Abstract
Available drugs have been used as an urgent attempt through clinical trials to minimize severe cases of hospitalizations with Coronavirus disease (COVID-19), however, there are limited data on common pharmacogenomics affecting concomitant medications response in patients with comorbidities. To identify the genomic determinants that influence COVID-19 susceptibility, we use a computational, statistical, and network biology approach to analyze relationships of ineffective concomitant medication with an adverse effect on patients. We statistically construct a pharmacogenetic/biomarker network with significant drug-gene interactions originating from gene-disease associations. Investigation of the predicted pharmacogenes encompassing the gene-disease-gene pharmacogenomics (PGx) network suggests that these genes could play a significant role in COVID-19 clinical manifestation due to their association with autoimmune, metabolic, neurological, cardiovascular, and degenerative disorders, some of which have been reported to be crucial comorbidities in a COVID-19 patient.
Subject terms: Predictive medicine, Gene regulatory networks
Introduction
Since the first outbreak of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection in Wuhan, Hubei, China in December 2019, more than 263 million people have been infected by SARS-CoV-2, leading to the COVID-19 pandemic with a total of 5.2 million deaths globally [1]. COVID-19 has been shown to be a multifactorial disease, with the scientific interest focused on the host genetic variants associated with critical illness aiming at identifying targets for efficient therapeutics development.
Drug repurposing is a cost-effective approach to quickly investigate whether current drugs could be used to potentially improve the clinical outcome of a patient [2–5]. To our knowledge, there are no approved drugs till today which have been shown to be safe and effective in terms of reducing the number of deaths and preventing severe COVID-19 [6–8].
Due to the urgency, scientists investigate the effect of different drug mixtures to propose an alternative, perhaps more effective, treatment, often at any cost [9]. However, initiating ineffective therapies and trying drug cocktails can be crucial, especially to patients with comorbidities on top of acute respiratory distress syndrome that require mechanical ventilation [10]. In this sense, co-medications can cause phenoconversion, a case where there is a discrepancy between drug-metabolizing genotype and phenotype status of an individual [11]. Inflammation and infection processes [12] in COVID-19 could constitute the causative ground for therapeutic failures that may be due to genetic interactions, yet to be explored [13, 14]. To this end, available reported drug-drug interactions (DDI) accompanied by a clinical outcome and information about possible adverse effects may help health professionals in treatment strategy decisions when comorbidities exist and co-administration of drugs is advised [15, 16].
Pharmacogenomics (PGx) approach uses an individual’s genetic information to predict drug response and guide optimal drug dosage for safer, and cost-effective treatments [17]. The recent advances in pharmacogenomics in combination with the rapid development of next-generation sequencing technology have led to remarkable findings such as the identification of various drug-related genetic loci variations associated with complex diseases [18]. By knowing the genetic interactions which affect the co-medication response, common genes could be considered as candidate biomarkers for optimum treatment and clinical management strategies. On the other hand, there are a limited number of methods to correctly estimate all possible drug-drug interactions (DDIs) by in silico approaches [19] and predict the successfulness of a COVID-19 treatment by just considering disease progression.
In this study, we use a computational approach to evaluate all possible pharmacogenes, with a recorded clinical outcome, which influences co-medication response in order to uncover the significant pharmacogenetic determinants that affect the drug response of COVID-19 patients [13]. We identify a cluster of genes, known risk factors for other diseases, and propose them as pharmacogenomic biomarkers of relevance in COVID-19 treatment and perhaps long-term outcomes. Our findings could help in clinical decision-making to combat COVID-19 pandemic with medication [13].
Materials and methods
Data-mining approach
The DDI datasets from the University of Liverpool [20–22] refer to drug interactions with three types of experimental therapies: (1) experimental COVID-19 antiviral therapies, such as Atazanavir (ATV), Hydroxychloroquine (HCLQ), Nitazoxanide (NTZ), Azithromycin (AZM), Interferon-beta (IFN-β), Remdesivir (RDV), Chloroquine (CLQ), Ivermectin (IVM), Ribavirin (RBV), Favipiravir (FAVI), Lopinavir/ritonavir (LPV/r); (2) experimental COVID-19 immune therapies, such as Anakinra (ANR), Baricitinib (BAR), Hydrocortisone (HC), COVID-19 Vaccines (VAC), Methylprednisolone (MP), Antibody Therapies: Convalescent plasma Bamlanivimab Casirivimab/Imdevimab (Ab Tx), Canakinumab (CAN), Ruxolitinib (RUX), Colchicine (COL), Sarilumab (SAR), Dexamethasone (DXM), Tocilizumab (TCZ); and (3) Experimental COVID-19 Adjunct Therapies, such as Aspirin, Dalteparin, and Enoxaparin. Each drug was tested for synonyms in the DrugBank database [23]. The final dataset included 375 drugs comprising 33 pharmaceutical activity groups. Interactions of these drugs within all three groups of experimental COVID-19 therapies fell in four categories [24–26] with relative scores as depicted in Supplementary File 1, sheet 1: (a) drugs should not be co-administered (score 3), (b) drugs with potential interaction which may require a dose adjustment or close monitoring (score 2), (c) drugs with potential interaction likely to be of weak intensity, additional action/monitoring or dosage adjustment unlikely to be required (score 1) and (d) drugs with no clinically significant interaction expected (score 0).
Drug-drug interactions (DDI)
All DDIs with recorded clinical adverse effects have been recorded in an excel file along with its pharmacogenomic evaluation through PharmGKB [27]. Using the annotation files from PharmaGKB along with the list of 375 drugs (from Liverpool datasets and driven by a data-mining algorithm), a drug-gene association list was created (Supplementary File 1, sheet 2). Importantly, each drug from various databases was tested for synonyms in the DrugBank database [23]. Drug-gene interactions extracted from PharmGKB for both drugs participating in a DDI were linked (Supplementary File 1, sheet 3). Drugs without PGx information were discarded from the analysis (Supplementary File 1, sheet 3).
Statistical assessment of DDIs
For each DDI, common (intersection) genes, the union of genes, and different genes were recorded. A statistical method was designed using the STATA package [28] to determine the significance of the overrepresentation of common genes (between two interacting drugs) that affect co-medication response. Briefly, a hypergeometric test (Fisher-EXACT) [29] was performed for each DDI using as background the total number of genes (998) associated with at least one drug. The test returned a score as the probability of randomly selecting successful common DDI pharmacogenes (PGx genes) in the set of union genes of a certain DDI compared with the finite population of the background genes (Supplementary File 1, sheet 4). The most significant DDIs are those enriched for common genes with a p-value < 0.05 and used as input for further analysis.
