Abstract
An essential protein translocation pathway in Escherichia coli and Bacillus subtilis involves the signal recognition particle (SRP), of which the 54-kDa homolog (Ffh) is an essential component. In a previous study, we found that a transposon insertion in the ylxM-ffh intergenic region of the designated secretion and acid tolerance (sat) operon of Streptococcus mutans resulted in an acid-sensitive phenotype. In the present study, we further characterized this genomic region in S. mutans after construction of bonafide sat operon mutants and confirmed the role of the SRP pathway in acid resistance. Northern blot and primer extension analyses identified an acid-inducible promoter upstream of ylxM that was responsible for upregulating the coordinate expression of all five genes of the sat operon when cells were grown at acid pH. Two constitutive promoters, one immediately upstream of satD and one just 3′ to the acid-inducible promoter, were also identified. Except for Ffh, the functions of the sat operon gene products are unknown. SatC, SatD, and SatE have no homology to proteins with known functions, although YlxM may function as a transcriptional regulator linked to genes encoding SRP pathway proteins. Nonpolar mutations created in each of the five genes of the sat locus resulted in viable mutants. Most striking, however, was the finding that a mutation in ffh did not result in loss of cell viability, as is the case in all other microbial species in which this pathway has been described. This mutant also lacked immunologically detectable Ffh and was severely affected in resistance to acid. Complementation of the mutation resulted in restoration of acid tolerance and reappearance of cytoplasmic Ffh. These data provide evidence that the SRP pathway plays an important role in acid tolerance in S. mutans.
Acid tolerance is regarded as an important determinant for survival of cariogenic bacteria in the biofilm on teeth (7, 37, 49.) The microorganisms in dental plaque are exposed to continual cycles of acid shock resulting from the formation of acid end products during metabolism of dietary carbohydrate by the acidogenic microflora. In vivo pH measurements have revealed that the intake of carbohydrates can drop the plaque pH from 7 to 4 in less than 20 min (21, 24, 52). In addition, frequent carbohydrate intake results in a lower pH of “resting plaque,” which has been associated with increased dental caries. Early studies (41) demonstrated that high caries activity is associated with rapid plaque acidification as well as tolerance of low pH by the plaque microorganisms.
Streptococcus mutans has been recognized as one of the major etiological agents in human dental caries (14, 27). In addition to its ability to colonize tooth surfaces and acidogenicity, S. mutans is acid tolerant. Glycolysis can occur at pH values as low as pH 4.4, whereas the minimum pH for growth is 4.8 (5). Svensäter et al. (42) demonstrated that a 2-h exposure of exponential-phase cells to pH 5.5 induced an acid tolerance response (ATR) that increased subsequent survival at pH 3.0. The mechanisms involved in acid tolerance of S. mutans are poorly understood. Protein synthesis is required for induction of an ATR, since it could be blocked by chloramphenicol in the medium (17). Sustained mildly acidic conditions increase H+/ATPase and glycolytic activities (2, 4, 15, 16). These metabolic changes permit S. mutans to maintain a constant transmembrane pH gradient of approximately 1.0 pH unit (8, 15).
Previous genetic studies in our laboratory focused on identification of genes that when inactivated by transposon Tn917 rendered the mutants acid sensitive (12). One of the mutants (AS17) affected in acid tolerance had the transposon integrated into the intergenic region between the ylxM and ffh homologs. This locus was subsequently designated sat for secretion and acid tolerance (12).
The ffh gene encodes a 54-kDa subunit homolog of the eucaryotic signal recognition particle (SRP) complex, which is part of a protein translocation pathway that is highly conserved throughout the domains of life (6, 9, 29, 54). Detailed studies of Escherichia coli and Bacillus subtilis have shown that this pathway is involved in protein export and membrane biogenesis. The protein encoded by ylxM is homologous to a protein of unknown function in Bacillus subtilis that is transcribed as an operon with ffh (19). YlxM contains helix-turn-helix motifs indicative of DNA-binding proteins, therefore suggesting a function in transcriptional regulation (35). Remarkably, the transposon integration in AS17 resulted in a viable mutant. In E. coli and B. subtilis, ffh is a gene essential for viability (19, 33, 51). Our previous data (11) suggested that AS17 had a reduction in rather than a complete loss of Ffh expression.
Further characterization of mutant AS17 revealed that it was incapable of an ATR (11). Furthermore, the H+/ATPase activity in membrane fractions of pH 5.0-shocked AS17 cells was comparable to that in pH 7.5-grown mutant cells, whereas it was twofold lower than the pH 5.0-induced activity in wild-type cell membranes (11). A lack of Ffh could result in an inability to rearrange the membrane in the way needed to generate an ATR, which would include the incorporation of additional H+/ATPase polypeptides. However, the phenotypic effects observed could be the result of polar effects of the insertion. In the present study, we addressed this issue by characterizing the transcriptional organization of the sat operon by Northern blotting and primer extension analysis. Subsequently, we created nonpolar kanamycin insertions in the individual genes of the sat operon. Growth experiments were performed to assess the involvement of each of the sat operon genes in acid tolerance of S. mutans. Our data show that the S. mutans sat operon consists of five open reading frames (ORFs) and confirm the functional role of ffh in acid tolerance.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. mutans strains were grown routinely in Todd-Hewitt broth (BBL, Becton Dickinson Microbiology Systems, Cockeysville, Md.) supplemented with 0.3% (wt/vol) yeast extract (THYE), aerobically at 37°C without agitation, or on THYE supplemented with 1.5% (wt/vol) agar, at 37°C in either candle jars or GasPak anaerobic chambers (BBL). When required, antibiotics were used at the following concentrations: erythromycin, 10 μg/ml; and kanamycin, 500 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMA-mcrBC) φ80lacZΔM15 ΔlacZ74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| INV110 | F−traD36 proAB+ lacIqZΔM15/rpsL (Strr) thr leu endA thi-1 lacYgalK galTara tonA tsx dam dcm supE44 Δ(lac-proAB) Δ(mcrC-mrr) 102::Tn10 (Tetr) | Invitrogen |
| INVαF′ | F−endA1 recA1hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1 φ80lacZΔM15 Δ(lacZYA-argF)U169 | Invitrogen |
| S. mutans | ||
| NG8 | Wild type | K. Knox |
| MK4 | ffh::Kanr; pMK14 in NG8 | This study |
| BK110 | satC::Kanr; pBK40 in NG8 | This study |
| BK120 | satD::Kanr; pBK41 in NG8 | This study |
| BK130 | satE::Kanr; pBK42 in NG8 | This study |
| BK140 | ΔylxM::Kanr; pBK45 in NG8 | This study |
| BK150 | gtfA::ylxM-ffh; pBK55 in NG8 | This study |
| BK151 | ffh::KanrgtfA::ylxM-ffh; pMK14 in BK150 | This study |
| BK152 | gtfA::pVA891; pVAgtfA in NG8 | This study |
| Plasmids | ||
| pALH124 | aphA3 Kanr cassette in pALH123 | A. L. Honeyman |
| pBAD-TOPO | Arabinose-inducible expression of His6 tag protein | Invitrogen |
| pCR2.1 | PCR product A/T cloning vector | Invitrogen |
| pPC185 | pUC18 derivative; Emr Apr | This study |
| pPC300 | Psat-ylxM-ffh in pDL278 | 11 |
| PUC18 | E. coli cloning vector | Invitrogen |
| pVA891 | Streptococcal suicide vector | 30 |
| pVAGTFA | gtfA in pVA891 | 26 |
| pBK8 | ffh in pBAD-TOPO | This study |
| pBK19 | Psat-ylxM in pCR2.1 | This study |
| pBK22 | satD in pCR2.1 | This study |
| pBK23 | satE in pCR2.1 | This study |
| pBK40 | satC::Kanr in pPC185 | This study |
| pBK41 | satD::Kanr in pPC185 | This study |
| pBK42 | satE::Kanr in pPC185 | This study |
| pBK43 | 5′ ylxM in pCR2.1 | This study |
| pBK44 | 3′ ylxM in pCR2.1 | This study |
| pBK45 | ΔylxM::Kanr in pUC18 | This study |
| pBK55 | Psat-ylxM-ffh (pPC300-derived) in pVAGTFA | This study |
| pMK2 | satD in pPC185 | This study |
| pMK4 | satC in pCR2.1 | This study |
| pMK6 | satE in pPC185 | This study |
| pMK8 | satC in pPC185 | This study |
| pMK10 | ffh in pCR2.1 | This study |
| pMK11 | ffh in pPC185 | This study |
| pMK14 | ffh::Kanr in pPC185 | This study |
Escherichia coli strains were cultured routinely aerobically at 37°C with vigorous shaking in Luria-Bertani (LB) broth or on LB agar (1.5% [wt/vol]) plates supplemented with the appropriate antibiotics: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; erythromycin, 300 μg/ml; or tetracycline, 15 μg/ml.
