Abstract
Oxygen starvation triggers the shiftdown of the obligate aerobe Mycobacterium bovis BCG to a state of dormancy. Two-dimensional electrophoresis showed a drastic up-regulation of the α-crystallin homolog, the putative response regulator Rv3133c, and the two conserved hypothetical proteins Rv2623 and Rv2626c in dormant bacilli.
Mycobacteria are obligate aerobes; i.e., they require oxygen for growth. However, it has been known for years that tubercle bacilli encounter hypoxic environments in acute disease, as well as in latent infection (32, 34, 39, 40, 45), and the capability of tubercle bacilli to adapt to hypoxic conditions appears to play a role in vivo (3, 29, 49). Wayne established a link between oxygen starvation and drug resistance. That author demonstrated that upon depletion of oxygen in culture, the bacillus terminates growth and develops into a defined dormant form (41–43, 47). Importantly, the dormant form of the bacterium was found to be resistant against conventional antimycobacterials (46, 47). Hence, hypoxic dormant bacteria could, at least in part, be responsible for the observed persistence of the pathogen during chemotherapy (11).
To study the dormancy response in tubercle bacilli, we applied the dormancy culture system that was developed by Wayne for Mycobacterium tuberculosis (47) to the attenuated BCG Pasteur ATCC 35734 strain of M. bovis (19, 20, 24, 31). Wayne's dormancy culture system is based on growth of the bacilli under oxygen-limited conditions in sealed tubes with stirring. Initially, the cultures grow exponentially and consume oxygen rapidly. A temporal oxygen gradient is generated, and the cultures terminate growth when the oxygen concentration reaches a hypoxic threshold level. The bacilli in the hypoxic stationary phase are in a reversible, apparently diploid, synchronized state of low metabolic activity, in which the cells maintain viability for extended periods without division (24, 47); i.e., the organisms are in a physiological state of dormancy, as defined by Kaprelyants et al. (22).
Our knowledge of the molecules involved in the mycobacterial dormancy response is fragmentary (5, 15, 17, 18, 26, 30, 37). Elegant biochemical work showed that dormant bacilli adapt their metabolism to anaerobiosis by switching to nitrate respiration (48) and reductive amination of glyoxylate (44). Genetic analysis demonstrated that the stringent response plays a crucial role in the adaptation to hypoxic stationary-phase survival (33). However, only a single polypeptide, the α-crystallin homolog, has been shown to be up-regulated in oxygen-starved tubercle bacilli (7, 38, 51, 52). The induction of this chaperon appears to be under control of the sigma factor SigF (4, 9, 10, 25). In this study, the dormancy response in BCG was analyzed using two-dimensional gel electrophoresis (21) to identify new dormancy-induced proteins.
Detection and identification of dormancy-induced proteins.