Functional enrichment analysis of the highly significant DDI PGx genes
The statistically significantly enriched genes (44) which commonly associated with both drug responses of a DDI were listed and used as input for pathway analysis (KEGG, Reactome) and functional (Gene Ontology) enrichment analysis (Supplementary File 1). The latter was done with the use of the FLAME web-tool [30]. Further functional enrichment analysis of the 44 PGx genes was performed with the use of STRING database at a protein-protein interaction level and by only selecting interactions which were curated and experimentally validated.
Gene-disease network
All 44 genes of statistically significant DDIs were further investigated in GWAS [24], OMIM [25], and GAD [26], to identify pathophysiological diseases (phenotypes) that are linked with their genetic variations using a recently developed bipartite network analysis methodology [31]. For this task, Cytoscape [32] network visualization tool was utilized.
Shared-disease gene–gene network
A shared-disease gene–gene network was constructed by directly connecting the 44 identified PGx genes (so-called PGx-biomarkers from now on) associated with a certain disease. Similarly to before, the network visualization was performed with Cytoscape [32].
Creation of the expanded PGx-biomarkers network
An expanded version of genetic interactions identified through the shared-disease gene-gene network and through PPI interactions (STRING) for the 44 (statistically significant genes of all DDIs) PGx-biomarkers was constructed by merging and overlaying both networks using the Dynet software [33] to visually evaluate common and different edges between nodes.
Results
Drug–drug interactions (DDI)
The Liverpool dataset [20–22] was used to retrieve all Drug–Drug Interactions (DDIs), from COVID-19 treatments where clinical adverse effects were reported. 375 drugs (or medication schemes) were found in total, 26 of which were repurposed drugs for COVID-19 treatment. In total 1989 DDIs were recovered according to the data mining workflow described below (see Material & methods along with Fig. 1A). DDIs are divided into four categories according to their clinical significance: (a) drugs that should not be co-administered (score 3), (b) drugs with potential interaction that may require a dose adjustment or close monitoring (score 2) and (c) drugs with potential interaction likely to be of weak intensity (score 1) (Supplementary File 1, sheet 1). Drugs with no clinically significant interaction were not considered in our analysis. From the significant interactions (DDIs), 15% belong to the first category, 70% to the second, and 15% to the third category.
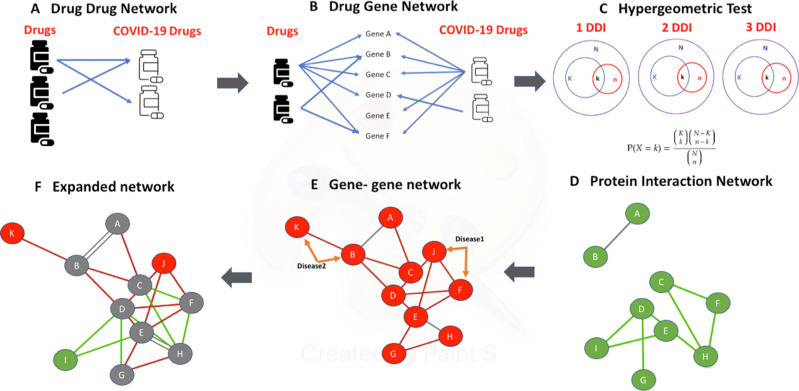
Fig. 1. Methodology workflow.
A A list of Drug-Drug Interactions (DDIs) with COVID-19 treatments and recorded clinical adverse effects from the Liverpool dataset was created (B) PGx information from PharmGKB was extracted for each drug participating in a DDI to build a drug-gene interaction list. C For each drug–drug interaction (DDI), a hypergeometric test was performed (Fisher test, p-value < 0.05 to determine the statistical significance of having overrepresented common genes in both drugs of the same co-medication with a clinical interaction. D Significant DDIs (p-value < 0.05) are selected, and their gene list has been used as input in the STRING database to find curated interactions between drug-interacting genes of each DDI. E Gene-disease associations of the above identified genes, based on GWAS, OMIM and GAD datasets, were retrieved and a new, sheared-disease gene-gene network was constructed with edges depicting genes associated with a common disease. F Networks of (D) and (E) are combined together providing an expanded PGx biomarker network that shows statistically significantly associated genes with COVID-19 treatment adverse effects.
Drug associated genes
Data mining in PharmaGKB using the 375 drugs as input, reported 998 pharmacogenes affecting the response of at least one drug (Fig. 1B). According to the PharmGKB level of significance, patients who carry variants in these genes should be prescribed with caution regarding drug safety and efficacy. However, nine out of the 26 repurposed COVID-19 treatments do not have PGx information in PharmaGKB (Supplementary File 1, sheet 3). Thus, we propose that more research is needed to identify PGx associations with such drugs.
Identification of PGx biomarkers
From the 1989 DDIs, only 1413 contained PGx information for both drugs. Hypergeometric tests were performed on each of the 1413 DDIs and resulted in 571 statistically significant DDIs. Interestingly, only seven interactions were found for COVID-19 Adjunct Therapies drugs, 127 for COVID-19 Immunotherapy drugs, and 443 DDIs were found for antiviral drugs (Table 1). A great deal of all these interactions fell into score category 2 i.e., drugs with potential interaction that may require a dose adjustment or close monitoring. Common genes of the two drugs in each DDI pair were considered if p-values < 0.05 according to the Fisher-Exact test. The sum of the common genes was 44 and presented the most significant association with drugs in DDIs (Supplementary File 1, sheet 4, Fig. 1C).
Table 1.
Number of drug-drug interactions by score (N = 571).
| Experimental COVID-19 therapies | DDI with score 1 | DDI with score 2 | DDI with score 3 |
|---|---|---|---|
| Adjunct Therapies | 1 (14%) | 6 (86%) | 0 (0%) |
| Immunotherapies | 2 (1%) | 110 (87%) | 15 (12%) |
| Antiviral | 35 (8%) | 305 (69%) | 103 (23%) |
Protein–Protein interactions of the PGx biomarkers
When these 44 genes were analyzed for their curated PPI interactions (co-expression, experiments, co-occurrence, databases, interaction) in the STRING database, a low-density interaction network was formed with four main interacting groups, such as xenobiotics metabolism partially overlapping with arachidonic-linoleic metabolism, cholesterol metabolism, immune response, and signal transducers some of which participate in tyrosine kinase pathways (Fig. 1D).
