Abstract
Inositol derivative compounds provide a nutrient source for soil bacteria that possess the ability to degrade such compounds. Rhizobium strains that are capable of utilizing certain inositol derivatives are better colonizers of their host plants. We have cloned and determined the nucleotide sequence of the myo-inositol dehydrogenase gene (idhA) of Sinorhizobium fredii USDA191, the first enzyme responsible for inositol catabolism. The deduced IdhA protein has a molecular mass of 34,648 Da and shows significant sequence similarity with protein sequences of Sinorhizobium meliloti IdhA and MocA; Bacillus subtilis IolG, YrbE, and YucG; and Streptomyces griseus StrI. S. fredii USDA191 idhA mutants revealed no detectable myo-inositol dehydrogenase activity and failed to grow on myo-inositol as a sole carbon source. Northern blot analysis and idhA-lacZ fusion expression studies indicate that idhA is inducible by myo-inositol. S. fredii USDA191 idhA mutant was drastically affected in its ability to reduce nitrogen and revealed deteriorating bacteroids inside the nodules. The number of bacteria recovered from such nodules was about threefold lower than the number of bacteria isolated from nodules initiated by S. fredii USDA191. In addition, the idhA mutant was also severely affected in its ability to compete with the wild-type strain in nodulating soybean. Under competitive conditions, nodules induced on soybean roots were predominantly occupied by the parent strain, even when the idhA mutant was applied at a 10-fold numerical advantage. Thus, we conclude that a functional idhA gene is required for efficient nitrogen fixation and for competitive nodulation of soybeans by S. fredii USDA191.
Sinorhizobium fredii and Bradyrhizobium spp. form nitrogen-fixing nodules on soybeans. On Midwestern soils, soybeans are predominantly nodulated by Bradyrhizobium japonicum serogroup 123 (34). However, serogroup 123 is a poor nitrogen fixer; hence, attempts have been made to overcome this problem. Commercial inoculants with better nitrogen-fixing ability than serogroup 123 are often utilized to enhance biological nitrogen fixation. However, serogroup 123 is extremely competitive on Midwestern soils and is able to exclude the introduced strains (17). Therefore, it becomes imperative that competitiveness of the commercial strains be improved to obtain beneficial effects from a commercial inoculum. We are interested in improving the nitrogen-fixing ability of S. fredii USDA191. This strain is a fast-growing Chinese isolate that is able to form nitrogen-fixing nodules on soybean and several other legumes (18, 21). S. fredii USDA191 is better suited for commercial inoculation production because it is a fast grower and produces fewer extracellular polysaccharides than the traditional symbiont B. japonicum. We are currently exploring strategies that will enhance USDA191 competitiveness relative to the indigenous rhizobia.
Symbiotic nitrogen fixation has been shown to be elevated in Rhizobium strains that have an increased acid tolerance, an uptake of hydrogenase, reduced levels of cytochrome o, increased expression of the nitrogen fixation gene (nifA) and C4-dicarboxylic acid transport genes (dctABD), and a flavonoid-independent hybrid NodD (8, 9, 10, 37, 39, 48). Ronson et al. (31) constructed rhizobial strains with additional copies of nifA and dctABD and demonstrated that such a genetically modified strain increased plant biomass under controlled environmental conditions. Bosworth and others (7) extended this study to field conditions. One of the recombinant Rhizobium meliloti strains (RMBPC-2) increased the alfalfa biomass by 13% when compared to the wild-type strain RMBPC on sites where soil nitrogen and organic matter content were lowest. However, on locations where soil nitrogen concentrations were high and native rhizobial populations were large, the recombinant rhizobium did not affect the yield of alfalfa (Medicago sativa [L.]) (7). In this study, the extra copies of nifA and dctABD genes were inserted in two symbiotically silent sites. The first site was referred to as the P3 region (4, 5) and the second as the ino region (46). The ino region is involved in inositol catabolism. Interestingly, when dctABD genes were inserted in the ino region, an increase in plant biomass was observed. However, insertion of the same genes in the P3 region resulted in a yield decrease (36). Based on this observation, it was suggested that the ino region may not be symbiotically silent and that inactivation of this region may actually benefit the host plants (36).
Plants secrete a wide array of compounds, some of which can be used as a carbon and/or nitrogen source by the rhizosphere bacteria. myo-Inositol is abundant in soil, and several microorganisms, including Rhizobium leguminosarum bv. viciae and Sinorhozobium meliloti, can grow using inositol as the sole carbon source (1, 14, 28). Rhizobium strains containing catabolism genes for the degradation of inositol may have a competitive advantage since inositol is abundant in the rhizosphere (47). Recently, Galbraith et al. (14) isolated a myo-inositol dehydrogenase gene (idhA) from S. meliloti and demonstrated that the activity of this gene product is essential for inositol catabolism as well as rhizopine utilization. A previous study established that bacterial strains that were capable of utilizing rhizopine had a fitness advantage and were able to nodulate their host plants more efficiently (15). These studies, along with the speculation that the inositol locus may have a role in symbiosis (36), prompted us to investigate the role of myo-inositol dehydrogenase in soybean-S. fredii USDA191 symbiosis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. Rhizobia were grown on a reciprocal shaker at 30°C in yeast extract mannitol (YEM) medium (44), and Escherichia coli was cultured in Luria-Bertani broth at 37°C (33). When appropriate, antibiotics were added at the following concentrations: kanamycin, 75 μg/ml; tetracycline, 10 μg/ml; spectinomycin, 50 μg/ml; gentamicin, 10 μg/ml; ampicillin, 100 μg/ml; and trimethoprim, 10 μg/ml (for counterselection against E. coli donor strains).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strain | ||