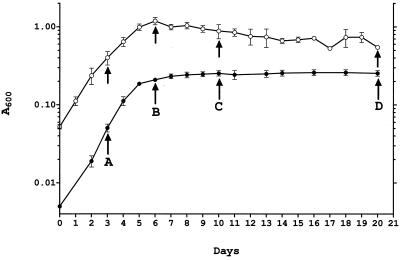
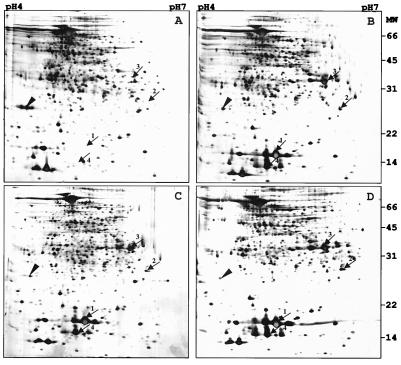
Figure 1 (solid circles) shows the growth curve of BCG in the Wayne dormancy culture system. Bacilli were harvested at various time points (Fig. 1, arrows A to D); washed twice in phosphate-buffered saline; resuspended in lysis buffer containing 9 M urea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 50 mM dithiothreitol, pefabloc at 1 mg ml−1, pepstatin at 1 μg ml−1, and leupeptin at 1 μg ml−1; and disrupted with 0.5-mm glass beads using a Mini Bead Beater (Biospec). Protein concentrations were determined using the Bio-Rad protein assay reagents and protocols. A 100-μg sample of total protein was subjected to isoelectric focusing using pH 4 to 7 Immobiline Dry Strips and an IPGphor isoelectric focusing unit as recommended by the manufacturer (Amersham Pharmacia) for 62,000 V-h. For separation in the second dimension, sodium dodecyl sulfate–12.5% polyacrylamide gels were used (Protean IIxi system; Bio-Rad) and proteins were detected by silver staining. Figure 2 shows a representative set of two-dimensional gels. Four proteins showed a drastic increase in their steady-state level in the hypoxic stationary phase (Fig. 2, arrows 1 to 4). The proteins were not (arrows 1, 2, and 4) or weakly (arrow 3) detectable in the extracts from exponentially growing cultures (Fig. 2A) and appeared as major spots immediately upon oxygen starvation-induced termination of growth (Fig. 2B). Elevated levels of the proteins were maintained throughout the hypoxic stationary phase (Fig. 2C and D). To identify the dormancy-induced proteins, the excised gel spots were subjected to in-gel digestion with trypsin to recover the peptides (36). Peptide sequence tags were generated from selected peptides by nanoelectrospray tandem mass spectrometry using a quadrupole–time-of-flight hybrid instrument (QSTAR; PE Sciex). Protein identity was revealed by searching sequence databases with a combination of the peptide sequence tags and the mass information (27, 50). Protein 1 was the α-crystallin homolog (Rv2031c) previously reported to be induced in oxygen-starved cultures of tubercle bacilli (51). Protein 2 was the 23-kDa putative response regulator Rv3133c. Proteins 3 and 4 were the 32-kDa conserved hypothetical protein Rv2623 and the 16-kDa conserved hypothetical protein Rv2626c, respectively. (Protein names and Rv numbers are according to the M. tuberculosis H37Rv genome annotation [6].)
FIG. 1.
Growth of BCG in the Wayne dormancy culture system and in aerated roller bottles. Solid circles show the growth curve of BCG under the oxygen-limited conditions of the Wayne dormancy culture system. Aerobic exponential precultures were diluted to an A600 of 0.005 with Dubos Tween-albumin broth (Difco) and grown in sealed tubes (2 by 12.5 cm; 17-ml culture volume) under gentle stirring at 170 rpm as described previously (12, 24). Anaerobiosis in the stationary phase was monitored using the redox indicator methylene blue (24). Empty circles show the growth curve of BCG under non-oxygen-limited conditions in roller bottles. Precultures were diluted to an A600 of 0.05 and grown in roller bottles (10 by 14 cm; 100-ml culture volume). Cultures were aerated in a roller apparatus at 1 rpm. The roller bottles were opened daily for turbidity measurements and to allow exchange of the air. Mean values and standard deviations from four independent experiments are shown. Arrows indicate the time points when samples were taken for protein and RNA analyses.
FIG. 2.
Temporal profile of protein contents of BCG grown in the Wayne dormancy culture system. Protein extracts were prepared at the time points (A to D) indicated by the arrows in Fig. 1. Samples (100 μg) of total protein were prepared from ≈ 100 ml of culture at time point A and ≈ 35 to 50 ml of culture at time points B to D and were subjected to two-dimensional electrophoresis. Panels A to D show silver-stained gels corresponding to the four time points. Arrows 1 to 4 indicate dormancy-induced protein spots. The arrowhead shows the 22-kDa alkyl hydroperoxide reductase (AhpC, Rv2428; 6, 35) which was found to be down-regulated in the hypoxic stationary phase. Molecular masses (MW) are indicated in kilodaltons. The experiment was carried out four times with the same results. Growth phase-dependent protein contents were also analyzed using isoelectric focusing strips at pHs 3 to 10. No additional dormancy-induced proteins were detected.
