Abstract
The class 1 integron integrase, IntI1, recognizes two distinct types of recombination sites, attI sites, found in integrons, and members of the 59-be family, found in gene cassettes. The efficiencies of the integrative version of the three possible reactions, i.e., between two 59-be, between attI1 and a 59-be, or between two attI1 sites, were compared. Recombination events involving two attI1 sites were significantly less efficient than the reactions in which a 59-be participated, and the attI1 × 59-be reaction was generally preferred over the 59-be × 59-be reaction. Recombination of attI1 with secondary sites was less efficient than the 59-be × secondary site reaction.
Gene cassettes are small mobile elements that generally include only a single gene and a downstream recombination site known as a 59-be (formerly 59-base element) (21, 31). Many of the antibiotic resistance genes found in gram-negative organisms are part of gene cassettes (19, 20), and cassettes that include genes for other functions have also been found in the small Vibrio cholerae chromosome (3, 28). The 59-be enable the recognition and mobilization of the cassettes by a site-specific recombinase (IntI integrase) that is encoded by an integron (11–13, 21, 27, 29). Gene cassettes are normally found inserted at a specific site (attI) in an integron, and this site is also recognized by the integrase (27, 33). Four classes of integron, each encoding a distinct IntI integrase, have been identified (see references 1, 10, 28, 31, and 36), and identical cassettes have been found residing in integrons of different classes, indicating that the pool of cassettes is shared. However, cassettes are also occasionally found integrated at secondary sites (2°rs) (32, 35).
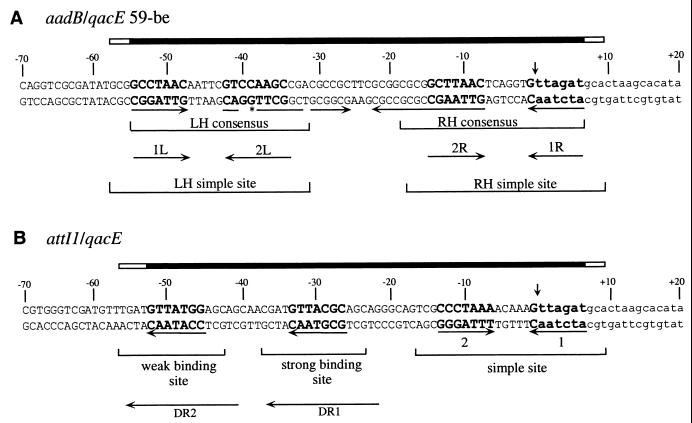
The attI-type sites and the 59-be-type sites have distinct architectures (Fig. 1). The 59-be sites have diverse sequences and lengths but share regions of about 25 bp at each end that conform to consensus sequences (13, 21, 37). The consensus regions are imperfect inverted repeats of one another, and each comprises a pair of inversely oriented integrase-binding domains, separated by a spacer of 7 or 8 bp (Fig. 1A) (37). The arrangement of each consensus region is similar to that of the simple sites recognized by other integrases. In contrast, the sequences of the attI sites are not closely related to each other and do not share most of these features (15, 22, 33). The attI1 site (Fig. 1B), the only attI site that has been examined in detail (22, 23, 30, 33), includes only one simple site structure, together with two further directly repeated integrase-binding domains located to the left (15, 18, 22). Since the 59-be and the attI sites represent two distinct types of recombination site, three formally distinct reactions, attI1 × 59-be, 59-be × 59-be, and attI1 × attI1, are all possible.
FIG. 1.
Structure of 59-be and attI1 sites. (A) Sequence of the aadB/qacE 59-be (bases 1810 to 1899 in reference 8) showing the extent of the 59-be defined by consensus (filled bar) and further bases predicted to be protected by bound IntI1 (15) and thus potentially involved in IntI1 recognition (open bar). Seven-base-pair putative core sites related to the core site consensus sequence (1L, 2L, 2R, and 1R; see reference 37) are in boldface type, and their relative orientations are indicated by arrows. An extra base in 2L is marked with an asterisk. Inverted repeats are underscored with arrows. The extents of the LH and RH 59-be consensus sequences and of the LH and RH potential simple sites (37) are indicated. The recombination crossover point is marked by a vertical arrow, and bases derived from the qacE cassette are in lowercase. (B) Sequence of attI1/qacE (bases 1219 to 1288 and bases 1880 to 1899 in reference 8), showing the extent of attI1 as defined by Partridge et al. (30). The core sites (1 and 2) of the simple site, the positions of weak and strong IntI1 binding sites determined experimentally (15) and of a pair of direct repeats DR1 and DR2 that overlap these binding sites, are shown. Core site sequences and the recombination crossover point are marked as for panel A.
The reactions catalyzed by the IntI1 integrase encoded by class 1 integrons have been studied most extensively. IntI1 can catalyze both integrative and excisive recombination events, and recombination between two 59-be, between attI1 and a 59-be, and between two attI1 sites has been documented (11–13, 21–23, 27, 30, 33, 37). In each of these reactions, the recombination crossover occurs between the G and TT (arrow in Fig. 1) (23, 37), indicating that only a single strand exchange is likely to be involved (37). IntI1 also recognizes the attI2 and attI3 sites from the integron classes 2 and 3, though the efficiency of recombination between attI2 or attI3 and a 59-be is much lower than that of the equivalent attI1 × 59-be reaction (22). Recombination of attI1 and 59-be with 2°rs has also been reported (16, 17, 23, 30, 33, 37).
The simplest potential route that leads to incorporation of gene cassettes into an integron involves IntI1-mediated conservative site-specific recombination between the 59-be in a circularized cassette and a site in the integron (21). In the wild, the circularized cassettes are presumably generated by the reverse reaction, namely, excision from an integron (13). If the recipient integron contains no cassettes, the cassette integration reaction is necessarily between attI1 and a 59-be, and this reaction has been demonstrated experimentally for IntI1 by transforming circular cassettes generated in vitro into cells containing a cassette-free integron (11). However, if, as is generally the case, one or more cassettes are already present in the integron, then recombination between two 59-be is also an option, and the incoming cassette can, in theory, be incorporated at a number of different positions. Thus, to understand cassette movement in general, it is necessary to know the relative efficiencies of the various possible reactions. Cassette integration events involving recombination between two 59-be were not observed in the assay for integration of circular cassettes (11), suggesting that recombination between attI1 and a 59-be is either far more efficient than recombination between two 59-be or that there is a mechanism that ensures preferential recognition of the attI1 site. However, in the experimental system developed by Martinez and de la Cruz (26, 27), which is the simplest, most widely used, and only quantifiable assay available, the preference for the attI1 × 59-be reaction over the 59-be × 59-be reaction is not consistently observed.