DNA manipulations.
PCRs were performed routinely with the High Fidelity PCR kit (Roche Molecular Biochemicals, Indianapolis, Ind.) using custom primers from Integrated DNA Laboratories, Inc. (Coralville, Iowa). All primers used in this study are listed in Table 2. Plasmid DNA preparations were obtained with the Qiaprep Spin Miniprep kit (Qiagen, Inc., Valencia, Calif.). S mutans chromosomal DNA was isolated as described previously (12). Natural transformation of S. mutans was performed as described previously (32). Transformed cells were spread on THYE agar supplemented with 10% (wt/vol) sucrose and the appropriate antibiotics, and the plates were incubated anaerobically at 37°C for 48 h.
TABLE 2.
Primers used in this study
| Primer no. | Sequencea | Positionb |
|---|---|---|
| BK5 | 5′-GCATTGAAATCCTACAGC-3′ | 19–36 |
| BK6 | 5′-CGTCCAATTGAACTTGTC-3′ | 294–277 |
| BK13 | 5′-ATGATTTACATCGCTATTATTGG-3′ | 3163–3185 |
| BK14 | 5′-GATTAAACAGATAGCATAAAT-3′ | 735–715c |
| BK15 | 5′-AAGGTAGCTACGACCTTA-3′ | 3781–3798 |
| BK18 | 5′-TTTTTTACGTTTTTTGGCTTT-3′ | 2516–2496 |
| BK23 | 5′-GACGACTATTGATAATAGAGAC-3′ | 832–853 |
| BK25 | 5′-GAGATCGCCAATAATAGCG-3′ | 3192–3174 |
| BK26 | 5′-GACAGGATTACCTGACAAAAAC-3′ | 3875–3854 |
| BK27 | 5′-TTATCTGTCAAAAGCGCCGCAT-3′ | 591–570 |
| BK28 | 5′-CATTTGCTTATCTGTCAA-3′ | 598–581 |
| BK36 | 5′-ggtaccATGGCTTTTGAAAGTTTA-3′ | 969–986 |
| BK39 | 5′-GTTTTTGACTTTTAGGGCGA-3′ | 3235–3216 |
| PC12 | 5′-ggatccGTTGTGGCTGCTTGGTC-3′ | 124–140 |
| PC18 | 5′-ggatccATGGCTTTTGAAAGTTTA-3′ | 969–986 |
| PC19 | 5′-ggtaccGGGAAGACCATTATAACG-3′ | 2595–2578 |
| PC20 | 5′-GCAACATTTGCGGCTTCT-3′ | 1669–1652 |
Sequence of primers in the sat locus based on S. mutans JH1005 sequence (GenBank accession no. U88582), except for primer BK14, which was based on the UAB159 genomic sequence (http://www.genomic.ou.edu/smutans.html); lowercase nucleotides indicate added restriction sites.
Position of primers based on S. mutans JH1005 sequence (GenBank accession no. U88582), unless noted otherwise.
The position indicated for BK14 is the position within the satE ORF, as based on the S. mutans UAB159 genomic sequence (http://www.genome.ou.edu /smutans.html).
Southern blot analyses were performed with digoxigenin (DIG)-labeled probes according to the DIG system manufacturer's protocols (Roche). Probes were generated by PCR with the PCR DIG labeling kit (Roche) according to the manufacturer's recommendations.
DNA sequencing was performed by the DNA Sequencing Core Laboratory of the University of Florida's Interdisciplinary Center for Biotechnology Research. Sequence analysis was carried out with MacVector V6.5.3 (Oxford Molecular, Ltd., Madison, Wis.) and BLAST, version 2.0, software (1) available from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
RNA isolation.
Total RNA was isolated from S. mutans NG8 with the RNeasy Mini or Midi kit (Qiagen), with several modifications. Overnight THYE cultures were subcultured 1:10 into prewarmed THYE at pH 7.0. Cells were grown to mid-exponential phase (optical density at 650 nm of ≈0.5), pelleted by centrifugation for 10 min at 4,000 × g, and resuspended in fresh THYE at pH 7.0 or 5.0. After a 1-h incubation at 37°C, bacteria were harvested by centrifugation for 10 min at 4,000 × g, washed once in ice-cold 10 mM Tris–1 mM EDTA (TE; pH 7.0), and resuspended in TE containing lysozyme (5 mg/ml; Sigma Chemical Co., St. Louis, Mo.) and mutanolysin (200 U/ml; Sigma Chemical Co.). The bacteria were incubated for 10 min at room temperature before proceeding with the Qiagen RNeasy kit protocol as provided by the manufacturer.
Northern blot analysis.
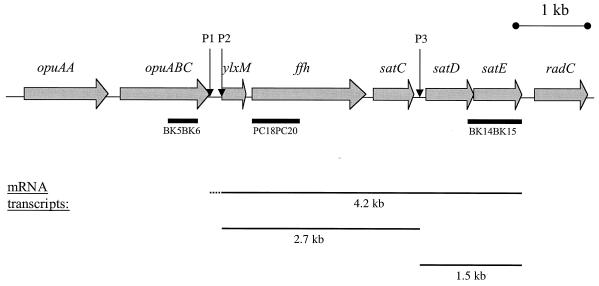
For Northern blot analysis, 10 μg of total RNA from both the pH 7.0- and pH 5.0-shocked S. mutans NG8 cultures was separated on a denaturing gel according to the Qiagen RNeasy kit protocol. Northern blotting and hybridization were performed according to the protocols provided with the Roche DIG system. DIG-labeled PCR probes were generated with the PCR DIG labeling kit (Roche) by using primers PC18 and PC20 for the ffh probe, BK15 and BK14 for the satE probe, and BK5 and BK6 for the opuABC probe. The position of the probes is indicated on the genomic map of the sat operon (Fig. 1).
FIG. 1.
Schematic representation of the sat operon locus in S. mutans. The sequences represented here that make up the sat operon were compiled from both our sequencing data (GenBank accession no. U88582) and the genome sequence of strain UAB159 (http://www.genome.ou.edu/smutans.html). Shaded arrows indicate ORFs; their designations are indicated above the sequence. Putative transcription start sites P1, P2, and P3, are indicated. The probes used in Northern blot experiments in this study are shown by bars below the sequence. sat operon mRNAs and their sizes as determined by Northern blot analysis are depicted below the sequence.
Primer extension.
To map the 5′ termini of sat operon mRNA transcripts, primer extension analysis was carried out with 2 pmol of [γ-32P]ATP (3,000 Ci/mmol; Amersham Pharmacia Biotech, Inc., Piscataway, N.J.)-labeled BK28 or BK39 on 15 μg of total RNA from pH 7.0- and pH 5.0-grown NG8 cells. After denaturing for 10 min at 70°C and subsequent cooling on ice, primer extension reactions were performed for 1 h at 42°C with 200 U of Superscript II reverse transcriptase (Gibco BRL, Grand Island, N.Y.). After heat inactivation, samples were treated with 2 U of Rnase H (Gibco BRL) for 20 min at 37°C. Following phenol extraction and ethanol precipitation, the DNA was resuspended in distilled water and diluted 1:1 with Sequenase stop buffer (U.S. Biochemicals Corp. Cleveland, Ohio). A sequencing ladder was generated with unlabeled BK28 and pBK19 as a template by using the Sequenase 7-deaza-dGTP DNA sequencing kit (U.S. Biochemicals Corp.) with [α-35S]dATP as a label, according to the manufacturer's protocol. The sequencing ladder and the primer extension reactions were run on an 8% denaturing polyacrylamide gel at 50 W for 3 h. The gel was dried and exposed to X-ray film.