3D visualization of the association of DDIs with PPIs
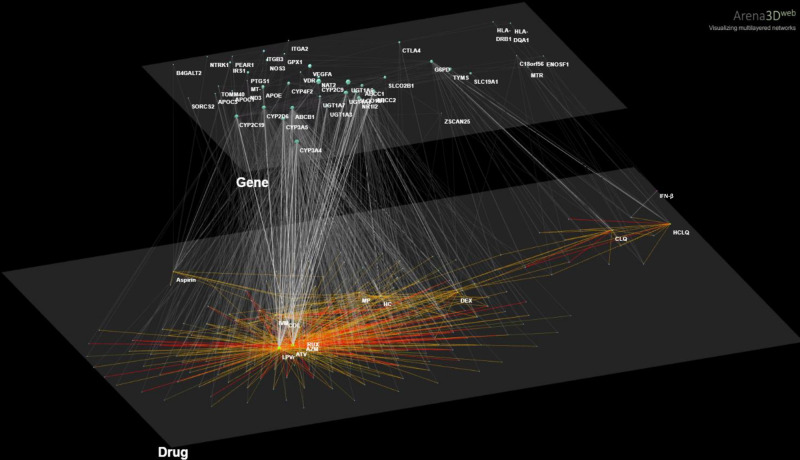
To visualize the proportion of COVID-19 drugs and other disease drugs that are associated with gene variants regulating drug response, a three-layer interaction network was created by Arena 3Dweb [34]. This 3D network connects the DDIs from Liverpool data with the PPIs (gene–gene interactions) from STRING with the aid of drug-gene interactions according to PharmGKB. The top layer shows the 571 drug-drug interactions (drugs combined in COVID-19 treatments) that have significant common PGx associations (Fisher Exact test, p-value < 0.05). The bottom layer represents the network between all 44 pharmacogenes significantly associated with toxicity due to drugs administered for COVID-19 treatment or co-administered for comorbidities. Edges in each layer represent the significant drug–drug interactions with a recorded clinical effect and the public protein interactions found in the STRING database (Fig. 2). In this way, we see which interacting drugs are associated with genes that they, themselves, interact with others.
Fig. 2. Arena3Dweb visualization.
A 3D visualization of a 2-layer network shows all the interactions retrieved from all sources described in the computational workflow (Fig. 1). DDIs are shown in the bottom layer with name annotations of COVID-19 treatments as the main nodes, whereas PPI interactions in the top layer with protein names. Drug-pharmacogene genes associations retrieved from PharmGKB are shown between layers.
Functional enrichment analysis of the 44 PGx biomarkers
Functional enrichment analysis of the 44-total common pharmacogenes with KEGG, and Reactome yielded significantly enriched pathways and biological terms that are highly involved. Pathways are sorted by significance (FDR < 0.05) and they are depicted in Supplementary File 4. All genes participate in the following main, commonly used, enriched pathways: (1) drug and xenobiotic/biological oxidation metabolism (CYP3A4, CYP2D6, NAT2, CYP2C9, UGT1A6, CYP3A5, UGT1A1, PTGS1, CYP2C19, UGT1A3, CYP4F2, MTR, UGT1A7), (2) arachidonic-linoleic metabolism and lipid metabolism (GPX1, ABCC1, CYP4F2, CYP2C9, PTGS1, CYP2C19, GPX1, CYP3A4, CYP2D6, VDR, SLCO1B1), (3) cholesterol/lipoprotein metabolism (KEGG and Reactome) (APOC1, APOC3, APOE), (4) tyrosine kinase pathways (NTRK1, ITGB3, VEGFA, ITGA2, NOS3, IRS1, APOE) and (5) metabolism of vitamins and cofactors (APOE, APOC3, NOS3, ABCC1, SLC19A1, MTR).
Upon enrichment, we then investigated whether certain drug categories participate in one or multiple DDIs. As shown in Table 2, there is no specific trend for any drug category to continuously present one type of adverse effect, apart from the fact that most DDIs in general (70%) are in score 2. Importantly, antiviral therapies present adverse effects mainly due to genes participating in xenobiotic/drug metabolic pathways (Table 2). Adverse effects from adjunct therapy drugs are attributed to genes responsible for Autoimmune, metabolic, and neurological diseases as well as to genes important for Drug Metabolism and related blood disorders.
Table 2.
Stratification genes by score and disease.
| COVID-19 treatments | Disease cluster | Genes involved in DDIs with score 1 | Genes involved in DDIs with score 2 | Genes involved in DDIs with score 3 |
|---|---|---|---|---|
| Adjunct therapies | 1 |
IRS1 ITGA2 ITGB3 NOS3 NTRK1 |
||
| 2 |
GPX1 B4GALT2 |
|||
| 3 |
CYP2C19 CYP2C9 CYP2D6 UGT1A6 |
CYP2C19 CYP2C9 CYP4F2 PTGS1 PEAR1 |
||
| 4 | NAT2 | NAT2 | ||
| Immunotherapies | 1 |
VEGFA CTLA4 |
||
| 2 | NR1I2 | NR1I2 | ||
| 3 | CYP3A4 |
CTP2D6 CYP3A4 UGT1A1 |
CYP2D6 UGTA1 |
|
| 4 |
ABCB1 TYMS VDR TYMSOS |
ABCB1 | ||
| Antiviral therapies | 1 |
HLA-DQA1 HLA-DRB1 MT-ND3 |
MT-ND3 | |
| 2 | NR1I2 |
NR1I2 APOE APOC1 APOC3 TOMM40 |
NR1I2 APOE APOC1 APOC3 TOMM40 SORCS2 |
|
| 3 |
CYP3A4 CYP2C19 CYP3A5 CYP2D6 UGT1A1 UGT1A3 ABCC2 SLCO1B1 |
CYP3A4 CYP2C19 CYP3A5 CYP2D6 UGT1A1 UGT1A3 ABCC2 SLCO1B1 G6PD UGT1A7 ENOSF1 SLC19A1 |
CYP3A4 CYP2C19 CYP3A5 CYP2D6 UGT1A1 UGT1A3 ABCC2 SLCO1B1 G6PD UGT1A7 |
|
| 4 |
ABCB1 ABCC1 |
ABCB1 ABCC1 TYMS TYMSOS MTR ZSCAN25 SLCO2B1 |
ABCB1 ABCC1 |
Genes are clustered in 4 disease groups. 1: Autoimmune, metabolic and neurological diseases, 2: Cardiovascular & other degenerativediseases, 3: Drug Metabolism and related blood disorders.