In the cointegration assay, the activity of any cloned recombination site can be tested by measuring cointegrate formation resulting from recombination between the cloned site and attI1 or either of the two 59-be, dfrB2/orfA (composite site at junction of first-named cassette/second-named cassette) and orfA/qacE, in the integron In3 found in the conjugative plasmid R388 (Fig. 2A). The site in In3 that participated in the reaction is determined by mapping the resultant cointegrates. When the activity of cloned 59-be was examined, recombination between the cloned site and attI1/dfrB2 in R388 was the predominant event in some cases (27), while in others only recombination between the cloned 59-be and the orfA/qacE 59-be was observed (21, 27, 37). The activity of the aadA1/qacE 59-be contained in fragments that include different lengths of the aadA1 cassette was measured in the study of Martinez and de la Cruz (27). With longer fragments 88% of the cointegrates were formed by recombination with the orfA/qacE 59-be of R388, but with shorter fragments the majority of the cointegrates were formed by recombination at attI1/dfrB2. The cloned orfA/qacE 59-be also recombined predominantly with attI1. These authors argued that the segment of aadA1 cassette DNA present in the larger cloned fragments was responsible for the change in recombination site specificity. However, in the study of Hall et al. (21), three different 59-be recombined exclusively with the R388 orfA/qacE 59-be, even though the relevant aadA1 region was not present. In a subsequent study, the cloned orfA/qacE 59-be also recombined exclusively with orfA/qacE in R388 (37). In addition, when the cloned site was attI1, the products of recombination between two attI1 sites were not detected (33), though such events have since been reported (23, 30).
FIG. 2.
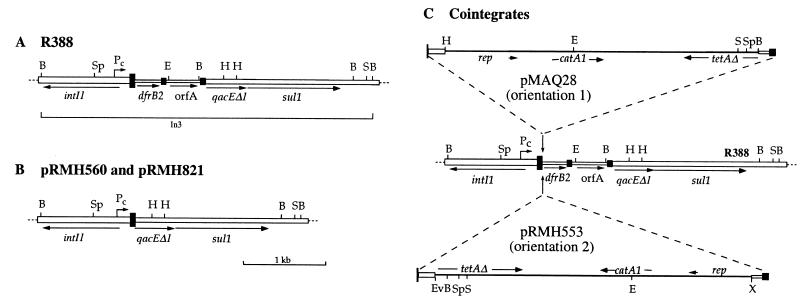
Structure of plasmids and cointegrates. (A) Integron region in R388. (B) Cassette-free integron region in pRMH560 and pRMH821. (C) Cointegrates formed with sites cloned in pACYC184. The genes are marked with horizontal arrows. The position of the Pc promoter is also marked with a bent arrow. The attI1 site is represented by a large filled box, and the 59-be are marked by smaller filled boxes. pMAQ28 and pRMH553 contain the aadB/qacE 59-be cloned in opposite orientations (see text). pACYC184 sequence is represented by a line, and open boxes represent cloned sequences adjacent to the 59-be. The replication (rep) region of pACYC184 and ori (arrowhead) are marked. Restriction sites: B, BamHI; E, EcoRI; Ev, EcoRV; H, HindIII; S, SalI; Sp, SphI; X, XbaI.
We have sought here to establish assay conditions which accurately reflect the relative efficiencies of the reactions catalyzed by IntI1. To achieve this end, the differences in the apparent specificity for the attI1 or 59-be sites in R388 sites observed in different laboratories have been examined and traced to an effect of the orientation of the cloned recombination site in the vector pACYC184. When the cloned site is in one orientation, cointegrates resulting from recombination at attI1 in R388 are not recovered. The relative efficiencies of the three main types of recombination event have been reexamined under conditions where this bias does not occur.
MATERIALS AND METHODS
Bacterial strains.
Escherichia coli UB5201 is F− pro met recA56 gyrA, E.coli UB1637 is F− his lys trp recA56 rpsL, and E. coli DH5α is supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1. Bacteria were routinely cultured in Luria-Bertani (LB) medium or LB agar supplemented as appropriate with ampicillin (100 μg/ml), chloramphenicol (25 μg/ml), nalidixic acid (NAL; 25 μg/ml), streptomycin (25 μg/ml), sulfamethoxazole (25μg/ml), or trimethoprim (TMP; 25 μg/ml). Antibiotics were obtained from Sigma. Mueller-Hinton agar (Bacto Laboratories) containing TMP (25 μg/ml) was used for screening for TMP sensitivity.
Plasmids.
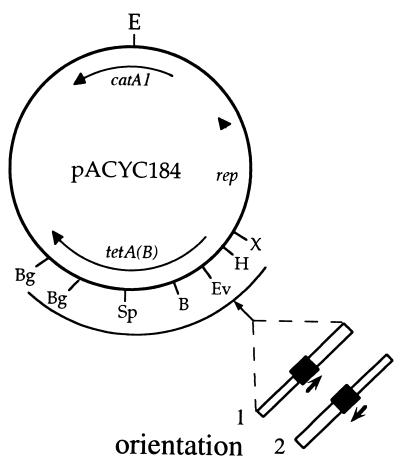
Plasmids are listed in Table 1. Short fragments containing 59-be or attI1 recombination sites were cloned into the same general region of pACYC184 (9), either into the unique EcoRV site (position 1680 in reference 34; accession no. X06403) or into pairs of sites located between positions 1425 (XbaI) and 2658 (BglI) (Table 1; see also Fig. 3). In some plasmids the promoter for the tetA(B) gene is absent or inactivated (see below), and in all cases the tetA(B) gene is interrupted. Orientation 1 (see text below and Fig. 3) refers to plasmids in which the 3′ end (righthand end in Fig. 1) of each composite site is closest to the origin of replication of pACYC184, while orientation 2 is the opposite.
TABLE 1.
