed 1.7-fold improved binding to BA.1 (median KD = 1.3 nM) and 2-fold reduced binding affinity to the WT RBD (median KD = 1.0 nM) relative to early antibodies, suggesting maturation towards Omicron BA.1 at the expense of WT affinity (Fig. 3A and 3B). These changes in binding recognition resulted in the late antibodies showing more balanced affinity profiles compared to the early antibodies (Fig 3B). For example, the majority of antibodies (73%) isolated at the late time point exhibited WT and BA.1 RBD affinities within two-fold of each other compared to only 24% of early antibodies (Fig. 3C).
Figure 3. Binding and neutralizing properties of RBD-directed antibodies induced by BA.1 breakthrough infection.
(A-B) Fab binding affinities of WT/BA.1 cross-reactive antibodies for recombinant WT and BA.1 RBD antigens, as measured by BLI, are plotted as bivariates for antibodies derived from (left) 1-month and (right) 5–6-month time points in (A) and summarized as a column dot plot in (B). Median affinities are indicated by black bars and shown below data points. (C) Proportions of WT/BA.1 cross-reactive antibodies at each time point that show increased affinity for the BA.1 RBD relative to WT (red shades) or increased affinity for WT RBD (blue shades). Values represent the percentage of antibodies belonging to each of the indicated categories. (D–E) Neutralizing activities of cross-binding antibodies against SARS-CoV-2 D614G and BA.1, as determined by an MLV-based pseudovirus neutralization assay. IC50 values are plotted in (D) as bivariates for antibodies isolated from (left) 1-month and (right) 5–6-month tie points and summarized as column dot plots in (E). Median IC50 values are indicated by black bars and shown below data points. (F) Proportions of WT/BA.1 cross-neutralizing antibodies at each time point that show increased neutralizing potency against BA.1 (red shades) or D614G (blue shades). Values represent the percentage of antibodies belonging to each of the indicated categories. Statistical comparisons were determined by (B and E) multiple Mann-Whitney U tests without adjustment for multiplicity across time points and Wilcoxon matched-pairs rank tests within each time point or (C and F) Mann-Whitney U test. IC50, 50% inhibitory concentration; KD, equilibrium dissociation constant; *P < 0.05; **P < 0.01; ****P < 0.0001.
To determine whether the improvement in binding affinity for BA.1 translated into enhanced neutralization potency, we assessed the antibodies for neutralizing activity against WT and BA.1 using a pseudovirus assay. Fifty-one percent and 42% of WT/BA.1 cross-binding antibodies isolated from the 1-month and 5–6-month time point, respectively, cross-neutralized D614G and BA.1 with IC50 < 2 μg/ml. Overall, the neutralizing antibodies displayed approximately 2-fold lower potency against D614G at the late time point relative to the acute time point, consistent with the observed reduction in WT RBD affinity over time (Fig. 3D and 3E). As expected, the improvement in BA.1 binding affinities over time translated into an overall improvement in neutralization potency (Fig. 3E). Approximately 16% of antibodies isolated at 5–6 months displayed neutralization IC50s <0.01 ug/ml compared to only 2% of antibodies isolated at the earlier time point (Fig. S5). As a result, forty-one percent of the neutralizing antibodies isolated at 6 months exhibited more potent activity against BA.1 relative to D614G, compared to only 7% of the acute neutralizing antibodies (Fig 3F). In summary, cross-reactive antibody responses induced following BA.1 breakthrough infection evolve toward increased BA.1 affinity and neutralization potency for at least 6 months post-infection.
Although the vast majority of antibodies isolated at the 5–6-month time point displayed WT/BA.1 cross-reactive binding, we identified a limited number of BA.1-specific antibodies in all four donors, comprising 1% to 15% of total RBD-specific antibodies (median = 4%) (Fig. S3). In contrast, we only detected BA.1-specific antibodies in a single donor at the acute time point (Fig. S3). Furthermore, unlike the BA.1-specific antibodies isolated at the early time point, which lacked somatic mutations, the BA.1-specific antibodies identified at 5–6 months displayed SHM levels similar to those of cross-reactive antibodies (median = 11 VH nucleotide substitutions) (Fig. 2F). Forty percent of BA.1-specific antibodies isolated at the late time point neutralized BA.1, with IC50s ranging from 0.002 to 0.089 μg/ml, and none of the antibodies displayed detectable neutralizing activity against D614G (Fig. S6). Thus, BA.1 breakthrough infection induces a limited and delayed de novo Omicron-specific B cell response that undergoes affinity maturation over time.
