Abstract
Population genetic studies suggest that Yersinia pestis, the cause of plague, is a clonal pathogen that has recently emerged from Yersinia pseudotuberculosis. Plasmid acquisition is likely to have been a key element in this evolutionary leap from an enteric to a flea-transmitted systemic pathogen. However, the origin of Y. pestis-specific plasmids remains obscure. We demonstrate specific plasmid rearrangements in different Y. pestis strains which distinguish Y. pestis bv. Orientalis strains from other biovars. We also present evidence for plasmid-associated DNA exchange between Y. pestis and the exclusively human pathogen Salmonella enterica serovar Typhi.
Yersinia pestis is primarily a rodent pathogen and is usually transmitted subcutaneously by the bite of an infected flea (13). The microorganism can spread to the regional lymph nodes of infected rodents and humans, where local replication can cause a swelling, or bubo (55). Subsequent bacteremia may sometimes result in pulmonary infection (secondary pneumonic plague) and direct respiratory spread (55). There are historical data suggesting that epidemic plague has on occasion been the leading cause of death in humans—a third of the population of Europe may have died in the 14th century during the second pandemic of plague known as the Black Death (79). The third pandemic resulting from the Hong Kong outbreak of 1894 killed millions of people (mainly in Asia) in the early years of the 20th century (55). Currently, most plague cases are reported from developing countries in Africa and Asia such as Madagascar, Tanzania, and Vietnam (75). Plague continues to have a high mortality rate: 20% of the 200 annual confirmed or presumptive cases (95% bubonic) in Madagascar (16) and 6 out of 19 cases in a recent series from the United States (18) were fatal infections. However, most of the countries reporting plague currently have much higher disease burdens associated with other infectious agents, including the exclusively human pathogen Salmonella enterica serovar Typhi (53).
The factors influencing the rise and fall of plague epidemics remain obscure (34), and it is vital to understand the genetic basis of the origins and evolution of such a potentially devastating pathogen as Y. pestis. The enteric pathogen Yersinia pseudotuberculosis is so closely related to Y. pestis in terms of chromosomal DNA hybridization that they could be classified as subspecies (5). On the basis of sequence analysis of multiple housekeeping genes, it has been proposed (1) that Y. pestis is a clone which has recently evolved from Y. pseudotuberculosis. Within the Y. pestis clone, distinct biovars (Antiqua, Medievalis, and Orientalis) have been associated with the three historically recognized pandemics (20). These biovars have been shown to be phylogenetically distinct by analysis of DNA restriction fragment polymorphisms (1) and ribotyping (29). It has been suggested that Y. pestis. bv. Orientalis has emerged most recently (1).
A role for key Y. pestis-specific gene mutations in evolution from Y. pseudotuberculosis was suggested by demonstration of increased virulence of Y. pseudotuberculosis for mice to a level comparable with that of Y. pestis when mutated in the chromosomal inv and plasmid-located yadA loci (61). These genes are intact open reading frames (ORFs) in Y. pseudotuberculosis but not in Y. pestis strains (61, 67). Conversely, introduction of a functional Y. pseudotuberculosis yadA gene into Y. pestis caused a significant decrease in virulence (61). However, the hypervirulent Y. pseudotuberculosis mutants were later found not to be isogenic with the wild-type strains (31), suggesting that the two single gene mutations were not the only factors involved.
The most obvious additional genetic complement present in Y. pestis compared with Y. pseudotuberculosis is in the form of plasmids. Both Y. pestis and Y. pseudotuberculosis possess a 70-kb plasmid (pCD1 in Y. pestis and pIB1 in Y. pseudotuberculosis) which encodes the Yop virulon, a type III secretion system essential for virulence in many hosts (9, 55). In addition, Y. pestis usually possesses two further plasmids which encode a variety of potentially virulence-associated determinants. A 9.5-kb plasmid called pPCP1 in Y. pestis KIM strains (alternatively known as pPst) encodes the plasminogen activator Pla (55). A 100- to 110-kb plasmid called pMT1 in KIM strains (alternatively known as pFra) encodes the murine toxin Ymt and the F1 capsular protein (55).
The key difference between Y. pseudotuberculosis and Y. pestis is the latter's capacity to colonize the flea and be transmitted to mammalian hosts by a subcutaneous route of infection. The chromosomal hms locus that is required (33) for blockage of the flea midgut by Y. pestis to maximize onward transmission is also present in Y. pseudotuberculosis (11). However, the Y. pestis-specific pMT1 or pFra plasmid encodes a murine toxin, which is apparently essential for flea colonization (35), and the F1 capsular antigen, which is an important protective immunogen although it is not essential for virulence in mammalian hosts (25). The Pla protease encoded by the pPCP1 or pPst plasmid is required (70) by some strains of Y. pestis for systemic spread after subcutaneous injection into a mammalian host. It seems likely that acquisition of these two plasmids was an essential step in the evolution of Y. pestis from Y. pseudotuberculosis. However, the limited sequence similarity between the Y. pestis plasmids (37, 42) and other bacterial plasmids gives few clues about their origin and capacity for mobility. In an effort to understand the pathogenicity and evolution of Y. pestis, we have undertaken the genome sequencing of this devastating pathogen, and plasmid DNA sequence comparison is an important application of these data. Genetic diversity of plasmids exceeds that of host chromosomes, largely due to a higher rate of recombination (6), and genetic variation associated with biovar emergence may be more easily revealed in plasmids than in the homogeneous chromosomal background (1).
Our data provide supporting evidence for the emergence of Y. pestis bv. Orientalis after biovars Medievalis and Antiqua. We present data demonstrating the common ancestry of more than 50% of the Y. pestis pMT1/pFra plasmid and a cryptic plasmid from a recent clinical isolate of S. enterica serovar Typhi, another species of Enterobacteriaceae which is an exclusively human pathogen.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study are listed in Table 1.
TABLE 1.
Bacterial strains used in this studya
| Strain | Properties | Source or reference |
|---|---|---|
| Y. pestis | ||
| A1122 | bv. Orientalis pFra | M. C. Chu, NCIDb |
| PEXU2 | bv. Orientalis pFra | M. C. Chu, NCID |
| PB6 | bv. Orientalis | M. C. Chu, NCID |
| F361/66 | bv. Orientalis pFra | M. C. Chu, NCID |
| 15–91 | bv. Orientalis pFra | M. C. Chu, NCID |
| 16–34 | bv. Orientalis pFra | M. C. Chu, NCID |
| AZ94-0666 | bv. Orientalis pFra | M. C. Chu, NCID |
| ZE94-2122 | bv. Orientalis pFra | M. C. Chu, NCID |
| TWS | bv. Orientalis pFra | P. Oyston, CBDEc |
| CO-92 | bv. Orientalis pFra | P. Oyston, CBDE (21) |
| GB | bv. Orientalis pFra | P. Oyston, CBDE (51) |
| JAVA 9 | bv. Orientalis | P. Oyston, CBDE |
| 195P | bv. Orientalis pFra | P. Oyston, CBDE |
| KIM | bv. Medievalis pMT1 | M. C. Chu, NCID |
| Harbin 35 | bv. Medievalis pFra | M. C. Chu, NCID |
| Nepal 516 | bv. Antiqua pFra | M. C. Chu, NCID |
| S. enterica serovar Typhi CT18 | pHCM1 pHCM2 | J. Wain, Centre for Tropical Diseasesd |
The presence of pFra/pMT1 was demonstrated by caf1 PCR (52).
NCID, National Center for Infectious Diseases, Fort Collins, Colo.
