Abstract
A derivative of SspC, a minor α/β-type, small, acid-soluble spore protein (SASP) from Bacillus subtilis, was generated that has a very high affinity for DNA. This protein (SspCΔ11-D13K) was able to confer UV resistance on spores lacking α/β-type SASP, and spores with SspCΔ11-D13K triggered germination normally. However, SspCΔ11-D13K blocked outgrowth of ≥90% of germinated spores, and SspCΔ11-D13K persisted in these germinated spores, whereas wild-type SspC was almost completely degraded. The outgrowth phenotype of spores with SspCΔ11-D13K is proposed to be due to the high stability of the SspCΔ11-D13K-DNA complex, which prevents rapid degradation of this α/β-type SASP early in germination. The persistence of this protein on spore DNA then interferes with transcription during spore outgrowth.
Spores of Bacillus and Clostridium species contain a number of small, acid-soluble spore proteins (SASP) which comprise 7 to 20% of total spore protein (25). One subset of these proteins, the α/β-type SASP, are encoded by multiple genes and comprise a large protein family whose amino acid sequences are very highly conserved within and between species (25). The α/β-type SASP are nonspecific DNA binding proteins which saturate the spore chromosome and protect spore DNA from damage caused by UV radiation, heat, and peroxides (2, 10, 19, 20; reviewed in references 23 and 24). In addition to the α/β-type SASP, another type of SASP, termed SASP-γ, is also found at very high levels within dormant spores. In contrast to the α/β-type SASP, the γ-type SASP are encoded by a single gene, do not bind to DNA, and also do not share extensive sequence homology with the α/β-type SASP (25). However, both types of SASP are cleaved during spore germination by the same sequence-specific endoproteinase, termed the germination protease, or GPR. This cleavage initiates the degradation of the α/β- and γ-type SASP to amino acids which support protein synthesis during this period of development (3, 16). The rapid degradation of α/β-type SASP during spore germination is essential to allow for DNA transcription and eventually DNA replication during spore outgrowth, the period between spore germination and the resumption of vegetative growth (16).
The α/β-type SASP are essentially unstructured in the absence of DNA and consequently are very sensitive to proteolysis (6, 21). However, α/β-type SASP are much more resistant to protease cleavage when bound to DNA and may be completely resistant to GPR cleavage while bound to DNA (21). Indeed, the current model of α/β-type SASP degradation during spore germination (16) suggests that the rapid rehydration and volume expansion of the spore core early in germination result in the partial dissociation of α/β-type SASP from the chromosome. The dissociated α/β-type SASP are then cleaved by GPR, and this cleavage depletes the pool of free α/β-type SASP and leads to further dissociation and further cleavage (16). One corollary of this model is that α/β-SASP should not bind to spore DNA too tightly, or their degradation during germination could be impaired and thus spore outgrowth inhibited.
While studying N-terminal deletion mutant forms of SspC, a minor α/β-type SASP from Bacillus subtilis, we generated a protein (SspCΔ11) which lacks amino acid residues Gln2 through Asn12 (Fig. 1) (4). Spores of B. subtilis that express SspCΔ11 as their major α/β-type SASP are more sensitive to UV radiation and heat than spores expressing wild-type SspC (4). SspCΔ11 also binds to pUC19 plasmid DNA with 30-fold-lower affinity and to poly(dA-dT) · poly(dA-dT) with >50-fold-lower affinity than does wild-type SspC (4). We sought to increase the affinity of SspCΔ11 for DNA by changing Asp13 to a lysine residue, thus generating SspCΔ11-D13K (Fig. 1). This additional change was chosen because α/β-type SASP with positively charged amino acid residues near the N terminus tend to bind to DNA with higher affinity (4). This effect is presumably due to previously identified protein-protein interactions that occur between adjacent DNA-bound α/β-type SASP, in which the positively charged N terminus of one α/β-type SASP interacts with the negatively charged GPR cleavage sequence of an adjacent DNA-bound α/β-type SASP (Fig. 1) (8). Based on the information noted above, we reasoned that SspCΔ11 may bind to DNA with low affinity due to unfavorable protein-protein interactions arising from electrostatic repulsion between Asp13 near the N terminus and the two glutamate residues in the GPR cleavage region (Fig. 1). The electrostatic repulsion model is also based upon data which indicate that SspCΔ14 (which lacks residues Gln2 through Leu16) binds to DNA with higher affinity than SspCΔ11 (4); note that SspCΔ14 has only uncharged residues in the N-terminal region (Fig. 1). Equilibrium binding studies indicated that as predicted, SspCΔ11-D13K has >350-fold-higher affinity for pUC19 than does SspCΔ11, and surprisingly, SspCΔ11-D13K was also found to bind to pUC19 with 12-fold-higher affinity than wild-type SspC does (4). In addition, a complex of SspCΔ11-D13K and poly(dG) · poly(dC) is remarkably more stable to thermal denaturation than an SspC-poly(dG) · poly(dC) complex (4), and the SspCΔ11-D13K-poly(dG) · poly(dC) complex only dissociates at temperatures slightly below the melting temperature of poly(dG) · poly(dC) (4).
