Abstract
Eight genes have been identified that function in the regulation, biosynthesis, and transport of rhizobactin 1021, a hydroxamate siderophore produced under iron stress by Sinorhizobium meliloti. The genes were sequenced, and transposon insertion mutants were constructed for phenotypic analysis. Six of the genes, named rhbABCDEF, function in the biosynthesis of the siderophore and were shown to constitute an operon that is repressed under iron-replete conditions. Another gene in the cluster, named rhtA, encodes the outer membrane receptor protein for rhizobactin 1021. It was shown to be regulated by iron and to encode a product having 61% similarity to IutA, the outer membrane receptor for aerobactin. Transcription of both the rhbABCDEF operon and the rhtA gene was found to be positively regulated by the product of the eighth gene in the cluster, named rhrA, which has characteristics of an AraC-type transcriptional activator. The six genes in the rhbABCDEF operon have interesting gene junctions with short base overlaps existing between the genes. Similarities between the protein products of the biosynthesis genes and other proteins suggest that rhizobactin 1021 is synthesized by the formation of a novel siderophore precursor, 1,3-diaminopropane, which is then modified and attached to citrate in steps resembling those of the aerobactin biosynthetic pathway. The cluster of genes is located on the pSyma megaplasmid of S. meliloti 2011. Reverse transcription-PCR with RNA isolated from mature alfalfa nodules yielded no products for rhbF or rhtA at a time when the nifH gene was strongly expressed, indicating that siderophore biosynthesis and transport genes are not strongly expressed when nitrogenase is being formed in root nodules. Mutants having transposon insertions in the biosynthesis or transport genes induced effective nitrogen-fixing nodules on alfalfa plants.
Root nodule bacteria, known collectively as the rhizobia, interact in a nitrogen-fixing symbiosis with leguminous plants. The bacteria induce the formation of nodules on the roots of specific plant hosts and differentiate within the nodules to form bacteroids, cells containing nitrogenase that are capable of converting atmospheric nitrogen to ammonia. There is a high demand for iron in the symbiotic interaction, although the mechanism by which it is provided in plant root nodules is not well understood. The nitrogenase enzyme complex contains iron, as do a number of proteins, such as ferrodoxin, that provide energy for nitrogen fixation. Furthermore, the oxygen-labile nitrogenase is protected in the root nodule by leghemoglobin, an iron-rich protein that buffers the oxygen level and is estimated to account for 30% of the total soluble protein in the nodule (2). The importance of iron in the symbiosis has been demonstrated by O'Hara et al. (39), who showed that iron deficiency limits nodule development in peanuts inoculated with Bradyrhizobium species.
Rhizobia are soil bacteria and as free-living organisms they must compete for the limited iron available, particularly in neutral and alkaline soils. Iron is the fourth most abundant element on Earth, yet organisms frequently face deprivation because iron in the Fe(III) oxidation state, present under aerobic conditions, forms insoluble oxyhydroxide polymers at neutral pH. Microorganisms have evolved a variety of strategies to acquire iron under limiting conditions (20, 12). They may reduce ferric iron and take up ferrous iron by a ferrous-iron-specific transport system. Some bacteria that infect eukaryotic hosts can acquire ferric iron bound to iron carriers such as transferrin and lactoferrin and, interestingly, Noya et al. (37) have shown that a Rhizobium strain can use leghemoglobin as an iron source. Alternatively, bacteria may produce high-affinity iron chelators, termed siderophores. Investigations suggest that, of these strategies, soil microorganisms rely predominantly on siderophore production for competitive acquisition of iron.
Siderophores are low-molecular-weight chelators having a high specificity for Fe(III). They are produced only under iron-limiting conditions, with production being negatively regulated by the Fur protein in conjunction with iron. A variety of structurally different siderophores from different strains of rhizobia have been characterized (6, 14, 29, 41, 42, 47, 50). The ability of Sinorhizobium (formerly Rhizobium) meliloti to take up ferrisiderophores has been correlated with the presence of specific siderophore receptor proteins in the outer membrane (46). Moreover, some bacteria may acquire iron by the uptake of exogenous siderophores, such as those produced by fungi (43). Many strains of root nodule bacteria from both fast- and slow-growing species do not produce high-affinity iron chelators. Strains of Bradyrhizobium, for example, have been shown to use citrate as a siderophore (21). This indicates that the production of a high-affinity iron-binding chelator, which may be of importance in iron-limiting soils, is not essential for inducing a nitrogen-fixing symbiosis, a conclusion supported by the report that a fhuB mutant of Rhizobium leguminosarum, defective in the uptake of the siderophore vicibactin, induced apparently normal nitrogen-fixing nodules on peas (51).
The siderophore produced by S. meliloti 1021, an endosymbiont of Medicago sativa, has been structurally characterized (42) and named rhizobactin 1021 to distinguish it from the unrelated rhizobactin, an amino polycarboxylic acid siderophore produced by S. meliloti DM4 (50). Mutants in the production, uptake, and regulation of rhizobactin 1021 have been isolated and tested for their nodulation ability and efficiency in nitrogen fixation. Gill et al. (18) investigated the nitrogenase activity of plants inoculated with wild-type and rhizobactin 1021 mutants over a 30-day period and found a minor increase in activity in the plants inoculated with the wild type. In a second experiment over a 70-day period, a significant increase in nitrogenase activity and total plant dry weight was observed with the wild type compared to some rhizobactin 1021 mutants. Barton et al. (7) also observed increased efficiency in nitrogen fixation by plants growing in low iron medium when inoculated with the wild type compared to plants inoculated with rhizobactin 1021 mutants. In a subsequent study it was shown that, in iron-rich medium, the regulation of rhizobactin synthesis is a factor in efficient nitrogen fixation (8). These studies suggest that rhizobactin 1021, while not essential for symbiosis, can contribute to the efficiency of nitrogen fixation under certain conditions of plant growth.
Genomic regions carrying some or all of the genes for rhizobactin 1021 regulation, biosynthesis, and transport have been cloned in separate studies (17, 46). We describe here the sequencing and functional analysis of a region of one of the cosmids encoding genes for the regulation, biosynthesis, and uptake of rhizobactin 1021.
MATERIALS AND METHODS
Materials.
All chemicals were analar grade. Chrome azurol S (CAS) was obtained from BDH Chemicals, Ltd., Poole, England. Sigma Chemical Co., St. Louis, Mo., supplied 2,2′-dipyridyl. Molecular biology reagents were from New England Biolabs, Beverly, Mass., unless otherwise indicated.