CBDE, Chemical & Biological Defence Establishment, Porton Down, United Kingdom.
The Centre for Tropical Diseases is in Ho Chi Minh City, Vietnam.
DNA library construction and sequencing.
A single colony of Y. pestis CO-92 was picked from Congo red agar and grown overnight at 28°C in BAB broth (50). A single colony of Salmonella enterica serovar Typhi CT18 was picked from Luria agar and grown overnight at 37°C in Luria broth. Total DNA was isolated as described previously (51). Random shotgun libraries of each genome were constructed and sequenced as previously described (54).
Sequence annotation and analysis.
The DNA was compared with sequences in the EMBL database using BLASTN and BLASTX (2). Transfer RNAs were predicted with tRNAscan-SE (43). Potential coding sequences (CDSs) were predicted using ORPHEUS (26) and GLIMMER (64) and also stop-to-stop prediction; the results were combined. The predicted protein sequences were searched against a nonredundant protein database using WUBLASTP and FASTA. The complete six-frame translation was used to search PROSITE, and the predicted proteins were compared against the PFAM (4) database of protein domain hidden Markov models. The results of all these analyses were assembled using the Artemis sequence viewer (63). Plasmids were defined as separate contiguous sequences from analysis of shotgun sequence data with no plasmid manipulations (separation or specific cloning). Statistical analysis of the frequency of occurrence of Chi sequences was performed using the RMES package (Unité de Biométrie, INRA, Jouy-en-Josas, France [http://www-bia.inra.fr/J/AB/genome/RMES/welcome.html]) to rank the number of Chi and Chi complement sequences against the frequency of occurrence of all octamers and their complements on one plasmid sequence strand.
PCR.
Reaction mixtures (20 μl) were prepared with 1 U of Taq polymerase (Roche Molecular Biochemicals, Lewes, United Kingdom), 1× Taq polymerase buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2), 0.2 mM (each) deoxynucleoside triphosphate and 0.2 μM (each) primer. DNA was obtained by boiling a single colony in 200 μl of distilled water for 5 min, using 1 μl of supernatant for the PCR template. For the 1,083-bp amplicon, primer P121 (5′-CCGTTCTACATCATCCATA-3′) and primer P221 (5′-TTATGGCTGGCAAATCTGA-3′) were used. PCR conditions were 94°C for 1 min and 25 cycles of 94°C for 30 s, 57.5°C for 30 s, and 72°C for 1 min, and then 72°C for 10 min. For the 1,865-bp amplicon, primer P126 (5′-GTTGAGCATTAGCGAGACC-3′) and primer P226 (5′-CTACCGCATTACTCCACTC-3′) were used. PCR conditions were 94°C for 1 min and 25 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, and then 72°C for 10 min.
Nucleotide sequence accession numbers.
The annotated Y. pestis plasmid sequences have been deposited in EMBL under accession numbers AL117211 (pFra), AL117189 (pYV), and AL109969 (pPst). The sequences of S. enterica serovar Typhi CT18 and the plasmid pHCM2 are available from the Sanger Centre (http://www.sanger.ac.uk/Projects/S_typhi).
RESULTS
Y. pestis interbiovar comparisons.
The plasmid sequences obtained for Y. pestis CO-92 were compared with those previously reported for Y. pestis KIM5. Y. pestis CO-92 is a recent clinical isolate of Y. pestis bv. Orientalis from a fatal case of primary plague pneumonia in the United States (21). Y. pestis KIM is a laboratory-passaged bv. Medievalis strain originally isolated in the Middle East (Kurdistan Iran man) (55). Two of the Y. pestis CO-92 plasmids, pPst and pCD1, show minor differences from their counterparts in Y. pestis KIM5. More extensive rearrangements and indels were seen for pMT1/pFra (Fig. 1; see Fig. 3).
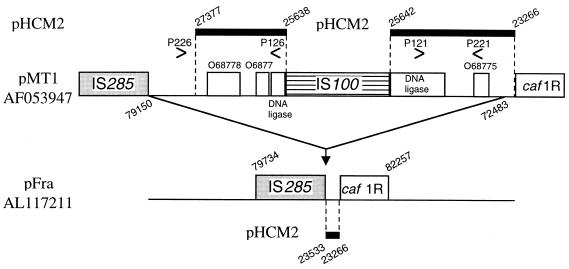
FIG. 1.
IS100-associated deletion in pFra of Y. pestis CO-92 compared with Y. pestis KIM5 pMT1 (GenBank accession no. AF053947) and S. enterica serovar Typhi CT18 plasmid pHCM2. A region of 6,668 bp present in KIM5 is shown above the corresponding region in CO-92, which lacks this insertion. Thick black lines above and below the two Y. pestis plasmids and linked to them by dotted lines indicate the regions of pHCM2 showing more than 95% identity to the adjacent Y. pestis sequence. Locations of PCR primers for the reactions shown in Fig. 2 are indicated by arrowheads. Numbers shown at an angle to the horizontal correspond to base-pair locations in the labeled plasmid. In one KIM5 sequence (GenBank accession no. AF074611), the regions shown on either side of this IS100 copy are separated by 24 kb due to IS100 recombination.
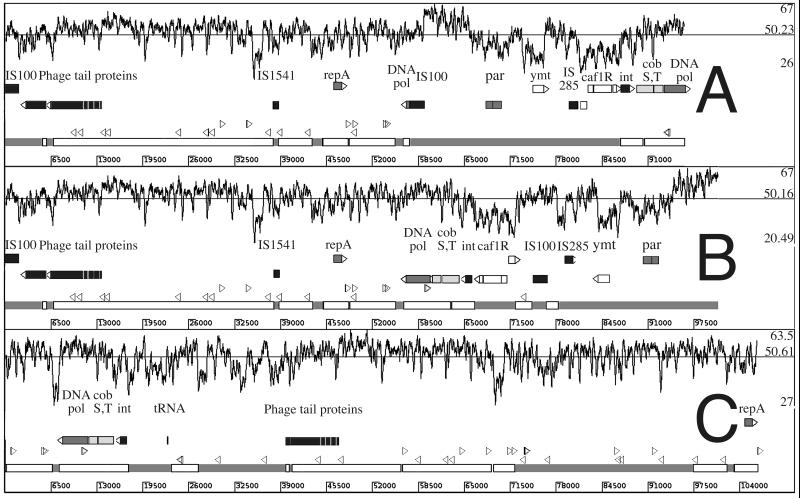
FIG. 3.
Comparison of the Y. pestis CO-92 pFra plasmid (A) with Y. pestis KIM5 pMT1 (GenBank accession no. AF053947) (B) and S. enterica serovar Typhi CT18 pHCM2 (C). Numbers on the horizontal scales are base pairs. Numbers on the vertical scale correspond to the plasmid GC percentage determined on a 200-bp window shown graphically along the top of each panel. The three numbers shown in each panel correspond to the mean percent GC for the whole plasmid and regional maxima and minima. Each panel contains the same features in the following order below the GC percentage graph. (i) Selected ORFs are shown as labeled blocks. Those transcribed to the right are shown above those transcribed to the left. ORF block shades are coded as follows: black, insertion sequence or phage related; dark gray, plasmid or DNA replication; light gray, non-DNA metabolism; white, virulence related. ORF abbreviations: DNA pol, DNA polymerase III alpha subunit; int, phage integrase; caf1R, F1 capsular operon and regulatory genes; ymt, murine toxin. (ii) Arrowheads corresponding to the location of E. coli Chi sequences are shown. Those pointing to the right are 5′-GCTGGTGG-3′, and those to the left are 5′-CCACCAGC-3′. (iii) Clear blocks in panels A and B correspond to regions of the Y. pestis plasmid sequence with sequence identity to S. enterica serovar Typhi pHCM2. The grey shading between the clear blocks corresponds to regions of the Y. pestis plasmids with no sequence identity to pHCM2. In panel C, the clear blocks represent regions of S. enterica serovar Typhi pHCM2 with significant sequence identity to Y. pestis (pFra and pMT1). The dark gray blocks represent regions with no sequence identity with Y. pestis pFra/pMT1. This figure is based on Artemis (63) views of individual annotated plasmid sequences arranged in a composite image using Adobe Photoshop.
pPst.