Plasmids
| Plasmid | Description | Cloned recombination sitea | Position in pACYC184b | Relevant phenotype | Source or reference |
|---|---|---|---|---|---|
| R388 | 33-kb IncW plasmid containing In3 (cassettes dfrB2-orfA) | NA | Tpr Sur Tra+ IntI1+ | 2 | |
| pRMH560 | Cassette-free derivative of R388 | NA | Sur Tra+ IntI1+ | 30 | |
| pRMH821 | pRMH560 derivative with mutated Pc sequence | NA | Sur Tra+ IntI1+ | This study | |
| pACYC184 | Cloning vector | NA | Tcr Cmr | 9 | |
| pSU2056 | 1,176-bp RsaI-BamHI fragment of In2 in pUC9 | NA | Apr IntI1+ | 27 | |
| pMAQ28 | 400-bp Sau3A-HindIII fragment | aadB/qacE 59-be | BamHI-HindIII (1) | Cmr | 21 |
| pMAQ34 | 806-bp HindIII-BamHI fragment | aadA2/cm/A 59-be | HindIII-BamHI (2) | Cmr | 21 |
| pMAQ53 | 344-bp BglII-BssHII fragment | aacC1/orfE 59-be | BamHI-EcoRV (1) | Cmr | This study |
| pMAQ74 | 317-bp BglI-HindIII fragment | orfD/qacE 59-be | BglI-HindIII (1) | Cmr | This study |
| pMAQ77 | 328-bp Sau3A-NlaIV fragment | oxa2/orfD 59-be | BamHI-EcoRV (1) | Cmr | This study |
| pMAQ93 | 360-bp HindIII fragment | aadA2/qacE 59-be | HindIII (1) | Cmr | This study |
| pMAQ541 | 203-bp SphI fragment | aadB/oxa2 59-be | SphI (1) | Cmr | This study |
| pMAQ542 | Same as pMAQ541 | aadB/oxa2 59-be | SphI (2) | Cmr | This study |
| pRMH173 | 321-bp Sau3A-HindIII fragment | oxa2/qacE 59-be | BamHI-HindIII (1) | Cmr | This study |
| pRMH251 | 262-bp TaqI-HindIII fragment | attIl/qacE | EcoRV (1) | Cmr | 33 |
| pRMH313 | Same as pRMH251 | attII/qacE | EcoRV (2) | Cmr | 30 |
| pRMH553 | 352-bp BstXI-HindIII fragment | aadB/qacE 59-be | EcoRV-XbaI (2) | Cmr | This study |
| pRMH555 | 976-bp EcoRV fragment | dfrA7/qacE 59-be | EcoRV (1) | Cmr | This study |
| pRMH556 | Same as pRMH555 | dfrA7/qacE 59-be | EcoRV (2) | Cmr | This study |
| pRMH638 | 395-bp NaeI-HindIII fragment | aadAI/qacE 59-be | EcoRV (1) | Cmr | This study |
| pRMH642 | 307-bp BamHI-HindIII fragment | orfA/qacE 59-be | EcoRV (1) | Cmr | This study |
| pRMH643 | Same as pRMH642 | orfA/qacE 59-be | EcoRV (2) | Cmr | This study |
| pRMH800 | Same as pRMH638 | aadAI/qacE 59-be | EcoRV (2) | Cmr | This study |
| pRMH814 | 493-bp BstNI-BamHI fragment | dfrB2/orfA 59-be | SalI-ClaI (1) | Cmr | This study |
| pRMH823 | 173-bp BstNI-EcoRI fragment | dfrB2/orfA 59-be | EcoRV (2) | Cmr | This study |
All sites were cloned into pACYC184. The sites found in integrons that contain gene cassettes are composite sites in which the bulk of the site comes from one module (5′-CS for attI1 or the first named cassette for 59-be) and the remainder (lowercase in Fig. 1) from the adjacent module (second-named cassette). The original attI1 site is found in the ancestral integron when it contains no cassettes, and each original 59-be is found in the free circular cassette (13).
Numbers in parentheses represent the orientation of the cloned fragment with respect to pACYC184 (see text for details). NA, not applicable.
FIG. 3.
Structure of pACYC184-based plasmids containing cloned recombination sites. The fragments containing recombination sites were cloned in the region between the BglI (Bg) and XbaI (X) sites (positions 2658 and 1425, respectively, under accession no. X06403), which is delineated by an arc. The orientation of the recombination sites (filled box) is shown by an arrow. Other symbols are as in Fig. 2.
The plasmid pRMH560 (Fig. 2B), a derivative of R388 that has lost both the dfrB2 and orfA cassettes, was generated by IntI1-mediated excision of cassettes as described previously (12). pRMH821 (Fig. 2B) is a derivative of pRMH560 in which the strong Pc promoter (−35 TTGACA, −10 TAAACT) found in R388 and pRMH560 is replaced by the 20-fold-weaker version (−35 TGGACA, −10 TAAGCT) known as the weak promoter. It was generated by IntI1-mediated resolution (21) of a cointegrate between pMAQ28 and pRMH560 that was found to contain the weak Pc (see Results). Plasmid pSU2056, present in the donor cells for cointegration assays as the source of IntI1, contains the weak version of Pc, and the substitutions found in some cointegrates may have arisen by RecA-independent recombination with pSU2056.
DNA procedures.
Plasmid DNA was isolated using an alkaline lysis method (4) and purified if necessary using Wizard Miniprep kits (Promega). For restriction mapping of cointegrates, digests were carried out according to the enzyme manufacturers' instructions, and fragments were separated on 1% (wt/vol) or 0.8% (wt/vol) agarose gels using EcoRI-digested bacteriophage SPP-1 DNA (Bresatec) and BamHI-digested R388 or pRMH560 as size markers. DNA fragments for cloning were isolated from agarose gels using a Geneclean II kit (Bio 101). Plasmid DNA for sequencing was prepared using a Wizard Miniprep kit. For manual sequencing, DNA was annealed to the primer according to the method of Jones and Schofield (24), and double-strand DNA sequencing was performed using a Sequenase kit, version 2.0 (U.S. Biochemicals), with reaction mixtures containing dITP. Alternatively, automated sequencing was carried out at the Macquarie Sequencing Facility, Macquarie University Sydney, using an ABI Prism 377 DNA sequencer (PE Biosystems) and Big Dye terminator mixes.
Conduction assays.
Recombination efficiency was determined using a conduction (mating-out) assay (21, 26, 27, 37) which measures conduction of a pACYC184-based plasmid, containing a cloned recombination site, from the donor strain UB1637 (RecA− Smr [streptomycin resistant]) to a recipient strain UB5201 (RecA− Nxr [NAL resistant]) or DH5α (RecA− Nxr). Donor cells also contained the Tra+ plasmid R388 (Tpr [TMP resistant] Sur [sulfamethoxazole resistant]) or one of its cassette-free derivatives pRMH560 (Sur) or pRMH821 (Sur) (Table 1; Fig. 2). IntI1 integrase was supplied in trans by the plasmid pSU2056 (Apr [ampicillin resistant]) (27). R388, pRMH560 or pRMH821 was introduced into the donor cells first, followed by the pACYC184-based plasmid containing the cloned recombination site and, finally, pSU2056. That each isolate had a full complement of plasmids after purification was confirmed by screening single colonies grown on LB agar for resistance to the relevant antibiotics.
