ABSTRACT
The duration of SARS-CoV-2 genomic RNA shedding is much longer than that of infectious SARS-CoV-2 in most COVID-19 patients. It is very important to determine the relationship between test results and infectivity for efficient isolation, contact tracing, and post-isolation. We characterized the duration of viable SARS-CoV-2, viral genomic and subgenomic RNA (gRNA and sgRNA), and rapid antigen test positivity in nasal washes, oropharyngeal swabs, and feces of experimentally infected Syrian hamsters. The duration of viral genomic RNA shedding is longer than that of viral subgenomic RNA, and far longer than those of rapid antigen test (RAgT) and viral culture positivity. The rapid antigen test results were strongly correlated with the viral culture results. The trend of subgenomic RNA is similar to that of genomic RNA, and furthermore, the subgenomic RNA load is highly correlated with the genomic RNA load.
IMPORTANCE Our findings highlight the high correlation between rapid antigen test and virus culture results. The rapid antigen test would be an important supplement to real-time reverse transcription-RCR (RT-PCR) in early COVID-19 screening and in shortening the isolation period of COVID-19 patients. Because the subgenomic RNA load can be predicted from the genomic RNA load, measuring sgRNA does not add more benefit to determining infectivity than a threshold determined for gRNA based on viral culture.
KEYWORDS: RT-PCR, SARS-CoV-2, Syrian hamsters, genomic and subgenomic RNA, rapid antigen test, viral culture
INTRODUCTION
The Coronavirus Infectious Disease 2019 (COVID-19) pandemic, caused by Severe Acute Respiratory Syndrome Virus 2 (SARS-CoV-2), has lasted for two and a half years and has severely affected global health and the world economy. In most COVID-19 patients, the duration of SARS-CoV-2 genomic RNA (gRNA) shedding is much longer than that of infectious SARS-CoV-2 (1). At present, guidelines for stopping COVID-19 patient isolation are mainly symptom-based, with isolation for 10 to 20 days depending on the condition (2) or viral nucleic acid test-based, with two consecutive negative or inconclusive results on real-time reverse transcription-PCR (RT-PCR) (3, 4). For the former, some asymptomatic carriers (5), immunocompromised patients (6), or symptom-resolved individuals (1) with infectivity may be improperly released from isolation too early. For the latter, a large number of recovered COVID-19 patients may be released from isolation too late (7), occupying a lot of medical resources which could have been used for other COVID-19 patients seeking treatment, because the presence of viral RNA in samples from COVID-19 patients does not necessarily mean viable SARS-CoV-2. While viral culture is the most reliable method for determining infectivity, it is time-consuming and labor-intensive, and it requires biosafety level 3 laboratories, precluding its wide use in most clinical labs. It is important to discover some other suitable index and find a reasonable threshold for infectivity determination. Subgenomic RNA (sgRNA) is an intermediate product of SARS-CoV-2 replication and transcription, and its presence indicates active SARS-CoV-2 replication in patients. The abundance of sgRNA in samples is closely related to the infectivity of SARS-CoV-2 (8) and may be used as diagnostic index for ending COVID-19 patient isolation. Rapid antigen-based testing is widely used for SARS-CoV-2 screening and diagnostics because it is readily available, is inexpensive, and returns results quickly. Some studies have shown that the rapid antigen test (RAgT) aligns better with viral culture results compared to RT-PCR (9). Here, we characterize the duration of viable SARS-CoV-2, viral gRNA and sgRNA, and RAgT positivity in different samples, including nasal washes, oropharyngeal swabs, and fresh feces, from experimentally infected Syrian hamsters. We found that RAgT results correlated strongly with viral culture results and that the viral sgRNA load is highly correlated with the gRNA load. Measuring sgRNA does not add more benefit to determining infectivity than a threshold determined for gRNA based on viral culture.
RESULTS
Viral shedding in nasal washes, oropharyngeal swabs, and feces of experimentally infected hamsters.
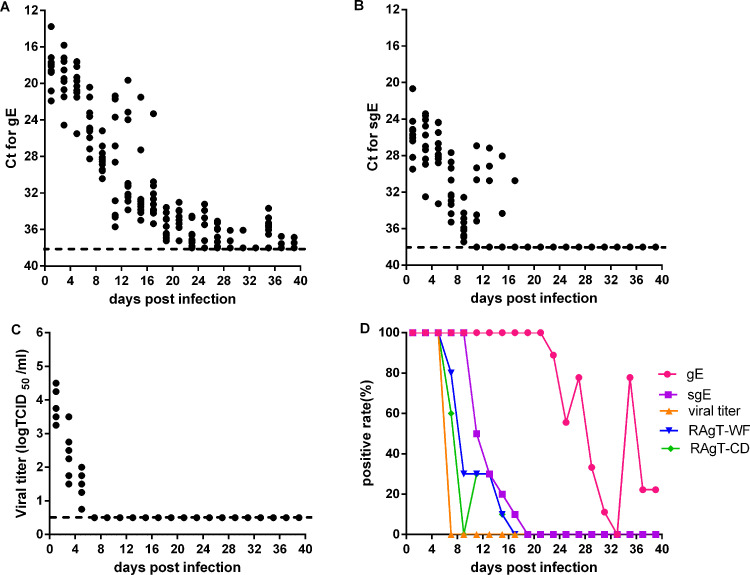
We first analyzed gRNA and sgRNA loads, viral titers, and RAgT results in nasal washes of experimentally infected Syrian hamsters. Nasal washes were collected from hamsters every 2 days for 39 days. The gRNA loads in nasal washes were maintained at a high level during the first five infection days, declined rapidly to a low level during the next week, and were finally maintained at low levels from 3 to 6 weeks (Fig. 1A). The positivity of gRNA was 100% during the first 3 weeks and gradually declined later (Fig. 1D). The sgRNA loads were also maintained at high levels during the first five infection days and then gradually decreased to undetectable levels by day 19 (Fig. 1B). The positivity of sgRNA was 100% during the first nine infection days and rapidly declined to 10% by day 17 (Fig. 1D). The viral titers decreased quickly from high to undetectable levels by day 7 (Fig. 1C). Viral culture positivity was 100% during the first five infection days and decreased to 0% at day 7 (Fig. 1D). The positivity of RAgT results was also 100% during the first five infection days, and later declined rapidly (Fig. 1D). The duration of gRNA shedding in nasal washes was longer than that of sgRNA, and far longer than those of RAgT and viral culture positivity.