Transcript levels of dormancy-induced proteins.
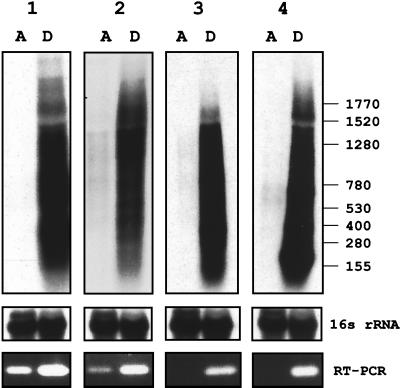
To determine whether the increase in the steady-state level of the dormancy-induced proteins correlates with an increase in the steady-state level of their mRNAs, total RNA was isolated from exponentially growing and hypoxic stationary-phase cultures and subjected to Northern blot analysis as previously described (19). Probes were isolated by PCR using BCG genomic DNA as the template. The primers were derived from the M. tuberculosis H37Rv genome sequence (6) and were as follows: 1, α-crystallin homolog (GCCACCACCCTTCCCGTTCAG [nucleotides 4 to 24] and ATGTCGTCCTCGTCAGCACCTACC [nucleotides 315 to 338]); 2, Rv3133c (TCGTAGGTGTAGGCGGGTTC [nucleotides 86 to 104] and CGGCGATCTGCTTGTTGGT [nucleotides 496 to 514]); 3, Rv2623 (GGCAGCCGTTCCCACATTG [nucleotides 294 to 312] and GGCTGATCGCGCACCACCAC [nucleotides 715 to 734]); 4, Rv2626c (CCACCGCACGCGACATCAT [nucleotides 5 to 23] and CGGAACACGGCGGACCTG [nucleotides 292 to 309]); 5, 16S rRNA (GCCTGGGAAACTGGGTCTAA [nucleotides 149 to 168] and TCTCCACCTACCGTCAATCC [nucleotides 467 to 486]). (The numbers in brackets specify the primer positions in the coding sequences of the respective genes.) The identities of the PCR fragments were confirmed by sequencing using a Perkin-Elmer ABI Prism 377 automated sequencer. Figure 3 shows high levels of the transcripts for all four proteins in dormant bacilli. In exponentially growing cultures, the transcripts were not or only weakly detectable. The dormancy-dependent increase in the mRNA levels was confirmed by reverse transcription-PCR analysis (Fig. 3). The primers used were the same as the primers employed to generate the probes for the Northern analysis. Reactions were performed using an Omniscript RT kit and a Taq polymerase kit (Qiagen) as recommended by the supplier. PCR amplification was carried out for 25 cycles. Taken together, these results show that the mRNA levels correlate with the protein levels and indicate that the level of the dormancy-induced proteins is regulated at the transcriptional level.
FIG. 3.
