Abstract
Neisseria meningitidis (the meningococcus) is a naturally competent bacterial species in which intra- and interspecific horizontal gene transfer is a major source of genetic diversity. In strains of the electrophoretic type 37 (ET-37) complex and of the A4 cluster, we identified genomic DNA coding for a novel restriction-modification system and for the tail of a previously unidentified prophage. Furthermore, a novel 7.2-kb DNA segment restricted to clones of the ET-37 complex and the A4 cluster was isolated and shown to occur both as a plasmid (pJS-B) and as a chromosomal integration. Neither the genomic loci nor pJS-B was present in ET-5 complex, lineage 3, or serogroup A meningococci. The differential distribution of the DNA segments described herein, as well as of opcA, porB, nmeAI, nmeBI, and nmeDI described previously, supports the concept of genetic isolation of hypervirulent lineages responsible for most cases of serogroup C disease worldwide.
The species Neisseria meningitidis is composed of a large variety of clones and clonal groupings (reviewed in reference 1). The majority of clones colonizing the nasopharynx of humans should be regarded as avirulent (7), and only a few hypervirulent clonal groupings are responsible for the majority of disease in humans. In Europe, serogroup B disease is caused mainly by the electrophoretic type 5 (ET-5) complex and lineage 3 whereas serogroup C disease is caused by the ET-37 complex and the A4 cluster (reviewed in reference 1). Horizontal gene transfer within the naturally competent species and with commensal neisseriae has resulted in genetic variability (2, 11–13, 27), but it has been demonstrated that hypervirulent lineages of both serogroups B and C can be readily distinguished by the presence or absence of several genomic loci; e.g., the ET-5 complex and lineage 3 are characterized by a class 3 porB gene, whereas a class 2 gene is found in the ET-37 complex and the A4 cluster (9, 43). Isotype I of the tbpB gene is restricted to meningococci of the ET-37 complex, while other hypervirulent lineages harbour isotype II (31). Furthermore, the latter lack the gene encoding the adhesin OpcA (32, 44).
We recently demonstrated a differential distribution of three novel restriction-modification (R-M) systems, nmeAI, nmeBI, and nmeDI (10), while a lineage 3-specific R-M system was described by Bart et al. (3). The differential distribution of meningococcal genes can be caused by rapid expansion of lineages with increased fitness, bottlenecking, periodic selection, and immune selection, which are mechanisms antagonizing the effects of recombination (2, 15, 16, 26, 33). In addition, we recently provided evidence that sexual isolation of clonal groupings is caused by differentially distributed R-M systems which partially disrupt genetic transformation (10). In the present study we have extended the list of currently known differentially distributed genes. We compared the ET-37 complex with the ET-5 complex by plasmid profiling and representational difference analysis (RDA). We identified a novel plasmid that is unique to the ET-37 complex and the A4 cluster and also identified two novel genomic loci of the ET-37 complex not present in the ET-5 complex, lineage 3, and serogroup A meningococci.
MATERIALS AND METHODS
Bacterial strains.
A total of 163 meningococcal strains were obtained from the following collections: Dominique A. Caugant (Oslo, Norway), Mark Achtman (Berlin, Germany), National Reference Centre for meningococci (NRZ; Heidelberg, Germany), Paula Kriz (Prague, Czech Republic), Manchester Public Health Laboratory (United Kingdom), E. R. Moxon (Oxford, United Kingdom), and Institute for Hygiene and Microbiology, University of Würzburg (our own isolates). Multilocus enzyme electrophoresis assignments were provided by D. A. Caugant (see also references 8, 9, and 14). Sequence types (STs) were obtained from the multilocus sequence typing (MLST) website (http://mlst.zoo.ox.ac.uk/) (28).
Recombinant DNA technology.
Restriction and DNA-modifying enzymes were purchased from New England BioLabs (Beverly, Mass.). Chromosomal DNA from N. meningitidis was purified on a CsCl gradient. For the extraction of meningococcal plasmid DNA, meningococci grown for 12 h on GC agar (Difco) were resuspended in phosphate-buffered saline. The pellet from 1 ml of a bacterial suspension with an optical density of 10 at 600 nm was used for plasmid preparation by the alkali lysis method (4). RDA was performed essentially as described previously (10). For DNA-DNA dot blot hybridizations, 20-μl volumes of suspensions containing 1010 CFU per ml of H2O were dotted onto nylon membranes (Macherey-Nagel, Düren, Germany). Alternatively, 200-ng portions of chromosomal DNA were spotted. Southern and colony blot hybridizations were performed as described previously (19) with digoxigenin-labeled probes. The probes 48/49 and 43/50, which were used for detection of pJS-B, comprised positions 1400 to 2382 and positions 5252 to 558, respectively (numbered according to EMBL database entry AJ277475). Probes 1–7 (positions 1455 to 2205, [AJ278708]) and 1–26 (positions 5193 to 5760 [AJ278707]) were used to detect chromosomal loci 37/1-7 and 37/1-26. PCR was performed on a thermal cycler obtained from Biometra (Göttingen, Germany). The thermostable DNA polymerase AmpliTaq was purchased from Perkin-Elmer (Weiterstadt, Germany).