For matings, 0.1 ml of stationary-phase cultures of donor and recipient cells were mixed, spread over the surface of an LB agar plate, and incubated overnight at 37°C. Cells were then harvested, and transconjugants were selected on agar containing TMP and NAL (R388) or sulfamethoxazole and NAL (pRMH560 and pRMH821), while cointegrates were selected on plates containing chloramphenicol and NAL. The conduction frequency was expressed as the ratio of Cmr (chloramphenicol-resistant) to Tpr or Sur transconjugants. Assays were repeated several times on at least two independent isolates obtained after transformation with pSU2056 and the mean value determined. For conduction frequencies below 10−6, where spontaneous mutation of the donor cells to Nxr can contribute significantly to the number of Cmr Nxr colonies detected, all colonies were screened for streptomycin resistance, which is characteristic of the donor.
Analysis of cointegrates.
In most cases, cointegrates resulting from recombination at attI1/dfrB2 in R388 could be differentiated from those at the dfrB2/orfA or the orfA/qacE 59-be by screening for TMP sensitivity, since in the former the dfrB2 gene conferring TMP resistance is separated from its promoter Pc, and cells containing the resulting cointegrates are Tps (Fig. 2C). In certain cases, however, the Ptet promoter was intact and correctly oriented to transcribe dfrB2 and confer Tpr even when the site of cointegration was attI1. In such cases, the site of cointegration was determined either by restriction mapping or by using E. coli DH5α as the recipient in the conduction assays. DH5α has a lower intrinsic resistance to TMP than the normal recipient, E. coli UB5201, and differentiation of the two levels of TMP resistance is possible in this background.
For pRMH560 and pRMH821 and for cointegrates where screening for TMP sensitivity was not applicable, the site of insertion of the test plasmid (attI1 or 2°rs) was determined by restriction mapping of cointegrate plasmid isolates with BamHI, BamHI-HindIII, or SphI as described previously (21, 33, 37). Cointegrates formed by recombination between pRMH560 and pACYC184 were initially mapped using BamHI. Isolates that appeared to be of the same map types were then mapped more precisely using combinations of the enzymes BamHI, EcoRI, HindIII, NcoI, and PvuII.
RESULTS
Activity of 59-be.
A number of different 59-be have been shown to be active IntI1-specific recombination sites by using a variety of assay systems (7, 12, 21, 27, 37). In this study, several different 59-be were cloned into the vector pACYC184, and their activities were assayed by measuring the frequency of recovery of cointegrates formed between the test plasmid (containing the 59-be) and any of the three sites in the conjugative plasmid R388 (Fig. 2A). The sites in R388 are the composite sites attI1/dfrB2 and the dfrB2/orfA and orfA/qacE 59-be, but for simplicity they will hereafter be referred to as attI1, dfrB2 59-be, and orfA 59-be, as the bulk of each site comes from the first-named module. All the 59-be tested here were also composite sites, and all of them were able to form cointegrates with R388 when IntI1 integrase was present (Table 2). However, the frequency varied over a large range from 8.5 × 10−5, which is only five fold higher than the background, to 1.1 × 10−2, indicating that 59-be differ markedly in their efficiency as IntI1 recombination sites.
TABLE 2.
Cointegration frequency of 59-be
| Plasmid | 59-be | Fragment lengtha | Orientation in vector | Cointegration frequency
|
R388 recombination site usedb | % Tpsc | ||
|---|---|---|---|---|---|---|---|---|
| No. of assays | Range | Mean | ||||||
| pACYC184 | 13 | 1.5 × 10−6–3.2 × 10−5 | 1.6 × 10−5 | |||||
| pMAQ28 | aadB/qacE | 202/198 | 1 | 5 | 4.5 × 10−3–1.6 × 10−2 | 1.1 × 10−2 | 60 orfA | <0.1 |
| pMAQ541 | aadB/oxa2 | 120/83 | 1 | 5 | 2.4 × 10−4–1.6 × 10−3 | 1.0 × 10−3 | 18 orfA | <0.4 |
| pRMH638 | aadAI1/qacE | 197/198 | 1 | 10 | 2.8 × 10−4–2.7 × 10−3 | 1.1 × 10−3 | 44 orfA | 2d |
| pMAQ93 | aadA2/qacE | 162/198 | 1 | 8 | 2.3 × 10−3–3.7 × 10−2 | 1.0 × 10−2 | 46 orfA, 4 2°rs | 1 |
| pMAQ34 | aadA2/cmlA | 162/644 | 2 | 12 | 3.4 × 10−4–2.3 × 10−3 | 1.2 × 10−3 | 16 attI1; 5 orfA | 87 |
| pRMH642 | orfA/qacE | 109/198 | 1 | 10 | 4.9 × 10−4–7.5 × 10−3 | 2.4 × 10−3 | 28 orfA | 8d |
| pRMH814 | dfrB2/orfA | 96/397 | 2 | 6 | 9.5 × 10−4–4.9 × 10−3 | 1.8 × 10−3 | 33 attI1; 3 2°rs | 89 |
| pRMH823 | dfrB2/orfA | 96/77 | 2 | 3 | 5.0 × 10−3–9.5 × 10−3 | 8.0 × 10−3 | 11 attI1 | 99 |
| pMAQ74 | orfD/qacE | 119/198 | 1 | 6 | 2.7 × 10−4–1.3 × 10−3 | 8.7 × 10−4 | 13 orfA | 0.3 |
| pRMH173 | oxa2/qacE | 123/198 | 1 | 5 | 1.1 × 10−4–5.2 × 10−4 | 2.8 × 10−4 | 15 orfA | <0.4 |
| pMAQ77 | oxa2/orfD | 123/205 | 1 | 10 | 5.3 × 10−5–1.0 × 10−3 | 8.9 × 10−4 | 3 orfA | 4 |
| pMAQ53 | aacC1/orfE | 317/27 | 1 | 7 | 1.6 × 10−5–7.2 × 10−4 | 2.9 × 10−4 | 49 orfA | <0.2 |
| pRMH555 | dfrA7/qacE | 221/755 | 1 | 12 | 1.9 × 10−5–2.7 × 10−4 | 8.5 × 10−5 | 27 orfA | NA |
Numbers refer to nucleotides in the cloned fragment to the left and right of the recombination crossover point.
Randomly selected cointegrates were mapped.
