Abstract
Staphylococcus epidermidis is a common pathogen in medical device-associated infections. Its major pathogenetic factor is the ability to form adherent biofilms. The polysaccharide intercellular adhesin (PIA), which is synthesized by the products of the icaADBC gene cluster, is essential for biofilm accumulation. In the present study, we characterized the gene locus inactivated by Tn917 insertions of two isogenic, icaADBC-independent, biofilm-negative mutants, M15 and M19, of the biofilm-producing bacterium S. epidermidis 1457. The insertion site was the same in both of the mutants and was located in the first gene, rsbU, of an operon highly homologous to the sigB operons of Staphylococcus aureus and Bacillus subtilis. Supplementation of Trypticase soy broth with NaCl (TSBNaCl) or ethanol (TSBEtOH), both of which are known activators of sigB, led to increased biofilm formation and PIA synthesis by S. epidermidis 1457. Insertion of Tn917 into rsbU, a positive regulator of alternative sigma factor ςB, led to a biofilm-negative phenotype and almost undetectable PIA production. Interestingly, in TSBEtOH, the mutants were enabled to form a biofilm again with phenotypes similar to those of the wild type. In TSBNaCl, the mutants still displayed a biofilm-negative phenotype. No difference in primary attachment between the mutants and the wild type was observed. Similar phenotypic changes were observed after transfer of the Tn917 insertion of mutant M15 to the independent and biofilm-producing strain S. epidermidis 8400. In 11 clinical S. epidermidis strains, a restriction fragment length polymorphism of the sigB operon was detected which was independent of the presence of the icaADBC locus and a biofilm-positive phenotype. Obviously, different mechanisms are operative in the regulation of PIA expression in stationary phase and under stress induced by salt or ethanol.
Staphylococcus epidermidis, a normal inhabitant of human skin and mucous membranes, is the predominant cause of foreign-body-associated infections (43). In addition, S. epidermidis is isolated with increasing frequency as the causative pathogen of nosocomial sepsis and other nosocomial infections, ranking among the five most frequent nosocomial pathogens (43, 49). The pathogenesis of S. epidermidis infections is correlated with the ability to form biofilms on polymer surfaces (5, 58).
Biofilm formation proceeds in two phases (23, 24). Primary attachment of bacterial cells to a polymer surface is a complex process influenced by a variety of factors, including hydrophobic interactions, presence of host proteins, and specific bacterial proteins and polysaccharides like the capsular polysaccharide adhesin, the autolysin AtlE, and other staphylococcal surface proteins (15, 17, 33, 38, 39, 50, 51). This is followed by the second phase leading to accumulation of bacteria in a multilayered biofilm embedded in an amorphous glycocalyx. Synthesis of the polysaccharide intercellular adhesin (PIA) is essential for bacterial cell accumulation because it mediates cell-to-cell adhesion of proliferating cells (26–28, 31, 32). PIA consists of two polysaccharide species which are composed of β-1,6-linked 2-deoxy-2-amino-d-glucopyranosyl residues containing non-N-acetylated amino groups, phosphate, and succinate (26) and is synthesized by the products of the icaADBC gene cluster (11, 16). In addition to having a function in intercellular adhesion, PIA is essential for hemagglutination mediated by S. epidermidis (10, 29, 42, 44). Recently, the significance of PIA as a virulence factor could be demonstrated by comparison of isogenic PIA-negative transposon mutant 1457-M10 and the corresponding wild-type strain in a central venous catheter rat infection model and a subcutaneous foreign-body mice infection model (45, 46).
By transposon mutagenesis, four unlinked gene loci were identified whose mutation leads to a biofilm-negative phenotype and abolished PIA synthesis and produces mutants that are classified, according to genotypic and phenotypic differences, as class I to IV mutants (30, 37). Class I mutants represent those in which icaADBC is inactivated. The other three genetic loci control expression of PIA synthesis and biofilm formation by directly or indirectly influencing expression of icaADBC on the level of transcription (30).
Understanding of the regulatory mechanisms regulating PIA synthesis and biofilm formation is of primary importance for the development of new preventive and therapeutic methods to combat S. epidermidis biomaterial-related infections. Therefore, in the present study, we characterized the genetic defect of the class III biofilm-negative mutants M15 and M19 at the molecular level.
(Part of this work will appear in the Ph.D. theses of J.K.-M.K., K.B., A.S., and H.R., Universitätsklinikum Hamburg-Eppendorf, Hamburg, Germany.)
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. epidermidis cells were grown in Trypticase soy broth (TSBBBL; Becton Dickinson, Cockeysville, Md.) at 37°C. For phenotypic characterization of the S. epidermidis strains, TSBBBL was supplemented with 4% NaCl (TSBNaCl) or 4% ethanol (TSBEtOH). E. coli cells were grown in Luria-Bertani (LB) broth or on LB agar at 37°C. Antibiotics were used at the following concentrations: erythromycin, 300 μg/ml; ampicillin, 150 μg/ml; and kanamycin, 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Reference or source | Comments |
|---|---|---|
| S. epidermidis | ||
| 1457 | 31 | Isolate from infected central venous catheter |
| M12 | 30 | Isogenic biofilm-negative Tn917 mutant of S. epidermidis 1457 (class II) |
| M15 | 30 | Isogenic biofilm-negative Tn917 mutant of S. epidermidis 1457 (class III) |
| M19 | 30 | Isogenic biofilm-negative Tn917 mutant of S. epidermidis 1457 (class III) |
| M16 | This study | Isogenic biofilm-positive control Tn917 mutant of S. epidermidis 1457 |
| 8400 | 31 | Blood culture isolate |
| 8400-M15 | This study | Transductant of S. epidermidis 8400 |
| 5179 | 31 | Blood culture isolate |
| RP62A | 7 | |
| SE5 | 42 | |
| 521 | 27 | Isolate from infected central venous catheter |
| 1057 | 27 | Isolate from infected central venous catheter |
| 939 | 27 | Blood culture isolate |
| 10333 | 31 | Blood culture isolate |
| 7837 | 31 | Blood culture isolate |
| 9225 | 31 | Blood culture isolate |
| 9896 | 31 | Blood culture isolate |
| S. aureus 35005 | This study | Clinical isolate |
| E. coli | ||
| MC1061 | 12 | |
| TOP10 | Invitrogen | |
| JKMK186-41 | This study | MC1061 containing pJKMK186-41 |
| JKMK186-46 | This study | MC1061 containing pJKMK186-46 |
| JKMK401 | This study | TOP10 containing pJKMK401 |
| JKMK402 | This study | TOP10 containing pJKMK402 |
| Plasmids | ||
| pBluescript II SK | Invitrogen | E. coli cloning vector for blue-white screening |
| pCRII | Invitrogen | E. coli cloning vector for direct cloning of PCR fragments |
| pJKMK186-41 | This study | pBluescript II SK containing HindIII/SalI fragment of 1457-M15 with chromosomal DNA flanking 5′ end (erm) of Tn917 |
| pJKMK186-46 | This study | pBluescript II SK containing HindIII/SalI fragment of 1457-M19 with chromosomal DNA flanking 5′ end (erm) of Tn917 |
| pJKMK401 | This study | pCRII containing sigB amplification product obtained from S. epidermidis 1457 with oligonucleotides JKMK4 and JKMK5 |
| pJKMK402 | This study | pCRII containing amplification product obtained with oligonucleotides 3R and 5L (sigB operon of S. epidermidis 1457) |
Phenotypic characterization.