To further explore the breadth of both WT/BA.1 cross-reactive and BA.1-specific neutralizing antibodies, we evaluated their binding reactivities with a panel of recombinant RBDs encoding mutations present in SARS-CoV-2 variants BA.2, BA.4/5, Beta, and Delta, and the more antigenically divergent SARS-CoV. D614G/BA.1 cross-neutralizing antibodies displayed 2.4-fold reduced affinity for the WT RBD and 3.4-fold improved affinity for the BA.1 RBD relative to early neutralizing antibodies, consistent with the pattern observed for all WT/BA.1 cross-binding antibodies (Fig. 4A, Fig. 3A–C). Furthermore, the WT/BA.1 cross-reactive antibodies isolated at 6 months broadly recognized other SARS-CoV-2 variants, except for BA.4/5, which was associated with a ≥5-fold loss in affinity for 57% (68/120) of the WT/BA.1 neutralizing antibodies (Fig. 4A, Fig. S7). Importantly, the 5–6-month antibodies displayed higher affinity binding to all Omicron sub-variants and Beta relative to the early antibodies, suggesting that the increased affinity to BA.1 also improved breadth of reactivity against other variants (Fig. 4A). In support of this finding, a significantly higher proportion (40%) of neutralizing antibodies isolated at 6 months displayed high affinity (KD < 10nM) binding to all five variants tested compared with early antibodies (22%) (Fig. 4B). Furthermore, antibodies isolated at the late time point displayed smaller differences in binding affinity against BA.1, BA.2, BA.4/5 and the early Beta and Delta variants relative to early antibodies (Fig. 4C). In contrast to the WT/BA.1 cross-reactive antibodies, the BA.1-specific neutralizing antibodies displayed limited breadth, with only 50% of these antibodies maintaining binding to BA.2 and none of the antibodies showing reactivity with WT, BA.4/5, Beta, or Delta (Fig. S6). We conclude that BA.1 breakthrough infection results in an overall broadening of the anti-SARS-CoV-2 neutralizing antibody repertoire.
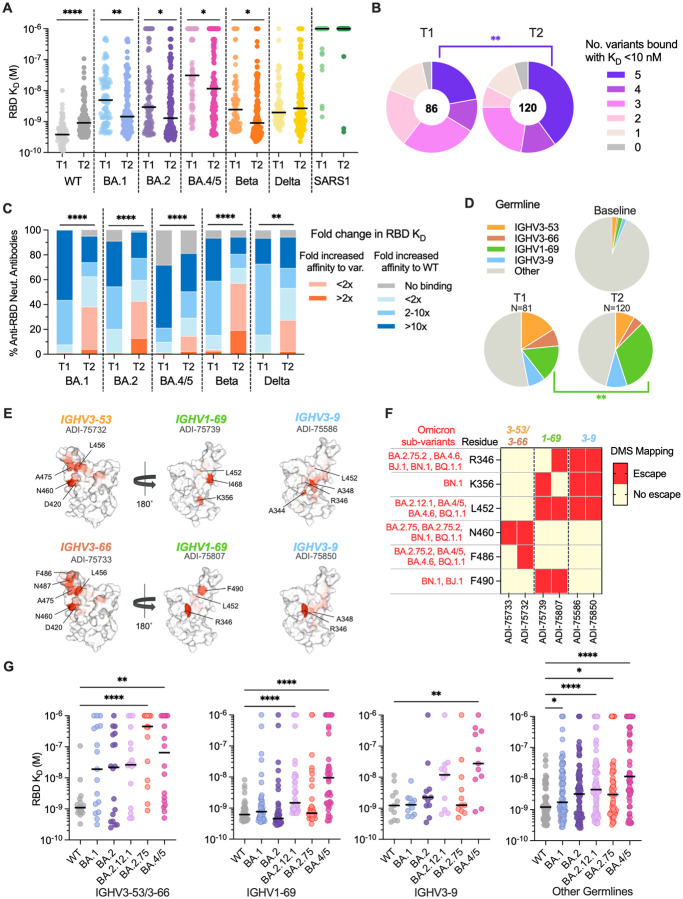
Figure 4. Breadth of D614G/BA.1 cross-neutralizing antibodies at early and late time points following BA.1 breakthrough infection.