| S. fredii USDA191 | Nod+ on soybean | 18 |
| S. meliloti USDA1002 | Nod+ on alfalfa | USDA-ARSa |
| S. terangae USDA4894 | Nod+ on Acacia spp. | USDA-ARS |
| S. medicae USDA1037 | Nod+ on Medicago spp. | USDA-ARS |
| S. saheli USDA4893 | Nod+ on Sesbania spp. | USDA-ARS |
| B. japonicum USDA6 | Nod+ on soybean | USDA-ARS |
| B. liaoningense USDA3622 | Nod+ on Arachis hypogaea | USDA-ARS |
| B. elkanii USDA76 | Nod+ on soybean | USDA-ARS |
| Rizobium sp. NGR234 | Broad host range | W. J. Broughton |
| R. etli USDA9032 | Nod+ on Phaseolus vulgaris | USDA-ARS |
| R. galegae USDA4128 | Nod+ on Galega spp. | USDA-ARS |
| R. mongolense USDA1844 | Nod+ on Medicago ruthenica | USDA-ARS |
| R. tropici USDA9030 | Nod+ on P. vulgaris | USDA-ARS |
| R. leguminosarum USDA2370 | Nod+ on peas | USDA-ARS |
| M. loti USDA3471 | Nod+ on Lotus japonicus | USDA-ARS |
| M. amorphae USDA10001 | Nod+ on Amorpha fruticosa | USDA-ARS |
| M. ciceri USDA3383 | Nod+ on Astragalus cicer | USDA-ARS |
| M. huakuii USDA4479 | Nod+ on Astragalus sinicus | USDA-ARS |
| M. mediterraneum USDA3392 | Nod+ on Cicer arietinum | USDA-ARS |
| M. plurifarium USDA3707 | Nod+ on Acacia spp. | USDA-ARS |
| M. tianshanense USDA3592 | Arid saline desert soil isolate | USDA-ARS |
| HBK101 | idhA mutant of USDA191, Kanr | This work |
| HBK102 | USDA191 containing pMP220 | This work |
| HBK103 | HBK101 carrying the idhA promoter-lacZ fusion | This work |
| HBK104 | HBK101 carrying pLAFRI | This work |
| HBK105 | HBK101 carrying pHBK191-4 | This work |
| HBK106 | idhA mutant of USDA191, Spcr | This work |
| E. coli DH5α | φ80lacZΔM15Δ(lacZYA-argF) U169 hsdR17 recA1 endA1 thi-1 | Gibco BRL |
| Plasmid | ||
| pGEM-7zf(+) | Apr | Promega |
| pBluescript II SK(+) | Apr | Stratagene |
| pGEM-T Easy | Apr | Promega |
| pMP220 | Tcr | 38 |
| pJQ200uc1 | Gmr | 29 |
| pUC-4K | Kmr | 43 |
| pHBK191-1 | Apr, 990-bp idhA PCR product in pGEM-T Easy | This work |
| pHBK191-2 | Apr, 2.1-kb EcoRI idhA-containing fragment in pGEM-7zf(+) | This work |
| pHBK191-3 | Apr, 2.4-kb XhoI idhA-containing fragment in pGEM-7zf(+) | This work |
| pHBK191-4 | Tcr, pLAFRI cosmid containing idhA | This work |
| pHBK191-5 | Tcr, 900-bp idhA promoter sequences in pMP220 | This work |
| pHBK191-6 | Apr, pBluescript II SK(+) modified to remove SmaI, EcoRI, and EcoRV restriction sites | This work |
| pHBK191-7 | Apr, 2.4-kb XhoI idhA-containing fragment in pHBK191-6 | This work |
| pHBK191-8 | Apr and Kmr, 1.3-kb Km cassette into EcoRI site of pHBK191-7 | This work |
| pHBK191-9 | Gmr and Kmr, 3.7-kb XhoI fragment from pHBK191-7 cloned in pJQ200uc1 | This work |
| pHBK191-10 | Apr and Spcr, 2.0-kb Ω fragment cloned into SalI site of pHBK191-7 | This work |
| pHBK191-11 | Spcr, 4.2-kb EcoRI fragment from pHBK191-7 cloned into the EcoRI site of pHBK191-7 | This work |
| pHBK191-12 | Gmr and Spcr, 4.4-kb XhoI fragment from pHBK191-11 cloned into the SmaI site of pJQ200uc1 | This work |
USDA-ARS, U.S. Department of Agriculture, Agricultural Research Service.
Molecular techniques.
Recombinant DNA techniques were performed according to standard protocols (33). Rhizobial genomic DNA was isolated according to the method described previously by Jagadish and Szalay (16), and DNA probes were labeled with [32P]dCTP by using a Multiprime DNA Labeling System (Amersham Life Science, Cleveland, Ohio). The construction of the cosmid clone bank of S. fredii USDA191 used in this study was described previously (3). DNA sequencing was performed with a Taq Dye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, Calif.) at the University of Missouri.
Cloning and nucleotide sequence analysis of idhA.
Based on the published sequence of idhA from S. meliloti (GenBank accession no. AF059313) we designed two primers: 5′-CAGTGATGACAGTGAGATTTG-3′ and 5′-TGCTGATCTGGCGCTTTTCC-3′. These primers were used to amplify a 990-bp DNA fragment from genomic DNA of S. fredii USDA191 by PCR. The PCR product was cloned into pGEM-T Easy vector (Promega, Madison, Wis.) to produce pHBK191–1. We used this 990-bp PCR fragment as a hybridization probe to screen a cosmid library of S. fredii USDA191 constructed in the vector pLAFRI (12). Three positive colonies were identified that yielded positive hybridization signals. Cosmid DNA was isolated from these three clones and digested with the restriction endonuclease EcoRI and separated by agarose gel electrophoresis. The DNA was transferred to a nylon membrane and hybridized with 32P-labeled 990-bp PCR product. All three positive cosmid clones revealed strong hybridization to a 2.1-kb EcoRI fragment. The 2.1-kb EcoRI fragment was subsequently cloned into pGEM-7zf(+) to produce pHBK191–2. A 1.3-kb region from pHBK191–2 was sequenced with appropriate primers synthesized by the DNA Core Facility at the University of Missouri.
Construction of strains and plasmids.
A 2.4-kb XhoI fragment (Fig. 1) from one of the cosmid clones was subcloned into pGEM-7zf(+) and pHBK191–6 to produce pHBK191–3 and pHBK191–7. For the construction of the idhA mutant (HBK101), a 1.3-kb EcoRI kanamycin cassette from pUC-4K (40) was inserted into the EcoRI site of pHBK191–7 to produce pHBK191–8. A 3.7-kb XhoI fragment from pHBK191–8 was purified from the gel and cloned into the SmaI site of the suicide plasmid pJQ200uc1 (29) to yield pHBK191–9. A second idhA mutant (HBK106) was created by digesting pHBK192 DNA with SalI, which resulted in a loss of a 120-bp fragment in the coding region of idhA (Fig. 1A). A 2.0-kb Ω fragment was inserted into the above SalI site to yield pHBK191–10. A 4.2-kb EcoRI fragment from pHBK191–10 was cloned into the EcoRI site of pHBK191–7 to yield pHBK191–11. A 4.4-kb XhoI fragment was isolated from pHBK191–11 and cloned into the SmaI site of pJQ200uc1 to yield pHBK191–12. pHBK191–9 and pHBK191–1 were individually transferred into S. fredii USDA191 by triparental mating using the helper plasmid pRK2013 (11). Marker exchange was achieved by selection on YEM plates containing 5% (wt/vol) sucrose. Mutants were confirmed by Southern blotting and hybridization with the wild-type region.