Biofilm production by S. epidermidis was measured by a semiquantitative adherence assay in the appropriate media in 96-well tissue culture plates (Nunclon Delta; Nunc, Roskilde, Denmark) as previously described (7, 31). For detection of PIA by immunofluorescence assay (IFA), S. epidermidis cells were grown in tissue culture dishes (Nunc) for 22 h in TSBBBL, TSBNaCl, or TSBEtOH, respectively. Cells were scraped off and diluted in phosphate-buffered saline to an optical density at 578 nm (OD578) of 0.3 to 0.5. The IFA procedure was then performed as previously described using a rabbit antiserum raised against purified PIA (16, 31).
For quantitation of PIA in bacterial extracts, S. epidermidis strains were grown in a 9-cm tissue culture dish (Nunc) for 22 h in TSBBBL, TSBNaCl, or TSBEtOH, respectively. Cells were scraped off and centrifuged (3,000 × g for 15 min). The culture supernatants were cleared by an additional centrifugation step (3,000 × g for 15 min), and NaN3 was added to a final concentration of 0.05%. The cell pellet was resuspended in 5 ml of phosphate-buffered saline containing 0.05% NaN3, and bacterial extracts were prepared by sonication (two 30-s cycles with a 3/16-in. tapered Microtip at 70% of the maximal amplitude) with Digital Sonifier 250-D (Branson, Danbury, Conn.). Cells were sedimented by centrifugation (3,000 × g for 15 min). The cell extracts were cleared by an additional centrifugation step (12,000 × g for 15 min). PIA concentrations in culture supernatants and cell extracts were determined by a specific coagglutination assay with PIA-specific antiserum (31).
For quantitation of primary attachment, bacterial strains were grown in TSBBBL, TSBNaCl, or TSBEtOH with shaking. Cells were harvested by centrifugation and resuspended and diluted in phosphate-buffered saline. Bacterial cell concentrations were determined by plating of appropriate dilutions. Bacterial dilutions (100 μl) at various concentrations were added in triplicate to the wells of 96-well cell culture plates (Nunclon Delta; Nunc) and incubated for 1 h at 37°C. After washing, attached cells were detected by enzyme-linked immunosorbent assay (ELISA) using rabbit anti-S. epidermidis 5179 serum and alkaline phosphatase-coupled anti-rabbit immunoglobulin G (Sigma) as previously described (25, 45).
Genetic methods.
Transduction of the Tn917 insertion of mutant M15 into the independent and biofilm-producing wild-type strain 8400 was performed essentially as described previously using S. epidermidis phage 71 (25, 37).
Chromosomal DNA of S. epidermidis was prepared as described previously (28). DNA was cleaved with restriction enzymes as suggested by the manufacturer (Pharmacia, Freiburg, Germany), and DNA fragments were separated by electrophoresis in 0.7% agarose gels in Tris-borate buffer (47).
DNA restriction fragments of the expected size were purified from agarose gels using the Gene Clean II kit (Bio 101, Inc., Vista, Calif.) and cloned in Escherichia coli MC1061 using pBluescript II SK (Stratagene, La Jolla, Calif.) as a vector. Selection was for ampicillin-resistant clones on LB agar plates. Positive clones were transferred to LB agar plates containing erythromycin selecting for the erythromycin resistance gene (erm) of Tn917 (12). Positive clones were further characterized.
For Southern blot analysis DNA fragments were transferred onto Zeta-Probe membranes (Bio-Rad, Munich, Germany) by alkaline capillary blotting (47). Hybridization was performed using probes labeled with [32P]dCTP (Amersham, Braunschweig, Germany) by the Ready to Go labeling kit (Pharmacia) as suggested by the manufacturer. The blots were exposed to Kodak X-Omat X-ray film.
Nucleotide sequence analysis was performed on an ABI Prism 310 sequencer by capillary electrophoresis using the ABI Prism dGTP BigDye Terminator Ready Reaction Kit (PE Applied Biosystems, Foster City, Calif.). Nucleotide sequences were analyzed subsequently with HUSAR software (DKFZ, Heidelberg, Germany).
Amplification of short DNA fragments (approximately 2 kb) was performed using the DyNazyme DNA Polymerase Kit (Finzyme, Espoo, Finnland) as described by the manufacturer. Oligonucleotides specific for rsbU (JKMK1 [5′-GTG GAA GAA TTT AAG CAA CA-3′] and JKMK2 [5′-GGA ATA TCT GTT TTT AAG CAT-3′]) and sigB (JKMK4 [5′-CTG AGC AAA TTA ACC AAT GG-3′] and JKMK5 [5′-TAA CTT TGT CCC ATT TCC AT-3′]) of S. aureus (57) and icaB (icaBforward [5′-TGG ATC AAA CGA TTT ATG ACA-3′] and icaBreverse [5′-ATG GGT AAG CAA GTG CGC-3′]) of S. epidermidis (11, 16) and oligonucleotides JK41.rev1 (5′-AGC GAA AAT ACC AAC CCA CG-3′), JKMK11 (5′-GAG GAA ATT GGT GTG CGA GG-3′), JKMK28 (5′-TGT GAA TGT CCA TAA GCA TCC-3′), and JKMK34 (5′-TTT CTT TTA GCC TCA GTT GC-3′) were synthesized by MWG Biotech (Munich, Germany).
For long-range PCR, the Expand Long Template PCR System (Boehringer, Mannheim, Germany) was used with oligonucleotides specific for the 5′ and 3′ junctions of Tn917 (5L [5′-CTC ACA ATA GAG AGA TGT CAC CG-3′] and 3R [5′-GGC CTT GAA ACA TTG GTT TAG TGG G-3′]) (48) as described by the manufacturer for an expected fragment length of 12 to 15 kb.
RESULTS
Cloning of DNA flanking Tn917 insertion sites.