FIG. 1.
Amino acid sequence alignment of wild-type and mutant SspC proteins. The amino acid sequences for wild-type SspC, SspCΔ11, and SspCΔ11-D13K are given in one-letter code (1, 25). Asterisks above amino acid residues indicate residues which are conserved in all α/β-type SASP identified from Bacillus, Sporosarcina, and Thermoactinomyces species (1, 25). The downward-pointing arrow indicates the peptide bond which is cleaved by GPR.
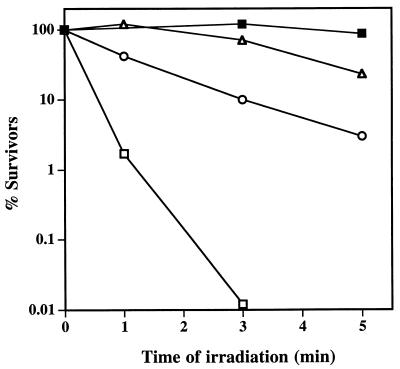
Spores which contain SspCΔ11 as their major α/β-type SASP are not as resistant to UV or heat as spores containing wild-type SspC, presumably at least in part because of the weaker binding of the variant protein to DNA compared to that of wild-type SspC (4). Although SspCΔ11-D13K binds to DNA with higher affinity than wild-type SspC in vitro, we wanted to determine if this protein is a functional α/β-type SASP in vivo. Therefore, we examined whether the D13K change increased the ability of SspCΔ11 to confer UV resistance to spores. SspC, SspCΔ11, and SspCΔ11-D13K were overexpressed to similar high levels in spores lacking the two major α/β-type SASP (termed α−β− spores) of B. subtilis (SspC, SspCΔ11, and SspCΔ11-D13K spores, respectively) as described previously (4), and the spores were purified as described previously (13). As predicted, α−β−SspCΔ11-D13K spores were more resistant to UV radiation than were α−β−SspCΔ11 spores and were almost as resistant as α−β−SspC spores (Fig. 2), indicating that the additional sequence change in SspCΔ11-D13K complements the slightly UV-sensitive phenotype of α−β−SspCΔ11 spores. However, we noted that α−β−SspCΔ11-D13K spore preparations consistently gave only 5 to 10% of the CFU per unit of optical density at 600 nm (OD600) on Luria-Bertani (LB) plates that α−β−, α−β−SspC, or α−β−SspCΔ11 spores gave. Resporulation of an α−β−SspCΔ11-D13K colony which had arisen from a spore and purification and analysis of the spores showed again that the colony-forming ability of these spores was 10- to 20-fold lower than expected. Expression of SspCΔ11-D13K in wild-type spores also conferred the same low-viability phenotype (data not shown).
FIG. 2.