FIG 1.
Viral load and test positivity in nasal washes of experimentally infected hamsters. Threshold cycle (CT) values for genomic (gRNA) (A), subgenomic RNA (sgRNA) (B), and viral titers (C) in nasal washes. (D) Test positivity of gRNA, sgRNA, viral culture, and two RAgT assays for SARS-CoV-2. Dotted lines represent the limits of detection of gRNA, sgRNA, and viral titers.
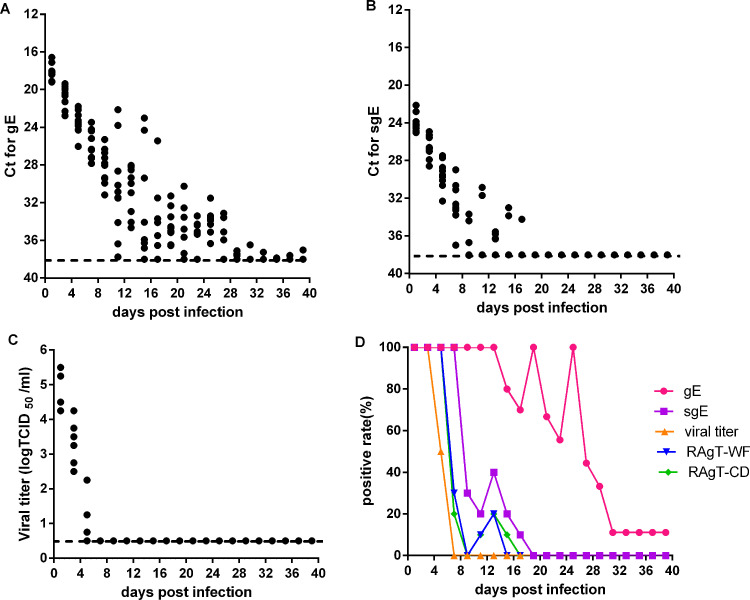
We then analyzed gRNA and sgRNA loads, viral titers, and RAgT results in oropharyngeal swabs of experimentally infected hamsters. Oropharyngeal swabs were collected every 2 days. gRNA loads gradually decreased to low levels during the first 2 weeks, were maintained at low levels during the next 2 weeks, and later decreased to almost undetectable levels (Fig. 2A). The positivity of gRNA was 100% during the first 13 infection days, then oscillated downward to 0% at day 31 (Fig. 2D). sgRNA loads quickly decreased to low levels during the first nine infection days (Fig. 2B), and the sgRNA positivity was 100% prior to day 7 and then quickly declined to 10% at day 17 (Fig. 2D). The viral titers decreased rapidly during the first 7 days (Fig. 2C). The positivity of viral culture was 100% on days 1 and 3, 50% on day 5, and 0% on day 7 (Fig. 2D). The positivity of RAgT was 100% during the first 5 days, then quickly declined to 30% or lower (Fig. 2D). The duration of gRNA shedding in oropharyngeal swabs was longer than that of sgRNA, and much longer than those of RAgT and viral culture positivity.
FIG 2.
Viral load and test positivity in oropharyngeal swabs of experimentally infected hamsters. CT values for gRNA (A) and sgRNA (B), and viral titers (C) in oropharyngeal swabs were shown. (D) The test positivity of gRNA, sgRNA, viral culture, and two RAgT assays for SARS-CoV-2. The dotted lines represented the limit of detection of gRNA, sgRNA and viral titer.