By genetic mapping, the Tn917 insertions of class III mutants M15 and M19 were shown to be closely linked (30). For identification of the inactivated chromosomal structures of these mutants, two identical 3.1-kb chromosomal SalI/HindIII fragments containing 0.8 kb of S. epidermidis chromosomal DNA flanking the 5′ end of Tn917, including the erythromycin resistance gene (erm) up to the first HindIII site of Tn917 (48), were cloned for both mutants in E. coli MC1061, resulting in plasmids pJKMK186-41 and pJKMK186-46. Nucleotide sequence analysis indicated that Tn917 was inserted at the same position in both mutants M15 and M19 (data not shown). The identities of the cloned fragments with the insertion site of Tn917 in the mutants were demonstrated by Southern blot hybridization using the cloned chromosomal fragment of mutant M15 as a probe (data not shown). The nucleotide sequence at the proximal end of the chromosomal fragment near the SalI site displayed an open reading frame (ORF) encoding 120 amino acids (ORF1), while the remaining sequence appeared to be noncoding. ORF1 was highly homologous (80.9% identical bases) to an ORF, designated ORF1 or ORF136, localized proximally to the sigB operon of S. aureus (21, 57). The noncoding region near the Tn917 insertion site of the mutants was homologous (67.6% identical bases) to the noncoding region directly preceding the S. aureus sigB operon.
Linkage of sigB of S. epidermidis and the Tn917 insertion site.
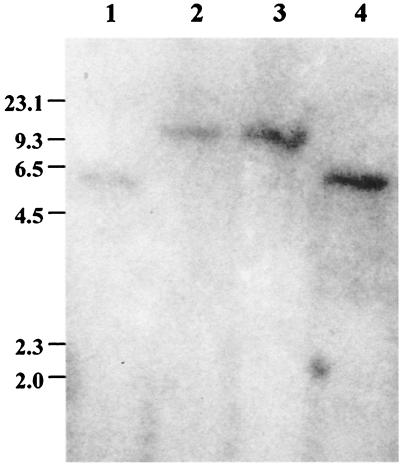
To ascertain the existence of a sigB homologue in S. epidermidis, we used oligonucleotides specific for the S. aureus rsbU (JKMK1 and JKMK3) and sigB (JKMK4 and JKMK6) genes for PCR amplification. Only with sigB-specific primers was a 696-bp fragment from S. epidermidis 1457 chromosomal DNA amplified which was similar in size to that amplified from a clinical S. aureus isolate. Nucleotide sequence analysis of the fragment obtained from S. epidermidis 1457(pJKMK401) revealed homology to sigB of S. aureus (21, 57). To determine the linkage of the S. epidermidis sigB homologue with the Tn917 insertion site of the class III mutants M15 and M19, the PCR fragment was used as a probe in a Southern blot assay (Fig. 1). Decreased mobility of the hybridizing EcoRI fragment of mutants M15 and M19, consistent with insertion of 5.2-kb Tn917, was observed, whereas no mobility change was detected with chromosomal DNA of class II mutant M12 (used as a control). Apparently, the sigB homologue of S. epidermidis 1457 is linked to the EcoRI fragment containing the Tn917 insertion of mutants M15 and M19. For chromosomal DNA of S. aureus, no hybridization signal was obtained when this probe was used under stringent hybridization conditions (data not shown).
FIG. 1.
Southern blot of EcoRI-digested chromosomal DNA using a 32P-labeled S. epidermidis sigB PCR fragment from pJKMK401 as a probe. Lanes 1, S. epidermidis 1457; 2, S. epidermidis M15; 3, S. epidermidis M19; 4, S. epidermidis M12 (class II mutant). The values on the left are sizes in kilobases.
Analysis of the sigB operon of S. epidermidis 1457.
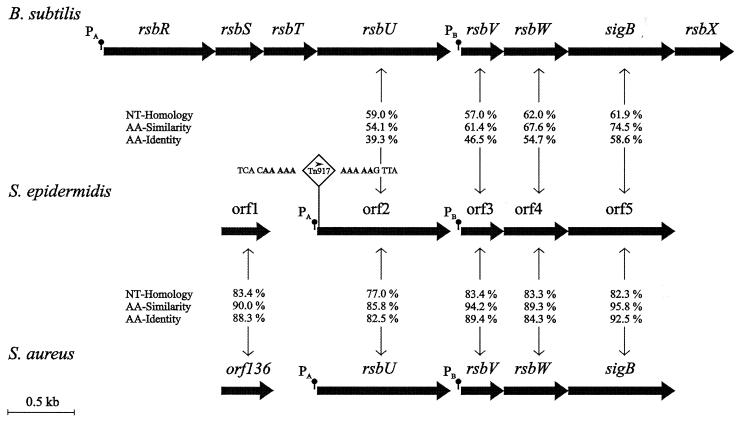
A 12.4-kb chromosomal EcoRI fragment containing the Tn917 insertion of mutant M15 was autoligated, and long-range PCR was performed using oligonucleotides 5L and 3R, which are complementary to the 5′ and 3′ junctions of Tn917. The resulting 7.2-kb PCR fragment was cloned, resulting in plasmid pJKMK402, which was sequenced by primer walking. Nucleotide sequences were completed by analysis of a DNA fragment generated by amplification of chromosomal DNA of wild-type S. epidermidis 1457 with primers JK41.rev1 and JKMK28, which overlap the transposon insertion site. Sequence analysis revealed an operon (2,700 nucleotides [nt]) consisting of four ORFs (ORF2 to ORF5) highly homologous to the sigB operons of S. aureus (21, 57) and Bacillus subtilis (18, 56). The four ORFs are organized in the same conserved order as their homologous counterparts rsbU, rsbV, rsbW, and sigB (Fig. 2). For each gene, a putative Shine-Dalgarno sequence could be detected (data not shown). As described for S. aureus and B. subtilis, ORF3, which is homologous to rsbV of S. epidermidis, is preceded by a putative ςB-dependent promoter with its −35 (TAGATTAA) and −10 (GGGTAT) promoter elements spaced by 14 nt, which conforms to the consensus sequence (13, 40). In the amino acid sequence of ORF4, which is homologous to RsbW, the residues thought to be important for ATP binding, and therefore kinase activity (19), are conserved in S. epidermidis, S. aureus, and B. subtilis (data not shown). These data permit the conclusion that these genes function similarly, and ORF2 to ORF5 were therefore named like their homologous counterparts in S. aureus, rsbU, rsbV, rsbW, and sigB (accession number AF274004).
FIG. 2.
Comparison of the organization of the sigB operon of S. epidermidis 1457 and that of the sigB operons of S. aureus and B. subtilis. In the physical maps, genes are indicated as arrows. Homology of nucleotide (NT) sequences and the identity and similarity of the deduced amino acid (AA) sequences between corresponding genes are shown. The positions of putative promoters are indicated. The Tn917 insertion site of mutants M15 and M19 is indicated. The transcriptional direction of the erm gene of Tn917 is shown by an arrowhead. The typical 5-nt duplication at the Tn917 insertion site is in boldface.
The Tn917 insertion sites in mutants M15 and M19 are localized 19 bp downstream of the translation start codon (GTG) of rsbU (Fig. 2).