Construction of nonpolar mutations in the sat operon genes.
To create a nonpolar mutation in ffh, a PCR product of BK36 and PC19 first was cloned into pCR2.1 T/A TOPO cloning vector (Invitrogen, Carlsbad, Calif.), to give pMK10. The KpnI insert fragment was subsequently cloned into streptococcal suicide vector pPC185, a pUC18 derivative in which the bla gene was replaced with an erythromycin cassette derived from pResEM749 (39). The resulting construct (pMK11) was HindIII digested, dephosphorylated, and ligated with HindIII-digested promoterless aphA3 Kanr cassette, which had been isolated from pALH124 (kindly provided by A. L. Honeyman, University of South Florida, Tampa), to result in pMK14 (ffh::Kanr). The approach for the satC, satD, and satE insertions was similar to that used for the ffh insertion. PCR products were generated with primers PC18 and BK25 for satC, BK13 and BK26 for satD, and BK15 and BK14 for satE and were cloned into pCR2.1 to give pMK4, pBK22, and pBK23, respectively. EcoRI insert fragments were subsequently cloned in pPC185, resulting in pMK8, pMK2, and pMK6, respectively. Thereafter, the PstI-digested Kanr cassette was cloned into the PstI site in satC in pMK8, the SphI-digested Kanr cassette was cloned into the SphI site in satD in pMK2, and the BamHI-digested Kanr cassette was cloned into the BclI site of satE in pMK6. These constructs were designated pBK40 (satC::Kanr), pBK41 (satD::Kanr), and pBK42 (satE::Kanr).
A different approach was taken to construct a kanamycin cassette insertion in ylxM. The promoter region and the 5′ 68 nucleotides (nt) of the ORF were amplified with primers PC12 and BK27 and then cloned into pCR2.1 to give pBK43. pBK44 contains a PCR fragment of the 3′ 22 nt plus 800 nt of downstream sequence that was amplified with primers BK23 and PC20. The inserts of pBK43 and pBK44 were isolated by digestion with BamHI-EcoRI and EcoRI-HindIII, respectively. These fragments were ligated into BamHI-HindIII-digested-pUC18 in the presence of the EcoRI-digested Kanr cassette, to generate a Kanr-inserted ylxM deletion construct, designated pBK45.
S. mutans strain NG8 was transformed with the Kanr insertion constructs after linearization by NarI. To verify the correct integration, Southern blotting was performed with chromosomal DNA isolated from Kanr Ems transformants, or Kanr transformants in the case of ΔylxM::Kanr. The resulting mutant strains were designated BK140 (ΔylxM::Kanr), MK4 (ffh::Kanr), BK110 (satC::Kanr), BK120 (satD::Kanr), and BK130 (satE::Kanr).
Complementation of the ffh mutation.
It was shown in previous work (11) that a mutation affecting ffh expression rendered the cell poorly competent for DNA uptake. In order to perform ffh complementation, we integrated another copy of ylxM-ffh, under the control of its own promoter, into the gtfA locus of the NG8 chromosome and then created a mutation in the native ffh locus by insertion of the Kanr cassette as follows. Integration vector pVAGTFA was created by insertion of a 2.5-kb gtfA fragment into PvuII-digested suicide vector pVA891 (26, 30). A 2.5-kb BamHI fragment, which contained the ylxM-ffh region, including its own putative promoters, was then isolated from pPC300 (11) and cloned into the BamHI site of pVAGTFA, to give pBK55. Single-crossover Emr mutants were isolated, and integration into the gtfA locus of NG8 was verified by Southern blotting. The resulting mutant, BK150, was subsequently transformed with NarI-linearized pMK14; DNA of the Kanr transformants was isolated and used in Southern blotting to verify the integration of the Kanr cassette into the original sat operon copy of ffh. The resulting complemented mutant was designated BK151. As a control in growth experiments, we used strain BK152, which has pVAGTFA without an insert integrated in the gtfA locus.
Acid sensitivity.
The effects of pH on the growth of the Kanr insertion mutants and complementation strains were evaluated in THYE media adjusted by addition of HCl to achieve pH 7.0, 6.0, 5.5, and 5.0. Sixteen-hour cultures of the strains were subcultured into THYE at 37°C at the experimental pH levels and grown to early exponential phase (Klett value of 50 U). The strains then were subcultured 1:4 into fresh medium at the experimental pH, and their growth at 37°C was recorded with a Klett colorimeter (Klett-Summerson, New York, N.Y.). The Klett colorimeter provides a simple means of performing growth curves on multiple small-volume (5 ml) cultures without removal of any of the culture to take measurements. Since the Klett instrument scale is logarithmic, growth curves are plotted with linear axes (1 scale increment = 1 U). Growth experiments were repeated three times with duplicate samples.
Western blot analysis.
To produce whole-cell extracts of recombinant E. coli, bacteria were harvested by centrifugation for 10 min at 4,000 × g, resuspended in TE, and diluted 1:1 in sample buffer (4% [wt/vol] sodium dodecyl sulfate [SDS], 2% [vol/vol] 2-mercaptoethanol, 20% [vol/vol] glycerol, 125 mM Tris-HCl [pH 6.8], 0.1 mg of bromophenol blue per ml). The samples were heated for 10 min at 100°C, and insoluble debris was removed by centrifugation for 10 min at 14,000 × g. S. mutans whole-cell extracts were obtained by mechanical homogenization of bacterial suspensions in TE with a Mini Beadbeater 8 (BiospecProducts, Inc., Bartlesville, Okla.) at maximum power with 0.1-mm-diameter glass beads (Biospec). The resulting cell lysate was diluted 1:1 in sample buffer and heated for 10 min at 100°C.
SDS-solubilized whole-cell proteins were separated on an SDS–10% polyacrylamide gel and transferred to a nitrocellulose membrane (pore size of 0.45 μm; Schleicher & Schuell, Inc., Keene, N.H.) by Western blotting (45). After blotting for 1 h at 100 V, the blots were incubated four times for 30 min at room temperature in phosphate-buffered saline (PBS) containing 0.3% (vol/vol) Tween 20 (TPBS). The blots were incubated overnight at room temperature with antisera appropriately diluted in TPBS. Subsequently, they were washed four times with TPBS and incubated for 2 h with horseradish peroxidase-labeled goat anti-mouse immunoglobulin G (1:1,000 in TPBS; Southern Biotechnology Associates, Inc., Birmingham, Ala.). After two washes with TPBS and two washes with PBS, the antibody-binding antigens were visualized with 4-chloro-1-naphthol and H2O2.
Ffh antiserum production.
To overproduce and purify Ffh, the ffh gene was amplified from the S. mutans NG8 genome with primers PC18 and BK18 and cloned into the pBAD-TOPO six-histidine tag (His6) expression vector (Invitrogen). The resulting construct was designated pBK8. Expression of Ffh (Ffh-V5-His6) from this construct was analyzed by Western blotting of whole-cell extracts with α-V5 antibody (1:5,000 dilution in TTBS; Invitrogen). Maximal induction of expression was achieved by adding 0.2% (wt/vol) arabinose to mid-exponential-phase E. coli TOP10 cells harboring pBK8 and subsequent incubation for 4 h at 37°C with vigorous shaking. Ffh-V5-His6 was semipurified with HisBind resin (Novagen, Madison, Wis.) according to the manufacturer's denaturing batch purification protocol. The Ffh-V5-His6 was separated on an SDS–10% polyacrylamide gel, the gel was stained with Coomassie brilliant blue R-250, and the 60 kDa-protein band of interest was excised. BALB/c mice were immunized with the isolated Ffh-V5-His6 gel slices followed by ascites production at the Hybridoma Core Laboratory of the University of Florida's Interdisciplinary Center for Biotechnology Research.