(A) Fab binding affinities of D614G/BA.1 cross-neutralizing antibodies isolated at 1-month (T1) and 5–6-month (T2) time points for recombinant SARS-CoV-2 variant RBDs and the SARS-CoV RBD, as determined by BLI. Black bars represent medians. (B) Pie charts showing the proportions of antibodies derived from (left) early and (right) late time points that bound the indicated number of SARS-CoV-2 variant RBDs with Fab KDs < 10 nM. The total number of antibodies is shown in the center of each pie. (C) Proportions of D614G/BA.1 cross-neutralizing antibodies with the indicated fold changes in Fab binding affinities for recombinant SARS-CoV-2 variant RBDs relative to the WT RBD. (D) Pie charts showing frequencies of the indicated convergent germline genes among D614G/BA.1 cross-neutralizing antibodies isolated at early (T1) and late (T2) timelines. Germline gene frequencies observed in baseline human antibody repertoires (upper right) are shown for comparison (25). (E) Structural projections of binding escape mutations determined for the indicated convergent antibodies using deep mutational scanning analysis of yeast-displayed SARS-CoV-2 BA.1 RBD mutant libraries. The RBD surface is colored by a gradient ranging from no escape (white) to strong escape (red) at each site. See Fig. S10 for additional details. (F) Heatmap summarizing convergent antibody escape mutations present in the indicated SARS-CoV-2 Omicron sub-lineages. (G) Fab binding affinities of convergent antibodies utilizing the indicated germline genes for SARS-CoV-2 WT and Omicron sub-variant RBD antigens, as measured by BLI. Black bars indicate median affinities. Statistical comparisons were determined by (A and C) Kruskal-Wallis test with Holms corrected multiple pairwise comparisons, (B and D) Fisher’s exact test, or (G) Kruskal-Wallis test with subsequent Dunn’s multiple comparisons with WT. KD, equilibrium dissociation constant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Among neutralizing antibodies isolated at both time points, we observed significant over-representation of four IGHV germline genes (IGHV1–69, IGHV3–53/3–66, and IGHV3–9) (8) (Fig S8A). At the 5–6-month time point, over half (54%) of the neutralizing antibodies were encoded by one of these four germlines, with one-third of these antibodies utilizing IGHV1–69 (Fig. 4D, Fig. S8). We previously found that BA.1-neutralizing IGHV1–69 antibodies isolated from the early time point preferentially paired with the light chain germline IGLV1–40 and targeted an antigenic site overlapping that of the class 3 antibody COV2–2130 and non-overlapping with the ACE2 binding site (8). Similarly, 69% of IGHV1–69 antibodies isolated at 5–6 months paired with the IGLV1–40 germline and the majority (80%) failed to compete with ACE2 for binding (Fig. S9A and C). Likewise, >90% of IGHV3–9 antibodies identified from both time points recognized a non-ACE2-competitive binding site, although unlike IGHV1–69 antibodies, IGHV3–9 antibodies recognize an epitope overlapping S309 and REGN10987 as well as COV2–2130, suggesting a distinct mode of binding from IGHV1–69 antibodies (Fig. S9C) (8). Lastly, IGHV3–53/66 antibodies isolated from both time points were characterized by short HCDR3s (median = 11 to 12 nucleotide substitutions) compared with baseline HCDR3 lengths (median = 15 substitutions) and displayed competitive binding with the ACE2 receptor (Fig. S9B and C). Thus, convergent antibody classes dominated the neutralizing antibody response at both early and late time points following BA.1 breakthrough infection, suggesting little to no change in B cell immunodominance hierarchy over time.