Gene-disease association network
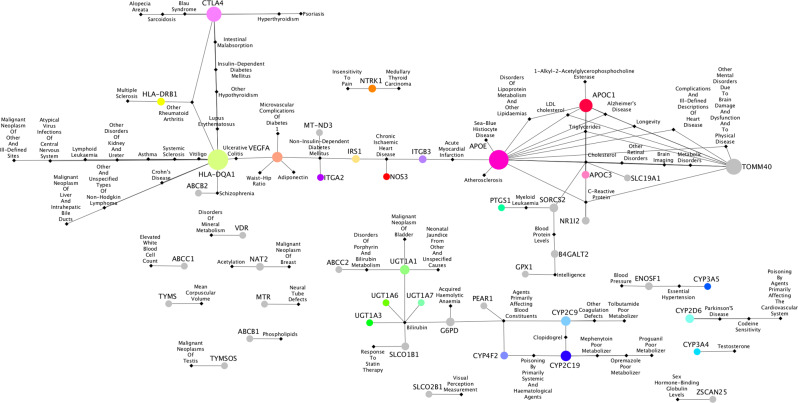
Next, a gene-disease network of the 44 pharmacogenes significantly associated with DDIs (Fig. 3, Supplementary File 3) was constructed depicting genes associated with diseases according to GWAS, OMIM, and GAD [31]. Visualization was performed with Cytoscape. Three major clusters of diseases became prominent: (a) Autoimmune, metabolic, and neurological diseases, (b) Cardiovascular and other degenerative diseases, (c) Drug Metabolism and related blood disorders. Additionally, a small number of gene-disease associations became prominent, however, they are disconnected from the major gene-disease clusters.
Fig. 3. The gene-disease association network.
Nodes in circles are PGx genes significantly predicted to affect clinical outcomes. Black nodes in rhombus represent the diseases. Edges between a gene and disease show the associations according to GWAS and Kontou et al. predictions. The size of a node indicates the higher connectivity with diseases. The top highly connected nodes with degree greater than 4 are: HLA-DQA1: 16, APOE: 15, TOMM40: 11, CTLA4: 10, APOC1: 8, CYP2C19: 5, VEGFA: 4, CYP2C9: 4. The full list of gene-disease interactions is provided in Supplementary File 3.
Shared-disease gene–gene network
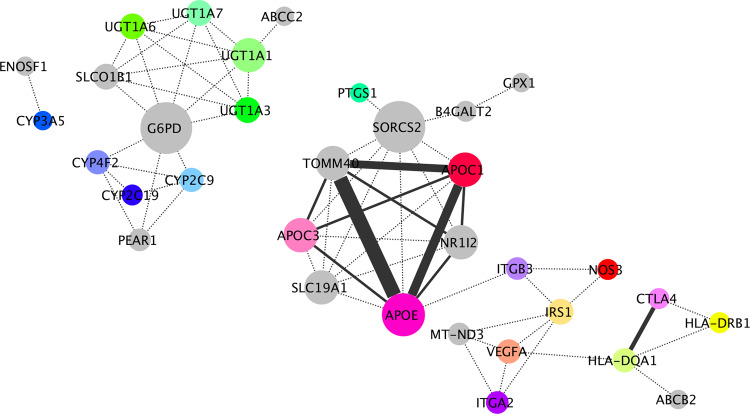
A shared-disease gene-gene network was then constructed by directly connecting the genes of the gene-disease network (associated with the same disease), with one edge (Fig. 4, and Fig. 1E, Supplementary File 3). From the three major disease clusters, three major gene clusters are formed. The importance of this network is that each gene of a cluster can be used as a predictor of a cluster of related diseases.
Fig. 4. The shared-disease gene–gene network predicted from the gene-disease network.
Edges between genes represent their common disease association that is extended from the gene-disease network. The size of a node indicates the higher connectivity with other genes. The line type indicates the number of common diseases (weight) between gene nodes (Supplementary File 3). Most genes shared only 1 common disease (dashed line). Thicker lines show interactions with the highest weight such as APOE-TOMM40: 11, APOE-APOC1: 7, APOC1-TOMM40: 6, CTLA4-HLA-DQA1: 4, APOC1-APOC3: 2, APOC1-NR1I2: 2, APOE-APOC3: 2, APOE-NR1I2: 2, TOMM40-APOC3: 2, TOMM40-NR1I2: 2.
Expanded PGx biomarker network
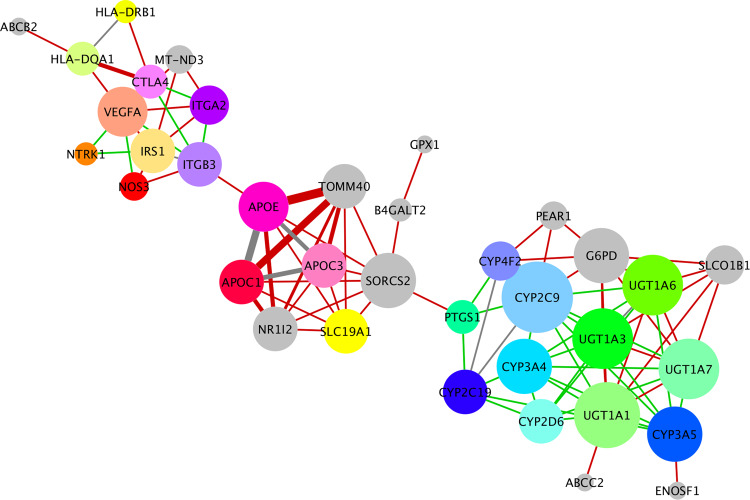
We then combined the information from the shared-disease gene-gene network and the gene-gene interaction network (PPI) generated by STRING and created the expanded PGx biomarker network (Fig. 5 and Fig. 1F, Supplementary File 3). Only 9 gene–gene interactions were found common between the two networks (APOC1:APOC3, APO3:APOC1, APOE: APOC3, CYP4F2:CYP2C19, CYP2C9:CYP2C19, CYP2C9:CYP4F2, UGT1A3:UGT1A6, HLA-DQA1: HLA-DRB1, ITGB3:IRS1). In this expanded network, the three gene clusters, representing the same three disease clusters were formed (Fig. 5). The first cluster includes 11 genes important risk factors for autoimmune, metabolic, and neurological diseases such as HLA-DRB1, HLA-DQA1, ITGA2, MT-ND3, VEGFA, CTL4, ABCB2. Some of them are involved in immune response (HLA-DRB1, HLA-DQA1) and some in-signal transduction (VEGFA, CTL4). The second cluster includes 6 apolipoproteins that are associated with cardiovascular and other degenerative diseases such as APOE, APOC1, APOC3, TOMM40, SLC19A1, NR1I2 SORCS2, PTGS1, ITGB3, NOS3, and IRS1. The last cluster includes 15 genes involved in drug metabolism pathways and is mainly members of cytochrome p450 (CYPs) and UDP-glucuronosyltransferase families (UGTs) (CYP2C19, CYP2C9, CYP4F2, PEAR1 G6PD, UGT1A1, ABCC2, SLCO1B1, UGT1A7, UGT1A6, UGT1A3). Remarkably, the extra information from the PPIs resulted in expanding each gene cluster rather than connecting the clusters, suggesting that the text-mining approach (from Liverpool data) we followed was of high specificity. The resulting expanded network expands, in a statistically significant manner, the number of genes that can be associated with a cluster of related diseases.