Nucleotide sequence accession number.
The updated nucleotide sequence of the sat operon is available from the GenBank database under accession no. U88582.
RESULTS
Genomic organization of the sat operon locus.
In this study, the nucleotide sequence downstream of the ylxM-ffh operon in S. mutans strain JH1005 (GenBank accession no. U88582), obtained from the DNA fragment in pJG301 containing ylxM-ffh followed by two complete ORFs and one partial putative ORF, was assessed. These three new ORFs were designated satC, satD, and satE (of which only the 5′ end was represented), with lengths of 525, 666, and 321 nt, respectively. Southern blotting confirmed that the genomic organization of the ylxM-satE region in strain NG8 was identical to that in JH1005 (data not shown). The nucleotide sequence of the sat locus in the S. mutans UAB159 genomic sequence (http://www.genome.ou.edu/smutans.html) was nearly identical (>99%) to the available JH1005 sequence. A schematic of the genomic organization of the sat operon based on a compilation of the two available S. mutans sequences is shown in Fig. 1.
Analysis of the genomic database sequence upstream of ylxM revealed the presence of two ORFs encoding homologs of Lactococcus lactis OpuAA (68% identity; GenBank accession no. AF37878) and OpuABC (66% identity; GenBank accession no. AF37879.) These opuA genes code for the two subunits of a glycine betaine ABC transport system involved in protection against osmotic stress conditions (48). The gene products of satC, satD, and satE have no homologs in the GenBank database (National Center for Biotechnology Information). In addition, their amino acid sequences contain no known protein motifs. Downstream of satE, a homolog of Bacillus subtilis RadC is encoded (43% identity; GenBank accession no. Q02170). Proteins of the RadC family are involved in DNA repair mechanisms (10).
Transcriptional organization of the sat operon.
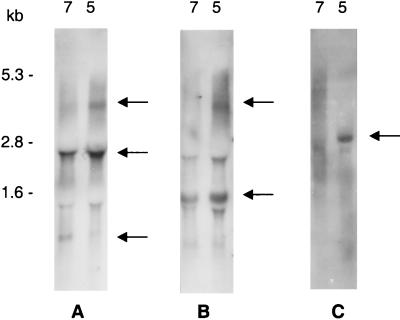
Northern blots on total RNA from mid-exponential-phase S. mutans NG8 cells that were exposed to pH 7.0 or 5.0 for 1 h following incubation in pH 7.0 medium were performed to identify the mRNAs transcribed from the sat locus under these conditions. A DIG-labeled ffh probe hybridized with three mRNA species, i.e., 4.2-, 2.7-, and 0.9-kb transcripts (Fig. 2A). The amount of the 4.2-kb transcript was significantly increased in pH 5.0-shocked cells compared to that in pH 7.0-shocked cells, whereas the 2.7-kb transcript appeared to be constitutively expressed. The amount of the 0.9-kb transcript was decreased under pH 5.0 conditions. An identical hybridization pattern was observed with a ylxM probe (data not shown).
FIG. 2.
Northern blot analysis of the sat operon. Ten micrograms of total RNA from pH 7- or 5-shocked S. mutans NG8 cells was hybridized with an ffh probe amplified with primers PC18 and PC20 (A), a satE probe amplified with primers BK14 and BK15 (B), and an opuABC probe amplified with primers BK5 and BK6 (C). Arrows indicate major mRNA transcripts. RNA size markers are indicated to the left of panel A.
Based on our sequencing data and the available S. mutans UAB159 genome sequence, probes homologous to the upstream opuABC sequence and the downstream satE sequence (Fig. 1) were generated. In addition to a constitutively expressed 1.5-kb transcript, the satE probe recognized a 4.2-kb transcript found in greater amounts in pH 5-shocked cells than in pH 7-shocked cells (Fig. 2B), as was the case with the ffh probe described above. Northern blots with total RNA isolated from the ffh::Kanr and satC::Kanr mutants revealed the existence of a 3.5-kb transcript (0.8 kb being contributed by the Kanr cassette) which hybridized only with the ylxM/ffh probes and not the satE probe (data not shown). These data confirm our conclusion that the 2.7-kb transcript in Fig. 2A is composed of ylxM, ffh, and satC only and does not include satE. A fainter 2.7-kb band in Fig. 2B is therefore likely an artifact due to its proximity to the 23S rRNA region. The opuABC probe hybridized with a 2.9-kb mRNA. This transcript also was strongly induced under pH 5.0 conditions (Fig. 2C).
Mapping of 5′ termini of sat operon mRNA.
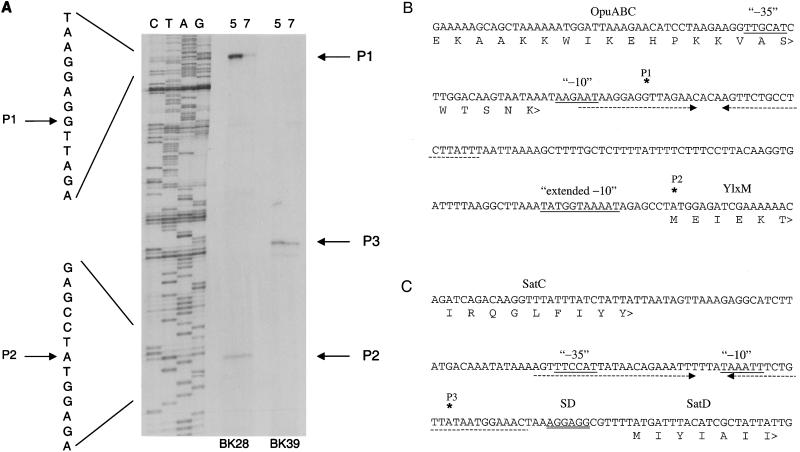
Putative transcription start sites in the sat operon were identified by primer extension mapping of the 5′ mRNA termini. Two major products were observed with primer BK28, which hybridizes within ylxM (Fig. 3A). The larger of the two was induced under acid conditions, whereas the smaller product was constitutively expressed. The 5′ terminus of the larger acid-inducible product mapped to a guanosine residue 104 nt upstream of ylxM (P1; Fig. 3B). This putative transcription start site is preceded by two hexameric sequences with similarities to consensus −10 and −35 promoter sequences. The 5′ terminus of the smaller constitutively expressed product mapped to the adenosine of the ylxM translation start codon (P2; Fig. 3B). This site is preceded by a putative −10 promoter sequence (TATGGTAAAAT) that is similar to the consensus extended −10 sequence (34, 50). No consensus −35 sequence was present upstream of this transcription start site. Primer extension with primer BK39, which hybridizes within satD, resulted in one major product that was constitutively expressed (Fig. 3A). The 5′ terminus of this product mapped to an adenosine residue 26 nt upstream of the satD gene (P3; Fig. 3C). Two hexameric sequences with similarities to consensus −10 and −35 promoter sequences were identified upstream of this putative transcription start site.
FIG. 3.
Primer extension mapping of the 5′ end points of transcripts expressing sat operon segments. (A) Primer extension reactions were performed with 15 μg of total RNA from pH 5- or 7-shocked S. mutans NG8. Reaction mixtures were primed with ylxM primer BK28 or with satD primer BK39. Lanes C, T, A, and G contain a sequencing ladder primed by primer BK28 on pBK19 plasmid DNA. Arrows indicate 5′ termini P1, P2, and P3. Nucleotide sequence for products of primer BK28 is indicated to the left of the products. (B) Sequence upstream of ylxM showing the two putative transcription start sites P1 and P2 (asterisks) and their putative promoter sites (underlined). The N-terminal amino acid sequence of YlxM is indicated. A putative inverted repeat terminator sequence for the upstream opu operon is indicated by opposite underlining arrows. (C) Intergenic sequence between satC and satD showing the putative transcription start site P3 as determined with primer BK39. Promoter sequences, inverted repeats, and SatC and SatD amino acid sequences are indicated as for panel B. SD, Shine-Dalgarno consensus ribosome binding site.