Resistance of α−β− spores overexpressing SspC, SspCΔ11, and SspCΔ11-D13K to UV radiation. Spores of various strains were purified, and their resistance to UV radiation at 254 nm was determined as described earlier (14) under conditions in which the spores were exposed to 45 J of UV radiation per m2 at 254 nm for 1 min. This experiment was performed twice with independent spore preparations. The relative resistances of the strains were the same in both experiments, with average D90 values (time to kill 90% of the initial spore population) of 30 s for α−β− spores, 3 min for SspCΔ11 spores, 8 min for SspCΔ11-D13K spores, and >20 min for SspC spores. Symbols: ■, α−β−SspC; ▵, α−β−SspCΔ11-D13K; ○, α−β−SspCΔ11; and □, α−β−pUB110.
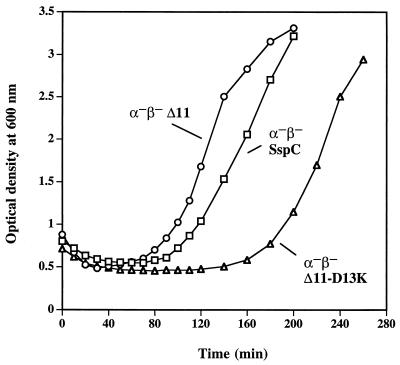
One explanation for the apparent low viability of α−β−SspCΔ11-D13K spores is that germination itself is defective in these spores. To test this explanation, α−β− spores expressing SspC, SspCΔ11, or SspCΔ11-D13K were examined for their ability to germinate after heat shock (70°C, 30 min) under a variety of conditions, including 20 mM Tris-HCl (pH 8.0)–100 mM KCl–8 mM l-alanine and rich medium (LB or 2× yeast-tryptone [YT]) (7) supplemented with 8 mM l-alanine. In general, α−β−SspCΔ11 spores germinated with greater efficiency (80 to 90%) than α−β−SspC or α−β−SspCΔ11-D13K spores (50 to 60%) as determined by the percentage of phase-dark spores present after 60 min of germination as observed by phase-contrast microscopy (data not shown). The germination efficiencies observed by microscopy were in good agreement with the percentages of total spore dipicolinic acid (DPA) released by each strain upon germination; α−β−SspCΔ11 spores released almost all their DPA, whereas α−β−SspC and α−β−SspCΔ11-D13K spores released only ∼50% of their total DPA after 1 h in 10 mM Tris-HCl (pH 8.0)–100 mM KCl–8 mM l-alanine at 37°C. The germination kinetics of α−β−SspC and α−β−SspCΔ11-D13K spores were also identical as measured by the initial decrease in OD600 after dilution into germination medium (Fig. 3 and data not shown). These data indicate that α−β−SspCΔ11 spores germinate more efficiently in liquid media than α−β−SspC and α−β−SspCΔ11-D13K spores for reasons not known; more importantly, these data also indicate that α−β−SspC and α−β−SspCΔ11-D13K spores germinate equally well, and thus that the low viability of α−β−SspCΔ11-D13K spores is not due to a germination defect. The germinated α−β−SspC and α−β−SspCΔ11-D13K spores also swell significantly as observed in a phase-contrast microscope, but the great majority of the germinated α−β−SspCΔ11-D13K spores remain round and never divide (data not shown). Consequently, the resumption of the vegetative growth of α−β−SspCΔ11-D13K spores at 37°C in 2× YT medium supplemented with 8 mM l-alanine is delayed by about 80 min compared to that of α−β−SspC spores (Fig. 3). This apparent difference in the times for the resumption of vegetative growth between α−β−SspC and α−β−SspCΔ11-D13K spores is ∼150 min when germination and outgrowth at 37°C are carried out in a slightly poorer medium, LB medium with 8 mM l-alanine (data not shown). The significant delay seen in the resumption of growth of α−β−SspCΔ11-D13K spores (Fig. 3 and data not shown) is consistent with the low viability of these spores; because only 5 to 10% of germinated α−β−SspCΔ11-D13K spores are able to successfully reinitiate vegetative growth, the time at which the OD600 of cultures of these spores can be seen to increase will be delayed relative to that for α−β−SspC spores, 10- to 20-fold more of which are viable.
FIG. 3.
