ABSTRACT
The receptor of the subgroup A avian leukosis virus (ALV-A) in chicken is Tva, which is the homologous protein of human CD320 (huCD320), contains a low-density lipoprotein (LDL-A) module and is involved in the uptake of transcobalamin bound vitamin B12/cobalamin (Cbl). To map the functional determinants of Tva responsible for ALV-A receptor activity, a series of chimeric receptors were created by swapping the LDL-A module fragments between huCD320 and Tva. These chimeric receptors were then used for virus entry and binding assays to map the minimal ALV-A functional domain of Tva. The results showed that Tva residues 49 to 71 constituted the minimal functional domain that directly interacted with the ALV-A gp85 protein to mediate ALV-A entry. Single-residue substitution analysis revealed that L55 and W69, which were spatially adjacent on the surface of the Tva structure, were key residues that mediate ALV-A entry. Structural alignment results indicated that L55 and W69 substitutions did not affect the Tva protein structure but abolished the interaction force between Tva and gp85. Furthermore, substituting the corresponding residues of huCD320 with L55 and W69 of Tva converted huCD320 into a functional receptor of ALV-A. Importantly, soluble huCD320 harboring Tva L55 and W69 blocked ALV-A entry. Finally, we constructed a Tva gene-edited cell line with L55R and W69L substitutions that could fully resist ALV-A entry, while Cbl uptake was not affected. Collectively, our findings suggested that amino acids L55 and W69 of Tva were key for mediating virus entry.
IMPORTANCE Retroviruses bind to cellular receptors through their envelope proteins, which is a crucial step in infection. While most retroviruses require two receptors for entry, ALV-A requires only one. Various Tva alleles conferring resistance to ALV-A, including Tvar1 (C40W substitution), Tvar2 (frame-shifting four-nucleotide insertion), Tvar3, Tvar4, Tvar5, and Tvar6 (deletion in the first intron), are known. However, the detailed entry mechanism of ALV-A in chickens remains to be explored. We demonstrated that Tva residues L55 and W69 were key for ALV-A entry and were important for correct interaction with ALV-A gp85. Soluble Tva and huCD320 harboring the Tva residues L55 and W69 effectively blocked ALV-A infection. Additionally, we constructed gene-edited cell lines targeting these two amino acids, which completely restricted ALV-A entry without affecting Cbl uptake. These findings contribute to a better understanding of the infection mechanism of ALV-A and provided novel insights into the prevention and control of ALV-A.
KEYWORDS: receptors, subgroup A avian leukosis virus, virus entry
INTRODUCTION
Enveloped virus infection is mediated by glycoproteins that protrude from the viral membrane. Paramyxoviruses, retroviruses, filoviruses, and coronaviruses contain envelope glycoproteins that are considered class I enveloped (Env) glycoproteins (1). The entry of enveloped viruses into host cells is initiated by binding the viral membrane with the host cell membrane. Retroviral Env glycoproteins are trimers of surface (SU) glycoproteins and transmembrane (TM) glycoprotein heterodimers, where SUs contain domains important for interaction with receptors (2, 3), whereas TMs contain proteins that are responsible for membrane fusion (4). The initial interaction of retroviral glycoproteins with a specific cell surface receptor results in a conformational change in the trimeric glycoprotein structure, exposing the TM glycoprotein domains responsible for membrane fusion. (5, 6). However, different retroviruses have different entry mechanisms. For some retroviruses that require dual receptors, such as the human immunodeficiency virus (HIV), the initial binding to CD4 on target cells triggers a change in the glycoprotein structure, allowing interaction with a second receptor (CCR5 or CXCR4) to complete the entry process (7). In contrast, the avian leukosis virus (ALV) is a simple retrovirus that requires only one receptor and uses an entry mechanism different from that of HIV; it interacts with a single functional receptor during entry, resulting in a conformational change in the viral glycoprotein (8, 9). The exploration of the ALV binding mechanism is of great significance as it may provide a deeper understanding of the entry mechanisms of retroviruses that require only one receptor.
The ALV Env protein is a heterodimer composed of an SU protein (gp85) and a TM protein (gp37). ALVs are divided into 11 subgroups, ALV-A to ALV-K, based on differences in five variable domains of their SU glycoproteins (10–14). The receptors used by the different subgroups of ALVs are not completely the same (8, 9). ALV-A and ALV-K use Tva (15–17), ALV-B, ALV-D, and ALV-E use Tvb (18–21), ALV-C uses Tvc (22, 23), and ALV-J uses chNHE1 (24). Tva, the homologous protein of human CD320 (huCD320), mediates the uptake of transcobalamin (TC)-bound vitaminB12 (cobalamin, Cbl) (25). Tva contains a low-density lipoprotein (LDL-A) module that is present in several proteins, such as the very low-density lipoprotein receptor, the LDL-A module of which mediates the entry of multiple alphaviruses into host cells (26). The LDL-A module is a cysteine-rich domain that relies on six cysteine residues to form a stable structure. Interestingly, this LDL-A structure is sufficient to mediate ALV-A entry into host cells (27). The LDL-A of Tva has a high affinity for gp85 of ALV-A and mediates the specific attachment of viruses to the host cell surface (28–30). Additionally, Tva is responsible for a series of conformational changes of ALV-A Env that are crucial for viral and host cell membrane fusion triggered by gp85 and Tva binding (31–33). Therefore, analysis of the interaction mechanism between ALV-A gp85 and its receptor Tva may elucidate the basic entry principles of retroviruses.
In the current study, we identified the amino acid residues of Tva that were critical for ALV-A entry. This study was part of ongoing studies that utilize ALV as a model to elucidate the mechanisms of retrovirus entry into host cells and resistance to ALV-A infection. We used domain swapping and single-residue substitutions between Tva and huCD320 to identify the functional domain and residues critical for the interaction with ALV-A gp85 and entry of ALV-A. The chimeric receptor proteins were evaluated for their ability to bind ALV-A gp85 and support virus entry into transduced Tva-knockout cells. Our analyses revealed that L55 and W69 of Tva were critical functional residues responsible for binding to ALV-A gp85 and mediating ALV-A entry into host cells. We further determined that their substitution did not affect the Tva structure or TC-Cbl uptake mediated by Tva.
RESULTS
C terminus of LDL-A played a vital role in the Tva-mediated ALV-A entry process.
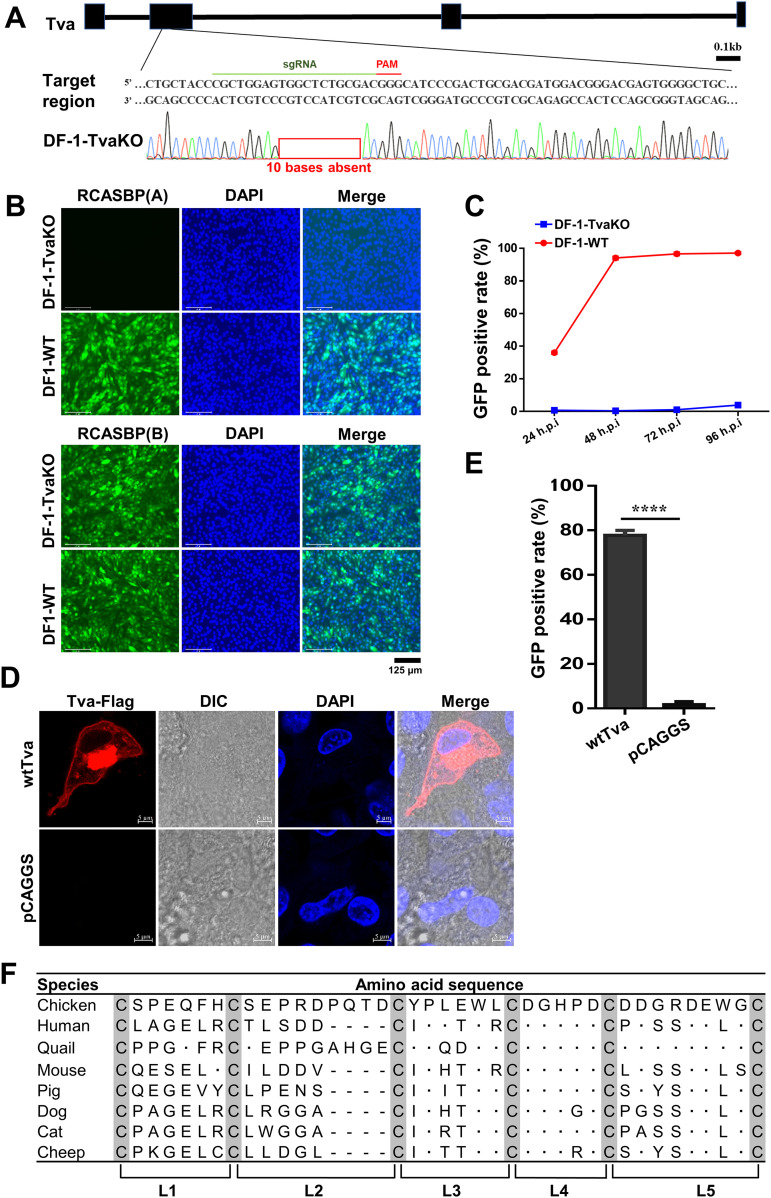
To map the key domain of Tva that mediates ALV-A entry into host cells, we expressed domain-swapped receptor proteins in Tva knocked out DF-1 (DF-1-TvaKO) cells and subjected the cells to virus entry assays. We first generated DF-1-TvaKO cells using CRISPR/Cas9 and then performed complement expression of different Tva substitutions in the DF-1-TvaKO cells by determining ALV-A entry levels using virus entry assays. Sequence analysis showed that the deletion of 10 bases in the Tva sequence resulted in a frameshift (Fig. 1A). Virus entry assay results showed that DF-1-TvaKO cells resisted the infection of RCASBP(A) (ALV-A enveloped replication-competent avian leukosis sarcoma virus harboring an eGFP reporter gene), without an effect on the infection of RCASBP(B) (Fig. 1B and C), indicating that the DF-1-TvaKO cells specifically resisted ALV-A entry. To establish the ALV-A entry assay based on DF-1-TvaKO cells, a membrane expression plasmid of wild-type Tva (wtTva) was constructed (Fig. 1D). DF-1-TvaKO cells transfected with the wtTva plasmid were infected with RCASBP(A). Flow cytometry results indicated that the entry level of RCASBP(A) in DF-1-TvaKO cells expressing wtTva was higher than 80% (Fig. 1E). These results demonstrated that the DF-1-TvaKO cell line could be employed for RCASBP(A) virus entry assays (referred to as virus entry assays) through Tva complement expression.