At least 100 cointegrates were tested. NA, not applicable.
All Tps recombinants examined had lost the dfrB2 cassette (insert order orfA-X, where X is the test plasmid) or duplicated the orfA cassette with integration occurring at the new orfA/dfrB2 59-be (orfA-X-dfrB2-orfA).
Recombinant types.
The proportion of cointegrates arising from each of the formally distinct attI1 × 59-be and 59-be × 59-be recombination events was examined both by screening for sensitivity to TMP and by mapping a number of cointegrates recovered from each cross as described previously (21). In most cases, cointegrates in which the test plasmid is integrated at attI1 can be identified by their Tps phenotype, since in these the dfrB2 gene, which confers resistance to TMP, is separated from its promoter Pc, and TMP resistance is not expressed (Fig. 2C). For most of the 59-be tested, >99% of the cointegrates were Tpr and had resulted from recombination of the test 59-be with the orfA 59-be in R388 (Table 2), indicating that this is the predominant event. Furthermore, in these cases, all of the rare Tps cointegrates examined had lost the dfrB2 cassette (Tpr) by IntI1-mediated excision from R388 or the cointegrate, and no recombinants arising from recombination of the test 59-be with attI1 or the dfrB2 59-be in R388 were detected. However, two of the sites tested here, the dfrB2/orfA and aadA2/cmlA 59-be, were exceptions; the majority of recombinants were Tps, diagnostic of recombination at attI1, and this was confirmed by restriction mapping.
Since others have shown that the orfA/qacE and aadA1/qacE 59-be are both able to recombine with the attI1 site in R388 (27), an explanation was sought for why cointegrates resulting from these events were not detected here when the same 59-be were tested. All of the 59-be examined here, except the dfrB2/orfA and aadA2/cmlA 59-be, are cloned in the same orientation (orientation 1 in Fig. 3) within a short region of the vector pACYC184. The dfrB2/orfA and aadA2/cmlA 59-be are cloned in the same region, but are in orientation 2. To examine the possibility that the orientation of the 59-be in the vector affects which of the recombination sites in R388 appear to be used, some of the other 59-be were recloned in orientation 2 in pACYC184 and retested. In all cases, a substantial proportion of the cointegrates recovered conferred a Tps phenotype (Table 3), indicating that recombination had now taken place at attI1 in R388, and this was confirmed by mapping randomly selected cointegrates. In two cases Tps recombinants also arose from events involving an R388 variant that had acquired an extra copy of the orfA cassette (orfA-dfrB2-orfA) and the orfA/dfrB2 site participated in the integrative recombination event. The Tpr cointegrates had all arisen from recombination between either the dfrB2 or orfA 59-be in R388 and the cloned 59-be. Thus, cointegrates arising from events involving the dfrB2 59-be in R388 were also now recovered (Table 3). It is clear that each of the 59-be tested in orientation 2 can recombine with attI1 and with at least one of the 59-be in R388.
TABLE 3.
Partner site preferences of 59-be cloned in orientation 2 in pACYC184
| Plasmid | 59-be | Fragment lengtha | Cointegration frequency
|
% Tpsb (range) | No. mappedc | R388 recombination site used | ||
|---|---|---|---|---|---|---|---|---|
| No. of assays | Range | Mean | ||||||
| pRMH553 | aadB/qacE | 154/198 | 9 | 2.6 × 10−3–1.0 × 10−2 | 8.4 × 10−3 | 88 (84–91) | 17 | 15 attI1; 2 orfA 59-be |
| 6 | 2.4 × 10−4–1.8 × 10−3 | 9.6 × 10−4 | 38 (32–45) | 13 | 6 attI1; 5 orfA 59-be; 2 2°rs | |||
| 4 | 3.8 × 10−3–2.4 × 10−2 | 1.1 × 10−2 | 99 (98–99) | 12 | 12 attI1 | |||
| pRMH643 | orfA/qacE | 109/98 | 9 | 7.3 × 10−5–2.0 × 10−3 | 5.1 × 10−4 | 48 (27–66) | 23 | 14 attI1; 5 dfrB2 59-be; 4 orfA 59-be |
| 4 | 1.8 × 10−4–9.9 × 10−3 | 3.6 × 10−3 | 85 (78–90) | 24 | 20 attI1; 3 orfA 5-be; 1 2°rs | |||
| pRMH800 | aadA1/qacE | 197/198 | 8 | 2.0 × 10−3–1.6 × 10−2 | 6.9 × 10−3 | 63 (45–79) | 32 | 19 attI1; 4 dfrB2 59-be; 4 orfA 59-be; 5 2°rs |
| pMAQ542 | aadB/oxa2 | 120/83 | 6 | 2.0 × 10−4–3.7 × 10−3 | 1.7 × 10−3 | NA | 18 | 5 attI1; 4 dfrB2 59-be; 9 orfA 59-be |
| pRMH556 | dfrA7/qacE | 221/755 | 9 | 2.8 × 10−4–9.2 × 10−4 | 6.3 × 10−4 | NA | 16 | 15 attI1; 1 orfA 59-be |
| 6 | 5.4 × 10−4–2.9 × 10−3 | 1.3 × 10−3 | 29 | 23 attI1; 3 orfA 59-be; 3 2°rs | ||||
Numbers refer to nucleotides in the cloned fragment to the left and right of the recombination crossover point.
Average with range of values from individual crosses shown in parentheses. NA, not applicable
Randomly selected recombinants were mapped.
To further examine this phenomenon, a derivative of R388 with no cassettes (pRMH560; Fig. 2B) and thus containing only the attI1/qacE recombination site (30) was constructed and substituted for R388 in the assay. Recombination with the aadB/qacE and orfA/qacE 59-be cloned in both orientations was measured (Table 4). For the 59-be in orientation 2 (pRMH553 and pRMH643), the frequencies were high, and all of the recombinants mapped (21 and 25, respectively) were the products of site-specific recombination with attI1 in pRMH560. When the 59-be were in orientation 1 (pMAQ28 and pRMH642), the frequencies of cointegrate formation were at least 1,000-fold lower than for orientation 2 and close to the background level for the vector (Table 4), indicating that no or a very low level of recombination involving the cloned 59-be sites had occurred. Thus, with the 59-be site in orientation 1 the products of recombination with attI1 are either not formed or not recovered efficiently.
TABLE 4.