The following oligonucleotides specific to 37/1-7 were used to analyze strains for the presence of the restriction-modification system and the insertion sequence IS1655: HC278 (5′-TTG CCC GTA ATT ATG GTT CTC-3′, positions 3,007 to 3,027 [AJ278708]), HC277 (5′-CCG TTG CTT TTG TCT TGG CG-3′, positions 3809 to 3790), and HC273 (5′-ATA CGC CAC AAT AGC TCA GC-3′, positions 4469 to 4450). The site of insertion of IS1655 (29) was determined by sequencing the HC278/HC273 PCR product with primer HC280 (5′-AAA GAG TTC TAC CAA CAC ACC-3′). Automated DNA sequencing was performed on a sequencer (model 377; Applied Biosystems, Foster City, Calif.) by the dye terminator cycle method with AmpliTaq. Nucleotide sequence data were analyzed with the Lasergene sequence analysis software (Dnastar, Madison, Wis.). DNA and protein sequences were compared with the GenBank and SWISS-PROT databases on the BLAST server hosted by the National Center for Biotechnology Information (NCBI), Bethesda, Md. Further DNA comparisons were made with sequence data released by the meningococcus genome-sequencing projects at the Sanger Centre (Cambridge, United Kingdom) (http://www.sanger.ac.uk/Projects/N_meningitidis/) and at The Institute for Genomic Research (TIGR) (http://www.tigr.org/).
Nucleotide sequence accession numbers.
The sequence of plasmid pJS-B has been submitted to the EMBL database and given accession number AJ277475. The sequences of loci 37/1-7 and 37/1-26 have been given accession numbers AJ278708 and AJ278707, respectively.
RESULTS
A novel plasmid, pJS-B (EMBL database accession number AJ277475), plus two novel genomic loci, i.e., 37/1-7 (AJ278708) and 37/1-26 (AJ278707), were identified in meningococci of the ET-37 complex and characterized further.
Plasmid pJS-B.
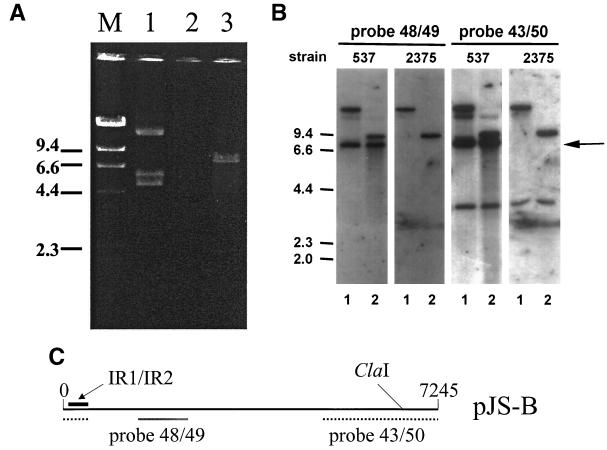
Representative strains of different clonal lineages including strains of the ET-37 complex were screened for the presence of plasmids. A 7,245-bp plasmid designated pJS-B was isolated from the ET-37 complex strain 537 (found in Norway) (original strain name P179; donation of D. A. Caugant) (Fig. 1). Eight open reading frames (ORFs) were identified on the plasmid. The nucleotide sequences of the ORFs, as well as the deduced amino acid sequences, did not reveal significant homologies to entries in public databases (GenBank, EMBL, SWISS-PROT). Therefore, we were not able to classify the plasmid type. Positions 20 to 307 of the plasmid were 93% identical to the IR1/IR2 inverted-repeat region upstream of the pivNG gene of Neisseria gonorrhoeae, which encodes the pilin gene-inverting protein homologue PivNG (accession number U65994) (5). Sequences homologous to the repeat region are also present in the genomes of meningococcal strains MC58 and Z2491 (29, 37). It has been speculated that the PivNG protein is involved in genomic rearrangements in gonococci (5). In the original description of IR1/IR2, the authors furthermore contended that the repeats might serve as recombination sites used by PivNG. This suggestion is interesting for the following reason: we could not isolate a plasmid from strain 2375 (ET-37 complex, ET-15 clone; Czech Republic) (Fig. 1), but by dot blot analysis with independent probes specific to pJS-B, which covered the whole plasmid, we could demonstrate that chromosomal DNA of 2375 carried a copy of the complete plasmid (data not shown). To further examine this finding, we conducted the following experiment. Chromosomal DNA of strains 537 and 2375 were digested either with BamHI plus ClaI or with EcoRV plus ClaI. These chromosomal DNA preparations contained small amounts of plasmid. ClaI cuts at a single site on the plasmid (position 6811), whereas EcoRV and BamHI do not cleave the plasmid. Southern blots of digested DNA with the independent probes 43/50 and 48/49 revealed identical patterns except for a 7.2-kb band representing the plasmid linearized by ClaI (Fig. 1). Taken together, these experiments showed that pJS-B appears to be a chromosomal locus, which most frequently can also be isolated as a plasmid. We hypothesize that the site of recombination is the IR1/IR2 region. Evidence for this hypothesis is lacking as several attempts to clone the chromosomal region containing the pJS-B insertion site have been unsuccessful.