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli cultures were grown at 37°C in Luria-Bertani (LB) medium (4). SOB medium used in the transformation of E. coli was as described by Inoue et al. (28). Sinorhizobium strains were grown at 30°C in tryptone-yeast extract (TY) medium (9). The CAS medium for the detection of siderophore production was prepared by the method of Schwyn and Neilands (48), with the modifications described by Reigh and O'Connell (46). The low-iron medium used in the preparation of siderophore and in iron nutrition bioassays consisted of TY medium with 200 μM 2,2′-dipyridyl. All glassware used in the preparation of low-iron and CAS media was initially washed in 2 N HCl and rinsed in distilled deionized water. Antibiotics were added to solid media at the following concentrations unless otherwise stated: ampicillin, 50 μg/ml; neomycin sulfate, 100 μg/ml; gentamicin sulfate, 30 μg/ml; rifampin, 100 μg/ml; chloramphenicol, 50 μg/ml; and tetracycline, 30 μg/ml.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmida | Relevant characteristic(s)b | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| S605 | met::Tn5 | W. Klipp, Bielefeld, Germany |
| A118 | Chromosomally located Tn5lac | 49 |
| DH5α | hsdR17 endA1 thi-1 gyrA96 relA1 recA1 supE44 ΔlacU169 (φ80dlacZΔM15) | Gibco-BRL |
| S. meliloti | ||
| 2011 | Wild typec | |
| 2011 rhbA62 | rhbA::Tn5lac, rif | This study |
| 2011 rhbC2 | rhbC::Tn5mob, rif | 46 |
| 2011 rhbE11 | rhbE::Tn5lac, rif | This study |
| 2011 rhbE9 | rhbE::Tn5, rif | This study |
| 2011 rhbF104 | rhbF::Tn5lac, rif | This study |
| 2011 rhbF107 | rhbF::Tn5lac, rif | This study |
| 2011 rhrA4 | rhrA::Tn5lac, rif | This study |
| 2011 rhrA6 | rhrA::Tn5, rif | This study |
| 2011 rhrA26 | rhrA::Tn5lac, rif | This study |
| 2011 rhtA45 | rhtA::Tn5lac, rif | This study |
| 2011 rhtA42 | rhtA::Tn5lac, rif | This study |
| 2011 rhtA1 | rhtA::Tn5, rif | This study |
| SmA818 | Cured of pSyma | 40 |
| SmA416BTW | Deletion in pSyma | 40 |
| Plasmids | ||
| pUC18 | Apr, lacZ′ | 36 |
| pJQ200ks | Gmr, mob sacB | 45 |
| pGR30 | Cosmid carrying rhizobactin 1021 synthesis, regulation, and uptake genes, Tcr | 46 |
| pRK600 | Tra+, Cmr | 15 |
| pDL80 | pJQ200ks carrying 8.0-kb genomic BamHI insert | This study |
| pUC18–52 | pUC18 carrying 5.2-kb genomic BamHI insert | This study |
| pDL30 | pJQ200ks carrying 3.0-kb XhoI-BamHI fragment from pUC18–52 | This study |
pUC18-based constructs used for sequencing are not included.
Apr, Cmr, Gmr, and Tcr, ampicillin, chloramphenicol, gentamicin, and tetracycline resistance, respectively; mob, mobilization; Tra+, transfer.
S. meliloti 2011 is the parent strain of 1021 (35) from which rhizobactin 1021 was purified and chemically characterized (42). A rifampin-resistant derivative (rif) was used in the construction of transposon insertion mutants. To facilitate the use of the npt probe to compare relative abundance of RNA in Tn5lac-induced mutants and wild type, a derivative with a Tn5 insertion in a rhamnose utilization gene (provided by Michael Hynes, University of Calgary) was used instead of wild type.
Fragment targeted transposon mutagenesis.
Transposon inserts in the restriction fragments of interest were selected in E. coli and then introduced into the S. meliloti genome. Selection for the transposition of Tn5 or Tn5lac from the chromosomes of E. coli S605 and E. coli A118, respectively, to plasmids carrying the restriction fragments to be mutated was carried out as follows. The mobilizable vector pJQ200ks was used to carry cloned genomic fragments making plasmids pDL80 and pDL30. These plasmids were introduced into E. coli S605 or A118 by transformation. Following growth to stationary phase the culture was concentrated 10-fold, and 0.1 ml was plated on LB medium containing 500 μg of neomycin per ml. The elevated level of neomycin enriches for cells in which a copy of the transposon has inserted into the multicopy plasmid. Colonies that arose were washed en masse from the plate, and plasmid DNA was prepared and used to transform E. coli DH5α, plating it on gentamicin and neomycin to select for plasmids carrying Tn5 or Tn5lac. Plasmid DNA was isolated from clones that grew and was analyzed by restriction digestion to confirm the presence of the transposon and to map the site of the insertion. The precise locations of Tn5lac insertions in the genomic insert were confirmed by DNA sequencing. Transposon inserts in regions of interest were directed into the genome of S. meliloti by triparental mating. Efficient mutagenesis was achieved when the transposon insert was flanked by at least 1 kb of cloned DNA on each side. The triparental mating was carried out by mixing equal volumes of logarithmic cultures of E. coli pRK600, the donor of the mobilizing plasmid, E. coli DH5α, carrying the cloned DNA with the transposon insert in the mobilizable plasmid, and S. meliloti 2011 rif, the recipient. The cells were incubated overnight on a 0.22 μm (pore-size) membrane (Millipore) on the surface of TY medium and then resuspended in sterile water. Selection was then made on TY medium with gentamicin, selecting for integration of the plasmid into the chromosome by a single-crossover recombination. Colonies that arose were purified and grown to stationary phase. The cells were then plated on TY medium containing neomycin and 5% sucrose. This medium selects for cells in which a second recombination event has excised the plasmid, leaving the transposon in the genome, since the plasmid carries the sacB gene, which confers sensitivity to sucrose. Correct insertion of the transposons into the genome was confirmed by Southern hybridization.
Rhizobactin bioassay.
Utilization of rhizobactin was measured in iron nutrition bioassays. Supernatant containing rhizobactin was prepared from wild-type strain 2011 grown to stationary phase in TY broth with 200 μM 2,2′-dipyridyl. The supernatant was concentrated 10-fold by vacuum centrifugation before use. Test strains were grown to stationary phase and then seeded (100 μl of culture into 25 ml of agar) in TY agar with 300 μM 2,2′-dipyridyl. A well was cut in the center of the agar; 100 μl of the concentrated supernatant, containing siderophore, was added; and the plates were incubated. The halo of growth observed in a positive reaction was optimally visualized 3 days after incubation.
Preparation and visualization of outer membrane proteins.
The method used for preparation of outer membrane proteins and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was as described previously (46) except that cultures were grown in the low-iron medium, described above, rather than low-iron minimal medium.
DNA manipulations.
Plasmid DNA preparation was by the rapid boiling method (25). Transformation of E. coli with plasmid DNA was done by the method of Inoue et al. (28). Cosmid DNA was prepared by the method of Little (31). Total genomic DNA was prepared as described by Meade et al. (35). Restriction enzyme digestion, DNA ligations, agarose gel electrophoresis, and Southern blotting were done by standard procedures (4). Nylon membranes (Boehringer Mannheim) were used for DNA transfer, and the probes were labeled by random priming using the DIG High Prime Labeling and detection Starter Kit II as directed by the manufacturer (Boehringer Mannheim). Restricted DNA fragments were prepared from agarose gels for cloning or labeling using essentially the method described by Mather et al. (34) as follows. After agarose gel electrophoresis and staining, the band of interest was cut from the gel and weighed. Two volumes of a saturated solution of sodium iodide were added followed by incubation at 55°C until the agarose had dissolved. Then, 5 μl of siliconized glass beads were added to the solution, and it was held on ice for 5 min. The solution was centrifuged for 5 min in a microfuge (Heraeus) at 13,000 rpm, and the pellet of beads was washed three times with ice-cold ethanol before elution of DNA from the beads by resuspension in 20 μl of Tris-EDTA buffer.
RNA isolation and RNase protection experiments.