Fig. 5. Expanded PGx biomarker network.
The 44 significant PGx risk factors affecting COVID-19 clinical outcome are predicted with their genetic interactions: This network combines overlay of curated PPIs (green unweighted edges) (Supplementary Fig. 1) and the gene-gene network predicted by Kontou et al. (red weighted edges) (Fig. 4, Supplementary File 3). Gray edges are common edges between the two networks. The line width indicates the number of common diseases between two genes as shown in Fig. 4. The size of a node indicates the degree connectivity.
Validation of our approach with existing data
Phenome enrichment analysis of the 44 genes using PheWeb 2019 yielded nine enriched diseases states with p-value < 0.05 ((1) lipoid metabolism, (2) hyperlipidemia, (3) multiple myeloma, (4) dementias, (5) hypothyroidism, (6) acute bronchospasm, (7) rheumatoid arthritis, (8) diabetic retinopathy with p-value < 0.05). GWAS enrichment analysis of the 44 PGx biomarkers returned five enriched phenotypes with a 5% level of significance: Plantar warts, Myositis, Pneumonia, Cervical cancer, Parental longevity (combined parental attained age), total bilirubin levels, Shingles, Neuromyelitis optica, Triglycerides, Serum metabolite levels. The above disease phenotypes show concordance with diseases in the gene-disease association network predicted herein, thus validating the present approach. In addition, our analysis could predict more gene-disease associations related to degenerative disease, cardiovascular, drug metabolism, blood disorders, and autoimmune diseases.
Genes found in PubMed regulate COVID-19 outcome
Our approach was validated with the help of all available relative literature published in PubMed. We searched for all studies of the articles containing one of the 44 pharmacogenes and their link to SARS-CoV-2 infection. We found at least one publication for only 30 of the 44 PGx biomarkers reporting involvement in COVID-19 disease outcome: HLA, ABCB2, CTLA4, VEGFA, MT-ND3, ITGA2, IRS1, ITGB3. NTRK1. GPX1, APOE, APOC, SLC19A1, PTGS1, CYPs, UGTs, G6PD (Table 3).
Table 3.
Gene Validation by related covid-19 literature.
| Autoimmune, metabolic, and neurological diseases | |
|
Verified HLA-DRB1 HLA-DQA1 CTLA4 VEGFA ITGB3 ITGA2 IRS1 MT-ND3 NOS3 ABCB2 |
HLA is associated with risk for severe COVID-19. CTLA4 expression levels were correlated with viral levels. VEGFA increases endothelial dysfunction and correlates with COVID-19 disease severity, stimulates sensory receptors in central and peripheral nervous systems, cooperates with sars-cov2 spike protein to induce analgesia. CTLA4: HLA: VEGFA are highly expressed and associated with progression, immune regulation, and symptoms in COVID patients. ITGB2/3 SARS-CoV-2 infection revealed changes in genes related to coagulation (ITGB3). IRS1 Sars-cov-2 viral infection activates stress response, which induce IRS-1 phosphorylation and insulin resistance. MT-MD3 mitochondrial gene downregulated in sars-cov-2. NOS3 constitutes an important endothelial protection mechanism. However, ARDS diffuse inflammatory process triggers a vasodilatation cascade in non-ventilated parts of the lungs (deregulating hypoxic vasoconstrictive mechanisms) and vasoconstriction in ventilated areas. The imbalance between vasoconstricting and vasodilating pathways leads to endothelium dysfunction. ABCB contributes to drug resistance, lysosomal accumulation of COVID-19 treatments and related to virus replications. |
| Unverified | NTRK1 |
| Cardiovascular & other degenerative diseases | |
|
Verified APOC1 APOE APOC3 SLC19A1 GPX1 |
APOC1: APOE: APOC3 are early prognostic biomarkers for progression to severe COVID-19. A blood proteome profiling analysis revealed distinct functional characteristics of plasma proteins between severe and non-severe COVID-19 patients showing that these regulators of lipid homeostasis increased over the course of the disease. SLC19A1. Lymphocyte Changes in Severe COVID-19: Delayed Over-Activation of STING and highly express the cGAMP importer SLC19A1. GPX1 role of selenium-dependent GPX1 in SARS-CoV-2 virulence as a molecular target. |
| Unverified | TOMM40, NR1I2, SORCS2, B4GALT2 |
| Drug Metabolism | |
|
Verified G6PD CYP4F2 CYP2C9 CYP2C19 CYP3A4 CYP2D6 CYP3A5 UTG1A1 UTG1A3 UTG1A6 UTG1A7 PTGS1 |
G6PD deficiency facilitates human coronavirus infection due to glutathione depletion and shows a potential link between inherited G6PD deficiency and the racial inequities in mortality. G6PD was significantly induced in the lungs in COVID-19 obese patients. G6PD deficient cells infected with human coronavirus show impaired cellular responses, viral proliferation and worsening oxidative damage. G6PD deficiency is a predisposing factor of COVID-19 and deficient individuals are at high risk of severe hemolysis and or thrombosis when given anti-malarial treatment. CYP4F2: CYP2C9: CYP2C19: CYP3A4: CYP2D6: CYP3A5. Many PGx studies had evaluated the Genetic polymorphisms in members of cytochrome p450 (CYPs) that complicate COVID-19 therapy. UTG1A1: UTG1A3: UTG1A6: UTG1A7 are associated with bilirubin, a compound that occurs in the normal catabolic pathway that breaks down heme. COVID-19 patients with elevated bilirubin levels had a higher mortality. Polymorphism in the UGT1A1 gene has been observed in a patient’s case with Gilbert Syndrome attenuating COVID-19 metabolic disturbances. PTGS1 inhibition with COVID-19 treatments triggers upregulation of IL10 gene expression and represses platelet aggregation. |
| Unverified | ENOSF1, ABCC2, SLCO1B1, PEAR1 |
| Disconnected genes | |
|
Verified VDR ZSCAN25 ABCB1 |
VDR exerts a critical role in prevention and protection of viral acute respiratory infection. VDR deficiency can aggravate respiratory syndrome by igniting a wounding response in stellate cells of the lung. VDR related genetic variants were implicated in severe COVID-19 in adults. ZSCAN25 associates with Sex Hormone-Binding Globulin (SHBG) Levels. Low SHBG were associated with mortality rate in patients with COVID-19, and low total and free testosterone levels were associated with mortality in men. ABCB1 has a possible role in lysosomal accumulation of COVID-19 treatments and related to virus replications. |
| Unverified | BCC1, TYMS, NAT2, MTR, TYMSOS, SLCO2B1 |
30 genes are found in literature that are related to COVID-19 susceptibility whereas 15 genes are not yet verified, however they have been predicted as candidate genes in relation to COVID-19 disease.