The 9,612-bp pesticin plasmid pPst showed two single-base-pair differences (one insertion and one deletion) and a single 2-bp insertion compared with the previously reported 9,610-bp KIM5 plasmid pPCP1 (GenBank accession no. AF053945) (37). All changes involved intergenic homopolymeric base-pair tracts. Only the single-base-pair differences were adjacent to a coding region: an insertion and a deletion 20 and 40 bp, respectively, upstream of the pst gene encoding pesticin. Neither involved the promoter or any proposed regulatory site (59). One 5-bp direct repeat flanking the IS100 insertion site in both the pPst and pPCP1 plasmids was also found, together with the next 5 bp of downstream sequence adjacent to one side of an IS100 insertion in Y. pestis bv. Medievalis KIM pMT1 and Y. pestis bv. Orientalis pFra (data not shown), suggesting that IS100 interplasmid recombination had occurred before divergence of these biovars.
pCD1.
Three of the four clusters of insertion elements identified in the 70,305-bp Y. pestis CO-92 pCD1 plasmid showed deletions or insertions compared with the two previously reported Y. pestis KIM5 sequences. These were 70,504 (37) and 70,509 (56) bp, respectively (the 5-bp indel accounting for the size difference between the two KIM sequences is not deleted from the CO-92 sequence). The major difference between the two KIM sequences and the CO-92 sequence was the location of the IS100 insertion element. In both biovars it was nested within another insertion element in a cluster of mobile genetic elements, but in the case of CO-92 it was inserted into a degenerate IS21-like element between sycE and sycH instead of a degenerate ISD1-like element between yscM and yopH as in Y. pestis KIM5. In both cases, the IS100 insertion site was flanked by a direct repeat of 5 bp. The absence of ambiguities on sequence assembly suggests that all copies of pCD1 in the cells of the colony used to make the sequence library had the same IS100 insertion site, distinct from that in KIM5 strains. However, PCR with primers from IS100 and both sets of flanking sequences gave products with the Y. pestis bv. Orientalis strains listed in Table 1 (data not shown), and we were unable to assign strain or biovar specificity to the pCD1 IS100 insertion site. Another insertion site for IS100 in pCD1 from Y. pestis bv. Orientalis strain EV76 has previously been demonstrated (GenBank accession no. X78302) (data not shown).
A 212-bp deletion of a partial IS285 sequence between yopD and yopM, apparently following recombination between two 3-bp direct repeats (located at 18,852 bp in CO-92), accounted for most of the difference in size between the KIM5 sequences and those of CO-92. It was located within one of three repeated sequences (R1-3) (60) forming part of an IS element which have apparently recombined to invert the yopM region in Y. enterocolitica pYVe227 (38) with respect to that in Y. pestis and Y. pseudotuberculosis. The deletion extended the length of R1 in Y. pestis CO-92 from 189 to 264 bp.
There were four other minor differences in non-insertion sequence ORFs from the KIM5 pCD1 sequences: three single-base-pair substitutions changing a single amino acid and an 11-bp insertion extending the Syc-like ORF7 reported upstream of ypkA (56) and rendering this ORF in CO-92 (ORF 1.73c) identical to that upstream of ypkA in Y. pseudotuberculosis pIB1 (28). There were no differences in the gene order in pCD1 between KIM5 and CO-92 other than the insertion sequence transposition. The point mutation in yadA which distinguishes Y. pestis pCD1 from its homologues in Y. enterocolitica and Y. pseudotuberculosis (56) was present.
pFra.
The 96,210-bp Y. pestis CO-92 pFra plasmid contained two IS100 elements present as inverted repeats, as described for pMT1 in KIM5 (37, 42). Deletions of 2,109 and 2,606 bp in CO-92 pFra compared with the published KIM5 pMT1 sequences were also found, corresponding to regions adjacent to an IS100 element (Fig. 1; see Fig. 3). In one of the published pMT1 sequences (GenBank accession no. AF053947) (37), these regions flanked a single IS100 element with a 5-bp direct repeat at the site of insertion and together with that element formed a single insertion of 6,669 bp compared with the CO-92 sequence (Fig. 1). In the other published KIM5 sequence (GenBank accession no. AF074611) (42), each was adjacent to a different IS100 copy, neither of which was flanked by a direct repeat. This reflected a previously reported inversion of the 24-kb region between the two IS100 copies in one KIM5 sequence compared with the other (37, 42). One of the two IS100 copies was inserted in a different location in CO-92 (bp 57267 to 59220) compared with the two KIM5 sequences (Fig. 1; see Fig. 3). Other differences in gene order between the two KIM5 sequences and CO-92 could be accounted for by insertion of this IS100 copy into a DNA polymerase III gene followed by recombination with the other IS100 sequence, inverting a 37-kb region in CO-92 (see Fig. 3).
In addition, a 63-bp deletion was detected in CO-92 pFra compared with the KIM5 pMT1 sequences, apparently following recombination between 12-bp imperfect direct repeats, resulting in the fusion of two unidentified KIM5 ORFs (O68739 and O68738 in one KIM5 sequence [37], Y1032 and Y1033 in the other [42]) to form ORF 1.22c. There were 12 sites showing differences with both KIM5 sequences. Four of these were conservative single-base-pair substitutions in coding regions, four were 1- or 2-bp indels in noncoding sequence, and four were single-base-pair substitutions in coding regions changing one or two amino acids. There were 12 sites where the CO-92 sequence showed identity with one but not both KIM5 sequences (i.e., KIM5 polymorphisms were as common as interbiovar differences). These comprised three single-base-pair substitutions (one conservative, two in noncoding regions), eight single-base-pair indels (three in noncoding regions), and one 5-bp polymorphism in a noncoding region. There were no differences in any known virulence-related factors.
PCR with primers internal to the two large deletions yielded no specific amplicons with a variety of Y. pestis bv. Orientalis strains, but they were present (Fig. 2) in a Y. pestis bv. Antiqua strain and a Y. pestis bv. Medievalis strain unrelated to KIM, suggesting that this deletion could be biovar specific. In addition to previously reported homologies, BLASTN searching showed that most of this deleted region (apart from the IS100 sequence) was present in pHCM2, one of two plasmids present in a multidrug-resistant strain of S. enterica serovar Typhi CT18 which originated in Vietnam (http://www.sanger.ac.uk/Projects/S_typhi). This plasmid also harbored 60 bp of the 63-bp deletion.
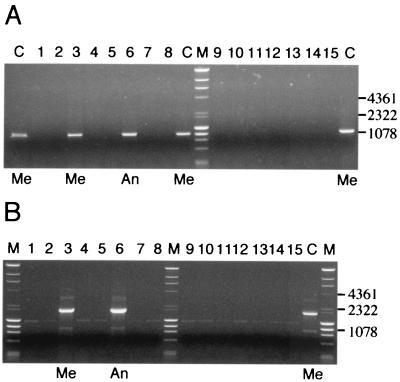
FIG. 2.