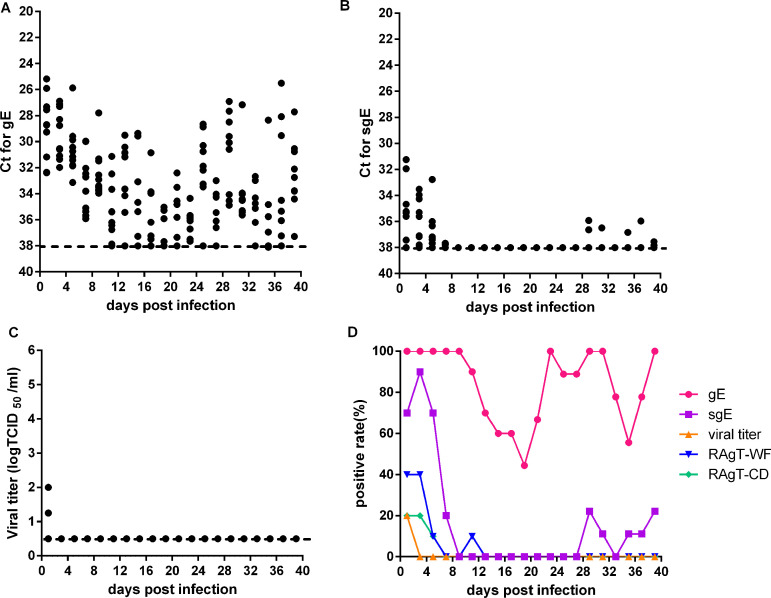
We also analyzed gRNA and sgRNA loads, viral titers, and RAgT results in the feces of experimentally infected hamsters. The gRNA loads in feces were much lower than those in nasal washes and oropharyngeal swabs, and gRNA was always oscillating at moderate levels (Fig. 3A). The positivity of gRNA was 100% during the first nine infection days, then declined during the next 2 weeks, rose again later, and was always higher than 40% (Fig. 3D). sgRNA loads were maintained at moderate levels in the first week and very low levels in the sixth week (Fig. 3B). The positivity of sgRNA was 70% to 90% during the first five infection days, and 10% to 30% during the sixth week (Fig. 3D). Only two fecal samples were positive for viral culture at day 1 (Fig. 3C). The positivity of RAgT was 10% to 40% during the first five infection days (Fig. 3D). The duration of gRNA positivity in feces was longer than that of sgRNA, and much longer than those of RAgT and viral culture positivity.
FIG 3.
Viral loads and test positivity in feces of experimentally infected hamsters. CT values for gRNA (A), sgRNA (B), and viral titers (C) in feces. (D) Test positivity of gRNA, sgRNA, viral culture, and the two RAgT assays for SARS-CoV-2. Dotted lines represent the limits of detection of gRNA, sgRNA and viral titer.
In summary, the duration of gRNA shedding is longer than that of sgRNA, and much longer than those of RAgT and viral culture positivity in nasal washes, oropharyngeal swabs, and feces. The RAgT results were more strongly correlated with viral culture results than with gRNA and sgRNA in experimentally infected hamsters.
Relationship between the CT value of gRNA and the positivity of sgRNA, RAgT, and viral culture.
For nasal washes, when the threshold cycle (CT) was >31, sgRNA of E gene (sgE) was undetectable. When CT < 26, the majority of RAgT results were positive; when CT was between 26 and 30, only a few RAgT results were positive; and when CT >30, the RAgT results were negative. When CT < 22, the majority of viral culture results were positive; when CT was between 22 and 26, only a small number of viral cultures were positive; and when CT > 26, viral cultures were all negative (Table 1). For oropharyngeal swabs, when CT > 30, sgE was undetectable. When CT < 26, the majority of RAgT results were positive; when CT was between 26 and 30, only a small number of RAgT results were positive; and when CT >30, all RAgT results were negative. When CT < 24, most of the virus culture results were positive, and when CT > 24, the viral culture results were all negative (Table 1). For feces, when CT > 32, sgE was undetectable; when CT > 30, the RAgT results were negative; and when CT > 28, the viral culture results were all negative. The incidence of viral culture and RAgT positivity decreased with increasing CT values (Table 1). When RAgT results were negative, viral culture were all negative, and the duration of RAgT positivity was slightly longer than that of viral culture positivity. Based on viral culture results, for nasal washes, Guangzhou Wondfo (WF) RAgT and Wuhan CDiagnosis (CD) RAgT had positive predictive values (PPV) of 61% (30/49) and 68% (30/44), respectively, and all had negative predictive values (NPV) of 100%. For oropharyngeal swabs, WF RAgT and CD RAgT had PPVs of 64% (25/39) and 68% (25/37), respectively, and all had NPVs of 100%. The WF RAgT results were highly concordant with the CD RAgT results. These results further confirmed that RAgT results were strongly correlated with viral culture results. The CT threshold for infectivity varied with specimen type, from 24 in oropharyngeal swabs to 28 in feces.
TABLE 1.
Viral culture and RAgT positivity rates according to CT values of gRNAa
| CT value for gE | Tested real-time RT-PCR assays (n) | Positive test results |
|||
|---|---|---|---|---|---|
| sgRNA | RAgT |
Viral culture | |||
| WF | CD | ||||
| Nasal washes | |||||
| <20 | 21 | 21/21 | 21/21 | 21/21 | 20/21 |
| 20−22 | 13 | 13/13 | 13/13 | 13/13 | 8/13 |
| 22−24 | 6 | 6/6 | 6/6 | 6/6 | 0/6 |
| 24−26 | 7 | 7/7 | 7/7 | 4/7 | 2/7 |
| 26−28 | 5 | 5/5 | 1/5 | 0/5 | 0/5 |
| 28−30 | 8 | 8/8 | 1/8 | 0/8 | 0/8 |
| 30−31 | 3 | 1/3 | 0/3 | 0/3 | 0/3 |
| 31−32 | 3 | 0/3 | 0/3 | 0/3 | 0/3 |
| 32−34 | 24 | 0/24 | 0/24 | 0/24 | 0/24 |
| Oropharyngeal swabs | |||||
| <20 | 13 | 13/13 | 13/13 | 13/13 | 13/13 |
| 20−22 | 6 | 6/6 | 6/6 | 6/6 | 6/6 |
| 22−24 | 12 | 12/12 | 11/12 | 11/12 | 6/12 |
| 24−26 | 8 | 8/8 | 5/8 | 4/8 | 0/8 |
| 26−28 | 10 | 9/10 | 2/10 | 1/10 | 0/10 |
| 28−30 | 12 | 5/12 | 2/12 | 2/12 | 0/12 |
| 30−32 | 9 | 0/9 | 0/9 | 0/9 | 0/9 |
| 32−34 | 15 | 0/15 | 0/15 | 0/15 | 0/15 |
| Feces | |||||
| 24−26 | 4 | 4/4 | 3/4 | 2/4 | 1/4 |
| 26−28 | 10 | 8/10 | 4/10 | 2/10 | 1/10 |
| 28−30 | 18 | 9/18 | 1/18 | 1/18 | 0/18 |
| 30−32 | 25 | 10/25 | 0/25 | 0/25 | 0/25 |
| 32−34 | 31 | 0/31 | 0/31 | 0/31 | 0/31 |
RAgT, rapid antigen test; CT, threshold cycle; gRNA, genomic RNA; gE, genomic RNA of gene E; RT-PCR, reverse transcription-PCR; sgRNA, subgenomic RNA. CD, Wuhan CDiagnosis; WF, Guangzhou Wondfo.