FIG. 1.
(A) Plasmid preparation by the alkali lysis method from strains 537, which carries pJS-B (lane 1, native plasmid; lane 3, plasmid cleaved with ClaI), and strain 2375, which is lacking the plasmid (lane 2). (B) Southern blot demonstrating plasmid pJS-B in its chromosomal and its plasmid forms in strains 537 and 2375. Chromosomal DNA was digested with ClaI-BamHI (lanes 1) or with ClaI-EcoRV (lanes 2). (C) Probe 48/49 covered positions 1400 through 2382 of the plasmid, and probe 43/50 covered positions 5252 through 558. Thus, only probe 43/50 contained the ClaI recognition site of the plasmid (position 6811). The hybridization patterns obtained with the two strains were identical, except for the linearized plasmid band, which appeared only in strain 537 (arrow). The remaining bands were due to hybridization of the probes to the chromosomal copy of pJS-B present in both strains. The position of the inverted repeat region IR1/IR2 of the plasmid is indicated.
Putative R-M system 37/1-7.
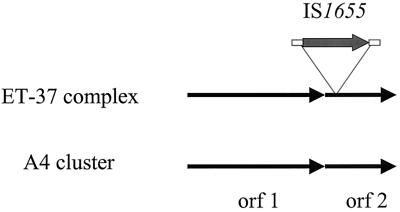
We and others have previously demonstrated the potential of RDA to identify R-M systems in meningococci (3, 10, 30, 38). We therefore performed RDA using strain 2120 (ET-37 complex) (40) as the tester and strain MC58 (ET-5 complex) (37) as the driver. From strain 2120, we isolated a novel hypothetical R-M system designated 37/1-7 with homology to a hypothetical R-M system (HP0054/HP0053) identified by genome sequencing of Helicobacter pylori (http://www.tigr.org) (39). The deduced amino acid sequence of ORF 1 of 37/1-7 exhibits 42% identity over 831 amino acids to the product of the coding sequence HP0054, i.e., a putative adenine/cytosine DNA methyltransferase. The partially sequenced deduced amino acid sequences of ORF 2 of 37/1-7 shows 46% identity over 263 amino acids to the product of HP0053, which is located immediately downstream of HP0054 in H. pylori. In contrast to methyltransferases (25), endonucleases are much less highly conserved and functional domains cannot be deduced from sequence data (21). Therefore, we can only imply that ORF 2 of 37/1-7 and HP0053 encode endonucleases, because the genes are located downstream of the genes encoding the modifying enzymes. This organization is frequently observed in bacterial R-M systems. In strain 2120, a copy of an insertion sequence (IS) belonging to the IS30 family was found to be inserted at position 260 of ORF 2. Therefore, ORF 2 must be considered to be disrupted and nonfunctional. The same IS occurs in 13 and 6 copies in the genomes of MC58 (ET-5 complex) (37) and Z2491 (subgroup IV-1) (29), respectively, and has been designated IS1655 (29). By PCR with primers HC278, HC277, and HC273, we confirmed that IS1655 was present in 59 of 59 ET-37 complex strains. The IS insertion, however, was not seen in A4 cluster strains, indicating that the IS insertion occurred after divergence of the ET-37 complex and the A4 cluster. It has apparently remained stable in ET-37 complex strains. We determined the site of insertion of IS1655 in 12 of 59 ET-37 complex strains. For this purpose, we sequenced the HC278/HC273 PCR product obtained from ET-37 complex strains by using the primer HC280. All 12 strains analyzed exhibited precisely the same site of insertion of IS1655. In one strain, which was kindly provided by D. A. Caugant (original strain designation, 401), we additionally observed the insertion of IS1301 (17, 19) into the left inverted repeat of IS1655. The insertion of IS1301 into IS1655 proved to be a rare exception, which might be explained by the fact that IS1301 only rarely occurs in ET-37 complex strains (19, 20). The stable insertion of IS1655 in ET-37 complex meningococci argues for a lack of mobility of this IS element. Taken together, we have demonstrated the occurrence of a novel putative R-M system in ET-37 and A4 cluster meningococci. Of the two clonal groupings, only the ET-37 complex exhibited an insertion of IS1655 in orf 2 (Fig. 2).
FIG. 2.
