Abstract
Six closely related N2-fixing bacterial strains were isolated from surface-sterilized roots and stems of four different rice varieties. The strains were identified as Serratia marcescens by 16S rRNA gene analysis. One strain, IRBG500, chosen for further analysis showed acetylene reduction activity (ARA) only when inoculated into media containing low levels of fixed nitrogen (yeast extract). Diazotrophy of IRBG500 was confirmed by measurement of 15N2 incorporation and by sequence analysis of the PCR-amplified fragment of nifH. To examine its interaction with rice, strain IRBG500 was marked with gusA fused to a constitutive promoter, and the marked strain was inoculated onto rice seedlings under axenic conditions. At 3 days after inoculation, the roots showed blue staining, which was most intense at the points of lateral root emergence and at the root tip. At 6 days, the blue precipitate also appeared in the leaves and stems. More detailed studies using light and transmission electron microscopy combined with immunogold labeling confirmed that IRBG500 was endophytically established within roots, stems, and leaves. Large numbers of bacteria were observed within intercellular spaces, senescing root cortical cells, aerenchyma, and xylem vessels. They were not observed within intact host cells. Inoculation of IRBG500 resulted in a significant increase in root length and root dry weight but not in total N content of rice variety IR72. The inoculated plants showed ARA, but only when external carbon (e.g., malate, succinate, or sucrose) was added to the rooting medium.
Rice (Oryza sativa) is arguably the most important cereal crop in the world, feeding more than 50% of the world's population (18, 38). However, the population is growing at a rapid rate; therefore, rice yields will need to be enhanced to match the increased consumption. Achieving these higher yields by 2020 will require at least double the amount of N fertilizers currently being used because, after water, N is the most limiting nutrient for rice growth (22). An alternative to the increased use of chemical fertilizers is to explore and improve the ability of rice to obtain N from biological N2 fixation (BNF) (37, 69).
It has long been known that rice can form natural associations with various N2-fixing bacteria, both phototrophs and heterotrophs (3, 4, 42, 53, 70). All or some of these may be responsible for supplying the plants with fixed N (27, 37). Moreover, in addition to the culturable diazotrophs associated with rice, a substantial molecular diversity of N2-fixing bacteria has been detected in field-grown rice based on retrieval of nifH or nifD gene fragments from root DNA (12, 63, 64). However, the contribution of the bacteria externally associated with rice is insufficient to sustain a high yield (39). It has been suggested that bacteria colonizing the plant interior might interact more closely with the host, with less competition for carbon sources and a more protected environment for N2 fixation (49, 51), such as that occurring in the relatively efficient N2-fixing symbioses between rhizobia and legumes (45).
In view of the above, a global frontier project which aims to transfer an N2 fixation capability to rice has begun (38). One of the approaches toward this goal is the use of natural N2-fixing endophytic bacteria associated with rice. It has been suggested that endophytic N2-fixing bacteria, particularly Acetobacter diazotrophicus and Herbaspirillum spp. (8, 26), may be responsible for the significant BNF observed in some Brazilian varieties of sugarcane (Saccharum spp.) (65). Similarly, Azoarcus spp. might be responsible for N2 fixation in Kallar grass (Leptochloa fusca) (42, 50, 51). Therefore, it is possible that rice varieties showing potential for N2 fixation (41, 57) could also harbor effective endophytic diazotrophs. Indeed, recent studies have shown that numerous and varied diazotrophic bacteria can be isolated from wetland rice after surface sterilization, thus indicating a potential endophytic location for them (6, 60). Surface sterilization in itself, however, is insufficient evidence to ascribe an endophytic location for these bacteria, and it is now considered that only direct localization using microscopy can provide such evidence (23, 26, 50). In view of the above, this study aimed to characterize the predominant endophytic diazotrophic bacteria that are present within various rice varieties grown under greenhouse conditions. Using light and electron microscopy, the colonization of rice seedlings by a selected isolate (Serratia marcescens IRBG500) was examined in detail. To the best of our knowledge, this is the first detailed ultrastructural study of a naturally occurring diazotrophic endophyte in rice.
MATERIALS AND METHODS
Isolation of endophytic bacteria and determination of diazotrophy.
Roots and stems of seven different rice varieties (Table 1) growing in nonsterile flooded soil in a greenhouse were collected and washed with tap water, blotted, and weighed. The roots were surface sterilized with 70% ethanol for 5 min and then treated with 0.2% mercuric chloride for 30 s. The stems were cut into small (approximately 5-cm) pieces and surface sterilized by dipping in 95% ethanol and flaming. Approximately 1 cm was then removed from each end. The root and the stem were checked for the efficacy of sterilization by rolling them on 0.1% tryptic soy agar (TSA) plates. They were then homogenized, under sterile conditions, with a mortar and pestle in phosphate-buffered saline, and different dilutions were placed on TSA plates to determine the total heterotrophic bacterial population. Serially diluted homogenate was also inoculated into tubes containing a semisolid N-free medium consisting of (per liter) malic acid (5 g), K2HPO4 (0.5 g), MgSO4 · 7H2O (0.2 g), NaCl (0.1 g), CaCl2 (0.02 g), and 0.5% bromothymol blue in 0.2 N KOH (2 ml), 1.64% Fe-EDTA solution (4 ml), and agar (2 g) (33). The final pH was adjusted to 7.0 by KOH. The medium was modified by adding yeast extract (0.02 g), as it is known that a trace amount of fixed nitrogen is required for the isolation of most diazotrophs from the rhizosphere of rice (67). The bacteria from the acetylene reduction activity (ARA)-positive tubes were further streaked onto agar plates (1.5% [wt/vol]) with the same medium containing 0.1 mM NH4Cl to obtain pure colonies.
TABLE 1.
Isolation of putative endophytic bacteria from seven varieties of rice grown under greenhouse conditions
| Rice variety | Mean log CFU g [fresh wt]−1 ± SD
|
|||
|---|---|---|---|---|
| Total no. of bacteria
|
ARA-positive bacteria
|
|||
| Root | Stem | Root | Stem | |
| IR387 | 5 ± 0.5 | 3 ± 1.0 | Nil | Nil |
| IR26578 | 6 ± 0.8 | 4 ± 0.75 | 3.5 ± 0.3 | 2 ± 1 |
| Palawan | 6 ± 0.3 | 3.5 ± 1.0 | Nil | Nil |
| Kinandang Patong | 5 ± 0.5 | 2 ± 1.0 | 3.2 ± 0.7 | Nil |
| Moroberekan | 6 ± 0.5 | 4 ± 1.5 | 2.8 ± 0.3 | 1 ± 0.5 |
| Oking Seroni | 6 ± 0.5 | 2 ± 1.0 | 2.0 ± 1.2 | Nil |
| IR72 | 5 ± 0.6 | 4.5 ± 1.0 | 2.8 ± 0.9 | 1 ± 0.8 |
Analysis of strain diversity and identification of diazotrophic bacteria in various rice varieties.
Diazotrophic bacteria isolated from different parts of rice were analyzed for diversity by fingerprinting using BOX-PCR amplification fragment length polymorphism as described by Verslovic et al. (66). A BOX A1R primer (5′-CTACGGCAAGGCGACGCTGACG-3′) was used at 50 pmol with 100 ng of template DNA in a 25-μl PCR mixture containing 1.25 mM each deoxynucleoside triphosphate and 2 U of AmpliTaq DNA polymerase (Perkin-Elmer) in a reaction buffer with 10% (vol/vol) dimethyl sulfoxide; 5× reaction buffer stock contained 83 mM ammonium acetate, 335 mM Tris-HCl, 33.5 mM MgCl2, 33.5 μM EDTA, 150 mM β-mercaptoethanol, and 850 μg of bovine serum albumin ml−1 (pH 8.8). PCR amplification was performed in a BIOMETRA Uno-Thermocycler with an initial denaturation (95°C, 7 min) followed by 30 cycles of denaturation (95°C, 30 s; 95°C, 1 min), annealing (52°C, 1 min), and extension (65°C, 8 min), with a single final extension (65°C, 16 min). After the reaction, the samples were separated on 1.5% agarose gels. The resulting amplification patterns were analyzed, and the strains were grouped according to their fingerprints.
