ABSTRACT
Duck Tembusu virus (DTMUV) is an emerging pathogenic flavivirus that mainly causes a decrease in egg production in infected waterfowl. Similar to other members of the Flaviviridae family, it can proliferate in most mammalian cells and may also pose a potential threat to nonavian animals. In previous studies, we found that DTMUV infection can upregulate suppressor of cytokine signaling 1 (SOCS1) to inhibit type I interferon (IFN) production and promote virus replication, but the specific mechanism is unclear. Furthermore, little is known about the regulatory role of ubiquitination during flavivirus infection. In this study, we found that activation of Toll-like receptor 3 (TLR3) signaling rather than type I IFN stimulation led to the upregulation of SOCS1 during DTMUV infection. Further studies revealed that JOSD1 stabilized SOCS1 expression by binding to the SH2 domain of SOCS1 and mediating its deubiquitination. In addition, JOSD1 also inhibited type I IFN production through SOCS1. Finally, SOCS1 acts as an E3 ubiquitin ligase that binds to IFN regulatory factor 7 (IRF7) through its SH2 domain and mediates K48-linked ubiquitination and proteasomal degradation of IRF7, ultimately inhibiting type I IFN production mediated by IRF7 and promoting viral proliferation. These results will enrich and deepen our understanding of the mechanism by which DTMUV antagonizes the host interferon system.
IMPORTANCE DTMUV is a newly discovered flavivirus that seriously harms the poultry industry. In recent years, there have been numerous studies on the involvement of ubiquitination in the regulation of innate immunity. However, little is known about the involvement of ubiquitination in the regulation of flavivirus-induced type I IFN signaling. In this study, we found that SOCS1 was induced by TLR3 signaling during DTMUV infection. Furthermore, we found for the first time that duck SOCS1 protein was also modified by K48-linked polyubiquitination, whereas our previous study found that SOCS1 was upregulated during DTMUV infection. Further studies showed that JOSD1 stabilized SOCS1 expression by mediating the deubiquitination of SOCS1. While SOCS1 acts as a negative regulator of cytokines, we found that DTMUV utilized SOCS1 to mediate the ubiquitination and proteasomal degradation of IRF7 and ultimately inhibit type I IFN production, thereby promoting its proliferation.
KEYWORDS: DTMUV, SOCS1, IRF7, ubiquitination, JOSD1
INTRODUCTION
Duck Tembusu virus (DTMUV) belongs to the Flaviviridae family of flaviviruses and is an emerging pathogen causing reduced egg production syndrome in infected ducks (1). Since its discovery in 2010, the rapid spread of the disease has caused substantial economic losses in the poultry industry of many Southeast Asian countries, including China, Malaysia, and Thailand (1–3). Immune organ defects and neurological dysfunction are the main clinical symptoms of DTMUV infection. Preinfection with DTMUV makes the virus impervious to later interferon (IFN) treatment, revealing that DTMUV has evolved some strategies to defend against host IFN-dependent antiviral responses (4).
The earliest response of the host to pathogen invasion is the recognition of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs), which triggers host countermeasures to respond to the cell physiology disturbance caused by pathogens (5, 6). PRRs mainly include Toll-like receptors (TLRs), retinoic acid-inducible gene I-like receptor (RLR), and nucleotide-binding oligomerization domain-like receptors (NLRs). Many studies have reported that the nonstructural (NS) protein of DTMUV can inhibit RIG-I-mediated type I IFN production. For example, DTMUV inhibits IFN-β production by disrupting RIG-I-mediated signaling through its NS protein 2A (NS2A) binding to duSTING (4). NS1 of DTMUV inhibits RIG-I-mediated type I IFN production by targeting the virus-induced signaling adaptor (VISA) (7). In addition, NS2B-3 of DTMUV also antagonized RIG-I-mediated IFN-β production by cleaving duSTING (8). Notably, none of the above-mentioned proteins of DTMUV could inhibit IFN regulatory factor 7 (IRF7)-mediated type I IFN production. However, previous studies have shown that DTMUV infection triggers TLR3-mediated innate immunity in the host, which promotes IRF7-mediated type I IFN production (9, 10). How DTMUV antagonizes IRF7-mediated type I IFN production is still an open area of investigation.
The signaling pathways that mediate the production of type I IFN are mainly divided into three categories: RLRs-MAVS-IFNs, DNA receptors-STING-IFNs, and TLRs-TRIF/MyD88-IFNs (11). Although the ligands, signal molecular composition, and signal transduction mechanisms recognized by these three types of signal pathways are quite different, they all eventually converge into IRF3/7. The activated IRF3/7 are incorporated into the nucleus through dimerization and promotes the production of type I IFN (12). Although IRF3/7 belong to the IRF3 subfamily and are similar in structure and function, there are many differences in their regulatory effects on type I IFN (13–15). IRF3 is constitutively expressed in mammalian cells, and its production is not affected by IFN stimulation or viral infection. In contrast, in most cells, IRF7 expression is low, and type I IFN signals strongly induce IRF7 production (11). In terms of type I IFN signal regulation, IRF3 mainly initiates the inducible expression of IFN-β, while IRF7 plays a role in the later stage of IFN-β production (16). Studies have found that IRF3 is missing in ducks and other poultry, so the production of type I IFN in ducks is mainly mediated by IRF7 (11, 17–19).
The suppressor of cytokine signaling (SOCS) family mainly contains eight members, namely, SOCS1 to SOCS7 and CIS. These eight members share a central SH2 domain, a carboxy-terminal SOCS BOX domain, an amino-terminal variable domain, and an extended SH2 subdomain (ESS) (20, 21). Among these, the SH2 domain of the SOCS protein is mainly involved in the recognition and binding of the substrate, and the ESS domain strengthens the binding of the SOCS protein to the substrate (21). The best-studied proteins in the SOCS family are SOCS1 and SOCS3, because they have a unique kinase-inhibitory region (KIR). SOCS1 and SOCS3 can bind to JAK through their KIR domains and inhibit JAK phosphorylation, thereby inhibiting JAK/STAT signal transduction (22). It is worth noting that in mammals, the SOCS BOX of the SOCS protein can recruit elongin B/C, Cullin-5, and RBX2 to form an E3 ubiquitin ligase complex (23). Studies have found that SOCS1 can bind to the p65 subunit of NF-κB and mediate the binding of the K48-linked polyubiquitin chain to p65 to promote the proteasomal degradation of p65 and negatively regulate NF-κB signaling (24). In addition, SOCS1 interacts with Mal (the adaptor protein of TLR2/4 downstream signals) and mediates its ubiquitination-dependent degradation to quickly terminate the innate immune response signal transduction (25). Therefore, SOCS1 is a key negative regulator of innate immunity. Although there are many studies on SOCS1-mediated ubiquitination of other proteins, there are few reports on the ubiquitination of SOCS1 itself. JOSD1 is a member of the deubiquitinase MJD family (26, 27). Studies have shown that JOSD1 can mediate the deubiquitination of SOCS1 and upregulate the expression of SOCS1 to inhibit the production of type I IFN (28). But does a similar thing happen with SOCS1 in birds? It is worth exploring.
Many viruses take advantage of SOCS1’s negative regulation of cytokine to suppress IFN signaling. For example, porcine reproductive and respiratory syndrome virus (PRRSV) upregulates SOCS1 to antagonize the production of IFN-β and IFN-stimulated genes (ISGs) and promote its proliferation (29). In addition, influenza A virus (IAV) infection can also promote the production of SOCS1 and SOCS3, while the upregulated SOCS1 and SOCS3 block the production of type I and II IFN by inhibiting the activation of JAK/STAT signals (30). Another example is that during hepatitis B virus (HBV) infection of plasmacytoid dendritic cells (pDCs), HBV exploits SOCS1 to restrict TLR9-mediated IFN-α production to promote its proliferation (31). Our previous study found that during DTMUV infection, SOCS1 is upregulated and can inhibit the production of type I IFN, but the specific mechanism is still unclear. In human pDCs, it was found that SOCS1 can interact with IRF7 to negatively regulate the production of type I IFN mediated by TLR7 (32). Still, the structure and function of the innate immune system of avians are far from those of mammals. Whether the viruses can also take advantage of duck SOCS1 to antagonize type I IFN production to promote self-proliferation during DTMUV infection has not been reported yet.
In this study, we found that the upregulation of SOCS1 during DTMUV infection was mediated by TLR3 activation rather than induced by type I IFN stimulation. Further experiments found that the upregulation of SOCS1 inhibited the production of type I IFN induced by DTMUV. As the critical regulator of type I IFN production is IRF7, we started with IRF7 and found that SOCS1 could mediate the ubiquitination of IRF7 and the subsequent proteasomal degradation, thereby inhibiting IRF7-mediated type I IFN production. In addition, we found that duck SOCS1 mainly undergoes K48-linked polyubiquitination, and duck JOSD1 can stabilize SOCS1 expression and inhibit type I IFN production by removing K48-linked polyubiquitination of SOCS1, thereby promoting viral replication. Overall, revealing the mechanism by which DTMUV inhibits IRF7-mediated type I IFN production through the JOSD1-SOCS1-IRF7 signaling axis will help us better understand the strategy of DTMUV to inhibit type I IFN production.
RESULTS
TLR3 plays a role in DTMUV induction of SOCS1 expression, and SOCS1 inhibits DTMUV-induced type I IFN production.
Previous studies have found that DTMUV infection of duck embryo fibroblasts (DEFs) could cause upregulation of SOCS1 mRNA levels, but the specific reason remained unclear (Fig. 1A) (33). In some studies, type I IFN secretion was found to induce SOCS1 upregulation (Fig. 1A), so we examined whether type I IFN secretion could stimulate SOCS1 upregulation during DTMUV infection. Type I IFN stimulation could induce the production of ISGs (such as MX, OASL, etc.) (Fig. 1A). Therefore, we detected the expression of SOCS1, MX, and OASL (positive control) after transfecting pCAGGS-IFN-α and pCAGGS-IFN-β overexpression plasmids into DEFs or stimulating DEFs with gradient doses of IFN-β. It can be seen from Fig. 1B and C that the stimulation of type I IFN could significantly upregulate the mRNA levels of SOCS1, MX, and OASL in the absence of DTMUV infection. In contrast, in the DTMUV-infected group, DTMUV induced upregulation of the SOCS1 mRNA level, but unlike MX and OASL, type I IFN did not further increase the SOCS1 mRNA level in DTMUV-infected cells (Fig. 1B and C). Some studies have found that activation of TLR signaling could also induce upregulation of SOCS1 (Fig. 1A) (32). And some studies have found that DTMUV infection can activate TLR3-mediated innate immunity (Fig. 1A) (10). First, changes in TLR3 mRNA level during DEF infection with DTMUV were detected, and it was found that the TLR3 mRNA level was significantly upregulated within 36 h compared with that in the control group (Fig. 1D). During DTMUV infection, knockdown of TLR3 significantly downregulated the mRNA level of SOCS1, while overexpression of TLR3 significantly upregulated the mRNA level of SOCS1 (Fig. 1E to G). In addition, the mRNA level of SOCS1 was significantly upregulated under either DTMUV infection or poly(I:C) stimulation (Fig. 1H). Therefore, the activation of TLR3 during DTMUV infection causes upregulation of SOCS1.