RNA was prepared from cultures grown in TY medium to mid exponential phase with 2,2′-dipyridyl (200 μM) added when iron-depleted growth was required. Cells were pelleted by centrifugation, RNA Whiz (Ambion) was added, and the total RNA was extracted as directed by the manufacturers. Riboprobes were prepared from PCR products amplified with a T7 phage promoter site incorporated into a 30-base sequence appended to the 5′ end of one of the primers for transcription of an antisense probe. In vitro transcription was carried out using the T7 MAXIscript kit from Ambion incorporating α-32P-labeled UTP (800 Ci/mmol). Probes were gel purified in 6% polyacrylamide and mixed with 15 μg of total RNA per reaction. Hybridization was at 45°C in 80% formamide. RNase protection was carried out with the RPA II Ribonuclease Protection Assay kit from Ambion. The RNA products were separated on a 6% polyacrylamide gel and exposed to X-ray film.
Reverse transcription.
RNA from free-living bacterial cultures was prepared as described above. RNA from root nodules was prepared by picking 20 nodules into 1 ml of RNA Whiz (Ambion), grinding the sample with a mortar and pestle, and then extracting it as directed by the manufacturer. Then, 1 μg of RNA was used for reverse transcription. It was mixed with 0.5 μg of the reverse primer for the fragment to be amplified, and the mixture was brought to 5 μl with sterile water. The priming reaction was incubated at 70°C for 10 min. To this was added 4 μl of 5× transcription buffer (supplied with the reverse transcriptase [RT] enzyme); 2 μl of 100 mM dithiothreitol; 1 μl of RNasin; 1 μl of a mix of dATP, dCTP, dGTP, and dTTP, each at a concentration of 10 mM; 6 μl of sterile water; and 1 μl of Moloney murine leukemia virus RT (Promega). The reaction mixture was incubated at 37°C for 1 h and was then heated to 95°C for 2 min to inactivate the enzyme. The synthesized cDNA was used for PCR.
Nodulation and nitrogen fixation assays.
Alfalfa (M. sativa) seeds were surface sterilized in ethanol and sodium hypochlorite. Seeds were grown in Jenson plant medium as described by Ogawa et al. (38) in test tubes and inoculated with approximately 108 rhizobia per tube. The nitrogen-fixing ability of whole plants was measured by the acetylene reduction assay (52).
DNA sequencing and sequence analysis.
Sequencing reactions were carried out using the dye terminator approach. The samples were loaded on ABI 377XL sequencers. Data processing was done with ABI's own sequence analysis software (version 3.1). Lane tracking was done with Perkin-Elmer's manual lane tracking kit. Gap4 (11) from the Staden package was used to assemble sequences into contigs. Gaps between contigs were closed by sequencing reactions using primers based on sequence from the ends of the contigs. Each base has been sequenced at least three times.
The Genemark program (http://genemark.biology.gatech.edu/GeneMark/) was used to identify open reading frames, and sequences were compared against database using BLASTN and BLASTP (1). Conserved regions within proteins were identified using the BLOCKS program (22). The CLUSTALW and Genedoc programs were used to perform global alignment of sequences.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been registered in the GenBank data base under accession no. AF110737.
RESULTS
Transposon mutagenesis and DNA sequencing of the region encoding rhizobactin 1021 regulation, biosynthesis, and transport.
The cosmid pGR30 carries all of the genes necessary for the regulation, biosynthesis, and transport of rhizobactin 1021 (46). The cosmid complemented a Tn5mob mutation in the genome of an S. meliloti mutant that was defective in siderophore biosynthesis. The Tn5mob insertion was mapped to a 1-kb BamHI fragment in the genome, a fragment flanked by BamHI fragments of 5.2 and 8.0 kb. These fragments were cloned in pJQ200ks and subjected to transposon insertion mutagenesis as described in Materials and Methods with Tn5 and Tn5lac. The Tn5lac transposon was used to create transcriptional fusions. However, none of the Tn5lac insertions that were successfully introduced into the genome of S. meliloti occurred in the correct orientation to facilitate analysis of transcription by this approach.
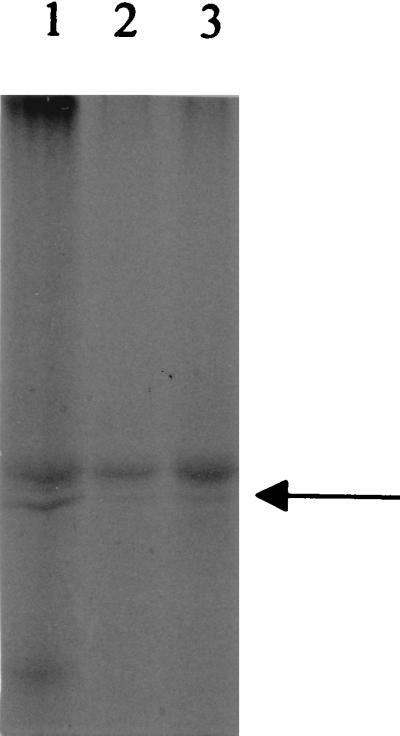
Twelve independent transposon insertions were made in the 5.2- and 8.0-kb BamHI fragments of the genome. The CAS assay for siderophore production, siderophore utilization as detected by the bioassay, and the profile of outer membrane proteins detected under low-iron growth conditions were then used to assign each of the 12 mutants to one of three phenotypic groups (Table 2). Mutants in the first group were defective in biosynthesis, producing no halo on CAS plates. They retained the ability to utilize the siderophore but yielded a much-reduced amount of the outer membrane receptor protein for rhizobactin 1021 (Fig. 1). Mutants in the second group were defective in transport. They did not produce the outer membrane receptor protein and were unable to utilize rhizobactin 1021 in the bioassay. They did produce rhizobactin and gave a slightly larger halo than the wild type when grown on CAS plates. Mutants in the third group were defective in both biosynthesis and transport. They did not produce a halo on CAS plates, did not utilize rhizobactin 1021 in the bioassay, and did not produce the outer membrane receptor.
TABLE 2.
Mutant phenotypes
| Mutant | Mutated gene | Halo on CAS mediuma | Rhizobactin uptake | Outer membrane receptor protein |
|---|---|---|---|---|
| 2011 | Wild type | ++ | Yes | Abundant |
| 2011 rhbA62 | rhbA | − | Yes | Reduced |
| 2011 rhbC2 | rhbC | − | Yes | NTb |
| 2011 rhbE9 | rhbE | − | Yes | Reduced |
| 2011 rhbE11 | rhbE | − | Yes | Reduced |
| 2011 rhbF104 | rhbF | − | Yes | Reduced |
| 2011 rhbF107 | rhbF | − | Yes | Reduced |
| 2011 rhrA4 | rhrA | − | No | None |
| 2011 rhrA6 | rhrA | − | No | None |
| 2011 rhrA26 | rhrA | − | No | None |
| 2011 rhtA45 | rhtA | +++ | No | None |
| 2011 rhtA42 | rhtA | +++ | No | None |
| 2011 rhtA1 | rhtA | +++ | No | None |
FIG. 1.
Separation by SDS-PAGE of outer membrane proteins isolated from cultures grown under iron-depleted conditions. Lane 1, wild-type strain 2011; lane 2, 2011 rhbE9; lane 3, rhbF107. The arrow indicates the position of the 72-kDa protein, RhtA.