Relationship between gRNA and sgRNA.
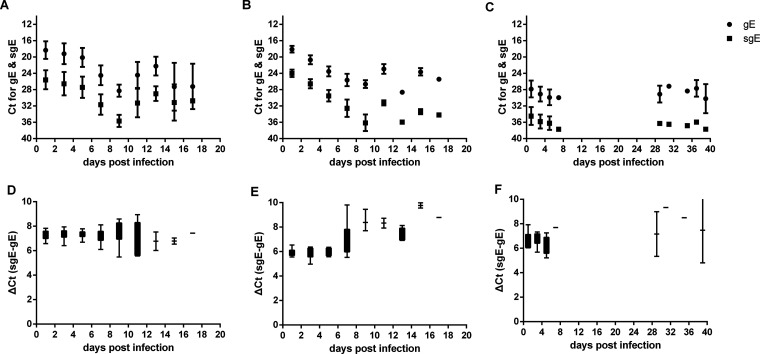
To understand the relationship between gRNA and sgRNA, we compared the CT and ΔCT values of gRNA and sgRNA in nasal washes, oropharyngeal swabs, and feces. The trend of CT for sgRNA over time was almost the same as that of CT for gRNA in nasal washes, oropharyngeal swabs, and feces (Fig. 4A to C). The ΔCT between sgRNA and gRNA is almost constant, with a mean value of 7.0 during the whole infection course (Fig. 4D to F). The gRNA load was about 128-fold higher than the sgRNA load. Hence, sgRNA is highly correlated with gRNA, and the sgRNA load can be predicted based on the gRNA load.
FIG 4.
Relationship between gRNA and sgRNA of E gene (gE and sgE) in experimentally infected hamsters. CT values for gRNA and sgRNA in nasal washes (A), oropharyngeal swabs (B), and feces (C) of experimentally infected hamsters. ΔCT values for gRNA and sgRNA in nasal washes (D), oropharyngeal swabs (E), and feces (F) of experimentally infected hamsters.
Relationship between gRNA, sgRNA, and infectious virus titer.
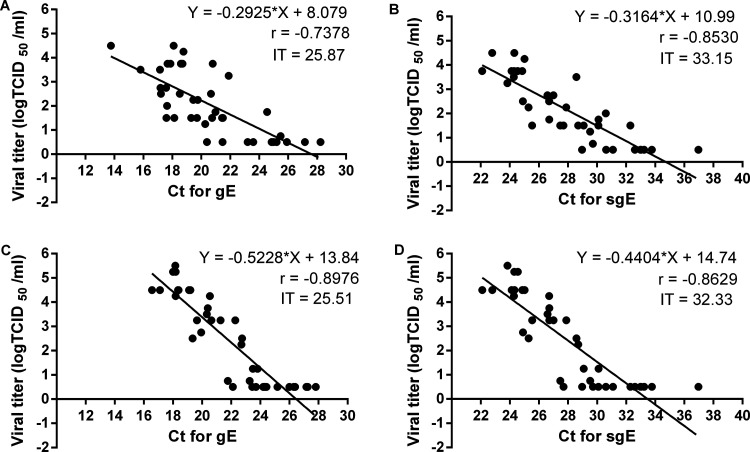
For nasal washes, viral titer gradually decreased with increasing CT values for genomic RNA of E gene (gE) and sgE. The slopes of the two regression lines for gE and sgE were similar. When the CT for gE was increased to 25.87 or the CT for sgE was increased to 33.15, infectious virus was no longer detectable (Fig. 5A and B). Therefore, the infection thresholds (IT) were 25.87 for gE and 33.15 for sgE in nasal washes. For oropharyngeal swabs, viral titers also decreased with increasing CT values for gE and sgE. The slope of the regression line for gE was slightly greater than that for sgE, and the slopes of the two regression lines in oropharyngeal swabs were obviously greater than those for nasal washes. The IT values were 25.51 for gE and 32.33 for sgE in oropharyngeal swabs (Fig. 5C and D).
FIG 5.
Infection threshold determination and correlation between gRNA, sgRNA, and viral titers in experimentally infected hamsters. Correlation and linear regression analysis between gRNA and viral titer in nasal washes (A) and oropharyngeal swabs (C). Correlation and linear regression analysis between sgRNA and viral titer in nasal washes (B) and oropharyngeal swabs (D). Infection threshold (IT) is the corresponding CT value in the linear regression equation when viral titer is 0.5 -log TCID50 (50% tissue culture infective dose). Linear regression equations, Pearson correlation coefficients (r), and infection thresholds are included in each panel.
Viral RNA load in a specific hamster.