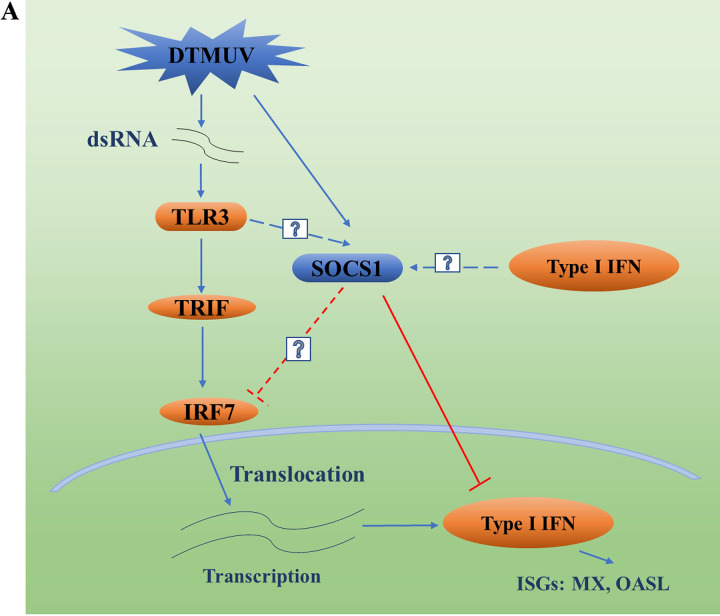
FIG 1.
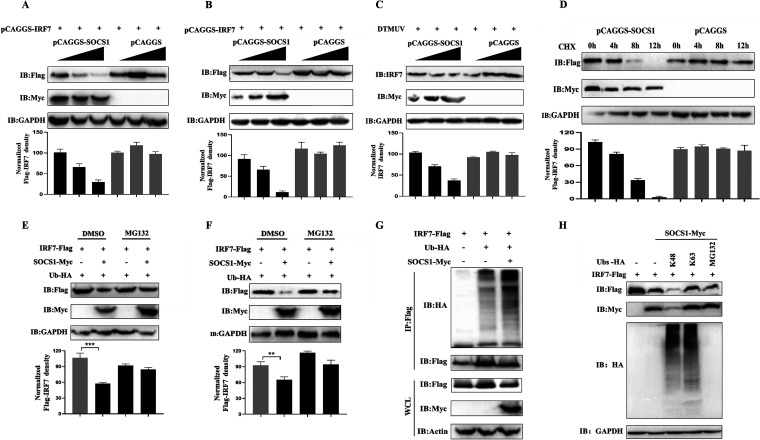
TLR3 plays a role in DTMUV induction of SOCS1 expression, and SOCS1 inhibits DTMUV-induced type I IFN production. (A) DTMUV infection activates TLR3-mediated innate immunity. Since birds lack IRF3, avian cells mainly mediate type I IFN production through IRF7, and the generated type I IFN continues to promote the production of ISGs such as MX and OASL to achieve antiviral responses. DTMUV infection has previously been found to upregulate SOCS1 to inhibit type I IFN production, thereby promoting the proliferation of the virus itself, but the mechanism of SOCS1 upregulation during DTMUV infection remains unclear. Some studies have shown that SOCS1 may be stimulated by type I IFN, and some studies have shown that the activation of TLR signaling can promote the production of SOCS1. Furthermore, the mechanism of how SOCS1 negatively regulates IRF7-mediated type I IFN during DTMUV infection remains unclear. (B) pCAGGS-IFNα, pCAGGS-IFNβ, and pCAGGS (1,250 ng/well) were transfected into DEFs for 24 h, and DEFs were infected with DTMUV at a multiplicity of infection (MOI) of 1 or left uninfected for 12, 24, 36, 48, 60 h, and then cell samples were collected. The mRNA levels of SOCS1, MX, and OASL (n = 3) were detected by RT-qPCR. (C) After DEFs were infected with DTMUV at an MOI of 1 or left uninfected for 36 h, they were stimulated with gradient doses of IFN-β supernatant (300 μL, 400 μL, and 500 μL) for 24 h and then samples were collected. The mRNA levels of SOCS1, MX, and OASL (n = 3) were detected by RT-qPCR. (D) After infecting DEFs with DTMUV at an MOI of 1 for different time periods (6 h, 12 h, 24 h, 36 h, 48 h, and 60 h), the cells were collected. The mRNA level of TLR3 (n = 3) was detected by RT-qPCR. (E) pCAGGS (1,250 ng/well), pCAGGS-TLR3 (1,250 ng/well), pCAGGS-TLR3 and shRNA-NC (1,250 ng/well), and pCAGGS-TLR3 and shRNA-TLR3 (1,250 ng/well) were transfected into DEFs for 36 h. The protein level of TLR3 (n = 3) was detected by Western blotting (IB). (F) pCAGGS-TLR3, pCAGGS, shRNA-TLR3, and shRNA-NC (1,250 ng/well) were transfected into DEFs for 36 h. Then the mRNA level of TLR3 (n = 3) was detected by RT-qPCR. (G) pCAGGS-TLR3, pCAGGS, shRNA-TLR3, and shRNA-NC (1,250 ng/well) were transfected into DEFs for 36 h. Then DEFs were infected with DTMUV at an MOI of 1 for 36 h. The mRNA level of SOCS1 (n = 3) was detected by RT-qPCR. (H) Cells were collected after infection of DEFs with DTMUV at an MOI of 1 or stimulating DEFs with poly(I:C) for 36 h. The mRNA level of SOCS1 (n = 3) was detected by RT-qPCR. (I and J) shRNA-NC, shRNA-SOCS1, pCAGGS, and pCAGGS-SOCS1 (1,250 ng/well) were transfected into DEFs for 24 h. Then DEFs were infected with DTMUV at an MOI of 1 or left uninfected for 36 h. The mRNA levels of IFN-α and IFN-β (n = 3) were detected by RT-qPCR. (K and L) NF-κB-Luc and IFN-β-Luc (400 ng/well) with 40 ng pRL-TK (40 ng/well) and gradient doses of pCAGGS or pCAGGS-SOCS1 (100, 200, and 400 ng/well) were cotransfected into DEFs for 24 h. Then DEFs were infected with DTMUV at an MOI of 1 for 36 h (n = 5). Promoter activity was measured by the dual-luciferase reporter system. The ratio of firefly luciferase activity to Renilla luciferase activity is the data obtained. Each experiment whose results are shown was repeated three times. The data are the mean ± SEM (n = 3). The significant differences between the groups were analyzed by t test. ns, not significant (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Studies have found that DTMUV can significantly activate TLR3-mediated innate immunity (9). Therefore, we examined whether SOCS1 knockdown or overexpression affected DTMUV-induced type I IFN mRNA levels. It was found that knockdown of SOCS1 resulted in upregulation of type I IFN induced by DTMUV and that ectopic expression of SOCS1 significantly inhibited the production of type I IFN induced by DTMUV (Fig. 1I and J). Furthermore, the dual-luciferase reporter system assay found that with increasing doses of pCAGGS-SOCS1 transfected, DTMUV-induced IFN-β signaling and NF-κB activation were inhibited in a dose-dependent manner (Fig. 1K and L). Therefore, SOCS1 downregulates type I IFN production during DTMUV infection.
SOCS1 affects type I IFN production via IRF7 to promote viral replication.
Because birds lack IRF3, the production of type I IFN during viral infection is mainly mediated by IRF7 (11). Therefore, we tested the changes in IRF7 mRNA and protein levels at different time points upon DTMUV infection of DEFs. As shown in Fig. 2A, compared with that in the control group, the mRNA level of IRF7 was significantly upregulated within 36 h after DTMUV infection but was significantly downregulated at 60 h of infection. Similarly, compared with that in the control group, the protein level of IRF7 was significantly upregulated within 48 h of DTMUV infection and was significantly downregulated at 60 h compared with those at other infection time points (Fig. 2B). In addition, compared with the control group, the mRNA level of type I IFN was significantly upregulated at 36 h after DTMUV infection. However, the type I IFN mRNA level was significantly downregulated at 48 h and 60 h of infection compared with that at 36 h of infection (Fig. 2C and D). Can SOCS1 negatively regulate type I IFN production through IRF7? First, we tested the effect of ectopic expression of SOCS1 or knockdown of SOCS1 on IRF7 mRNA levels. The results showed that knockdown of SOCS1 caused upregulation of the IRF7 mRNA level, and the overexpression of SOCS1 significantly downregulated the mRNA level of IRF7 (Fig. 2E). Next, we tested whether SOCS1 affected IRF7-mediated type I IFN production through the dual-luciferase reporter system and quantitative real-time PCR (RT-qPCR). The results of the dual-luciferase reporter system showed that SOCS1 inhibited IRF7-mediated IFN-β signaling in a dose-dependent manner (Fig. 2F). Consistent with the results of the dual-luciferase test, the overexpression of SOCS1 significantly inhibited IRF7-mediated IFN-β production during DTMUV infection (Fig. 2G). Therefore, SOCS1 inhibits type I IFN production through IRF7 during DTMUV infection.
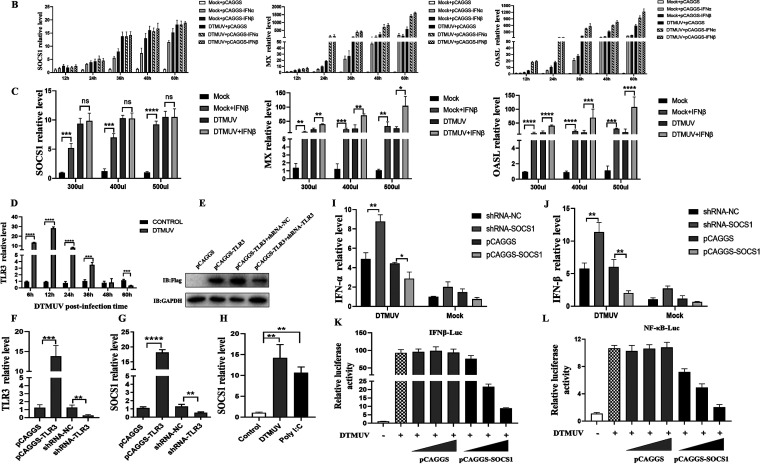
FIG 2.