Phenotypic characterization.
In S. aureus and B. subtilis, ςB is known to regulate specific genes in stationary phase and under different stress conditions like an osmotic shift and the presence of ethanol (14, 21, 22). Therefore, we compared the phenotypic properties of mutant M15 and the corresponding wild-type strain, S. epidermidis 1457, using different stress conditions.
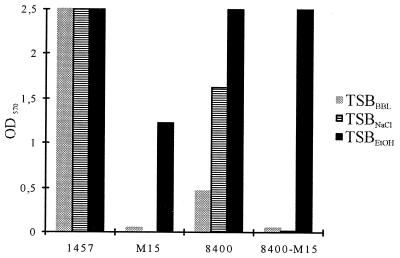
Under standard biofilm assay conditions, mutant M15 exhibited a biofilm-negative phenotype in TSBBBL whereas S. epidermidis 1457 was a strong biofilm producer (Fig. 3). In TSBNaCl, mutant M15 was also biofilm negative. In this medium, the wild-type strain produced even more biofilm, which could not be quantified, however, because the respective OD values were outside the detection range of the spectrophotometer used. However, wild-type S. epidermidis 8400 produced less biofilm in TSBBBL than did S. epidermidis 1457. With this strain, the increased biofilm production in TSBNaCl could be quantified (Fig. 3). As with osmotic stress, both wild-type strains S. epidermidis 1457 and 8400 displayed increased biofilm formation in TSBEtOH (Fig. 3). Interestingly, in contrast to the response of mutant M15 to osmotic stress, this mutant was a strong biofilm producer in TSBEtOH (Fig. 3). To investigate the phenotypic differences in biofilm formation resulting from insertion of Tn917 from mutant M15 into a different genetic background, the Tn917 insertion of this mutant was transduced into independent wild-type strain 8400, resulting in mutant 8400-M15. Insertion of Tn917 into the transductant 8400-M15 at the expected site was demonstrated using PCR with primers JK41rev1 plus 5L and 3R plus JKMK28, respectively, followed by nucleotide sequence analysis (data not shown). With this transductant, similar phenotypic properties regarding biofilm formation in the presence of ethanol and osmotic stress were obtained (Fig. 3). Similar results were obtained for mutants M19 and 8400-M19 (data not shown).
FIG. 3.
Biofilm formation under different stress conditions. S. epidermidis 1457, isogenic mutant M15, independent wild-type S. epidermidis 8400, and its transductant 8400-M15 were analyzed using TSBBBL, TSBNaCl, and TSBEtOH as the growth media. Results of a representative experiment are shown.
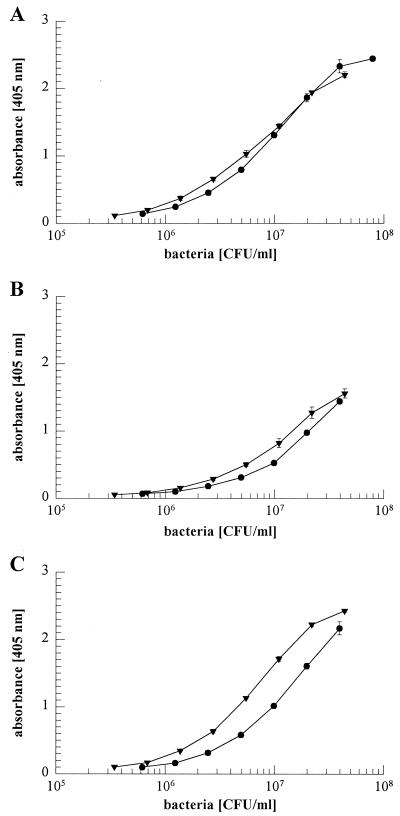
To exclude the possibility that inactivation of rsbU affected the primary attachment of S. epidermidis to polymer surfaces under the different growth conditions analyzed, mutant M15 and S. epidermidis 1457 were compared for primary attachment. Staphylococcal cells attached to cell culture plates also used for the biofilm assay after 60 min of incubation were detected by ELISA with an antiserum raised against biofilm-negative S. epidermidis 5179 (25, 45, 45). There were no significant differences in primary attachment between mutant and wild-type cells grown in TSBBBL, TSBNaCl, and TSBEtOH (Fig. 4). Similar results were obtained with mutant M19 (data not shown).
FIG. 4.
Primary attachment of S. epidermidis 1457 (●) and isogenic mutant M15 (▾) grown in TSBBBL (A), TSBNaCl (B), or TSBEtOH (C) to polystyrene cell culture plates (Nunclon Delta; Nunc). Bacteria were inoculated into the plates at various concentrations and incubated for 1 h at 37°C. Attached bacterial cells were detected by ELISA as described in Materials and Methods. Results of a representative experiment are shown.
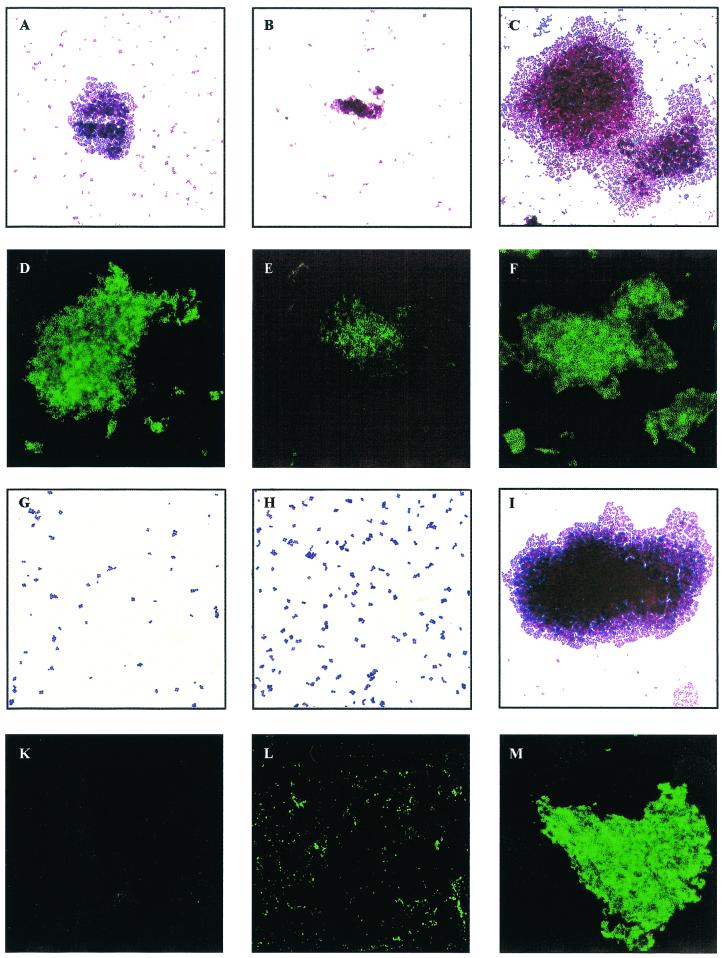
Wild-type S. epidermidis 1457 formed smaller but more compact cell clusters in TSBNaCl compared to the standard medium, while in TSBEtOH, significantly larger cell clusters were observed (Fig. 5A to C). In an indirect IFA using PIA-specific antiserum, expression of PIA by S. epidermidis 1457 was observed in all of the growth media used (Fig. 5D to F). Despite the formation of smaller cell clusters by S. epidermidis 1457 grown in TSBNaCl, no significant difference in intensity of fluorescence was observed (Fig. 5B and D to F). In contrast, mutant M15 did not produce detectable cell clusters in TSBBBL and TSBNaCl (Fig. 5G and H). In parallel with reconstituted biofilm production in TSBEtOH, mutant M15 was located in large cell clusters (Fig. 5I). As detected by the specific IFA, expression of PIA in TSBEtOH by mutant M15 was comparable to that of wild-type cells (Fig. 5M). With mutant M15 grown in TSBNaCl, only irregular, speckled fluorescence could be detected (Fig. 5L), whereas in TSBBBL, PIA expression was not detected (Fig. 5K). Similar results were obtained with mutant M19 (data not shown).