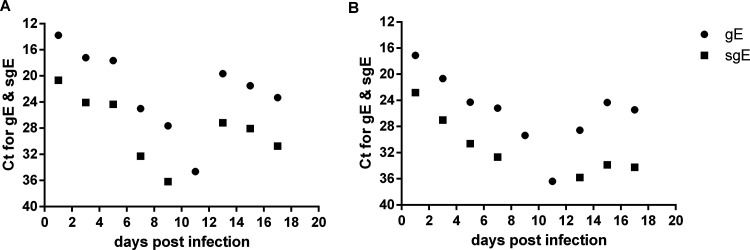
The young Syrian hamster is not a lethal disease model for SARS-CoV-2. In our study, an infected hamster died unexpectedly on day 18. We determined viral gRNA and sgRNA levels in nasal washes and oropharyngeal swabs. gRNA and sgRNA in nasal washes and oropharyngeal swabs gradually decreased to low levels before day 11, but by day 13 and later, gRNA and sgRNA loads again increased to high levels, and the gRNA and sgRNA in oropharyngeal swabs increased to moderate levels (Fig. 6A and B).
FIG 6.
Viral load of an infected hamster which died at day 18. CT values for gRNA and sgRNA in nasal washes (A) and oropharyngeal swabs (B).
DISCUSSION
RT-PCR has been the gold standard for SARS-CoV-2 diagnostics because of its high sensitivity, high specificity, and high throughout. However, RT-PCR does not determine infectivity. The duration of viral gRNA shedding is much longer than that of infectivity. It is very important to determine the relationship between test results and infectivity for efficient isolation, contact tracing, and release from isolation.
In this study, we first characterized viral gRNA and sgRNA loads and viral culture and RAgT positivity in a time series of sequentially sampled samples, including nasal washes, oropharyngeal swabs, and feces, of experimentally infected hamsters. We found that the duration of viral gRNA shedding was longer than that of sgRNA, and far longer those that of RAgT and viral culture positivit. Phuphuakrat et al. (10) reported that the median times to negativity of gRNA, sgRNA and viable SARS-CoV-2 in non-critically ill COVID-19 patients were 18, 11, and 7 days, and Immergluck et al. (11) reported that sgRNA was concordant with antigen detection in symptomatic outpatients, but remained detectable in 13 of 16 (82.1%) antigen-negative persons. The results of these two studies strongly validated our findings. Additionally, we found that the RAgT results correlated more strongly with viral culture results than with gRNA and sgRNA. Pekosz et al. also reported similar results in COVID-19 patients (12).
RAgT is a very practical method for SARS-CoV-2 diagnosis in the field and clinical lab. It is readily accessible, inexpensive, and can return results 15 min after sample preparation, which allows rapid isolation of infected individuals. The high PPV of RAgT suggested that most RAgT-positive individuals are infectious, which aids in early initiation of isolation and contact tracing action for COVID-19 cases. However, compared with RT-PCR, RAgT is less sensitive. Some COVID-19 patients with low viral loads in the early disease stage may be missed by the RAgT assay. Through serial specimen collection, the three-rapid antigen test can increase the sensitivity of RAgT from 68.5% to an enough high level of 95.8% (13). The very high NPV suggests that individuals with negative RAgT results are temporarily not infectious, and whether the nucleic acid based-test is positive or negative, these recovered COVID-19 patients should be allowed to stop isolation immediately. RAgT may be useful in guiding isolation decisions and contact tracing, shortening isolation times, and reducing waste of medical resources. Consistent with our results, a recent study reported that RAgT reduced isolation times by 2 days compared with the standard isolation time of 10 days for health care personnel (14). RAgT would be an important supplement to RT-PCR in SARS-CoV-2 diagnostics and COVID-19 prevention and control. This was strongly confirmed during the fifth wave of COVID-19 in Hong Kong, China (15).
CT values were inversely proportion to viral RNA loads in the collected specimens. In this study, we found that viral culture positivity decreased with increasing CT values. Viral RNA shedding in COVID-19 patients can persist for several weeks, but detectable viral RNA does not necessarily mean infectious virus. A suitable CT threshold for gRNA is necessary for infectivity determination. When CT > 26, only one sample was positive for viable SARS-CoV-2, and when CT > 28, all viral culture results in samples from infected hamsters were negative. Kim et al. (1) reported that when CT > 28.4, and Bullard et al. (16) reported that when CT > 24, viral culture results were all negative. Because CT values in the same specimen differed with RT-PCR assays (17), the differences between CT thresholds for gRNA in these three studies are acceptable. Correlation and linear regression analysis between gRNA and viral titers showed that infection thresholds for gE were 25.87 in nasal washes and 25.51 in oropharyngeal swabs. To be conservative, the CT threshold for infectivity can be set to be 28 to 30. Because the infectious virus shedding time in feces is very short and viral titers in very few positive fecal samples were also very low, we think that it may not be necessary to sample anal swabs or feces for daily SARS-CoV-2 monitoring and diagnostics.