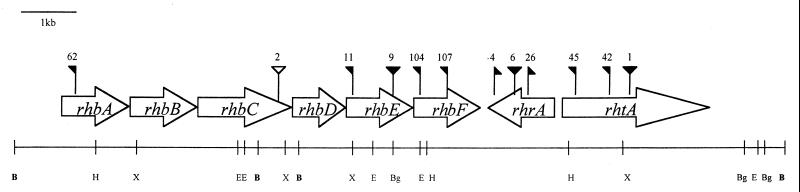
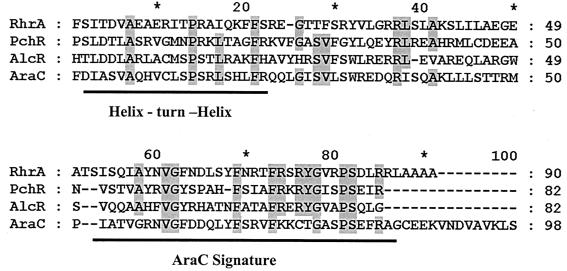
The DNA sequence of the 14.2-kb region comprising the three BamHI fragments was determined. Eight open reading frames were identified. The precise positions of the transposon insertion mutations were confirmed by sequencing, and genes were named according to the phenotypes of the mutants (Fig. 2). Six of the genes were named rhbABCDEF (rhizobactin 1021 biosynthesis), another was named rhtA (rhizobactin 1021 transport), and the eighth was named rhrA (rhizobactin 1021 regulation). The deduction of functions from the mutant phenotypes correlated well with the prediction of function based on similarities shown by the protein products of the genes when subjected to a BLASTP database search (Table 3). The products of four of the six biosynthetic genes were found to have similarity with, among others, the four proteins IucABCD involved in aerobactin biosynthesis. The products of the remaining two biosynthetic genes show similarity to the products of a pair of adjacent genes, dat and dcc, involved in the synthesis of 1,3-diaminopropane in Acinetobacter baumannii. The product of the rhtA gene has 61% similarity with IutA the receptor protein that functions in the transport of aerobactin. The rhrA gene encodes a regulator that controls both the biosynthesis and the transport of rhizobactin 1021 (see below) and has motifs found in AraC-type transcriptional activators (Fig. 3).
FIG. 2.
Organization of genes and locations of transposon insertions in the rhizobactin 1021 regulon. Symbols: , Tn5lac insertion, flag indicates orientation of lac gene; , Tn5 insertion; , Tn5mob insertion; B, BamHI; H, HindIII; X, XhoI; E, EcoRI; Bg, BglII.
TABLE 3.
Amino acid identity and similarity to proteins of the rhizobactin regulon
| Protein | Homologue
|
Identity (%) | Similarity (%)b | ||
|---|---|---|---|---|---|
| Protein | Sourcea | Accession no. | |||
| RhbA | Dat | A. baumannii | AB001599 | 49 | 63 |
| RhbB | Dcc | A. baumannii | D55724 | 34 | 54 |
| RhbC | IucA | E. coli | X76100 | 24 | 42 |
| RhbD | IucB | E. coli | X76100 | 21 | 40 |
| RhbE | AlcA | B. bronchiseptica | V61153 | 35 | 56 |
| IucD | E. coli | V90207 | 28 | 51 | |
| RhbF | IucC | E. coli | X76100 | 33 | 54 |
| AlcC | B. bronchiseptica | V61153 | 27 | 46 | |
| RhrA | AlcR | B. pertussis | AF018255 | 19 | 33 |
| PchR | P. aeruginosa | L11657 | 18 | 31 | |
| AraC | E. coli | V00259 | 14 | 29 | |
| RhtA | IutA | E. coli | P14542 | 40 | 61 |
B., Bordetella.
Similarity to uncharacterized sequences from genome sequencing is not included.
FIG. 3.
Amino acid sequence alignments of RhrA with AraC-type transcriptional regulators in the carboxy terminus. The helix-turn-helix motif and the AraC signature motif, as identified using the BLOCKS programme, are identified.
The rhbABCDEF genes comprise an operon.
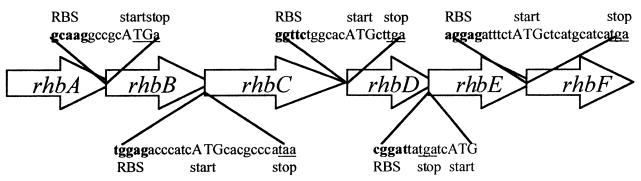
The rhbABCDEF genes show a remarkable absence of intervening sequence, with the ribosome-binding sites for rhbB, rhbC, rhbD, rhbE, and rhbF all occurring within the upstream open reading frame (Fig. 4). Furthermore for rhbB, rhbC, rhbD and rhbF the start codon occurs upstream within the preceding gene. The organization of the genes and the overlaps that exist at the gene junctions suggested that the six genes comprise an operon. An RNase protection experiment using riboprobes for rhbA and rhbF, the first and last of the six genes, confirmed the operon structure. The presence of transcripts in the S. meliloti wild type and mutant rhbA62 was investigated. Mutant rhbA62 has a transposon insertion downstream of the rhbA riboprobe sequence and should protect the probe. However, it would be expected to have a polar effect on downstream genes, including rhbF, within an operon. A comparison of RNase protection of the rhbA and rhbF probes by the wild type and mutant rhbA62 revealed the rhbF transcript in the wild type only (Fig. 5).
FIG. 4.
DNA sequences at the gene junctions in the rhizobactin 1021 biosynthesis operon.
FIG. 5.
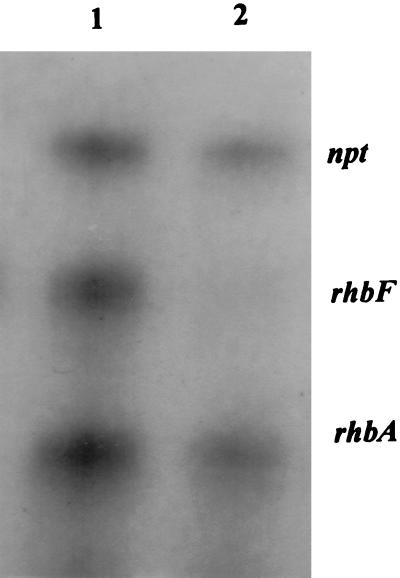
RNase protection of a 148-nucleotide riboprobe for rhbF and a 100-nucleotide riboprobe for rhbA. Lane 1, RNA from 2011::Tn5 in which the Tn5 insertion is outside the rhizobactin regulon and does not affect rhizobactin 1021 regulation, biosynthesis, or transport; lane 2, RNA from mutant 2011 rhbA62. The Tn5lac insertion in mutant rhbA62 is located downstream from the 100-nucleotide region protected by the rhbA riboprobe, and therefore a signal for rhbA is observed. A 200-nucleotide riboprobe for the npt gene of Tn5 allows comparison of RNA levels in the reactions. RNA was prepared from cultures grown under iron-depleted conditions.
In a separate RNase protection experiment (data not shown), we examined the effect of a transposon insertion in the open reading frame immediately upstream of rhbA on the transcription of rhbA. A transcript for rhbA was observed in this case, indicating that a promoter must be located immediately in front of rhbA. The open reading frame located upstream of rhbA showed no similarity to genes involved in siderophore biosynthesis, but we do not exclude the possibility that there may be readthrough from it into the rhbABCDEF operon.
Iron and RhrA regulate transcription of rhizobactin 1021 biosynthesis and transport.