Germination and outgrowth of α−β− spores overexpressing SspC, SspCΔ11, and SspCΔ11-D13K. Spores were heat shocked for 30 min at 70°C followed by resuspension at an OD600 of 0.75 to 0.9 in 2× YT medium supplemented with 8 mM l-alanine and cultured at 37°C with shaking. This experiment was performed twice with independent spore preparations, with similar results. Symbols: ○, α−β−SspCΔ11; □, α−β−SspC; and ▵, α−β−SspCΔ11-D13K.
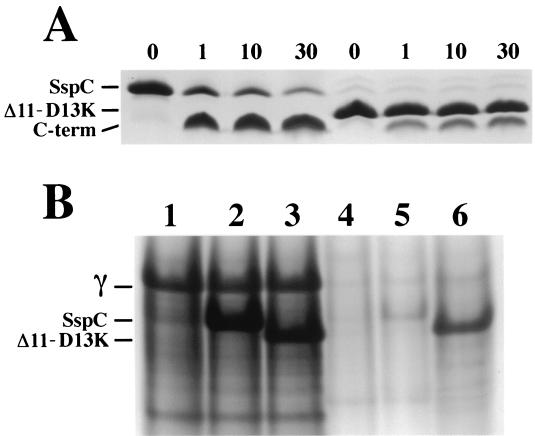
Because the SspCΔ11-D13K protein has a higher affinity for DNA than does wild-type SspC in vitro, a second possible explanation for the apparent low viability of SspCΔ11-D13K spores is that SspCΔ11-D13K is not efficiently degraded by GPR during spore germination, and thus, the germinated spore dies because it cannot reinitiate vegetative growth appropriately. Indeed, it is known that the overexpression of α/β-type SASP in Escherichia coli results in cell death most likely due to the interruption of transcription and DNA metabolism (18), and possibly the same phenomenon occurs in germinating SspCΔ11-D13K spores. Previous work has shown that DNA-bound α/β-type SASP are very resistant to digestion by GPR compared to free α/β-type SASP (21). To determine whether the increased affinity of SspCΔ11-D13K for DNA results in reduced degradation by GPR in vitro compared to that of wild-type SspC, purified protein-DNA complexes were made of each α/β-type SASP and supercoiled pUC19 plasmid DNA followed by the addition of partially purified recombinant Bacillus megaterium GPR (9). Under these conditions (10 mM Tris-HCl [pH 7.4]–150 mM NaCl–2 mM CaCl2 at 37°C), approximately 90% of wild-type SspC is digested in 30 min, while less than 75% of SspCΔ11-D13K is cleaved in the same time (Fig. 4A). That the difference in GPR cleavage of the two proteins is due to differences in their DNA binding affinity was shown by both the similar rates of cleavage and the complete cleavage (within 1 min) of both SspC and SspCΔ11-D13K by GPR in the absence of added plasmid DNA (data not shown).
FIG. 4.
Degradation of SspC and SspCΔ11-D13K in vitro and in vivo. (A) SspC and SspCΔ11-D13K (50 μM) were equilibrated with supercoiled pUC19 plasmid DNA (0.13 mg/ml) in 10 mM Tris-HCl (pH 7.4)–150 mM NaCl–2 mM CaCl2 at 37°C for 90 min prior to addition of partially purified B. megaterium GPR to 20 μg/ml. Samples were removed before (0 min) and after addition of GPR (1, 10, and 30 min) for analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on Tris-tricine gels (17). The positions of full-length proteins and the C-terminal GPR cleavage peptide (C-term) are indicated on the left of the figure, and the minutes of incubation with GPR are given above the lanes. (B) Total SASP were acid extracted from dry, ruptured dormant spores (lanes 1 through 3) and purified germinated spores (germinated for 100 min as described in the text) (lanes 4 through 6), and samples from equivalent amounts of spores and germinated spores were run on polyacrylamide gel electrophoresis at low pH (15). The samples in each lane are as follows: lane 1, α−β− dormant spores; lane 2, α−β−SspC dormant spores; lane 3, α−β−SspCΔ11-D13K dormant spores; lane 4, α−β− germinated spores; lane 5, α−β−SspC germinated spores; and lane 6, α−β−SspCΔ11-D13K germinated spores. The positions of SASP-γ (γ) SspC, and SspCΔ11-D13K are indicated on the left.