Construction of nonpolar insertions in sat operon genes.
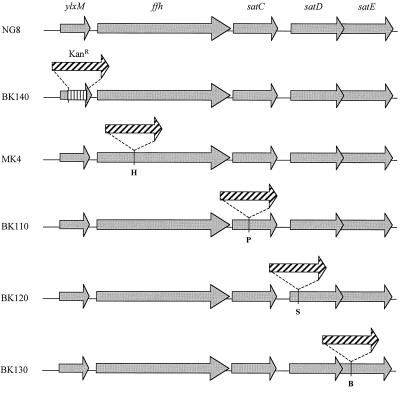
To determine their function in acid tolerance, insertional mutations were made in each of the individual genes of the sat operon with a nonpolar kanamycin cassette. This cassette contains a promoterless aphA3 kanamycin resistance gene with upstream stop codons in all three reading frames and without transcription termination sequences to allow transcription readthrough into downstream sequences (A. L. Honeyman, personal communication). In the cases of the ffh, satC, satD, and satE genes, the insertion was made in a unique restriction site within the gene. The Kanr cassette was inserted at the HindIII site at position 629 of ffh, the PstI site at position 396 in satC, the SphI site at position 322 in satD, and the BclI site at position 321 in satE. In the case of ylxM, no useful restriction sites were present within the ORF. Therefore, a 5′ fragment and a 3′ fragment of this gene were amplified and cloned with the Kanr cassette inserted between the PCR fragments. In this construct (pBK45), the Kanr cassette replaces residues 69 to 308 of the ylxM ORF.
In all cases, transformation with the NarI-linearized Kanr insertion constructs resulted in viable mutations. Transformants were visible on THYES plates after 24 h under anaerobic conditions in all cases except for the ffh::Kanr mutation, in which case colonies only became visible after 48 h. Southern blotting confirmed that integration in the chromosome had occurred as schematically depicted in Fig. 4.
FIG. 4.
Nonpolar insertional inactivation of sat operon genes by a promoterless Kanr cassette. Schematic representation of the genomic organization in the wild-type NG8 and mutants BK140 (ΔylxM::Kanr), MK4 (ffh::Kanr), BK110 (satC::Kanr), BK120 (satD::Kanr), and BK130 (satE::Kanr). Restriction sites used for insertion in MK4, BK110, BK120, and BK130 are indicated. H, HindIII; P, PstI; S, SphI; B, BclI.
Effect of insertional inactivations on growth and acid tolerance.
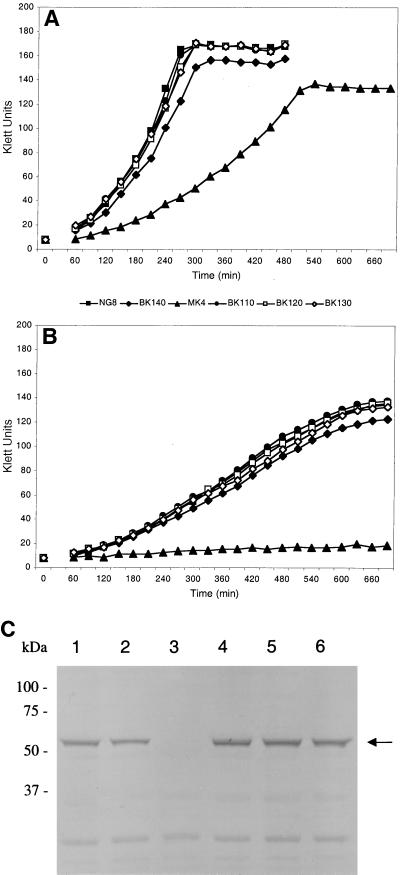
The Kanr insertion mutants were grown in THYE at pH 5.0 to 7.0 to assess the importance of the sat operon genes in S. mutans for normal growth and acid tolerance. None of the mutations in the genes of the sat operon caused a complete inability to grow in THYE at pH 7.0 (Fig. 5A). The wild-type strain NG8 had a mean maximum growth rate of 55.4 Klett units h−1 and a final Klett value of 167 ± 3 U. The ffh mutant MK4 had a greatly reduced growth rate of 24.4 Klett units h−1 and a final Klett value of 135 ± 4 U. A mutation in ylxM (BK140) had a small effect on the growth rate (47.5 Klett units h−1) and yield (155 ± 3 U). The satC (BK110), satD (BK120), and satE (BK130) mutants had growth rates and yields similar to those of parent strain NG8. The same growth patterns were observed at pH 6.0 and 5.5, with gradually reduced growth rates and yields for all strains compared to those at pH 7.0 (data not shown). At pH 5.0 (Fig. 5B), the growth rate of NG8 was reduced to 15.8 Klett units h−1, with a final Klett value of 123 ± 7 U. The ffh mutant was incapable of growth at this pH. The growth rate and yield of BK140 were not significantly lower than those of the wild type: 15.0 Klett units h−1 and 118 ± 6 U, respectively. As at the other pHs, growth of BK110, BK120, and BK130 was similar to that of NG8.
FIG. 5.
Effect of Kanr insertions on growth and Ffh expression. The growth capacities of mutants MK4, BK110, BK120, BK130, and BK140 were compared to that of wild-type strain NG8 in THYE adjusted to pH 7 (A) and 5 (B). The growth experiments were performed in duplicate in three separate experiments. The results of a typical experiment are shown. (C) Ffh expression was determined by Western blot analysis with polyclonal mouse anti-Ffh serum. Fifty micrograms of SDS-soluble whole-cell protein was separated on an SDS–10% polyacrylamide gel and blotted onto nitrocellulose. The full-size Ffh protein band is indicated (arrow). Lanes: 1, wild-type strain NG8; 2, BK140; 3, MK4; 4, BK110; 5, BK120; 6, BK130.
Complementation of ffh
Complementation of the ffh mutation was performed to confirm that the acid-sensitive phenotype of mutant MK4 was caused by the disruption of this gene. We tried to complement this mutant directly by using pBK55; however, as was shown previously with a Tn917 mutant (11), ffh mutants in S. mutans are not naturally competent. Therefore, a second copy of ffh, including its putative promoter region and ylxM, was integrated in the gtfA locus of NG8, before interruption of the native ffh locus with construct pMK14, to generate complemented ffh-knockout mutant strain BK151.
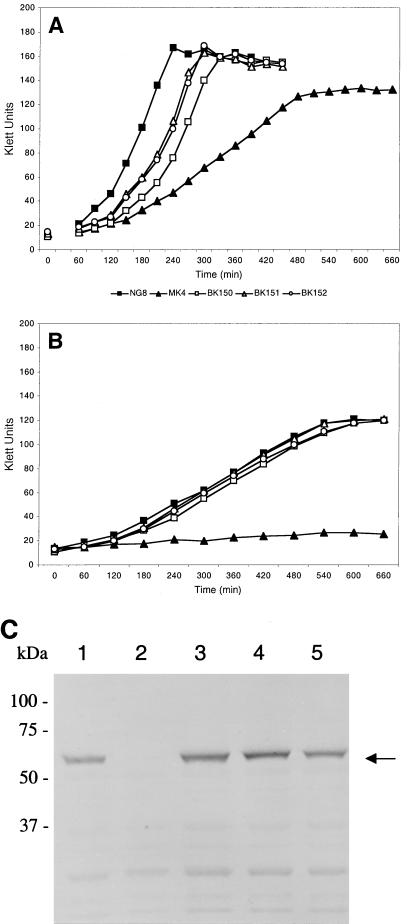
In growth experiments, the intact second copy of ffh in the gtfA locus in BK150 had no marked effect on growth rate and yield compared to that in wild-type cells (Fig. 6A and B). In contrast to mutant MK4, mutant BK151, in which the ffh gene of the sat locus in BK150 had been interrupted, grew at growth rates and final yields similar to those of wild-type NG8 at both pH 7.0 (Fig. 6A) and 5.0 (Fig. 6B). These data indicate that a copy of ffh in the gtfA locus complemented the ffh mutation, causing growth defects in MK4. Alteration of the gtfA locus did not affect growth rate and yield, as shown by integration of vector pVAGTFA alone (BK152).