FIG 1.
Establishment of a virus entry assay using Tva-knockout DF-1 (DF-1-TvaKO) cells. (A) Schematic diagram of the generation of DF-1-TvaKO cells and sequence analysis of the DF-1-TvaKO cells. (B) DF-1-WT and DF-1-TvaKO cells were challenged with RCASBP(A) or RCASBP(B) at an MOI of 1 and analyzed by fluorescence microscopy at 120 hpi. Scale bar: 125 μm. (C) DF-1-WT and DF-1-TvaKO cells were infected with RCASBP(A) at an MOI of 1 and analyzed by flow cytometry at 24, 48, 72, and 96 hpi. (D) Confocal analysis of Tva expression on the DF-1-TvaKO cell membrane. Nuclei were counterstained with DAPI. Scale bar: 2 μm. (E) The proportion of GFP-positive cells in DF-1-TvaKO cells transfected with a wtTva expression plasmid or pCAGGS (negative control) and infected with RCASBP(A) as analyzed by flow cytometry at 72 hpi. (F) Amino acid sequence comparison of the LDL-A module of CD320 proteins of different species. Data from three independent experiments are shown as means ± standard deviations of triplicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significant difference.
Tva contained an LDL-A module that was highly homologous to the second LDL-A module of huCD320. The LDL-A module was a cysteine-rich domain that relied on six cysteines to form a stable structure. To allow sequence comparison of the LDL-A modules of different species, we split LDL-A into five fragments based on the six conserved cysteine residues (L1, L2, L3, L4, and L5) (Fig. 1F). The amino acids in L1 and L2 were not conserved between Tva and huCD320, whereas those in L3 to L5 were relatively conserved (14 out of 21 residues were conserved) (Fig. 1F).
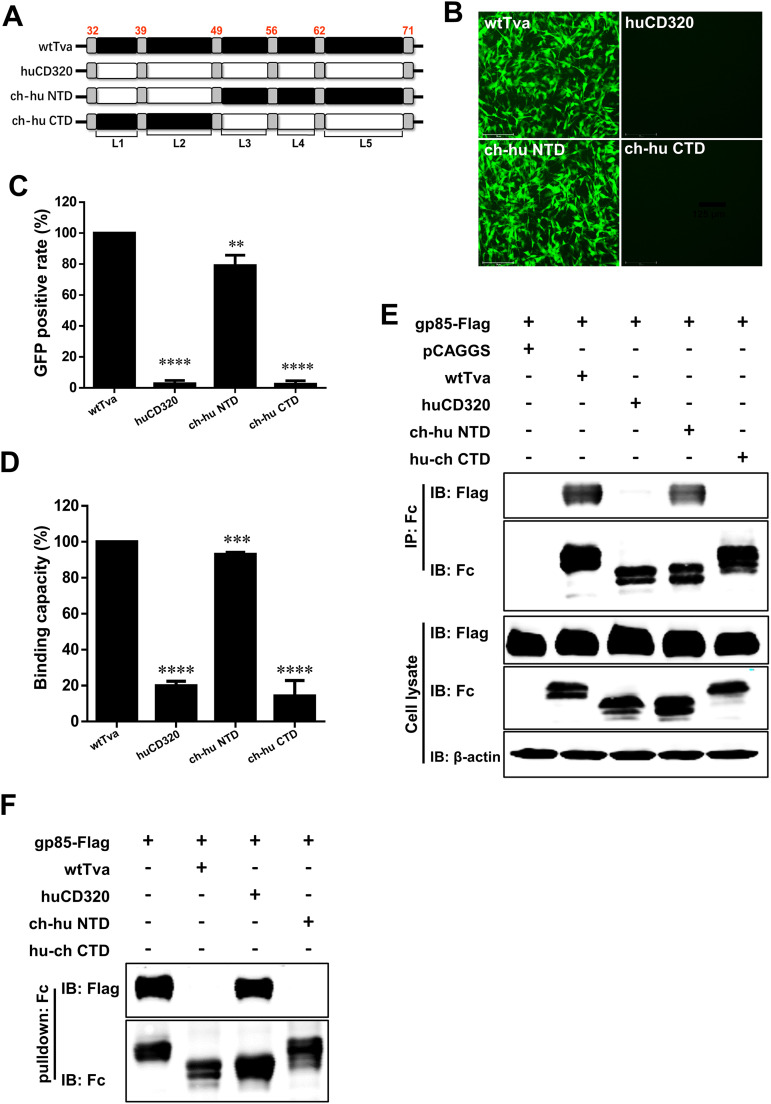
To determine the key domain of Tva interacting with ALV-A gp85, we adopted strategies for complementary domain exchange between the Tva and huCD320 LDL-A modules. We substituted the N-terminal domain (NTD) (containing L1 and L2) or the C-terminal domain (CTD) (containing L3, L4, and L5) of wtTva with the corresponding domains of huCD320 (Fig. 2A) and evaluated the chimeric receptors for their ability to mediate ALV-A entry. Virus entry assay results showed that the relative level of RCASBP(A) entry mediated by chimeric Tva containing the NTD of huCD320 was higher than 70%, whereas the level of virus entry mediated by chimeric Tva containing the CTD of huCD320 was only 10% (Fig. 2B and C). This result indicated that the C terminus of the LDL-A module was crucial for Tva to mediate ALV-A entry.
FIG 2.
The C terminus of the LDL-A module in Tva was involved in binding between the Tva receptor and ALV-A gp85. (A) Schematic diagram of the strategy used for constructing chimeric Tva with the N terminus or C terminus of the LDL-A module substituted with the corresponding fragments of huCD320. (B and C) Entry of RCASBP(A) virus into DF-1-TvaKO cells expressing different chimeric Tva receptors. (B) Virus entry levels as analyzed by fluorescence microscopy at 72 hpi. Scale bar: 125 μm. (C) Virus entry levels as analyzed by counting the proportion of GFP-positive cells using flow cytometry at 72 hpi. The entry level of RCASBP(A) virus into DF-1-TvaKO cells expressing wtTva was set to 100% and the values for the chimeric Tva receptors were calculated as its proportions. (D) gp85-binding abilities of different chimeric Tva receptors expressed in transfected DF-1-TvaKO cells as evaluated by receptor binding assays. The binding capacity of wtTva was set to 100% and the values for the chimeric Tva receptors were calculated as its proportions. (E) Physical interactions between the chimeric Tva receptors (with an Fc tag) and gp85 (with a Flag tag) as determined by Co-IP assays in 293T cells. (F) Validation of the interaction of different chimeric Tva receptors (with an Fc tag) with gp85 (with a Flag tag) as assessed by pulldown assays. Data from three independent experiments are shown as means ± standard deviations of triplicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significant difference.
To further confirm the above results, a gp85 protein-cell binding assay was performed using soluble forms of ALV-A gp85 (referred to as gp85) tagged with human IgG-Fc and produced in 293T cells (34). The relative gp85-binding capacity of chimeric Tva containing the NTD of the huCD320 LDL-A module (ch-hu NTD) was similar to that of wtTva (>80%), whereas the binding capacity of chimeric Tva containing the CTD of the huCD320 LDL-A module (ch-hu CTD) was only 20% (Fig. 2D). This result indicated that the C terminus of the LDL-A module of Tva directly binds to gp85.
To assess whether the C terminus of the LDL-A module can interact with gp85, the ch-hu NTD and ch-hu CTD plasmids were engineered to replace their transmembrane domains with Fc tags for soluble expression. Co-immunoprecipitation (Co-IP) assays were performed by transiently coexpressing gp85 and ch-hu NTD or ch-hu CTD, respectively, in 293T cells. The results showed that ch-hu NTD formed a complex with gp85, whereas ch-hu CTD did not (Fig. 2E). A pulldown assay was performed to confirm the Co-IP results. As shown in Fig. 2F, ch-hu NTD pulled down gp85, whereas ch-hu CTD did not. This further indicated that the C terminus of the LDL-A module of Tva played a vital role in interacting with ALV-A gp85.
Taken together, these results demonstrated that the C terminus of the LDL-A module of Tva played a crucial role in mediating ALV-A entry and binding with ALV-A gp85.
L55 and W69 of Tva were required for mediating ALV-A entry.