The data on the thermal stability and in vitro GPR digestion of α/β-type SASP-DNA complexes indicate that SspCΔ11-D13K has a significantly higher affinity for DNA than do SspCΔ11 and wild-type SspC, and that this tight binding to DNA can result in less than complete cleavage of SspCΔ11-D13K by GPR. If DNA-bound SspCΔ11-D13K is indeed preventing spore outgrowth in α−β−SspCΔ11-D13K spores, then significant amounts of uncleaved SspCΔ11-D13K protein should be present in these germinated spores. To test this prediction, purified α−β−, α−β−SspC, and α−β−SspCΔ11-D13K spores were germinated at 37°C in LB medium supplemented with 8 mM l-alanine for 100 min; this time was chosen to give maximum spore germination without significant spore outgrowth. Because α−β− spores overexpressing wild-type SspC and SspCΔ11-D13K germinate less efficiently than α−β− spores, germinated spores were purified from ungerminated spores by centrifugation through a solution of 50% metrizoic acid as described earlier (13), and total SASP were extracted from disrupted dormant spores and purified germinated spores as described previously (5). Proteins from equivalent amounts of dormant spores and germinated spores (based on the OD600 and taking into account the 50% decrease in spore OD600 upon germination) were resolved by polyacrylamide gel electrophoresis at low pH (Fig. 4B). Dormant spores of all three strains had roughly the same levels of SASP-γ, and 100 min after initiating spore germination virtually all the SASP-γ had been degraded in all three strains (Fig. 4B). The α−β−SspC and α−β−SspCΔ11-D13K dormant spores also contained the same level of these α/β-type SASP, and the great majority of wild-type SspC (and SspCΔ11; data not shown) was degraded during germination (Fig. 4B, lanes 2, 3, and 5). In contrast, substantial amounts of SspCΔ11-D13K (∼30 to 40%) remained long after germination had initiated (Fig. 4B, lane 6). Presumably the SspCΔ11-D13K remaining in germinated spores is bound to DNA and therefore is resistant to GPR as well as degradation by other proteases (see below). Purified germinated spores were also plated on LB agar with kanamycin to determine plating efficiency, and these spores again gave only ∼8% of the colonies per unit of OD600 of germinated spores as compared to α−β− or α−β−SspC germinated spores (data not shown).
The continued presence of significant levels of SspCΔ11-D13K in germinated spores expressing this protein and this protein's high affinity for DNA in vitro strongly suggests that outgrowth of these spores is inhibited due to the persistence of this protein on germinated spore DNA, although some of these germinated spores (5 to 10%) are able to degrade SspCΔ11-D13K sufficiently to allow vegetative growth to resume. However, successful degradation of SspCΔ11-D13K and resumption of vegetative growth appears to be a stochastic event, because resporulated clones of SspCΔ11-D13K spore survivors still show the low-spore-viability phenotype.
In contrast to the decrease in spore viability due to undegraded SspCΔ11-D13K, slowing degradation of wild-type α/β-type SASP during spore germination by inactivation of GPR causes no noticeable decrease in spore viability (16). While the reason(s) for this difference between the effects of undegraded SspCΔ11-D13K and wild-type α/β-type SASP on spore viability is not clear, there are at least two possible explanations, which are not mutually exclusive. First, wild-type α/β-type SASP bind relatively weakly to DNA; in fact, the major α/β-type SASP of B. subtilis (α and β) have a much lower affinity than does SspC for DNA (11, 22). Consequently, even though 50% of the genome remains complexed with α/β-type SASP early in germination of gpr spores (16), the proteins are likely rapidly dissociating and reassociating with the DNA such that essentially all regions of the genome are available for transcription at least some of the time. In contrast, the much tighter binding of SspCΔ11-D13K may keep some regions of the germinated spore genome constantly covered with protein, thus precluding their transcription and resulting in spore death. This may especially be the case if some regions of the genome must be transcribed at an appropriate time relative to other regions to ensure an orderly progression through spore outgrowth. A second possible explanation is based on the fact that while degradation of wild-type α/β-type SASP is slowed during germination of gpr spores, this degradation still takes place, presumably due to other nonspecific proteases (16); indeed, degradation of SASP-α and -β is largely complete after 90 min of germination of gpr spores in a rich medium (16). In contrast, SspCΔ11-D13K persists in wild-type spores germinated for >100 min (Fig. 4B, lane 6). Since the viability of α−β−SspCΔ11-D13K spores is not further reduced by introduction of a gpr mutation (data not shown), presumably SspCΔ11-D13K bound to DNA is also resistant to other nonspecific proteases that can degrade α/β-type SASP. Consequently, SspCΔ11-D13K persists much longer in germinated spores than do wild-type α/β-type SASP in germinated gpr spores, and thus SspCΔ11-D13K causes significant spore death.