FIG. 6.
Effect of complementation of ffh::Kanr into the gtfA locus on growth and Ffh expression. Growth capacities of complementation strains BK150 (gtfA::ffh), BK151 (ffh::Kanr gtfA::ffh), and BK152 (gtfA::pVAGTFA), were compared with those of mutant MK4 and wild-type strain NG8 in THYE adjusted to pH 7 (A) and 5 (B). The growth experiments were performed in duplicate in three separate experiments. The results of a typical experiment are shown. (C) Ffh expression was determined by Western blot analysis with polyclonal mouse anti-Ffh serum. Fifty micrograms of SDS-soluble whole-cell protein was separated on an SDS–10% polyacrylamide gel and blotted onto nitrocellulose. The full-size Ffh protein band is indicated (arrow). Lanes: 1, wild-type strain NG8; 2, MK4; 3, BK151; 4, BK150; 5, BK152.
Ffh protein in mutants.
The calculated molecular mass of Ffh is 56,963 Da. The mouse polyclonal antiserum raised against the Ffh-V5-His6 protein specifically recognized a protein of the expected size, i.e., 57,000 Da, in whole-cell protein extracts of S. mutans NG8 (Fig. 5C, lane 1). Therefore, this serum was used to verify the effects of the Kanr insertions in the individual genes of the sat operon on Ffh protein expression. As shown in Fig. 5C, the ffh mutant MK4 (lane 3) did not express any full-length Ffh. Moreover, the anti-Ffh antibodies did not bind to smaller antigens representing truncated products of the insertionally inactivated ffh gene. In the other insertion mutants (lanes 2 and 4 to 6), full-length Ffh was expressed at levels similar to those in the wild type (lane 1).
The anti-Ffh serum was also used to verify complementation in BK151. As shown in Fig. 6C, integration of the ylxM-ffh region under the control of its own promoter into the gtfA locus (BK150 [lane 4]) did not affect Ffh expression. Subsequent interruption of ffh in the sat locus with a Kanr cassette (BK151 [lane 3]) did not result in the loss of Ffh expression, in contrast to MK4 (lane 2). These data indicate that complementation of Ffh expression has been achieved in strain BK151. As a control, the insertion of vector pVAGTFA by itself had no effect on Ffh expression (lane 5).
DISCUSSION
The ylxM-ffh operon is highly conserved in gram-positive eubacteria. This gene cluster was first reported in B. subtilis (19), where primer extension analysis implied that these genes form an operon. Members of our group previously reported the same organization in S. mutans (12). The ylxM-ffh gene cluster can also be found in several other gram-positive species, such as Streptococcus pneumoniae, Streptococcus pyogenes, Staphylococcus aureus, and Enterococcus faecalis (Microbial Genome Blast Database; http://www.ncbi.nlm.nih.gov/), whereas the gram-negative species contain no ylxM homologs and thus lack the ylxM-ffh operon. In the Mycoplasma mycoides and Mycoplasma pneumoniae genomes, a ylxM homolog is located immediately downstream of ftsY, the gene encoding the Ffh docking protein (35). The tight linkage with the components of the SRP translocation pathway implies a role for YlxM in this pathway in gram-positive species. However, although YlxM has characteristics of a transcriptional regulator (35), the function of this protein remains to be elucidated.
In S. mutans, preliminary primer extension data suggested that ylxM and ffh are cotranscribed (13). In the present study, we determined that S. mutans ffh is transcribed in polycistronic mRNAs containing ylxM-ffh as well as putative ORFs downstream of ffh (Fig. 1). A 4.2-kb transcript was composed of ylxM-ffh-satC-satD-satE, whereas a 2.7-kb mRNA contained the first three genes and a 1.5-kb mRNA contained the last two genes. Putative terminator structures are found upstream of ylxM, between satC and satD, and downstream of satE and are consistent with the transcript sizes. The small 0.9-kb transcript (Fig. 2A) could be the result of preliminary termination or truncation of a larger transcript. The 5′ mRNA termini as determined by primer extension were consistent with the transcriptional organization. The three termini were located upstream of ylxM (P1), at the adenosine of the translation start codon of ylxM (P2), and in the satC-satD intergenic region (P3). Unlike the P1 and P3 termini, the P2 terminus results in a leaderless mRNA. Although rare, this type of transcript has been reported several times in other species, such as S. pneumoniae (28, 34, 53), E. coli and streptomycetes (22). Although leaderless transcripts lack upstream Shine-Dalgarno ribosome binding sites, they are functional in translation. It has been suggested that a sequence downstream of the translation start site plays a role in enhancement of ribosome binding (38, 40). Recently, O'Connor et al. (31) proved by mutation analysis of the proposed downstream ribosome binding site sequence that this region was not involved in the initiation of translation. The mechanism of translation of these transcripts, therefore, is still unclear.
Transcription of ffh in S. mutans is amplified in response to acid stress (11). The data presented here are consistent with these findings. Although the amounts of the 2.7- and 1.5-kb mRNA appear to be constitutive, upregulation of the 4.2-kb full-length sat operon transcript could account for an increase in total ffh transcript (Fig. 2A and B). The upregulation of transcription appears to be driven by an acid-inducible promoter upstream of the P1 5′ terminus, whereas transcription starting at P2 and P3 is constitutive, as determined by primer extension (Fig. 3). Preliminary data from experiments employing modified rapid amplification of cDNA ends confirmed that P2 and P3 are genuine transcription start sites with 5′ cap structures (data not shown). However, no further experimental evidence has been provided yet for the authenticity of P1 as a true transcription start site, suggesting this terminus could have been generated by truncation of a larger mRNA. This implies that the acid-inducible transcription is driven from a promoter further upstream, possibly by the opuA operon promoter. Induction of opuA promoter activity under pH 5 conditions is supported by our Northern blot data indicating upregulation of a 2.9-kb transcript containing opuABC (Fig. 2C). Although transcription of the opuA genes in L. lactis and B. subtilis is regulated by environmental signals, especially osmotic stress, a link with acid stress has not been reported. A coordinated upregulation of transcription of both SRP translocation and osmoprotection genes might be indicative of a general stress response induced by low pH in batch culture. Recent cross-protection studies in S. mutans revealed a slight cross-protective effect of osmotic stress adaptation on resistance to acid stress (43). However, acid adaptation did not render S. mutans resistant to osmotic stress. Further research is needed to reveal the nature of the linkage, if any, between the sat operon and the opuA operon, as well as their respective roles in acid tolerance of S. mutans.
Induction of full-length mRNA upon acid stress could result in an increased protein translation of all of the sat operon ORFs, implying a possible role for all of these gene products in acid tolerance. In acid-sensitive mutant AS17, the transposon insertion in the ylxM-ffh intergenic region decreased ffh expression (11). However, polar effects on the transcription of downstream genes cannot be ruled out, since satC, satD, and satE are cotranscribed with ylxM-ffh. Therefore, it was unclear which of the sat operon genes was actually associated with the acid-sensitive phenotype, especially since the function of SatC, SatD, and SatE is unknown. Homologs of the overlapping ORFs satD and satE are also found in S. pyogenes, but they have no assigned function (Microbial Genomes Blast Database; http://www.ncbi.nlm.nih.gov/). In that species, these genes are linked, but they are not located near ffh. Interestingly, satC has no homology to any known sequence or motifs. In the present study, we provide evidence that only ffh is involved in acid tolerance. The ffh mutant was unable to initiate growth at pH 5.0 (Fig. 5B). Complementation of the ffh mutant with an intact copy of the gene in the gtfA locus restored Ffh expression (Fig. 6C) and abolished any growth defects (Fig. 6A and B). Mutations in satC, satD, or satE did not affect growth at acidic conditions, nor did they result in defects in Ffh expression (Fig. 5C). Finally, although its mutant was slightly affected in growth, ylxM expression is not essential for Ffh expression.