SOCS1 affects type I IFN production via IRF7 to promote viral replication. After infection of DEFs with DTMUV at an MOI of 1 for different time periods (6 h, 12 h, 24 h, 36 h, 48 h, and 60 h), the cells were collected. The mRNA levels of IRF7, IFN-α, and IFN-β (n = 3) were detected by RT-qPCR (A, C, and D). The protein level of IRF7 (n = 3) was detected by Western blotting (B). (E) shRNA-NC, shRNA-SOCS1, pCAGGS and pCAGGS-SOCS1 (1,250 ng/well) were transfected into DEFs for 24 h. Then DEFs were infected with DTMUV for 36 h. The mRNA level of IRF7 (n = 3) was detected by RT-qPCR. (F) IFN-β-Luc (400 ng/well) and 40 ng pRL-TK (40 ng/well), pCAGGS-IRF7 (100 ng), and gradient doses of pCAGGS or pCAGGS-SOCS1 (100, 200, and 400 ng/well) were cotransfected into DEFs for 24 h. Then DEFs were infected with DTMUV for 36 h (n = 5). Promoter activity was measured by the dual-luciferase reporter system. The ratio of firefly luciferase activity to Renilla luciferase activity is the data obtained. (G) pCAGGS (625 ng/well), pCAGGS-IRF7 (625 ng/well), pCAGGS-IRF7 and pCAGGS, and pCAGGS-IRF7 and pCAGGS-SOCS1 (625 ng/well) were transfected into DEFs for 24 h. Then DEFs were infected with DTMUV or left uninfected for 36 h. The mRNA level of IFN-β (n = 3) was detected by RT-qPCR. (H to K) pCAGGS, pCAGGS-IRF7, pCAGGS-IRF7 and pCAGGS, pCAGGS-IRF7 and pCAGGS-SOCS1, shRNA-NC, shRNA-IRF7, shRNA-IRF7 and pCAGGS, and shRNA-IRF7 and pCAGGS-SOCS1 (625 ng/well) were transfected into DEFs for 24 h. Then DEFs were infected with DTMUV for 36 h. RT-qPCR (H and J) and Western blotting (I) were used to detect the mRNA and protein levels of IRF7 and the viral E protein (n = 3). The cells were harvested for titer detection (K). The viral E protein densitometric values in panel I was quantified and analyzed. Each experiment was repeated three times. The data are the means ± SEM (n = 3). The significant differences between the groups were analyzed by t test. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
If SOCS1 affects type I IFN production through IRF7, does SOCS1 also affect virus replication through IRF7? As shown in Fig. 2H and I, pCAGGS-IRF7 and shRNA-IRF7 could effectively overexpress and knock down the mRNA and protein levels of IRF7 in DEFs, respectively. Notably, when pCAGGS-SOCS1 and pCAGGS-IRF7 were cotransfected into DEFs, the mRNA and protein levels of IRF7 were downregulated (Fig. 2H and I). Moreover, as shown in Fig. 2I to K, when IRF7 was overexpressed in DEFs, the viral E protein expression level, viral copy number, and viral titer of DTMUV were downregulated. When IRF7 was knocked down, the viral E protein expression level, viral copy number, and viral titer were upregulated (Fig. 2I to K). However, when SOCS1 and IRF7 were coexpressed in DEFs, the viral E protein expression level, viral copy number, and viral titer were significantly upregulated (Fig. 2I to K), indicating that overexpression of SOCS1 interfered with the inhibitory effect of IRF7 on the proliferation of DTMUV. Taken together, these results show that SOCS1 can affect the proliferation of DTMUV through IRF7.
SOCS1 binds to IRF7 through its SH2 domain.
Does SOCS1 have direct interaction with IRF7? By indirect immunofluorescence, we found that in the control group, SOCS1 colocalized with IRF7 in the cytoplasm, while in the DTMUV infection group, SOCS1 colocalized with IRF7 in the nucleus (Fig. 3A). We further conducted anti-Flag and anti-Myc antibody (Ab) immunoprecipitation experiments and found that SOCS1 and IRF7 could bind directly (Fig. 3B and C).
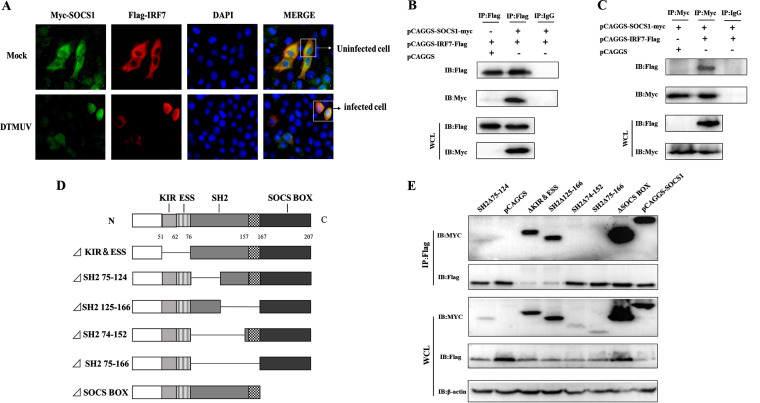
FIG 3.
SOCS1 binds to IRF7 through its SH2 domain. (A) pCAGGS-IRF7-Flag and pCAGGS-SOCS1-Myc (625 ng/well) were transfected into DEFs for 24 h. Then DEFs were stimulated with DTMUV or left unstimulated for 24 h, and cell samples were collected. The cellular localization of IRF7 and SOCS1 was observed by indirect immunofluorescence (original magnification, ×600). (B and C) pCAGGS-IRF7-Flag (1,500 ng/well) and pCAGGS-SOCS1-Myc (1,000 ng/well), pCAGGS-SOCS1-Myc and pCAGGS (1,500 ng/well), and pCAGGS-IRF7-Flag and pCAGGS were cotransfected into 293T cells for 36 h. Cell lysates were immunoprecipitated with anti-Myc or anti-Flag or control mouse IgG and then anti-Flag and anti-Myc for IB. (D) The various domains of SOCS1 were deleted sequentially. (E) The wild-type and mutant plasmids of SOCS1 were transfected together with the IRF7 plasmid into 293T cells for 36 h. The interaction between SOCS1 and IRF7 was detected by anti-Flag Ab for IP, and then anti-Myc was used for IB detection. Each experiment was repeated three times. The data are the means ± SEM (n = 3).
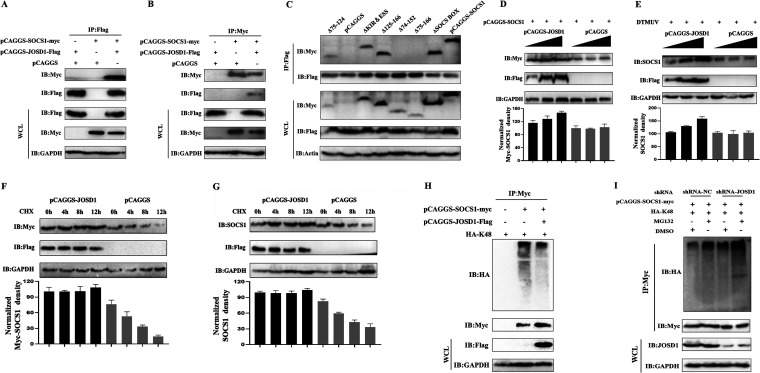
To know which domain of SOCS1 is involved in IRF7 interaction, Myc tag plasmids with deletion of each domain of SOCS1 were constructed (Fig. 3D). The immunoprecipitation test was used to detect whether they bind to IRF7. The results showed that the mutants lacking KIR and ESS (ΔKIR&ESS) and the mutants lacking SOCS BOX (ΔSOCS BOX) could interact with IRF7. In addition, the mutants with the N-terminal truncation of SH2 (ΔSH2 75–124) and the mutants with the C-terminal truncation (ΔSH2 125–166) could both bind to IRF7. It is worth noting that when we deleted part of the SH2 domain and the amino acid sequence close to the SOCS BOX (ΔSH2 74–152) or completely deleted the SH2 domain (ΔSH2 75–166), SOCS1 lost its ability to bind to IRF7 (Fig. 3E). In summary, the above results indicate that the SH2 domain of SOCS1 is the critical region for binding to IRF7.
SOCS1 mediates the ubiquitination of IRF7 and subsequent proteasomal degradation.
SOCS1 is a protein with ubiquitin ligase activity, and it can be seen from Fig. 2I that SOCS1 inhibited the expression of IRF7, so could SOCS1 mediate the ubiquitination of IRF7 and its proteasomal degradation? We transfected gradient amounts of pCAGGS-SOCS1-Myc and fixed amounts of pCAGGS-IRF7-Flag into DEFs or 293T cells and found that as the transfection amount of SOCS1 increased, the expression of IRF7 was downregulated in a dose-dependent manner (Fig. 4A and B). In addition, when gradient doses of pCAGGS-SOCS1-Myc were transfected into DEFs, SOCS1 also inhibited the expression of endogenous IRF7 in a dose-dependent manner (Fig. 4C). Next, pCAGGS-SOCS1-Myc and pCAGGS-IRF7-Flag were cotransfected into 293T cells. Then the cells were treated with protein synthesis inhibitor cycloheximide (CHX) for different periods to determine the half-life of IRF7. It was found that when SOCS1 and IRF7 were cotransfected, the conversion rate of IRF7 increased (Fig. 4D).
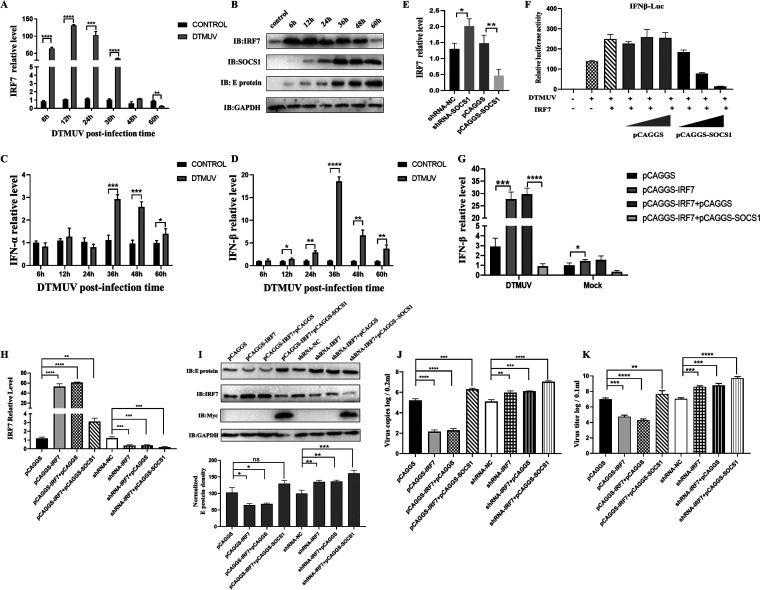
FIG 4.