Effect of 59-be site orientation on recombination with attI1
| Plasmida | Recombination site | Orientation | Mean cointegration frequencyb
|
||
|---|---|---|---|---|---|
| R388c | pRMH560 | pRMH821d | |||
| pMAQ28 | aadB/qacE 59-be | 1 | 1.1 × 10−2 (5) | 1.2 × 10−7 (7) | 5.3 × 10−2 (4) |
| pRMH553 | aadB/qacE 59-be | 2 | 9.2 × 10−3 (13) | 6.3 × 10−2 (6) | 4.4 × 10−2 (4) |
| pRMH642 | orfA/qacE 59-be | 1 | 2.4 × 10−3 (10) | 5.5 × 10−6 (6) | ND |
| pRMH643 | orfA/qacE 59-be | 2 | 1.5 × 10−3 (13) | 3.6 × 10−3 (4) | ND |
| pRMH251 | attI1/qacE | 1 | 7.3 × 10−3 (14) | 4.3 × 10−8 (11) | ND |
| pRMH313 | attI1/qacE | 2 | 3.5 × 10−3 (9) | 2.5 × 10−5 (6) | ND |
| pACYC184 | 1.6 × 10−5 (13) | 5.4 × 10−7 (14) | ND | ||
The strength of the Pc promoter in R388 influences recovery of cointegrates.
To determine if any recombination had occurred between attI1 in pRMH560 and the 59-be in orientation 1, the rare recombinants recovered were mapped. A total of 23 cointegrates, three to four recombinants from each of six independent crosses, formed between pMAQ28 and pRMH560 were examined. All appeared to have arisen by recombination between attI1 and the aadB/qacE 59-be. However, for five cointegrates (from two crosses) the predicted 7-kb fragment generated by cleavage at the BamHI sites in the 5′- and 3′-CS (see Fig. 2B) was slightly smaller than expected. Further mapping revealed that either a 0.4-kb (two cointegrates from one cross) or a 0.7-kb (three cointegrates from another cross) segment of the 5′-CS that includes the SphI site was missing. This suggested that the deletion of Pc, which is located 0.3 kb to the right of this SphI site, might be the factor allowing the recovery of cointegrates.
Although the remaining 18 of 23 cointegrates were indistinguishable from the predicted product of recombination at attI1, when the 5′-CS of one recombinant from each of the six crosses was sequenced, mutations in Pc were detected. In all cases the −35 region had undergone a change from TTGACA to TGGACA, and in four of the six, the −10 sequence was also changed from TAAACT to TAAGCT. Different class 1 integrons contain variants of Pc which differ in strength over a 20- to 30-fold range (6, 14, 25), and the version normally present in R388 (TTGACA with TAAACT) is the strongest. The observed substitutions reduce the strength of Pc by 10- to 20-fold (6, 14, 25), and it is possible that survival of these particular cointegrates might be a result of this.
To further examine this hypothesis, a derivative of pRMH560 with the weak Pc configuration TGGACA (17 bp) TAAGCT was recovered by IntI1-mediated resolution of one of the cointegrates, and this plasmid, pRMH821, was substituted for pRMH560 in the assay. With the aadB/qacE 59-be cloned in orientation 1, the cointegration frequency was 5.3 × 10−2 (Table 4), which is substantially higher than that obtained with pRMH560 and close to the value observed for orientation 2 using pRMH560. Restriction mapping of four cointegrates from each of four crosses showed that they were the products of recombination at attI1 in pRMH821. Taken together, these results show that, for 59-be sites cloned in orientation 1, cointegrates resulting from recombination at attI1 are not recovered when Pc is strong but can be recovered when Pc is weak.
Efficiencies of recombination of a 59-be with a 59-be or attI1 partner.
Using data obtained exclusively with sites cloned in orientation 2 in pACYC184, it is possible to assess the relative efficiencies of the various reactions catalysed by IntI1. When the cloned 59-be had a choice of partner sites in R388, the distribution of cointegrate types recovered from individual crosses varied over a considerable range (Tables 2 and 3). Generally, the fraction of Tps recombinants was reproducible for individual donor strain isolates but differed between isolates, and this may indicate that much of the recombination occurs during a short period after the introduction of the plasmid encoding IntI1. However, cointegrates resulting from recombination at attI1 were generally recovered at a higher frequency than cointegrates resulting from recombination at the dfrB2 or orfA 59-be. Thus, each of the 59-be is not only able to recombine with attI1 but attI1 is also likely to be the overall preferred partner site for the 59-be tested here. In the case of the dfrB2/orfA 59-be (pRMH814 and pRMH823), the bias toward recombination with attI1 was consistently extremely high, and no recombinants formed with the orfA 59-be were recovered (Table 2). It is therefore possible to conclude that, though the properties of individual 59-be may influence the outcome, overall the attI1 × 59-be recombination events occur at a higher efficiency than 59-be × 59-be events.
Recombination between two attI1 sites.
In a previous study (33), which examined recombination of cloned attI1/qacE fragments with R388, no cointegrates arising from recombination of attI1/qacE with the attI1 site in R388 were found. However, the attI1/qacE fragments used were all in orientation 1 in pACYC184. One of these attI1/qacE fragments was therefore recloned in orientation 2 (pRMH313) and retested. The recombination frequency was similar to that with the orientation 1 clone (Table 4). However, 1 cointegrate arising from recombination at attI1 in R388 was now found among 18 mapped. The fact that the majority of recombination events involved the orfA 59-be in R388 indicates that the efficiency of attI1 × attI1 recombination is at least 10-fold lower than the attI1 × 59-be reaction. Thus, though recombination between two attI1 sites can occur, the attI1 × 59-be recombination reaction is more efficient and is thus highly preferred if a choice of sites is available.
This conclusion was confirmed by testing the ability of the attI1/qacE recombination site to form cointegrates with the attI1/qacE site in pRMH560. In orientation 2, the observed recombination frequency was low (2.5 × 10−5), but most of the cointegrates mapped (22 of 27) had resulted from legitimate recombination events involving the two attI1 sites. When attI1/qacE was in orientation 1 the level of recombination was close to the background level (Table 4), and all 24 of the cointegrates mapped had arisen by recombination between the attI1 site in one plasmid and a 2°rs in the other. The absence of attI1 × attI1 recombination confirms that the products of this reaction are not recovered when the cloned site is in orientation 1.
Recombination of 59-be and attI1 with 2°rs.
The frequency of recombination between pRMH560 and pACYC184, which provides a direct measure of attI1 × 2°rs recombination, was 30-fold lower than the corresponding value for R388 and pACYC184 (Table 4). The frequency of recombination of R388 with pACYC184 measures recombination of 59-be with 2°rs (17, 33), since all of the cointegrates analyzed had arisen by recombination between the orfA 59-be and a 2°rs in pACYC184. Products of recombination between attI1 and a 2°rs were not found among 51 recombinants examined by Francia et al. (17) or 13 recombinants examined by Recchia et al. (33). Thus, recombination of the orfA 59-be with 2°rs is more efficient than recombination of attI1 with 2°rs.