Although significantly affected in its growth, our ffh mutant was viable, whereas in other organisms, ffh knockout attempts failed, thereby suggesting that Ffh expression is essential for viability of the cells (19, 33, 51). It can be argued that insertion of a Kanr cassette at position 629 in ffh results in a truncated but functional Ffh fragment containing the NH2-terminal 209 amino acid residues. However, based on homology to E. coli (23, 36) and B. subtilis Ffh (19, 25, 44), this putative truncated protein would only contain the first two GTP-binding domains of the G domain. The domains essential for binding to both hydrophobic signal sequences of transported proteins (44) and small cytoplasmic RNA (25) are encompassed in the C-terminal (residues 297 to 516) M domain (11). A BLAST search of the S. mutans genomic database, using the E. coli ffh sequence as the query, identified some partial ORFs that showed statistically significant sequence similarity to Ffh; however, they appear to be related to the SRP receptor FtsY, the membrane docking protein for Ffh. Although we cannot rule out the possibility that this protein can take over part of the function of Ffh in ffh knockout mutants, it is unlikely, because this protein lacks significant homology to the essential signal sequence-binding domain of Ffh. Taking these results together, we conclude that MK4 is a genuine ffh mutant that expresses no functional Ffh.
The underlying defects in the acid-sensitive phenotype of MK4 remain to be elucidated. Tn917 mutant AS17 was unable to rapidly increase H+/ATPase activity in the membrane upon acid shock (11), as part of an overall acid tolerance response (2, 3, 15). In other S. mutans strains, exposure to acid conditions induces an upregulation of synthesis of at least 36 proteins, albeit transient in nature for most of them (17). At present, the identity and localization of these proteins are unknown. The SRP pathway could be involved in the ATR in these strains by translocating the upregulated membrane and extracellular proteins. The strain (NG8) used in the present study, however, did not display a classical ATR (data not shown). This strain is atypical in that preincubation at pH 5.0 to 5.5 does not reduce the number of cells killed by subsequent incubation at pH 3.5. The genetic regulatory factors responsible for the ATR in S. mutans would appear, therefore, to be distinct from the regulatory genes controlling expression of the sat locus. Instead, as mentioned above, upregulation of the sat operon may be part of a more generalized stress response involving the osmoprotection (opuA) locus and possibly other loci. Further studies will explore the genetic basis of this generalized stress response.
In contrast to the mechanism of translocation, the repertoire of proteins that are translocated by the SRP pathway has barely been studied in the model organisms E. coli and B. subtilis. The main SRP substrates in E. coli appear to be a subset of polytopic inner membrane proteins (46, 47). In B. subtilis, the requirement of SRP for the localization of membrane proteins has not been analyzed. However, Ffh depletion inhibits the secretion of over 80% of extracellular proteins, as determined by two-dimensional gel electrophoresis (18). To gain insight into the role of the SRP pathway in S. mutans, analysis of the SRP-dependent proteins is of great importance. Future research will therefore focus on the differential protein patterns in membranes and culture supernatants of our ffh mutant and the wild-type strain, grown under identical conditions.
ACKNOWLEDGMENTS
We thank M. Swanson and R. Hector for excellent technical advice on primer extension; A. L. Honeyman, University of South Florida, Tampa, for providing pALH124; and D. G. Cvitkovitch, University of Toronto, Toronto, Canada, for providing pVAGTFA.
This study was supported by NIH grant DE08007–14.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Belli W A, Marquis R E. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl Environ Microbiol. 1991;57:1134–1138. doi: 10.1128/aem.57.4.1134-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belli W A, Marquis R E. Catabolite modification of acid tolerance of Streptococcus mutans GS5. Oral Microbiol Immunol. 1994;9:29–34. doi: 10.1111/j.1399-302x.1994.tb00211.x. [DOI] [PubMed] [Google Scholar]
- 4.Bender G R, Sutton S V W, Marquis R E. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infect Immun. 1986;53:331–338. doi: 10.1128/iai.53.2.331-338.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender G R, Thibodeau E A, Marquis R E. Reduction of acidurance of streptococcal growth and glycolysis by fluoride and gramicidin. J Dent Res. 1985;64:90–95. doi: 10.1177/00220345850640021701. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein H D. Protein targeting: getting into the groove. Curr Biol. 1998;8:R715–R718. doi: 10.1016/s0960-9822(98)70456-7. [DOI] [PubMed] [Google Scholar]
- 7.Bowden G H W. Which bacteria are cariogenic in humans? In: Johnson N M, editor. Dental caries. 1. Markers of high and low risk groups and individuals. Cambridge, United Kingdom: Cambridge University Press; 1991. pp. 266–286. [Google Scholar]
- 8.Dashper S G, Reynolds E C. pH regulation by Streptococcus mutans. J Dent Res. 1992;71:1159–1165. doi: 10.1177/00220345920710050601. [DOI] [PubMed] [Google Scholar]
- 9.De Gier J-W L, Valent Q A, Von Heijne G, Luirink J. The E. coli SRP: preferences of a targeting factor. FEBS Lett. 1997;408:1–4. doi: 10.1016/s0014-5793(97)00402-x. [DOI] [PubMed] [Google Scholar]
- 10.Felsenszwalb I, Sargentini N J, Smith K C. Characterization of a new radiation-sensitive mutant, Escherichia coli K12 radC102. Radiat Res. 1984;97:615–625. [PubMed] [Google Scholar]
- 11.Gutierrez J A, Crowley P J, Cvitkovitch D G, Brady L J, Hamilton I R, Hillman J D, Bleiweis A S. Streptococcus mutans ffh, a gene encoding a homolog of the 54 kDa subunit of the signal recognition particle, is involved in resistance to acid stress. Microbiology. 1999;145:357–366. doi: 10.1099/13500872-145-2-357. [DOI] [PubMed] [Google Scholar]
- 12.Gutierrez J A, Crowley P J, Brown D P, Hillman J D, Youngman P, Bleiweis A S. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J Bacteriol. 1996;178:4166–4175. doi: 10.1128/jb.178.14.4166-4175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez J A, Cvitkovitch D G, Hamilton I R, Hillman J D, Bleiweis A S. The Streptococcus mutans ffh gene is involved in acidurance. J Dent Res. 1997;76:102. . (Abstract.) [Google Scholar]
- 14.Hamada S, Slade H D. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev. 1980;44:331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamilton I R, Buckley N D. Adaptation by Streptococcus mutans to acid tolerance. Oral Microbiol Immunol. 1991;6:65–71. doi: 10.1111/j.1399-302x.1991.tb00453.x. [DOI] [PubMed] [Google Scholar]
- 16.Hamilton I R, Ellwood D C. Effects of fluoride on carbohydrate metabolism by washed cells of Streptococcus mutans grown at various pH values in a chemostat. Infect Immun. 1978;19:434–442. doi: 10.1128/iai.19.2.434-442.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton I R, Svensäter G. Acid-regulated proteins induced by Streptococcus mutans and other oral bacteria during acid shock. Oral Microbiol Immunol. 1998;13:292–300. doi: 10.1111/j.1399-302x.1998.tb00710.x. [DOI] [PubMed] [Google Scholar]
- 18.Hirose I, Sano K, Shioda I, Kumano M, Nakamura K, Yamane K. Proteome analysis of Bacillus subtilis extracellular proteins: a two-dimensional protein electrophoretic study. Microbiology. 2000;146:65–75. doi: 10.1099/00221287-146-1-65. [DOI] [PubMed] [Google Scholar]