SOCS1 mediates the ubiquitination of IRF7 and subsequent proteasomal degradation. (A and B) pCAGGS-IRF7-Flag (1,000 ng/well) and gradient doses of pCAGGS-SOCS1-Myc (500, 1,000, and 1,500 ng/well) were cotransfected into DEFs (A) or 293T cells (B) for 36 h, and then cell samples were collected. The protein levels of IRF7 and SOCS1 were detected by Western blotting. (C) Gradient doses of pCAGGS-SOCS1-Myc (500, 1,000, and 1,500 ng/well) were transfected into DEFs for 24 h. After DEFs were infected with DTMUV at an MOI of 1 for 36 h, cell samples were collected. Western blotting was used to detect endogenous IRF7 protein level. (D) pCAGGS-SOCS1-Myc (1,000 ng/well) and pCAGGS-IRF7-Flag (1,500 ng/well) were transfected into 293T cells for 36 h. The cells were then treated with CHX (200 μg/mL), a protein synthesis inhibitor. Then the cells were collected at different time points (0 h, 4 h, 8 h, and 12 h). The protein level of IRF7 was detected by Western blotting. (E and F) pCAGGS-IRF7-Flag (1,000 ng/well), pCAGGS-SOCS1-Myc (1,200 ng/well), and Ub-HA (800 ng/well) were cotransfected into DEFs (E) or 293T cells (F) for 36 h. Cells were harvested by treatment with proteasome inhibitor MG132 (10 μM) or control DMSO for 6 h. The protein level of IRF7 was detected by Western blotting. (G) pCAGGS-IRF7-Flag (1,000 ng/well), pCAGGS-SOCS1-Myc (1,200 ng/well), and HA-Ub (800 ng/well) were cotransfected into 293T cells for 36h. Immunoprecipitation of cell lysates was detected with anti-Flag, and then anti-HA was used for IB to detect ubiquitinated IRF7. (H) HA-Ub-K48 and HA-Ub-K63 (800 ng/well) with pCAGGS-IRF7-Flag (1,000 ng/well) and pCAGGS-SOCS1-Myc (1,200 ng/well) were cotransfected into 293T cells for 36 h. At the same time, a group of cells was treated with MG132 for 6 h. Then the cells were collected, and the protein level of IRF7 was detected by Western blotting. The IRF7 protein densitometric values were quantified and analyzed. Each experiment was repeated three times. The data are the means ± SEM (n = 3).
To examine whether SOCS1 mediates the proteasomal degradation of IRF7, pCAGGS-SOCS1-Myc and pCAGGS-IRF7-Flag were cotransfected into 293T cells or DEFs and treated with the proteasome inhibitor (MG132) to examine the expression level of IRF7 protein. It was found that compared with that in the dimethyl sulfoxide (DMSO) treatment group, the IRF7 protein level in 293T cells or DEFs in the MG132 treatment group was replenished (Fig. 4E and F). Therefore, SOCS1 mediates the proteasomal degradation of IRF7.
Furthermore, the immunoprecipitation test found that when SOCS1 was overexpressed, the ubiquitination level of IRF7 increased (Fig. 4G). However, what type of ubiquitination modification does SOCS1 induce on IRF7? We used the ubiquitin mutant plasmids HA-Ub-K63 (only the lysine at position 63 was retained, and the lysines at the remaining positions were mutated to arginine) and HA-Ub-K48 (only the lysine at position 48 was retained, and the lysines at the remaining positions were mutated to arginine) to identify the type of ubiquitination that SOCS1 induced on IRF7. The results showed that compared with the case with the control group, when HA-Ub-K48 was cotransfected with IRF7 and SOCS1, IRF7 was significantly degraded (Fig. 4H). However, in the group in which HA-Ub-K63 was cotransfected with IRF7 and SOCS1 and MG132 treatment was used, the expression of IRF7 was significantly increased (Fig. 4H). Therefore, SOCS1 mediated the K48-linked polyubiquitination of IRF7. Taken together, the above-described experimental results indicate that SOCS1 mediates K48-linked ubiquitination and proteasomal degradation of IRF7.
SOCS1 mainly undergoes K48-linked polyubiquitination.
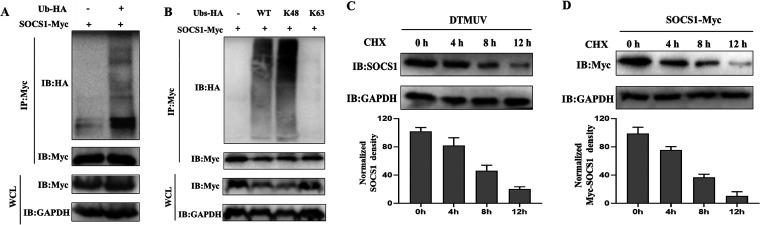
As shown in Fig. 4H, we found that SOCS1 was degraded when pCAGGS-SOCS1-Myc was cotransfected with HA-Ub-K48, so we speculated that SOCS1 might also undergo ubiquitination-mediated degradation. To test this conjecture, we first cotransfected pCAGGS-SOCS1-Myc with ubiquitin plasmids and then performed immunoprecipitation experiments with anti-Myc Ab. The results showed that SOCS1 was indeed ubiquitinated compared with the case with the control group (Fig. 5A). What type of ubiquitination modification mainly occurs on SOCS1? We cotransfected pCAGGS-SOCS1-Myc with HA-Ub-K48 or HA-Ub-K63 and found that SOCS1 mainly underwent K48-linked ubiquitination (Fig. 5B). Since the K48-linked ubiquitination modification is primarily associated with protein degradation events, we transfected pCAGGS-SOCS1-Myc into 293T cells and treated them with CHX for various time periods. The results showed that both endogenous and exogenous SOCS1 were downregulated over time (Fig. 5C and D). Therefore, SOCS1 mainly undergoes K48-linked polyubiquitination, and this ubiquitination modification leads to the degradation of SOCS1.
FIG 5.
SOCS1 mainly undergoes K48-linked polyubiquitination. (A) pCAGGS-SOCS1 (1,500 ng/well) and HA-Ub (1,500 ng/well) were cotransfected into 293T cells for 36 h. Immunoprecipitation of cell lysates was detected with anti-Myc, and then anti-HA was used for IB to detect ubiquitinated SOCS1. (B) HA-Ub-K48 and HA-Ub-K63 or HA-Ub (1,500 ng/well) with pCAGGS-SOCS1 (1,500 ng/well) were cotransfected into 293T cells for 36 h. Immunoprecipitation of cell lysates was detected with anti-Myc, and then anti-HA was used for IB to detect ubiquitinated SOCS1. (C) After infection of DEFs with DTMUV for 36 h, the cells were treated with CHX (200 μg/mL). Then the cells were collected at different time points (0 h, 4 h, 8 h, and 12 h). Western blotting was used to detect endogenous SOCS1 protein level. (D) pCAGGS-SOCS1 (2,000 ng/well) was transfected into 293T cells for 36 h. Cells were collected by treatment with CHX (200 μg/mL) for different time periods (0 h, 4 h, 8 h, and 12 h). The protein level of SOCS1 was detected by Western blotting. The SOCS1 protein densitometric values were quantified and analyzed. Each experiment was repeated three times. The data are the means ± SEM (n = 3).
JOSD1 binds to SOCS1 and removes its K48-linked polyubiquitination.
In previous studies, we found that the expression of SOCS1 was upregulated during DTMUV infection and SOCS1 would undergo K48-linked polyubiquitination, so we speculated that there might be deubiquitinating enzymes to mediate the deubiquitination of SOCS1 during DTMUV infection. It has been previously reported that human JOSD1 can mediate the deubiquitination of SOCS1. Is there a similar situation in ducks? To test this, we used anti-Flag and anti-Myc Ab immunoprecipitation experiments and found that SOCS1 and JOSD1 could bind directly (Fig. 6A and B). Further screening revealed that the SH2 domain of SOCS1 was mainly involved in the binding with JOSD1 (Fig. 6C). If JOSD1 could attenuate K48-linked ubiquitination of SOCS1, could JOSD1 upregulate SOCS1 expression? After cotransfecting the gradient dose of pCAGGS-JOSD1-Flag or pCAGGS with a certain amount of pCAGGS-SOCS1-Myc, we found that the expression of exogenous SOCS1 protein was upregulated with the increase of JOSD1 transfection dose (Fig. 6D). Similarly, the expression of endogenous SOCS1 protein was upregulated with the increase of JOSD1 transfection dose during DTMUV infection (Fig. 6E). To further verify the above conclusions, we cotransfected pCAGGS-JOSD1-Flag and pCAGGS-SOCS1-Myc into DEFs and treated them with CHX for different periods. It was found that the half-life of SOCS1 protein in the cotransfection with the JOSD1 group was significantly longer than that in the control group (Fig. 6F). In addition, during DTMUV infection, the half-life of endogenous SOCS1 was also prolonged under JOSD1 overexpression (Fig. 6G). Therefore, JOSD1 could indeed stabilize SOCS1 expression. Could JOSD1 remove the K48-linked polyubiquitin chain of SOCS1? As shown in Fig. 6H, the results showed that the ubiquitination level of SOCS1 in the JOSD1 overexpression group was significantly downregulated compared with that in the control group. In addition, the ubiquitination level of SOCS1 treated with MG132 in the shRNA-NC group (control group for shRNA-JOSD1) was significantly higher than that in the DMSO treatment group (Fig. 6I). Under the same DMSO or MG132 treatment, when JOSD1 was knocked down, the ubiquitination level of SOCS1 was significantly higher than that in the shRNA-NC group (Fig. 6I). Collectively, the results show that JOSD1 upregulates SOCS1 expression by removing K48-linked polyubiquitin chains of SOCS1.
FIG 6.
JOSD1 binds to SOCS1 and removes its K48-linked polyubiquitination. (A and B) pCAGGS-JOSD1-Flag (1,500 ng/well) and pCAGGS-SOCS1-Myc (1,500 ng/well), pCAGGS-SOCS1-Myc and pCAGGS (1,500 ng/well), and pCAGGS-JOSD1-Flag and pCAGGS were cotransfected into 293T cells for 36 h. Immunoprecipitation of cell lysates was detected with anti-Myc or anti-Flag, and then anti-Flag and anti-Myc were used for IB. (C) The wild-type and mutant plasmids of SOCS1 were transfected together with the JOSD1-Flag plasmid into 293T cells for 36 h. The interaction between SOCS1 and JOSD1 was detected by anti-Flag Ab for IP, and then anti-Myc was used for IB detection. (D) pCAGGS-SOCS1-Myc (1,000 ng/well) and gradient doses of pCAGGS-JOSD1-Flag (500, 1,000, and 1,500 ng/well) were cotransfected into 293T cells for 36 h, and then the cell samples were collected. The protein level of exogenous SOCS1 was detected by Western blotting. (E) Gradient doses of pCAGGS-JOSD1-Flag (500, 1,000, and 1,500 ng/well) were transfected into DEFs for 24 h. After DEFs were infected with DTMUV at an MOI of 1 for 36 h, cell samples were collected. Western blotting was used to detect endogenous SOCS1 protein level. (F) pCAGGS-SOCS1-Myc (1,500 ng/well) and pCAGGS-JOSD1-Flag (1,000 ng/well) were transfected into 293T cells for 36 h. Cells were collected by treatment with CHX (200 μg/mL) for different time periods (0 h, 4 h, 8 h, and 12 h). The protein level of exogenous SOCS1 was detected by Western blotting. (G) pCAGGS-JOSD1-Flag or pCAGGS (1,000 ng/well) was transfected into DEFs for 24 h. Then DEFs were infected with DTMUV for 36 h. Cells were collected by treatment with CHX (200 μg/mL) for different time periods (0 h, 4 h, 8 h, and 12 h). The protein level of endogenous SOCS1 was detected by Western blotting. (H) HA-Ub-K48 (1,000 ng/well), pCAGGS-SOCS1-Myc (1,000 ng/well) and HA-Ub-K48, pCAGGS-JOSD1-Flag (1,000 ng/well), and pCAGGS-SOCS1-Myc and HA-Ub-K48 were cotransfected into 293T cells for 36 h. Immunoprecipitation of cell lysates was detected with anti-Myc, and then anti-HA was used for IB to detect ubiquitinated SOCS1. (I) HA-Ub-K48 (800 ng/well) and pCAGGS-SOCS1-Myc (1,000 ng/well) with shRNA-JOSD1 (1,200 ng/well) or shRNA-NC (1,200 ng/well) were cotransfected into DEFs cells for 36 h. Cells were treated with DMSO or MG132 for 6 h. Immunoprecipitation of cell lysates was detected with anti-Myc, and then anti-HA was used for IB to detect ubiquitinated SOCS1. The SOCS1 protein densitometric values were quantified and analyzed. Each experiment was repeated three times. The data are the means ± SEM (n = 3).