FIG. 5.
Cell cluster formation and PIA expression by S. epidermidis 1457 and mutant M15 under different stress conditions. S. epidermidis 1457 (A to F) and isogenic mutant M15 (G to M) were grown in tissue culture plates as a biofilm in TSBBBL (A, D, G, and K), TSBNaCl (B, E, H, and L), or TSBEtOH (C, F, I, and M) for 22 h at 37°C. Cells were scraped from the surface, and appropriate dilutions in phosphate-buffered saline were applied to microscope slides or immunofluorescence slides. Microphotographs of representative fields are shown after Gram staining (A to C and G to I) or after IFA using a PIA-specific antiserum as described in Materials and Methods (D to F and K to M). Results of a representative experiment are shown.
As IFA allows only qualitative PIA detection, synthesis of PIA was quantified in bacterial cell extracts and culture supernatants of the cells grown as biofilms on tissue culture plates in the respective media. With wild-type S. epidermidis 1457, a significant eightfold increase in the PIA concentration was observed in the culture supernatant of cells grown in TSBNaCl and TSBEtOH compared to that of TSBBBL-grown cells (Table 2). Only a minor increase in cell-associated PIA was detected with cells grown in TSBNaCl or TSBEtOH (Table 2). With mutant M15, consistent with its biofilm-negative phenotype in TSBBBL, PIA production was hardly detectable (Table 2). In TSBNaCl, a small but significant increase in the PIA concentration in the culture supernatant was detected, which could correspond to the irregular speckled fluorescence seen in these cells. In contrast, the concentrations of cell-associated PIA and PIA detected in culture supernatants of mutant M15 grown in TSBEtOH were not significantly different from those of S. epidermidis 1457 grown in the same medium, which is consistent with reconstituted biofilm production of the mutant (Table 2). Similar results were obtained with S. epidermidis 8400, its transductant 8400-M15 (Table 2), and mutant M19 (data not shown).
TABLE 2.
Quantitation of PIA produced using osmotic and ethanol stress
| Strain and PIA location | TSBBBL | TSBNaCl | TSBEtOH |
|---|---|---|---|
| 1457 | |||
| Supernatant | 32a | 256 | 512 |
| Cell associated | 256 | 512 | 512 |
| M15 | |||
| Supernatant | 4 | 64 | 256 |
| Cell associated | 4 | 16 | 256 |
| 8400 | |||
| Supernatant | 2 | 128 | 128 |
| Cell associated | 64 | 256 | 256 |
| 8400-M15 | |||
| Supernatant | 2 | 4 | 64 |
| Cell associated | 2 | 2 | 256 |
Reciprocal titer of PIA determined by coagglutination using a PIA-specific coagglutination assay as described in Materials and Methods.
Characterization of a sigB RFLP type of S. epidermidis wild-type strains.
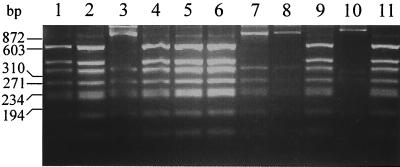
As the activity of sigB apparently influences biofilm formation by S. epidermidis, the sigB operons of different S. epidermidis clinical isolates, including reference strains RP62A (6) and SE5 (42), were amplified by PCR using oligonucleotides JKMK11 and JKMK34, spanning the region from rsbU to the noncoding region downstream sigB. A restriction fragment length polymorphism (RFLP) of the resulting fragments (approximately 2.6 kb) was analyzed after cleavage with HinfI. Two different fragment patterns, designated A and B, were detected (Fig. 6). The respective sigB RFLP type did not correlate with the icaADBC genotype characterized by PCR using icaB-specific oligonucleotides (icaBforward and icaBreverse) or with the expression phenotype of biofilm formation (Table 3).
FIG. 6.
sigB RFLP of different clinical S. epidermidis isolates. PCR fragments containing almost the complete sigB operon were cleaved with HinfI. Two different sigB RFLP types, A and B, were detected. The S. epidermidis strains which display sigB RFLP type A were 1457 (lane 1), RP62A (lane 2), 521 (lane 4), 1057 (lane 5), 10333 (lane 6), 9225 (lane 9), and 939 (lane 11). The S. epidermidis strains which display sigB RFLP type B were SE5 (lane 3), 5179 (lane 7), 7837 (lane 8), and 9896 (lane 10).
TABLE 3.
Phenotypic and genotypic characterization of S. epidermidis wild-type strains
| Strain | icaB PCR | sigB RFLP type | Biofilm formation (OD570) |
|---|---|---|---|
| 1457 | + | A | 2.500 |
| RP62A | + | A | 2.446 |
| SE5 | + | B | 1.800 |
| 521 | + | A | 1.643 |
| 1057 | + | A | 0.798 |
| 939 | + | A | 0.724 |
| 10333 | + | A | 0.035 |
| 5179 | + | B | 0.014 |
| 7837 | + | B | 0.009 |
| 9225 | − | A | 0.004 |
| 9896 | − | B | 0.027 |
DISCUSSION
In this study, we characterized the genetic defect of two isogenic PIA- and biofilm-negative Tn917 mutants, M15 and M19 (class III), with transposon insertion sites distinct from the icaADBC locus (30). Sequence analysis of chromosomal DNA flanking the Tn917 insertion sites of M15 and M19 demonstrated identical insertions at nt 19 of rsbU, which is the first gene of an operon of S. epidermidis with high homology to the sigB operon of S. aureus, B. subtilis, and Listeria monocytogenes (2, 21, 55, 57). In S. epidermidis, the same order of the genes, rsbU, rsbV, rsbW, and sigB, coding for alternative sigma factor ςB and three regulatory proteins was detected as described for S. aureus (21, 57). As with S. aureus, no evidence of additional regulatory proteins, like RsbRST and RsbX, flanking the sigB operon of S. epidermidis was obtained (data not shown). In addition to a putative housekeeping promoter in front of rsbU, a ςB-like recognition sequence was observed preceding the gene rsbV, as described for S. aureus and B. subtilis (21, 57). The homology of the nucleotide sequence and the organization of this operon in S. epidermidis suggest a general function similar to that observed for S. aureus or B. subtilis.