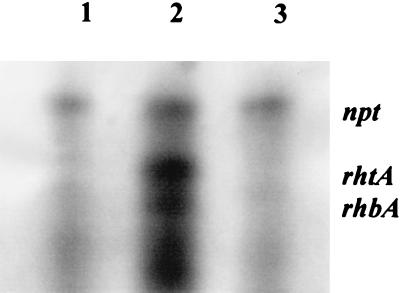
Repression of transcription of both the biosynthesis operon and the rhtA transport gene in the presence of iron was shown by RNase protection. Riboprobes for rhbA and rhtA were not protected by RNA from the wild-type strain grown under iron-replete conditions in contrast to the protection observed with RNA from a culture grown under iron-depleted conditions. Moreover, when mutant rhrA26, carrying a transposon insertion in the rhrA gene, was grown under iron-depleted conditions the rhbABCDEF and rhtA transcripts were not detected, indicating that RhrA is a transcriptional activator of both the biosynthesis operon and the outer membrane receptor gene (Fig. 6). The rhtA transcript was present in greater abundance than the rhbA transcript in the wild type under iron-depleted conditions. This may result from differential activation of transcription by RhrA or may arise from different stabilities of the transcripts.
FIG. 6.
RNase protection of a 150-nucleotide riboprobe for rhtA and a 100-nucleotide riboprobe for rhbA. Lane 1, RNA from mutant 2011 rhrA26 grown under iron-depleted conditions; lane 2, RNA from 2011::Tn5 grown under iron-depleted conditions; lane 3, RNA from 2011::Tn5 grown under iron-replete conditions. The Tn5 insertion in 2011::Tn5 does not affect rhizobactin 1021 regulation, biosynthesis, or transport. A 200-nucleotide riboprobe for the npt gene of Tn5 allows comparison of RNA levels in the reactions.
The rhizobactin 1021 regulon is located on the pSyma megaplasmid.
Two derivatives of S. meliloti 2011 (kindly provided prior to publication by Michael Hynes, University of Calgary), namely, SmA818, a strain that has been cured of the pSyma megaplasmid and strain SmA416 BTW that has a deletion in pSyma encompassing the symbiotic genes, were tested for the production of rhizobactin 1021 on CAS medium. The cured strain did not produce rhizobactin 1021, while the deleted strain produced amounts similar to that produced by the wild type. The conclusion from this result, that the regulon involved in rhizobactin 1021 biosynthesis and transport is located on pSyma, is in agreement with the report of Barloy-Hubler et al. (5) who located rhbF on the pSyma megaplasmid when constructing a high-resolution physical map of pSyma.
Symbiotic properties of mutants.
M. sativa (alfalfa) seeds were germinated and infected with the wild type and the mutants, rhbA62 and rhtA45, representatives of the biosynthetic and transport mutant phenotypes. No differences were observed in the time of onset of nodulation for plants inoculated with the wild-type or mutant strains. No differences in the patterns of nodulation were detected for the mutant and wild-type strains. In each case the numbers of nodules per plant varied between one and five. After 35 days whole plants were assayed for acetylene reduction as a measure of nitrogenase activity, and similar levels of activity were detected in plants inoculated with the wild type and each of the two mutant strains. The effect of a mutation in the rhizobactin 1021 receptor gene on symbiotic properties has not previously been investigated, and it was considered possible that the receptor transports an iron carrier, other than rhizobactin 1021, that may be essential for nitrogen fixation. However, it is clear from these results that the rhizobactin 1021 receptor is not essential for nitrogen fixation.
rhbF and rhtA transcripts are not detectable by RT-PCR in mature nodules.
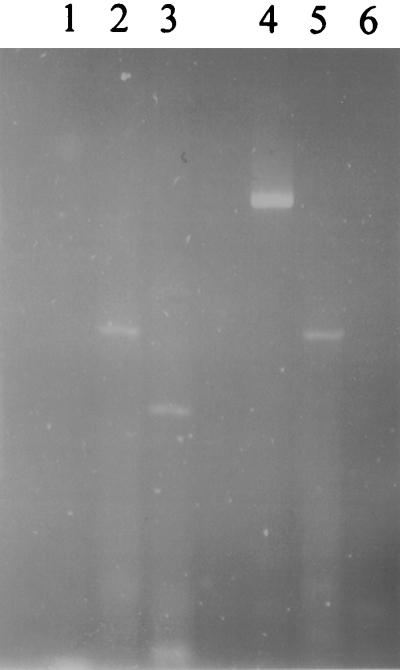
rhbF and rhtA transcripts were not detectable by RT-PCR in mature nodules. RNA isolated from nodules 35 days after inoculation with a strain carrying Tn5 inserted outside the siderophore biosynthesis-transport cluster was used in RT-PCR. The reactions were primed for rhbF and rhtA transcripts and were amplified with the same primers as were used to produce the DNA probes for RNase protection. No products were observed. A product was obtained for a 400-nucleotide nifH fragment that was primed and amplified as a positive control with the same template RNA. A transcript for npt from Tn5 was also detected. In contrast to the results with nodule RNA, products for rhbF and rhtA, as well as npt, were obtained in reactions conducted under the same conditions with an equal amount of RNA template from a free-living wild-type culture grown under iron-depleted conditions (Fig. 7).
FIG. 7.
Products from RT-PCR separated on a 4% agarose gel. The RNA template for the reactions in lanes 1, 2, and 3 was from free-living cells grown under iron-depleted conditions. The RNA template for the reactions in lanes 4, 5, and 6 was from nodules. Lanes 1 and 4, reactions primed for a 400-nucleotide nifH fragment; lanes 2 and 5; reactions primed for a 200-nucleotide npt fragment; lanes 3 and 6; reactions primed for a 148-nucleotide rhbF fragment.
DISCUSSION
The demonstration that the transcription of rhizobactin 1021 biosynthesis and transport genes is iron dependent is in keeping with the expected Fur type regulation of siderophore genes. Potential Fur box sequences CAGAAGCAGAGTGATTGTA (underlined bases are shared with the 19-base E. coli Fur box consensus) upstream of rhbA and GATAATGGCGAACCTCATT in the intergenic region between rhrA and rhtA were identified. In addition to its regulation by iron we have shown that rhizobactin 1021 production and transport are under the control of the AraC-like transcriptional activator, RhrA. In RhrA mutants we detected no siderophore production by the CAS assay, and we were unable to detect the outer membrane receptor, RhtA. These results correlate with the negligible detection of rhbA and rhtA mRNA by RNase protection and imply that under iron stress the biosynthesis and transport of rhizobactin 1021 are reliant on transcriptional activation by RhrA. RhrA has similarities to PchR, YbtA, and AlcR, three AraC-type regulators that have been shown to activate the expression of biosynthesis and transport genes for pyochelin in Pseudomonas aeruginosa (23, 24), yersiniabactin in Yersinia pestis (16), and alcaligin in Bordetella species (10, 44), respectively. PchR and YbtA also repress their own expression. For PchR and YbtA, repeat sequences were identified upstream of the regulated genes. We identified a seven-base repeat (GGCAATT) and a six-base repeat (GTTCGC) in the sequence upstream of rhtA. One or both of these may function in an interaction between RhrA and DNA.