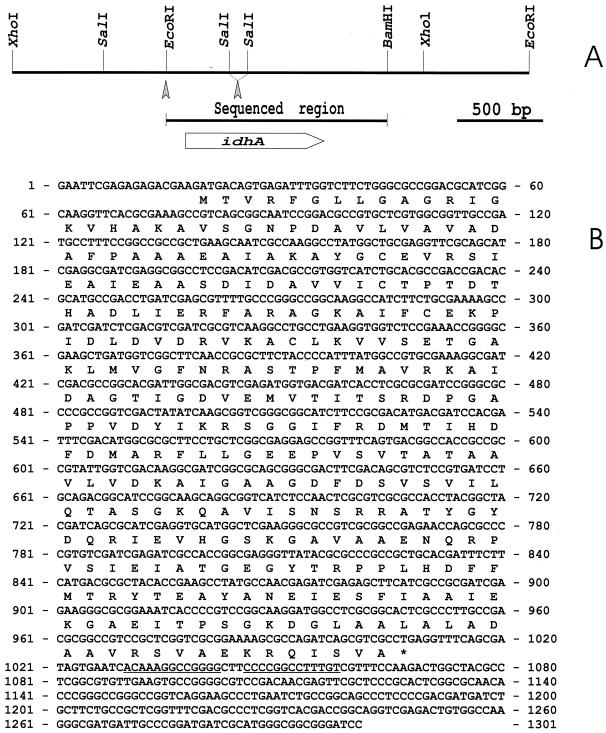
FIG. 1.
(A) Physical map of a 4-kb DNA segment of S. fredii USDA191 containing the idhA gene. An empty arrow indicates the location and the direction of the idhA gene. The arrowheads point to the sites into which a kanamycin or spectinomycin cassette was inserted to inactivate the idhA gene. (B) Nucleotide sequence of the idhA gene of S. fredii USDA191. The sequenced region covers 1,301 nucleotides. The ORF for idhA begins at position 20 and ends at position 1009. The predicted amino acid sequence is given below the nucleotide sequence. Transcriptional termination sequences represented by an inverted repeat are underlined. These sequences appear in the GenBank database as accession no. AF274867.
Northern analysis.
Five-milliliter starter cultures of S. fredii USDA191, HBK101, and HBK106 were grown at 30°C in YEM overnight. The cells were harvested by centrifugation and washed with minimal media. The cells were transferred to a fresh 20 ml of minimal media and grown for another 8 h either in the presence or absence of 0.2% myo-inositol. Total RNA was isolated by the hot-phenol method (45). Fifteen micrograms of RNA was resolved on a 1.5% agarose gel containing formaldehyde and transferred to a nylon membrane by capillary blotting. After transfer, the membrane was baked at 80°C for 2 h and hybridized with the S. fredii USDA191 idhA gene that had been labeled with [32P]dCTP. Prehybridization (4 h) and hybridization (18 h) were at 68°C in a solution containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 10× Denhardt's solution, 0.1% (wt/vol) sodium dodecyl sulfate (SDS), and 100 μg of denatured salmon sperm DNA/ml. After hybridization, membranes were washed twice for 20 min at 68°C in 2× SSC–0.5% SDS and were then washed for 30 min at 68°C in 0.5× SSC–0.5% SDS.
Enzyme assays.
Bacteria were grown in 250 ml of minimal medium with 0.2% succinate as the carbon source and 0.02% myo-inositol as the inducer. The specific myo-inositol dehydrogenase activity was measured according to the method described previously by Poole et al. (28) and was expressed as nanomoles of NADH reduced min−1/milligram of protein. The values represent the mean of three determinations.
Nodulation and nitrogenase assays.
Nodulation assays were performed as described previously by Balatti et al. (2). Acetylene reduction rates were determined by the method of Schwinghamer et al. (35).
Assessment of competition for nodulation.
Soybean cv. McCall seeds were surface sterilized and germinated on 1% water agar at 30°C for 3 days (19). Rhizobial strains HBK102 and HBK103 (Table 1) were pelleted from log-phase cultures, washed in YEM, and resuspended in YEM to 108 cells/ml. Bacterial growth in liquid cultures was estimated turbidimetrically relative to a standard curve that had been validated by bacterial counts with a Petroff-Hauser counter. For assessing the competition for soybean nodulation, strains HBK102 and HBK103 were mixed to provide ratios of 1:0, 1:1; 1:5, 1:10, 0:1, 5:1, and 10:1. Three-day-old soybean seedlings were dipped into bacterial suspensions, and the seedlings were transferred to autoclaved, plastic growth pouches that had been premoistened with nitrogen-free nutrient solution. The pouches were incubated in a growth chamber at 400 μmol of photons/m2/s with a 12-h photoperiod. Twenty-five days after inoculation, 100 nodules were randomly selected from a total of 30 plants per treatment and were histochemically stained for β-galactosidase activity.
β-Galactosidase assay.
The β-galactosidase assays were performed as described previously by Miller (24). In planta expression of idhA-lacZ was measured histochemically as described previously by Krishnan and Pueppke (20). Briefly, soybean nodules were sliced in two, and halves were fixed in glutaraldehyde and treated with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), essentially as described by Boivin et al. (6). Each nodule was viewed by bright-field microscopy for the presence of blue, an indication of β-galactosidase activity driven by the idhA promoter. Photographs representative of these nodules were recorded on Kodak Gold 200 film (Eastman Kodak Company, Rochester, N.Y.) with the help of an Olympus SZH microscope (Olympus Optical Co., Ltd., Tokyo, Japan).
Ultrastructural analysis.
Soybean nodules induced by S. fredii USDA191 and HBK101 were harvested at 15 and 25 days after inoculation. They were cut into small cubes with a razor blade and were immediately fixed in 2.5% glutaraldehyde buffered at pH 7.2 with 50 mM sodium phosphate for 4 h at room temperature. After the primary fixation, the samples were washed extensively with several changes of phosphate buffer. Tissue samples were then postfixed with 2% aqueous osmium tetroxide and embedded in Spurr's resin as described previously by Krishnan et al. (22). Thin sections were cut with a diamond knife on an ultramicrotome, mounted on uncoated 200-mesh nickel grids, and stained with uranyl acetate and lead citrate. Sections were examined with a JEOL JEM 100B (JEOL USA, Inc., Peabody, Mass.) electron microscope at 80 kV.
RESULTS
Cloning the S. fredii USDA191 idhA gene region.
To isolate the idhA gene of S. fredii USDA191, we synthesized primers corresponding to the 5′ and 3′ of the published S. meliloti idhA gene sequence (13). These primers were utilized to amplify a 990-bp fragment from S. fredii USDA191 genomic DNA. Using this PCR product as a hybridization probe, we screened a genomic cosmid library of S. fredii USDA191. Three positive cosmids were identified by colony hybridization. Southern blot analysis revealed that all three cosmids contained a 2.1-kb EcoRI hybridization DNA fragment. This fragment was subcloned in pGEM-7zf(+) to produce the plasmid pHBK191–2. Subsequently, a 2.4-kb XhoI fragment that overlaps the 2.1-kb EcoRI fragment was also cloned. The physical map of this DNA region is shown in Fig. 1A.