Given the dominance of these public clonotypes in BA.1 breakthrough infection donors, we sought to determine their escape mutations in the BA.1 background. We randomly selected one to two antibodies belonging to each convergent germline and performed deep mutational scanning (DMS) analysis using a library encoding all possible amino acid substitutions from BA.1 (Fig. S10A) (13). Antibodies encoded by IGHV3–53 (ADI-75733) and IGHV3–66 (ADI-75732) displayed similar escape profiles, consistent with their shared sequence features and competitive binding profiles (Fig. 4E and Fig. S10C) (8). RBD positions N460 and F486, which are mutated in emergent variants (N460K in B.2.75, BA.2.75.2, BN.1, and BQ.1; F486S in BA.2.75.2; and F486V in BA.4/5, BA.4.6, and BQ.1.1), were associated with binding escape from IGHV3–53/66 antibodies (Fig. 4F and Fig. 10C). IGHV1–69 and IGHV3–9 antibodies both showed reduced binding to RBDs incorporating mutations at positions 344–349, 356, 452–453, 468, and 490. Notably, residues R346, K356, L452, and F490 are mutated across evolutionarily diverse Omicron sub-lineages, including BA.4.6 (R346T, L452R), BA.4/5 (L452R), BA.2.12.1 (L452Q), BJ.1 (R346T, F490V), BN.1 (R346T, K356T, F490S), and BQ.1.1 (R346T, L452R) (Fig. 4F and Fig. S10C). Consistent with these escape profiles, IGHV1–69 and IGHV3–9 class antibodies displayed reduced binding to BA.2.12.1 and BA.4/5 relative to early Omicron variants, likely due to the unique L452Q/R mutations present in these variants compared with BA.1 and BA.2 (Fig. 4G). Consistent with DMS-based predictions, both BA.2.75 and BA.4/5 RBDs displayed increased binding resistance to IGHV3–53/66 antibodies (Fig. 4F and 4G). Thus, convergent D614G/BA.1 cross-neutralizing antibodies recognize epitopes commonly mutated in recently emerging Omicron sub-variants, providing a molecular explanation for the high degree of antigenic convergence observed in recent Omicron sub-variant evolution and their increased level of immune evasion relative to BA.1.
In summary, BA.1 breakthrough infection in mRNA-vaccinated individuals induces broadly neutralizing serological and MBC responses that persist for at least six months after infection, supporting real-world studies showing that BA.1 breakthrough infection provides protection against symptomatic BA.1, BA.2, and BA.5 infection for at least 5–6 months (14–16). Furthermore, although the acute B cell response following breakthrough infection is primarily mediated by recall of cross-reactive vaccine-induced MBCs, these MBC clones accumulate somatic mutations and evolve increased breadth and potency for at least 6 months following infection. Although this enhanced neutralization breadth and potency was not reflected in the serum antibody response, it is possible that a second heterologous exposure may broaden the serological repertoire by activating these affinity matured MBCs, akin to the improved serum neutralization breadth observed following mRNA booster vaccination (17, 18). Nevertheless, our data indicate that infection or vaccination with antigenically divergent SARS-CoV-2 variants may provide long-term benefits by broadening pre-existing anti-SARS-CoV-2 B cell memory.
Finally, we found that convergent classes of neutralizing antibodies dominated the BA.1 breakthrough response at both early and late time points, reminiscent of the antibody response elicited following primarily infection or vaccination with early ancestral SARS-CoV-2 strains (19–21). The sustained prevalence of public clones that target residues frequently mutated in emerging Omicron subvariants suggests that this response is the driving force behind the continued antigenic drift of Omicron. Thus, in contrast to current approaches to the design of universal vaccines for certain highly antigenically variable viruses, such as HIV and influenza, which aim to focus the neutralizing response on a limited number of relatively conserved epitopes, the development of “variant-proof” COVID-19 vaccines may require a different strategy: engineering of spike-based immunogens that induce a diversity of neutralizing antibodies targeting numerous co-dominant epitopes, with the goal of limiting convergent immune pressure and therefore constraining viral evolution (22–24).
Supplementary Material
Acknowledgements:
We thank T. Boland for assistance with sequence analysis. We acknowledge E. Krauland, J. Nett, M. Vasquez, and C.G. Rappazzo for helpful comments on the manuscript. We also thank the Flow Cytometry and Genomics Shared Facilities at Fred Hutchinson Cancer Center. All IgGs were sequenced by Adimab’s Molecular Core and produced by the High-Throughput Expression group.
Funding:
T.N.S. is supported by the NIAID/NIH (K99AI166250). D.M. is funded by the NCI Cancer Center Support Grant (5P30 CA023108-41). D.R.B. is funded by the Bill and Melinda Gates Foundation INV-004923 and by the James B. Pendleton Charitable Trust. J.D.B. is supported by the NIH/NIAID (R01AI141707) and is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Competing interests: C.I.K. is a former employee and holds shares in Adimab. LLC. P.K., H.L.D., E.R.C., and J.C.G. are current employees and hold shares in Adimab LLC. L.M.W. is an employee and holds shares in Invivyd Inc. T.N.S. and J.D.B. consult with Apriori Bio. J.D.B. has consulted for Moderna and Merck on viral evolution and epidemiology. D.R.B. is a consultant for IAVI, Invivyd, Adimab, Mabloc, VosBio, Nonigenex, and Radiant. C.I.K. and L.M.W. are inventors on a provisional patent application describing the SARS-CoV-2 antibodies reported in this work. T.N.S. and J.D.B. may receive a share of intellectual property revenue as inventors on Fred Hutchinson Cancer Center–optioned technology and patents related to deep mutational scanning of viral proteins. The other authors declare that they have no competing interests.