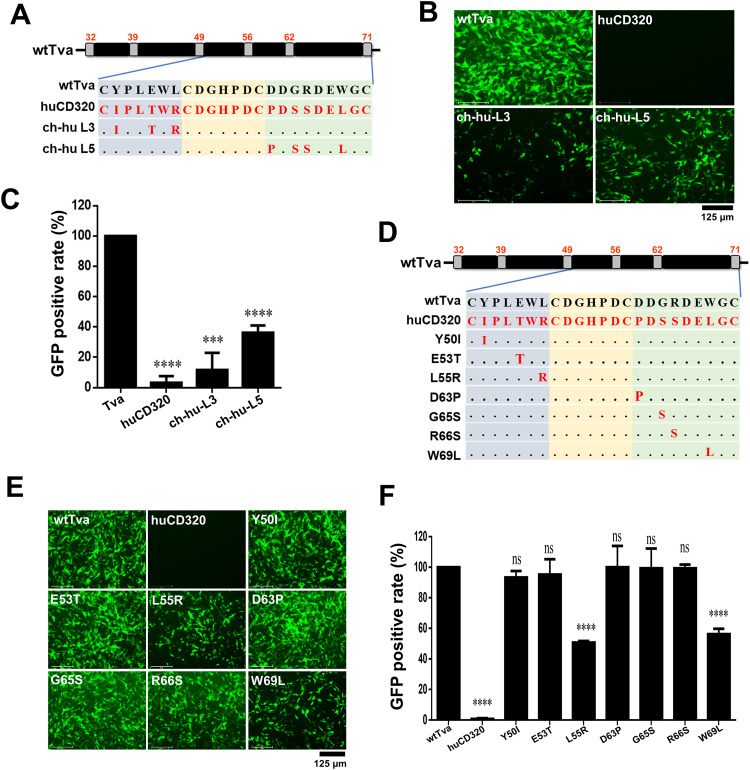
To extensively identify the CTD fragments of the LDL-A module of Tva responsible for the receptor function, we first comparatively analyzed the sequences of the C termini of the LDL-A modules of Tva and huCD320 (Fig. 3A). The L4 sequence was identical between huCD320 and Tva, whereas the L3 and L5 sequences showed some differences. Next, we constructed chimeric plasmids based on the wtTva backbone harboring the L3 (ch-hu-L3) or L5 (ch-hu-L5) domains of huCD320 (Fig. 3A). Virus entry assay results showed that the abilities of ch-hu-L3 and ch-hu-L5 to mediate viral infection were decreased to 12% and 36%, respectively, compared with that of wtTva (Fig. 3B and C). Combined with the results shown in Fig. 1, which suggested that the C terminus of the LDL-A module was crucial for RCASBP(A) entry, these findings indicated that both L3 and L5 in the C terminus of the LDL-A module participated in Tva-mediated ALV-A entry.
FIG 3.
L55 and W69 of Tva were the key residues for virus entry. (A) Schematic diagram of the strategy used for constructing chimeric Tva receptors with the L3 or L5 fragment of the LDL-A module substituted with the corresponding fragment of huCD320. (B and C) Entry of RCASBP(A) virus into DF-1-TvaKO cells expressing the chimeric Tva receptors. (B) Virus entry levels as analyzed by fluorescence microscopy at 72 hpi. Scale bar: 125 μm. (C) Virus entry levels as analyzed by counting the proportion of GFP-positive cells using flow cytometry at 72 hpi. (D) Schematic diagram of the strategy used for constructing chimeric Tva receptors with seven different single-residue substitutions. (E and F) Entry of RCASBP(A) virus into DF-1-TvaKO cells expressing the chimeric Tva receptors. (E) Virus entry levels as analyzed by fluorescence microscopy at 72 hpi. Scale bar: 125 μm. (F) Virus entry levels as analyzed by counting the proportion of GFP-positive cells using flow cytometry at 72 hpi. Data from three independent experiments are shown as means ± standard deviations of triplicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significant difference.
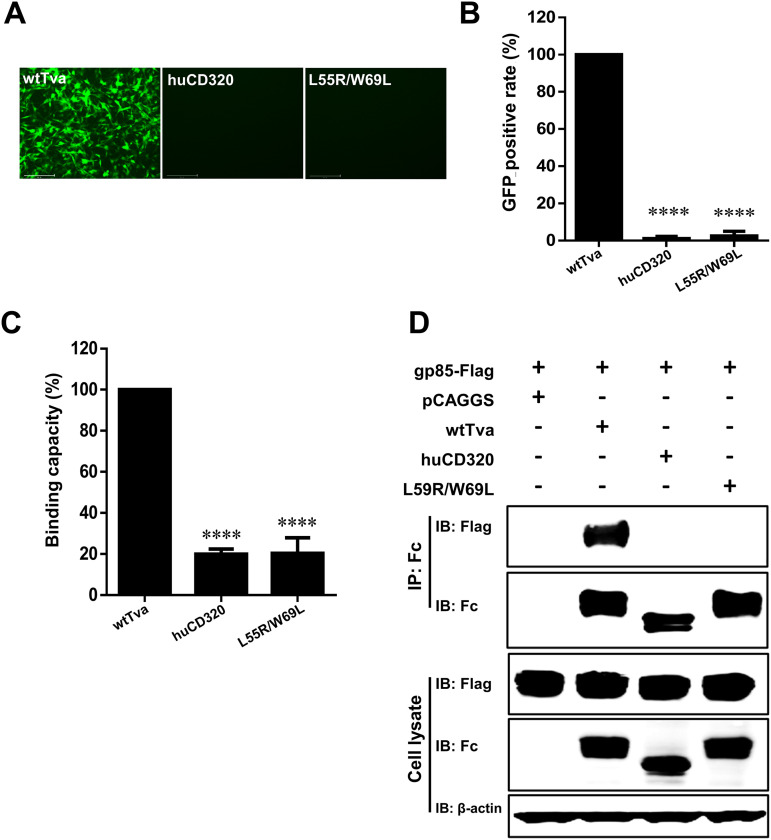
There were seven residues in L3 and L5 that differed between Tva and huCD320: Y50, E53, L55, D63, G65, R66, and W69. To pinpoint the key amino acids responsible for Tva-mediated ALV-A entry, we constructed seven chimeric plasmids in which a single residue in the wtTva backbone was substituted with the corresponding residue of huCD320 (Fig. 3D). Virus entry assay results revealed that L55R and W69L substitutions decreased the entry level of RCASBP(A) to 51% and 56%, respectively, compared with that of wtTva (Fig. 3E and F). Next, we evaluated the effect of simultaneous substitution of L55 and W69 in the wtTva backbone with the corresponding huCD320 residues (L55R/W69L). Virus entry assay results showed that simultaneous L55 and W69 substitutions completely abolished ALV-A entry (Fig. 4A and B), indicating that these two residues played a crucial role in mediating ALV-A entry.
FIG 4.
Tva with L55/W69 substitution lost its ability to mediate ALV-A infection. (A and B) Entry of RCASBP(A) virus into DF-1-TvaKO cells expressing Tva with L55/W69 substitution. wtTva was used as a positive control, and huCD320 was used as a negative control. (A) Virus entry levels as analyzed by fluorescence microscopy at 72 hpi. Scale bar: 125 μm. (B) Virus entry levels as analyzed by counting the proportion of GFP-positive cells using a flow cytometric at 72 hpi. (C) gp85-Binding abilities of the chimeric Tva receptors expressed on DF-1-TvaKO cells as evaluated by receptor binding assays. (D) Physical interactions between the chimeric Tva receptors (with an Fc tag) and gp85 (with a Flag tag) as determined by Co-IP assays in 293T cells. Data from three independent experiments are shown as means ± standard deviations of triplicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significant difference.
A protein-cell binding assay showed that the relative gp85-binding capacity of Tva harboring the L55R/W69L substitutions was decreased to 20% compared with that of wtTva (Fig. 4C). Furthermore, Co-IP assay results showed that L55R/W69L Tva did not interact with ALV-A gp85 (Fig. 4D). These observations indicated that L55 and W69 of Tva were the key residues involved in binding and interaction of the receptor with ALV-A gp85.
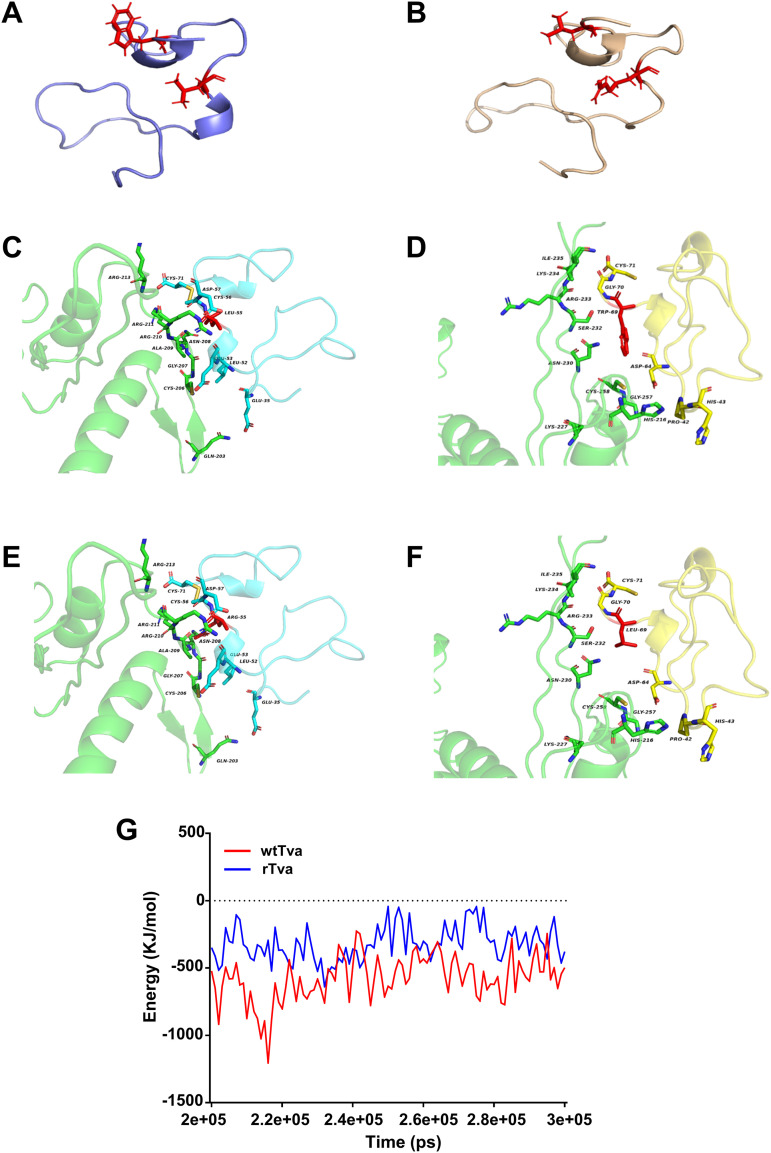
To analyze the molecular details of Tva binding to gp85 through structural analysis, we predicted and analyzed the three-dimensional structures of wtTva and L55R/W69L Tva (rTva). The wtTva structural model showed that L55 and W69 were exposed on the surface of wtTva and spatially adjacent (Fig. 5A). Structural analysis further indicated that the substitution of L55 and W69 did not destroy the Tva structure (Fig. 5A and B). Docking analysis results of Tva and gp85 showed that Tva interacted with multiple amino acids of gp85 via the C terminus of the LDL-A module and that L55 (Fig. 5C) and W69 (Fig. 5D) were in different docking surfaces formed between Tva and gp85. Although the interaction interface did not change considerably after substitution (Fig. 5E and F), the binding free energy between Tva and gp85 increased (Fig. 5G), indicating that the interaction force between Tva and gp85 was weakened after substitution.