In biofilm-producing S. epidermidis 1457, inactivation of rsbU, which is a positive regulator of ςB (53), led to a biofilm-negative phenotype, which is caused by severely decreased PIA synthesis, indicating a defect in biofilm accumulation of rsbU mutant M15. This was confirmed directly, as no significant differences in primary attachment to polystyrene tissue culture plates were observed between the mutant and wild-type strains (Fig. 4). Apparently, RsbU activity is essential for expression of biofilm formation and PIA synthesis in S. epidermidis. As the Tn917 insertion of mutant M15 transduced into the independent genetic background of biofilm-producing S. epidermidis 8400 resulted in similar phenotypic properties, it is extremely unlikely that the observed phenotypic changes are caused not by the transposon insertion into rsbU but by nonspontaneous mutations in association with the Tn917 insertion, as was observed with inactivation of xpr in S. aureus KSI9051, where, in parallel with exchange of the mutated allele by transduction, independent point mutations in agrC occurred with high frequency (35).
Abolished PIA synthesis due to insertion of Tn917 into rsbU of S. epidermidis suggests ςB-dependent expression of PIA. This is corroborated by the observations that the spontaneous rsbU deletion mutant S. aureus 8325 has a phenotype similar to that of experimentally induced sigB deletion mutants and that its phenotype can be complemented by expression of ςB in trans from an independent promoter (22). Similar to sigB and rsbU mutants of S. aureus, mutants M15 and M19 exhibited significant differences in colony morphology compared to wild-type S. epidermidis 1457 (22, 30). Recently, it was reported that a sigB-null mutant of a clinical biofilm-producing S. aureus strain was biofilm negative (41). However, the ways in which biofilm expression by S. epidermidis and S. aureus are regulated seem to be fundamentally different, as most icaADBC-positive S. epidermidis strains produce a biofilm in vitro but almost all clinical S. aureus strains are biofilm negative in vitro despite the presence of icaADBC (8, 34; M. A. Horstkotte, J. K.-M. Knobloch, and D. Mack, unpublished results).
Mutant M15 could be complemented by expression of icaADBC from a xylose-dependent promoter to a biofilm-producing phenotype, indicating a defect on the level of transcription of icaADBC leading to the biofilm-negative phenotype of this mutant (30). Indeed, no icaADBC-specific transcript was observed in the mutant compared with biofilm-producing wild-type S. epidermidis 1457 in the mid-exponential growth phase (30). The transcriptional start site of the icaADBC mRNA was determined at nt 732 of the published sequence (accession number SE43366), 29 nt proximal to the start codon of the icaA gene (11, 16). Surprisingly, using a consensus-directed search strategy, only a putative ςA-dependent promoter starting at nt 701 was detected. A consensus-directed search strategy for ςB-dependent promoters (40) revealed a possible promoter (AGTGTAGT N18 GGAAAA) with low homology to the consensus sequence starting at nt 529 of this sequence (11, 16), which probably is not active for transcription of the icaADBC mRNA transcribed downstream from nt 732 (11, 13). In addition, use of the same approach to search the nucleotide sequence of icaR, the putative regulator of icaADBC (59), revealed no sequence homologous to ςB-like recognition sequences. In the sequences of icaR and the icaADBC locus (accession number AF086783) of S. aureus (8), no sequence homologous to ςB-dependent promoters preceding these genes could be detected. These observations strongly suggest that there is no direct ςB-dependent regulation of icaADBC transcription. However, the possibility cannot be completely excluded that ςB-dependent transcription of icaADBC starting from the putative ςB-dependent promoter occurs under special physiological conditions.
As in S. epidermidis, in the closely related species S. aureus, the ςB operon consists of four genes, rsbU, rsbV, rsbW, and sigB, with the same identified or predicted functions as the homologous downstream module in B. subtilis (21, 36, 57). The central module of ςB regulation consists of anti-sigma factor RsbW and anti-anti-sigma factor RsbV (1, 3, 9, 20, 54). This module is additionally regulated by RsbU, an RsbV-specific phosphatase activating RsbV (53, 54, 56). For B. subtilis, a second RsbV-specific phosphatase, RsbP, was described (52).
Phenotypic characterization clearly shows that PIA synthesis and biofilm formation by S. epidermidis are significantly increased by environmental stresses like high osmolarity and the presence of ethanol. At least two different pathways of induction exist. Apparently, induction of PIA synthesis by NaCl depends on a functional rsbU gene, as mutants M15 and 8400-M15 were completely biofilm negative in the presence of NaCl. However, ethanol stress leads to induction of PIA synthesis and biofilm formation independent of rsbU. This is in contrast to observations on B. subtilis, where induction of ςB by ethanol or salt stress depends on rsbU (53, 54). These results suggest that in S. epidermidis, additional pathways could exist, as suggested for S. aureus (4, 22), which activate ςB by regulators substituting for RsbU or activate PIA expression by ςB- independent pathways. Additionally, the possibility cannot be completely excluded that RsbU has two different roles in S. epidermidis and activates icaADBC transcription by a ςB-independent mechanism.
A HinfI RFLP with two different patterns (A and B) was observed in a PCR fragment containing almost the complete sigB operon of S. epidermidis in 11 clinical isolates. However, the sigB operon was detected in all strains and the respective sigB RFLP type did not correlate with the icaADBC genotype (Table 3). In addition, there was no correlation of the sigB RFLP type with the observed biofilm formation phenotype (Table 3), indicating no obvious functional genetic defects correlating with the sigB-RFLP types, although the possibility of point mutations or very small deletions of only a few base pairs cannot be completely excluded.
The regulatory mechanisms controlling expression of biofilm formation and PIA synthesis in S. epidermidis are only basically known. At least three unlinked genetic loci control expression of icaADBC on the level of transcription (30). One of these loci is RsbU, a positive regulator of alternative sigma factor ςB. As is apparent from the data presented here, ςB may act only indirectly via an additional, unknown, factor or RsbU may, by itself, be a regulator of icaADBC transcription. Activation of PIA expression by different stress stimuli apparently uses different pathways. It is of primary importance to further characterize the molecular mechanisms controlling expression of icaADBC and biofilm formation, as it is reasonable to anticipate that interference with these mechanisms will improve the therapy and prevention of biomaterial-related S. epidermidis infections.