Steady-state levels of mRNAs encoding dormancy-induced proteins in growing and dormant cultures. Autoradiograms of Northern blots of total RNA from exponentially growing (lanes A, from time point A in Fig. 1) and hypoxic stationary-phase cultures (lanes D, from time point D in Fig. 1) grown in the Wayne dormancy culture system are shown. The blots were hybridized with [α-32P]dATP-labeled probes specific to the transcripts encoding the dormancy-induced proteins: 1, α-crystallin homolog; 2, 23-kDa response regulator; 3, 32-kDa conserved hypothetical protein; 4, 16-kDa conserved hypothetical protein. X-ray films were exposed for 1 day. Sizes are indicated in bases. 16S rRNA shows the blots after rehybridization with a probe specific to 16S rRNA, demonstrating equal loading of rRNA. Five micrograms of total RNA (prepared from ≈20 ml of culture at time point A and from ≈35 ml of culture at time point D) was loaded. At the bottom, the agarose gel electrophoretic analysis of reverse transcriptase (RT) PCRs carried out with primers specific to the dormancy-induced genes and 150 ng of total RNA is shown. Carrying out the identical reactions but without the reverse transcription step did not yield any bands, demonstrating that the bands generated by reverse transcriptase PCR were derived from RNA and not from contaminating genomic DNA. A control PCR with genomic DNA yielded bands of the same size observed for the reverse transcriptase PCRs. Reverse transcriptase PCRs with primers specific to 16S rRNA yielded bands of the same intensity independent of the RNA sample; thus confirming equal loading of total RNA into the reverse transcriptase reactions (data not shown). All experiments were repeated once, yielding the same results. The values on the right are numbers of nucleotides.
It should be noted that the up-regulation of the steady-state level of the α-crystallin homolog mRNA in an oxygen-starved dormant culture is in apparent contradiction to the results obtained by Hu and Coates (16). Those authors observed a down-regulation of the transcript in nonagitated cultures of the tubercle bacillus. This discrepancy might be due to the different culture system used by those investigators (14).
Analysis of dormancy-induced proteins in aerated stationary-phase cultures.
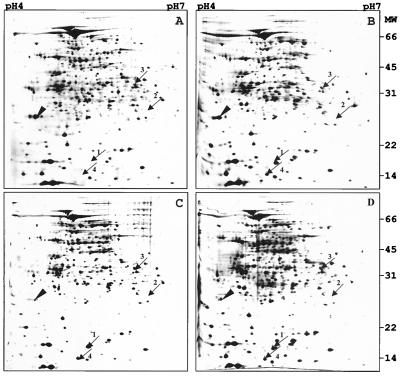
Next, we determined whether the induction of the four proteins is specific to the hypoxic stationary-phase cultures generated in the Wayne dormancy culture model, as opposed to aerated stationary-phase cultures. Figure 1 (empty circles) shows the growth curve of BCG in agitated roller bottles. Bacilli were harvested at various time points (Fig. 1, arrows A to D), and 100 μg of total protein was subjected to two-dimensional gel electrophoresis as described above. Figure 4 shows a representative set of gels. The α-crystallin homolog and the 16-kDa conserved hypothetical protein were found to be induced in aerated stationary-phase cultures. The two proteins were not detectable in exponentially growing roller bottle cultures and appeared as major spots immediately upon termination of growth (Fig. 4, arrows 1 and 4). Thus, induction of the α-crystallin homolog and the 16-kDa conserved hypothetical protein was not specific to the hypoxic dormant culture. In contrast, the 23-kDa response regulator was not detectable in aerated stationary-phase cultures (Fig. 4, arrow 2) and the 32-kDa hypothetical protein maintained a level that was similar to its level in an exponentially growing culture (Fig. 4, arrow 3). Thus, the induction of the 23-kDa response regulator and the 32-kDa hypothetical protein appears to be specific to the dormancy response. These results are consistent with a previous analysis of the protein contents of aerated stationary-phase cultures employing two-dimensional gel electrophoresis carried out by Barry and coworkers (51). Those authors found one protein, the α-crystallin homolog, to be strongly up-regulated in stationary-phase cultures of the tubercle bacillus grown in roller bottles. However, up-regulation of the16-kDa conserved hypothetical protein was not reported. This might be due to the parameters used by the investigators for separation of the proteins in the second dimension. The α-crystallin homolog migrated at the bottom of their gels. Our studies show that the 16-kDa conserved hypothetical protein migrates below the α-crystallin homolog (Fig. 4, arrows 1 and 4). Hence, the 16-kDa conserved hypothetical protein might have migrated out of the gel under the running conditions employed by those authors.
FIG. 4.