PCR for IS100-flanking regions from Y. pestis KIM5 pMT1 (bv. Medievalis) showing absence in Y. pestis bv. Orientalis strains and presence in other biovar Medievalis and Antiqua strains. DNA fragment sizes are given in base pairs. (A) PCR with primers P121 and P221. (B) PCR with primers P226 and P126. Lanes: 1, strain A1122; 2, PEXU2; 3, Harbin 35; 4, PB6; 5, F361/66; 6, Nepal 516; 7, 15–91; 8, 16–34; 9, AZ94-0666; 10, ZE94-2122; 11, TWS; 12, CO-92; 13, GB; 14, JAVA 9; 15, 195P; C, KIM (control); M, molecular weight marker. Additional text labels indicate lanes containing DNA templates from Y. pestis bv. Antiqua (An) and bv. Medievalis (Me) strains. All other numbered lanes contain DNA from Y. pestis bv. Orientalis strains. A 0.9% agarose gel was used. The digital image was obtained with GelDoc (Bio-Rad) and was cropped and resized in Adobe Photoshop.
pFra and pHCM2 comparison.
More than 52% of pFra from Y. pestis CO-92 was highly similar to pHCM2 (Table 2). Because some of the shared DNA sequence was in regions absent in Y. pestis bv. Orientalis strains, the percentage of KIM5 pMT1 resembling pHCM2 was higher than that of CO-92 pFra at 56.2%, with 57.6% of pHCM2 showing similarity with pMT1. More than 90% of the similar regions showed more than 96% DNA identity. The G+C content of the regions in common (51.9%) was closer to the overall S. enterica serovar Typhi G+C content of 52.05% than the Yersinia background level of 47.6%, while the unique regions of both plasmids showed a lower G+C content.
TABLE 2.
Overall sequence features of pFra, pMT1, and pHCM2
| Parameters | Value for plasmid or specific hosta
|
||
|---|---|---|---|
| Y. pestis CO-92 pFra | Y. pestis KIM5 pMTIb | S. enterica serovar Typhi CT18 pHCM2 | |
| Chromosomal G+C (%) | 47.6 | 47.6 | 52.05 |
| Plasmid size (bp) | 96,210 | 100984 | 106515 |
| Plasmid G+C (%) | 50.23 | 50.16 | 50.61 |
| Proportion of plasmid with cross-species similarity (%) | 58.6 | 59.6 | 52.6 (pFra) |
| 56.2 (pMT1) | |||
| G+C content of plasmid regions with cross-species similarity (%) | 51.9 | 51.9 | 51.9 |
| G+C content of nonhomologous regions (%) | 47.8 | 47.8 | 49.1 non-pFra |
| 48.9 non-pMT1 | |||
| Number of Chi and Chi complement sites on one plasmid sequence strand | 19 | 20 | 32 |
| Expectedc Chi frequency (in same length sequence of random base order) | 2.96 | 3.06 | 3.36 |
| Probabilityd (rank) of random Chi occurrence in plasmid based on dinucleotide frequency | <10−38 (1) | <10−44 (1) | <10−116 (1) |
Sixty-two ORFs in Y. pestis CO-92 pFra had their closest homologue outside Y. pestis in predicted ORFs in pHCM2. For a further four predicted ORFs in pHCM2, homologues in the region of Y. pestis KIM5 pMT1 were absent in CO-92 (Fig. 1). The DNA sequences sharing significant homology between pFra and pHCM2 included those encoding the repA gene (42) and the surrounding iteron-based replication control system in the ori region (Fig. 3). pFra encodes a parABS-like system (ORFs 1.68c and 1.67c) (Fig. 3) resembling that of bacteriophages P1 and P7 (42) which is functional in Escherichia coli (78), but this region is absent in pHCM2. The partial repeats of fragments of the S. enterica serovar Typhimurium parAB loci previously noted (42) adjacent to the murine toxin gene in Y. pestis KIM5 pMT1 and the adjacent resolvase homologue were present in CO-92 (ORF 1.70) but not in pHCM2. Neither pFra/pMT1 nor pHCM2 had an obvious set of transfer or mobilization genes.
There is only one significant segment of low G+C composition in the regions common to pFra/pMT1 and pHCM2. This is adjacent to an IS200-like sequence, IS1541, in pFra (Fig. 3) and contains the pFra ORF 1.34 with no significant similarity on BLAST searching other than in pHCM2. The blocks of sequence similarity between pFra and pHCM2 occur in the same order in both plasmids. Most of the sequence present in Y. pestis KIM5 but not in Y. pestis CO-92 is present in pHCM2, and there are no inversions of any homologous sequence in pHCM2 compared with Y. pestis KIM5, while there is one such inversion in CO-92, reflecting the 37-kb inversion compared with KIM5 (Fig. 3).
Repeated elements.
The three insertion elements encoded by pFra (Two IS100 copies and a single IS1541) were outside the regions of sequence similarity with pHCM2 (Fig. 3). No insertion elements were apparent in the pHCM2 sequence. The E. coli Chi site GCTGGTGG and the complement CCACCAGC occur 19 times in pFra (Table 2) and are restricted to the regions also found in pHCM2 (Fig. 3). This sequence is not found in the other two Y. pestis plasmids. The R'MES program (23) showed it to be the most overrepresented of the 65,536 possible octanucleotides in pFra, pMT1, and pHCM2 (Table 2), based on the dinucleotide composition of each plasmid. A novel 57-bp imperfect repeat is present upstream of three unidentified ORFs in both pHCM2 and pFra (data not shown). Another five copies of this repeat are present between unidentified ORFs in pHCM2. These repeat sequences were not found elsewhere in the Y. pestis or S. enterica serovar Typhi genome sequence and were not deposited in GenBank. They do not show the palindromic symmetry typical of integrons or super integrons (62).
DISCUSSION
Y. pestis biovar-associated plasmid differences.
The most obvious differences between the Y. pestis CO-92 and KIM5 plasmid sequences were those related to recombination between IS100 elements or IS100 transposition. No IS-related transposition differences were apparent in CO-92 pPst compared with KIM pPCP1 despite the presence of an IS100 sequence, possibly due to the limited number of potential insertion sites in this small plasmid. A 24-kb inversion between two IS100 copies in pMT1 was previously noted on comparison of two sequences of the same strain of Y. pestis KIM5 (37, 42). From the loss of direct repeats flanking IS100 in one of these sequences, this inversion can be attributed to IS100 recombination, presumably during 30 years of laboratory storage (8) or during the E. coli subcloning of Y. pestis plasmids that was carried out to obtain the KIM plasmid sequences (37, 42). Laboratory strains of bacteria that are stored for many years are particularly subject to IS-related rearrangements (48). In contrast, Y. pestis CO-92 is a recent clinical isolate (1992) (21) with fewer laboratory passages, and the genomic shotgun method employed in this study did not require subcloning of the entire pFra plasmid, reducing the scope for in vitro rearrangement that could influence sequence results.