Genomic organization of the putative R-M system 37/1-7 in the ET-37 complex and the A4 cluster. A stable insertion of IS1655 in ORF 2 was found in the ET-37 complex but not in the A4 cluster. Note that the 3′ end of ORF 2 was not sequenced.
Putative prophage 37/1-26.
The RDA described above comparing the ET-37 complex with the ET-5 complex resulted in isolation of a chromosomal fragment of the ET-37 complex strain containing prophage DNA. The genomic fragment was designated 37/1-26. We have currently sequenced 6,764 bp of the putative meningococcal prophage. Four ORFs with identical orientation have been identified in 37/1-26. The deduced amino acids of ORF 1 (partial ORF coding for 195 amino acids), ORF 2 (252 amino acids), ORF 3 (241 amino acids), and ORF 4 (1,422 amino acids) exhibit 54, 44, 36, and 34% identity over 192, 194, 200, and 1,001 amino acids to the structural proteins L (SWISS-PROT accession number P03738), K (P03729), I (P03730), and J (P03749), respectively, of the Escherichia coli bacteriophage lambda. Proteins L, K, I, and J are structural proteins involved in tail assembly of phage lambda. The database comparisons therefore suggest that we have found a fragment of a novel meningococcal lambdoid prophage. Using the BLAST servers of the Sanger Centre and of TIGR, respectively, no significant homologies to any of the coding sequences of the genomes of the serogroup A strain Z2491 and of the serogroup B strain MC58 could be detected. This finding demonstrates that 37/1-26 is not related to prophages identified by the recent meningococcal genome sequencing efforts (29, 37).
Distribution of pJS-B, 37/1-7 and 37/1-26.
We performed DNA-DNA hybridizations in a dot blot format to investigate the distribution of pJS-B, 37/1-7, and 37/1-26. A total of 163 meningococcal strains were included in the analysis (ET-37 complex, n = 59; A4 cluster, n = 36; ET-5 complex, n = 19; lineage 3, n = 5; serogroup A [ST-1-7], n = 7; other electrophoretic or sequence types not belonging to hypervirulent lineages, n = 37). The data are summarized in Table 1. Epidemiologically related isolates gave similar results. For example, 28 ET-37 complex strains belonging to clone ET-15, which were isolated in the Czech Republic or Bavaria between 1993 and 1998 (18, 23, 24), were positive for all probes used. The genomic loci 37/1-7 (putative R-M system) and 37/1-26 (putative prophage) were present in all ET-37 complex strains and in more than 90% of the A4 cluster strains. pJS-B was identified in most of the ET-37 complex strains (88%) and in many of the A4 cluster strains (58.3%). Among the ET-37 complex strains negative for pJS-B was strain FAM18, which is currently under genome sequence investigation at the Sanger Centre. The plasmid, 37/1-7, and 37/1-26 were not found in the ET-5 complex, lineage 3, and serogroup A meningococci. 37/1-7 and 37/1-26 were infrequently found in strains not belonging to the hypervirulent lineages shown in Table 1. This finding probably suggests sporadic horizontal transfer of 37/1-7 and 37/1-26 to these lineages. Taken together, the data demonstrate a differential distribution of the novel DNA fragments identified in this study, which proved to be rather specific for the ET-37 complex and the A4 cluster.
TABLE 1.
Differential distribution of 37/1-7, 37/1-26, and pJS-Ba
| Clonal lineage | No. of strains | No. of countries | No. (%) of:
|
||
|---|---|---|---|---|---|
| 37/1-7 | 37/1-26 | pJS-B | |||
| ET-37 complex | 59 | 10 | 59 (100) | 59 (100) | 52 (88.1) |
| A4 cluster | 36 | 14 | 34 (92.0) | 35 (97.2) | 21 (58.3) |
| ET-5 complex | 19 | 4 | 0 | 0 | 0 |
| Lineage 3 | 5 | 5 | 0 | 0 | 0 |
| Serogroup A (ST-1-7) | 7 | 5 | 0 | 0 | 0 |
| Otherb | 37 | 11 | 4 (10.8) | 6 (16.2) | 0 |
The presence of DNA fragments was determined by DNA-DNA hybridization using dot blots.
Electrophoretic or sequence types not belonging to hypervirulent linages.