DISCUSSION
When the various reactions catalyzed by IntI1 were assayed under conditions where cointegrates formed by all potential recombination events could be recovered, it was found that the fraction of recombination events between a 59-be and either a 59-be or the attI1 site was highly variable between independent isolates of the donor strain. However, the 59-be × attI1 reaction was generally most efficient, and this reflects the results obtained by assaying the incorporation of free gene cassettes into an integron that contains a cassette (11). In the case of the dfrB2/orfA 59-be, this bias was consistently very large, and it is likely therefore that 59-be differ in their preferences for partner sites. Recombination events involving two 59-be were overall less efficient than recombination between attI1 and a 59-be, but far more efficient than recombination between two attI1 sites.
The data presented here have resolved the discrepancy between experimental studies of IntIl-mediated cassette integration using two different experimental systems. When circularized gene cassettes were transformed into cells containing a plasmid with an integron carrying one or more integrated cassettes, the incoming cassette was always inserted at the attI1 site to take up the first position in the cassette array (11). In contrast, in the cointegration assay, where the cassette is replaced by a plasmid carrying a 59-be, the outcome was variable and integration of such plasmids at attI1 was frequently not detectable (21, 27, 33, 37). In these cases, only the 59-be in the promoter-distal cassette of the recipient conjugative plasmid (R388) participated in IntI1-mediated recombination events. This effect has been traced to two factors in the cointegration assay: the orientation of the cloned recombination site with respect to the vector and the strength of the Pc promoter in the recipient integron in R388. The possibility that transcription from Pc disrupts recombination at attI1 is not consistent with the finding that recombinants are recovered when the site is cloned in orientation 2. A possible explanation lies in the fact that when the 59-be is cloned in orientation 1, the Pc promoter in cointegrates will face toward the rep region of the pACYC184 vector (Fig. 2C). Transcription into the rep region of plasmids is known to reduce the copy number (5, 38). It appears that when Pc is strong, as in In3 in R388, and not separated from rep by appropriate sequences (the dfrB2 and orfA cassettes which presumably attenuate transcription readthrough), replication of the cointegrate is disrupted, leading to failure to recover cointegrates formed by recombination with either the attI1 site or the dfrB2 59-be in R388. The presence of the R388 rep region in the cointegrates does not appear to compensate for this effect.
In the light of these findings it is also possible to reinterpret the data from the study of Martinez and de la Cruz (27), which used the same assay. The longer fragments containing the aadA1/qacE 59-be are cloned in pACYC184 in orientation 1, and with these plasmids the recombination outcomes were the same as those obtained in both past and present studies from our laboratory using sites (attI1 and 59-be) cloned in pACYC184 in orientation 1. The shift to preferential recombination of the aadA1/qacE 59-be with attI1, observed when shorter fragments were tested (27), coincides not with the loss of a critical sequence element as suggested by these authors but with a change in the cloning vector from pACYC184 to pSU2718/9. The orfA/qacE 59-be was also cloned in the closely related vector pSU19. In these vectors, the multicloning site, and hence the cloned fragment, is located on the opposite side of the rep region, and our preliminary data indicate that the orientation of a cloned recombination site with respect to pSU2718/9 does not influence the recombination outcomes (data not shown). The data obtained with these shorter clones (27) are consistent with that presented here and support the conclusion that 59-be prefer to recombine with attI1.
IntI1 also catalyzes quite substantial levels of recombination between a 59-be and 2°rs (17, 33). In these studies and in the present study, the frequency of such events, assayed by recombination with the pACYC184 vector alone, was at least 2 orders of magnitude above the background levels seen when the IntI1-producing plasmid, pSU2056, was not present. This significant level of IntI1-mediated recombination between a 59-be and a 2°rs is likely to be important in the evolution of bacteria because it disseminates gene cassettes to new locations in bacterial chromosomes or plasmids (see references 32 and 35). It may also explain why the strains containing pSU2056, which is used to supply IntI1 for the cointegration assays, are unstable. Recombination of attI1 with 2°rs was detected here and also by Hansson et al. (23), but the frequency of such events was at least an order of magnitude lower than for 59-be with 2°rs. However, the relevance of attI1 recombination with 2°rs is not obvious, particularly as it has the potential to inactivate the attI1 site and thus prevent the incorporation of gene cassettes.
The integron-gene cassette site-specific recombination system is unusual in that there are two distinct types of sites. Both attI1 and the 59-be type sites potentially bind four molecules of IntI1, but the configuration of these binding sites is different (Fig. 1). The fact that the architecture of attI1 differs from that of 59-be appears to play a role in ensuring that cassettes are integrated preferentially adjacent to the attI site of the integron. This location is optimal for expression of the gene contained in the incoming cassette (14). Whether deletion of cassettes (the excision reaction) also preferentially involves the attI1 site remains to be established.
ACKNOWLEDGMENTS
We thank Maryam Parsekhian for the construction and assay of some of the plasmids and Finn Grey and Rachelle Ellis for technical assistance.
M.-J.K. was the recipient of an Australian Postgraduate award. This work was supported by a grant from the Australian National Health and Medical Research Council.