Nucleotide sequence of idhA
In order to characterize the idhA gene, the DNA sequence of a 1,301-bp EcoRI and BamHI fragment was determined (Fig. 1B). Analysis of the DNA sequence using the open reading frame (ORF) finder program identified a 990-bp-long ORF. Twenty base pairs downstream of the stop codon, a palindromic structure with the potential to form a stem-loop structure was identified (Fig. 1B). The predicted ORF encodes a protein of 329 amino acids with a molecular weight of 34,648. The theoretical isoelectric point of the protein was estimated to be 5.39. The amino acid sequences of S. fredii USDA191 that were submitted to a search of the SwissProt data bank revealed striking sequence similarity to the deduced protein sequences of S. meliloti idhA (79% identity, 82% similarity), B. subtilis yrbE (36% identity, 51% similarity), B. subtilis yisS (34% identity, 47% similarity), Streptomyces griseus strI (27% identity, 43% similarity), S. meliloti mocA (26% identity, 40% similarity), and B. subtilis iolG (22% identity, 40% similarity). The idhA, iolG, and strI genes encode myo-inositol dehydrogenase. The mocA gene product is involved in rhizopine catabolism. The yrbE and yisS genes encode hypothetical proteins of unknown function. Conserved among all these sequences is an N-terminal NADH-binding motif that is involved in the binding of the ADP moiety to NADH in dehydrogenases (41).
S. fredii idhA mutant is unable to utilize myo-inositol.
In order to verify if the cloned region contains the inositol catabolism region, we mutated the cloned region by inserting a kanamycin cassette into the EcoRI site or a spectinomycin cassette into the SalI site, respectively (Fig. 1A). These cassettes were recombined into the genome to create the idhA mutants. When inositol mutants were grown on minimal media containing myo-inositol as the sole carbon source, no noticeable growth was observed. However, when a cosmid containing the wild-type gene was introduced into the inositol mutants, it restored the ability of these mutants to utilize myo-inositol (data not shown). In order to demonstrate that the cloned region encodes an enzyme with myo-inositol dehydrogenase activity, we performed an enzyme assay utilizing cell extracts from S. fredii USDA191 grown in minimal media with and without 0.02% myo-inositol. S. fredii USDA191 grown in the absence of myo-inositol had a specific activity of myo-inositol dehydrogenase of 5 nmol of NADH reduced min−1/mg of protein. When the medium was supplemented with 0.02% myo-inositol, the enzyme activity was elevated to 70 nmol of NADH reduced min−1/mg of protein, indicating that the myo-inositol dehydrogenase activity is inducible. We were also able to detect myo-inositol dehydrogenase activity from soybean nodule extracts, although at considerably lower levels than those observed from Rhizobium grown in culture media. The inositol mutants, HBK101 and HBK106, did not display any detectable myo-inositol dehydrogenase activity either in the presence or absence of myo-inositol. HBK105, which harbors the cloned idhA gene, displayed specific myo-inositol dehydrogenase activity of 4 nmol of NADH reduced min−1/mg of protein and 209 nmol of NADH reduced min−1/mg of protein when grown in the absence and presence of myo-inositol, respectively.
S. fredii idhA is inducible by myo-inositol.
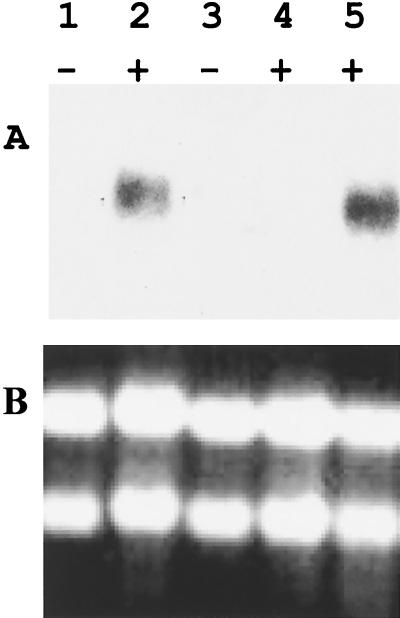
In order to determine if the idhA gene is inducible by myo-inositol, we cloned a 900-bp upstream sequence (XhoI-EcoRI fragment) (Fig. 1) into pMP220, a promoter probe vector (36). This construct was mobilized into S. fredii USDA191 by triparental mating to produce the strain HBK191–5. The promoter activity was measured by assaying β-galactosidase activity. HBK191–5 grown in minimal media exhibited β-galactosidase activity of 579 Miller units. The activity was elevated about fourfold (2,743 Miller units) when the cultures were grown in the presence of 0.2% myo-inositol, indicating that the idhA promoter is inducible by myo-inositol. The relatively high levels of β-galactosidase activity observed in HBK191–5 even in the absence of myo-inositol could be related to the copy number of the promoter probe vector. The induction of idhA by myo-inositol was further verified by Northern blot analysis. No RNA transcripts were detected using idhA-specific probe from USDA191 and HBK101 grown in the absence of myo-inositol. Supplementation of the culture medium with 0.2% myo-inositol clearly induced the transcription of USDA191 idhA (Fig. 2). The probe hybridized to a 1.0-kb RNA transcript (Fig. 2). RNA from the inositol mutant grown either in the presence or absence of myo-inositol revealed no hybridizing signal (Fig. 2).
FIG. 2.
(A) Effect of myo-inositol on the transcription of idhA of S. fredii USDA191. The bacteria initially grown in YEM media were harvested, washed, and grown in minimal slat medium in the presence or absence of 0.2% myo-inositol to an A660 of 1.0. Total RNA was isolated and used for Northern blot analysis. The RNA (15 μg) was resolved on a 1.5% formaldehyde gel and probed with a 1-kb EcoRI fragment containing the idhA gene. Lanes 1 and 2, USDA191; lanes 3 and 4, HBK101; lane 5, HBK104. The + and − signs indicate the presence and absence of myo-inositol in the culture media. (B) Ethidium bromide-stained gel picture showing uniform loading and integrity of RNA samples.
Nodulation and nitrogen fixation by idhA mutant of S. fredii
The symbiotic phenotypes of parental strain USDA191 and the idhA mutant (HBK101) were examined on McCall soybean. Both strains formed nodules on soybean tap and lateral roots. The nitrogen fixation levels were estimated by measuring acetylene reduction to assay nitrogenase activity. A comparison of nitrogen-fixing ability of these strains at 15 days after inoculation showed that the nitrogenase activity of the idhA mutant was several times lower than that of the wild type. This difference was particularly pronounced 25 days after inoculation, at which point the acetylene reduction rates of the USDA191 and the idhA mutant were 7.9 and 0.06 μmol/mg/h, respectively. Plants inoculated with HBK101 were stunted with yellow leaves. In order to rule out the possibility that the idhA mutant was delayed in its ability to fix nitrogen, we measured the nitrogenase activity in 35- and 50-day-old soybean nodules induced by the idhA mutant. The acetylene reduction was barely detectable (less than 0.02 μmol/mg/h) in 35- and 50-day-old nodules, suggesting that the idhA mutant was defective in its ability to fix nitrogen. Furthermore, we isolated bacteria from the nodules and found that about threefold fewer bacteria were present in the nodules initiated by the idhA mutant than in the wild-type nodules.
Soybean nodules initiated by S. fredii idhA mutant have aberrant ultrastructure.