FIG 5.
L55R/W69L substitution abrogated the interaction force between Tva and gp85. (A and B) Three-dimensional models of wtTva (shown in purple) and rTva (shown in brown) as predicted. Residues 55 and 69 are highlighted in red. (C) Docking results of wtTva and gp85 showed that Tva residue 55 was located on the docking surface. (D) Docking results of wtTva and gp85 showed that Tva residue 69 was located on the docking surface. (E) Docking results of Tva with residue 55 substituted and gp85 showed that Tva residue 55 was located on the docking surface. (F) Docking results of Tva with residue 69 substituted and gp85 showed that Tva residue 69 was located on the docking surface. (G) The binding energy between gp85 and wtTva or rTva.
Taken together, these results demonstrated that L55 and W69 of Tva interacted directly with gp85 to mediate ALV-A entry into host cells.
HuCD320 harboring L55 and W69 of Tva acted as a functional receptor and mediated ALV-A entry.
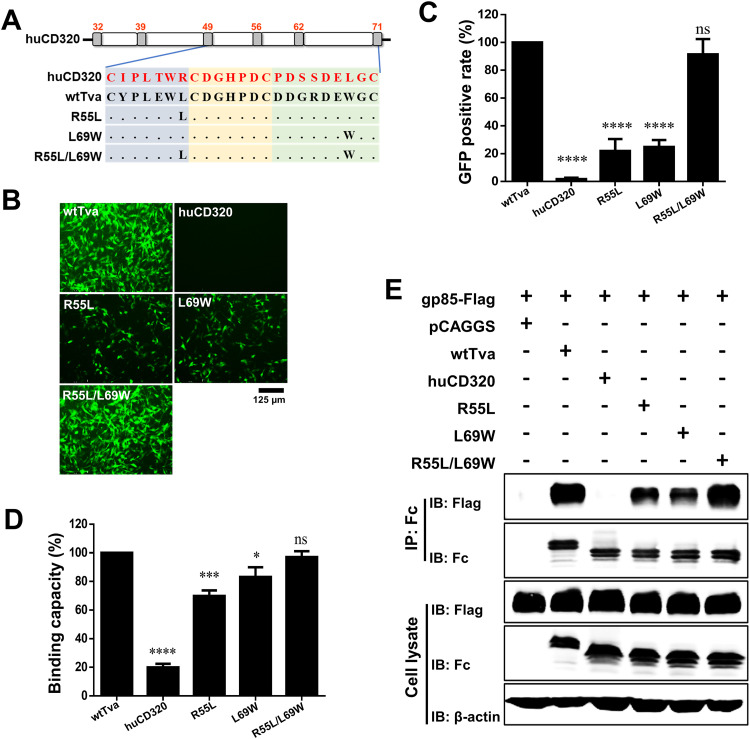
To further explore the roles of L55 and W69 of Tva in receptor function, three chimeric huCD320 proteins containing L55 (R55L) or W69 (L69W) single-residue substitution or simultaneous L55 and W69 (R55L/L69W) substitutions with the corresponding amino acids of Tva were constructed (Fig. 6A). Virus entry assay results showed that the RCASBP(A) entry level in DF-1-TvaKO cells with R55L or L69W substitution was only 22% and 25%, respectively (Fig. 6B and C). In contrast, the RCASBP(A) entry level in DF-1-TvaKO cells with R55L/L69W was higher than 90%. (Fig. 6B and C). These results demonstrated that the simultaneous substitution of L55 and W69 enabled the naturally nonfunctional ALV-A receptor to effectively mediate ALV-A entry.
FIG 6.
Substituting R55 and L69 of huCD320 with L55 and W69 of Tva converted huCD320 into a functional ALV-A receptor that mediated virus entry. (A) Schematic diagram of the strategy used for constructing chimeric huCD320 proteins substituted with L55, W69, or L55 and W69 of Tva. (B and C) Entry of RCASBP(A) virus into DF-1-TvaKO cells expressing the chimeric huCD320 proteins. (B) Virus entry levels as analyzed by fluorescence microscopy at 72 hpi. Scale bar: 125 μm. (C) Virus entry levels as analyzed by counting the proportion of GFP-positive cells using flow cytometry at 72 hpi. (D) gp85-Binding abilities of the chimeric Tva proteins expressed on DF-1-TvaKO cells as evaluated by receptor binding assays. (E) Physical interactions between the chimeric huCD320 proteins (with an Fc tag) and gp85 (with a Flag tag) as determined by Co-IP assays in 293T cells. Data from three independent experiments are shown as means ± standard deviations of triplicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significant difference.
Protein-cell binding and Co-IP assays revealed that R55L/L69W substitution enabled huCD320 to effectively bind and interact with gp85 (Fig. 6D and E), whereas single-residue substitution did not, with relative binding recovery rates of only 70% (R55L) and 80% (L69W) (Fig. 6D and E).
Taken together, these results demonstrated that substituting R55 and L69 of huCD320 with L55 and W69 of Tva converted huCD320 into a functional receptor that mediates ALV-A entry.
Soluble huCD320 harboring L55 and W69 of Tva effectively blocked ALV-A entry.
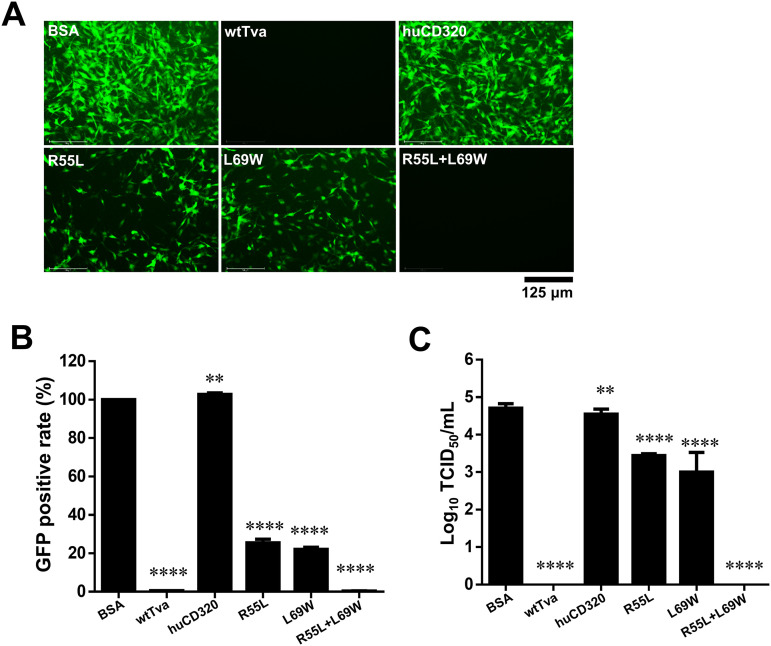
To verify the roles of residues L55 and W69 of Tva in the soluble form of the receptor and to evaluate their potential application in blocking virus entry, three soluble chimeric huCD320 proteins with R55L, L69W, or R55L/L69W substitution were constructed and successfully expressed in 293T cells for virus blocking assays. RCASBP(A) was first incubated with the different soluble chimeric huCD320 proteins and then used to infect DF-1 cells. Flow cytometry results indicated that huCD320 proteins with L55/W69 substitution nearly completely blocked RCASBP(A) entry (Fig. 7A and B). In contrast, chimeric huCD320 proteins with L55 or W69 substitution exhibited a weak blockade effect, decreasing the RCASBP(A) infection rate to 60% and 70%, respectively (Fig. 7A and B). Next, the ALV-A strain RAV-1 was incubated with the different soluble chimeric huCD320 proteins and used to infect DF-1 cells. Cell culture supernatants were collected at 7 days postinfection (dpi) for viral titer detection using a median 50% tissue culture infectious dose (TCID50) assay. The TCID50 assay results showed that the released viral titers were decreased 18.2- and 50-fold after incubation with chimeric huCD320 protein with L55 or W69 substitution, respectively (Fig. 7C). No viral titer was detected after incubation with chimeric huCD320 protein with L55/W69 substitution.
FIG 7.
Soluble huCD320 harboring L55 and W69 of Tva effectively blocked viral entry. (A and B) Entry of RCASBP(A) virus into DF-1 cells incubated with different soluble chimeric huCD320 or Tva proteins. Bovine serum albumin (BSA) was used as a negative control. (A) Virus entry levels as analyzed by fluorescence microscopy at 7 days postinfection (dpi). Scale bar: 125 μm. (B) Virus entry levels as analyzed by counting the proportion of GFP-positive cells using flow cytometry at 7 dpi. The entry level of RCASBP(A) virus incubated with BSA was set to 100% and the values for the soluble chimeric huCD320 and Tva proteins were calculated as their proportions. (C) Entry of RAV-1 virus into DF-1 cells incubated with the soluble chimeric huCD320 or Tva proteins as assessed by measuring virus titers in cell supernatants collected 7 dpi. BSA was used as a negative control. Data from three independent experiments are shown as means ± standard deviations of triplicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significant difference.