The deletions we have detected adjacent to one of the IS100 elements in Y. pestis CO-92 pFra compared to KIM pMT1 (Fig. 1) are demonstrable in other Y. pestis bv. Orientalis strains but not in those of Y. pestis bv. Antiqua or Medievalis (Fig. 2). Consistent biovar-specific deletions are unlikely to be in vitro phenomena. These deletions are largely responsible for the 4.8-kb difference in size between Y. pestis CO-92 pFra and KIM5 pMT1. It is well recognized that pFra differs in size in strains of different origins (24). In particular, the pFra of two Y. pestis bv. Orientalis strains has previously been noted on restriction mapping to be 3 kb smaller than the pFra of two Y. pestis bv. Antiqua strains (57). However, not only is this sequence present in Y. pestis bv. Medievalis and Antiqua strains, but most of it forms part of the pFra plasmid-related sequence which is encoded by the S. enterica serovar Typhi plasmid. This suggests that the interbiovar difference is indeed due to a unique deletion in Y. pestis bv. Orientalis pFra strains rather than an insertion in Y. pestis bv. Medievalis and Antiqua strains. This is consistent with a reported Orientalis-specific chromosomal point mutation (11) and supports phylogenetic analysis based on restriction patterns on chromosomal hybridization with IS100, which suggested that this biovar had arisen after the other two (1). The definition of these deleted segments offers the first plasmid-based test for Y. pestis biovar differentiation by molecular means.
Horizontal DNA transfer between Yersinia and other Enterobacteriaceae.
Salmonella species are clearly distinct from Yersinia species in 16S rRNA (15, 72) and other sequenced loci (46). Therefore, the presence of such a high degree of sequence identity on a potentially mobile element implies recent horizontal genetic exchange. There is some evidence of chromosomal DNA exchange between S. enterica serovar Typhimurium and Yersinia species other than Y. pestis. Intervening sequences of 100 bp in the rrl genes (23S rRNA genes) of highly pathogenic Yersinia enterocolitica strains are more closely related to sequences in S. enterica serovar Typhimurium than those of other Yersinia strains (68). This intervening sequence is not present in Y. pestis (68) or S. enterica serovar Typhi (44).
Short regions of similarity in nonstructural genes have been reported between the virulence-associated plasmids of S. enterica serovar Dublin and Y. pseudotuberculosis (40). Numerous short stretches of pMT1 (maximum length, 1,150 bp) over a 30-kb region have been previously noted to show high levels of DNA similarity with other plasmid and DNA sequences (42). The IS100 and IS285 insertion sequences found on Y. pestis plasmids and in particular pFra are present in the enteropathogenic E. coli adherence factor plasmid (74). IS285 incorporates a 117-bp region with more than 76% identity to the gene encoding the 38-amino-acid enteroaggregative E. coli heat-stable enterotoxin 1 (EAST1) which is found in a variety of diarrhea-associated E. coli strains (76). The 225-amino-acid ORF pMT1.64c is 89% identical to a 227-amino-acid ORF from the virulence plasmid of E. coli O157:H7 (12).
The region of pFra/pMT1 forming a mosaic of DNA identities and insertion sequences is also the region that encodes the known Y. pestis virulence-associated factors (capsule and the murine toxin). Interestingly, these genes have already been noted to be composed of a lower GC percentage than the remainder of the plasmid (42) and the Y. pestis host. Strikingly, similarity to the pHCM2 plasmid is almost absent from this region, apart from the IS100-flanking sequence not present in Y. pestis bv. Orientalis strains.
The overrepresentation of the canonical E. coli Chi sequence seen on pHCM2 and the corresponding regions of pFra has not previously been reported for naturally occurring plasmids. R'MES analysis of the 23 complete plasmid sequences from Enterobacteriaceae represented on the Entrez Genomes database (http://www.ncbi.nlm.nih.gov:80/PMGifs/Genomes/eub_p.html) revealed only one further plasmid with a similar degree of Chi overrepresentation: S. enterica serovar Typhi plasmid pR27 (66) contains Chi as the 4th most overrepresented octanucleotide. This plasmid shows extensive sequence similarity to pHCM1, the other plasmid present in S. enterica serovar Typhi CT18 (http://www.sanger.ac.uk/Projects/S_typhi) in which Chi is the 5th most overrepresented octanucleotide. The contrasting complete absence of Chi sequences in the other two Y. pestis plasmids and the virulence-associated regions of pFra is intriguing. In E. coli, Chi sequences regulate the different functions of the RecBCD complex (exonuclease-recombinase) in repairing double-stranded breaks in host DNA and destroying foreign DNA cleaved by host endonucleases (71, 73). Different sequences may fulfill the function of Chi sequences in different bacterial hosts (71), and the presence of a specific Chi sequence in plasmid genes may be a signature of origin from a specific bacterial genome. Phylogenetic analysis suggests that multiple horizontal transfers of chromosomal bacterial genes to plasmids commonly occur (47), and these genes would necessarily include the respective specific bacterial Chi sequence, because Chi sequences tend to be in coding DNA (41). RecBCD-induced recombination due to Chi sequences may promote the survival of horizontally transferred DNA between closely related bacteria (32). It has been suggested that Chi sequences overlapping the start codons of plasmid replication proteins may promote recombination in replicon evolution (49). None of the Chi sequences identified in pFra or pHCM2 overlap start codons in this way. Related Enterobacteriaceae may use the same Chi sequence; e.g., S. enterica serovar Typhimurium has RecBCD-like enzymes active at E. coli Chi sites (45). Specific Chi sequence activity in Yersinia species and S. enterica serovar Typhi remains to be determined.
pFra is known to integrate into the Y. pestis chromosome (58), and the plasmid regions shared with pHCM2 contain Chi sequences and apparent metabolic genes (cobS and cobT) not found on plasmids in other organisms. Another recently described Y. pestis plasmid containing ORFs highly homologous to E. coli chromosomal genes (22) does not contain Chi sequences. The pla gene on pPst encodes a serine protease whose amino acid sequence is 70% identical (69) to those of products of chromosomal genes from E. coli and S. enterica serovar Typhimurium.
Homologous recombination between identical sequence on the plasmid and the chromosome (e.g., copies of IS100) is the suggested mechanism for the observed phenomenon of chromosomal integration of pFra (58). Other site-specific integration processes may have been functional in the acquisition of chromosomal genes by pFra and pHCM2. A predicted asparagine tRNA gene is present in the pHCM2-specific region at one boundary of the sequence common to both plasmids (Fig. 3) upstream of an integrase gene. tRNA genes are known to be integration sites of pathogenicity islands in the bacterial chromosome (30), and the Yersinia high-pathogenicity island is apparently inserted into an asparagine tRNA (10). There are no direct repeats of portions of the tRNA gene elsewhere in pHCM2 to suggest a currently excisable pathogenicity island. The integrase, cobS, and cobT genes are also present in pFra with no obvious interrupted genes at the boundaries of shared sequence in either plasmid.
Mechanism of transfer.
A transferable plasmid, pIPI202, conferring multiple drug resistances has recently been found in a clinical Y. pestis isolate (27). This 150-kb plasmid has an Inc6-C origin of replication (unlike pFra and pHCM2). Although there are no obvious mobilization genes on the pFra/pMT1 plasmid, cointegration (19) with another conjugative plasmid (27) or an ori-T-independent method of transfer, such as that mediated by the Constin element SXT from Vibrio cholerae (36), remain possibilities. The presence of endogenous transducing phages in Y. pestis has not been demonstrated (55), but phage genes are represented on the common regions of pFra and pHCM2. Both Y. pestis and S. enterica serovar Typhi are among the seven species not known to be naturally competent which contain homologues of the HP0333 gene involved in natural transformation in Helicobacter pylori (3). They may have been naturally competent in the past or require specific conditions to show this property. Recently it has been shown that genes on another nonconjugative plasmid (E. coli pO157) may be transferred from E. coli to other enteric bacteria via membrane vesicles (77). This has not been shown for Salmonella or Yersinia species.