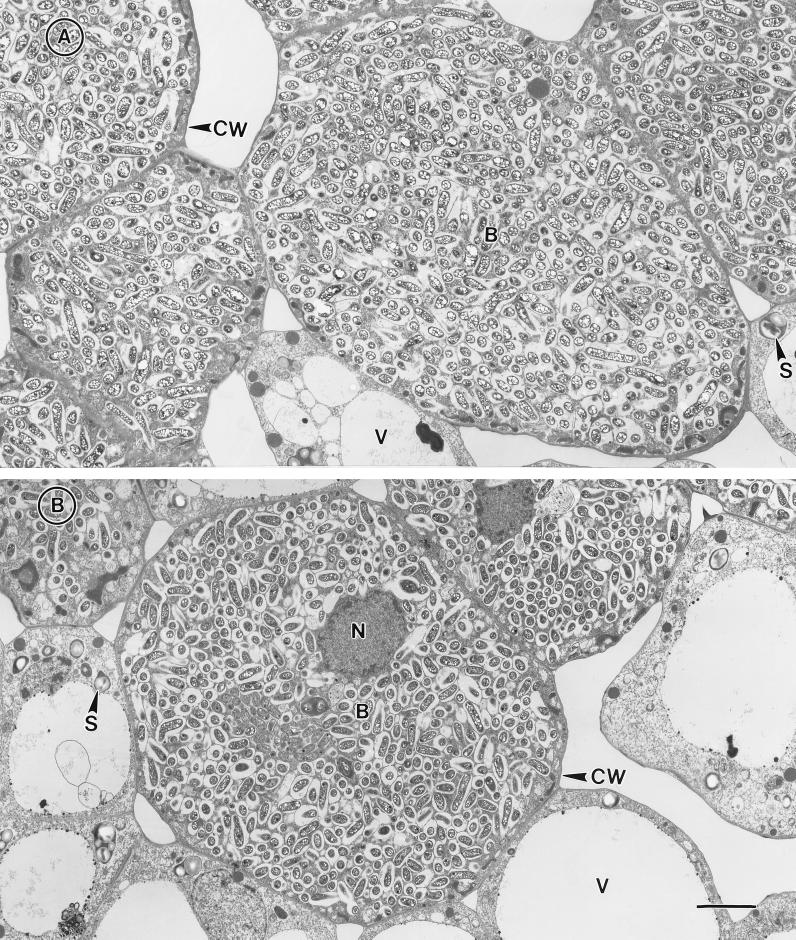
Since the inositol mutant had reduced nitrogen-fixing capacity when compared with the parent strain, we examined whether the mutant-induced nodules were altered in their ultrastructure. Figure 3 compares the ultrastructure of 15-day-old McCall nodules induced by the wild-type S. fredii USDA257 with the ultrastructure of those initiated by the inositol mutant. Nodules produced by the parental strain exhibited the typical internal structure of soybean nodules (Fig. 3A). Cells that contained numerous bacteria and a few noninfected cells predominantly occupied the central region of the nodules. The noninfected cells contained prominent vacuoles, and the cytoplasm of these cells contained several peroxisomes and starch granules. In contrast, the infected cells contained bacteroids that were enclosed by peribacteroid membranes. Most of the symbiosome contained more than one bacteroid (Fig. 3A). In contrast the nodules initiated by the inositol mutant revealed a symbiosome that contained only one bacteriod (compare Fig. 3A and B). A closer examination of the nodules at higher magnification revealed abnormal structural features, including the occurrence of whorls of loosely arranged membranes and bacteroids showing signs of senescence (data not shown).
FIG. 3.
Low-magnification transmission electron micrographs of 15-day-old soybean nodules initiated by S. fredii USDA191 (A) and the idhA mutant (B). The infected cells contain numerous bacteria that are surrounded by peribacteroid membranes. Note the presence of densely packed poly-β-hydroxybutyrate granules within the bacteroids. Bar, 5 μm; CW, cell wall; B, bacteroids; S, starch; V, vacuole; and N, nucleus.
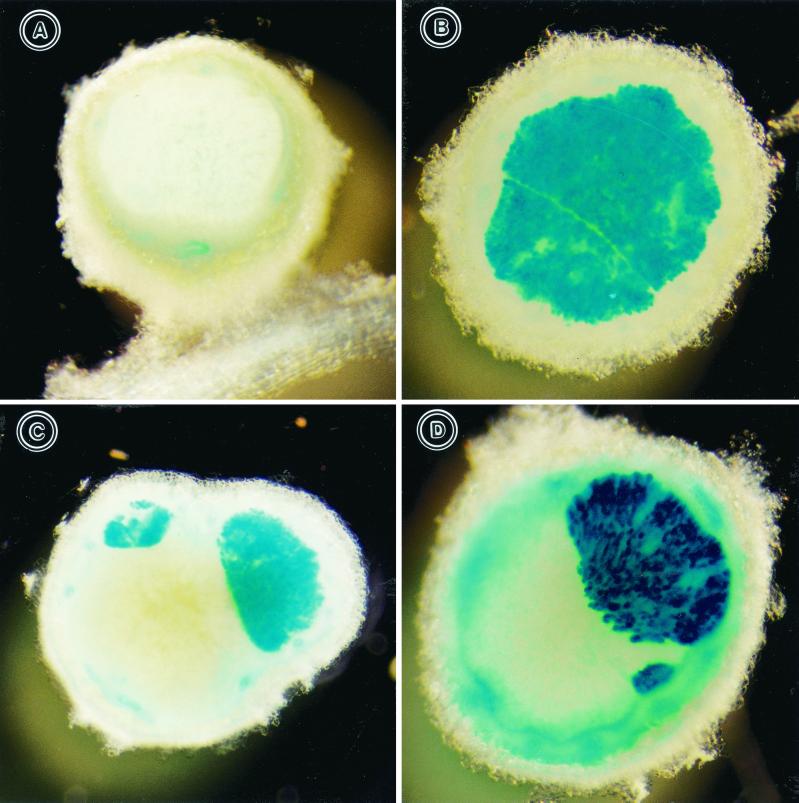
S. fredii idhA mutant is impaired in its ability to compete for soybean nodulation.
We also compared the competitiveness of the S. fredii USDA257 inositol mutant with that of its parent strain. Since the idhA-lacZ fusions were strongly expressed in culture, even in the absence of induction, we introduced the idhA-lacZ constructs into the USDA191 inositol mutant. Our preliminary experiments indicated that the idhA-lacZ fusion was expressed strongly in nodules when visualized by histochemical staining with X-Gal, a chromogenic substrate for the lacZ-encoded β-galactosidase. Nodules occupied by USDA191 containing the vector pMP220 (HBK102) were unstained, while the nodules occupied by USDA191 carrying the idhA promoter-lacZ fusion (HBK103) stained blue (Fig. 4A and B). Nodules occupied by both the competitors contained blue and white sectors (Fig. 4C and D). Table 2 gives the results of competition experiments in which strain HBK102 was paired with HBK103 at various ratios. Controls inoculated with HBK102 or HBK103 yielded nodules that were 100% unstained or 100% blue, respectively (Table 2). Under competitive conditions, USDA191 always dominated nodule occupancy, even when the competitor strain had been applied at a greater numerical advantage (Table 2). At a 1:1 ratio in the inoculum, the inositol mutant occupied only 11% of the nodules, while USDA191 was found in 59% of the nodules. Interestingly, both the competitors occupied 30% of the nodules. A fivefold increase in inoculum concentration of USDA191 in comparison with the idhA mutant almost completely prevented the latter strain from forming nodules (Table 2). In contrast, increasing the idhA mutant concentration by 5- to 10-fold over that of USDA191 only marginally improved its ability to compete for soybean nodulation. In spite of increased numbers, the idhA mutant was found only in 29% of the nodules, while the USDA191, in spite of a 10-fold-lower concentration, was found in 33% of the nodules (Table 2). These results clearly indicate that the inositol mutant is severely affected in competitiveness.