Steady-state levels of dormancy-induced proteins in aerated roller bottle cultures. Protein extracts were prepared at the time points (A to D) indicated by the arrows in Fig. 1. Samples of total protein (100 μg) were prepared from ≈25 ml of culture at time point A and ≈10 to 15 ml of culture at time points B to D and were subjected to two-dimensional electrophoresis. Panels A to D show silver-stained gels corresponding to the four time points. Arrows 1 to 4 indicate the migration areas of the dormancy-induced protein spots shown in Fig. 2. The arrowhead shows the 22-kDa alkyl hydroperoxide reductase found to be down-regulated in the aerated stationary phase. Molecular masses (MW) are indicated in kilodaltons. The experiment was carried out four times with the same results.
Conclusions.
In the present work, we identified four proteins that were markedly up-regulated in hypoxic stationary-phase cultures of BCG generated in the Wayne dormancy model. The increase in the steady-state level of the dormancy-induced proteins correlated with an increase in the steady-state level of their transcripts, suggesting transcriptional control of gene expression. To determine whether up-regulation of the four proteins was specific to the oxygen depletion-induced stationary phase, the protein contents of aerated stationary-phase cultures were analyzed. The α-crystallin homolog and the 16-kDa conserved hypothetical protein were found to be induced in aerated stationary-phase cultures. This indicates that induction of the two proteins is not specific to dormant bacilli. In contrast, the 23-kDa putative response regulator was not detectable in aerated stationary-phase cultures. The 32-kDa conserved hypothetical protein was found to maintain a level that was similar to its level observed in exponentially growing culture. This suggests that the expression of the two molecules is not up-regulated upon termination of growth per se. Rather, low oxygen tension appears to be required to increase the level of these proteins. To determine whether hypoxic conditions alone (without concurrent termination of growth) are sufficient to induce the level of the 23-kDa putative response regulator and the 32-kDa conserved hypothetical protein, contents of cultures grown under mild hypoxic conditions that allow ongoing replication have to be analyzed.
The up-regulation of the four proteins in dormant bacilli suggests that they play roles in development of the dormant state and/or maintenance of viability during dormancy. What could their functions be? The α-crystallin homolog belongs to the 20-kDa small heat shock protein family and was shown to possess a chaperon function ascribed to other members of the family. Hence, the α-crystallin homolog could play a role in enhancing long-term protein stability and, therefore, long-term survival of bacilli (51). The three new dormancy-induced proteins were predicted by the M. tuberculosis H37Rv genome project (6). However, biochemical or genetic data for these proteins are not available. Two of the three new dormancy-induced proteins are annotated as “conserved hypothetical proteins” (6). To predict the domain architecture of these proteins, their sequence (derived from M. tuberculosis H37Rv genome database TubercuList [Institut Pasteur, Paris, France; http://genolist.pasteur.fr/TubercuList/] data release R2) was searched against the protein domain families database Pfam (Sanger Centre [http://www.sanger.ac.uk/Software/Pfam/]; version 5.5; 2). The similarity search suggested that the 32-kDa conserved hypothetical protein contains two universal stress protein domains (accession number PF00582). This domain is found in the UspA protein in Escherichia coli. UspA is up-regulated in stationary-phase cultures and plays a role in the survival of growth-arrested cells (13). Thus, it is tempting to speculate that the 32-kDa mycobacterial protein plays a role in survival during dormancy in tubercle bacilli. However, it is important to note that the 32-kDa protein is different from UspA in that it is twice the size and the universal stress protein domain is repeated twice within the molecule. The 16-kDa conserved hypothetical protein was predicted to contain two cystathionine beta synthase (CBS) domains (accession number PF00571). This recently discovered domain is named after the enzyme where it was originally identified (1). The domain is usually present as a pair, and this CBS domain dimer associates to form a single compact structure. Pairs of CBS domains are found in a large number of functionally diverse proteins, such as inosine-monophosphate dehydrogenases and chloride channels (1). Although the role of the CBS domain in these proteins is unclear, it may be involved in protein-protein interaction and protein regulation. In contrast to these proteins that contain a pair of CBS domains in the context of other, unrelated domains, the 16-kDa conserved hypothetical mycobacterial protein appears to consist of only a pair of CBS domains not fused to other domains.