Given the structural similarity between rhizobactin 1021 and aerobactin the strong homology between the receptor proteins for the two siderophores, RhtA and IutA respectively, is not surprising. Mutants carrying transposon insertions in the rhtA gene did not produce the 72-kDa iron-regulated outer membrane protein and did not transport rhizobactin 1021, confirming that this is the receptor protein for the siderophore. These mutants also produced larger, more-intense haloes than the wild type on CAS plates, suggesting that the inability to transport rhizobactin 1021 results in its accumulation in the medium. The amount of the 72-kDa protein was reduced in biosynthesis mutants (Table 2), indicating that in the absence of rhizobactin 1021, the synthesis of the receptor protein is downregulated. The basis for the downregulation may be related to the observation by Koster et al. (30) that the outer membrane ferric-pseudobactin receptor of Pseudomonas putida WCS358 was expressed only in cells cultured in iron-deficient medium containing pseudobactin BN8 or pseudobactin BN7. We found homology within RhtA to motifs conserved among TonB-dependent outer membrane proteins, indicating that RhtA may be a TonB-dependent receptor.
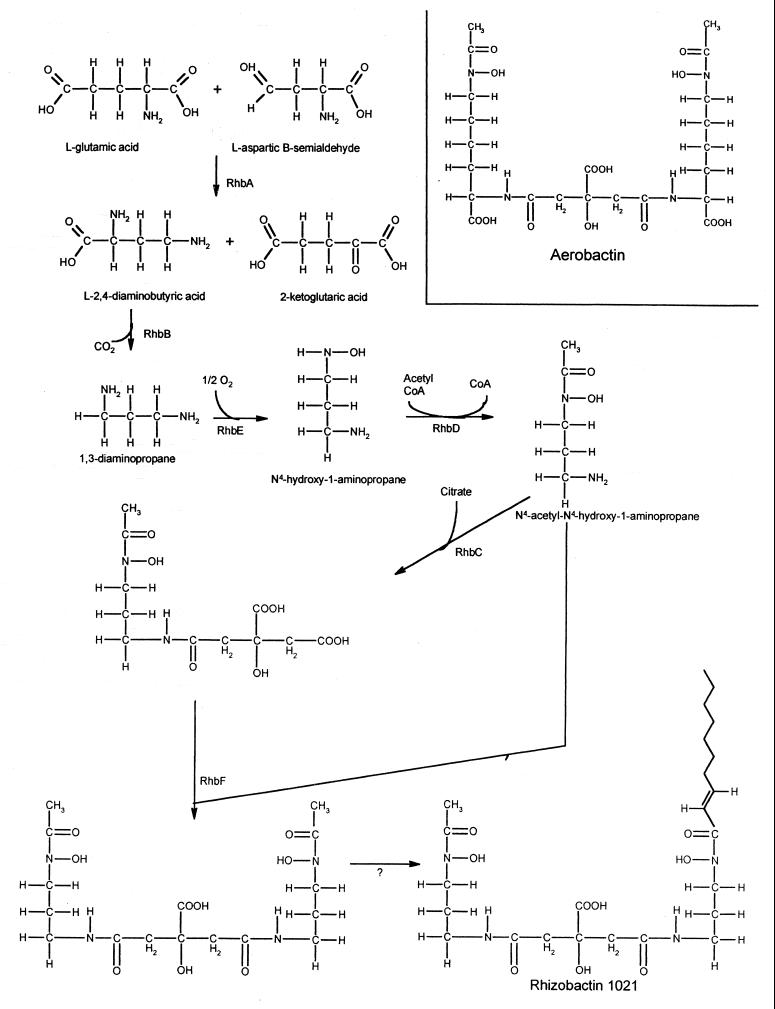
Siderophores are synthesized from common cell metabolites, frequently amino acids. The evidence from the present study suggests that rhizobactin 1021 is synthesized from a novel compound 1,3-diaminopropane, which has previously only been detected in A. baumannii. The phenotypes of transposon induced mutations in the rhbABCDEF operon and the predicted functions of the gene products, coupled to the knowledge of the chemical structure of rhizobactin 1021 (42), allows a biosynthetic pathway for rhizobactin 1021 to be proposed. Two of the predicted protein products of the biosynthetic genes, RhbA and RhbB, are homologous to the DABA AT and DABA DC enzymes of A. baumannii involved in the production of 1,3-diaminopropane (26, 27). The products of the remaining four biosynthetic genes, rhbCDEF, are homologous to products of the four genes, iucABCD, involved in the biosynthesis of aerobactin. Both aerobactin and rhizobactin 1021 are citrate-based hydroxymate siderophores with similar structures, differing only in the presence of a fatty acid moiety in rhizobactin 1021 and an additional carboxylic acid moiety and two saturated carbons in each of the symmetrical arms of aerobactin. The aerobactin molecule is built on lysine residues and, significantly, if a similar pathway were to use 1,3-diaminopropane instead of lysine, the resulting siderophore would have the exact structure determined for the core of rhizobactin 1021. It seems reasonable to propose therefore that rhizobactin 1021 is formed by the synthesis of 1,3-diaminopropane, which is subsequently incorporated into rhizobactin 1021 by steps similar to those that incorporate lysine into aerobactin. A diagram for the proposed pathway is shown in Fig. 8. The strong protein homologies and the similarity between the organization of the rhbAB genes of S. meliloti and the dat and dcc genes of A. baumannii suggest that 1,3-diaminopropane is made in S. meliloti. No function for 1,3-diaminopropane has been identified in A. baumannii but, interestingly, we have identified a sequence with homology to the Fur box consensus upstream from the dat gene in the published sequence. The presence of a strong transcriptional terminator downstream from dcc in A. baumannii indicates that there are no additional genes in the operon (26). Based on the homologies between the products of the four genes that follow in the rhizobactin 1021 biosynthetic operon and the corresponding enzymes involved in aerobactin biosynthesis, it can be proposed that 1,3-diaminopropane may be hydroxylated by RhsE to form N4-hydroxy-1-aminopropane, which is then acetylated by RhsD to form N4-acetyl-N4-hydroxy-1-aminopropane. The similarity between RhsC and RhsF and the subunits of the aerobactin synthetase complex, IucA and IucC (13), suggests that the S. meliloti proteins may act as a synthetase complex adding two molecules of N4-acetyl-N4-hydroxy-1-aminopropane to citrate, forming the core of rhizobactin 1021.
FIG. 8.
Proposed pathway for the synthesis of rhizobactin 1021. The structure of aerobactin is shown for comparison.
Unexpected overlaps exist in the DNA sequence at the junctions of the biosynthesis genes. The overlapping nature of the biosynthesis genes is unusual but not without precedent. Interestingly, the four aerobactin biosynthesis genes also show this intercistronic organization. Martinez et al. (32) proposed that the reason for the overlapping gene junctions may be to bring about posttranscriptional control that would vary the levels of different translation products from within the same mRNA transcript. These authors predicted a strong secondary structure in the mRNA transcript for the aerobactin biosynthesis genes. Folded structures in mRNA are known to influence ribosome programming (3). Secondary structure may prevent ribosome binding within the transcript but would be relieved by translation. This suggests coupling of translation in which the translation of one region of the sequence is dependent on the opening of the mRNA secondary structure by translation of another region. Whether differential gene expression and translational coupling occur in the rhizobactin biosynthetic operon is not yet established. Analysis of the sizes and relative abundance of the gene products and the effects when premature stop codons are introduced may help in the elucidation of the functional significance of the sequences at the gene junctions.