FIG. 4.
Histochemical staining of nodules produced by S. fredii HBK102 and HBK103. HBK102 carries the cloning vector pMP220, and HBK103 is an idhA mutant carrying the idhA promoter cloned in pMP220. Twenty-five-day-old nodules were sectioned into two halves and stained with X-Gal as described in Materials and Methods. Inocula are as follows: HBK102 (A), HBK103 (B), and a mixture of HBK102 and HBK103 (C and D).
TABLE 2.
Nodule occupancy in dually inoculated McCall soybeana
| USDA191/USDA191 idhA mutant ratio | No. of nodules examined | % Nodules occupied by:
|
||
|---|---|---|---|---|
| USDA191 | USDA191 idhA mutant | Both | ||
| 1:0 | 108 | 100 | 0 | 0 |
| 0:1 | 101 | 0 | 100 | 0 |
| 1:1 | 112 | 59 | 11 | 30 |
| 5:1 | 102 | 80 | 4 | 16 |
| 10:1 | 112 | 85 | 0 | 15 |
| 1:5 | 108 | 29 | 33 | 38 |
| 1:10 | 122 | 28 | 33 | 39 |
Nodules were harvested 25 days after inoculation and were stained histochemically with X-Gal. Results are from replicated experiments.
Sequences similar to S. fredii idhA are widely distributed in rhizobia.
Earlier studies have shown that the occurrence of rhizopine synthesis (mos) and catabolism (moc) genes is restricted in their distribution (32). Since the idhA gene of S. fredii USDA191 has significant sequence similarity to the S. meliloti mocA gene, we examined the occurrence of the idhA gene among 24 different Rhizobium species. Unlike the moc genes, idhA is widely distributed in different Rhizobium species from diverse geographical locations. Under stringent hybridization conditions, we were able to detect sequences similar to idhA in Rhizobium sp. strain NGR234, Rhizobium etli, Rhizobium galegae, Rhizobium mongolense, Rhizobium tropici, R. leguminosarum, Mesorhizobium amorphae, Mesorhizobium ciceri, Mesorhizobium huakuii, Mesorhizobium loti, Mesorhizobium mediterraneum, Mesorhizobium plurifarium, Mesorhizobium tianshanense, S. meliloti, Sinorhizobium terangae, Sinorhizobium medicae, and Sinorhizobium saheli. However, under similar hybridization conditions we were unable to detect any sequences similar to idhA in Rhizobium huautlaense, Azorhizobium caulinodans, B. japonicum, Bradyrhizobium liaoningense, and Bradyrhizobium elkanii. We also examined the occurrence of the idhA gene in 10 different S. fredii strains obtained from the U.S. Department of Agriculture collection. All S. fredii strains revealed strong hybridization to a 2.1-kb EcoRI DNA fragment.
DISCUSSION
myo-Inositol, which is abundantly present in soil, is widely used by soil bacteria as a carbon source. myo-Inositol dehydrogenase is the first enzyme responsible for the catabolism of myo-inositol in microorganisms such as Cryptococcus melibiosum (42), B. subtilis (30), R. leguminosarum bv. viciae (28), and S. meliloti (14). We have demonstrated that S. fredii USDA191, a soybean symbiont, also contains a myo-inositol dehydrogenase that is required for growth on inositol as the sole carbon source. The nucleotide sequence of the S. fredii USDA191 idhA gene, which consists of 329 amino acids and has a molecular weight of 34,648, is very similar to those for the S. meliloti idhA (14) and B. subtilis yrbE (23) and iolG (13) genes. The B. subtilis iolG gene has been identified as the myo-inositol 2-dehydrogenase gene (idh) and is a part of an operon consisting of 10 iol genes (26, 47). Mutation analysis has shown that the iol operon is probably transcribed as an 11.5-kb mRNA that encodes all the 10 iol genes (47). In case of S. fredii USDA191, the idhA does not appear to be a part of an operon. Our Northern blot analysis indicates that the idhA of S. fredii USDA191 is transcribed as a 1-kb mRNA. Our results are consistent with the findings from S. meliloti, where the entire inositol degradation gene was localized within a 2-kb BamHI fragment and no other adjacent DNA region was required for growth on myo-inositol as the sole carbon source (14).
The IdhA of S. fredii USDA191 also reveals amino acid similarity to MocA of S. meliloti (32). Both IdhA and MocA contain, in their N-terminal region, a NADH-binding motif, which has been suggested to bind to the ADP moiety of NADH in dehydrogenases (41). The MocA is required for rhizopine utilization (32). Rhizopine (l-3-O-methyl-scyllo-inosamine), which occurs in alfalfa nodules induced by specific S. meliloti strains, can serve as a nutrient source for bacterial growth. Soybean nodules also accumulate several inositol derivatives. myo-Inositol, d-chiro-inositol, 3-O-methyl-d-chiro-inositol, and 4-O-methyl-myo-inositol are the most abundant water-soluble forms of carbon in the soybean nodules (40). Even though these compounds have been proposed to function as osmotic protectants, their precise role in symbiosis still needs to be elucidated. Our ultrastructural studies of soybean nodules initiated by idhA mutants indicate that the bacteroids undergo structural alterations. In the absence of myo-inositol dehdrogenase activity, the bacteroids within soybean nodules will be unable to catabolize myo-inositol. The accumulation of myo-inositol may result in toxicity, leading to a reduction in the number of viable bacteroids within the nodules. The occurrence of nonfunctional bacteroids within soybean nodules will lead to lower nitrogen-fixing capacity, as in the case of the idhA mutant of S. fredii USDA191. The other possibility is that inositol may be essential for the growth and maturation of the bacterioids prior to nitrogen fixation. If the maturation is blocked, as in the case of the idhA mutant, then senescence may follow rapidly, resulting in the loss of nitrogen fixation. This possibility appears to be more likely since dicarboxylic acids rather than sugars are needed for nitrogen fixation.