JOSD1 inhibits type I IFN production through SOCS1.
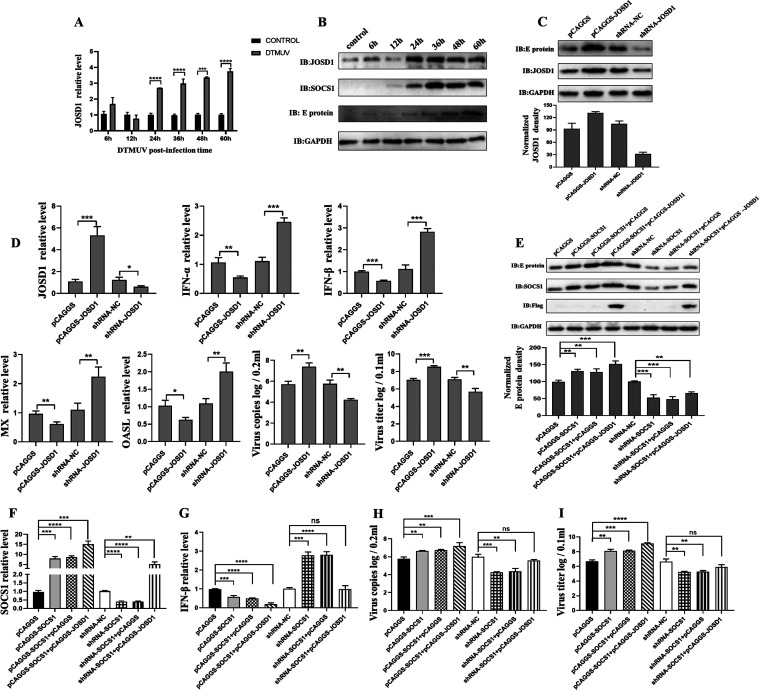
Previously, we found that JOSD1 could mediate the deubiquitination of SOCS1 to stabilize SOCS1, so did JOSD1 regulate type I IFN production through SOCS1 during DTMUV infection, thereby affecting virus replication? First, we examined the expression changes of JOSD1 mRNA level and protein level during DTMUV infection. The results showed that JOSD1 expression was upregulated over time during DTMUV infection (Fig. 7A and B), which also explained why SOCS1 was upregulated during DTMUV infection despite ubiquitination-mediated degradation. Next, the effect of JOSD1 on viral E protein expression was examined by knockdown or overexpression of JOSD1. The results showed that the expression of viral E protein was upregulated when JOSD1 was overexpressed, while the expression of viral E protein was downregulated when JOSD1 was knocked down (Fig. 7C), which indicated that JOSD1 promoted DTMUV replication. The effects of knockdown or overexpression of JOSD1 on type I IFN, ISGs, viral copy number, and viral titer were further examined. The data showed that the mRNA levels of type I IFN, MX, and OASL were all downregulated when JOSD1 was overexpressed, while the mRNA levels of type I IFN, MX, and OASL were all upregulated when JOSD1 was knocked down (Fig. 7D). In contrast, the viral copy number and viral titer were upregulated when JOSD1 was overexpressed, while the viral copy number and viral titer were downregulated when JOSD1 was knocked down (Fig. 7D). Therefore, JOSD1 promotes viral replication by affecting the production of type I IFN and downstream ISGs.
FIG 7.
JOSD1 inhibits type I IFN production through SOCS1. (A and B) After infection of DEFs with DTMUV at an MOI of 1 for different time periods (6 h, 12 h, 24 h, 36 h, 48 h, and 60 h), the cells were collected. The mRNA level of JOSD1 (n = 3) was detected by RT-qPCR. The protein level of JOSD1 (n = 3) was detected by Western blotting. (C) shRNA-NC, shRNA-JOSD1, pCAGGS, and pCAGGS-JOSD1 (2,500 ng/well) were transfected into DEFs for 24 h. Then DEFs were infected with DTMUV for 36 h. The protein levels of the viral E protein and JOSD1 (n = 3) were detected by Western blotting. (D) shRNA-NC, shRNA-JOSD1, pCAGGS, and pCAGGS-JOSD1 (1,250 ng/well) were transfected into DEFs for 24 h. Then DEFs were infected with DTMUV for 36 h. The mRNA levels of JOSD1, IFN-α, IFN-β, MX, and OASL and the viral copy number (n = 3) were detected by RT-qPCR. The cells were harvested for titer detection. pCAGGS, pCAGGS-SOCS1, pCAGGS-SOCS1 and pCAGGS, pCAGGS-SOCS1 and pCAGGS-JOSD1-Flag, shRNA-NC, shRNA-SOCS1, shRNA-SOCS1 and pCAGGS, and shRNA-SOCS1 and pCAGGS-JOSD1-Flag (625 ng/well) were transfected into DEFs for 24 h, respectively. Then DEFs were infected with DTMUV for 36 h. RT-qPCR (F, G, and H) and Western blotting (E) were used to detect the mRNA or protein levels of IFN-β, SOCS1, and the viral E protein (n = 3). The cells were harvested for titer detection (I). The viral E protein densitometric values in panel E were quantified and analyzed. Each experiment was repeated three times. The data are the means ± SEM (n = 3). The significant differences between the groups were analyzed by t test. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To verify that JOSD1 affects type I IFN production through SOCS1 and promotes virus replication. JOSD1 was overexpressed in the case of knockdown or overexpression of SOCS1 and the changes in the mRNA level of type I IFN, the expression of viral E protein, and the viral titer were observed. The results showed that overexpression of SOCS1 inhibited IFN-β production, while cotransfection of JOSD1 with SOCS1 enhanced the inhibitory effect of SOCS1 on IFN-β production (Fig. 7F and G). IFN-β mRNA levels were upregulated upon SOCS1 knockdown, whereas IFN-β production was downregulated when JOSD1 was cotransfected with shRNA-NC compared with that in the control group (Fig. 7F and G). On the contrary, viral E protein expression, viral copy number, and viral titer were upregulated when SOCS1 was overexpressed, and viral E protein, viral copy number, and viral titer were all downregulated when SOCS1 was knocked down (Fig. 7E, H and I). Notably, when JOSD1 and SOCS1 were cotransfected, viral E protein expression, viral copy number, and viral titer were upregulated significantly compared with those in the control group (Fig. 7E, H, and I). Therefore, JOSD1 antagonizes type I IFN production by stabilizing SOCS1 to promote viral replication.
DISCUSSION
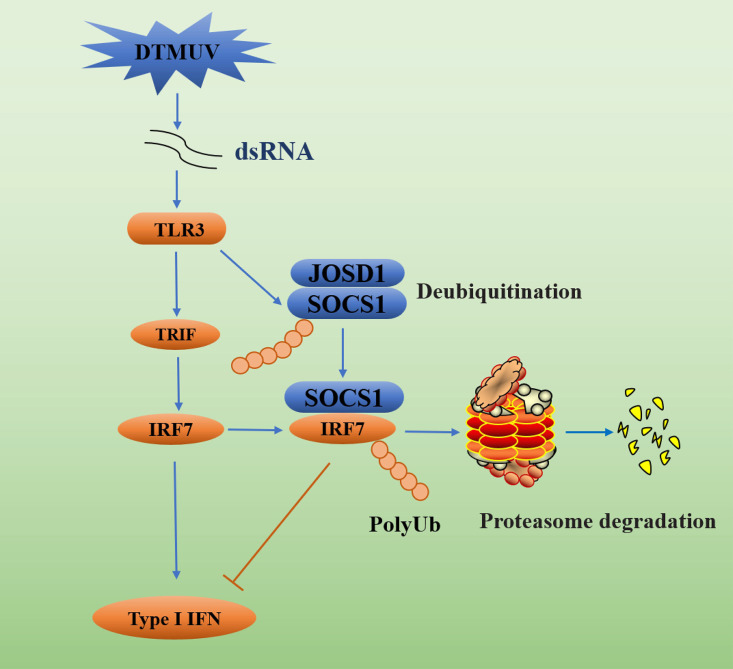
Collectively, we found that activation of TLR3 signaling during DTMUV infection promoted SOCS1 production. However, upregulated SOCS1 inhibited IRF7-mediated type I IFN production by mediating IRF7 ubiquitination and proteasomal degradation to promote viral replication. Furthermore, we found that SOCS1 itself undergoes K48-linked polyubiquitination and degradation. Further exploration revealed that duck JOSD1 stabilized SOCS1 expression by deubiquitinating SOCS1, ultimately inhibiting the production of type I IFN and downstream ISGs (Fig. 8).
FIG 8.
DTMUV antagonizes IRF7-mediated type I IFN signaling through the SOCS1-JOSD1-IRF7 signaling axis. Although activation of TLR3-mediated type I IFN production by DTMUV infection inhibits viral replication, activation of TLR3 signaling also upregulates SOCS1, and JOSD1 stabilizes SOCS1 expression by deubiquitinating SOCS1. Ultimately, SOCS1 inhibits type I IFN production and promotes viral replication by mediating IRF7 ubiquitination and proteasomal degradation.
In previous studies, we found that DTMUV infection could cause the upregulation of SOCS1 and SOCS1 could inhibit the production of type I IFN during DTMUV infection, but the specific mechanism is still unclear. Studies have shown that the activation of TLR signals can induce the upregulation of SOCS1. For example, in human pDCs, TLR7 can quickly boost the upregulation of SOCS1 and SOCS3 after being stimulated by natural or synthetic ligands (32). In addition, in macrophages, stimulation by the TLR ligand CpG-DNA or lipopolysaccharide (LPS) can promote the expression of SOCS1 and SOCS3, and their expression prevents the lethal LPS response from harming the host (34–36). Similarly, TLR-mediated activation of IFNAR-STAT1 signaling promotes the production of SOCS1 and SOCS3 by inducing the upregulation of TAM (Tyro3, Axl, and Mer) receptor tyrosine kinases, which ultimately antagonize TLR signaling and cytokine production (37). However, whether there is a similar mechanism during DTMUV infection has not been reported. In our study, we found that DTMUV could induce TLR3 upregulation (Fig. 1D) and that stimulation of TLR3 signal instead of type I IFN stimulation promoted the expression of SOCS1 (Fig. 1B, C, and E to H). Furthermore, upregulation of SOCS1 inhibited DTMUV-induced type I IFN production (Fig. 1I to L).