ACKNOWLEDGMENTS
We thank Rainer Laufs for his continuous support. For the kind gift of E. coli MC1061, we thank J. A. Gutierrez, Department of Oral Biology, University of Florida, Gainesville.
This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Paul Gerson Unna-Forschungszentrum für Experimentelle Dermatologie, Beiersdorf AG, Hamburg, Germany, to D.M.
REFERENCES
- 1.Alper S, Dufour A, Garsin D A, Duncan L, Losick R. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J Mol Biol. 1996;260:165–177. doi: 10.1006/jmbi.1996.0390. [DOI] [PubMed] [Google Scholar]
- 2.Becker L A, Çetin M S, Hutkins R W, Benson A K. Identification of the gene encoding the alternative sigma factor ςB from Listeria monocytogenes and its role in osmotolerance. J Bacteriol. 1998;180:4547–4554. doi: 10.1128/jb.180.17.4547-4554.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boylan S A, Rutherford A, Thomas S M, Price C W. Activation of Bacillus subtilis transcription factor ςB by a regulatory pathway responsive to stationary-phase signals. J Bacteriol. 1992;174:3695–3706. doi: 10.1128/jb.174.11.3695-3706.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan P F, Foster S J, Ingham E, Clements M O. The Staphylococcus aureus alternative sigma factor ςB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J Bacteriol. 1998;180:6082–6089. doi: 10.1128/jb.180.23.6082-6089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen G D, Baldassarri L, Simpson W A. Colonization of medical devices by coagulase-negative staphylococci. In: Bisno A L, Waldvogel F A, editors. Infections associated with indwelling medical devices. Washington, D.C.: American Society for Microbiology; 1994. pp. 45–78. [Google Scholar]
- 6.Christensen G D, Simpson W A, Bisno A L, Beachey E H. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun. 1982;37:318–326. doi: 10.1128/iai.37.1.318-326.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen G D, Simpson W A, Younger J J, Baddour L M, Barrett F F, Melton D M, Beachey E H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cramton S E, Gerke C, Schnell N F, Nichols W W, Götz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dufour A, Haldenwang W G. Interactions between a Bacillus subtilis anti-ς factor (RsbW) and its antagonist (RsbV) J Bacteriol. 1994;176:1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fey P D, Ulphani J S, Götz F, Heilmann C, Mack D, Rupp M E. Characterization of the relationship between polysaccharide intercellular adhesin and hemagglutination in Staphylococcus epidermidis. J Infect Dis. 1999;179:1561–1564. doi: 10.1086/314762. [DOI] [PubMed] [Google Scholar]
- 11.Gerke C, Kraft A, Süssmuth R, Schweitzer O, Götz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem. 1998;273:18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 12.Gutierrez J A, Crowley P J, Brown D P, Hillman J D, Youngman P, Bleiweis A S. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J Bacteriol. 1996;178:4166–4175. doi: 10.1128/jb.178.14.4166-4175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haldenwang W G. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hecker M, Völker A. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigmaB regulon. Mol Microbiol. 1998;29:1129–1136. doi: 10.1046/j.1365-2958.1998.00977.x. [DOI] [PubMed] [Google Scholar]
- 15.Heilmann C, Hussain M, Peters G, Götz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 16.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 17.Herrmann M, Vaudaux P E, Pittet D, Auckenthaler R, Lew P D, Schumacher-Perdreau F, Peters G, Waldvogel F A. Fibronectin, fibrinogen, and laminin act as mediators of adherence of clinical staphylococcal isolates to foreign material. J Infect Dis. 1988;158:693–701. doi: 10.1093/infdis/158.4.693. [DOI] [PubMed] [Google Scholar]
- 18.Kalman S, Duncan M L, Thomas S M, Price C W. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J Bacteriol. 1990;172:5575–5585. doi: 10.1128/jb.172.10.5575-5585.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang C M, Brody M S, Akbar S, Yang X, Price C W. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor ςB in response to environmental stress. J Bacteriol. 1996;178:3846–3853. doi: 10.1128/jb.178.13.3846-3853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang C M, Vijay K, Price C W. Serine kinase activity of a Bacillus subtilis switch protein is required to transduce environmental stress signals but not to activate its target PP2C phosphatase. Mol Microbiol. 1998;30:189–196. doi: 10.1046/j.1365-2958.1998.01052.x. [DOI] [PubMed] [Google Scholar]
- 21.Kullik I, Giachino P. The alternative sigma factor sigmaB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch Microbiol. 1997;167:151–159. doi: 10.1007/s002030050428. [DOI] [PubMed] [Google Scholar]
- 22.Kullik I, Giachino P, Fuchs T. Deletion of the alternative sigma factor ςB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol. 1998;180:4814–4820. doi: 10.1128/jb.180.18.4814-4820.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mack D. Molecular mechanisms of Staphylococcus epidermidis biofilm formation. J Hosp Infect. 1999;43(Suppl.):S113–S125. doi: 10.1016/s0195-6701(99)90074-9. [DOI] [PubMed] [Google Scholar]
- 24.Mack D, Bartscht K, Dobinsky S, Horstkotte M A, Kiel K, Knobloch J K M, Schäfer P. Staphylococcal factors involved in adhesion and biofilm formation on biomaterials. In: An Y H, Friedman R J, editors. Handbook for studying bacterial adhesion: principles, methods, and applications. Totowa, N.J: Humana Press; 2000. pp. 307–330. [Google Scholar]
- 25.Mack D, Bartscht K, Fischer C, Rohde H, de Grahl C, Dobinsky S, Horstkotte M A, Kiel K, Knobloch J K M. Genetic and biochemical analysis of Staphylococcus epidermidis biofilm accumulation. Methods Enzymol. 2001;336:215–239. doi: 10.1016/s0076-6879(01)36592-8. [DOI] [PubMed] [Google Scholar]
- 26.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, Laufs R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178:175–183. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mack D, Haeder M, Siemssen N, Laufs R. Association of biofilm production of coagulase-negative staphylococci with expression of a specific polysaccharide intercellular adhesin. J Infect Dis. 1996;174:881–884. doi: 10.1093/infdis/174.4.881. [DOI] [PubMed] [Google Scholar]