The strains showing similar fingerprints were further identified by direct sequencing of PCR-amplified 16S rRNA gene (rDNA) fragments as described earlier (21). The PCR primers 25f (5′-AACTKAAGAGTTTGATCCTGGCTC-3′) and 1492r (5′-ACGGYTACCTTGTTACGACTT-3′) were used at 50 pmol each with 1 to 2 ng of template DNA in PCR buffer containing 20 mM Tris-HCl (pH 8.7), 10 mM KCl, 0.005% Tween 20, 100 μg of nuclease-free bovine serum albumin ml−1, 7.5 mM MgCl2, 1.25 mM each deoxynucleoside triphosphate, and 2 U of Taq polymerase (Beckman Instruments, Munich, Germany), in a total volume of 25 μl. The DNA was amplified in a Techne Progene thermocycler (Thermo-Dux, Wertheim, Germany) after initial denaturation at 95°C with 27 cycles of 1 min of denaturation at 95°C, 2 min of annealing at 59°C, and 2 min of extension at 72°C, with a final extension step of 4 min at 72°C. The amplification product was treated with exonuclease I (10 U) and shrimp alkaline phosphatase (2 U; Amersham International, Little Chalfont, United Kingdom) for 15 min at 37°C and then for 15 min at 80°C. The DNA template was dialyzed against distilled water for 15 min before direct sequencing with 7-deaza-dGTP (catalog no. 2438; Amersham International) according to the manufacturer's instructions. A Techne Cyclogene thermocycler (Thermo-Dux) was used with 25 cycles of denaturation at 95°C for 30 s and annealing and extension at 60°C for 30 s, after an initial denaturation at 97°C for 1 min. Sequencing primers were labeled at the 5′ end with the fluorescent dye Cy-5 in order to allow automated sequencing with an ALFexpress apparatus (Pharmacia). Primers used were 35fC (5′-CTKAAGAGTTTGATCMTGGCTCAGATTGAACG-3′), 342fC (5′-CTCCTACGGGAGGCAGCAG-3′), and 530mfC (5′-CTACGTGCCAGCMGCCGCGG-3′). The resulting sequences were compared with known sequences by using the BLAST program.
Diazotrophy of S. marcescens IRBG500.
Diazotrophy of S. marcescens IRBG500 and S. marcescens IRBG500-gusA was determined on the basis of ARA (5) and 15N2 incorporation (2, 32) and by sequence analysis of the nifH fragment. As glucose is the preferred carbon source for growth of diazotrophic strains of enterobacteriaceae (10), nitrogen fixation was characterized using nitrogen-free dextrose medium (10), which was modified by addition of yeast extract (100 mg liter−1), with or without 10 mM NH4Cl as an additional N source. The bacteria were grown in Luria broth to stationary phase, and then 200 μl of each culture was inoculated into 10 ml of nitrogen-free dextrose medium. The cells were incubated on a orbital shaker (100 rpm) at 30°C overnight, and then 10% (vol/vol) of the headspace was replaced with 15N2 (99.5% 15N; Monsanto Research Corporation, Miamisburg, Ohio) before being incubated for another 3 days. After this time, 1 ml of acetylene was injected into the flasks, which were then incubated for a further 4 to 6 hours at 30°C. Ethylene produced by the bacteria was detected on a Hitachi 164F gas chromatograph. After determination of the ARA, a subsample was removed for protein estimations by the dye-binding method of Bradford (9), and the remainder of the cultures were freeze-dried. A 10-mg fine subsample of the freeze-dried material was used for analysis of 15N by a mass spectrometer (VG model 903) equipped with a Dumas elemental analyzer (Roboprep-CN 7001; Europa Scientific Ltd., Crewe, United Kingdom) at the Analytical Services Laboratory, International Rice Research Institute (IRRI). Uninoculated flasks served as controls. For evaluation in the context of related bacteria, the nitrogenase activity (ARA) of IRBG500 was compared with those of the type strains of S. marcescens (LMG2792) and S. rubidaea (LMG5019), as well as two strains of diazotrophic enterobacteria previously isolated from the rhizosphere of rice (35), Klebsiella planticola and Enterobacter cloacae.
The presence of nif genes in IRBG500 was confirmed by amplification of nifH gene segments by PCR. Plasmid DNA was prepared according to the method of Kado and Liu (30), and 1 to 5 ng of this DNA was used as the template for amplification of nifH by PCR with primers 19B (5′-CGGGATCCGCIWTYTAYGGIAARGGIGG-3′) and 407B (5′-CGGGATCCAAICCRCCRCAIACIACRTC-3′). PCR was carried out in a BIOMETRA Uno-Thermocycler with an initial denaturation of 4 min, then 35 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C, with a final extension of 4 min at 72°C (20), using PCR buffer as described above for BOX-PCR analysis. The amplified DNA fragment was sequenced by the dideoxy-chain termination method using an automated DNA sequencer (model 377; Applied Biosystems). Both strands were sequenced entirely.
Marking of a rice endophytic diazotroph (IRBG500) with gusA.
Escherichia coli S17.1 containing transposon-based β-glucuronidase (GUS) marker pCAM120 (Tn5ssgusA20), which has the gusA gene under the control of a constitutive kanamycin resistance gene promoter (68), was maintained on Luria agar plates with 100 μg of ampicillin ml−1. pCAM120 was transferred to IRBG500 by conjugation using a filter-mating technique (58). Because the E. coli strains used in conjugation are auxotrophs, the transconjugants were selected on minimal medium (M9) (56) plates containing 10 mM glucose, 10 mM potassium nitrate, and 0.2 mg of spectinomycin ml−1. Color reagent 5-bromo-4-chloro-3-indolyl-β-d-glucuronate (X-Gluc; Biosynth AG) was added to the medium at 40 μg ml−1, and the blue colonies were selected for further analysis.
Inoculation of rice with IRBG500 marked with gusA.
Dehulled seeds of rice variety IR72 were surface sterilized with 70% ethanol followed by 0.2% mercuric chloride for 30 s and were washed thrice with sterile water. The seeds were germinated on 0.1% TSA plates, and seedlings free of visual bacterial and fungal contamination were used for inoculation with the gusA-marked strain. The latter were grown in Luria broth supplemented with spectinomycin (100 μg ml−1) until they reached an optical density of 0.6. The cells were then harvested by centrifugation, washed twice with normal saline, and resuspended in phosphate-buffered saline. Surface-sterilized seedlings were grown in 80-ml glass tubes with 20 ml of Fahraeus medium without nitrogen (13). They were inoculated with a bacterial suspension containing approximately 107 bacteria and grown in a growth chamber (14-h light/10-h dark cycle) at 27°C (day) and 25°C (night). Uninoculated plants served as controls.
Enumeration of IRBG500 colonizing rice roots and stems.
Plants were sampled at various times, from 1 to 14 days after inoculation (DAI). Loosely attached bacteria were removed by washing the roots in excess sterile water, and the roots were then immersed in sterile water and vortexed for 30 s. The resulting solution was serially diluted and placed on Luria broth agar plates containing spectinomycin (100 μg ml−1) and X-Gluc (40 μg ml−1). Bacterial colonies showing blue coloration were then counted, and these counts were assumed to be those bacteria that were closely associated with the root surface. In another set, the roots were surface sterilized by immersion in 95% ethanol for 5 min followed by treatment with 3% calcium hypochlorite containing 0.1% sodium dodecyl sulfate for 1 min. After three washes with sterile distilled water followed by maceration in saline, the homogenate was serially diluted and plated on Luria agar as described above. The saline solution before maceration was plated to determine the efficiency of surface sterilization, and the number of bacteria present in this solution, if any, was subtracted from the total count after maceration.
Preparation and specificity of a polyclonal antibody raised against IRBG500.
A polyclonal antibody was raised against IRBG500 as described by Hurek et al. (19). Briefly, the bacteria were grown in Luria broth to log phase, centrifuged, and washed in normal saline. The washed, centrifuged cells were then resuspended in 3 volumes of normal saline and were killed by treatment with UV for 30 min before injection into rabbits. The cross-reaction of the antibody against various bacteria commonly isolated from the rhizosphere or interior of rice (27) (Azoarcus communis, Azospirillum spp., Enterobacter spp., K. planticola, E. coli, Pseudomonas spp., Rhizobium strain IRBG74, and Herbaspirillum seropedicae Z67) was determined by enzyme-linked immunosorbent assay (ELISA) (1). Briefly, log-phase cultures of the bacteria were normalized according to protein concentration, and various dilutions were loaded into 96-well polyvinyl chloride microtiter plates (Nunc, Roskilde, Denmark) and then probed with the anti-IRBG500 antibody. Anti-rabbit antibody conjugated to alkaline phosphatase was used as the secondary antibody, and p-nitrophenyl phosphate (Sigma Chemical Co., St. Louis, Mo.) was the substrate. The hydrolyzed p-nitrophenol was monitored using a plate reader (Minireader II; Dynatech Laboratories Inc., Chantilly, Va.) at a wavelength of 405 nm (1).
Microscopic studies of infection and colonization by IRBG500.
At least three seedlings from three independent inoculations were collected at 2, 5, and 7 DAI and stained for GUS activity in 50 mM potassium phosphate buffer (pH 7.0) containing 400 μg of X-Gluc ml−1 for 4 to 6 h as described by Jefferson et al. (28). Roots and shoots showing blue coloration were cut into small (1- to 2-mm) pieces and fixed in 4% glutaraldehyde (in 50 mM phosphate buffer [pH 7.0] containing 0.1% [vol/vol] Triton X-100) (61) under vacuum for 30 min and then at atmospheric pressure overnight. The fixed samples were prepared for light and transmission electron microscopy (TEM) by being rinsed in 50 mM phosphate buffer, dehydrated in an ethanol series, and embedded in LR white acrylic resin (London Resin Company, London, United Kingdom) (24).