Most intriguing is the apparently dormancy-specific up-regulation of the 23-kDa putative response regulator. This molecule contains a helix-turn-helix DNA binding domain of the LuxR family and is thus likely to act as a phosphorylation-dependent transcription factor (6). Response regulators, together with their respective sensor histidine protein kinase, are part of two-component signal transduction systems and play key roles in a variety of developmental and adaptive processes in bacteria (23). Thus, it is conceivable that the 23-kDa response regulator plays a role in the control of the mycobacterial dormancy response. Inspection of the genomic locus surrounding the gene encoding the 23-kDa response regulator in M. tuberculosis H37Rv showed that the gene appears to form an operon with the gene for Rv3132c, which is a putative sensor histidine protein kinase (6). A recent transcriptional analysis of the gene locus suggests that the two genes are indeed cotranscribed (8). The genomic organization in BCG is the same (C.B. and T.D., unpublished data), which could indicate that this putative kinase represents the sensor involved in the control of the activity of the dormancy-induced 23-kDa response regulator. Based on the likely cotranscription of the two genes, one might expect dormancy-dependent coinduction of the two proteins. However, inspection of the predicted migration area of the histidine kinase based on its theoretical isoelectric point (pH 4.8) and molecular mass (62 kDa) (6) did not reveal any dormancy-dependent protein spot (Fig. 2). Whether the failure to detect up-regulation of the histidine kinase on two-dimensional gels might be due to masking of the kinase by other proteins remains to be elucidated by the employment of higher-resolution two-dimensional gels or kinase-specific antibodies.
Two-component systems work by phosphorylation of the response regulator. Why is it, then, that the steady-state level of the 23-kDa response regulator is increased upon entry into dormancy? This phenomenon is found in numerous two-component systems. Conditions that lead to elevated phosphorylation levels of the response regulator cause a concomitant increase in its expression. One example is Spo0A, the master regulator of sporulation in Bacillus subtilis (28). The response regulator mediates its own up-regulation when the environment signals initiation of this developmental process. It appears that the high phosphorylated response regulator levels needed to initiate the developmental process are not present until a threshold level of the stimulus signals autoactivation and thus amplification of the signal transduction apparatus. In this manner, autoactivation ensures that the program is not initiated inappropriately and allows a rapid response once the appropriate conditions prevail. Whether positive autoregulation is also the mechanism underlying the dormancy-dependent up-regulation of the 23-kDa response regulator remains to be elucidated. Genetic analyses are under way to determine the functional roles of the response regulator, its candidate histidine kinase, and the two conserved hypothetical proteins in the dormancy response of tubercle bacilli.
Acknowledgments
We thank Alice Tay, Institute of Molecular and Cell Biology (IMCB) Genome Analysis and Sequencing Laboratory: for the DNA sequence analysis. We thank Bernadette Murugasu-Oei and Amanda Lim for comments on the manuscript.
This study was supported by the IMCB.