An intriguing feature of the rhizobactin 1021 structure is the lipid moiety that gives the siderophore an asymmetric structure and is responsible for its amphiphilic properties. We did not conclusively identify the gene or genes for the addition of the lipid group. However, immediately downstream from rhtA we sequenced an open reading frame with 38% similarity to iucB and low similarity to a malonyl-coenzyme A decarboxylase. A transposon insertion in this open reading frame did not affect siderophore production as measured by the CAS assay or rhizobactin 1021 uptake as measured in the bioassay. However, given that rhizobactin 1021 without the lipid group would be identical to schizokinen, a functional siderophore, it would not be surprising that the siderophore would still be active in the absence of the lipid. Until recently, mycobactins were the only other siderophores reported to have a lipid moiety. The lipid of mycobactin may anchor the siderophore to the cell, and it has been suggested that mycobacteria sequester iron in a two step process in which exochelins are excreted to bind iron and deliver it to the anchored mycobactin (19). Recently, lipids have also been detected in aquachelins and marinobactins of marine bacteria (33). It was suggested that the lipid in these siderophores may facilitate micelle formation, providing protection for the siderophore against degradative cleavage. The lipids of different siderophores may have different biological functions.
We did not detect expression of rhbF or rhtA by RT-PCR with RNA isolated from 35-day-old nodules. Expression of the nitrogenase structural gene, nifH, was readily detectable by this approach with the same RNA. The npt gene was also detectable in nodules infected with a Tn5lac-induced mutant. In control experiments we detected rhbF and rhtA by RT-PCR under similar experimental conditions with RNA isolated from the wild-type strain grown under iron-depleted conditions. These experiments indicate that rhbF and rhtA are not expressed in mature nodules in which nitrogenase genes are actively expressed and when nitrogenase is active, as measured by acetylene reduction. The absence of any difference in acetylene reduction by plants infected with siderophore biosynthesis mutants and the wild type concurs with this observation. Yeoman et al. (54) used a reporter gene fusion to investigate an iron-regulated fhuA gene in nodules induced by R. leguminosarum and found the gene product in the meristematic zone but not in the bacteroids. Our results indicate that active expression of biosynthesis or transport genes for rhizobactin 1021 is not occurring in the nodule after 35 days. At this time bacteria in the meristematic zone may be relatively inactive. However, although we observed no difference in the number of nodules or the time of onset of nodulation in comparisons between mutants and wild type, we cannot rule out the expression of the genes in the early stages of nodulation. We also cannot rule out the expression of the genes late in nodule development. Gill et al. (18) reported a difference in the effectiveness of nodules induced by mutants and wild type 70 days after infection.
Rhizobactin 1021 is likely to be a factor that contributes to the competitive ability of free-living S. meliloti in iron-depleted soils. A more extensive investigation of gene expression throughout the course of nodulation will be necessary to assess whether any contribution is made to the effectiveness of S. meliloti by rhizobactin 1021 in planta. However, we can conclude that siderophore biosynthesis or transport genes are not expressed when nitrogenase genes are actively expressed. How the supply of iron is provided to form nitrogenase, at a stage in nodulation when it is highly active, remains to be resolved.
ACKNOWLEDGMENTS
We are grateful to Sabine Thamm for technical assistance and to Jiping Li for preliminary analysis of the pGR30 clone.
This work was partially supported by Public Health Service grant AI19018 from the National Institutes of Health to J.H.C.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appleby C A. Leghemoglobin and Rhizobiumrespiration. Annu Rev Plant Physiol. 1984;35:443–478. [Google Scholar]
- 3.Atkins J F, Gesteland R F. Intricacies of ribosomal frameshifting. Nat Struct Biol. 1999;6:206–207. doi: 10.1038/6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1996. [Google Scholar]
- 5.Barloy-Hubler F, Capela D, Barnett M J, Kalman S, Federspeil N A, Long S R, Galibert F. High resolution physical map of the Sinorhizobium meliloti1021 pSyma megaplasmid. J Bacteriol. 2000;182:1185–1189. doi: 10.1128/jb.182.4.1185-1189.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barsomian G D, Urzainqui A, Lohman K, Walker G C. Rhizobium melilotimutants unable to synthesize anthranilate display a novel symbiotic phenotype. J Bacteriol. 1992;174:4416–4426. doi: 10.1128/jb.174.13.4416-4426.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton L L, Fekete F A, Vester C R, Gill P R, Neilands J B. Physiological characteristics of Rhizobium meliloti 1021 Tn5mutants with altered rhizobactin activities. J Plant Nutr. 1992;15:2145–2156. [Google Scholar]
- 8.Barton L L, Johnson G V, Schitoskey K, Wertz M. Siderophore-mediated iron metabolism in growth and nitrogen fixation by alfalfa nodulated with Rhizobium meliloti. J Plant Nutr. 1996;19:1201–1210. [Google Scholar]
- 9.Beringer J E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 10.Beaumont F C, Kang H Y, Brickman T J, Armstrong S K. Identification and characterization of alcR, a gene encoding an AraC-like regulator of alcaligin siderophore biosynthesis and transport in Bordetella pertusis and Bordetella bronchiseptica. J Bacteriol. 1998;180:862–870. doi: 10.1128/jb.180.4.862-870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonfield J K, Smith K F, Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;24:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Lorenzo V, Neilands J B. Characterization of iucA and iucC genes of the aerobactin system of plasmid ColV-K30 in Escherichia coli. J Bacteriol. 1986;167:350–355. doi: 10.1128/jb.167.1.350-355.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dilworth M J, Carson K C, Giles R G F, Byrne L T, Glenn A R. Rhizobium leguminosarum bv. viciaeproduces a novel cyclic trihydroxamate siderophore, vicibactin. Microbiology. 1998;144:781–791. doi: 10.1099/00221287-144-3-781. [DOI] [PubMed] [Google Scholar]
- 15.Finan T M, Kunkel B, De Vos G F, Signer E R. Second symbiotic megaplasmid in Rhizobium meliloticarrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fetherston J D, Bearden S W, Perry R D. YbtA, an AraC-type regulator of Yersinia pestispesticin/yersiniabactin receptor. Mol Microbiol. 1996;22:315–325. doi: 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- 17.Gill P R, Neilands J B. Cloning a genomic region required for a high-affinity iron-uptake system in Rhizobium meliloti1021. Mol Microbiol. 1989;3:1183–1189. doi: 10.1111/j.1365-2958.1989.tb00268.x. [DOI] [PubMed] [Google Scholar]
- 18.Gill P R, Barton L L, Scoble M D, Neilands J B. A high affinity iron transport system of Rhizobium meliloti may be required for efficient nitrogen fixation in planta. Plant Soil. 1991;130:211–217. [Google Scholar]
- 19.Gobin J, Horwitz M A. Exochelins of Mycobacterium tuberculosis remove iron from human iron-binding proteins and donate iron to mycobactins in the M. tuberculosiscell wall. J Exp Med. 1996;183:1527–1532. doi: 10.1084/jem.183.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerinot M L. Microbial iron transport. Annu Rev Microbiol. 1994;48:743–772. doi: 10.1146/annurev.mi.48.100194.003523. [DOI] [PubMed] [Google Scholar]