DISCUSSION
N. meningitidis is one of the most variable bacterial species colonizing the human host. This is reflected by the fact that over 1,000 STs have been identified by MLST since publication of the method in 1998 (28). The genetic variability is due in large part to recombination (11, 12). However, despite frequent recombination, the population structure of N. meningitidis is not panmictic. Only a few clonal groupings, which have been known for decades, have been demonstrated to be responsible for majority of cases of serogroup A, B, and C meningococcal disease. Accordingly, measurement of the frequency of recombination by using the homoplasy test revealed that recombination was much less obvious in meningococci than in H. pylori (34–36). In particular, hypervirulent clonal lineages of meningococci causing serogroup C disease have been genetically uniform over decades, because the vast majority of ET-37 complex and A4 cluster strains belong to only four STs and because many specific genetic markers, including the ones demonstrated in this study, are found in most strains of the lineages isolated from many countries over many years. These findings may be due to increased fitness of strains of these STs and their rapid expansion, or other purifying mechanisms such as bottlenecking and immune selection. Furthermore, genetic isolation should be taken into consideration: although ET-37 complex meningococci are easily transformable by plasmids and genomic DNA in the laboratory, it is unclear how actively this clonal grouping participates in horizontal gene transfer in nature. Linz et al. (27) recently made the striking observation that 95 ET-37 complex strains isolated in Mali between 1990 and 1992 showed no sequence variation in the tbpB gene whereas 98 subgroup IV-1 serogroup A strains isolated in the Gambia in the mid-1980s exhibited independent import of 17 foreign tbpB alleles. This finding suggests that ET-37 complex meningococci incorporate foreign DNA less frequently in nature than do serogroup A meningococci.
One possible mechanism of genetic isolation of meningococcal lineages may be the result of a differential distribution of R-M systems, as we recently demonstrated for nmeBI (10). This R-M system was found in ET-5 complex meningococci but not in ET-37 complex meningococci. We showed that ET-5 complex meningococci were moderately protected from uptake of DNA derived from the ET-37 complex, if NmeBI restriction sites were placed adjacent to the selective marker in the transforming DNA (10). It remains unclear, however, whether similar mechanisms act on transformation of ET-37 complex meningococci.
Does the hypothesis that genetic isolation separates ET-5 and ET-37 complex meningococci also hold true for housekeeping genes, which are abundant in the neisserial gene pool? This question can be easily addressed by using the information deposited in the MLST database. The prototype of the ET-5 complex is ST-32, and that of the ET-37 complex is ST-11. ST-32 and ST-11 differ in all alleles of housekeeping genes analyzed by MLST. We retrieved from the MLST database all STs which differed from ST-32 or ST-11 in only one of seven housekeeping gene alleles. We found that several alleles were shared by these closely related derivatives of the ST-32 and the ST-11, i.e., abcZ8, adk-3, gdh-6, and pdhC3. This finding demonstrates that alleles of the ST-32 complex enter the ST-11 complex and vice versa. How does this finding fit into the hypothesis of genetic isolation of the ET-37 (ST-11) complex suggested by the differential distribution of pJS-B, 37/1-7, and 37/1-26? The reasons for this discrepancy are unclear. One might speculate that selective pressures account for the finding. On the other hand, it has to be taken into account that the size of the MLST database far exceeds the number of strains analyzed in this study, which makes a comparison of the results difficult. However, a reason for the lack of mobility of DNA islands such as 37/1-7 within the neisserial gene pool may be that recombination of DNA islands, in contrast to recombination of housekeeping genes, is difficult due to their heterologous nature.
Plasmid profiles, to our knowledge, have not been systematically related to clonal groupings before. We recently described the identification of plasmid pJS-A which was found only in serogroup A meningococci of subgroup VI (EMBL database accession number AJ238491, [20]). This plasmid provided an example of a differentially distributed plasmid, which had been taken up by a defined subset of clones within subgroup VI. As described for pJS-A, pJS-B is restricted to a defined subset of meningococcal clones, i.e., the A4 cluster and the ET-37 complex. These examples illustrate that plasmid profiling in meningococci must take into account the genetic diversity of this species. The most interesting feature of plasmid pJS-B was the presence of a conserved IR1/IR2 region, which was described for gonococci and which has also been identified in meningococcal genomes (37). We suggest that this plasmid integrates chromosomally by recombination via this IR1/IR2 region. We identified strains which harbor a chromosomal version of the plasmid but do not release an extrachromosomal copy. It is unclear how plasmid liberalization is regulated and whether piv genes are involved. piv genes in gonococci are located adjacent to IR1/IR2, and their putative role in recombination via the repeats has been discussed previously (5). We also do not know yet whether pJS-B can replicate autonomously. Initial attempts to transfer the plasmid to pJS-B-negative strains failed. Nevertheless, pJS-B is another of the few examples for bacterial plasmids released from chromosomal copies, as described before in Vibrio cholerae for the CTXφ bacteriophage, which integrates chromosomally or replicates as a plasmid (41, 42). It is unclear whether pJS-B contributes to the pathogenicity of the ET-37 complex. No homology to virulence genes could be found by comparison with public databases. Furthermore, a role in pathogenicity is questionable, because not all of the pathogenic ET-37 strains, e.g., strain 2120 (40), carried pJS-B.