REFERENCES
- 1.Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci. 1997;22:12–13. doi: 10.1016/s0968-0004(96)30046-7. [DOI] [PubMed] [Google Scholar]
- 2.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonnhammer E L L. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishai W R. Lipid lunch for a persistent pathogen. Nature. 2000;406:683–685. doi: 10.1038/35021159. [DOI] [PubMed] [Google Scholar]
- 4.Chen P, Ruiz R E, Li Q, Silver R F, Bishai W R. Construction and characterization of a Mycobacterium tuberculosis mutant lacking the alternate sigma factor gene, sigF. Infect Immun. 2000;68:5575–5580. doi: 10.1128/iai.68.10.5575-5580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen P, Gomez J, Bishai W R. Mycobacterial transcription regulation in stationary phase. In: Hatful G F, Jacobs W R Jr, editors. Molecular genetics of mycobacteria. Washington, D.C.: American Society for Microbiology; 2000. pp. 149–156. [Google Scholar]
- 6.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton J, Squares S, Squares R, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham A F, Spreadbury C L. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton α-crystallin homolog. J Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dasgupta N, Kapur V, Singh K K, Das T K, Sachdeva S, Jyothisri K, Tyagi J S. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber Lung Dis. 2000;803:141–159. doi: 10.1054/tuld.2000.0240. [DOI] [PubMed] [Google Scholar]
- 9.DeMaio J, Zhang Y, Ko C, Young D B, Bishai W R. A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1996;93:2790–2794. doi: 10.1073/pnas.93.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeMaio J, Zhang Y, Ko C, Bishai W R. Mycobacterium tuberculosis sigF is part of a gene cluster with similarities to the Bacillus subtilis sigF and sigB operons. Tuber Lung Dis. 1997;78:3–12. doi: 10.1016/s0962-8479(97)90010-1. [DOI] [PubMed] [Google Scholar]
- 11.Dick T. Dormant tubercle bacilli: the key to more effective TB chemotherapy? J Antimicrob Chemother. 2001;47:117–118. doi: 10.1093/jac/47.1.117. [DOI] [PubMed] [Google Scholar]
- 12.Dick T, Lee B H, Murugasu-Oei B. Oxygen depletion induced dormancy in Mycobacterium smegmatis. FEMS Microbiol Lett. 1998;163:159–164. doi: 10.1111/j.1574-6968.1998.tb13040.x. [DOI] [PubMed] [Google Scholar]
- 13.Diez A, Gustavsson N, Nystrom T. The universal stress protein A of Escherichia coli is required for resistance to DNA damaging agents and is regulated by a RecA/FtsK-dependent regulatory pathway. Mol Microbiol. 2000;36:1494–1503. doi: 10.1046/j.1365-2958.2000.01979.x. [DOI] [PubMed] [Google Scholar]
- 14.Hu Y M, Butcher P D, Sole K, Mitchison D A, Coates A R M. Protein synthesis is shutdown in dormant Mycobacterium tuberculosis and is reversed by oxygen or heat shock. FEMS Microbiol Lett. 1998;158:139–145. doi: 10.1111/j.1574-6968.1998.tb12813.x. [DOI] [PubMed] [Google Scholar]
- 15.Hu Y, Coates A R M. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J Bacteriol. 1999;181:469–476. doi: 10.1128/jb.181.2.469-476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Y, Coates A R M. Transcription of the stationary-phase-associated hspX gene of Mycobacterium tuberculosis is inversely related to synthesis of the 16-kilodalton protein. J Bacteriol. 1999;181:1380–1387. doi: 10.1128/jb.181.5.1380-1387.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y, Butcher P D, Mangan J A, Rajandream M A, Coates A R M. Regulation of hmp gene transcription in Mycobacterium tuberculosis: effects of oxygen limitation and nitrosative and oxidative stress. J Bacteriol. 1999;181:3486–3493. doi: 10.1128/jb.181.11.3486-3493.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Y, Mangan J A, Dhillon J, Sole K M, Mitchison D A, Butcher P D, Coates A R. Detection of mRNA transcripts and active transcription in persistent Mycobacterium tuberculosis induced by exposure to rifampin or pyrazinamide. J Bacteriol. 2000;182:6358–6365. doi: 10.1128/jb.182.22.6358-6365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutter B, Dick T. Upregulation of narX, encoding a putative ‘fused nitrate reductase’ in anaerobic dormant Mycobacterium bovis BCG. FEMS Microbiol Lett. 1999;178:63–69. doi: 10.1111/j.1574-6968.1999.tb13760.x. [DOI] [PubMed] [Google Scholar]