- 28.Mack D, Nedelmann M, Krokotsch A, Schwarzkopf A, Heesemann J, Laufs R. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect Immun. 1994;62:3244–3253. doi: 10.1128/iai.62.8.3244-3253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mack D, Riedewald J, Rohde H, Magnus T, Feucht H H, Elsner H A, Laufs R, Rupp M E. Essential functional role of the polysaccharide intercellular adhesin of Staphylococcus epidermidis in hemagglutination. Infect Immun. 1999;67:1004–1008. doi: 10.1128/iai.67.2.1004-1008.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mack D, Rohde H, Dobinsky S, Riedewald J, Nedelmann M, Knobloch J K M, Elsner H-A, Feucht H H. Identification of three essential regulatory gene loci governing expression of the Staphylococcus epidermidis polysaccharide intercellular adhesin and biofilm formation. Infect Immun. 2000;68:3799–3807. doi: 10.1128/iai.68.7.3799-3807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mack D, Siemssen N, Laufs R. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect Immun. 1992;60:2048–2057. doi: 10.1128/iai.60.5.2048-2057.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mack D, Siemssen N, Laufs R. Identification of a cell cluster associated antigen specific for plastic-adherent Staphylococcus epidermidis which is functionally related to intercellular adhesion. Zentbl Bakteriol Suppl. 1994;26:411–413. [Google Scholar]
- 33.McKenney D, Hübner J, Muller E, Wang Y, Goldmann D A, Pier G B. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect Immun. 1998;66:4711–4720. doi: 10.1128/iai.66.10.4711-4720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenney D, Pouliot K L, Wang Y, Murthy V, Ulrich M, Doring G, Lee J C, Goldmann D A, Pier G B. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science. 1999;284:1523–1527. doi: 10.1126/science.284.5419.1523. [DOI] [PubMed] [Google Scholar]
- 35.McNamara P J, Iandolo J J. Genetic instability of the global regulator agr explains the phenotype of the xpr mutation in Staphylococcus aureus KSI9051. J Bacteriol. 1998;180:2609–2615. doi: 10.1128/jb.180.10.2609-2615.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyazaki E, Chen J-M, Ko C, Bishai W R. The Staphylococcus aureus rsbW (orf159) gene encodes an anti-sigma factor of SigB. J Bacteriol. 1999;181:2846–2851. doi: 10.1128/jb.181.9.2846-2851.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nedelmann M, Sabottke A, Laufs R, Mack D. Generalized transduction for genetic linkage analysis and transfer of transposon insertions in different Staphylococcus epidermidis strains. Zentbl Bakteriol. 1998;287:85–92. doi: 10.1016/s0934-8840(98)80151-5. [DOI] [PubMed] [Google Scholar]
- 38.Nilsson M, Frykberg L, Flock J I, Pei L, Lindberg M, Guss B. A fibrinogen-binding protein of Staphylococcus epidermidis. Infect Immun. 1998;66:2666–2673. doi: 10.1128/iai.66.6.2666-2673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pascual A, Fleer A, Westerdaal N A, Verhoef J. Modulation of adherence of coagulase-negative staphylococci to Teflon catheters in vitro. Eur J Clin Microbiol. 1986;5:518–522. doi: 10.1007/BF02017694. [DOI] [PubMed] [Google Scholar]
- 40.Petersohn A, Bernhardt J, Gerth U, Höper D, Koburger T, Völker U, Hecker M. Identification of ς(B)-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J Bacteriol. 1999;181:5718–5724. doi: 10.1128/jb.181.18.5718-5724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rachid S, Ohlsen K, Wallner U, Hacker J, Hecker M, Ziebuhr W. Alternative transcription factor ςB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J Bacteriol. 2000;182:6824–6826. doi: 10.1128/jb.182.23.6824-6826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rupp M E, Archer G L. Hemagglutination and adherence to plastic by Staphylococcus epidermidis. Infect Immun. 1992;60:4322–4327. doi: 10.1128/iai.60.10.4322-4327.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rupp M E, Archer G L. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin Infect Dis. 1994;19:231–243. doi: 10.1093/clinids/19.2.231. [DOI] [PubMed] [Google Scholar]
- 44.Rupp M E, Sloot N, Meyer H G, Han J, Gatermann S. Characterization of the hemagglutinin of Staphylococcus epidermidis. J Infect Dis. 1995;172:1509–1518. doi: 10.1093/infdis/172.6.1509. [DOI] [PubMed] [Google Scholar]
- 45.Rupp M E, Ulphani J S, Fey P D, Bartscht K, Mack D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun. 1999;67:2627–2632. doi: 10.1128/iai.67.5.2627-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rupp M E, Ulphani J S, Fey P D, Mack D. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect Immun. 1999;67:2656–2659. doi: 10.1128/iai.67.5.2656-2659.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Shaw J H, Clewell D B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985;164:782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thylefors J D, Harbarth S, Pittet D. Increasing bacteremia due to coagulase-negative staphylococci: fiction or reality? Infect Control Hosp Epidemiol. 1998;19:581–589. doi: 10.1086/647878. [DOI] [PubMed] [Google Scholar]
- 50.Tojo M, Yamashita N, Goldmann D A, Pier G B. Isolation and characterization of a capsular polysaccharide adhesin from Staphylococcus epidermidis. Infect Dis. 1988;157:713–722. doi: 10.1093/infdis/157.4.713. . (Erratum, 158:268, 1988.) [DOI] [PubMed] [Google Scholar]
- 51.Veenstra G J, Cremers F F, van Dijk H, Fleer A. Ultrastructural organization and regulation of a biomaterial adhesin of Staphylococcus epidermidis. J Bacteriol. 1996;178:537–541. doi: 10.1128/jb.178.2.537-541.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vijay K, Brody M S, Fredlund E, Price C W. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigmaB transcription factor of Bacillus subtilis. Mol Microbiol. 2000;35:180–188. doi: 10.1046/j.1365-2958.2000.01697.x. [DOI] [PubMed] [Google Scholar]
- 53.Voelker U, Dufour A, Haldenwang W G. The Bacillus subtilis rsbU gene product is necessary for RsbX-dependent regulation of ςB. J Bacteriol. 1995;177:114–122. doi: 10.1128/jb.177.1.114-122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voelker U, Voelker A, Haldenwang W G. Reactivation of the Bacillus subtilis anti-ςB antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J Bacteriol. 1996;178:5456–5463. doi: 10.1128/jb.178.18.5456-5463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiedmann M, Arvik T J, Hurley R J, Boor K J. General stress transcription factor ςB and its role in acid tolerance and virulence of Listeria monocytogenes. J Bacteriol. 1998;180:3650–3656. doi: 10.1128/jb.180.14.3650-3656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wise A A, Price C W. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor ςB in response to environmental signals. J Bacteriol. 1995;177:123–133. doi: 10.1128/jb.177.1.123-133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu S, de Lencastre H, Tomasz A. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziebuhr W, Heilmann C, Götz F, Meyer P, Wilms K, Straube E, Hacker J. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect Immun. 1997;65:890–896. doi: 10.1128/iai.65.3.890-896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ziebuhr W, Krimmer V, Rachid S, Lößner I, Götz F, Hacker J. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32:345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]