Members of the SOCS family are well-known negative regulators of cytokines, especially for playing an indispensable role in the negative regulation of TLR signaling (38–40). The purpose of the production of SOCS protein by the host is to prevent the excessive immune response from causing damage to the body. However, with the evolution of pathogens, many viruses such as PRRSV, IAV, herpes simplex virus (41), Japanese encephalitis virus (42), Zika virus (ZIKV), and hepatitis C virus (43) all use SOCS protein to promote their proliferation. The above examples also prove the crucial negative regulatory role of SOCS protein in innate immunity. There have been many reports about viruses using SOCS1 to negatively regulate the production of IFN. For example, during respiratory syncytial virus infection, its nonstructural protein 1 suppresses the type I IFN-mediated antiviral response and chemokine production by upregulating SOCS1 and SOCS3 (44). There are similar reports for flaviviruses. ZIKV infection leads to the upregulation of SOCS1 and SOCS3 expression, thereby repressing RLR-mediated secretion of type I and III IFN. This indicates that during ZIKV infection, SOCS protein may negatively regulate the innate antiviral immune response to promote virus replication (45). SOCS1 usually inhibits the phosphorylation of JAK through its KIR and negatively regulates JAK/STAT signals. When SOCS1 is overexpressed, it will reduce the phosphorylation level of JAK1, TYK2, and STAT1, thereby inhibiting the antiviral response induced by type I IFN (46). In recent years, it has been newly discovered that SOCS1 can also negatively regulate innate immune signaling by acting as an E3 ubiquitin ligase, and the IRF family appears to be a new target of SOCS1. For example, human T cell leukemia virus type 1 infection can induce SOCS1, while SOCS1 ubiquitinates IRF3 to cause the proteasomal degradation of IRF3 and negatively regulate IFN-β signaling to promote virus proliferation (47). It has also been found in human pDCs that both SOCS1 and SOCS3 play the role of endogenous E3 ubiquitin ligase to mediate the ubiquitination and proteasomal degradation of IRF7, thereby negatively regulating the production of type I IFN mediated by TLR7 (32). However, the relevant mechanism of duck SOCS1 regulation of IFN during DTMUV infection is still undetermined.
IRF3/7 are essential factors that regulate the production of type I IFN, and both RLR and TLR signal activation will converge on IRF3/7 to promote the transcription of type I IFN. There are two main mechanisms for destroying IRF3/7-mediated type I IFN production: (i) repression of the dimerization and nuclear translocation of IRF3/7 and (ii) direct labeling of IRF3/7 through ubiquitination and mediation of their proteasomal degradation to terminate the type I IFN signal quickly. For example, peptidyl-prolyl isomerase Pin1 (48), E3 ubiquitin ligase HOIL-1 (49), and transcription factor FoxO1 (50) promote ubiquitination and proteasomal degradation of IRF3 to terminate IRF3-mediated type I IFN signaling quickly. In addition, E3 ubiquitin ligases such as RAUL, triple motif 21 (TRIM21), and TRIM25 can promote the ubiquitination of IRF3 or IRF7 and subsequent proteasomal degradation, thereby negatively regulating the innate immunity mediated by TLR signals (51–54). However, some studies have found that due to the lack of IRF3 in poultry, the replacement of IRF3 with IRF7 plays a crucial role in producing type I IFN (11). Therefore, we started with IRF7 and found that DTMUV infection resulted in downregulation of IRF7 mRNA and protein levels at 60 h (Fig. 2A and B). But the mechanism of IRF7 downregulation during DTMUV infection is unknown. In this study, we found that SOCS1 can inhibit IRF7-mediated type I IFN production to promote DTMUV replication (Fig. 2E to K). Further mechanistic studies found that SOCS1 bound to IRF7 through its SH2 domain (Fig. 3). Additionally, SOCS1 led to the downregulation of IRF7 expression during DTMUV infection by promoting K48-linked ubiquitination and subsequent proteasomal degradation of IRF7 (Fig. 4).
In addition, we found that when SOCS1 was cotransfected with HA-K48 plasmid, the protein level of SOCS1 was downregulated (Fig. 4H). And some studies have shown that SOCS1 does undergo K48-linked polyubiquitination modification (28), so why is SOCS1 upregulated during DTMUV infection? Is there a deubiquitination enzyme targeting duck SOCS1 for deubiquitination? And is there a similar situation in poultry? The results indicated that duck SOCS1 indeed bound to K48-linked polyubiquitin chains (Fig. 5A and B). Further studies found that duck JOSD1 directly bound to SOCS1 and mediated its deubiquitination to stabilize the expression of SOCS1 (Fig. 6). In addition, JOSD1 promoted viral replication by inhibiting the production of type I IFN and downstream ISGs through SOCS1 (Fig. 7). But what enzymes mediate the ubiquitination of SOCS1? The SH2 domain of SOCS1 is involved in binding JOSD1 and IRF7 at the same time. Is there a competitive relationship between them? This needs further clarification. Although we found for the first time in this study that JOSD1 stabilizes the expression of SOCS1 by deubiquitination, thereby inhibiting type I IFN production and promoting viral replication, the upregulation mechanism of JOSD1 during DTMUV infection is still a research gap. However, the upregulation of JOSD1 may be related to the induction of type I IFN or the activation of the innate immune pathway, and it is also possible that the entry of viral components into the body induces the upregulation of JOSD1, but the specific mechanism remains to be further explored. Furthermore, although our study elucidates the molecular mechanism by which the JOSD1-SOCS1-IRF7 pathway negatively regulates type I IFN production in DTMUV-infected cells, the specific role of this pathway in viral pathogenesis needs to be further determined in vivo.
In conclusion, our study revealed a novel mechanism by which DTMUV antagonizes IRF7-mediated type I IFN via the JOSD1-SOCS1-IRF7 negative-feedback signaling axis (Fig. 8). This also suggests that the JOSD1-SOCS1-IRF7 signaling axis may be a possible therapeutic target for future DTMUV vaccine development.
MATERIALS AND METHODS
Ethics approval and consent to participate.
The Animal Ethics Committee of Sichuan Agricultural University (approval no. 2015-016) approved the use of duck embryos in this study. Duck embryos were used in accordance with the National Institutes of Health’s guidelines for performing animal experiments.
Cells and virus.
Primary duck embryo fibroblasts (DEFs) were obtained from duck embryos aged 9 to 12 days. We used Dulbecco’s modified Eagle’s medium (DMEM) (12800-058; Gibco, USA) supplemented with 10% newborn calf serum (Gibco) to cultivate DEFs for 12 to 24 h at 37°C and 5% CO2. We used RPMI 1640 (31800-014; Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) to cultivate human embryonic kidney (HEK) 293T cells for 12 to 24 h at 37°C and 5% CO2. The Institute of Preventive Veterinary Medicine, Sichuan Agricultural University, provided the DTMUV CQW1 strain (GenBank accession no. KM233707.1). The viral titer of the DTMUV CQW1 strain was 6.1 × 10−6 50% tissue culture infective doses (TCID50)/100 μL.
Plasmids and reagents.
pCAGGS-IRF7-Flag, pCAGGS-IFN-β-Flag, shRNA-IRF7, HA-ubiquitin (HA-Ub), NF-κB-Luc, IFN-β-Luc, pRL-TK, and shRNA-TLR3 were provided by the Institute of Preventive Veterinary Medicine, Sichuan Agricultural University. shRNA-SOCS1 and shRNA-JOSD1 were synthesized by GenePharma (Shanghai, China). HA-Ub-K48 and HA-Ub-K63 were purchased from Addgene. According to the sequence published in GenBank and the pCAGGS plasmid of our laboratory, we designed specific primers for the overexpression vectors of duck SOCS1, JOSD1, IFN-α, TLR3, and SOCS1 domain deletion mutants (as shown in Table 1). Then the duck template cDNA was used to amplify the corresponding gene. The amplified SOCS1, JOSD1, IFN-α, TLR3, and SOCS1 domain deletion mutants were cloned into the pCAGGS vector by restriction enzyme digestion and ligation. The double enzyme digestion test verifies whether the plasmid is constructed successfully. The integrity and fidelity of inserts were verified by sequencing (Wangke, China). The restriction enzymes EcoRI, KpnI, XhoI, and SmaI were purchased from TaKaRa (Japan). Xuedong Wu (Sichuan Agricultural University, China) kindly provided the anti-E monoclonal antibody. The primary antibodies used in this study were anti-Myc (71D10; CST, USA), anti-Flag (D6W5B; CST), anti-HA (MBL, Japan), anti-β-actin (Proteintech, China), anti-GAPDH (Proteintech, China), and anti-JOSD1 (ABclone, Wuhan, China). Anti-duck IRF7 polyclonal antibody was developed by ABclone. Anti-duck SOCS1 polyclonal antibody was developed in our laboratory. HRP-conjugated secondary antibody was purchased from Proteintech.
TABLE 1.
Primers used to amplify duck SOCS1, JOSD1, TLR3, and IFN-α
| Primer name | Sequence (5′–3′) |
|---|---|
| pCAGGS-SOCS1-Myc F | CCGGAATTCGCCACCATGGAGCAGAAACTCATCTCTGAAGAGGATCTGATGGTAGCGCACAGCAAG |
| pCAGGS-SOCS1-Myc R | CGGGGTACCTTACAGATCCTCTTCAGAGATGAGTTTCTGCTCGATCTGAAATGGGAATGATTTCAGG |
| pCAGGS-JOSD-Flag F | CCGGAATTCGCCACCATGGGCATGAGTTGCGTGCCATGGAAAG |
| pCAGGS-JOSD-Flag R | CCGCTCGAGTTACTTATCGTCGTCATCCTTGTAATCCACGTCAGCTCGCCAGCT |
| pCAGGS-IFNα-Flag F | CCGGAATTCGCCACCATGGCTGGGCCATCAGCC |
| pCAGGS-IFNα-Flag R | CCGGGTACCTTACTTATCGTCGTCATCCTTGTAATCGCGCATGGTGCGGGTGAG |
| pCAGGS-TLR3-Flag F | CCGCCCGGGGCCACCATGGGAAGTGATATTCTT |
| pCAGGS-TLR3-Flag R | CCGCTCGAGTTACTTATCGTCGTCATCCTTGTAATCCCGTGCTTTACTATTAGA |
| pCAGGS-ΔKIR&ESS-Myc F | CAGAGCAACACGCACGGCTTTTACTGGGGACCT |
| pCAGGS-ΔKIR&ESS-Myc R | TCCCCAGTAAAAGCCGTGCGTGTTGCTCTGTGC |
| pCAGGS-ΔSH2 75-124-Myc F | CTGGATGCCTGTGGCAACTTTCAGACTGGGCGT |
| pCAGGS-ΔSH2 75-124-Myc R | CCCAGTCTGAAAGTTGCCACAGGCATCCAGCAA |
| pCAGGS-ΔSH2 125-166-Myc F | ACCAGCATCCGGATAGTGCAGCCCTTGCAGGA |
| pCAGGS-ΔSH2 125-166-Myc R | CTGCAAGGGCTGCACTATCCGGATGCTGGTGGG |
| pCAGGS-ΔSH2 74-152-Myc F | TTGCTGGATGCCTGTTCCCCGAGGAAGGTACTGGTT |
| pCAGGS-ΔSH2 74-152-Myc R | TACCTTCCTCGGGGAACAGGCATCCAGCAAGCT |
| pCAGGS-ΔSH2 75-166-Myc F | CTGGATGCCTGTGGCGTGCAGCCCTTGCAGGA |
| pCAGGS-ΔSH2 75-166-Myc R | CTGCAAGGGCTGCACGCCACAGGCATCCAGCAA |
| pCAGGS-ΔSOCS BOX-Myc R | CGGGGTACCTTACAGATCCTCTTCAGAGATGAGTTTCTGCTCACGGACTTTGCGCAGGG |
Type I IFN collection and stimulation.