We recently identified three novel R-M systems differentially distributed among clonal groupings of meningococci (10). Identification of meningococcal R-M systems by RDA was also reported by others (3, 30, 38). The potential of RDA to identify bacterial R-M systems was demonstrated again in this study by the identification of 37/1-7. We recently suggested that meningococcal R-M systems might contribute to sexual isolation, as demonstrated by a reduction of the uptake of double-stranded DNA by NmeBI (10). However, 37/1-7 is probably not involved in restriction of horizontal gene transfer, because the endonuclease gene is disrupted by IS insertion in ET-37 complex strains. It is nevertheless interesting to observe the large number of R-M systems specific to distinct clonal groupings, which suggests that R-M systems frequently entered subsets of clones of meningococci. A systematic analysis of the distribution of such genes, as performed in this and our recent study (10), will be very helpful to the development of rapid molecular identification schemes for hypervirulent lineages based on PCR. Such approaches are desirable for routine typing of meningococci, because although MLST has definitly become the “gold standard” of meningococcal typing, it is still rather time-consuming and expensive.
Apart from a 1967 report of bacteriophages isolated from N. meningitidis (6), there is only one description of a complete prophage genome in meningococci by Klee et al. (22), who performed a detailed analysis of meningococcal loci not present in gonococci. One of these regions, region 3 contained a prophage designated pmn1, which consisted of 52 ORFs, of which 16 were homologous to phage Mu. pmn1 was found in group A meningococci, and analysis of the MC58 genome revealed that the phage was not present in this ET-5 complex strain. Despite the small number of reports on meningococcal prophages and bacteriophages, the serogroup A genome (subgroup IV-1) was shown to carry five partial or complete prophages (pmn1 to pmn5) (29). Furthermore, 16 ORFs derived from prophages or with phage-related functions were recovered from the serogroup B genome (ET-5 complex) (37). Therefore, one might assume that several bacteriophages entered meningococci in the course of their evolution and that at least cryptic versions of prophages are still present. In this study, we detected prophage DNA in the ET-37 complex. Homology searches suggest that the four ORFs are structural genes involved in tail assembly of a lambdoid bacteriophage. We did not sequence the entire phage genome, because the genome of the ET-37 complex strain FAM18, which carries 37/1-26, is currently being sequenced at the Sanger Centre. The genome sequence data will shortly reveal whether 37/1-26 is part of a complete or cryptic prophage. With the genome data available, we will be able to determine, in future experiments, whether the prophage is functional, whether it carries genes related to pathogenicity, and which conditions result in phage liberalization.
Taken together, plasmid profiling and RDA allowed us to identify previously unknown DNA specific to the ET-37 complex and A4 cluster. The distribution of the fragments underlines the close relationship of these clonal groupings. More importantly, it provides evidence for the presence of effective barriers to genetic exchange between hypervirulent lineages. This barrier affects several groups of genes, e.g., genes encoding R-M systems (nmeAI, nmeBI, nmeDI, and 37/1-7), genes encoding outer membrane proteins (opcA and porB), prophage DNA (37/1-26), and plasmids (pJS-B). Future studies should focus on the basic mechanisms conferring this barrier to transformation.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (grant VO-718/3-2).
We thank D. A. Caugant, M. Achtman, R. Borrow, I. Ehrhard, and E. R. Moxon for providing us with strains. M. Achtman is also gratefully acknowledged for the donation of chromosomal DNA and for helpful discussions. H. Schmidt is thanked for comments on prophages, and Ken Haynes is thanked for critically reading the manuscript. We are grateful to Gabi Heinze and Anja Suhl for their skilled technical assistance. This publication made use of the Multi Locus Sequence Typing website (http://mlst.zoo.ox.ac.uk) developed by Man-Suen Chan and sited at the Wellcome Trust Centre for the Epidemiology of Infectious Disease, University of Oxford. The development of this site is funded by the Wellcome Trust. We acknowledge the very helpful comments of the anonymous reviewers of the initial version of the manuscript.