Subgenomic RNA is a transcriptional intermediate and a marker of actively replicating virus (18). A previous study showed that sgRNA correlates strongly with viral infectivity (8). We determined viral gRNA and sgRNA loads in experimentally infected hamsters and found that viral sgRNA loads were obviously lower than viral gRNA loads, and the duration of viral sgRNA shedding is obviously shorter than that of viral gRNA. Perera et al. (7) also showed that the duration of sgRNA shedding is shorter than that of gRNA in 35 mild COVID-19 patients. However, the trend of sgRNA is highly similar to that of gRNA. Furthermore, the ΔCT values between sgE and gE were almost constant, about 7.0, during the whole infection process, suggesting that sgRNA is highly correlated with viral gRNA. This result is consistent with correlation and linear analysis which showed that the slopes of the two regression lines were almost the same. We can easily predict the viral sgRNA load from the viral gRNA load, and the shorter sgRNA shedding time is due to the lower abundance of sgRNA. Dimcheff et al. (19) and Verma et al. (20) reported a similarly high correlation between gRNA and sgRNA. Therefore, sgRNA did not provide more useful information than viral gRNA in infectivity determination. The CT threshold for sgE to determine infectivity in all samples can be predicted as 35. Indeed, when CT for sgE > 35, all viral culture results were negative. The infection threshold for sgE can be predicted to be 33 in nasal washes and 31 in oropharyngeal swabs (Fig. 4). This was consistent with the results of linear regression analysis between sgRNA and viral titers, showing that ITs for sgE were 33.15 in nasal washes and 32.33 in oropharyngeal swabs. Indeed, when CT for sgE > 33, viral culture results were all negative in non-fecal samples.
The gRNA shedding pattern varied greatly in feces, nasal washes, and oropharyngeal swabs. gRNA was continuously detectable over all 40 experimental days in most fecal samples, and was maintained at relatively high levels. However, gRNA in nasal washes and oropharyngeal swabs gradually decreased with infection time, was only detected in a small number of samples, and was maintained at very low levels after 28 days. The gRNA shedding time in feces was much longer than those in nasal washes and oropharyngeal swabs. Similarly, in a few clinically recovered COVID-19 patients, the median clearance time of viral RNA in stool (22 days) was much longer than that in nasopharyngeal secretions (12 days) (21). Prolonged viral RNA shedding in rectal swabs (22) or feces (23) has also been found in pediatric patients with COVID-19. Therefore, the continued detection of gRNA in feces of infected hamsters may mirror SARS-CoV-2 shedding in stools or rectal swabs of some COVID-19 patients. Compared with that in the respiratory tract, SARS-CoV-2 receptor ACE2 and the priming protease TMPRSS2 were highly expressed in the intestine, especially the small intestine, of hamsters (24) and humans (25, 26), forming a suitable environment for SARS-CoV-2 replication and spread. In our study, low levels of sgRNA were detected in feces during the last 10 days, indicating weakly or moderately active replication in the intestines of hamsters. The underlying mechanism behind long SARS-CoV-2 shedding in human and hamster feces remains to be explored in the future.
Viral shedding in immunocompromised COVID-19 patients can persist for several weeks or even several months (27). In our study, most of the time, the gRNA and sgRNA loads in nasal washes of the dead hamster were maintained at relatively high levels. Usually, SARS-CoV-2 is not lethal to young hamsters. We speculated that the virus may have adapted to this hamster or that the dead hamster may have been compromised or had another unknown disease. The true factors leading to this hamster’s death have yet to be explored.
In conclusion, the duration of viral gRNA shedding was far longer than those of RAgT and viral culture positivity, and RAgT results correlated strongly with viral culture results. RAgT may be an important supplement to RT-PCR in guiding isolation periods for COVID-19 patients and early COVID-19 screening. sgRNA was highly correlated with gRNA and may be no more useful for identifying infectivity than a CT threshold determined for gRNA based on viral culture results. For gRNA, the CT threshold for infectivity can be conservatively set to be 28 to 30.
MATERIALS AND METHODS
Ethics and biosecurity statement.
All animal experiments were approved by the Animal Care and Use Committee of Changchun Veterinary Research Institute. All experiments involving infectious SARS-CoV-2 were performed in the Biosafety Level 3 Laboratories of Changchun Veterinary Research Institute.
Study design and sample processing.
Ten male Syrian hamsters, 5 weeks old, were anesthetized with isoflurane and intranasally inoculated with 105 50% tissue culture infective dose (TCID50) of the virus (BetaCoV/Beijing/IME-BJ05-2020). Every other day, nasal washes were collected from all animals with 1 mL phosphate-buffered saline (PBS) for 39 days, oropharyngeal swabs were collected and immersed into 1 mL PBS, and fresh feces were collected into 1 mL PBS. All samples were vibrated and centrifuged. The sample supernatants were used for viral culture, genomic and subgenomic RNA quantification, and rapid antigen testing.
Viral genomic RNA and subgenomic RNA quantification.
Two hundred μL of the sample supernatants of nasal washes, oropharyngeal swabs, and feces from hamsters was used to extract RNA using viral RNA minikits (Qiagen, Hilden, Germany) according to the manufacturers’ instructions. Viral gRNA and sgRNA were quantified using RT-PCR as described previously (28, 29). The cutoff values for gRNA and sgRNA were 38, and a CT value of <38 was considered to be positive.
Rapid antigen test.
Two rapid antigen testing kits (Wuhan CDIAGNOSIS and Guangzhou Wondfo Biotech Company) were used for SARS-CoV-2 testing. RAgT was performed according to the manufacturers’ instruction with slight modification. Here, 100 μL of the sample supernatants was mixed with 100 μL of the antigen extraction buffer and incubated for 2 min. The mixed solution was added into the sample pad of the rapid antigen test strip, and the test results were read at 15 min.
SARS-CoV-2 virus culture.