Data and materials availability:
Omicron BA.1 yeast-display deep mutational scanning libraries are available from Addgene (accession # 1000000187). Complete computational pipeline with intermediate and final data files is available from GitHub: https://github.com/jbloomlab/SARS-CoV-2RBD_Omicron_MAP_Adimab. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. IgGs are available from L.M.W. under a material transfer agreement from Invivyd Inc.
References and Notes:
- 1.WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data, (available at https://covid19.who.int/).
- 2.Tseng H. F. et al. , Nat. Med. 28, 1063–1071 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tartof S. Y. et al. , Lancet Respir. Med. 10, 689–699 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q. et al. , Nature. 608, 603–608 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuekprakhon A. et al. , Cell. 185, 2422–2433.e13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qu P. et al. , BioRxiv (2022), doi: 10.1101/2022.08.14.503921. [DOI] [Google Scholar]
- 7.Coronavirus (COVID-19) Update: FDA Authorizes Moderna, Pfizer-BioNTech Bivalent COVID-19 Vaccines for Use as a Booster Dose | FDA, (available at https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-moderna-pfizer-biontech-bivalent-covid-19-vaccines-use).
- 8.Kaku C. I. et al. , Sci. Immunol. 7, eabq3511 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quandt J. et al. , Sci. Immunol. 7, eabq2427 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z. et al. , BioRxiv (2022), doi: 10.1101/2022.08.11.503601. [DOI] [Google Scholar]
- 11.Cao Y. et al. , BioRxiv (2022), doi: 10.1101/2022.09.15.507787. [DOI] [Google Scholar]
- 12.Chalkias S. et al. , N. Engl. J. Med. (2022), doi: 10.1056/NEJMoa2208343. [DOI] [Google Scholar]
- 13.Starr T. N. et al. , BioRxiv (2022), doi: 10.1101/2022.09.20.508745. [DOI] [Google Scholar]
- 14.Chemaitelly H. et al. , medRxiv (2022), doi: 10.1101/2022.02.24.22271440. [DOI] [Google Scholar]
- 15.Malato J. et al. , N. Engl. J. Med. 387, 953–954 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin D.-Y. et al. , N. Engl. J. Med. (2022), doi: 10.1056/NEJMc2209371. [DOI] [Google Scholar]
- 17.Muecksch F. et al. , Nature. 607, 128–134 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Beltran W. F. et al. , Cell. 185, 457–466.e4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robbiani D. F. et al. , Nature. 584, 437–442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnes C. O. et al. , Nature. 588, 682–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z. et al. , Nature. 592, 616–622 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanzavecchia A., Frühwirth A., Perez L., Corti D., Curr. Opin. Immunol. 41, 62–67 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Ward A. B., Wilson I. A., Curr. Opin. Immunol. 65, 50–56 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwong P. D., Mascola J. R., Immunity. 48, 855–871 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Briney B., Inderbitzin A., Joyce C., Burton D. R., Nature. 566, 393–397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers T. F. et al. , Science. 369, 956–963 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wec A. Z. et al. , Proc Natl Acad Sci USA. 117, 6675–6685 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gietz R. D., Schiestl R. H., Nat. Protoc. 2, 31–34 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Raybould M. I. J., Kovaltsuk A., Marks C., Deane C. M., Bioinformatics. 37, 734–735 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakharkar M. et al. , Sci. Immunol. 6 (2021), doi: 10.1126/sciimmunol.abg6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Omicron BA.1 yeast-display deep mutational scanning libraries are available from Addgene (accession # 1000000187). Complete computational pipeline with intermediate and final data files is available from GitHub: https://github.com/jbloomlab/SARS-CoV-2RBD_Omicron_MAP_Adimab. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. IgGs are available from L.M.W. under a material transfer agreement from Invivyd Inc.