A major conclusion drawn from the data in this communication is that to be effective, α/β-type SASP must bind to DNA tightly enough to effect the global change in chromosome DNA conformation required for spore resistance to UV radiation and other damaging treatments (10–12) but not so tightly that dissociation of α/β-type SASP from all parts of the spore chromosome does not occur efficiently and rapidly during spore germination. Interestingly, many of the minor α/β-type SASP (such as SspC) studied to date have higher affinity for DNA than do the major α/β-type SASP which comprise ∼75 to 85% of the total α/β-type SASP pool (6, 22). It may be that the low-affinity major α/β-type SASP are sufficient to bind to most of the chromosome in spores, but that certain regions of the chromosome may require higher-affinity minor α/β-type SASP for efficient binding. This might explain why spores contain several different α/β-type SASP. In this manner, the spore chromosome could be saturated with these proteins without the need for an excessive amount of high-affinity α/β-type SASP, as the latter proteins could persist after initiation of spore germination and significantly retard or even prevent spore outgrowth.
Acknowledgments
This work was supported by National Institutes of Health grant GM19698.
REFERENCES
- 1.Driks A, Setlow P. Morphogenesis and properties of the bacterial spore. In: Brun Y V, Shimkets L J, editors. Prokaryotic development. Washington, D.C.: American Society for Microbiology; 1999. pp. 191–218. [Google Scholar]
- 2.Fairhead H, Setlow B, Setlow P. Prevention of DNA damage in spores and in vitro by small, acid-soluble proteins from Bacillus species. J Bacteriol. 1993;175:1367–1374. doi: 10.1128/jb.175.5.1367-1374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hackett R H, Setlow P. Properties of spores of Bacillus subtilis strains which lack the major small, acid-soluble protein. J Bacteriol. 1988;170:1403–1404. doi: 10.1128/jb.170.3.1403-1404.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayes C S, Hernandez-Alarcon E, Setlow P. N-terminal amino acid residues mediate protein-protein interactions between DNA-bound α/β-type small, acid-soluble spore proteins from Bacillus species. J Biol Chem. 2001;276:2267–2275. doi: 10.1074/jbc.M007858200. [DOI] [PubMed] [Google Scholar]
- 5.Hayes C S, Illades-Aguiar B, Casillas-Martinez L, Setlow P. In vitro and in vivo oxidation of methionine residues in small, acid-soluble spore proteins from Bacillus species. J Bacteriol. 1998;180:2694–2700. doi: 10.1128/jb.180.10.2694-2700.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayes C S, Peng Z-Y, Setlow P. Equilibrium and kinetic interactions between DNA and a group of novel, non-specific DNA binding proteins from spores of Bacillus and Clostridium species. J Biol Chem. 2000;275:35040–35050. doi: 10.1074/jbc.M005669200. [DOI] [PubMed] [Google Scholar]
- 7.Hayes C S, Setlow P. Analysis of deamidation of small, acid-soluble spore proteins from Bacillus subtilis in vitro and in vivo. J Bacteriol. 1997;179:6020–6027. doi: 10.1128/jb.179.19.6020-6027.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayes C S, Setlow P. Identification of protein-protein contacts between α/β-type small, acid-soluble spore proteins of Bacillus species bound to DNA. J Biol Chem. 1998;273:17326–17332. doi: 10.1074/jbc.273.28.17326. [DOI] [PubMed] [Google Scholar]