Discussion
Bioinformatics has been very helpful in identifying potential molecular mechanisms and key genes involved in COVID-19 [35]. Due to the limited data on common PGx affecting concomitant medications response, to our knowledge, this is the first attempt which tries to combine bioinformatics, data mining, and network biology approaches to mine and analyze drug interactions of repurposed COVID-19 treatments. This effort aims at predicting the most significant genes which minimize the effectiveness and/or safety of drugs and could be possible early prognostic markers in relation to COVID-19. The predicted PGx network could initiate the development of clinically applicable diagnostic and prognostic tests for patients at high risk of COVID-19 and those who cannot be vaccinated due to health conditions and allergies [36].
Extended network analysis of these markers’ genetic interactions indicates that patients who carry variants of these genes may have long-term cardiovascular, immune, or neurological effects after infection with SARS-CoV2 as they are found to be known risk factors of heart, neuron, liver, and metabolic diseases. Several predicted genes in the network are currently used as prognostic biomarkers (Table 3), however, some others (14 genes) are not yet verified in their relation to COVID-19, and thus, through this work, they could be of great benefit for the prevention and assessment of disease severity and could play a role for the disease outcome. The receptor neurotrophic tyrosine kinase (NTRK1) is involved in the development and the maturation of the central and peripheral nervous system, and it is the only unverified gene in the first cluster of the network. NTRK1 is associated with insensitivity to pain and thyroid carcinoma but it doesn’t share common disease with VEGFA. However, there is a curated interaction between NTRK1 and VEGFA gene suggesting that both are participating in the same signaling pathway. A recent study suggests that SARS-CoV-2 spike protein interferes with endogenous ligand VEGFA, promotes a signal arriving at the central nervous system, and stimulates specialized sensory receptors in the peripheral nervous system inducing analgesia [37, 38]. Thus, small-molecule inhibitors of this signaling for the treatment of neuropathic pain and cancer are being tested and could have added potential of inhibiting SARS-CoV-2 virus entry [39].
Network analysis reveals some genes that are central and tend to connect different clusters of diseases (bottlenecks) that may play a significant role in disease outcome in patients with comorbidities. Such genes in the first cluster are the ITGA2/ITGB3 and IRS1 that connect pathways of the neurological system with cardiovascular respectively. Moreover, IRS1 has a central node that interacts with NTRK1 and ITGB3 via curation suggesting that it is an immediate connector of genes related to cardiovascular and degenerative diseases. All genes of the second cluster seem to be central with dense connectivity with apolipoproteins APOC1, APOE, APOC3 which are early prognostic biomarkers for severe COVID-19 and share common diseases of cholesterol and mental disorders with genes such as TOMM40, the mitochondrial import receptor subunit. TOMM40 is an unverified gene in relation to COVID-19 but it is known as a risk factor for metabolic or neurological and mental disorders.
Interestingly, dementia was among the common comorbidities and was associated with higher mortality due to the APOE homozygous genotyping in hospitalized COVID-19 patients [40–42]. These findings strongly highlight the possible cognitive impact of COVID-19 due to the APOE/APOC cluster with TOMM40 and their interactions [43–45] and could be also potential genetic biomarkers of COVID-19 severity. In addition, network analysis shows SORCS2 as a bottleneck that connects the 2nd and the 3rd cluster of drug metabolic genes through PTGS1 and shares common diseases of myeloid leukemia and cholesterol. PTGS1 according to literature has an unknown role in COVID-19, but its inhibition with COVID-19 treatments triggers upregulation of IL10 gene expression and represses platelet aggregation [46]. A distant connected sub-cluster of SORCS2:B4GALT2:GPX1 genes is associated with blood protein levels and intelligence. SORCS2 and B4GALT2 genes were not found as verified in relation to COVID-19 however, they are found as risk factors of diseases in cholesterol and protein levels, and they may indirectly increase the susceptibility of patients to SARS-CoV-2 and increase the risk of death. The NR1I2 gene, a member of the nuclear receptor subfamily 1 Group, is associated with disease of C-reactive protein, maybe another significant node as in COVID-19 the elevated levels of CRP might be linked to the overproduction of inflammatory cytokines in severe patients [47]. We suggest that elevated levels of C-reactive protein may be an early marker to predict risk for severity of COVID-19 and NR1I2 may be a prognostic biomarker as well as its neighbors [47]. In the third cluster, several CYP genes are known as PGx biomarkers affecting the metabolism of repurposed drugs and known risk of liver disease. Our analysis has predicted multiple bilirubin-associated genes including ABCC2, SLCO1B1, G6PD, UGT1A1/3/6/7, and other essential hypertension and blood pressure-associated genes PEAR1, G6PD, SLO1B1, ABCC2, ENOSF1 densely interacting with CYPs.