Sections for light microscopy and TEM were immunogold labeled with an antibody raised against IRBG500 (see above), which was diluted 1:800 with blocking (immunogold labeling [IGL]) buffer (24). To reduce cross-reactions with plant material, the diluted IRBG500 antibody was first cross-absorbed overnight with a rice seedling homogenate prepared as described by da Rocha et al. (12). The homogenate was prepared by grinding surface-sterilized seedlings in liquid nitrogen and then washing with acetone to remove the chlorophyll.
Semithin (1- to 2-μm) sections for light microscopy were collected on gelatin-coated glass slides and incubated for 1 h in IGL buffer and then for 1 h in the cross-absorbed antibody. After being washed, the slides were incubated for 1 h in a 1:50 dilution of 5-nm gold-labeled goat anti-rabbit antibody (BioCell, Cardiff, United Kingdom) in IGL buffer. The gold labeling was then visualized for light microscopy using a BioCell silver enhancement kit. Toluidine blue (0.01%) was used to lightly counterstain the gold-labeled sections. In parallel with the sections used for immunogold silver enhancement, serial sections were collected on uncoated slides and stained with 1% toluidine blue. The protocol for immunogold labeling ultrathin (50- to 70-nm) sections for TEM was as described above except that the sections were collected on pioloform-coated nickel grids and were labeled with 15-nm gold-labeled goat anti-rabbit antibody (Amersham International). The sections for light microscopy were viewed under a Zeiss Axiophot 2 optical microscope, and the ultrathin sections were viewed by TEM using a JEOL 1200 EX.
In addition to immunogold labeling with the IRBG500 antibody, serial sections were also labeled with polyclonal antibodies raised against GUS (Clontech, Basingstoke, United Kingdom) or against the Fe protein of the nitrogenase enzyme complex originally isolated from Rhodospirillum rubrum (a gift from P. W. Ludden, Madison, Wis.). Both antibodies were diluted 1:100 in IGL buffer.
For each immunogold assay, the following controls were performed on serial sections: (i) omission of the primary antibody and (ii) replacement of the primary antibody with either preimmune or normal serum (Sigma) diluted appropriately (1:800 or 1:100) in IGL buffer.
Nitrogenase activity of rice inoculated with IRBG500.
The plants were taken from the tubes at 20 DAI and washed with sterile distilled water three times to remove the loosely associated bacteria. They were then transferred to fresh Fahraeus liquid medium without N and containing 10 mM malate, sucrose, or citrate as a carbon source. The tubes were incubated in a 14-h light/10-h dark cycle as described earlier, and ARA was determined at 12 and 14 h after injection of acetylene. Uninoculated plants served as controls. After measurement of nitrogenase activity, the number of bacteria in the tubes was determined by culturable counts on Luria agar plates.
Plant growth-promoting activity of IRBG500.
The plant growth-promoting activity of IRBG500 was determined by measuring the length of roots and shoots as well as by comparing the dry weights of the inoculated plants with those of the uninoculated control plants at 20 DAI. Total N content of the plants was estimated by a Perkin-Elmer 2400 CHN analyzer (Perkin-Elmer Corp., Norwalk, Conn.) as described earlier (29). For statistical analysis, the data were subjected to analysis of variance and the means (± standard deviations [SD]) were compared using Duncan's multiple range test.
RESULTS
Isolation and enumeration of diazotrophic endophytes from rice.
Diazotrophic bacteria were isolated from surface-sterilized roots and stems of five of the seven varieties of rice (Table 1). The population of the diazotrophic endophytes was in the range of 102 to 103 g (fresh weight)−1 of the roots and the stems, whereas the total heterotrophic bacteria recovered were on the order of 105 to 106 g (fresh weight)−1 (Table 1). After further purification of the diazotrophic isolates, one predominant morphotype was obtained in four of the varieties tested. BOX-PCR analysis showed that all four of these isolates gave similar fingerprints (data not shown). One of these isolates, designated IRBG500, was chosen for further studies. A 400-bp 16S rDNA fragment was amplified from IRBG500 (accession no. AF286867), and the sequence was aligned with reference sequences from the GenBank data library. The sequence showed highest identity of 99.5% to S. marcescens (accession no. M59160).
Nitrogenase activity of IRBG500.
Diazotrophy in IRBG500 and its gusA-linked transformant was confirmed on the basis of ARA and incorporation of 15N2 (Table 2). The presence of small amounts of fixed nitrogen (100 mg liter of yeast extract−1) was found to be essential to induce ARA by IRBG500, although activity (both ARA and 15N2 incorporation) was abolished or inhibited in the presence of 10 mM ammonium chloride (Table 2). The nitrogenase activity (ARA) of IRBG500 was less than half of that of K. planticola IRBG185 and E. cloacae IRBG236 (Table 2). No nitrogenase activity was shown by the type strains of S. marcescens (LMG2792) and S. rubidaea (LMG5019) (Table 2).
TABLE 2.
Nitrogenase activity (ARA) and 15N2 incorporation by S. marcescens IRBG500 and ARA of related bacteriaa
| Bacterial strain | ARA (nmol of C2H4 h−1 mg of protein−1) | 15N atom% excess |
|---|---|---|
| S. marcescens | ||
| IRBG500 | 65 ± 15 | 0.154 ± 0.006 |
| IRBG500-gusA | 70 ± 12 | 0.175 ± 0.008 |
| IRBG500 + 10 mM NH4Cl | 0 | 0.002 ± 0.001 |
| K. planticola | 150 ± 24 | ND |
| E. cloacae | 135 ± 30 | ND |
| S. marcescens LMG2792 | 0 | ND |
| S. rubidaea LMG5019 | 0 | ND |
Results are means ± SD of four independent experiments for ARA and two for 15N2 incorporation. ND, not determined.
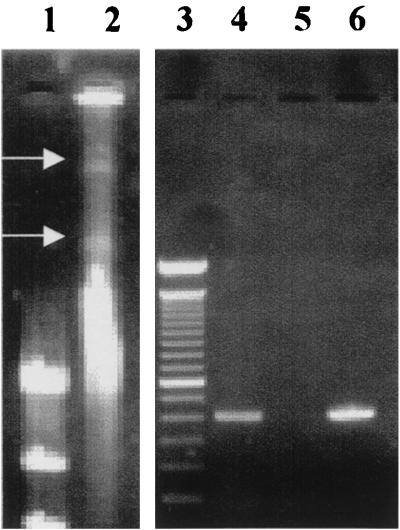
IRBG500 showed the presence of high-molecular-weight extrachromosomal elements when extracted with the method of Kado and Liu (30) (Fig. 1, lane 2). Using primers designed to amplify the fragment between positions 19 and 407 of the nifH gene of Azotobacter vinelandii M20568 (20), a product of approximately 400 bp was amplified with plasmid DNA isolated from IRBG500 (Fig. 1, lane 4). No such fragment was amplified from the type strain of S. marcescens (Fig. 1, lane 5). The nucleotide sequence of the amplified 390-bp fragment showed 60 to 73% homology with the other nifH sequences from the DNA database.
FIG. 1.
Isolation of high-molecular-weight plasmids from S. marcescens IRBG500 and amplification of the nifH fragment. Lane 1, λ-HindIII molecular weight marker; lane 2, DNA extract showing two high-molecular-weight DNA bands (arrows) (the intense and diffuse band is the chromosomal DNA); lane 3, 100-bp molecular weight marker; lane 4, PCR product from IRBG500; lane 5, PCR product from S. marcescensT; lane 6, PCR product from the nifH gene of Azoarcus sp. strain BH72.
Specificity of the IRBG500 antibody.
In ELISAs, the polyclonal antibody showed strong reaction with IRBG500 and its gusA-marked derivative up to a dilution of 1:15,000 (data not shown) but did not show a reaction at any dilutions with any of the other bacteria tested. There were also no reactions observed with any of the bacteria, including IRBG500, when the ELISAs were performed with preimmune serum used as the primary antibody (data not shown).
Colonization of rice seedlings by gusA-marked IRBG500.
Transconjugants containing gusA were obtained at a frequency of 10−6 and showed a growth rate similar to that of the wild type, IRBG500. IRBG500-gusA colonized the exterior and interior of roots, stems, and leaves of rice variety IR72, as suggested by reisolation of the inoculated strain. Both the external and internal populations, particularly of the roots, increased up to 14 DAI (Table 3). The bacterial population then remained stable for a further 20 days of observation (data not shown). Although the bacteria could be reisolated from surface-sterilized roots and stems at 3 DAI, they could be isolated from leaves only after 7 DAI, and numbers in the leaves remained relatively low throughout the experiment (Table 3). No bacteria could be isolated from the control plants under the test conditions. Throughout the study, both control and inoculated plants were without any visible disease symptoms.
TABLE 3.