Inactivating idhA in S. meliloti and S. fredii produced different symbiotic effects. myo-Inositol dehydrogenase mutants of S. meliloti were reported to have no observable symbiotic phenotype (14). These mutants were able to form nitrogen-fixing nodules on alfalfa that were indistinguishable from those induced by the parental strain (14). Based on this observation, it was concluded that the idhA gene of S. meliloti is not essential for the establishment of nitrogen-fixing symbiosis (14). Another study showed that inactivation of the inositol site by insertion of dctABD genes resulted in increased plant biomass (36). In our study, we found that the idhA mutant of S. fredii USDA191, even though it formed pink, nitrogen-fixing nodules on soybean roots, exhibited a considerably lower acetylene reduction rate than that of the parental strain. In addition, the idhA mutant also revealed developmental abnormalities in the nodule ultrastructure. Our results indicate that the idhA gene may have an important function in soybean symbiosis.
It has been suggested that bacterial strains that can synthesize and degrade rhizopine have a competitive advantage in the rhizosphere (15). Since rhizopine is an inositol derivative, it was suggested that genes involved in inositol catabolism might be involved in rhizopine degradation. It is now known that the idhA gene of S. meliloti is essential for rhizopine utilization (14). However, the competitive ability of S. meliloti idhA mutants has not been examined. Our study clearly shows the importance of an intact idhA gene for S. fredii USDA191's ability to successfully compete for soybean nodulation. Presumably idhA plays an important role in utilizing myo-inositol and inositol derivatives that are abundantly found in the rhizosphere. Soil bacteria containing an active idhA gene will have a competitive advantage in the rhizosphere. Attempts are currently being made to establish “biased rhizospheres” by creating transgenic plants expressing mos genes and beneficial soil bacteria expressing moc genes (32). A similar approach can also be taken to create “biased rhizospheres” by expressing myo-inositol synthesis genes in transgenic plants and expressing the idhA gene in desirable soil bacteria.
ACKNOWLEDGMENTS
We thank Larry Darrah and Jerry White for critical reading of this manuscript.
This work was supported in part by funds from Urbana Laboratories, St. Joseph, Mo.
REFERENCES
- 1.Bahar M, de Majnik J, Wexler M, Fry J, Poole P S, Murphy P J. A model for the catabolism of rhizopine in Rhizobium leguminosarum involves a ferredoxin oxygenase complex and the inositol degradative pathway. Mol Plant-Microbe Interact. 1998;11:1057–1068. doi: 10.1094/MPMI.1998.11.11.1057. [DOI] [PubMed] [Google Scholar]
- 2.Balatti P A, Krishnan H B, Pueppke S G. Calcium regulates growth of Rhizobium fredii and its ability to nodulate soybean cv. Peking. Can J Microbiol. 1991;37:542–548. [Google Scholar]
- 3.Bellato C, Krishnan H B, Cubo T, Temprano F, Pueppke S G. The soybean cultivar specificity gene nolX is present, expressed in a nodD-dependent manner, and of symbiotic significance in cultivar-nonspecific strains of Rhizobium (Sinorhizobium) fredii. Microbiology. 1997;143:1381–1388. doi: 10.1099/00221287-143-4-1381. [DOI] [PubMed] [Google Scholar]
- 4.Better M, Ditta G, Helinski D R. Deletion analysis of Rhizobium meliloti symbiotic promoters. EMBO J. 1985;4:2419–2424. doi: 10.1002/j.1460-2075.1985.tb03950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Better M, Lewis B, Corbin D, Ditta G, Helinski D R. Structural relationships among Rhizobium meliloti symbiotic promoters. Cell. 1983;35:479–485. doi: 10.1016/0092-8674(83)90181-2. [DOI] [PubMed] [Google Scholar]
- 6.Boivin C, Carnut S, Malpica C A, Truchet G, Rosenberg C. Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell. 1990;2:1157–1170. doi: 10.1105/tpc.2.12.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosworth A H, Williams M K, Albrecht K A, Hankinson T R, Kwiatkowski R, Beynon J, Ronson C W, Cannon F, Wacek T J, Triplett E W. Alfalfa yield response to inoculation with recombinant strains of Rhizobium meliloti carrying extra copy of dct and/or modified nifA expression. Appl Environ Microbiol. 1994;60:3815–3832. doi: 10.1128/aem.60.10.3815-3832.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannon F C, Beynon J, Hankinson T, Kwiatkowski R, Legocki R P, Ratcliffe H, Ronson C, Szeto W, Williams M. Increasing biological nitrogen fixation by genetic manipulation. In: Bothe H, de Bruijn F J, Newton W E, editors. Nitrogen fixation: a hundred years after. Stuttgart, Germany: Gustav Fisher; 1988. pp. 735–740. [Google Scholar]
- 9.Chen H, Richardson A E, Gartner E, Djordjevic M A, Roughley R J, Rolfe B J. Construction of an acid-tolerant Rhizobium leguminosarum bv. trifolii strain with enhanced capacity for nitrogen fixation. Appl Environ Microbiol. 1991;57:2005–2011. doi: 10.1128/aem.57.7.2005-2011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans H J, Harker A R, Papen H, Russell S A, Hanus F J, Zuber M. Physiology, biochemistry, and genetics of the uptake hydrogenase in rhizobia. Annu Rev Microbiol. 1987;41:335–361. doi: 10.1146/annurev.mi.41.100187.002003. [DOI] [PubMed] [Google Scholar]
- 11.Figurski D, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 13.Fujita Y, Shindo K, Miwa Y, Yoshida K. Bacillus subtilis inositol dehydrogenase-encoding gene (idh): sequence and expression in Escherichia coli. Gene. 1991;108:121–125. doi: 10.1016/0378-1119(91)90496-x. [DOI] [PubMed] [Google Scholar]
- 14.Galbraith M P, Feng S F, Borneman J, Triplett E W, de Bruijn F J, Rossbach S. A functional myo-inositol catabolism pathway is essential for rhizopine utilization by Sinorhizobium meliloti. Microbiology. 1998;144:2915–2924. doi: 10.1099/00221287-144-10-2915. [DOI] [PubMed] [Google Scholar]
- 15.Gordon D M, Ryder M H, Heinrich K H, Murphy P J. An experimental test of the rhizopine concept in Rhizobium meliloti. Appl Environ Microbiol. 1996;62:3991–3996. doi: 10.1128/aem.62.11.3991-3996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jagadish M N, Szalay A A. Directed transposon Tn5 mutagenesis and complementation in slow-growing, broad host range cowpea Rhizobium. Mol Gen Genet. 1984;196:290–300. [Google Scholar]
- 17.Kapusta G, Rouwenhorst D L. Influence of inoculum size on Rhizobium japonicum serogroup distribution frequency in soybean nodules. Agron J. 1973;65:916–919. [Google Scholar]