Since we were unable to purchase commercial duck IFN-α or IFN-β, we collected the supernatant of cells transfected with pCAGGS-IFNβ to stimulate DEFs. The specific operation was as follows. When the cell confluence reached about 70%, the IFN-β overexpression plasmids were transfected into 293T cells for 36 h, and the cell supernatant was collected after freezing and thawing three times. The transfection step was carried out in accordance with the Lipofectamine 3000 transfection reagent instructions. The collected supernatant was used to stimulate DEFs with a gradient dose for 24 h, and then DEF samples were collected.
RNA isolation and RT-qPCR analysis.
Based on the manufacturer’s instructions (TaKaRa, Japan), the total RNA of the cells in each well was separately extracted with RNAiso plus reagent. After the total RNA is prepared, we first used PrimeScript RT master mix (TaKaRa, Japan) to reverse transcribe the RNA into a cDNA strand according to the manufacturer’s instructions. Then quantitative real-time PCR (RT-qPCR) (Bio-Rad, USA) was used to detect the mRNA level of the target gene. The primers used in this study are listed in Table 2. β-Actin was used as an internal reference for the target gene. All experiments included three sets of repetitions. The relative expression of the target gene was calculated by the threshold cycle (2−ΔΔCT) method.
TABLE 2.
RT-qPCR primers used in the present study
| Primer name | Sequence (5′–3′) |
|---|---|
| SOCS1 F | CTTGCTGGATGCCTGTGG |
| SOCS1 R | CTGCGTGCTGTCCCTGAT |
| JOSD1 F | ATTTACCACGAGAAGCAG |
| JOSD1 R | TCACATCATAGTTCCCATT |
| DTMUV-E F | AATGGCTGTGGCTTGTTTGG |
| DTMUV-E R | GGGCGTTATCACGAATCTA |
| β-actin F | CCGGGCATCGCTGACA |
| β-actin R | GGATTCATCATACTCCTGCTTTGCT |
| IFN-α F | CCACCATGCCTGGGCCATCAG |
| IFN-α R | AGGAGAAGGCGTTGGCGGGAG |
| IFN-β F | CGCCTGGACACGCTAATA |
| IFN-β R | AGCTGGTGCCTCTTGCTC |
| IRF7 F | CGCCACCCGCCTGAAGAAGT |
| IRF7 R | CTGCCCGAAGCAGAGGAAGAT |
| TLR3 F | ATGTCATGCAAACCTGACCA |
| TLR3 R | CCAGGGTCTTGAAAGGATCA |
| MX F | CCTAAGGGAGAAAGGACACT |
| MX R | GACCACGACACTTCACAACC |
| OASL F | GCAGGCAGAGGCTGTCGTTC |
| OASL R | ATGGACTCGCCGTTGGAGGA |
Transfection and luciferase activity assays.
After inoculation of DEFs into 24-well plates for 12 to 24 h, transfection was performed when the cell confluence reached about 70%. Lipofectamine 3000 (Invitrogen) was used to combine 400 ng of luciferase reporter plasmid (NF-κB-Luc or IFN-β-Luc) and 40 ng of Renilla luciferase expression plasmid pRL-TK (Promega) with a gradient dose of empty plasmid pCAGGS or specific expression plasmids (as shown in Table 1) for cotransfection into cells. Then the dual-luciferase detection kit (Promega) was used to detect the reporter gene activity according to the manufacturer's instructions. The ratio of firefly luciferase activity to Renilla luciferase activity is the data obtained.
Virus titer determination.
Specific plasmids were cotransfected into DEFs for 24 h. DEFs were then infected with DTMUV at a multiplicity of infection (MOI) of 1 for 36 h. Then cell cultures were freeze-thawed three times and centrifuged at 2,000 × g for 10 min. The viral contents in the supernatants were titrated using TCID50 in DEFs. Tissue culture wells with a cytopathic effect (CPE) were deemed positive. The titer was calculated on the basis of a previously described method (55).
Indirect immunofluorescence.
DEFs were inoculated into slides in 12-well plates. When the cells reach about 70 to 90% confluence, the designated plasmids were cotransfected into the cells. At 24 h after the plasmid was transfected into the cells, the cells were first fixed with 4% paraformaldehyde at 4°C overnight, then washed three times with phosphate-buffered saline (PBS), and permeabilized with 0.25% Triton X-100 at 4°C for 1 h. After the cells were washed three times with PBS, they were blocked with 5% bovine serum albumin (BSA) diluted in PBS for 1 h and then incubated and washed with the primary antibody and the secondary antibody in turn. Finally, the slides were fixed with 90% glycerol buffer and observed under a microscope (80i; Nikon).
Coimmunoprecipitation and Western blot analysis.
Specific plasmids were cotransfected into 293T cells or DEFs for 36 h. After harvesting of the cells, the cells were lysed with immunoprecipitation (IP) lysis buffer (Thermo Fisher Scientific). For the IP test, 0.5 mg of designated Ab or control IgG was added to 0.55 mL of the lysate and incubated for 12 to 24 h at 4°C. After incubation, protein G magnetic beads (Bio-Rad) were added to pull down at 4°C for 4 to 6 h and washed three times with precooled PBS with Tween 20 (PBST). Next, SDS-PAGE was performed to separate equal amounts of cell samples, the separated protein was transferred to a polyvinylidene fluoride membrane, and then the primary antibody and secondary antibody were incubated sequentially; ECL reagent (Bio-Rad) was used to visualize the target protein.
Ubiquitination assay.
After 36 h of transient cotransfection of the specified plasmid into the 293T cells or DEFs, the cells were treated with the proteasome inhibitor MG132 for 6 h before harvesting. The total protein was purified with IP lysis buffer containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and denatured with 1% SDS at 95°C for 5 min to dissociate any noncovalently bound proteins. Part of the processed cell lysate was taken as a positive control, and a designated Ab was added to the remaining part. After IP, the magnetic beads were washed three times with precooled PBST. The magnetic beads were gently eluted with PBS, 5× loading buffer was added to boil for 10 min, and the ubiquitinated protein was detected by Western blotting.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (32172833/31872475), Sichuan Province Program (2022NSFSC0078), and the Program Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (CARS-SVDIP), and the earmarked fund for China Agriculture Research System (CARS-42-17).
Contributor Information
Anchun Cheng, Email: chenganchun@vip.163.com.
Renyong Jia, Email: jiary@sicau.edu.cn.
Bryan R. G. Williams, Hudson Institute of Medical Research
REFERENCES
- 1.Su J, Li S, Hu X, Yu X, Wang Y, Liu P, Lu X, Zhang G, Hu X, Liu D, Li X, Su W, Lu H, Mok NS, Wang P, Wang M, Tian K, Gao GF. 2011. Duck egg-drop syndrome caused by BYD virus, a new Tembusu-related flavivirus. PLoS One 6:e18106. 10.1371/journal.pone.0018106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Z, Fu Y, Ji Y, Wei J, Cai X, Zhu Q. 2013. Development and validation of one-step SYBR green real-time RT-PCR for the rapid detection of newly emerged duck Tembusu virus. Avian Dis 57:595–601. 10.1637/10484-010713-Reg.1. [DOI] [PubMed] [Google Scholar]
- 3.Yan L, Yan P, Zhou J, Teng Q, Li Z. 2011. Establishing a TaqMan-based real-time PCR assay for the rapid detection and quantification of the newly emerged duck Tembusu virus. Virol J 8:464. 10.1186/1743-422X-8-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang W, Jiang B, Zeng M, Duan Y, Wu Z, Wu Y, Wang T, Wang M, Jia R, Zhu D, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Liu Y, Zhang L, Yu Y, Pan L, Chen S, Cheng A. 2020. Binding of duck Tembusu virus nonstructural protein 2A to duck STING disrupts induction of its signal transduction cascade to inhibit beta interferon induction. J Virol 94:e01850-19. 10.1128/JVI.01850-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saito T, Gale M, Jr.. 2007. Principles of intracellular viral recognition. Curr Opin Immunol 19:17–23. 10.1016/j.coi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Thompson AJ, Locarnini SA. 2007. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol Cell Biol 85:435–445. 10.1038/sj.icb.7100100. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Lei CQ, Ji Y, Zhou H, Ren Y, Peng Q, Zeng Y, Jia Y, Ge J, Zhong B, Li Y, Wei J, Shu HB, Zhu Q. 2016. Duck Tembusu virus nonstructural protein 1 antagonizes IFN-β signaling pathways by targeting VISA. J Immunol 197:4704–4713. 10.4049/jimmunol.1502317. [DOI] [PubMed] [Google Scholar]
- 8.Wu Z, Zhang W, Wu Y, Wang T, Wu S, Wang M, Jia R, Zhu D, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Liu Y, Zhang L, Yu Y, Pan L, Merits A, Chen S, Cheng A. 2019. Binding of the duck Tembusu virus protease to STING is mediated by NS2B and is crucial for STING cleavage and for impaired induction of IFN-β. J Immunol 203:3374–3385. 10.4049/jimmunol.1900956. [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Luo G, Yang Z, Lin S, Chen S, Wang S, Goraya MU, Chi X, Zeng X, Chen JL. 2016. Avian Tembusu virus infection effectively triggers host innate immune response through MDA5 and TLR3-dependent signaling pathways. Vet Res 47:74. 10.1186/s13567-016-0358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo H, Cheng A, Zhang X, Pan Y, Wang M, Huang J, Zhu D, Chen S, Liu M, Zhao X, Wu Y, Yang Q, Zhang S, Yu Y, Pan L, Tian B, Rehman MU, Chen X, Liu Y, Zhang L, Yin Z, Jing B, Jia R. 2020. DEF cell-derived exosomal miR-148a-5p promotes DTMUV replication by negative regulating TLR3 expression. Viruses 12:94. 10.3390/v12010094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng Y, Zhu W, Ding C, Niu Q, Wang H, Yan Y, Sun J. 2019. IRF7 is involved in both STING and MAVS mediating IFN-β signaling in IRF3-lacking chickens. J Immunol 203:1930–1942. 10.4049/jimmunol.1900293. [DOI] [PubMed] [Google Scholar]
- 12.Ramos HJ, Gale M, Jr.. 2011. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr Opin Virol 1:167–176. 10.1016/j.coviro.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikushima H, Negishi H, Taniguchi T. 2013. The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harbor Symp Quant Biol 78:105–116. 10.1101/sqb.2013.78.020321. [DOI] [PubMed] [Google Scholar]