These findings demonstrated that soluble huCD320 with simultaneous substitution with L55 and W69 of Tva effectively blocked ALV-A infection.
Precise gene editing of both L55 and W69 of chicken Tva conferred resistance to ALV-A entry.
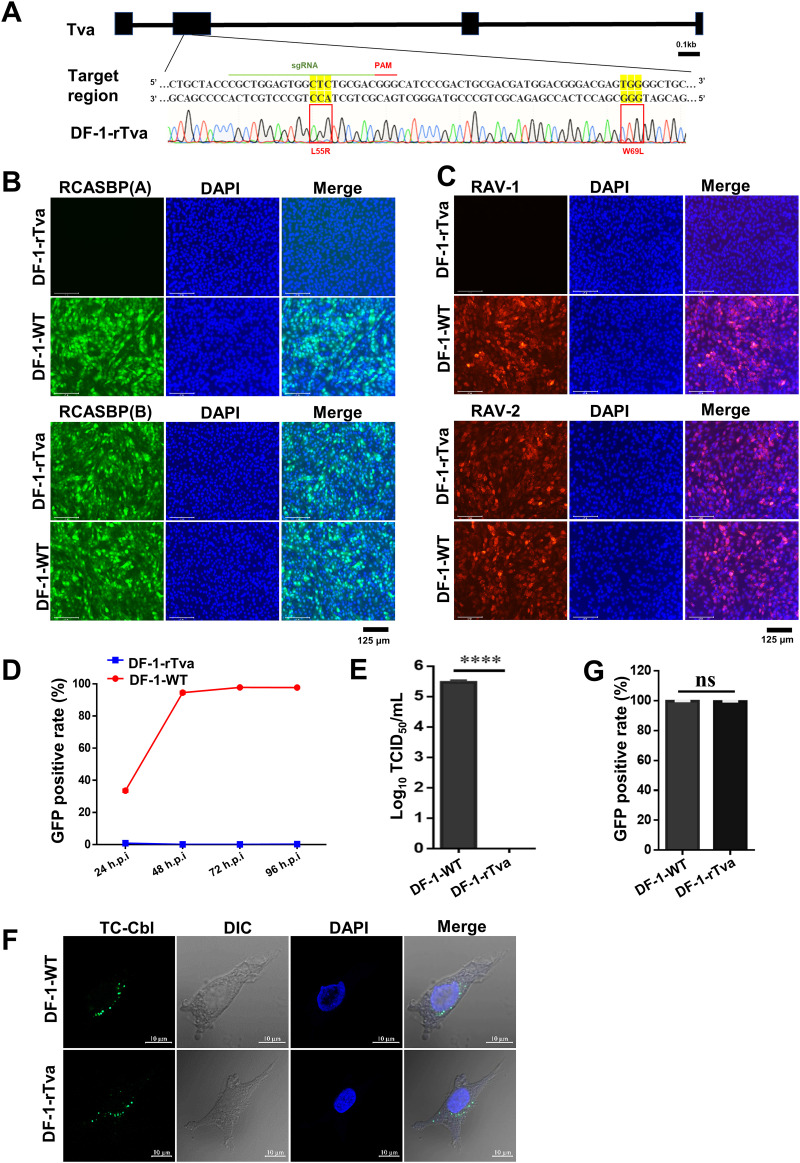
To validate the receptor function of L55 and W69 of Tva on ALV-A entry in vivo, we carried out precise L55 and W69 amino-acid editing of endogenous Tva in DF-1 cells to obtain L55R and W69L substitutions (35, 36). The editing efficiency was approximately 20%. Sequence analysis revealed that we obtained a Tva-modified DF-1 cell line (DF-1-rTva) with both L55R and W69L substitutions (Fig. 8A). The cell growth rate of DF-1-rTva cells as evaluated using a CCK8 kit was not remarkably different from that of wild-type DF-1 (DF-1-WT) cells, indicating that Tva modification did not affect cell growth.
FIG 8.
Precise gene editing of both L55 and W69 of chicken Tva conferred resistance to ALV-A. (A) Schematic diagram of the construction of Tva gene-edited DF-1 cells and sequence analysis of the DF-1-rTva cells. (B) DF-1-WT and DF-1-rTva cells were challenged with RCASBP(A) or RCASBP(B) at an MOI of 1 and analyzed by fluorescence microscopy at 120 hpi. Scale bar: 125 μm. (C) DF-1-WT and DF-1-rTva cells were challenged with RAV-1 or RAV-2 (wild-type ALV-B strains) at an MOI of 1 and analyzed by fluorescence microscopy using an anti-ALV-A monoclonal antibody at 120 hpi. Scale bar: 125 μm. (D) DF-1-WT and DF-1-rTva cells were infected with RCASBP(A) at an MOI of 1 and analyzed by flow cytometry at 24, 48, 72, and 96 hpi. (E) Entry of RAV-1 virus into DF-1-WT or DF-1-rTva cells as analyzed by measuring virus titers in cell supernatants collected 7 dpi. (F) Confocal microscopy analysis of TC-Cbl uptake in DF-1-WT and DF-1-rTva cells. Nuclei were counterstained with DAPI. Scale bar: 2 μm. (G) TC-Cbl uptake in DF-1-WT and DF-1-rTva cells as analyzed by flow cytometry. Data from three independent experiments are shown as means ± standard deviations of triplicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significant difference.
Virus entry assay results showed that DF-1-rTva fully resisted the entry of RCASBP(A) and RAV-1 but did not affect the infection of RCASBP(B) and RAV-2 (ALV-B strain) (Fig. 8B and C). Flow cytometry results showed that DF-1-rTva cells resisted RCASBP(A) entry, whereas the entry level in DF-1-WT cells gradually increased over time (Fig. 8D). Next, DF-1-rTva and DF-1-WT cells were infected with the RAV-1 strain. TCID50 assay results showed that the viral titer in DF-1-WT cells was 105.47 TCID50/mL, whereas no viral titer was detected in DF-1-rTva cells (Fig. 8E). Thus, endogenous Tva with L55/W69 substitution can fully resist ALV-A entry into cells, indicating that L55 and W69 were the key amino acid sites for Tva-mediated ALV-A entry.
Tva mediates the uptake of Cbl in chickens. Cbl binds to the circulating transporter TC to form a TC-Cbl complex that binds to the membrane receptor Tva to enter cells (25). To investigate the effect of L55R/W69L substitution on Cbl uptake, a Cbl uptake experiment was performed using DF-1-rTva cells. Confocal microscopy revealed that the amount of Cbl signal in DF-1-rTva cells was similar to that in DF-1-WT cells (Fig. 8F). Flow cytometry results showed that the TC-Cbl-positive cell rate was highly similar between DF-1-rTva cells and DF-1-WT cells (Fig. 8G). These results suggested that simultaneous L55R and W69L substitution did not affect Cbl uptake by Tva.
Taken together, these results demonstrated that Tva with simultaneous L55R and W69L substitution lost its ability to mediate ALV-A infection but not its Cbl uptake ability.
DISCUSSION
The first and most crucial step in the infection process of enveloped viruses is the binding of Env glycoproteins to receptor proteins on the host cell. Because of differences in their gp85 proteins, different subgroups of ALVs depend on different host receptor proteins to enter cells. Therefore, the identification of receptor amino acids that are key for virus entry is of great significance in exploring the entry mechanisms of different subgroups of ALVs. In this study, by swapping domains between Tva and huCD320, we discovered that the key Tva domain mediating ALV-A entry into host cells is in the C terminus of the LDL-A module and comprises residues 49 to 71. More specifically, L55 and W69 are the key amino acids of Tva in mediating ALV-A entry. These two residues of L55 and W69 of Tva can convert huCD320, the nonfunctional receptor of ALV-A, into a functional receptor to mediate the entry of ALV-A. More importantly, precise editing of Tva to obtain L55R and W69L substitutions in DF-1 cells resulted in blockade of ALV-A infection, without affecting TC-Cbl uptake.
Tva contained an LDL-A module that was essential for maintaining its structure. The C terminus of LDL-A is relatively conservative among different species, whereas its N terminus is not. Our results showed that the C terminus of the LDL-A module is critical for Tva interaction with gp85 and ALV-A entry. Moreover, structural prediction showed that Tva forms an interaction interface with gp85 through the C terminus of its LDL-A module, which includes fragments L3, L4, and L5, as shown in Fig. 5. L4 is conserved in most LDL-A modules, whereas L3 and L5 are not well conserved. Virus entry assay results demonstrated that L3 and L5 are important for Tva-mediated ALV-A entry. Thus, although L4 was involved in the interaction interface between Tva and gp85, L3 and L5 played a more critical role in this interaction.
Next, we mapped the key amino acids in L3 and L5 that mediate virus entry. L55 is a key amino acid in L3 involved in ALV-A entry. The function of L55 in mediating ALV-A entry in quail is controversial (37). One study suggested that L55 is present on the surface of the LDL-A structure and is directly involved in the interaction between Tva and gp85 (38). Another study suggested that L55 interacts with a few spatially adjacent amino acids of Tva to form a hydrophobic domain and is buried into the core of LDL-A to maintain the correct conformation of Tva (39). Our results showed that L55 was in the interaction interface between Tva and gp85 and could interact with gp85 and partially mediate ALV-A entry. These results, which are in line with the first hypothesis above (38), further demonstrated that L55 may not play a role in maintaining LDL-A structure but mediated ALV-A entry through direct interaction with gp85. L55 is an uncharged amino acid, whereas arginine (R) is a positively charged amino acid. Therefore, we speculate that when L55 is substituted with R55, the positive charge of this residue may result in repulsive interactions with the positively charged R210, R211, and R213 residues on the interaction surface of gp85, which weakens the interaction between Tva and gp85 and reduces the former’s ability to mediate ALV-A entry.