- 19.Honda K, Nakamura K, Nishiguchi M, Yamane K. Cloning and characterization of a Bacillus subtilis gene encoding a homolog of the 54-kilodalton subunit of mammalian signal recognition particle and Escherichia coli Ffh. J Bacteriol. 1993;175:4885–4894. doi: 10.1128/jb.175.15.4885-4894.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutchinson C A, Peterson S N, Gill S R, Cline R T, White O, Fraser C M, Smith H O, Venter J C. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 21.Imfeld T, Lutz F. Intraplaque acid formation assessed in children and young adults. Pediatr Dent. 1980;2:87–93. [Google Scholar]
- 22.Jannsen G R. Eubacterial, archaebacterial, and eucaryotic genes that encode leaderless mRNA. In: Baltz R H, Hegeman G D, Skatrud P L, editors. Industrial microorganisms: basic and applied molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 59–67. [Google Scholar]
- 23.Jensen C G, Brown S, Pederson S. Effect of 4.5S RNA depletion on Escherichia coli protein synthesis and secretion. J Bacteriol. 1994;176:2502–2506. doi: 10.1128/jb.176.9.2502-2506.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen E M, Wefel J S. Human dental plaque responses to meals and the effects of chewing gum. Br Dent J. 1989;167:204–208. doi: 10.1038/sj.bdj.4806971. [DOI] [PubMed] [Google Scholar]
- 25.Kurita K, Honda K, Suzuma S, Takamatsu H, Nakamura K, Yamane K. Identification of a region of Bacillus subtilis Ffh, a homolog of mammalian SRP54 protein, that is essential for binding to small cytoplasmic RNA. J Biol Chem. 1996;271:13140–13146. doi: 10.1074/jbc.271.22.13140. [DOI] [PubMed] [Google Scholar]
- 26.Li Y-H, Lau P C Y, Lee J H, Ellen R P, Cvitkovitch D G. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol. 2001;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loesche W J. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez P, Martinez S, Diaz A, Espinosa M, Lacks S A. Characterization of the polA gene of Streptococcus pneumoniae and comparison of the DNA polymerase I it encodes to homologous enzymes from Escherichia coli and phage T7. J Biol Chem. 1989;264:4255–4263. [PubMed] [Google Scholar]
- 29.Luirink J, Dobberstein B. Mammalian and Escherichia coli signal recognition particles. Mol Microbiol. 1994;11:9–13. doi: 10.1111/j.1365-2958.1994.tb00284.x. [DOI] [PubMed] [Google Scholar]
- 30.Macrina F L, Evans R P, Tobian J A, Hartley D L, Clewell D B, Jones H R. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene. 1983;25:145–150. doi: 10.1016/0378-1119(83)90176-2. [DOI] [PubMed] [Google Scholar]
- 31.O'Connor M, Asai T, Squires C L, Dahlberg A E. Enhancement of translation by the downstream box does not involve base pairing of mRNA with the penultimate stem sequence of 16S rRNA. Proc Natl Acad Sci USA. 1999;96:8973–8978. doi: 10.1073/pnas.96.16.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perry D, Kuramitsu H K. Genetic transformation of Streptococcus mutans. Infect Immun. 1981;32:1295–1297. doi: 10.1128/iai.32.3.1295-1297.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Philips G J, Silhavy T J. The E. coli ffh gene is necessary for viability and efficient protein export. Nature. 1992;359:744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- 34.Sabelnikov A G, Greenberg B, Lacks S A. An extended −10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J Mol Biol. 1995;250:144–155. doi: 10.1006/jmbi.1995.0366. [DOI] [PubMed] [Google Scholar]
- 35.Samuelsson T, Macao B, Bölske G. A 13-kDA protein with a helix-turn-helix motif is encoded by bacterial operons related to the SRP pathway. Biochem Biophys Res Commun. 1997;231:839–843. doi: 10.1006/bbrc.1997.6199. [DOI] [PubMed] [Google Scholar]
- 36.Samuelsson T, Olsen M, Wikström P M, Johansson B R. The GTPase activity of Escherichia coli Ffh protein is important for normal cell growth. Biochim Biophys Acta. 1995;1267:83–91. doi: 10.1016/0167-4889(95)00034-p. [DOI] [PubMed] [Google Scholar]
- 37.Sansone C, van Houte J, Joshipura K, Kent R, Margolis H C. The association of mutans streptococci and non-mutans streptococci capable of acidogenesis at a low pH with dental caries on enamel and root surfaces. J Dent Res. 1993;72:508–516. doi: 10.1177/00220345930720020701. [DOI] [PubMed] [Google Scholar]
- 38.Shean C S, Gottesman M E. Translation of the prophage λ cI transcript. Cell. 1992;70:513–522. doi: 10.1016/0092-8674(92)90175-c. [DOI] [PubMed] [Google Scholar]
- 39.Shiroza T, Kuramitsu H K. Construction of a model secretion system for oral streptococci. Infect Immun. 1993;61:3745–3755. doi: 10.1128/iai.61.9.3745-3755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sprengart L M, Fatscher H P, Fuchs E. The initiation of translation in E. coli: apparent base pairing between the 16S rRNA and downstream sequences of the mRNA. Nucleic Acids Res. 1990;18:1719–1723. doi: 10.1093/nar/18.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephan R M. Intra-oral hydrogen-ion concentration associated with dental caries activity. J Dent Res. 1944;23:257–266. [Google Scholar]
- 42.Svensäter G, Larsson U-B, Greif E C G, Cvitkovitch D G, Hamilton I R. Acid tolerance response and survival by oral streptococci. Oral Microbiol Immunol. 1997;12:266–273. doi: 10.1111/j.1399-302x.1997.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 43.Svensäter G, Sjögreen B, Hamilton I R. Multiple stress responses in Streptococcus mutans and the induction of general and stress-specific proteins. Microbiology. 2000;146:107–117. doi: 10.1099/00221287-146-1-107. [DOI] [PubMed] [Google Scholar]
- 44.Takamatsu H, Bunai K, Horinaka T, Oguro A, Nakamura K, Watabe K, Yamane K. Identification of a region for binding to presecretory protein in Bacillus subtilis Ffh, a homolog of the 54-kDa subunit of the mammalian signal recognition particle. Eur J Biochem. 1997;248:575–582. doi: 10.1111/j.1432-1033.1997.00575.x. [DOI] [PubMed] [Google Scholar]
- 45.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ulbrandt N D, Newitt J A, Bernstein H D. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 47.Valent Q A, Scotti P A, High S, de Gier J-W, von Heijne G, Lentzen G, Wintermayer W, Oudega B, Luirink J. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Heide T, Poolman B. Osmoregulated ABC-transport system of Lactococcus lactis senses water stress via changes in the physical state of the membrane. Proc Natl Acad Sci USA. 2000;97:7102–7106. doi: 10.1073/pnas.97.13.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Houte J, Lopman J, Kent R. The final pH of bacteria comprising the predominant flora of sound and carious root and enamel surfaces. J Dent Res. 1996;75:1008–1014. doi: 10.1177/00220345960750040201. [DOI] [PubMed] [Google Scholar]
- 50.Voskuil I M, Chambliss G H. The −16 region of Bacillus subtilis and other Gram-positive bacterial promoters. Nucleic Acids Res. 1998;26:3584–3590. doi: 10.1093/nar/26.15.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walter P, Johnson A E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 52.Yamada T, Igarashi K, Mitsutomi M. Evaluation of cariogenicity of glycosylsucrose by a new method of measuring pH under human dental plaque in situ. J Dent Res. 1980;59:2157–2162. [Google Scholar]
- 53.Zhang Y-B, Ayalew S, Lacks S A. The rnhB gene encoding RNase HII of Streptococcus pneumoniae and evidence of conserved motifs in eucaryotic genes. J Bacteriol. 1997;179:3828–3836. doi: 10.1128/jb.179.12.3828-3836.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zweib C, Samuelsson T. SRPDB (signal recognition particle database) Nucleic Acids Res. 2000;28:171–172. doi: 10.1093/nar/28.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]