Vero-E6 cells (CRL1586, ATCC, Manassas, VA) were cultured in growth medium (GM) containing high-glucose Dulbecco’s modified Eagle’s medium (DMEM; HyClone, USA), supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C with 5% CO2. Cells were grown to 80% confluence in 96-well plates and the GM was removed and replaced with maintenance medium, which is identical to the GM, supplemented with 2% FBS. Next, 100 μL of serial 10-fold dilutions of the sample supernatants was added to Vero-E6 cells. The cytopathic effect was observed under a microscope. Viral titers were determined by the Reed-Muench method.
Statistical analysis.
All data were analyzed with GraphPad Prism v6.01. A Pearson correlation was used to analyze data correlation between gRNA, sgRNA, and viral titers, and a linear regression analysis was used to fit the linear regression curves. The infection threshold is the corresponding CT value in the linear regression equation when the viral titer is 0.5-log TCID50.
ACKNOWLEDGMENTS
This research was supported by the National Natural Science Foundation of China (32000134) and the National Major Research and Development Program (2020YFC0840800).
Chunmao Z. designed the project, and Chunmao Z., C.Z., H.C., Z.G., Z.C., F.Y., and Y.L. performed the experiments. Chunmao Z., C.Z., H.C., and Z.G. analyzed the data. Chunmao Z. drafted the manuscript, and Y.G., J.L., and Z.G. critically revised the manuscript.
All authors have declared no conflicting interests in this study.
Contributor Information
Chunmao Zhang, Email: jk704715@sina.com.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Kim M-C, Cui C, Shin K-R, Bae J-Y, Kweon O-J, Lee M-K, Choi S-H, Jung S-Y, Park M-S, Chung J-W. 2021. Duration of culturable SARS-CoV-2 in hospitalized patients with COVID-19. N Engl J Med 384:671–673. 10.1056/NEJMc2027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC. 2020. Discontinuation of isolation for persons with COVID-19 not in healthcare settings (interim guidance). Available from https://stacks.cdc.gov/view/cdc/88539. Accessed 6 May. CDC, Atlanta, GA. [Google Scholar]
- 3.Hong KH, Lee SW, Kim TS, Huh HJ, Lee J, Kim SY, Park JS, Kim GJ, Sung H, Roh KH, Kim JS, Kim HS, Lee ST, Seong MW, Ryoo N, Lee H, Kwon KC, Yoo CK. 2020. Guidelines for laboratory diagnosis of coronavirus disease 2019 (COVID-19) in Korea. Ann Lab Med 40:351–360. 10.3343/alm.2020.40.5.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Health Commission of China (CNHC). 2022. The ninth edition of Chinese diagnosis and treatment plan for COVID-19. Available from http://www.nhc.gov.cn/yzygj/s7653p/202203/b74ade1ba4494583805a3d2e40093d88.shtml [Chinese]. Accessed 15 March. CNHC, Beijing, China. [Google Scholar]
- 5.Avanzato VA, Matson MJ, Seifert SN, Pryce R, Williamson BN, Anzick SL, Barbian K, Judson SD, Fischer ER, Martens C, Bowden TA, de Wit E, Riedo FX, Munster VJ. 2020. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 183:1901–1912.e9. 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baang JH, Smith C, Mirabelli C, Valesano AL, Manthei DM, Bachman MA, Wobus CE, Adams M, Washer L, Martin ET, Lauring AS. 2021. Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J Infect Dis 223:23–27. 10.1093/infdis/jiaa666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perera RAPM, Tso E, Tsang OTY, Tsang DNC, Fung K, Leung YWY, Chin AWH, Chu DKW, Cheng SMS, Poon LLM, Chuang VWM, Peiris M. 2020. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis 26:2701–2704. 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santos Bravo M, Berengua C, Marín P, Esteban M, Rodriguez C, del Cuerpo M, Miró E, Cuesta G, Mosquera M, Sánchez-Palomino S, Vila J, Rabella N, Marcos MÁ. 2022. Viral culture confirmed SARS-CoV-2 subgenomic RNA value as a good surrogate marker of infectivity. J Clin Microbiol 60:e01609-21. 10.1128/JCM.01609-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young S, Taylor SN, Cammarata CL, Varnado KG, Roger-Dalbert C, Montano A, Griego-Fullbright C, Burgard C, Fernandez C, Eckert K, Andrews JC, Ren H, Allen J, Ackerman R, Cooper CK. 2020. Clinical evaluation of BD Veritor SARS-CoV-2 point-of-care test performance compared to PCR-based testing and versus the Sofia 2 SARS Antigen point-of-care test. J Clin Microbiol 59:e02338-20. 10.1128/JCM.02338-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phuphuakrat A, Pasomsub E, Srichatrapimuk S, Kirdlarp S, Suksatu A, Srisaowakarn C, Manopwisedjaroen S, Ludowyke N, Purwono PB, Priengprom T, Wongsa A, Thakkinstian A, Hongeng S, Malathum K, Thitithanyanont A, Tassaneetrithep B. 2022. Detectable duration of viable SARS-CoV-2, total and subgenomic SARS-CoV-2 RNA in noncritically ill COVID-19 patients: a prospective cohort study. Microbiol Spectr: 10:e00503-22. 10.1128/spectrum.00503-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Immergluck K, Gonzalez MD, Frediani JK, Levy JM, Figueroa J, Wood A, Rogers BB, O'Neal J, Elias-Marcellin R, Suessmith A, Sullivan J, Schinazi RF, Babiker A, Piantadosi A, Vos MB, Martin GS, Lam WA, Waggoner JJ. 2021. Correlation of SARS-CoV-2 subgenomic RNA with antigen detection in nasal midturbinate swab specimens. Emerg Infect Dis 27:2887–2891. 