We found that W69 in L5 was another important amino acid in mediating ALV-A entry. W69 is an aromatic amino acid with a relatively large side chain. Interestingly, aromatic amino acids have been identified as key amino acids in receptors for other subgroups of ALVs. For example, Y42 of Tvb is crucial for the binding of ALV-B gp85 (40), W48 of Tvc is involved in the binding of ALV-C gp85 (23), and W38 of chNHE1 is key for ALV-J entry (34). These studies demonstrated that aromatic amino acids play important roles in other retroviral receptors that mediate virus entry. Studies have shown that the HIV receptor CD4 contains an aromatic amino acid, F43, located in a protein loop formed by two cysteines involved in virus entry (41). Residue 375 of gp120 is in the cavity formed by F43 of CD4. This cavity allows CD4 to make numerous contacts with conserved gp120 residues, which are critical for gp120 binding to CD4 (42). Thus, aromatic amino acids in receptors play important roles in viral Env protein binding. However, the role of W69 in the interaction between Tva and gp85 remains unclear. In this study, the structural analysis showed that W69 is in the loop formed by C56 and C71 and is exposed on the surface of the LDL-A structure and located in the Tva-gp85 interaction interface. The binding and Co-IP results demonstrated that W69 can directly interact with gp85. Tryptophan (W) has a relatively large side chain, whereas leucine (L) is a smaller amino acid. Substitution of the larger W69 with the smaller L69 leads to a reduction in the contact area between Tva and gp85, which may be accounted for the weakened interaction between Tva and gp85 and the reduction in Tva-mediated ALV-A entry.
The binding interface between proteins relies on multiple intermolecular contacts, involving 10 to 30 side chains per protein (43, 44). This is consistent with reports stating that multiple receptor residues are involved in the efficient binding of receptors to viruses. For example, L36, Q37, L41, and Y42 of Tvb are key amino acids for ALV-B binding (45), W48 and Y105 of Tvc are key amino acids for ALV-C binding (23), and A30, V33, W38, and E39 of chNHE1 are key amino acids for ALV-J binding (34). The receptors for these viruses have multiple amino acids that are involved in the binding to the viral SU. In addition, protein activation is usually involved in different activation pathways with multiple metastable states (46). Our results showed that L55 or W69 substitution alone did not completely inhibit virus entry, whereas simultaneous L55/W69 substitution did, suggesting two key metastable states between Tva and gp85 in the entry pathways. Therefore, when L55 is substituted with R55, Tva can still bind to gp85 through W69-dependent interaction and vice versa.
The avian receptor Tva is a homolog of mammalian CD320 and is one of the cellular receptor molecules involved in TC-mediated Cbl uptake (25). Previous studies have shown that Tva also acts as a cellular receptor molecule for TC-mediated Cbl uptake in chickens. ALV-A infection can partially inhibit Cbl uptake (25), possibly because of partial interaction of Tva with TC-Cbl, the binding sites of which only partially overlap those of ALV. In this study, although ALV-A entry was inhibited after substitution of L55 and W69 of Tva, TC-Cbl could still enter host cells. Our results demonstrated that ALV-A and TC-Cbl entry into host cells relied on different amino acids of Tva.
Although Tva-deficient gene-edited chickens have constructed and could completely resist ALV-A infection, they exhibit specific Cbl metabolic disturbances due to a lack of Tva (47). In this study, we successfully constructed DF-1-rTva cells harboring L55R and W69L substitutions. DF-1-rTva cells resisted ALV-A entry, whereas their TC-Cbl uptake ability was not affected. These results corroborated that L55R and W69L of Tva were crucial for gp85 binding and ALV-A entry and suggested that ALV-A and TC-Cbl bound with different amino acids of Tva to enter host cells in vivo. These findings contributed to a better understanding of the molecular mechanisms of the interactions of Tva with ALV-A and Cbl and provided gene-editing target sites for the development of chickens resistant to ALV-A infection.
MATERIALS AND METHODS
Cells and viruses.
The 293T cells and DF-1 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) and 100 μg/mL of penicillin and streptomycin in a humidified incubator at 37°C, containing 5% CO2. The ALV-A prototype strains RAV-1 and ALV-B prototype strains RAV-2 were kindly provided by Venugopal Nair (Pirbright Institute, Pirbright, UK) and were propagated in DF-1 cells. The fluorescently tagged ALV strain RCASBP(A) (ALV-A enveloped replication-competent avian leukosis sarcoma virus vector harboring an eGFP reporter gene) was kindly provided by Stephen H. Hughes (National Cancer Institute, Frederick, MD) and was propagated in DF-1 cells.
Expression of soluble gp85 (sgp85) protein.
The gp85 gene (GenBank accession number AYN55358.1) was PCR-amplified from proviral DNA of RAV-1 and fused with an N-terminal signal peptide sequence and a C-terminal Fc tag of human IgG or a Flag tag inserted into the pCAGGS vector to generate pCAGGS-sgp85-Fc and pCAGGS-sgp85-Flag plasmids. Sgp85 protein was prepared by transfecting the pCAGGS-sgp85-Fc plasmid into 293T cells using X-Treme gene-HP-DNA transfection reagent (06366236001; Roche). At 48 h postinfection (hpi), the cell culture medium was collected and purified using protein A affinity matrix beads (L00210; GenScript) according to the manufacturer’s instructions. The concentration of sgp85 was determined using a bicinchoninic acid protein assay kit (23227; Thermo Scientific), following the manufacturer’s instructions.
Construction of various chimeric receptors.
Tva was cloned from chicken embryo fibroblasts by reverse transcription-PCR (RT-PCR) using primers based on the published chicken Tva sequence (accession number: NC_052600). huCD320 was cloned from 293T cells by RT-PCR using primers based on the published huCD320 sequence (accession number NG_028124). Constructs harboring Tva and huCD320 domain exchange substitutions or single/multiple-residue substitutions were generated by overlap PCR and inserted into the pCAGGS vector for transmembrane expression. All these chimeric constructs contained the signal peptide sequence, transmembrane domain, and cytoplasmic domain with a Flag tag. Likewise, constructs for soluble expression of Tva and huCD320 domain exchange mutants or point mutations were generated by overlap PCR and inserted into the pCAGGS vector. All these chimeric constructs contained the signal peptide sequence at the N terminus, the Fc region of human IgG at the N terminus, and a cytoplasmic domain with a HA tag. All constructs were verified by DNA sequencing. The primer sequences of all oligonucleotides used in this study are available upon request.
Generation of DF-1-TvaKO cells.
DF-1-TvaKO cells were generated using the CRISPR/Cas9 technology. Guide (g)RNA target sites (GATCGTGCGGCTCCGAACAG) were designed using E-CRISPR (http://www.e-crisp.org/E-CRISP/designcrispr.html) (48, 49). DNA fragments containing the U6 promoter, target gRNA, and the gRNA scaffold were fused by overlap PCR and then inserted into the pMD-18T vector (6011; TaKaRa Bio). DF-1 cells were transfected with a plasmid carrying gRNA and pMJ920 (number 42234; Addgene) using the TransIT-X2 delivery system (MIR 6000; Mirus) according to the manufacturer’s instructions. Cells showing green fluorescence were sorted into 96-well plates using flow cytometry, and monoclonal cells were identified by sequence analysis and tested for resistance to RCASBP(A), RCASBP(B), or RAV-1 infection.
Receptor binding assay.
A receptor binding assay was performed to evaluate the binding ability between Tva and gp85 (34, 50). DF-1-TvaKO cells were transfected with a plasmid encoding wtTva or different chimeric receptors. At 24 h posttransfection, the cells were resuspended in phosphate-buffered saline (PBS) and pelleted (1,000 × g, 10 min). Then, the cells were washed three times with ice-cold PBS containing 5% (wt/vol) FBS and incubated with 500 μL of sgp85 protein (200 ng/μL) on ice for 1 h. After three washes with ice-cold PBS containing 5% (wt/vol) FBS, anti-human IgG (Fc specific)-FITC antibody (diluted 1:200, F9512; Sigma) was added to the cells for another 1 h. After three washes with ice-cold PBS containing 5% (wt/vol) FBS, the cells were fixed using 4% paraformaldehyde (PFA) (P1110; Solarbio) at room temperature for 15 min. After washing, the cells were incubated in a 1:200 dilution of anti-Flag antibody produced in mice (F1804; Sigma) on ice for 1 h. After washing, the cells were incubated in a 1:200 dilution of anti-mouse IgG-TRITC antibody (T2402; Sigma) on ice for 1 h. After three washes, the stained cells were analyzed by fluorescence-activated cell sorting (FACS) using a FACS ARIA II flow cytometer (Cytomics FC 500; BD Biosciences).
Virus entry assay.
DF-1-TvaKO cells were transfected with plasmids encoding wtTva or different chimeric receptors as described above. At 24 h posttransfection, the transfected cells were infected with RCASBP(A) virus at a multiplicity of infection (MOI) of 1 at 37°C in the presence of 5% CO2 for 2 h. Then, the cells were washed three times with PBS at room temperature and maintained in DMEM containing 2% (wt/vol) FBS at 37°C in the presence of 5% CO2. Virus entry levels were determined at 72 h postinfection by measuring the percentage of virus-positive cells showing green fluorescence by flow cytometry. Briefly, the cells were washed with PBS at room temperature, digested with trypsin, and pelleted (1,000 × g) for 10 min. The cells were fixed in 4% PFA at room temperature for 15 min. After washing, the cells were first incubated with an anti-Flag antibody and then stained with an anti-mouse IgG-TRITC antibody. The stained cells were analyzed by FACS as described above.
Co-IP assay.