REFERENCES
- 1.Achtman M. Global epidemiology of meningococcal disease. In: Cartwright K, editor. Meningococcal disease. Chichester, England: John Wiley & Sons, Ltd.; 1995. pp. 159–175. [Google Scholar]
- 2.Achtman M. Microevolution and epidemic spread of serogroup A Neisseria meningitidis—a review. Gene. 1997;192:135–140. doi: 10.1016/s0378-1119(97)00083-8. [DOI] [PubMed] [Google Scholar]
- 3.Bart A, Dankert J, van Der Ende A. Representational difference analysis of Neisseria meningitidis identifies sequences that are specific for the hyper-virulent lineage III clone. FEMS Microbiol Lett. 2000;188:111–114. doi: 10.1111/j.1574-6968.2000.tb09179.x. [DOI] [PubMed] [Google Scholar]
- 4.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrick C S, Fyfe J A, Davies J K. Neisseria gonorrhoeae contains multiple copies of a gene that may encode a site-specific recombinase and is associated with DNA rearrangements. Gene. 1998;220:21–29. doi: 10.1016/s0378-1119(98)00424-7. [DOI] [PubMed] [Google Scholar]
- 6.Cary S G, Hunter D H. Isolation of bacteriophages active against Neisseria meningitidis. J Virol. 1967;1:538–542. doi: 10.1128/jvi.1.3.538-542.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caugant D A, Bovre K, Gaustad P, Bryn K, Holten E, Hoiby E A, Froholm L O. Multilocus genotypes determined by enzyme electrophoresis of Neisseria meningitidis isolated from patients with systemic disease and from healthy carriers. J Gen Microbiol. 1986;132:641–652. doi: 10.1099/00221287-132-3-641. [DOI] [PubMed] [Google Scholar]
- 8.Caugant D A, Froholm L O, Bovre K, Holten E, Frasch C E, Mocca L F, Zollinger W D, Selander R K. Intercontinental spread of a genetically distinctive complex of clones of Neisseria meningitidis causing epidemic disease. Proc Natl Acad Sci USA. 1986;83:4927–4931. doi: 10.1073/pnas.83.13.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caugant D A, Mocca L F, Frasch C E, Froholm L O, Zollinger W D, Selander R K. Genetic structure of Neisseria meningitidis populations in relation to serogroup, serotype, and outer membrane protein pattern. J Bacteriol. 1987;169:2781–2792. doi: 10.1128/jb.169.6.2781-2792.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claus H, Friedrich A, Frosch M, Vogel U. Differential distribution of two novel restriction-modification systems in clonal lineages of Neisseria meningitidis. J Bacteriol. 2000;182:1296–1303. doi: 10.1128/jb.182.5.1296-1303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feil E J, Enright M C, Spratt B G. Estimating the relative contributions of mutation and recombination to clonal diversification: a comparison between Neisseria meningitidis and Streptococcus pneumoniae. Res Microbiol. 2000;151:465–469. doi: 10.1016/s0923-2508(00)00168-6. [DOI] [PubMed] [Google Scholar]
- 12.Feil E J, Maiden M C, Achtman M, Spratt B G. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol Biol Evol. 1999;16:1496–1502. doi: 10.1093/oxfordjournals.molbev.a026061. [DOI] [PubMed] [Google Scholar]
- 13.Goodman S D, Scocca J J. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1988;85:6982–6986. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grahlow W D, Caugant D A, Hoiby E A, Selander R K. Occurrence of clones of the ET-5 complex of Neisseria meningitidis in the German Democratic Republic. Z Klin Med. 1990;45:947–950. [Google Scholar]
- 15.Gupta S, Anderson R M. Population structure of pathogens: the role of immune selection. Parasitol Today. 1999;15:497–501. doi: 10.1016/s0169-4758(99)01559-8. [DOI] [PubMed] [Google Scholar]
- 16.Gupta S, Maiden M C, Feavers I M, Nee S, May R M, Anderson R M. The maintenance of strain structure in populations of recombining infectious agents. Nat Med. 1996;2:437–442. doi: 10.1038/nm0496-437. [DOI] [PubMed] [Google Scholar]
- 17.Hammerschmidt S, Hilse R, van Putten J P, Gerardy-Schahn R, Unkmeir A, Frosch M. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 1996;15:192–198. [PMC free article] [PubMed] [Google Scholar]
- 18.Hauri A M, Ehrhard I, Frank U, Ammer J, Fell G, Hamouda O, Petersen L. Serogroup C meningococcal disease outbreak associated with discotheque attendance during carnival. Epidemiol Infect. 2000;124:69–73. doi: 10.1017/s0950268899003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilse R, Hammerschmidt S, Bautsch W, Frosch M. Site-specific insertion of IS1301 and distribution in Neisseria meningitidis strains. J Bacteriol. 1996;178:2527–2532. doi: 10.1128/jb.178.9.2527-2532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilse R, Stoevesandt J, Caugant D A, Claus H, Frosch M, Vogel U. Distribution of the meningococcal insertion sequence IS1301 in clonal lineages of Neisseria meningitidis. Epidemiol Infect. 2000;124:337–340. doi: 10.1017/s0950268899003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeltsch A, Kröger M, Pingoud A. Evidence for an evolutionary relationship among type-II restriction endonucleases. Gene. 1995;160:7–16. doi: 10.1016/0378-1119(95)00181-5. [DOI] [PubMed] [Google Scholar]