Infection and spread of S. marcescens IRBG500 in rice variety IR72
| Time of harvesting (DAI) | Bacterial population (log CFU g [fresh wt]−1)a
|
|||||
|---|---|---|---|---|---|---|
| Root
|
Stem
|
Leaf
|
||||
| Surface | Internal | Surface | Internal | Surface | Internal | |
| 1 | 2.0 ± 1 | ND | ND | ND | ND | ND |
| 3 | 3.0 ± 1 | 1.0 ± 0.5 | 1.5 ± 0.5 | 1.0 ± 0.6 | ND | ND |
| 7 | 6.7 ± 1 | 3.5 ± 1.4 | 2.0 ± 1.0 | 3.0 ± 0.8 | 2.0 ± 1.0 | 1.0 ± 0.8 |
| 14 | 7.5 ± 1.8 | 6.4 ± 1.6 | 1.0 ± 0.7 | 5.0 ± 1.2 | 2.0 ± 1.2 | 2.2 ± 1.8 |
Results are means ± SD of at least three independent observations. ND, not detected.
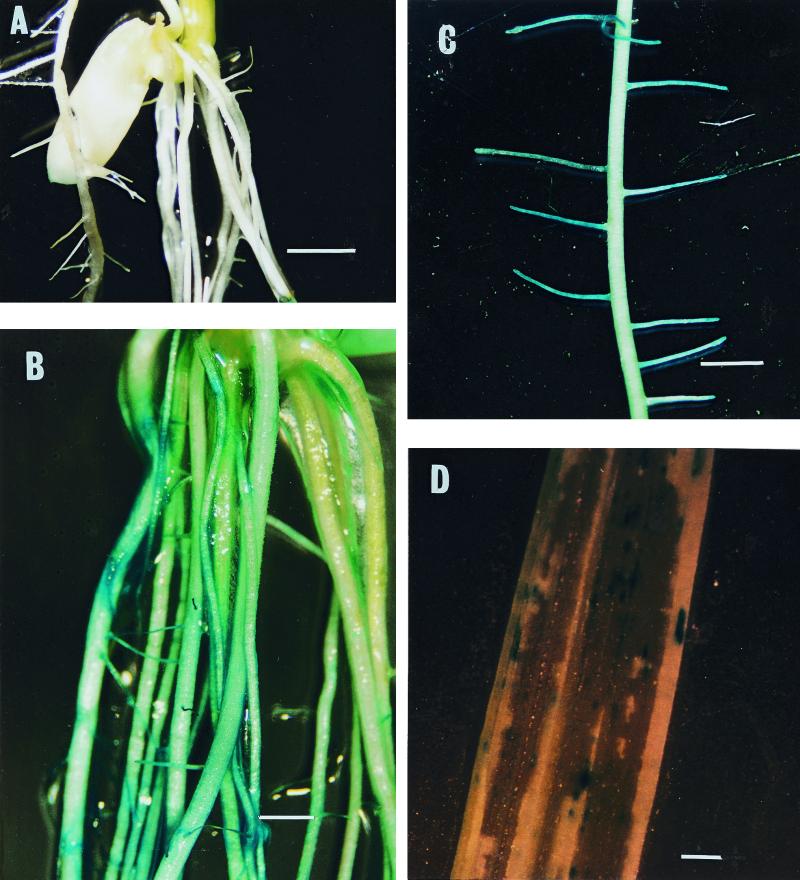
At 5 DAI, uninoculated control plants did not show any blue coloration with X-Gluc (Fig. 2A). However, roots and the basal part of the stem of the inoculated plants showed blue coloration when stained for GUS activity (Fig. 2B), with that on the lateral roots being most intense (Fig. 2C). Blue coloration could also be observed on the lower part of the stem as well as on leaves at 7 DAI (Fig. 2D).
FIG. 2.
Light micrographs of GUS-stained roots, stems, and leaves of rice variety IR72 at 5 days after inoculation with S. marcescens IRBG500 marked with gusA. Control uninoculated plants showed no GUS activity (A). GUS activity was observed on all roots (B), with the most intense color development on the lateral roots (C). GUS staining was also observed on the stems (D). Bars: 2 mm (A and B) and 0.5 mm (C and D).
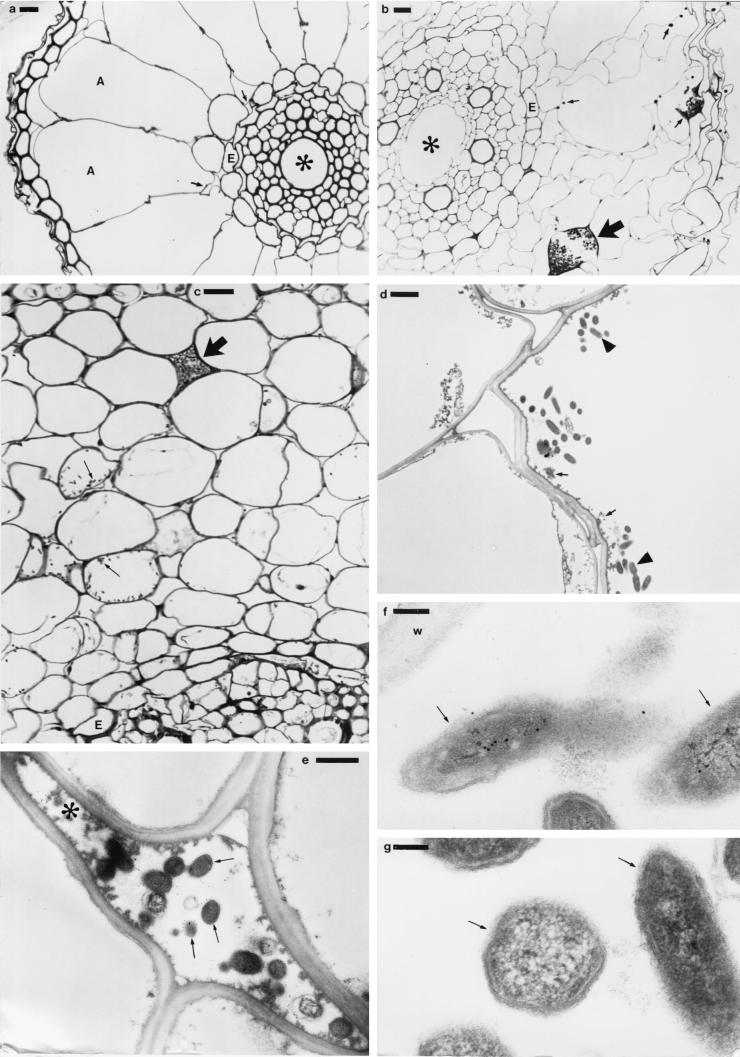
More detailed studies of root material sectioned for light microscopy confirmed that there was no internal or surface colonization of uninoculated control plants (Fig. 3a). However, light and electron microscopy of inoculated plants showed that by 5 DAI, in addition to the surface colonization (Fig. 2b to d) there was also considerable internal colonization of the roots, stems, and leaves. Transverse sections of roots showing GUS activity revealed bacteria within many of the cortical intercellular spaces next to the stele and within the aerenchyma (Fig. 3b and c), although there was no evidence that the bacteria had penetrated the endodermis to colonize the root vascular system (Fig. 3b and c). Interestingly, many of the root cortical cells contained bacteria (Fig. 3c), and by TEM, both intracellular bacteria (Fig. 3d) and those in the intercellular spaces (Fig. 3e) were healthy in appearance and were immunogold labeled with the antibody raised against IRBG500 (Fig. 3e). On the other hand, the host plant cells containing IRBG500 appeared not to have intact cytoplasm (Fig. 3d), and the intercellular bacteria were often surrounded by electron-dense material (Fig. 3e). In addition to reacting to the IRBG500-specific antibody (Fig. 3e), the bacteria in the roots also reacted with an anti-GUS antibody (Fig. 3f), but there was no reaction when serial sections were incubated in nonimmune serum (Fig. 3g).
FIG. 3.
(a) Light micrograph of a toluidine blue (1%)-stained transverse section of a root from an uninoculated control plant. No bacteria are visible within the intercellular spaces (arrows), aerenchyma (A), or vascular cylinder (∗). Light (b to c) and transmission electron (d to g) micrographs show sections of rice roots at 5 days after inoculation with S. marcescens strain IRBG500. Transverse sections of infected roots were stained with 1% toluidine blue (b to c). (b) Bacteria have colonized intercellular spaces and aerenchyma (small arrows), sometimes very densely (large arrow), but none have penetrated the endodermis (E) to colonize the vascular cylinder (∗). (c) Higher magnification showing bacteria within the cortical cells (small arrows), as well as a dense colony of intercellular bacteria (large arrow). (d) Bacteria (arrowheads) colonizing a cortical cell (see panel c). Note that the host cytoplasm is no longer intact and exists only as fragments (small arrows). (e) Bacteria colonizing an intercellular space in the cortex (see panel c). The bacteria (arrows) were immunogold labeled using a primary antibody raised against S. marcescens strain IRBG500 and a secondary goat anti-rabbit antibody conjugated to 15-nm gold particles. Note that electron-dense material has accumulated close to the bacteria (∗). (f and g) Transmission electron micrographs of serial sections to panels d and e. The bacteria in these sections (arrows) were immunogold labeled with a primary antibody raised against GUS (f) or with normal rabbit serum (g), followed by a secondary goat anti-rabbit antibody conjugated to 15-nm gold particles. (f) Bacteria showing internal immunogold labeling (arrows), indicating the expression of GUS protein. w, plant cell wall. (g) Bacteria with no significant internal immunogold labeling (arrows). Bars: 10 μm (a to c), 2 μm (d), 1 μm (e), and 100 nm (f, g).