Future work is needed to identify PGx associations with repurposed COVID-19 treatments that do not have PGx information in PharmaGKB and include more interactions in the network. A potential solution to this problem would be to include variants and genes having predicted effects derived from existing publicly available computational prediction methods [ref. 1–2]. However, such an approach has many problems related to the large number of potential “targets” that need to be evaluated and added to the model. Nevertheless, it is a possible extension of our approach that needs to be investigated in future studies. In addition, evaluation of population-specific PGx landscape genetic analysis of the genes proposed as biomarkers is needed that might aid in a better understanding of the inconsistency in therapy response [48, 49].
Supplementary information
Acknowledgements
GAP would like to acknowledge support from the Hellenic Foundation for Research and Innovation (H.F.R.I) under the “First Call for H.F.R.I Research Projects to support faculty members and researchers and the procurement of high-cost research equipment grant”, Grant ID: 1855-BOLOGNA.
Author contributions
PGB participated in the conception of the study, in data collection, in the analysis and in the interpretation of the results. TC did most of data collection, performed most of the analysis, participated in the interpretation of the results, and drafted the initial version of the manuscript. GCB participated in data collection, in the analysis and in the interpretation of the results. PIK participated in data collection, in the analysis and in the interpretation of the results. IT and GAP participated in the interpretation of the results. All authors participated in drafting the manuscript. All authors read and approved the final version of the manuscript.
Funding
GAP was supported by the project ‘The Greek Research Infrastructure for Personalised Medicine (pMedGR)’ (MIS 5002802), which is implemented under the Action ‘Reinforcement of the Research and Innovation Infrastructure’, funded by the Operational Program ‘Competitiveness, Entrepreneurship and Innovation’ (NSRF 2014–2020) and co-financed by Greece and the European Union (Eu-ropean Regional Development Fund).
Data availability
All data generated or analyzed during this study are included in this published article and its Supplementary Information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41397-022-00289-1.
References
- 1.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal A, Rochwerg B, Lamontagne F, Siemieniuk RA, Agoritsas T, Askie L, et al. A living WHO guideline on drugs for covid-19. Bmj. 2020;370:m3379. doi: 10.1136/bmj.m3379. [DOI] [PubMed] [Google Scholar]
- 3.Han N, Hwang W, Tzelepis K, Schmerer P, Yankova E, MacMahon M, et al. Identification of SARS-CoV-2-induced pathways reveals drug repurposing strategies. Sci Adv. 2021;7:eabh3032. doi: 10.1126/sciadv.abh3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lovato ECW, Barboza LN, Wietzikoski S, de Souza ANV, Auth PA, Junior AG, et al. Repurposing drugs for the management of patients with confirmed coronavirus disease 2019 (COVID-19) Curr Pharm Des. 2021;27:115–26.. doi: 10.2174/1381612826666200707121636. [DOI] [PubMed] [Google Scholar]
- 5.Santos J, Brierley S, Gandhi MJ, Cohen MA, Moschella PC, Declan ABL, et al. Repurposing therapeutics for potential treatment of SARS-CoV-2: a review. Viruses. 2020;12:705. doi: 10.3390/v12070705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gyanwali P, Sharma S, Pant S, Koirala P, Adhikari K, Koirala J, et al. Safety and efficacy of different therapeutic interventions on prevention and treatment of COVID-19. J Nepal Health Res Counc. 2020;18:151–8. doi: 10.33314/jnhrc.v18i2.2806. [DOI] [PubMed] [Google Scholar]
- 7.Mehraeen E, Najafi Z, Hayati B, Javaherian M, Rahimi S, Dadras O, et al. Current treatments and therapeutic options for COVID-19 patients: a systematic review. Infect Disord Drug Targets. 2021;22:e260721194968. doi: 10.2174/1871526521666210726150435. [DOI] [PubMed] [Google Scholar]
- 8.Stasi C, Fallani S, Voller F, Silvestri C. Treatment for COVID-19: an overview. Eur J Pharm. 2020;889:173644. doi: 10.1016/j.ejphar.2020.173644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang WD, Jeon S, Kim S, Lee SY. Drugs repurposed for COVID-19 by virtual screening of 6,218 drugs and cell-based assay. Proc Natl Acad Sci USA. 2021;118:e2024302118. [DOI] [PMC free article] [PubMed]
- 10.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–81.. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah RR, Smith RL. Addressing phenoconversion: the Achilles’ heel of personalized medicine. Br J Clin Pharm. 2015;79:222–40. doi: 10.1111/bcp.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah RR. Pharmacogenetics and precision medicine: Is inflammation a covert threat to effective genotype-based therapy? Ther Adv Drug Saf. 2017;8:267–72.. doi: 10.1177/2042098617712657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fricke-Galindo I, Falfán-Valencia R. Pharmacogenetics approach for the improvement of COVID-19 treatment. Viruses. 2021;13:413. [DOI] [PMC free article] [PubMed]
- 14.Via M, Tcheurekdjian H, González Burchard E. Role of interactions in pharmacogenetic studies: leukotrienes in asthma. Pharmacogenomics. 2013;14:923–9. doi: 10.2217/pgs.13.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodge C, Marra F, Marzolini C, Boyle A, Gibbons S, Siccardi M, et al. Drug interactions: a review of the unseen danger of experimental COVID-19 therapies. J Antimicrob Chemother. 2020;75:3417–24. doi: 10.1093/jac/dkaa340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seden K, Gibbons S, Marzolini C, Schapiro JM, Burger DM, Back DJ, et al. Development of an evidence evaluation and synthesis system for drug–drug interactions, and its application to a systematic review of HIV and malaria co-infection. PLoS One. 2017;12:e0173509. doi: 10.1371/journal.pone.0173509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein DB, Tate SK, Sisodiya SM. Pharmacogenetics goes genomic. Nat Rev Genet. 2003;4:937–47. doi: 10.1038/nrg1229. [DOI] [PubMed] [Google Scholar]
- 18.Haga SB, Burke W. Using pharmacogenetics to improve drug safety and efficacy. Jama. 2004;291:2869–71. doi: 10.1001/jama.291.23.2869. [DOI] [PubMed] [Google Scholar]
- 19.Telbisz Á, Ambrus C, Mózner O, Szabó E, Várady G, Bakos É, et al. Interactions of Potential Anti-COVID-19 Compounds with Multispecific ABC and OATP Drug Transporters. Pharmaceutics. 2021;13:81. [DOI] [PMC free article] [PubMed]
- 20.Detailed recommendations for interactions with experimental COVID-19 antiviral therapies: University of Liverpool; 2021 [Available from: www.covid19-druginteractions.org].