- 21.Guerinot M L, Meidl E J, Plessner O. Citrate as a siderophore in Bradyrhizobium japonicum. J Bacteriol. 1990;172:3298–3303. doi: 10.1128/jb.172.6.3298-3303.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henikoff S, Henikoff J G. Protein family classification based on searching a database of blocks. Genomics. 1994;19:97–107. doi: 10.1006/geno.1994.1018. [DOI] [PubMed] [Google Scholar]
- 23.Heinrichs D E, Poole K. Cloning and sequence analysis of a gene (pchR) encoding an AraC family activator of pyochelin and ferripyochelin receptor synthesis in Pseudomonas aeruginosa. J Bacteriol. 1993;175:5882–5889. doi: 10.1128/jb.175.18.5882-5889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinrichs D E, Poole K. PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J Bacteriol. 1996;178:2586–2592. doi: 10.1128/jb.178.9.2586-2592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes D S, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 26.Ikai H, Yamamoto S. Sequence analysis of the gene encoding a novel l-2,4-diaminobutyrate decarboxylase of Acinetobacter baumannii: similarity to the group II amino acid decarboxylases. Arch Microbiol. 1996;166:128–131. doi: 10.1007/s002030050366. [DOI] [PubMed] [Google Scholar]
- 27.Ikai H, Yamamoto S. Identification and analysis of a gene encoding l-2,4-diaminobutyrate:2-ketoglutarate 4-aminotransferase involved in the 1,3-diaminopropane production pathway in Acinetobacter baumannii. J Bacteriol. 1997;179:5118–5125. doi: 10.1128/jb.179.16.5118-5125.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coliwith plasmids. Gene. 1991;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 29.Jadhav R S, Desai A J. Isolation and characterisation of siderophore from cowpea Rhizobium(peanut isolate) Curr Microbiol. 1992;24:537–542. [Google Scholar]
- 30.Koster M, Van De Vossenberg J, Leong J, Weisbeek P J. Identification and characterization of the pupB gene encoding an inducible ferric-pseudobactin receptor of Pseudomonas putidaWCS358. Mol Microbiol. 1993;8:591–601. doi: 10.1111/j.1365-2958.1993.tb01603.x. [DOI] [PubMed] [Google Scholar]
- 31.Little P F R. Choice and use of cosmid vectors. In: Glover D M, editor. DNA cloning. III. Washington, D.C.: IRL Press; 1987. pp. 33–34. [Google Scholar]
- 32.Martinez J L, Herrero M, de Lorenzo V. The organisation of intercistronic regions of the aerobactin operon of pColV-K30 may account for the differential expression of the iucABCD iutAgenes. J Mol Biol. 1994;238:288–293. doi: 10.1006/jmbi.1994.1290. [DOI] [PubMed] [Google Scholar]
- 33.Martinez J S, Zhang G P, Holt P D, Jung H-T, Carrano C J, Haygood M G, Butler A. Self-assembling amphiphilic siderophores from marine bacteria. Science. 2000;287:1245–1247. doi: 10.1126/science.287.5456.1245. [DOI] [PubMed] [Google Scholar]
- 34.Mather M W, Keightly J A, Fee J A. Recovery and cloning of genomic DNA fragments from dried agarose gels. Methods Enzymol. 1993;218:695–704. doi: 10.1016/0076-6879(93)18052-e. [DOI] [PubMed] [Google Scholar]
- 35.Meade H M, Long S R, Ruvkun G B, Brown S E, Ausubel F M. Physical and genetic characterisation of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- 37.Noya F, Arias A, Fabiano E. Heme compounds as iron sources for nonpathogenic Rhizobiumbacteria. J Bacteriol. 1997;179:3076–3078. doi: 10.1128/jb.179.9.3076-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogawa J, Brierley H I, Long S R. Analysis of Rhizobium meliloti nodulation mutant WL131: novel insertion sequence ISRm3 in nodG and altered nodHprotein product. J Bacteriol. 1991;173:3060–3065. doi: 10.1128/jb.173.10.3060-3065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Hara G W, Dilworth M J, Boonkerd N, Parkpian P. Iron-deficiency specifically limits nodule development in peanut inoculated with Bradyrhizobiumsp. New Phytol. 1988;108:51–57. doi: 10.1111/j.1469-8137.1988.tb00203.x. [DOI] [PubMed] [Google Scholar]
- 40.Oresnik I J, Liu S-L, Yost C K, Hynes M F. Megaplasmid pRme2011a of Sinorhizobium melilotiis not required for viability. J Bacteriol. 2000;182:3582–3586. doi: 10.1128/jb.182.12.3582-3586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patel H N, Chakraborty R N, Desai S B. Isolation and partial characterisation of phenolate siderophore from Rhizobium leguminosarumIARI102. FEMS Microbiol Lett. 1988;56:131–134. [Google Scholar]
- 42.Persmark M, Pittman P, Buyer J S, Schwyn B, Gill P R, Neilands J B. Isolation and structure of rhizobactin 1021, a siderophore from the alfalfa symbiont Rhizobium meliloti1021. J Am Chem Soc. 1993;115:3950–3956. [Google Scholar]
- 43.Plessner O, Klapatch T, Guerinot M L. Siderophore utilization by Bradyrhizobium japonicum. Appl Environ Microbiol. 1993;59:1688–1690. doi: 10.1128/aem.59.5.1688-1690.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pradel E, Guiso N, Locht C. Identification of AlcR, an AraC-type regulator of alcaligin siderophore synthesis in Bordetella bronchiseptica and Bordetella pertussis. J Bacteriol. 1998;180:871–880. doi: 10.1128/jb.180.4.871-880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 46.Reigh G, O'Connell M. Siderophore-mediated iron transport correlates with the presence of specific iron regulated proteins in the outer membrane of Rhizobium meliloti. J Bacteriol. 1993;175:94–102. doi: 10.1128/jb.175.1.94-102.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rioux C R, Jordan D C, Rattray J B M. Iron requirement of Rhizobium leguminosarumand secretion of anthranilic acid during growth on an iron-deficient medium. Arch Biochem. 1986;248:175–182. doi: 10.1016/0003-9861(86)90414-5. [DOI] [PubMed] [Google Scholar]
- 48.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 49.Simon R, Quandt J, Klipp W. New derivatives of transposon Tn5suitable for mobilisation of replicons, generation of operon fusions and induction of genes in gram-negative bacteria. Gene. 1989;80:161–169. doi: 10.1016/0378-1119(89)90262-x. [DOI] [PubMed] [Google Scholar]
- 50.Smith M J, Schoolery J N, Schwyn B, Holden I, Neilands J B. Rhizobactin, a structurally novel siderophore from Rhizobium meliloti. J Am Chem Soc. 1985;107:1739–1743. [Google Scholar]
- 51.Stevens J B, Carter R A, Hussain H, Carson K C, Dilworth M J, Johnston A W B. The fhu genes of Rhizobium leguminosarum, specifying siderophore uptake proteins: fhuDCB are adjacent to a pseudogene version of fhuA. Microbiology. 1999;145:593–601. doi: 10.1099/13500872-145-3-593. [DOI] [PubMed] [Google Scholar]
- 52.Wacek T, Brill W J. Simple, rapid assay for screening nitrogen fixing ability in soybean. Crop Sci. 1976;16:519–522. [Google Scholar]
- 53.Welch T J, Chai S, Crosa J H. The overlapping angB and angG genes are encoded within the trans-acting factor region of the virulence plasmid in Vibrio anguillarum: essential role in siderophore biosynthesis. J Bacteriol. 2000;182:6762–6773. doi: 10.1128/jb.182.23.6762-6773.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeoman K H, Wisniewski-Dye F, Timoney C, Stevans J B, deLuca N G, Downie J A, Johnston A W B. Analysis of the Rhizobium leguminosarum siderophore-uptake gene fhuA: differential expression in free-living bacteria and nitrogen-fixing bacteroids and distribution of fhuApseudogenes in different strains. Microbiology. 2000;146:829–837. doi: 10.1099/00221287-146-4-829. [DOI] [PubMed] [Google Scholar]