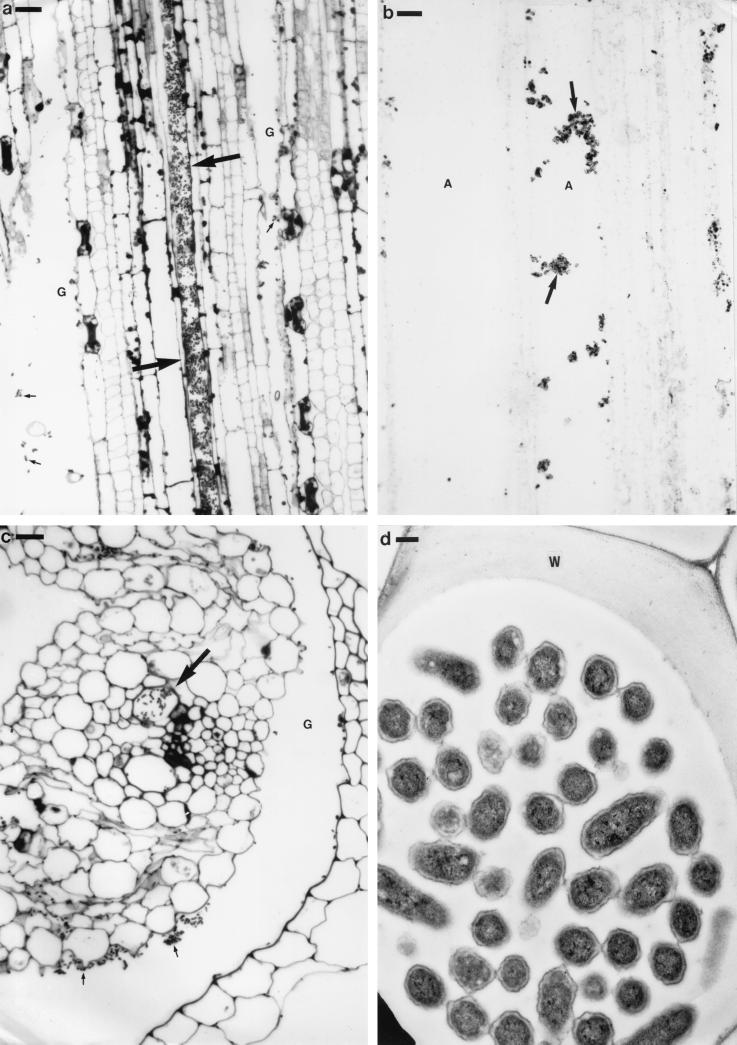
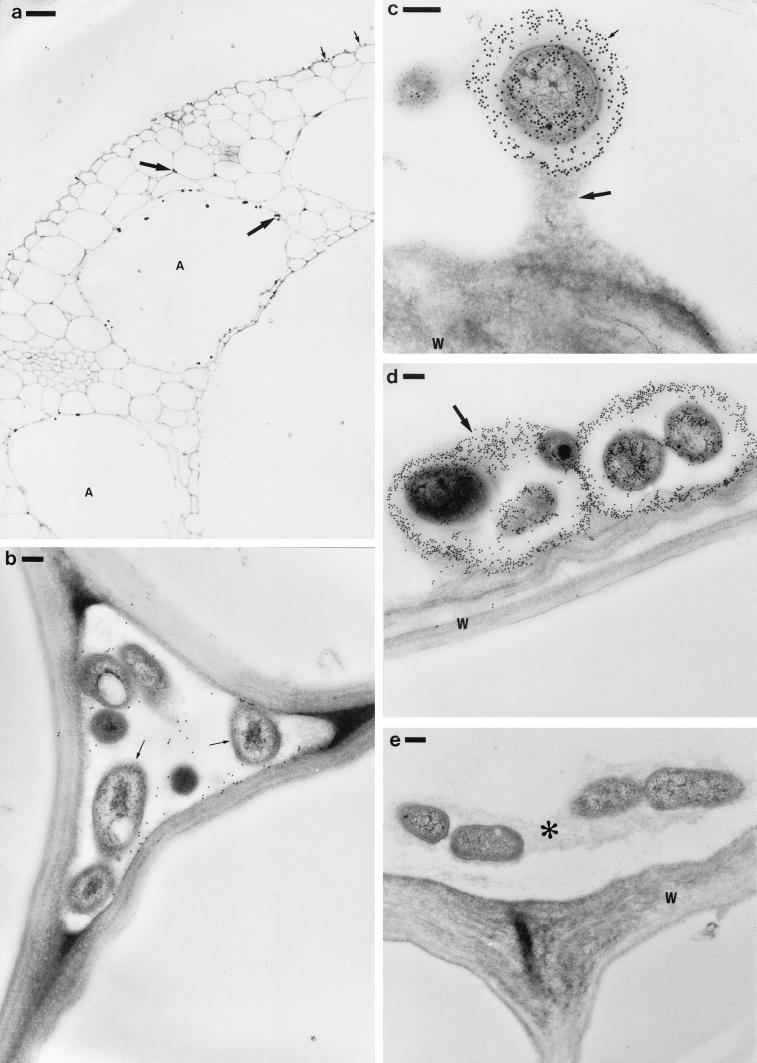
In the stems, large numbers of bacteria were observed within the aerenchyma and intercellular spaces and in the gaps between the leaf sheaths (Fig. 4a to c). Again, these bacteria were recognized by immunogold labeling with an antibody raised against IRBG500 (Fig. 4b). Unlike in the roots, high numbers of bacteria were also observed within some of the xylem vessels (Fig. 4a, c, and d), and these appeared not to have provoked any obvious host defense response. In the leaves, although the bacteria were fewer in number, some had readily colonized the aerenchyma and the intercellular spaces (Fig. 5). They were also occasionally seen within the xylem (not shown). The bacteria in the aerenchyma often showed high levels of immunogold labeling, with a dense halo of gold particles around individual bacteria or around bacterial microcolonies (Fig. 5c and d). These bacteria appeared to be attached to the host cell walls via the gold-labeled material that surrounded them. In contrast to the bacteria in the aerenchyma, those within the intercellular spaces were much less densely gold labeled (Fig. 5b).
FIG. 4.
Longitudinal (a to b) and transverse (c to d) sections of rice stems at 5 days after inoculation with S. marcescens strain IRBG500. (a) Infected stem stained with 1% toluidine blue. Two leaf sheaths are visible, and bacteria (small arrows) can be seen in the gaps (G) between them. Note that the xylem in one of the leaf sheaths is particularly densely colonized (large arrows). (b) Bacteria colonizing the stem aerenchyma (A). The bacterial colonies (arrows) were immunogold labeled as for Fig. 3e (followed by silver enhancement). (c) Infected stem stained with 1% toluidine blue. A vascular bundle in one of the leaf sheaths is shown, and one of the metaxylem vessels is heavily infected with bacteria (large arrow). As in panel b, bacteria (small arrows) can also be seen within the gaps (G) between the leaf sheaths. (d) Transmission electron micrograph of a stem protoxylem vessel containing numerous bacteria. Note that there is no obvious host defense material surrounding the bacteria. W, cell wall. Bars: 20 μm (a), 10 μm (b to c), and 250 nm (d).
FIG. 5.
Light (a) and transmission electron (b to e) micrographs of rice leaves at 7 days after inoculation with S. marcescens strain IRBG500. (a) Transverse section of bacteria (arrows) colonizing the leaf aerenchyma (A) and intercellular spaces. Some bacteria can also be seen on the leaf surface (small arrows). The bacteria were immunogold labeled (followed by silver enhancement) as for Fig. 4b. (b) Bacteria (arrows) within an intercellular space. The section was immunogold labeled as for Fig. 4b. (c) A bacterium attached to the cell wall (W) of the aerenchyma by a stalk of electron-dense material (large arrow). The section was immunogold labeled as for Fig. 4b; note that the bacterium is very densely gold labeled compared to the intercellular bacteria in panel b, and also that the majority of the labeling is of extracellular material surrounding it (small arrow). (d) Microcolony of five bacteria attached to the cell wall (W) of the aerenchyma. The section was immunogold labeled as for Fig. 4b. Again, note that the extracellular material surrounding the bacteria is very densely labeled (arrow). (e) Microcolony of bacteria attached to the cell wall (W) of the aerenchyma. This section was treated as for Fig. 4b except that the primary antibody was omitted. There is no gold labeling visible on the bacteria or on the material surrounding them (∗). Bars: 20 μm (a) and 200 nm (b to e).