293T cells cultured in a 6-well plate were transfected with the corresponding plasmid using PolyJet transfection reagent (SL100688, SignaGen) according to the manufacturer’s instructions. After 48 h, the cells were lysed in lysis buffer (P0013F; Beyotime). The lysates were collected by centrifugation. The supernatants were incubated with 20 μL of protein A affinity matrix beads at 4°C overnight. The samples were then washed with ice-cold PBS five times. The immunoprecipitated proteins were boiled at 100°C for 10 min, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and detected by Western blotting. In brief, after being separated on 12.5% SDS-PAGE, the proteins were transferred onto a nitrocellulose membrane (HATF00010; Merck-Millipore). The membranes were incubated with HA monoclonal antibody (H9658; Sigma) at room temperature for 1 h. After three washes with PBST (PBS containing 0.05% Tween 20), the membranes were incubated with IRDye 800CW anti-mouse IgG antibody (926-32212; LI-COR Biosciences) or IRDye 800CW anti-Human IgG antibodies (925-32232, LI-COR Biosciences) at room temperature for 1 h. After three washes with PBST, the membrane blots were scanned using an Odyssey infrared imaging system (LI-COR Biosciences).
Pulldown assay.
For the in vitro binding assay, human IgG-Fc fusions of wtTva or different chimeric Tva proteins were expressed in 293T cells. The cell culture medium was collected, and the proteins were purified using a protein A column. The protein A column was then incubated with 20 μg of human IgG-Fc fused with different chimeric proteins at 4°C for 2 h under gentle agitation. After five washes with ice-cold PBS, the column was incubated with a lysate of 293T cells expressing gp85 with a Flag tag at 4°C for 2 h under gentle agitation. After five washes with ice-cold PBS, the bound proteins were separated by SDS-PAGE and detected by Western blotting.
Structural analysis.
The chicken Tva sequence has a high similarity with the quail domain sequence, and the co-crystal structure of the quail Tva domain has been reported. Based on the quail Tva co-crystal structure (Protein Data Bank code: 1k7b) (https://www.uniprot.org/uniprotkb/P98162/entry), wtTva and substituted Tva models were built using modeller 9.23. The ALV-A gp85 protein structure was predicted using the open-source server trRosseta (https://yanglab.nankai.edu.cn/trRosetta/). The model was optimized by molecular dynamics simulation. All simulations and analyses were done using AMBER 20. Initial docking models of Tva and ALV-A gp85 were constructed using hdock (http://hdock.phys.hust.edu.cn/). The highest-scoring model was further optimized using Rosetta 2020 to obtain a final docking model. The free energy of Tva–ALV-A gp85 binding was obtained by molecular dynamics simulation using GROMACS software based on the final docking model.
Blocking assay.
RCASBP(A) or RAV-1 was incubated with various Tva proteins at 4°C for 1 h and then used for DF-1 cell infection at 4°C for 2 h. Unbound viruses were then removed by washing three times with PBS. The infected cells were cultured for 7 days and then harvested. Viral infection levels were quantified based on the percentage of GFP-positive cells as determined by FACS as described above. Viral titers were determined by the TCID50 assay, as described previously (50).
Confocal microscopy.
DF-1 cells were transfected with various Tva plasmids using the TransIT-X2 delivery system according to the manufacturer’s instructions. At 24 h posttransfusion, the cells were fixed in 4% PFA at room temperature for 30 min. The cells were blocked in PBS with 5% bovine albumin fraction V (10735078001; Sigma) and processed as described previously. The cells were incubated with a 1:1000 dilution of Flag antibody produced in mice for 1 h. After washing with PBS, the cells were incubated with Alexa Fluor 594-conjugated anti-mouse IgG (A11012; Invitrogen) for another 1 h. After staining with 4′,6-diamidino-2-phenylindole (DAPI) (C0065-50; Solarbio) for 30 min, the cells were examined using a laser confocal microscope (LSM980; Zeiss).
CRISPR/Cas9-mediated homologous recombination.
We prepared CRISPR/Cas9 constructs by cloning the Tva-specific gRNA sequence (GATCGTGCGGCTCCGAACAG) into the gRNA scaffold of a pMD18-T vector. Single-stranded oligodeoxynucleotides (ssODN) carrying L55R and W69L substitutions were used as homologous recombination templates. DF-1 cells were transfected with plasmid pMJ920, sgRNA, and ssODN using the TransIT-X2 delivery system according to the manufacturer’s instructions. Cells showing green fluorescence were sorted into 96-well plates using flow cytometry, and monoclonal cells were identified by sequence analysis and evaluated for resistance to RCASBP(A)GFP, RCASBP(B)GFP, RAV-1, or RAV-2 infection.
TC-Cbl uptake assay.
DF-1-WT cells and DF-1-rTva cells were seeded on microscope cover glasses and 24 h later, they were incubated with TC-Cbl (25) at 37°C for 1 h. Then, the cells were washed with PBS, fixed in 4% PFA, stained with DAPI for 10 min, and examined using a laser confocal microscope. In addition, cells incubated with TC-Cbl were digested and resuspended in PBS containing 5% (wt/vol) FBS for FACS analysis as described above.
Statistical analysis.
GraphPad Prism software (version 7.03; GraphPad Software, San Diego, CA) was employed for statistical analysis. Student's t test was used to assess differences between groups. Statistical significance was set at P < 0.05.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31872482), the Heilongjiang Touyan Innovation Team Program (TD2019C003), and China's Agricultural Research System (CARS-41-G15).
We declare no conflict of interest.
Contributor Information
Xiaomei Wang, Email: wangxiaomei@caas.cn.
Yulong Gao, Email: gaoyulong@caas.cn.
Viviana Simon, Icahn School of Medicine at Mount Sinai.
REFERENCES
- 1.Weissenhorn W, Hinz A, Gaudin Y. 2007. Virus membrane fusion. FEBS Lett 581:2150–2155. 10.1016/j.febslet.2007.01.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delos SE, Burdick MJ, White JM. 2002. A single glycosylation site within the receptor-binding domain of the avian sarcoma/leukosis virus glycoprotein is critical for receptor binding. Virology 294:354–363. 10.1006/viro.2001.1339. [DOI] [PubMed] [Google Scholar]
- 3.Li X, Zhu H, Wang Q, Sun J, Gao Y, Qi X, Wang Y, Gao H, Gao Y, Wang X. 2015. Identification of a novel B-cell epitope specific for avian leukosis virus subgroup J gp85 protein. Arch Virol 160:995–1004. 10.1007/s00705-014-2318-6. [DOI] [PubMed] [Google Scholar]
- 4.Xu M, Qian K, Shao H, Yao Y, Nair V, Ye J, Qin A. 2022. Glycosylation of ALV-J envelope protein at sites 17 and 193 is pivotal in the virus infection. J Virol 96:e0154921. 10.1128/JVI.01549-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao Y, Wang L, Gu C, Herschhorn A, Désormeaux A, Finzi A, Xiang SH, Sodroski JG. 2013. Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer. Proc Natl Acad Sci USA 110:12438–12443. 10.1073/pnas.1307382110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Barnes CO, Yang Z, Nussenzweig MC, Bjorkman PJ. 2018. Partially open HIV-1 envelope structures exhibit conformational changes relevant for coreceptor binding and fusion. Cell Host Microbe 24:579–592.e4. 10.1016/j.chom.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knipping F, Newby GA, Eide CR, McElroy AN, Nielsen SC, Smith K, Fang Y, Cornu TI, Costa C, Gutierrez-Guerrero A, Bingea SP, Feser CJ, Steinbeck B, Hippen KL, Blazar BR, McCaffrey A, Mussolino C, Verhoeyen E, Tolar J, Liu DR, Osborn MJ. 2022. Disruption of HIV-1 co-receptors CCR5 and CXCR4 in primary human T cells and hematopoietic stem and progenitor cells using base editing. Mol Ther 30:130–144. 10.1016/j.ymthe.2021.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnard RJ, Elleder D, Young JA. 2006. Avian sarcoma and leukosis virus-receptor interactions: from classical genetics to novel insights into virus-cell membrane fusion. Virology 344:25–29. 10.1016/j.virol.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 9.Federspiel MJ. 2019. Reverse engineering provides insights on the evolution of subgroups A to E avian sarcoma and leukosis virus receptor specificity. Viruses 11:497. 10.3390/v11060497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai J, Howes K, Payne LN, Skinner MA. 1995. Sequence of host-range determinants in the env gene of a full-length, infectious proviral clone of exogenous avian leukosis virus HPRS-103 confirms that it represents a new subgroup (designated J). J Gen Virol 76:181–187. 10.1099/0022-1317-76-1-181. [DOI] [PubMed] [Google Scholar]
- 11.Cui N, Su S, Chen Z, Zhao X, Cui Z. 2014. Genomic sequence analysis and biological characteristics of a rescued clone of avian leukosis virus strain JS11C1, isolated from indigenous chickens. J Gen Virol 95:2512–2522. 10.1099/vir.0.067264-0. [DOI] [PubMed] [Google Scholar]
- 12.Huebner RJ. 1966. Diseases of the avian leukosis complex. A commentary on the conference deliberations. Avian Dis 10:441–452. 10.2307/1588252. [DOI] [PubMed] [Google Scholar]
- 13.Langlois AJ, Beard D, Beard JW. 1970. Strain MC29, an avian leukosis virus of unique properties. Bibl Haematol:95–105. 10.1159/000391697. [DOI] [PubMed] [Google Scholar]
- 14.Payne LN, Nair V. 2012. The long view: 40 years of avian leukosis research. Avian Pathol 41:11–19. 10.1080/03079457.2011.646237. [DOI] [PubMed] [Google Scholar]