10.3201/eid2711.211135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pekosz A, Parvu V, Li M, Andrews JC, Manabe YC, Kodsi S, Gary DS, Roger-Dalbert C, Leitch J, Cooper CK. 2021. Antigen-based testing but not real-time polymerase chain reaction correlates with severe acute respiratory syndrome coronavirus 2 viral culture. Clin Infect Dis 73:e2861–e2866. 10.1093/cid/ciaa1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lind ML, Schultes OL, Robertson AJ, Houde AJ, Cummings DAT, Ko AI, Kennedy BS, Richeson RP. 2022. Testing frequency matters: an evaluation of the diagnostic performance of a SARS-CoV-2 rapid antigen test in United States correctional facilities. Clin Infect Dis ciac450. 10.1093/cid/ciac450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tande AJ, Swift MD, Challener DW, Berbari EF, Tommaso CP, Christopherson DR, Binnicker MJ, Breeher LE. 2022. Utility of follow-up coronavirus disease 2019 (COVID-19) antigen tests after acute severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection among healthcare personnel. Clin Infect Dis ciac235. 10.1093/cid/ciac235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung P-HH, Chan C-P, Jin D-Y. 2022. Lessons learned from the fifth wave of COVID-19 in Hong Kong in early 2022. Emerg Microbes Infect 11:1072–1078. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9004509/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, Boodman C, Bello A, Hedley A, Schiffman Z, Doan K, Bastien N, Li Y, Van Caeseele PG, Poliquin G. 2020. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clinical Infectious Diseases 71:2663–2666. 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nalla AK, Casto AM, Huang M-LW, Perchetti GA, Sampoleo R, Shrestha L, Wei Y, Zhu H, Jerome KR, Greninger AL. 2020. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J Clin Microbiol 58:e00557-20. 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sola I, Almazan F, Zúñiga S, Enjuanes L. 2015. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol 2:265–288. 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimcheff DE, Valesano AL, Rumfelt KE, Fitzsimmons WJ, Blair C, Mirabelli C, Petrie JG, Martin ET, Bhambhani C, Tewari M, Lauring AS. 2021. Severe acute respiratory syndrome coronavirus 2 total and subgenomic RNA viral load in hospitalized patients. J Infect Dis 224:1287–1293. 10.1093/infdis/jiab215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verma R, Kim E, Martínez-Colón GJ, Jagannathan P, Rustagi A, Parsonnet J, Bonilla H, Khosla C, Holubar M, Subramanian A, Singh U, Maldonado Y, Blish CA, Andrews JR. 2021. SARS-CoV-2 subgenomic RNA kinetics in longitudinal clinical samples, p ofab310. Open Forum Infect Dis 8:ofab310. 10.1093/ofid/ofab310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Díaz LA, García-Salum T, Fuentes-López E, Reyes D, Ortiz J, Chahuan J, Levican J, Almonacid LI, Valenzuela GH, Serrano E, Budnik S, Gandara V, Gallardo A, Seydewitz MF, Ferrés M, Cofré C, Álvarez M, Pavez C, Candia R, Monrroy H, Espino A, Rada G, Ortiz L, Valderrama S, Salinas E, Toro A, Ortega M, Pizarro M, Medina RA, Riquelme A. 2022. High prevalence of SARS-CoV-2 detection and prolonged viral shedding in stools: a systematic review and cohort study. Gastroenterol Hepatol S0210-5705(22)00001-2. 10.1016/j.gastrohep.2021.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, Guo Q, Sun X, Zhao D, Shen J, Zhang H, Liu H, Xia H, Tang J, Zhang K, Gong S. 2020. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med 26:502–505. 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xing Y-H, Ni W, Wu Q, Li W-J, Li G-J, Wang W-D, Tong J-N, Song X-F, Wong GW-K, Xing Q-S. 2020. Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J Microbiol Immunol Infect 53:473–480. 10.1016/j.jmii.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suresh V, Parida D, Minz AP, Sethi M, Sahoo BS, Senapati S. 2020. Tissue distribution of ACE2 protein in Syrian golden hamster (Mesocricetus auratus) and its possible implications in SARS-CoV-2 related studies. Front Pharmacol 11:579330. 10.3389/fphar.2020.579330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M-Y, Li L, Zhang Y, Wang X-S. 2020. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty 9:23–29. 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao W, Feng Q, Wang X. 2021. Computational analysis of TMPRSS2 expression in normal and SARS-CoV-2-infected human tissues. Chem Biol Interact 346:109583. 10.1016/j.cbi.2021.109583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi B, Choudhary MC, Regan J, Sparks JA, Padera RF, Qiu X, Solomon IH, Kuo H-H, Boucau J, Bowman K, Adhikari UD, Winkler ML, Mueller AA, Hsu TY-T, Desjardins M, Baden LR, Chan BT, Walker BD, Lichterfeld M, Brigl M, Kwon DS, Kanjilal S, Richardson ET, Jonsson AH, Alter G, Barczak AK, Hanage WP, Yu XG, Gaiha GD, Seaman MS, Cernadas M, Li JZ. 2020. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 383:2291–2293. 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 29.Zhang C, Cui H, Li E, Guo Z, Wang T, Yan F, Liu L, Li Y, Chen D, Meng K, Li N, Qin C, Liu J, Gao Y, Zhang C. 2022. The SARS-CoV-2 B. 1.351 variant can transmit in rats but not in mice. Front Immunol 13:869809. 10.3389/fimmu.2022.869809. [DOI] [PMC free article] [PubMed] [Google Scholar]