- 14.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. 2001. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol 19:623–655. 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 15.Honda K, Takaoka A, Taniguchi T. 2006. Type I inteferon [sic] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25:349–360. 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett 441:106–110. 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 17.Magor KE, Miranzo Navarro D, Barber MR, Petkau K, Fleming-Canepa X, Blyth GA, Blaine AH. 2013. Defense genes missing from the flight division. Dev Comp Immunol 41:377–388. 10.1016/j.dci.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang B, Qi ZT, Xu Z, Nie P. 2010. Global characterization of interferon regulatory factor (IRF) genes in vertebrates: glimpse of the diversification in evolution. BMC Immunol 11:22. 10.1186/1471-2172-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cormican P, Lloyd AT, Downing T, Connell SJ, Bradley D, O’Farrelly C. 2009. The avian Toll-like receptor pathway—subtle differences amidst general conformity. Dev Comp Immunol 33:967–973. 10.1016/j.dci.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Elliott J, Johnston JA. 2004. SOCS: role in inflammation, allergy and homeostasis. Trends Immunol 25:434–440. 10.1016/j.it.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimura A, Naka T, Kubo M. 2007. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol 7:454–465. 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 22.Yoshimura A, Ito M, Chikuma S, Akanuma T, Nakatsukasa H. 2018. Negative regulation of cytokine signaling in immunity. Cold Spring Harb Perspect Biol 10:a028571. 10.1101/cshperspect.a028571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piessevaux J, Lavens D, Peelman F, Tavernier J. 2008. The many faces of the SOCS box. Cytokine Growth Factor Rev 19:371–381. 10.1016/j.cytogfr.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Strebovsky J, Walker P, Lang R, Dalpke AH. 2011. Suppressor of cytokine signaling 1 (SOCS1) limits NFkappaB signaling by decreasing p65 stability within the cell nucleus. FASEB J 25:863–874. 10.1096/fj.10-170597. [DOI] [PubMed] [Google Scholar]
- 25.Mansell A, Smith R, Doyle SL, Gray P, Fenner JE, Crack PJ, Nicholson SE, Hilton DJ, O’Neill LA, Hertzog PJ. 2006. Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat Immunol 7:148–155. 10.1038/ni1299. [DOI] [PubMed] [Google Scholar]
- 26.Komander D, Clague MJ, Urbé S. 2009. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10:550–563. 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 27.Seki T, Gong L, Williams AJ, Sakai N, Todi SV, Paulson HL. 2013. JosD1, a membrane-targeted deubiquitinating enzyme, is activated by ubiquitination and regulates membrane dynamics, cell motility, and endocytosis. J Biol Chem 288:17145–17155. 10.1074/jbc.M113.463406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Zhang L, Zhang Y, Zhao P, Qian L, Yuan Y, Liu J, Cheng Q, Xu W, Zuo Y, Guo T, Yu Z, Zheng H. 2017. JOSD1 negatively regulates type-I interferon antiviral activity by deubiquitinating and stabilizing SOCS1. Viral Immunol 30:342–349. 10.1089/vim.2017.0015. [DOI] [PubMed] [Google Scholar]
- 29.Luo X, Chen XX, Qiao S, Li R, Xie S, Zhou X, Deng R, Zhou EM, Zhang G. 2020. Porcine reproductive and respiratory syndrome virus enhances self-replication via AP-1-dependent induction of SOCS1. J Immunol 204:394–407. 10.4049/jimmunol.1900731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uetani K, Hiroi M, Meguro T, Ogawa H, Kamisako T, Ohmori Y, Erzurum SC. 2008. Influenza A virus abrogates IFN-gamma response in respiratory epithelial cells by disruption of the Jak/Stat pathway. Eur J Immunol 38:1559–1573. 10.1002/eji.200737045. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y, Hu Y, Shi B, Zhang X, Wang J, Zhang Z, Shen F, Zhang Q, Sun S, Yuan Z. 2009. HBsAg inhibits TLR9-mediated activation and IFN-alpha production in plasmacytoid dendritic cells. Mol Immunol 46:2640–2646. 10.1016/j.molimm.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 32.Yu CF, Peng WM, Schlee M, Barchet W, Eis-Hübinger AM, Kolanus W, Geyer M, Schmitt S, Steinhagen F, Oldenburg J, Novak N. 2018. SOCS1 and SOCS3 target IRF7 degradation to suppress TLR7-mediated type I IFN production of human plasmacytoid dendritic cells. J Immunol 200:4024–4035. 10.4049/jimmunol.1700510. [DOI] [PubMed] [Google Scholar]
- 33.Huang S, Cheng A, Cui M, Pan Y, Wang M, Huang J, Zhu D, Chen S, Liu M, Zhao X, Wu Y, Yang Q, Zhang S, Ou X, Mao S, Yu Y, Tian B, Liu Y, Zhang L, Yin Z, Jing B, Chen X, Jia R. 2020. Duck Tembusu virus promotes the expression of suppressor of cytokine signaling 1 by downregulating miR-148a-5p to facilitate virus replication. Infect Genet Evol 85:104392. 10.1016/j.meegid.2020.104392. [DOI] [PubMed] [Google Scholar]
- 34.Dalpke AH, Opper S, Zimmermann S, Heeg K. 2001. Suppressors of cytokine signaling (SOCS)-1 and SOCS-3 are induced by CpG-DNA and modulate cytokine responses in APCs. J Immunol 166:7082–7089. 10.4049/jimmunol.166.12.7082. [DOI] [PubMed] [Google Scholar]
- 35.Crespo A, Filla MB, Russell SW, Murphy WJ. 2000. Indirect induction of suppressor of cytokine signalling-1 in macrophages stimulated with bacterial lipopolysaccharide: partial role of autocrine/paracrine interferon-alpha/beta. Biochem J 349:99–104. 10.1042/0264-6021:3490099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagawa R, Naka T, Tsutsui H, Fujimoto M, Kimura A, Abe T, Seki E, Sato S, Takeuchi O, Takeda K, Akira S, Yamanishi K, Kawase I, Nakanishi K, Kishimoto T. 2002. SOCS-1 participates in negative regulation of LPS responses. Immunity 17:677–687. 10.1016/s1074-7613(02)00449-1. [DOI] [PubMed] [Google Scholar]
- 37.Rothlin CV, Ghosh S, Zuniga EI, Oldstone MB, Lemke G. 2007. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell 131:1124–1136. 10.1016/j.cell.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 38.Alston CI, Dix RD. 2019. SOCS and herpesviruses, with emphasis on cytomegalovirus retinitis. Front Immunol 10:732. 10.3389/fimmu.2019.00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinjyo I, Hanada T, Inagaki-Ohara K, Mori H, Aki D, Ohishi M, Yoshida H, Kubo M, Yoshimura A. 2002. SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity 17:583–591. 10.1016/s1074-7613(02)00446-6. [DOI] [PubMed] [Google Scholar]
- 40.Kimura A, Naka T, Muta T, Takeuchi O, Akira S, Kawase I, Kishimoto T. 2005. Suppressor of cytokine signaling-1 selectively inhibits LPS-induced IL-6 production by regulating JAK-STAT. Proc Natl Acad Sci USA 102:17089–17094. 10.1073/pnas.0508517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frey KG, Ahmed CM, Dabelic R, Jager LD, Noon-Song EN, Haider SM, Johnson HM, Bigley NJ. 2009. HSV-1-induced SOCS-1 expression in keratinocytes: use of a SOCS-1 antagonist to block a novel mechanism of viral immune evasion. J Immunol 183:1253–1262. 10.4049/jimmunol.0900570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kundu K, Dutta K, Nazmi A, Basu A. 2013. Japanese encephalitis virus infection modulates the expression of suppressors of cytokine signaling (SOCS) in macrophages: implications for the hosts' innate immune response. Cell Immunol 285:100–110. 10.1016/j.cellimm.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Ma CJ, Ni L, Zhang CL, Wu XY, Kumaraguru U, Li CF, Moorman JP, Yao ZQ. 2011. Cross-talk between programmed death-1 and suppressor of cytokine signaling-1 in inhibition of IL-12 production by monocytes/macrophages in hepatitis C virus infection. J Immunol 186:3093–3103. 10.4049/jimmunol.1002006. [DOI] [PubMed] [Google Scholar]
- 44.Zheng J, Yang P, Tang Y, Pan Z, Zhao D. 2015. Respiratory syncytial virus nonstructural proteins upregulate SOCS1 and SOCS3 in the different manner from endogenous IFN signaling. J Immunol Res 2015:738547. 10.1155/2015/738547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seong RK, Lee JK, Shin OS. 2020. Zika virus-induction of the suppressor of cytokine signaling 1/3 contributes to the modulation of viral replication. Pathogens 9:163. 10.3390/pathogens9030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song MM, Shuai K. 1998. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. J Biol Chem 273:35056–35062. 10.1074/jbc.273.52.35056. [DOI] [PubMed] [Google Scholar]
- 47.Olière S, Hernandez E, Lézin A, Arguello M, Douville R, Nguyen TL, Olindo S, Panelatti G, Kazanji M, Wilkinson P, Sékaly RP, Césaire R, Hiscott J. 2010. HTLV-1 evades type I interferon antiviral signaling by inducing the suppressor of cytokine signaling 1 (SOCS1). PLoS Pathog 6:e1001177. 10.1371/journal.ppat.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saitoh T, Tun-Kyi A, Ryo A, Yamamoto M, Finn G, Fujita T, Akira S, Yamamoto N, Lu KP, Yamaoka S. 2006. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat Immunol 7:598–605. 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- 49.Zhang M, Tian Y, Wang RP, Gao D, Zhang Y, Diao FC, Chen DY, Zhai ZH, Shu HB. 2008. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res 18:1096–1104. 10.1038/cr.2008.277. [DOI] [PubMed] [Google Scholar]
- 50.Lei CQ, Zhang Y, Xia T, Jiang LQ, Zhong B, Shu HB. 2013. FoxO1 negatively regulates cellular antiviral response by promoting degradation of IRF3. J Biol Chem 288:12596–12604. 10.1074/jbc.M112.444794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Y, Hayward GS. 2010. The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity 33:863–877. 10.1016/j.immuni.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Higgs R, J NG, Ben Larbi N, Breen EP, Fitzgerald KA, Jefferies CA. 2008. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J Immunol 181:1780–1786. 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Higgs R, Lazzari E, Wynne C, J NG, Espinosa A, Wahren-Herlenius M, Jefferies CA. 2010. Self protection from anti-viral responses–Ro52 promotes degradation of the transcription factor IRF7 downstream of the viral Toll-Like receptors. PLoS One 5:e11776. 10.1371/journal.pone.0011776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Yan S, Yang B, Wang Y, Zhou H, Lian Q, Sun B. 2015. TRIM35 negatively regulates TLR7- and TLR9-mediated type I interferon production by targeting IRF7. FEBS Lett 589:1322–1330. 10.1016/j.febslet.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 55.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hyg 27:493–497. [Google Scholar]