- 22.Klee S R, Nassif X, Kusecek B, Merker P, Beretti J L, Achtman M, Tinsley C R. Molecular and biological analysis of eight genetic islands that distinguish Neisseria meningitidis from the closely related pathogen Neisseria gonorrhoeae. Infect Immun. 2000;68:2082–2095. doi: 10.1128/iai.68.4.2082-2095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krizova P, Musilek M. Changing epidemiology of meningococcal invasive disease in the Czech Republic caused by new clone Neisseria meningitidis C:2a: P1.2(P1.5), ET-15/37. Cent Eur J Public Health. 1995;3:189–194. [PubMed] [Google Scholar]
- 24.Krizova P, Musilek M, Kalmusova J. Development of the epidemiological situation in invasive meningococcal disease in the Czech Republic caused by emerging Neisseria meningitidis clone ET-15/37. Cent Eur J Public Health. 1997;5:214–218. [PubMed] [Google Scholar]
- 25.Kumar S, Cheng X, Klimasauskas S, Mi S, Posfai J, Roberts R J, Wilson G G. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994;22:1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine B R. Periodic selection, infectious gene exchange and the genetic structure of E. coli populations. Genetics. 1981;99:1–23. doi: 10.1093/genetics/99.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linz B, Schenker M, Zhu P, Achtman M. Frequent interspecific genetic exchange between commensal neisseriae and Neisseria meningitidis. Mol Microbiol. 2000;36:1049–1058. doi: 10.1046/j.1365-2958.2000.01932.x. [DOI] [PubMed] [Google Scholar]
- 28.Maiden M C, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B G. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkhill J, Achtman M, James K D, et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 30.Perrin A, Nassif X, Tinsley C. Identification of regions of the chromosome of Neisseria meningitidis and Neisseria gonorrhoeae which are specific to the pathogenic Neisseria species. Infect Immun. 1999;67:6119–6129. doi: 10.1128/iai.67.11.6119-6129.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rokbi B, Renauld-Mongenie G, Mignon M, Danve B, Poncet D, Chabanel C, Caugant D A, Quentin-Millet M-J. Allelic diversity of the two transferrin binding protein B gene isotypes among a collection of Neisseria meningitidis strains representative of serogroup B disease: implication for the composition of a recombinant TbpB-based vaccine. Infect Immun. 2000;68:4938–4947. doi: 10.1128/iai.68.9.4938-4947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seiler A, Reinhardt R, Sarkari J, Caugant D A, Achtman M. Allelic polymorphism and site-specific recombination in the opc locus of Neisseria meningitidis. Mol Microbiol. 1996;19:841–856. doi: 10.1046/j.1365-2958.1996.437970.x. [DOI] [PubMed] [Google Scholar]
- 33.Spratt B G, Maiden M C. Bacterial population genetics, evolution and epidemiology. Philos Trans R Soc London Ser B. 1999;354:701–710. doi: 10.1098/rstb.1999.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suerbaum S. Genetic variability within Helicobacter pylori. Int J Med Microbiol. 2000;290:175–181. doi: 10.1016/S1438-4221(00)80087-9. [DOI] [PubMed] [Google Scholar]
- 35.Suerbaum S, Achtman M. Evolution of Helicobacter pylori: the role of recombination. Trends Microbiol. 1999;7:182–184. doi: 10.1016/s0966-842x(99)01505-x. [DOI] [PubMed] [Google Scholar]
- 36.Suerbaum S, Smith J M, Bapumia K, Morelli G, Smith N H, Kunstmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tettelin H, Saunders N J, Heidelberg J, et al. Complete genome sequence of Neisseria meningitidis serogroup B Strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 38.Tinsley C R, Nassif X. Analysis of the genetic differences between Neisseria meningitidis and Neisseria gonorrhoeae: two closely related bacteria expressing two different pathogenicities. Proc Natl Acad Sci USA. 1996;93:11109–11114. doi: 10.1073/pnas.93.20.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomb J F, White O, Kerlavage A R, Clayton R A, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. . (Erratum, 389:412, 1997.) [DOI] [PubMed] [Google Scholar]
- 40.Vogel U, Morelli G, Zurth K, Claus H, Kriener E, Achtman M, Frosch M. Necessity of molecular techniques to distinguish between Neisseria meningitidis strains isolated from patients with meningococcal disease and from their healthy contacts. J Clin Microbiol. 1998;36:2465–2470. doi: 10.1128/jcm.36.9.2465-2470.1998. . (Erratum, 37:882, 1999.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 42.Waldor M K, Rubin E J, Pearson G D, Kimsey H, Mekalanos J J. Regulation, replication, and integration functions of the Vibrio cholerae CTXphi are encoded by region RS2. Mol Microbiol. 1997;24:917–926. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]
- 43.Wang J F, Caugant D A, Morelli G, Koumare B, Achtman M. Antigenic and epidemiologic properties of the ET-37 complex of Neisseria meningitidis. J Infect Dis. 1993;167:1320–1329. doi: 10.1093/infdis/167.6.1320. [DOI] [PubMed] [Google Scholar]
- 44.Zhu P, Morelli G, Achtman M. The opcA and ψopcB regions in Neisseria: genes, pseudogenes, deletions, insertion elements and DNA islands. Mol Microbiol. 1999;33:635–650. doi: 10.1046/j.1365-2958.1999.01514.x. [DOI] [PubMed] [Google Scholar]