- 9.Illades-Aguiar B, Setlow P. Studies of the processing of the protease which initiates degradation of small, acid-soluble proteins during germination of spores of Bacillus species. J Bacteriol. 1994;176:2788–2795. doi: 10.1128/jb.176.10.2788-2795.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mason J M, Setlow P. Essential role of small, acid-soluble spore proteins in resistance of Bacillus subtilis spores to UV light. J Bacteriol. 1986;167:174–178. doi: 10.1128/jb.167.1.174-178.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohr S C, Sokolov N V H A, He C, Setlow P. Binding of small acid-soluble spore proteins from Bacillus subtilis changes the conformation of DNA from B to A. Proc Natl Acad Sci USA. 1991;88:77–81. doi: 10.1073/pnas.88.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholson W L, Setlow B, Setlow P. Ultraviolet irradiation of DNA complexed with α/β-type small, acid-soluble proteins from spores of Bacillus or Clostridium species makes spore photoproduct but not thymine dimers. Proc Natl Acad Sci USA. 1991;88:8288–8292. doi: 10.1073/pnas.88.19.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholson W L, Setlow P. Sporulation, germination, and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons Ltd.; 1990. pp. 391–450. [Google Scholar]
- 14.Popham D L, Sengupta S, Setlow P. Heat, hydrogen peroxide, and UV resistance of Bacillus subtilis spores with increased core water content and with or without major DNA-binding proteins. Appl Environ Microbiol. 1995;61:3633–3638. doi: 10.1128/aem.61.10.3633-3638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reisfield R A, Lewis V J, Williams D E. Disk electrophoresis of basic proteins and peptides on polyacrylamide gels. Nature. 1962;195:281–283. doi: 10.1038/195281a0. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez-Salas J-L, Santiago-Lara M L, Setlow B, Sussman M D, Setlow P. Properties of Bacillus megaterium and Bacillus subtilis mutants which lack the protease that degrades small, acid-soluble proteins during spore germination. J Bacteriol. 1992;174:807–814. doi: 10.1128/jb.174.3.807-814.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 18.Setlow B, Hand A R, Setlow P. Synthesis of a Bacillus subtilis small, acid-soluble spore protein in Escherichia coli causes DNA to assume some characteristics of spore DNA. J Bacteriol. 1991;173:1642–1653. doi: 10.1128/jb.173.5.1642-1653.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Setlow B, Setlow C A, Setlow P. Killing bacterial spores by organic hydroperoxides. J Ind Microbiol. 1997;18:384–388. [Google Scholar]
- 20.Setlow B, Setlow P. Binding of small, acid-soluble spore proteins to DNA plays a significant role in the resistance of Bacillus subtilis spores to hydrogen peroxide. Appl Environ Microbiol. 1993;59:3418–3423. doi: 10.1128/aem.59.10.3418-3423.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Setlow B, Setlow P. Binding to DNA protects α/β-type small, acid-soluble spore proteins of Bacillus and Clostridium species against digestion by their specific protease as well as other proteases. J Bacteriol. 1995;177:4149–4151. doi: 10.1128/jb.177.14.4149-4151.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Setlow B, Sun D, Setlow P. Interaction between DNA and α/β-type small, acid-soluble spore proteins: a new class of DNA-binding protein. J Bacteriol. 1992;174:2312–2322. doi: 10.1128/jb.174.7.2312-2322.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Setlow P. Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu Rev Microbiol. 1995;49:29–54. doi: 10.1146/annurev.mi.49.100195.000333. [DOI] [PubMed] [Google Scholar]
- 24.Setlow P. Mechanisms which contribute to the long-term survival of spores of Bacillus species. J Appl Bacteriol Symp Suppl. 1994;76:49S–60S. doi: 10.1111/j.1365-2672.1994.tb04357.x. [DOI] [PubMed] [Google Scholar]
- 25.Setlow P. Small, acid-soluble spore proteins of Bacillus species: structure, synthesis, genetics, function and degradation. Annu Rev Microbiol. 1988;42:319–338. doi: 10.1146/annurev.mi.42.100188.001535. [DOI] [PubMed] [Google Scholar]