- 18.Keyser H H, Bohlool B B, Hu T S, Weber D F. Fast-growing rhizobia isolated from root nodules of soybean. Science. 1982;215:1631–1632. doi: 10.1126/science.215.4540.1631. [DOI] [PubMed] [Google Scholar]
- 19.Krishnan H B, Pueppke S G. Sequence and analysis of the nodABC region of Rhizobium fredii USDA257, a nitrogen-fixing symbiont of soybean and other legumes. Mol Plant-Microbe Interact. 1991;5:512–520. doi: 10.1094/mpmi-4-512. [DOI] [PubMed] [Google Scholar]
- 20.Krishnan H B, Pueppke S G. A nolC-lacZ fusion in Rhizobium fredii facilitates direct assessment of competition for nodulation of soybean. Can J Microbiol. 1992;38:515–519. [Google Scholar]
- 21.Krishnan H B, Pueppke S G. Host range, RFLP, and antigenic relationships between Rhizobium fredii strains and Rhizobium sp. NGR234. Plant Soil. 1994;161:21–29. [Google Scholar]
- 22.Krishnan H B, Franceschi V R, Okita T W. Immunocytochemical studies on the role of the Golgi complex in protein-body formation in rice seeds. Planta. 1986;169:471–480. doi: 10.1007/BF00392095. [DOI] [PubMed] [Google Scholar]
- 23.Kunst F, Ogasawara N, Moszer I, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 25.Murphy P J, Heycke N, Banfalvi Z, Tate M E, de Bruijn F J, Kondorosi A, Tempe J, Schell J. Genes for the catabolism and synthesis of an opine-like compound in Rhizobium meliloti are closely linked and on the Sym plasmid. Proc Natl Acad Sci USA. 1987;84:493–497. doi: 10.1073/pnas.84.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nihashi J, Fujita Y. Catabolite repression of inositol dehydrogenase and gluconate kinase syntheses in Bacillus subtilis. Biochim Biophys Acta. 1984;798:88–95. doi: 10.1016/0304-4165(84)90014-x. [DOI] [PubMed] [Google Scholar]
- 27.Perret X, Fellay R, Bjourson A J, Cooper J E, Broughton W J. Subtraction hybridization and shot-gun sequencing: a new approach to identify symbiotic loci. Nucleic Acids Res. 1994;22:1335–1341. doi: 10.1093/nar/22.8.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poole P S, Blyth A, Reid C J, Walters K. Myo-inositol catabolism and catabolite regulation in Rhizobium leguminosarum bv. viciae. Microbiology. 1994;140:2787–2795. [Google Scholar]
- 29.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 30.Ramaley R, Fujita Y, Freese E. Purification and properties of Bacillus subtilis inositol dehydrogenase. J Biol Chem. 1979;254:7684–7690. [PubMed] [Google Scholar]
- 31.Ronson C W, Bosworth A, Genova M, Gudbrandsen S, Hankinson T, Kwiatkowaski R, Ratcliffe H, Robie C, Sweeney P, Szeto W, Williams M, Zablotowicz R. Field release of genetically engineered Rhizobium meliloti and Bradyrhizobium japonicum strains. In: Gresshoff P M, Roth L E, Newton W E, editors. Nitrogen fixation: achievements and objectives. New York, N.Y: Chapman and Hall; 1990. pp. 397–403. [Google Scholar]
- 32.Rossbach S, Kulpa D A, Rossbach U, de Bruijn F J. Molecular and genetic characterization of the rhizopine catabolism (mocABCR) genes of Rhizobium meliloti L5–30. Mol Gen Genet. 1994;245:11–24. doi: 10.1007/BF00279746. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Schmidt E L, Zidwick M J, Abebe H M. Bradyrhizobium japonicum serocluster 123 and diversity among member isolates. Appl Environ Microbiol. 1986;51:1212–1215. doi: 10.1128/aem.51.6.1212-1215.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwinghamer E A, Evans H J, Dawson M D. Evaluation of effectiveness in mutant strains of Rhizobium by acetylene reduction relative to other criteria for N2 fixation. Plant Soil. 1970;33:192–212. [Google Scholar]
- 36.Scupham A J, Bosworth A H, Ellis W R, Wacek T J, Albrecht K A, Triplett E W. Inoculation with Sinorhizobium meliloti RMBPC-2 increases alfalfa yield compared with inoculation with a nonengineered wild-type strain. Appl Environ Microbiol. 1996;62:4260–4262. doi: 10.1128/aem.62.11.4260-4262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soberon M, Williams H D, Poole R K, Escamilla E. Isolation of a Rhizobium phaseoli cytochrome mutant with enhanced respiration and symbiotic nitrogen fixation. J Bacteriol. 1989;171:465–472. doi: 10.1128/jb.171.1.465-472.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spaink H P, Okker R J H, Wijffelman C A, Pees E, Lugtenberg B J J. Promoters in the nodulation region of the Rhizobium leguminosarum sym plasmid pRL1JI. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 39.Spaink H P, Okker R J H, Wijffelman C A, Tak T, Roo L G, Pees E, van Brussel A A N, Lugtenberg B J J. Symbiotic properties of rhizobia containing a flavonoid-independent hybrid nodD product. J Bacteriol. 1989;171:4045–4053. doi: 10.1128/jb.171.7.4045-4053.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Streeter J G. Carbohydrate, organic acid, and amino acid compostion of bacterioids and cytosol from soybean nodules. Plant Physiol. 1987;85:768–773. doi: 10.1104/pp.85.3.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson J, Donkersloot J A. N-(carboxyalkyl) amino acids: occurrence, synthesis, and functions. Annu Rev Biochem. 1992;61:517–557. doi: 10.1146/annurev.bi.61.070192.002505. [DOI] [PubMed] [Google Scholar]
- 42.Vidal-Leiria M, van Uden N. Inositol dehydrogenase from the yeast Cryptococcus melibiosum. Biochim Biophys Acta. 1973;293:295–303. doi: 10.1016/0005-2744(73)90337-9. [DOI] [PubMed] [Google Scholar]
- 43.Vieira J, Messing J. The pUC plasmids, and M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 44.Vincent J M. A manual for the practical study of root-nodule bacteria. Oxford, United Kingdom: Blackwell Scientific Publications; 1970. [Google Scholar]
- 45.Wang S, Stacey G. Studies of the Bradyrhizobium japonicum nodD1 promoter: a repeated structure for the nod box. J Bacteriol. 1991;173:3356–3365. doi: 10.1128/jb.173.11.3356-3365.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams M K, Cannon F, McLean P, Beyon J. Vector for integration of genes into a defined site in the Rhizobium meliloti genome. In: Palacios R, Verma D P S, editors. Molecular genetics of plant-microbe interactions. St. Paul, Minn: APS Press; 1988. pp. 198–199. [Google Scholar]
- 47.Yoshida K, Aoyama D, Ishio I, Shibayama T, Fujita Y. Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis. J Bacteriol. 1997;179:4591–4598. doi: 10.1128/jb.179.14.4591-4598.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yurgel S N, Soberon M, Sharypova L A, Miranda J, Morera C, Simarov B V. Isolation of Sinorhizobium meliloti Tn5 mutants with altered cytochrome terminal oxidase expression and improved symbiotic performance. FEMS Microbiol Lett. 1998;165:167–173. doi: 10.1111/j.1574-6968.1998.tb13142.x. [DOI] [PubMed] [Google Scholar]