Whatever the mode or direction of transfer, extensive recombination with chromosomal or other plasmid DNA has occurred in at least one of the new hosts after the transfer event to achieve the sharp delineations observed between plasmid regions common to both replicons and those unique to each plasmid (Fig. 3). Of the two plasmids, pFra shows more mosaicism, evidenced by variability in G+C composition and Chi sequence distribution, suggesting that it has recombined more than pHCM2, probably due to the presence of multiple insertion sequences. This implies that pFra originally resembled pHCM2 even more closely than it does now. The G+C composition of the regions common to both plasmids is much closer to the S. enterica serovar Typhi chromosomal background than that of Y. pestis, and insertion sequences present in pFra are found elsewhere in Y. pestis and not in pHCM2 or the S. enterica serovar Typhi chromosome, suggesting that any direct transfer was from S. enterica serovar Typhi to Y. pestis or Y. pseudotuberculosis and not in the other direction. Experiments are planned to search for conjugative plasmids related to pFra and pHCM2 in other S. typhi strains. The source for the virulence-related and lifestyle-specific genes on the unique portions of Y. pestis pFra remains obscure, but the G+C content and other sequence features (lack of Chi sequences) suggest an origin outside Enterobacteriaceae.
Date of plasmid transfer.
The opportunities for direct contact between S. enterica serovar Typhi and Y. pestis are limited. Host adaptation limits typhoid fever to humans and a few higher primates (39). Although Y. pestis is invasive for epithelial cells in vitro (17) and by the oral route in animal models (14), it does not colonize the gut (14) and has a restricted capacity for survival in the environment (55). This would confine contact between the two organisms to a dually infected human host or the gut of a flea vector which had fed on multiple hosts. Y. pseudotuberculosis however, is, a gastrointestinal pathogen with a wide host range likely to bring it into contact with Salmonella species in the environment or vertebrate gut. Like Y. pestis, S. enterica serovar Typhi is homogenous in population genetic analysis (65), forming only two clones. However, unlike the Y. pestis relationship with Y. pseudotuberculosis, phylogenetic analysis does not place it as a typical subclone of another S. enterica serovar (7, 65)—there are distinct serovar Typhi-specific polymorphisms not falling within the allelic variation seen within other currently identifiable Salmonella serovars. The very recent origin proposed for the Y. pestis clone (1,500 to 20,000 years ago) (1) is not therefore applicable to S. enterica serovar Typhi, and it is likely to have been in existence before Y. pestis.
The presence of most of the pFra biovar-specific deletion in the cryptic S. enterica serovar Typhi plasmid pHCM2 suggests that horizontal transfer of the plasmid occurred before emergence of the Orientalis biovar or involved a non-Orientalis biovar, which would be unusual in Vietnam. Plague arrived in Vietnam during the third pandemic in 1898, and most current isolates reported are Y. pestis bv. Orientalis, although some Y. pestis bv. Medievalis strains have been reported from the High-Plateaux (T. Mai, A. Guiyoule, D. Vinh, D. Tuyet, D. Thung, and E. Carniel, Ned. Tidjschr. Med. Microbiol. 6, Suppl. II, Abstr. P-70, 1998). The simplest route for direct plasmid transfer would be between an ancestral Y. pseudotuberculosis strain that gave rise to Y. pestis and an S. enterica serovar Typhi strain. The degree of identity retained by the plasmids is compatible with the hypothesis (1) that pFra plasmid acquisition and Y. pestis emergence could have been as recent as 2,000 years ago.
ACKNOWLEDGMENTS
Pathogen sequencing at the Sanger Centre is funded by the Wellcome Trust through its Beowulf Genomics initiative. M.B.P. acknowledges support from the Trustees of St. Bartholomew's Hospital.
We thank Nick Loman and Alex Lam for assistance with software installation and database searches and Sophie Schbath-Grammagnat for information on RMES utilization.
REFERENCES
- 1.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Ando T, Israel D A, Kusugami K, Blaser M J. HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori. J Bacteriol. 1999;181:5572–5580. doi: 10.1128/jb.181.18.5572-5580.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonnhammer E L. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bercovier H, Mollaret H. Intra and interspecies relatedness of Y. pestis by DNA hybridisation and its relationship with Yersinia pseudotuberculosis. Curr Microbiol. 1980;4:225. [Google Scholar]
- 6.Boyd E F, Hill C W, Rich S M, Hartl D L. Mosaic structure of plasmids from natural populations of Escherichia coli. Genetics. 1996;143:1091–1100. doi: 10.1093/genetics/143.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd E F, Wang F S, Beltran P, Plock S A, Nelson K, Selander R K. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J Gen Microbiol. 1993;139:1125–1132. doi: 10.1099/00221287-139-6-1125. [DOI] [PubMed] [Google Scholar]
- 8.Brubaker R. Interconversion of purine mononucleotides in Pasteurella pestis. Infect Immun. 1970;1:446–454. doi: 10.1128/iai.1.5.446-454.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brubaker R R. Factors promoting acute and chronic diseases caused by yersiniae. Clin Microbiol Rev. 1991;4:309–324. doi: 10.1128/cmr.4.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchrieser C, Brosch R, Bach S, Guiyoule A, Carniel E. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asntRNA genes. Mol Microbiol. 1998;30:965–978. doi: 10.1046/j.1365-2958.1998.01124.x. [DOI] [PubMed] [Google Scholar]
- 11.Buchrieser C, Rusniok C, Frangeul L, Couve E, Billault A, Kunst F, Carniel E, Glaser P. The 102-kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect Immun. 1999;67:4851–4861. doi: 10.1128/iai.67.9.4851-4861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burland V, Shao Y, Perna N T, Plunkett G, Sofia H J, Blattner F R. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli 0157:H7. Nucleic Acids Res. 1998;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler T. Plague and other Yersinia infections. New York, N.Y: Plenum Press; 1983. [Google Scholar]
- 14.Butler T, Fu Y S, Furman L, Almeida C, Almeida A. Experimental Yersinia pestis infection in rodents after intragastric inoculation and ingestion of bacteria. Infect Immun. 1982;36:1160–1167. doi: 10.1128/iai.36.3.1160-1167.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang H R, Loo L H, Jeyaseelan K, Earnest L, Stackebrandt E. Phylogenetic relationships of Salmonella typhi and Salmonella typhimurium based on 16S rRNA sequence analysis. Int J Syst Bacteriol. 1997;47:1253–1254. doi: 10.1099/00207713-47-4-1253. [DOI] [PubMed] [Google Scholar]