Control sections incubated in nonimmune serum (Fig. 3g) or with the primary antibody omitted (Fig. 5e) showed little or no gold labeling. Similarly, none of the bacteria in the roots, stems, or leaves reacted significantly with an antibody raised against the Fe protein of nitrogenase (data not shown).
Nitrogenase activity of plants inoculated with IRBG500.
Significant nitrogenase activity (ARA) by the inoculated plants could be detected at 20 DAI, although only when carbon sources were added to the medium, with organic acids, such as citrate and malate, giving the maximum activity (12.5 ± 1.5 and 14.8 ± 2.8 nmol of C2H4 h−1 106 bacteria−1, respectively, versus 5.5 ± 1.5 with sucrose). ARA was abolished when the roots were surface sterilized (data not shown).
Growth promotion of IR72 by IRBG500.
Inoculation of IR72 with IRBG500 resulted in increased root length and dry weight of these plants compared with the uninoculated control plants (Table 4). There was, however, no significant difference in the total N of the inoculated plants compared to uninoculated seedlings, it typically being 0.2 to 0.3 mg plant−1 (data not shown).
TABLE 4.
Growth promotion activity of S. marcescens IRBG500a
| Strain | Length (cm)
|
Dry wt (mg)
|
||
|---|---|---|---|---|
| Root | Shoot | Root | Shoot | |
| IR72 (control) | 11.2 | 18.5 | 4.62 | 5.85 |
| IR72 + IRBG500 | 14.8b | 19.2 | 6.40b | 6.46 |
| LSD (0.05) | 1.5 | 2.3 | 0.47 | 1.22 |
Results are means of three independent experiments with at least 10 plants in each experiment. Plants were examined at 20 DAI. The data were subjected to analysis of variance, and the means were compared by Duncan's multiple range test.
Significantly different from the control at 5% level.
DISCUSSION
Isolation and characterization of predominant endophytic diazotrophs.
Sterilization by treating with ethanol and mild flaming was effective in removing most of the microbes from the plant surfaces. Diverse diazotrophic bacteria were then isolated from the surface-sterilized roots and stems of five rice varieties growing in nonsterile soil in greenhouse conditions. Interestingly, no diazotrophs could be isolated from two other varieties examined (IR387 and Palawan). Within the five varieties that contained diazotrophs (Table 1), the total diazotrophic bacteria present in the roots and stems were only 5 to 10% of the total heterotrophic bacteria that could be cultured, consistent with the earlier findings of Barraquio et al. (6).
A group of N2-fixing bacteria isolated from the surface-sterilized stems and roots of these five varieties showed similar fingerprints with BOX-PCR and were identified as S. marcescens based on a 400-bp 16S rDNA sequence. Although S. marcescens is generally regarded as a potential human and animal pathogen, various species of Serratia have been isolated from cotton (Gossypium hirsutum) and sweet corn (Zea mays) (43), as well as the rhizosphere of rice (54) and rice seeds (44). Indeed, nonclinical isolates of S. marcescens have been used as agricultural biocontrol agents due to their chitinase activity (31, 43, 54) and ability to induce systemic resistance in plants (48).
Although this is the first report of diazotrophy by S. marcescens, Krishnapillai and Postgate (34) previously transferred Klebsiella genes involved in N2 fixation to S. marcescens and showed that they were functional. The only other report of diazotrophy in the genus Serratia is that of an S. rubidea strain isolated from the endorhizosphere of wheat (Triticum aestivum) and Ammophila arenaria (55).
The diazotrophy of IRBG500 was confirmed via the significant incorporation of 15N2, but the 15N atom percent excess was approximately half of that reported for other plant-associated diazotrophs, such as Agrobacterium tumefaciens (32) and Herbaspirillum spp. (2). Both ARA incorporation and 15N2 incorporation were inhibited by the presence of ammonium, indicating that the regulation of nitrogenase of IRBG500 was similar to that of many other diazotrophs (17). Finally, the S. marcescens strains in the present study (including IRBG500) were observed to have reduced nitrogenase activity (ARA) as a result of repeated subculturing (data not shown). The reason for this is not known but is currently under investigation at IRRI. It is possible that the N2-fixing ability of IRBG500 is localized on extrachromosomal elements present. Such extrachromosomal nitrogenase has been detected in other enteric bacteria such as Enterobacter agglomerans (59) and Rahnella aquatilis (7). Although an expected fragment of 400 bp was amplified when the plasmid DNA was used as the template, it remains to be confirmed if the nitrogenase genes are localized on the plasmid because the plasmid DNA isolated also had contaminating genomic DNA (Fig. 1, lane 2).
Infection and colonization of rice seedlings by S. marcescens IRBG500.
To study the colonization of rice by diazotrophic S. marcescens, strain IRBG500 was marked with gusA and inoculated onto rice seedlings under gnotobiotic conditions. The gusA-marked strain could be reisolated from surface-sterilized roots and stems of the inoculated seedlings, indicating internal colonization and systemic spreading within the plants. Staining of the whole roots showed that the GUS activity was most intense on some of the younger lateral roots, and it is possible that the bacteria entered these at their junctions with the primary roots. This type of infection has been observed with other diazotrophic endophytes, such as A. diazotrophicus in sugarcane (24), Azoarcus spp. in rice and Kallar grass (28), and Herbaspirillum spp. in sugarcane and rice (25–27).
A major drawback of in situ GUS staining is that the presence of blue color does not unequivocally confirm the location, or even the presence, of the GUS-labeled bacteria because the color can diffuse into bacterium-free plant material (19, 51). Therefore, to prove that S. marcescens strain IRBG500 is genuinely endophytic in rice, tissues stained for GUS activity were fixed with glutaraldehyde, embedded in resin, and sectioned for optical and electron microscopy. This also reduced the possibility that the bacteria observed within the plants had accidentally moved into them from the external population (27, 50). The bacteria in the sections reacted well with a polyclonal antibody raised against IRBG500, which was specific to IRBG500 because it did not react with other bacteria that are commonly isolated from the interior of wetland rice. The bacteria also showed reaction with an anti-GUS antibody, confirming that the bacteria inside rice tissues were the inoculated strain of gusA-marked IRBG500 (Fig. 3f).
Inside the plant, bacteria were mainly localized within the aerenchyma and the intercellular spaces. These apoplastic locations seem to be the preferred sites of the few endophytic diazotrophs so far examined in rice. For example, Hurek et al. (19) and Egener et al. (13) have shown dense colonies of Azoarcus in the aerenchyma of rice (and Kallar grass) roots, and James et al. (27) have presented micrographs showing H. seropedicae extensively colonizing the intercellular spaces and aerenchyma of roots, stems, and leaves. In addition to the apoplastic localization, the cortical cells in some of the roots also showed intracellular bacteria. However, TEM studies showed that these cortical cells did not have intact cytoplasm. It is possible that the bacterium-colonized cortical cells were preprogramed to senesce as part of the processes involved in the formation of the lysigenous aerenchyma that characterizes the roots of wetland rice (50), and hence, these cells may already have been dead or dying prior to their colonization by the bacteria. In contrast to the root cortical and aerenchyma cells, the stems and leaves showed little or no sign of degradation associated with bacterial colonization, even though bacterial numbers were high in the localized areas that were observed microscopically (Fig. 4 and 5). A possible reason for the lack of an obvious defense response from the stems and leaves could be that the bacteria were primarily localized within intercellular spaces or within already dead cells, such as xylem and aerenchyma, and were not observed penetrating intact host cells.
In the present study, the bacteria were not found within the xylem of the roots, although the xylem of the stem was extensively colonized. This contrasts with work on Azoarcus and H. seropedicae that has shown colonization of root xylem vessels, albeit at a relatively low frequency (19, 27). In the stems, the bacteria were present in the xylem vessels in fairly high concentrations, and apparently unoccluded xylem may have allowed spreading of the bacteria through the stems to the leaves via the transpiration stream. This has also been suggested for rice infected with Azoarcus (19) and H. seropedicae (27). Colonization of xylem vessels by endophytic diazotrophs, such as A. diazotrophicus and Herbaspirillum spp., is commonly reported in other tropical grasses, particularly sugarcane and sorghum (23–25, 47). James and Olivares (26) have suggested that xylem could be a suitable nonnodular niche for N2 fixation because it can provide the low pO2 required for the expression and function of nitrogenase and also allow for the exchange of fixed nitrogen. Indeed, James et al. (25) have shown that the phytopathogen H. rubrisubalbicans can express nitrogenase protein within sorghum leaf protoxylem. On the other hand, N2 fixation is an energetically demanding process, and carbon sources tend to be low in xylem (26). Although IRBG500 colonized stem xylem vessels in high numbers, it could not be labeled with an antinitrogenase antibody, suggesting that conditions in the xylem were not conducive for N2 fixation by IRBG500.