- 15.Bates P, Young JA, Varmus HE. 1993. A receptor for subgroup A rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74:1043–1051. 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 16.Přikryl D, Plachý J, Kučerová D, Koslová A, Reinišová M, Šenigl F, Hejnar J. 2019. The novel avian leukosis virus subgroup K shares its cellular receptor with subgroup A. J Virol 93:e00580-19. 10.1128/JVI.00580-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young JA, Bates P, Varmus HE. 1993. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J Virol 67:1811–1816. 10.1128/JVI.67.4.1811-1816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adkins HB, Brojatsch J, Naughton J, Rolls MM, Pesola JM, Young JA. 1997. Identification of a cellular receptor for subgroup E avian leukosis virus. Proc Natl Acad Sci USA 94:11617–11622. 10.1073/pnas.94.21.11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adkins HB, Brojatsch J, Young JA. 2000. Identification and characterization of a shared TNFR-related receptor for subgroup B, D, and E avian leukosis viruses reveal cysteine residues required specifically for subgroup E viral entry. J Virol 74:3572–3578. 10.1128/jvi.74.8.3572-3578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brojatsch J, Naughton J, Adkins HB, Young JA. 2000. TVB receptors for cytopathic and noncytopathic subgroups of avian leukosis viruses are functional death receptors. J Virol 74:11490–11494. 10.1128/jvi.74.24.11490-11494.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brojatsch J, Naughton J, Rolls MM, Zingler K, Young JA. 1996. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell 87:845–855. 10.1016/S0092-8674(00)81992-3. [DOI] [PubMed] [Google Scholar]
- 22.Elleder D, Stepanets V, Melder DC, Senigl F, Geryk J, Pajer P, Plachy J, Hejnar J, Svoboda J, Federspiel MJ. 2005. The receptor for the subgroup C avian sarcoma and leukosis viruses, Tvc, is related to mammalian butyrophilins, members of the immunoglobulin superfamily. J Virol 79:10408–10419. 10.1128/JVI.79.16.10408-10419.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munguia A, Federspiel MJ. 2008. Efficient subgroup C avian sarcoma and leukosis virus receptor activity requires the IgV domain of the Tvc receptor and proper display on the cell membrane. J Virol 82:11419–11428. 10.1128/JVI.01408-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chai N, Bates P. 2006. Na+/H+ exchanger type 1 is a receptor for pathogenic subgroup J avian leukosis virus. Proc Natl Acad Sci USA 103:5531–5536. 10.1073/pnas.0509785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krchlíková V, Mikešová J, Geryk J, Bařinka C, Nexo E, Fedosov SN, Kosla J, Kučerová D, Reinišová M, Hejnar J, Elleder D. 2021. The avian retroviral receptor Tva mediates the uptake of transcobalamin bound vitamin B12 (cobalamin). J Virol 95:e02136-20. 10.1128/JVI.02136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark LE, Clark SA, Lin C, Liu J, Coscia A, Nabel KG, Yang P, Neel DV, Lee H, Brusic V, Stryapunina I, Plante KS, Ahmed AA, Catteruccia F, Young-Pearse TL, Chiu IM, Llopis PM, Weaver SC, Abraham J. 2022. VLDLR and ApoER2 are receptors for multiple alphaviruses. Nature 602:475–480. 10.1038/s41586-021-04326-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rong L, Gendron K, Bates P. 1998. Conversion of a human low-density lipoprotein receptor ligandbinding repeat to a virus receptor: identification of residues important for ligand specificity. Proc Natl Acad Sci USA 95:8467–8472. 10.1073/pnas.95.15.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balliet JW, Berson J, D'Cruz CM, Huang J, Crane J, Gilbert JM, Bates P. 1999. Production and characterization of a soluble, active form of Tva, the subgroup A avian sarcoma and leukosis virus receptor. J Virol 73:3054–3061. 10.1128/JVI.73.4.3054-3061.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilbert JM, Bates P, Varmus HE, White JM. 1994. The receptor for the subgroup A avian leukosis-sarcoma viruses binds to subgroup A but not to subgroup C envelope glycoprotein. J Virol 68:5623–5628. 10.1128/JVI.68.9.5623-5628.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rong L, Edinger A, Bates P. 1997. Role of basic residues in the subgroup-determining region of the subgroup A avian sarcoma and leukosis virus envelope in receptor binding and infection. J Virol 71:3458–3465. 10.1128/JVI.71.5.3458-3465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Damico RL, Crane J, Bates P. 1998. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc Natl Acad Sci USA 95:2580–2585. 10.1073/pnas.95.5.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilbert JM, Hernandez LD, Balliet JW, Bates P, White JM. 1995. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J Virol 69:7410–7415. 10.1128/JVI.69.12.7410-7415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernandez LD, Peters RJ, Delos SE, Young JA, Agard DA, White JM. 1997. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J Cell Biol 139:1455–1464. 10.1083/jcb.139.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guan X, Zhang Y, Yu M, Ren C, Gao Y, Yun B, Liu Y, Wang Y, Qi X, Liu C, Cui H, Zhang Y, Gao L, Li K, Pan Q, Zhang B, Wang X, Gao Y. 2018. Residues 28 to 39 of the extracellular loop 1 of Chicken Na(+)/H(+) exchanger type I mediate cell binding and entry of subgroup J avian leukosis virus. J Virol 92:e01627-17. 10.1128/JVI.01627-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koslová A, Trefil P, Mucksová J, Reinišová M, Plachý J, Kalina J, Kučerová D, Geryk J, Krchlíková V, Lejčková B, Hejnar J. 2020. Precise CRISPR/Cas9 editing of the NHE1 gene renders chickens resistant to the J subgroup of avian leukosis virus. Proc Natl Acad Sci USA 117:2108–2112. 10.1073/pnas.1913827117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuo Q, Wang Y, Cheng S, Lian C, Tang B, Wang F, Lu Z, Ji Y, Zhao R, Zhang W, Jin K, Song J, Zhang Y, Li B. 2016. Site-directed genome knockout in chicken cell line and embryos can use CRISPR/Cas gene editing technology. G3 (Bethesda) 6:1787–1792. 10.1534/g3.116.028803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melder DC, Pike GM, VanBrocklin MW, Federspiel MJ. 2015. Model of the TVA receptor determinants required for efficient infection by subgroup A avian sarcoma and leukosis viruses. J Virol 89:2136–2148. 10.1128/JVI.02339-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tonelli M, Peters RJ, James TL, Agard DA. 2001. The solution structure of the viral binding domain of Tva, the cellular receptor for subgroup A avian leukosis and sarcoma virus. FEBS Lett 509:161–168. 10.1016/S0014-5793(01)03086-1. [DOI] [PubMed] [Google Scholar]
- 39.Wang QY, Huang W, Dolmer K, Gettins PG, Rong L. 2002. Solution structure of the viral receptor domain of Tva and its implications in viral entry. J Virol 76:2848–2856. 10.1128/jvi.76.6.2848-2856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reinisova M, Senigl F, Yin X, Plachy J, Geryk J, Elleder D, Svoboda J, Federspiel MJ, Hejnar J. 2008. A single-amino-acid substitution in the TvbS1 receptor results in decreased susceptibility to infection by avian sarcoma and leukosis virus subgroups B and D and resistance to infection by subgroup E in vitro and in vivo. J Virol 82:2097–2105. 10.1128/JVI.02206-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prévost J, Tolbert WD, Medjahed H, Sherburn RT, Madani N, Zoubchenok D, Gendron-Lepage G, Gaffney AE, Grenier MC, Kirk S, Vergara N, Han C, Mann BT, Chénine AL, Ahmed A, Chaiken I, Kirchhoff F, Hahn BH, Haim H, Abrams CF, Smith AB, 3rd, Sodroski J, Pazgier M, Finzi A. 2020. The HIV-1 env gp120 inner domain shapes the Phe43 cavity and the CD4 binding site. mBio 11:e00280-20. 10.1128/mBio.00280-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark AJ, Gindin T, Zhang B, Wang L, Abel R, Murret CS, Xu F, Bao A, Lu NJ, Zhou T, Kwong PD, Shapiro L, Honig B, Friesner RA. 2017. Free energy perturbation calculation of relative binding free energy between broadly neutralizing antibodies and the gp120 glycoprotein of HIV-1. J Mol Biol 429:930–947. 10.1016/j.jmb.2016.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lo Conte L, Chothia C, Janin J. 1999. The atomic structure of protein-protein recognition sites. J Mol Biol 285:2177–2198. 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 45.Knauss DJ, Young JA. 2002. A fifteen-amino-acid TVB peptide serves as a minimal soluble receptor for subgroup B avian leukosis and sarcoma viruses. J Virol 76:5404–5410. 10.1128/jvi.76.11.5404-5410.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J, Lee IH, Joung I, Lee J, Brooks BR. 2017. Finding multiple reaction pathways via global optimization of action. Nat Commun 8:15443. 10.1038/ncomms15443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koslová A, Trefil P, Mucksová J, Krchlíková V, Plachý J, Krijt J, Reinišová M, Kučerová D, Geryk J, Kalina J, Šenigl F, Elleder D, Kožich V, Hejnar J. 2021. Knock-out of retrovirus receptor gene Tva in the chicken confers resistance to avian leukosis virus subgroups A and K and affects Cobalamin (Vitamin B(12))-dependent level of methylmalonic Acid. Viruses 13:2504. 10.3390/v13122504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang S, Yu M, Liu A, Bao Y, Qi X, Gao L, Chen Y, Liu P, Wang Y, Xing L, Meng L, Zhang Y, Fan L, Li X, Pan Q, Zhang Y, Cui H, Li K, Liu C, He X, Gao Y, Wang X. 2021. TRIM25 inhibits infectious bursal disease virus replication by targeting VP3 for ubiquitination and degradation. PLoS Pathog 17:e1009900. 10.1371/journal.ppat.1009900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Yu M, Xing L, Liu P, Chen Y, Chang F, Wang S, Bao Y, Farooque M, Li X, Guan X, Liu Y, Liu A, Qi X, Pan Q, Zhang Y, Gao L, Li K, Liu C, Cui H, Wang X, Gao Y. 2020. The bipartite sequence motif in the N and C termini of gp85 of subgroup J avian leukosis virus plays a crucial role in receptor binding and viral entry. J Virol 94:e01232-20. 10.1128/JVI.01232-20. [DOI] [PMC free article] [PubMed] [Google Scholar]