- 16.Chanteau S, Ratsifasoamanana L, Rasoamanana B, Rahalison L, Randriambelosoa J, Roux J, Rabeson D. Plague, a reemerging disease in Madagascar. Emerg Infect Dis. 1998;4:101–104. doi: 10.3201/eid0401.980114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cowan C, Jones H A, Kaya Y H, Perry R D, Straley S C. Invasion of epithelial cells by Yersinia pestis: evidence for a Y. pestis-specific invasin. Infect Immun. 2000;68:4523–4530. doi: 10.1128/iai.68.8.4523-4530.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crook L D, Tempest B. Plague. A clinical review of 27 cases. Arch Intern Med. 1992;152:1253–1256. doi: 10.1001/archinte.152.6.1253. [DOI] [PubMed] [Google Scholar]
- 19.Davison J. Genetic exchange between bacteria in the environment. Plasmid. 1999;42:73–91. doi: 10.1006/plas.1999.1421. [DOI] [PubMed] [Google Scholar]
- 20.Devignat R. Varietés de l'espèce Pasteurella pestis. Nouvelle hyphothèse. Bull WHO. 1951;4:247–263. [PMC free article] [PubMed] [Google Scholar]
- 21.Doll J M, Zeitz P S, Ettestad P, Bucholtz A L, Davis T, Gage K. Cat-transmitted fatal pneumonic plague in a person who traveled from Colorado to Arizona. Am J Trop Med Hyg. 1994;51:109–114. doi: 10.4269/ajtmh.1994.51.109. [DOI] [PubMed] [Google Scholar]
- 22.Dong X Q, Lindler L E, Chu M C. Complete DNA sequence and analysis of an emerging cryptic plasmid isolated from Yersinia pestis. Plasmid. 2000;43:144–148. doi: 10.1006/plas.1999.1432. [DOI] [PubMed] [Google Scholar]
- 23.El Karoui M, Biaudet V, Schbath S, Gruss A. Characteristics of Chi distribution on different bacterial genomes. Res Microbiol. 1999;150:579–587. doi: 10.1016/s0923-2508(99)00132-1. [DOI] [PubMed] [Google Scholar]
- 24.Filippov A A, Solodovnikov N S, Kookleva L M, Protsenko O A. Plasmid content in Yersinia pestis strains of different origin. FEMS Microbiol Lett. 1990;55:45–48. doi: 10.1016/0378-1097(90)90165-m. [DOI] [PubMed] [Google Scholar]
- 25.Friedlander A M, Welkos S L, Worsham P L, Andrews G P, Heath D G, Anderson G W, Pitt M L M, Estep J, Davis K. Relationship between virulence and immunity as revealed in recent studies of the F1 capsule of Yersinia pestis. Clin Infect Dis. 1995;21:S178–S181. doi: 10.1093/clinids/21.supplement_2.s178. [DOI] [PubMed] [Google Scholar]
- 26.Frishman D, Mironov A, Mewes H W, Gelfand M. Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res. 1998;26:2941–2947. doi: 10.1093/nar/26.12.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galimand M, Guiyoule A, Gerbaud G, Rasoamanana B, Chanteau S, Carniel E, Courvalin P. Multidrug resistance in Yersinia pestis mediated by a transferable plasmid. N Engl J Med. 1997;337:677–680. doi: 10.1056/NEJM199709043371004. [DOI] [PubMed] [Google Scholar]
- 28.Galyov E E, Hakansson S, Wolf-Watz H. Characterization of the operon encoding the YpkA Ser/Thr protein kinase and the YopJ protein of Yersinia pseudotuberculosis. J Bacteriol. 1994;176:4543–4548. doi: 10.1128/jb.176.15.4543-4548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guiyoule A, Grimont F, Iteman I, Grimont P A, Lefevre M, Carniel E. Plague pandemics investigated by ribotyping of Yersinia pestis strains. J Clin Microbiol. 1994;32:634–641. doi: 10.1128/jcm.32.3.634-641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 31.Han Y W, Miller V L. Reevaluation of the virulence phenotype of the inv yadA double mutants of Yersinia pseudotuberculosis. Infect Immun. 1997;65:327–330. doi: 10.1128/iai.65.1.327-330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Handa N, Ohashi S, Kobayashi I. Clustering of chi sequence in Escherichia coli genome. Microb Comp Genomics. 1997;2:287–298. doi: 10.1089/omi.1.1997.2.287. [DOI] [PubMed] [Google Scholar]
- 33.Hinnebusch B, Perry R, Schwan T. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- 34.Hinnebusch B J. Bubonic plague: a molecular genetic case history of the emergence of an infectious disease. J Mol Med. 1997;75:645–652. doi: 10.1007/s001090050148. [DOI] [PubMed] [Google Scholar]
- 35.Hinnebusch J, Cherepanov P, Du Y, Rudolph A, Dixon J D, Schwan T, Forsberg A. Murine toxin of Yersinia pestis shows phospholipase D activity but is not required for virulence in mice. Int J Med Microbiol. 2000;290:483–487. doi: 10.1016/S1438-4221(00)80070-3. [DOI] [PubMed] [Google Scholar]
- 36.Hochhut B, Marrero J, Waldor M K. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae 0139. J Bacteriol. 2000;182:2043–2047. doi: 10.1128/jb.182.7.2043-2047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu P, Elliott J, McCready P, Skowronski E, Garnes J, Kobayashi A, Brubaker R R, Garcia E. Structural organization of virulence-associated plasmids of Yersinia pestis. J Bacteriol. 1998;180:5192–5202. doi: 10.1128/jb.180.19.5192-5202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iriarte M, Cornelis G. The 70-kilobase virulence plasmid of yersiniae. In: Kaper J, Hacker J, editors. Pathogenicity islands and other mobile virulence elements. Washington, D.C.: ASM Press; 1999. pp. 91–126. [Google Scholar]
- 39.Kingsley R A, Baumler A J. MicroReview: host adaptation and the emergence of infectious disease: the Salmonella paradigm. Mol Microbiol. 2000;36:1006–1014. doi: 10.1046/j.1365-2958.2000.01907.x. [DOI] [PubMed] [Google Scholar]
- 40.Krause M, Harwood J, Fierer J, Guiney D. Genetic analysis of homology between the virulence plasmids of Salmonella dublin and Yersinia pseudotuberculosis. Infect Immun. 1991;59:1860–1863. doi: 10.1128/iai.59.5.1860-1863.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lao P J, Forsdyke D R. Crossover hot-spot instigator (Chi) sequences in Escherichia coli occupy distinct recombination/transcription islands. Gene. 2000;243:47–57. doi: 10.1016/s0378-1119(99)00564-8. [DOI] [PubMed] [Google Scholar]
- 42.Lindler L E, Plano G V, Burland V, Mayhew G F, Blattner F R. Complete DNA sequence and detailed analysis of the Yersinia pestis KIM5 plasmid encoding murine toxin and capsular antigen. Infect Immun. 1998;66:5731–5742. doi: 10.1128/iai.66.12.5731-5742.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowe T M, Eddy S R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattatall N R, Daines D A, Liu S L, Sanderson K E. Salmonella typhi contains identical intervening sequences in all seven rrl genes. J Bacteriol. 1996;178:5323–5326. doi: 10.1128/jb.178.17.5323-5326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKittrick N H, Smith G R. Activation of Chi recombinational hotspots by RecBCD-like enzymes from enteric bacteria. J Mol Biol. 1989;210:485–495. doi: 10.1016/0022-2836(89)90125-3. [DOI] [PubMed] [Google Scholar]
- 46.Mollet C, Drancourt M, Raoult D. rpoB sequence analysis as a novel basis for bacterial identification. Mol Microbiol. 1997;26:1005–1011. doi: 10.1046/j.1365-2958.1997.6382009.x. [DOI] [PubMed] [Google Scholar]
- 47.Moreira D. Multiple independent horizontal transfers of informational genes from bacteria to plasmids and phages: implications for the origin of bacterial replication machinery. Mol Microbiol. 2000;35:1–5. doi: 10.1046/j.1365-2958.2000.01692.x. [DOI] [PubMed] [Google Scholar]
- 48.Naas T, Blot M, Fitch W M, Arber W. Dynamics of IS-related genetic rearrangements in resting Escherichia coli K-12. Mol Biol Evol. 1995;12:198–207. doi: 10.1093/oxfordjournals.molbev.a040198. [DOI] [PubMed] [Google Scholar]