Effect of S. marcescens IRBG500 on growth, nitrogenase activity, and total N content of infected rice seedlings.
Although IRBG500 could colonize both the roots and the stems, ARA could not be detected in the plants inoculated with IRBG500, and the bacteria within the rice seedlings also showed no immunogold reaction with an antibody against the Fe protein of the nitrogenase complex. When carbon was added to the rooting medium, however, significant ARA could be detected, suggesting that the bacteria associated with the rice seedlings were limited by the low availability of carbon and energy. This is consistent with recent reports that supplementation of carbon was essential for nitrogenase activity of Azorhizobium caulinodans in association with rice (46) and also for expression of dinitrogenase reductase by Klebsiella pneumoniae in association with maize (11). However, an additional factor causing the carbon limitation on rice-associated BNF in the present study could be that the plants analyzed were in the early stages of growth (20 DAI). In field-grown rice, significant ARA was obtained only after 40 days of transplantation (36). Although IRBG500 did not show ARA in association with rice, it promoted the growth of the inoculated seedlings but led to no enhancement of their N content, suggesting that the growth promotion was probably due to mechanisms other than N2 fixation.
The results presented here show that S. marcescens IRBG500 could become endophytically established in roots, stems, and leaves and could also increase the root length and root dry weight of the inoculated plants. Although endophytes have been confirmed and localized in various plants and are postulated to play an important role in sustainable crop production (62), the mechanisms of their endophytic establishment are not well known. It has been suggested that the bacteria could enter through the fissures created by the emergence of lateral roots or could actively dissolve the cell wall components to gain entry (26, 50). Therefore, it would be interesting to determine the biochemical and molecular mechanisms of entry into rice by S. marcescens IRBG500, especially as some strains possess hydrolytic enzymes, such as chitinase (14), lipase (40), and ligninase (52). Finally, IRBG500 could not be isolated from two of the seven rice varieties examined, which indicates that there may be some host specificity in the interaction between IRBG500 and rice; this also warrants further exploration.
ACKNOWLEDGMENTS
We are thankful to W. L. Barraquio for useful discussions and to J. I. Sprent for use of TEM facilities at the University of Dundee.
This work was supported by grants from the German Agency for Technical Cooperation (GTZ) to IRRI in collaboration with the Max Planck Institute, Marburg, Germany, and the University of Bremen, Bremen, Germany.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1992. [Google Scholar]
- 2.Baldani V L D, Baldani J I, Olivares F, Döbereiner J. Identification and ecology of Herbaspirillum seropedicae and the closely related Pseudomonas rubrisubalbicans. Symbiosis. 1992;13:65–73. [Google Scholar]
- 3.Barraquio W L, de Guzman M R, Barrion M, Watanabe I. Population of aerobic heterotrophic nitrogen-fixing bacteria associated with wetland and dryland rice. Appl Environ Microbiol. 1982;42:124–128. doi: 10.1128/aem.43.1.124-128.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barraquio W L, Ladha J K, Watanabe I. Isolation and identification of a N2 fixing Pseudomonas associated with wetland rice. Can J Microbiol. 1983;29:867–873. doi: 10.1139/m83-141. [DOI] [PubMed] [Google Scholar]
- 5.Barraquio W L, Padre B C, Watanabe I, Knowles R. Nitrogen fixation by Pseudomonas saccharophila Doudoroff ATCC 15946. J Gen Microbiol. 1986;132:237–241. [Google Scholar]
- 6.Barraquio W L, Revilla L, Ladha J K. Isolation of endophytic diazotrophic bacteria from wetland rice. Plant Soil. 1997;194:15–24. [Google Scholar]
- 7.Berge O, Heulin T, Achouak W. Rahnella aquatilis, a nitrogen-fixing enteric bacterium associated with the rhizosphere of wheat and maize. Can J Microbiol. 1990;37:195–203. [Google Scholar]
- 8.Boddey R M, de Oliveira O C, Urquiaga S, Reis V M, Olivares F L, Baldani V L D, Dobereiner J. Biological nitrogen fixation associated with sugar cane and rice: contribution and prospects for improvement. Plant Soil. 1995;174:195–209. [Google Scholar]
- 9.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Cannon F C, Dixon R A, Postgate J R, Primrose S B. Chromosomal integration of Klebsiella nitrogen fixation genes in Escherichia coli. J Gen Microbiol. 1974;80:227–239. doi: 10.1099/00221287-80-1-227. [DOI] [PubMed] [Google Scholar]
- 11.Chelius M K, Triplett E W. Immunolocalization of dinitrogenase reductase produced by Klebsiella pneumoniae in association with Zea mays. Appl Environ Microbiol. 2000;66:783–787. doi: 10.1128/aem.66.2.783-787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.da Rocha A, Ohki S T, Hiruki C. Detection of mycoplasm like organisms in situ by indirect immunofluorescence microscopy. Phytopathology. 1986;76:864–868. [Google Scholar]
- 13.Egener T, Hurek T, Reinhold-Hurek B. Endophytic expression of nif genes of Azoarcus sp. strain BH72 in rice roots. Mol Plant-Microbe Interact. 1999;12:813–819. doi: 10.1094/MPMI.1998.11.1.71. [DOI] [PubMed] [Google Scholar]
- 14.Engelhard M, Hurek T, Reinhold-Hurek B. Preferential occurrence of diazotrophic endophyte, Azoarcus sp. in wild rice species and land races of Oryza sativa in comparison with modern races. Environ Microbiol. 2000;2:131–141. doi: 10.1046/j.1462-2920.2000.00078.x. [DOI] [PubMed] [Google Scholar]
- 15.Fahraeus A. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol. 1957;16:374–381. doi: 10.1099/00221287-16-2-374. [DOI] [PubMed] [Google Scholar]
- 16.Grimont P A D, Grimont F. The genus Serratia. Annu Rev Microbiol. 1978;32:221–248. doi: 10.1146/annurev.mi.32.100178.001253. [DOI] [PubMed] [Google Scholar]
- 17.Hill S. Physiology of nitrogen fixation in free-living heterotrophs. In: Stacey G, Evans H J, Burris R H, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 87–135. [Google Scholar]
- 18.Hossain M, Fischer K S. Rice research for food security and sustainable agricultural development in Asia: achievements and future challenges. GeoJournal. 1995;35:286–295. [Google Scholar]
- 19.Hurek T, Reinhold-Hurek B, Van Montagu M, Kellenberger E. Root colonization and systemic spreading of Azoarcus sp. strain BH72 in grasses. J Bacteriol. 1994;176:1913–1923. doi: 10.1128/jb.176.7.1913-1923.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurek T, Egener T, Reinhold-Hurek B. Divergence in nitrogenases of Azoarcus spp., proteobacteria of the β subclass. J Bacteriol. 1997;179:4172–4178. doi: 10.1128/jb.179.13.4172-4178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurek T, Wagner B, Reinhold-Hurek B. Identification of N2-fixing plant-and-fungus-associated Azoarcus species by PCR-based genomic fingerprints. Appl Environ Microbiol. 1997;63:4331–4339. doi: 10.1128/aem.63.11.4331-4339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International Rice Research Institute. Rice research in a time of change. International Rice Research Institute's medium term plan for 1994–1998. Makati City, Philippines: Internal Rice Research Institute; 1993. [Google Scholar]
- 23.James E K. Nitrogen fixation in endophytic and associative symbiosis. Field Crops Res. 2000;65:197–209. [Google Scholar]
- 24.James E K, Reis V M, Olivares F L, Baldani J I, Dobereiner J. Infection of sugar cane by the nitrogen-fixing bacterium Acetobacter diazotrophicus. J Exp Bot. 1994;275:757–766. [Google Scholar]
- 25.James E K, Olivares F L, Baldani J I, Dobereiner J. Herbaspirillum, an endophytic diazotroph colonizing vascular tissue in the leaves of Sorghum bicolor L. Moench. J Exp Bot. 1997;48:785–797. [Google Scholar]
- 26.James E K, Olivares F L. Infection and colonization of sugarcane and other graminaceous plants by endophytic diazotrophs. Crit Rev Plant Sci. 1998;17:77–119. [Google Scholar]
- 27.James E K, Gyaneshwar P, Mathan N, Barraquio W L, Ladha J K. Endophytic diazotrophs associated with rice. In: Ladha J K, Reddy P M, editors. The quest for nitrogen fixation in rice. Makati City, Philippines: International Rice Research Institute; 2000. pp. 119–140. [Google Scholar]