REFERENCES
- 1.Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankun R, Ohsuka S, Kato N, Ohta M. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avila P, de la Cruz F. Physical and genetic map of the IncW plasmid R388. Plasmid. 1988;20:155–157. doi: 10.1016/0147-619x(88)90019-4. [DOI] [PubMed] [Google Scholar]
- 3.Barker A, Clark C A, Manning P A. Identification of VCR, a repeated sequence associated with a locus encoding a hemagglutinin in Vibrio cholerae O1. J Bacteriol. 1994;176:5450–5458. doi: 10.1128/jb.176.17.5450-5458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bujard H, Stueber D, Gentz R, Deuschle U, Peschke U. Insertion of transcriptional elements outside the replication region can interfere with the replication, maintenance, and stability of ColE1-derived plasmids. In: Helinski D R, Cohen S N, Clewell D B, Jackson D A, Hollaender A, editors. Plasmids in bacteria. New York, N.Y: Plenum Press; 1985. pp. 45–52. [DOI] [PubMed] [Google Scholar]
- 6.Bunny K L. Expression of cassette-associated antibiotic resistance genes in class 1 integrons. Ph. D. thesis. Australia: Macquarie University Sydney; 1997. [Google Scholar]
- 7.Bunny K L, Hall R M, Stokes H W. New mobile gene cassettes containing an aminoglycoside resistance gene, aacA7, and a chloramphenicol resistance gene, catB3, in an integron in pBWH301. Antimicrob Agents Chemother. 1995;39:686–693. doi: 10.1128/AAC.39.3.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron F H, Groot Obbink D J, Ackerman V P, Hall R M. Nucleotide sequence of the AAD(2") aminoglycoside adenylyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538–1 and dhfrII in R388. Nucleic Acids Res. 1986;14:8625–8635. doi: 10.1093/nar/14.21.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark C A, Purins L, Kaewrakon P, Manning P A. VCR repetitive sequence elements in the Vibrio cholerae chromosome constitute a mega-integron. Mol Microbiol. 1997;26:1137–1143. doi: 10.1046/j.1365-2958.1997.d01-5533.x. [DOI] [PubMed] [Google Scholar]
- 11.Collis C M, Grammaticopoulos G, Briton J, Stokes H W, Hall R M. Site-specific insertion of gene cassettes into integrons. Mol Microbiol. 1993;9:41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x. [DOI] [PubMed] [Google Scholar]
- 12.Collis C M, Hall R M. Site-specific deletion and rearrangement of integron insert genes catalysed by the integron DNA integrase. J Bacteriol. 1992;174:1574–1585. doi: 10.1128/jb.174.5.1574-1585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collis C M, Hall R M. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol Microbiol. 1992;6:2875–2885. doi: 10.1111/j.1365-2958.1992.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 14.Collis C M, Hall R M. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–162. doi: 10.1128/aac.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collis C M, Kim M-J, Stokes H W, Hall R M. Binding of the purified integron DNA integrase IntI1 to integron- and cassette-associated recombination sites. Mol Microbiol. 1998;29:477–490. doi: 10.1046/j.1365-2958.1998.00936.x. [DOI] [PubMed] [Google Scholar]
- 16.Francia M V, Avila P, de la Cruz F, García Lobo M. A hot spot in plasmid F for site-specific recombination mediated by Tn21 integron integrase. J Bacteriol. 1997;179:4419–4425. doi: 10.1128/jb.179.13.4419-4425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francia M V, de la Cruz F, García Lobo M. Secondary sites for integration mediated by the Tn21 integrase. Mol Microbiol. 1993;10:823–828. doi: 10.1111/j.1365-2958.1993.tb00952.x. [DOI] [PubMed] [Google Scholar]
- 18.Gravel A, Fournier B, Roy P H. DNA complexes obtained with the integron integrase IntI1 at the attI1 site. Nucleic Acids Res. 1998;26:4347–4355. doi: 10.1093/nar/26.19.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall R M, Collis C M. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 20.Hall R M, Collis C M. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist Updates. 1998;1:109–119. doi: 10.1016/s1368-7646(98)80026-5. [DOI] [PubMed] [Google Scholar]
- 21.Hall R M, Brookes D E, Stokes H W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 22.Hall R M, Collis C M, Kim M-J, Partridge S R, Recchia G D, Stokes H W. Mobile gene cassettes in evolution. Ann NY Acad Sci. 1999;87:68–80. doi: 10.1111/j.1749-6632.1999.tb08866.x. [DOI] [PubMed] [Google Scholar]
- 23.Hansson K, Sköld O, Sundström L. Non-palindromic attI sites of integrons are capable of site-specific recombination with one another and with secondary targets. Mol Microbiol. 1997;26:441–453. doi: 10.1046/j.1365-2958.1997.5401964.x. [DOI] [PubMed] [Google Scholar]
- 24.Jones D S C, Schofield J P. A rapid method for isolating high-quality plasmid DNA suitable for DNA sequencing. Nucleic Acids Res. 1990;18:7463–7464. doi: 10.1093/nar/18.24.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lévesque C, Brassard S, Lapointe J, Roy P H. Diversity and relative strength of tandem promoters for the antibiotic resistance genes of several integrons. Gene. 1994;142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 26.Martinez E, de la Cruz F. Transposon Tn21 encodes a RecA-independent site-specific integration system. Mol Gen Genet. 1988;211:320–325. doi: 10.1007/BF00330610. [DOI] [PubMed] [Google Scholar]
- 27.Martinez E, de la Cruz F. Genetic elements involved in Tn21 site-specific integration, a novel mechanism for the dissemination of antibiotic resistance genes. EMBO J. 1990;9:1275–1281. doi: 10.1002/j.1460-2075.1990.tb08236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazel D, Dychinco B, Webb V, Davies J. A distinctive class of integron in the Vibrio cholerae genome. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 29.Ouellette M, Roy P H. Homology of ORFs from Tn2603 and from R46 to site-specific recombinases. Nucleic Acids Res. 1987;15:10055. doi: 10.1093/nar/15.23.10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Partridge S R, Recchia G D, Scaramuzzi C, Collis C M, Stokes H W, Hall R M. Definition of the attI1 site of class 1 integrons. Microbiology. 2000;146:2855–2864. doi: 10.1099/00221287-146-11-2855. [DOI] [PubMed] [Google Scholar]
- 31.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 32.Recchia G D, Hall R M. Plasmid evolution by acquisition of mobile gene cassettes: plasmid pIE723 contains the aadB gene cassette precisely inserted at a secondary site in the IncQ plasmid RSF1010. Mol Microbiol. 1995;15:179–187. doi: 10.1111/j.1365-2958.1995.tb02232.x. [DOI] [PubMed] [Google Scholar]
- 33.Recchia G D, Stokes H W, Hall R M. Characterisation of specific and secondary recombination sites recognised by the integron DNA integrase. Nucleic Acids Res. 1994;22:2071–2078. doi: 10.1093/nar/22.11.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose R E. The nucleotide sequence of pACYC184. Nucleic Acids Res. 1988;16:355. doi: 10.1093/nar/16.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segal H, Elisha B G. Identification and characterization of an aadB gene cassette at a secondary site in a plasmid from Acinetobacter. FEMS Microbiol Lett. 1997;153:321–326. doi: 10.1111/j.1574-6968.1997.tb12591.x. [DOI] [PubMed] [Google Scholar]
- 36.Stokes H W, Hall R M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 37.Stokes H W, O'Gorman D B, Recchia G D, Parsekhian M, Hall R M. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
- 38.Stueber D, Bujard H. Transcription from efficient promoters can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1982;1:1399–1404. doi: 10.1002/j.1460-2075.1982.tb01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]