- 49.Osborn A M, da Silva Tatley F M, Steyn L M, Pickup R W, Saunders J R. Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology. 2000;146:2267–2275. doi: 10.1099/00221287-146-9-2267. [DOI] [PubMed] [Google Scholar]
- 50.Oyston P C, Dorrell N, Williams K, Li S R, Green M, Titball R W, Wren B W. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect Immun. 2000;68:3419–3425. doi: 10.1128/iai.68.6.3419-3425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oyston P C, Russell P, Williamson E D, Titball R W. An aroA mutant of Yersinia pestis is attenuated in guinea pigs, but virulent in mice. Microbiology. 1996;142:1847–1853. doi: 10.1099/13500872-142-7-1847. [DOI] [PubMed] [Google Scholar]
- 52.Oyston P C, Williamson E D, Leary S E, Eley S M, Griffin K F, Titball R W. Immunization with live recombinant Salmonella typhimurium aroA producing F1 antigen protects against plague. Infect Immun. 1995;63:563–568. doi: 10.1128/iai.63.2.563-568.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pang T, Levine M M, Ivanoff B, Wain J, Finlay B B. Typhoid fever—important issues still remain. Trends Microbiol. 1998;6:131–133. doi: 10.1016/s0966-842x(98)01236-0. [DOI] [PubMed] [Google Scholar]
- 54.Parkhill J, Wren B W, Mungall K, Ketley J M, Churcher C, Basham D, Chillingworth T, Davies R M, Feltwell T, Holroyd S, Jagels K, Karlyshev A V, Moule S, Pallen M J, Penn C W, Quail M A, Rajandream M A, Rutherford K M, van Vliet A H, Whitehead S, Barrell B G. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 55.Perry R D, Fetherston J D. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perry R D, Straley S C, Fetherston J D, Rose D J, Gregor J, Blattner F R. DNA sequencing and analysis of the low-Ca2+-response plasmid pCD1 of Yersinia pestis KIM5. Infect Immun. 1998;66:4611–4623. doi: 10.1128/iai.66.10.4611-4623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Popov Iu A, Iashechkin Iu I, Drozdov I G. Molecular genetic analysis of DNA structure of pFra plasmids in plague pathogen of varying biovar. Genetika. 1998;34:198–205. [PubMed] [Google Scholar]
- 58.Protsenko O A, Filippov A A, Kutyrev V V. Integration of the plasmid encoding the synthesis of capsular antigen and murine toxin into Yersinia pestis chromosome. Microb Pathog. 1991;11:123–128. doi: 10.1016/0882-4010(91)90005-u. [DOI] [PubMed] [Google Scholar]
- 59.Rakin A, Boolgakowa E, Heesemann J. Structural and functional organization of the Yersinia pestis bacteriocin pesticin gene cluster. Microbiology. 1996;142:3415–3424. doi: 10.1099/13500872-142-12-3415. [DOI] [PubMed] [Google Scholar]
- 60.Reisner B S, Straley S C. Yersinia pestis YopM: thrombin binding and overexpression. Infect Immun. 1992;60:5242–5252. doi: 10.1128/iai.60.12.5242-5252.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosqvist R, Skurnik M, Wolf-Watz H. Increased virulence of Yersinia pseudotuberculosis by two independent mutations. Nature. 1988;334:522–524. doi: 10.1038/334522a0. [DOI] [PubMed] [Google Scholar]
- 62.Rowe-Magnus D A, Mazel D. Resistance gene capture. Curr Opin Microbiol. 1999;2:483–488. doi: 10.1016/s1369-5274(99)00004-1. [DOI] [PubMed] [Google Scholar]
- 63.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M A, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 64.Salzberg S L, Delcher A L, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Selander R K, Beltran P, Smith N H, Helmuth R, Rubin F A, Kopecko D J, Ferris K, Tall B D, Cravioto A, Musser J M. Evolutionary genetic relationships of clones of Salmonella serovars that cause human typhoid and other enteric fevers. Infect Immun. 1990;58:2262–2275. doi: 10.1128/iai.58.7.2262-2275.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sherburne C K, Lawley T D, Gilmour M W, Blattner F R, Burland V, Grotbeck E, Rose D J, Taylor D E. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 2000;28:2177–2186. doi: 10.1093/nar/28.10.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simonet M, Riot B, Fortineau N, Berche P. Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect Immun. 1996;64:375–379. doi: 10.1128/iai.64.1.375-379.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Skurnik M, Toivanen P. Intervening sequences (IVSs) in the 23S ribosomal RNA genes of pathogenic Yersinia enterocolitica strains. The IVSs in Y. enterocolitica and Salmonella typhimurium have a common origin. Mol Microbiol. 1991;5:585–593. doi: 10.1111/j.1365-2958.1991.tb00729.x. [DOI] [PubMed] [Google Scholar]
- 69.Sodeinde O A, Goguen J D. Nucleotide sequence of the plasminogen activator gene of Yersinia pestis: relationship to ompT of Escherichia coli and gene E of Salmonella typhimurium. Infect Immun. 1989;57:1517–1523. doi: 10.1128/iai.57.5.1517-1523.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sodeinde O A, Subrahmanyam Y V, Stark K, Quan T, Bao Y, Goguen J D. A surface protease and the invasive character of plague. Science. 1992;258:1004–1007. doi: 10.1126/science.1439793. [DOI] [PubMed] [Google Scholar]
- 71.Sourice S, Biaudet V, El Karoui M, Ehrlich S D, Gruss A. Identification of the Chi site of Haemophilus influenzae as several sequences related to the Escherichia coli Chi site. Mol Microbiol. 1998;27:1021–1029. doi: 10.1046/j.1365-2958.1998.00749.x. [DOI] [PubMed] [Google Scholar]
- 72.Sproer C, Mendrock U, Swiderski J, Lang E, Stackebrandt E. The phylogenetic position of Serratia, Buttiauxella and some other genera of the family Enterobacteriaceae. Int J Syst Bacteriol. 1999;49:1433–1438. doi: 10.1099/00207713-49-4-1433. [DOI] [PubMed] [Google Scholar]
- 73.Taylor A F, Smith G R. Regulation of homologous recombination: Chi inactivates RecBCD enzyme by disassembly of the three subunits. Genes Dev. 1999;13:890–900. doi: 10.1101/gad.13.7.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tobe T, Hayashi T, Han C G, Schoolnik G K, Ohtsubo E, Sasakawa C. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect Immun. 1999;67:5455–5462. doi: 10.1128/iai.67.10.5455-5462.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.World Health Organization. Human plague in 1997. Wkly Epidemiol Rec. 1999;74:340–344. [PubMed] [Google Scholar]
- 76.Yamamoto T, Taneike I. The sequences of enterohemorrhagic Escherichia coli and Yersinia pestis that are homologous to the enteroaggregative E. coli heat-stable enterotoxin gene: cross-species transfer in evolution. FEBS Lett. 2000;472:22–26. doi: 10.1016/s0014-5793(00)01414-9. [DOI] [PubMed] [Google Scholar]
- 77.Yaron S, Kolling G L, Simon L, Matthews K R. Vesicle-mediated transfer of virulence genes from Escherichia coli 0157:H7 to other enteric bacteria. Appl Environ Microbiol. 2000;66:4414–4420. doi: 10.1128/aem.66.10.4414-4420.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Youngren B, Radnedge L, Hu P, Garcia E, Austin S. A plasmid partition system of the P1–P7par family from the pMT1 virulence plasmid of Yersinia pestis. J Bacteriol. 2000;182:3924–3928. doi: 10.1128/jb.182.14.3924-3928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ziegler P. The black death. London, United Kingdom: Penguin; 1982. [Google Scholar]