- 21.Detailed recommendations for interactions with experimental COVID-19 immune therapies: University of Liverpool; 2021 [Available from: www.covid19-druginteractions.org].
- 22.Detailed recommendations for interactions with experimental COVID-19 adjunct therapies University of Liverpool 2021 [Available from: www.covid19-druginteractions.org].
- 23.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–d82.. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, et al. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamosh A, Scott AF, Amberger J, Bocchini C, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic acids Res. 2002;30:52–5. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet. 2004;36:431–2. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 27.Whirl-Carrillo M, Huddart R, Gong L, Sangkuhl K, Thorn CF, Whaley R, et al. An evidence-based framework for evaluating pharmacogenomics knowledge for personalized medicine. Clin Pharm Ther. 2021;110:563–72. doi: 10.1002/cpt.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.StataCorp L. Stata Statistical Software: Release 13. Stata Press College Station, TX. 2013.
- 29.Kim HY. Statistical notes for clinical researchers: Chi-squared test and Fisher’s exact test. Restor Dent Endod. 2017;42:152–5. doi: 10.5395/rde.2017.42.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thanati F, Karatzas E, Baltoumas FA, Stravopodis DJ, Eliopoulos AG, Pavlopoulos GA. FLAME: a web tool for functional and literature enrichment analysis of multiple gene lists. Biology (Basel). 2021;10:665. [DOI] [PMC free article] [PubMed]
- 31.Kontou PI, Pavlopoulou A, Dimou NL, Pavlopoulos GA, Bagos PG. Network analysis of genes and their association with diseases. Gene. 2016;590:68–78. doi: 10.1016/j.gene.2016.05.044. [DOI] [PubMed] [Google Scholar]
- 32.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goenawan IH, Bryan K, Lynn DJ. DyNet: visualization and analysis of dynamic molecular interaction networks. Bioinformatics. 2016;32:2713–5. doi: 10.1093/bioinformatics/btw187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karatzas E, Baltoumas FA, Panayiotou NA, Schneider R, Pavlopoulos GA. Arena3Dweb: interactive 3D visualization of multilayered networks. Nucleic Acids Res. 2021;49:W36–w45. doi: 10.1093/nar/gkab278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ceylan H. A bioinformatics approach for identifying potential molecular mechanisms and key genes involved in COVID-19 associated cardiac remodeling. Gene Rep. 2021;24:101246. doi: 10.1016/j.genrep.2021.101246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizzi C, Dalabira E, Kumuthini J, Dzimiri N, Balogh I, Başak N, et al. A European spectrum of pharmacogenomic biomarkers: implications for clinical pharmacogenomics. PLoS One. 2016;11:e0162866. doi: 10.1371/journal.pone.0162866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jobe A, Vijayan R. Neuropilins: C-end rule peptides and their association with nociception and COVID-19. Comput Struct Biotechnol J. 2021;19:1889–95. doi: 10.1016/j.csbj.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moutal A, Martin LF, Boinon L, Gomez K, Ran D, Zhou Y, et al. SARS-CoV-2 spike protein co-opts VEGF-A/neuropilin-1 receptor signaling to induce analgesia. Pain. 2021;162:243–52.. doi: 10.1097/j.pain.0000000000002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Miller S, Patek M, Moutal A, Cabel CR, Thorne CA, Campos SK, et al. In silico identification and validation of inhibitors of the interaction between neuropilin receptor 1 and SARS-CoV-2 Spike protein. bioRxiv. 2020:2020.09.22.308783.
- 40.Kuo CL, Pilling LC, Atkins JL, Fortinsky RH, Kuchel GA, Melzer D. APOE e4 genotypes increase risk of delirium during COVID-19 related hospitalizations: evidence from a large United Kingdom cohort. J Gerontol A Biol Sci Med Sci. 2021;77:879–80. [DOI] [PMC free article] [PubMed]
- 41.Kuo CL, Pilling LC, Atkins JL, Masoli JAH, Delgado J, Kuchel GA, et al. APOE e4 genotype predicts severe COVID-19 in the UK Biobank Community Cohort. J Gerontol A Biol Sci Med Sci. 2020;75:2231–2. doi: 10.1093/gerona/glaa131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodrigues JCG, Pinto P, Leitão LPC, Vinagre L, Monte N, Fernandes MR, et al. Influence of APOE locus on poor prognosis of COVID-19. Heliyon. 2021;7:e07379. doi: 10.1016/j.heliyon.2021.e07379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miners S, Kehoe PG, Love S. Cognitive impact of COVID-19: looking beyond the short term. Alzheimers Res Ther. 2020;12:170. doi: 10.1186/s13195-020-00744-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Numbers K, Brodaty H. The effects of the COVID-19 pandemic on people with dementia. Nat Rev Neurol. 2021;17:69–70. doi: 10.1038/s41582-020-00450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang C, Zhang M, Garcia G, Jr., Tian E, Cui Q, Chen X, et al. ApoE-Isoform-dependent SARS-CoV-2 neurotropism and cellular response. Cell Stem Cell. 2021;28:331–42.e5. doi: 10.1016/j.stem.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao J, Tian S, Lu D, Yang J, Zeng H, Zhang F, et al. Systems pharmacological study illustrates the immune regulation, anti-infection, anti-inflammation, and multi-organ protection mechanism of Qing-Fei-Pai-Du decoction in the treatment of COVID-19. Phytomedicine. 2021;85:153315. doi: 10.1016/j.phymed.2020.153315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ali N. Elevated level of C-reactive protein may be an early marker to predict risk for severity of COVID-19. J Med Virol. 2020;92:2409–11. doi: 10.1002/jmv.26097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sahana S, Sivadas A, Mangla M, Jain A, Bhoyar RC, Pandhare K, et al. Pharmacogenomic landscape of COVID-19 therapies from Indian population genomes. Pharmacogenomics. 2021;22:603–18. doi: 10.2217/pgs-2021-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stanković B, Kotur N, Gašić V, Klaassen K, Ristivojević B, Stojiljković M, et al. Pharmacogenomics landscape of COVID-19 therapy response in Serbian population and comparison with worldwide populations. J Med Biochem. 2020;39:488–99. doi: 10.5937/jomb0-26725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Information files.