- 28.Jefferson R A, Kavanagh T A, Bevan M W. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jimenez R R, Ladha J K. Automated elemental analysis: a rapid and reliable but expensive measurement of total carbon and nitrogen in plant and soil samples. Commun Soil Sci Plant Anal. 1993;24:1897–1924. [Google Scholar]
- 30.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:373–378. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalbe C, Marten P, Berg G. Strains of genus Serratia as beneficial rhizobacteria of oilseed rape with antifungal properties. Microbiol Res. 1996;151:433–439. doi: 10.1016/S0944-5013(96)80014-0. [DOI] [PubMed] [Google Scholar]
- 32.Kanvinde L, Sastry G R K. Agrobacterium tumefaciens is a diazotrophic bacterium. Appl Environ Microbiol. 1990;56:2087–2092. doi: 10.1128/aem.56.7.2087-2092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirchhof G, Reis V M, Baldani J I, Eckart B, Dobereiner J, Hartman A. Occurrence, physiological and molecular analysis of endophytic diazotrophic bacteria in gramineous energy plants. Plant Soil. 1997;194:45–56. [Google Scholar]
- 34.Krishnapillai V, Postgate J R. Expression of Klebsiella his and nif genes in Serratia marcescens, Erwinia herbicola and Proteus mirabilis. Arch Microbiol. 1980;127:115–118. doi: 10.1007/BF00428014. [DOI] [PubMed] [Google Scholar]
- 35.Ladha J K, Barraquio W L, Watanabe I. Isolation and identification of nitrogen-fixing Enterobacter cloacae and Klebsiella planticola associated with rice plants. Can J Microbiol. 1983;29:1301–1308. doi: 10.1139/m83-141. [DOI] [PubMed] [Google Scholar]
- 36.Ladha J K, Triol A C, Daroy L G, Caldo G, Ventura W, Watanabe I. Plant associated N2 fixation (C2H2) reduction by five rice varieties, and relationship with plant growth characters as affected by straw incorporation. Soil Sci Plant Nutr. 1986;32:91–106. [Google Scholar]
- 37.Ladha J K, Reddy P M. Extension of nitrogen fixation to rice—necessity and possibilities. GeoJournal. 1995;35:363–372. [Google Scholar]
- 38.Ladha J K, de Bruijn F J, Malik K A. Assessing opportunities for nitrogen fixation in rice: a frontier project. Plant Soil. 1997;194:1–10. [Google Scholar]
- 39.Ladha J K, Kirk G J D, Bennett J, Peng S, Reddy C K, Reddy P M, Singh U. Opportunities for increased nitrogen use efficiency from improved lowland rice germplasm. Field Crops Res. 1998;56:41–47. [Google Scholar]
- 40.Li X, Tetling S, Winkler U K, Jaeger K, Benedick M J. Gene cloning, sequence analysis, purification, and secretion by Escherichia coli of and extracellular lipase from Serratia marcescens. Appl Environ Microbiol. 1995;61:2674–2680. doi: 10.1128/aem.61.7.2674-2680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malarvizhi P, Ladha J K. Influence of available N and rice genotype on associative nitrogen fixation. Soil Sci Soc Am J. 1999;63:93–99. [Google Scholar]
- 42.Malik K A, Rakhshanda B, Mehnaz S, Rasul G, Mirza M S, Ali S. Association of nitrogen-fixing, plant growth promoting rhizobacteria (PGPR) with Kallar grass and rice. Plant Soil. 1997;194:37–44. [Google Scholar]
- 43.McInroy J A, Kloepper J W. Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant Soil. 1995;173:337–349. [Google Scholar]
- 44.Mukhopadhyay K, Garrison N K, Hinton D M, Bacon C W, Khush G S, Peck H D, Datta N. Identification and characterization of bacterial endophytes of rice. Mycopathologia. 1996;134:151–159. doi: 10.1007/BF00436723. [DOI] [PubMed] [Google Scholar]
- 45.Mylona P, Pawlowski K, Bisseling T. Symbiotic nitrogen fixation. Plant Cell. 1995;7:869–885. doi: 10.1105/tpc.7.7.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nieuwenhove V C, Holm L V, Kulasooriya S A, Vlassak K. Establishment of Azorhizobium caulinodans in the rhizosphere of wetland rice (Oryza sativa L.) Biol Fertile Soils. 2000;31:143–149. [Google Scholar]
- 47.Olivares F L, James E K, Baldani J I, Dobereiner J. Infection of mottled stripe disease susceptible and resistant varieties of sugarcane by the endophytic diazotroph Herbaspirillum. New Phytol. 1997;35:723–737. [Google Scholar]
- 48.Press C M, Wilson M, Tuzun S, Kloepper J W. Salicylic acid produced by S. marcescens 90–166 is not the primary determinant of induced systemic resistance in cucumber or tobacco. Mol Plant-Microbe Interact. 1997;10:761–768. [Google Scholar]
- 49.Quispel A. A critical evaluation of the prospects of nitrogen fixation with non-legumes. Plant Soil. 1991;137:1–11. [Google Scholar]
- 50.Reinhold-Hurek B, Hurek T. Interactions of gramineous plants with Azoarcus spp. and other diazotrophs: identification, localization and perspectives to study their function. Crit Rev Plant Sci. 1998;17:29–54. [Google Scholar]
- 51.Reinhold-Hurek B, Hurek T. Life in grasses: diazotrophic endophytes. Trends Microbiol. 1998;6:139–144. doi: 10.1016/s0966-842x(98)01229-3. [DOI] [PubMed] [Google Scholar]
- 52.Rhoads T L, Mikell A T, Eley M H. Investigation of the lignin degrading activity of Serratia marcescens: biochemical screening and ultrastructural evidence. Can J Microbiol. 1995;41:592–600. doi: 10.1139/m95-079. [DOI] [PubMed] [Google Scholar]
- 53.Roger P A, Ladha J K. Biological N2 fixation in wetland rice fields: estimation and contribution to nitrogen balance. Plant Soil. 1992;141:41–55. [Google Scholar]
- 54.Rosales A M, Vantomme R, Swings J, Deley J, Mew T W. Identification of some bacteria from paddy antagonistic to several rice fungal pathogens. J Phytopathol. 1993;138:189–208. [Google Scholar]
- 55.Ruppel S. Isolation and characterization of dinitrogen-fixing bacteria from the rhizosphere of Triticum aestivum and Ammophila arenaria. Dev Soil Sci. 1988;18:253–262. [Google Scholar]
- 56.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 57.Shrestha R K, Ladha J K. Genotypic variation in promotion of rice nitrogen fixation as determined by 15N dilution. Soil Sci Am J. 1996;60:1815–1821. [Google Scholar]
- 58.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis of Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 59.Singh M, Kleeberger A, Klingmüller W. Location of nitrogen fixation (nif) genes on indigenous plasmids of Enterobacter agglomerans. Mol Gen Genet. 1983;190:373–378. [Google Scholar]
- 60.Stoltzfus J R, So R, Malarvizhi P P, Ladha J K, de Bruijn F J. Isolation of endophytic bacteria from rice and assessment of their potential for supplying rice with biologically fixed nitrogen. Plant Soil. 1997;194:25–36. [Google Scholar]
- 61.Stomp A. Histochemical localization of β-glucuronidase. In: Gallagher S R, editor. GUS protocols. New York, N.Y: Academic Press Inc.; 1992. pp. 103–113. [Google Scholar]
- 62.Sturz A V, Christie B R, Nowak J. Bacterial endophytes: potential role in developing sustainable system of crop production. Crit Rev Plant Sci. 2000;19:1–30. [Google Scholar]
- 63.Ueda T, Suga Y, Yahiro N, Matsuguchi T. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol. 1995;177:1414–1417. doi: 10.1128/jb.177.5.1414-1417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ueda T, Suga Y, Yahiro N, Matsuguchi T. Genetic diversity of N2-fixing bacteria associated with rice roots by molecular evolutionary analysis of nifD library. Can J Microbiol. 1995;41:235–240. doi: 10.1139/m95-032. [DOI] [PubMed] [Google Scholar]
- 65.Urquiaga S, Cruz K H S, Boddey R M. Contribution of nitrogen fixation to sugar cane: nitrogen-15 and nitrogen balance estimates. Soil Sci Soc Am J. 1992;56:105–114. [Google Scholar]
- 66.Versalovic J, Schneider M, de Bruijn F J, Lupski J R. Genomic fingerprinting of bacteria using repetitive sequences based polymerase chain reaction. Methods Mol Cell Biol. 1994;5:25–40. [Google Scholar]
- 67.Watanabe I, Barraquio W L. Low levels of fixed nitrogen are required for isolation of free-living N2-fixing organisms from rice roots. Nature. 1979;277:565–566. [Google Scholar]
- 68.Wilson K J, Sessitch A, Carbo J C, Giller K E, Akkermans A D L, Jefferson R A. β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other Gram negative bacteria. Microbiology. 1995;141:1691–1705. doi: 10.1099/13500872-141-7-1691. [DOI] [PubMed] [Google Scholar]
- 69.Wu P, Zhang G, Ladha J K, McCouch S R, Huang N. Molecular-marker-facilitated investigation on the ability to stimulate N2 fixation in the rhizosphere by irrigated rice plants. Theor Appl Genet. 1995;91:1171–1183. doi: 10.1007/BF00220926. [DOI] [PubMed] [Google Scholar]
- 70.You C, Zhou F. Non-nodular endorhizospheric nitrogen fixation in wetland rice. Can J Microbiol. 1989;35:403–408. [Google Scholar]