- 20.Hutter B, Dick T. Analysis of the dormancy-inducible narK2 promoter in Mycobacterium bovis BCG. FEMS Microbiol Lett. 2000;188:141–146. doi: 10.1111/j.1574-6968.2000.tb09185.x. [DOI] [PubMed] [Google Scholar]
- 21.Jungblut P R, Schaible U E, Mollenkopf H-J, Zimny-Arndt U, Raupach B, Mattow J, Halada P, Lamer S, Hagens K, Kaufman S H E. Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol Microbiol. 1999;33:1103–1117. doi: 10.1046/j.1365-2958.1999.01549.x. [DOI] [PubMed] [Google Scholar]
- 22.Kaprelyants A S, Gottschal J C, Kell D B. Dormancy in non-sporulating bacteria. FEMS Microbiol Rev. 1993;104:271–286. doi: 10.1111/j.1574-6968.1993.tb05871.x. [DOI] [PubMed] [Google Scholar]
- 23.Lengeler J W, Postma P W. Global regulatory networks and signal transduction pathways. In: Lengeler J W, Drews G, Schlegel H G, editors. Biology of the prokaryotes. Stuttgart, Germany: Thieme; 1999. pp. 491–523. [Google Scholar]
- 24.Lim A, Eleuterio M, Hutter B, Murugasu-Oei B, Dick T. Oxygen depletion induced dormancy in Mycobacterium bovis BCG. J Bacteriol. 1999;181:2252–2256. doi: 10.1128/jb.181.7.2252-2256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manabe Y C, Chen J M, Ko C G, Chen P, Bishai W R. Conditional sigma factor expression, using the inducible acetamidase promoter, reveals that the Mycobacterium tuberculosis sigF gene modulates expression of the 16-kilodalton alpha-crystallin homologue. J Bacteriol. 1999;181:7629–7633. doi: 10.1128/jb.181.24.7629-7633.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manganelli R, Dubnau E, Tyagi S, Kramer F R, Smith I. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol Microbiol. 1999;31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- 27.Mann M, Wilm M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 28.Marahiel M A, Zuber P. Sporulation and cell differentiation. In: Lengeler J W, Drews G, Schlegel H G, editors. Biology of the prokaryotes. Stuttgart, Germany: Thieme; 1999. pp. 586–601. [Google Scholar]
- 29.McKinney J D, Hoener zu Bentrup K, Munoz-Elias E J, Miczak A, Chen B, Chan W T, Swenson D, Sacchettini J C, Jacobs W R, Jr, Russell D G. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 30.Mizrahi V, Dawes S S, Rubin H. DNA replication. In: Hatful G F, Jacobs Jr W R, editors. Molecular genetics of mycobacteria. Washington, D.C.: American Society for Microbiology; 2000. pp. 159–172. [Google Scholar]
- 31.Murugasu-Oei B, Dick T. Bactericidal activity of nitrofurans on growing and dormant Mycobacterium bovis BCG. J Antimicrob Chemother. 2000;46:917–919. doi: 10.1093/jac/46.6.917. [DOI] [PubMed] [Google Scholar]
- 32.Parrish N M, Dick J D, Bishai W R. Mechanisms of latency in Mycobacterium tuberculosis. Trends Microbiol. 1998;6:107–112. doi: 10.1016/s0966-842x(98)01216-5. [DOI] [PubMed] [Google Scholar]
- 33.Primm T P, Andersen S J, Mizrahi V, Avarbock D, Rubin H, Barry C E., 3rd The stringent response of Mycobacterium tuberculosis is required for long-term survival. J Bacteriol. 2000;182:4889–4898. doi: 10.1128/jb.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segal W. Growth dynamics of in vivo and in vitro grown mycobacterial pathogens. In: Kubica G P, Wayne LG L G, editors. The mycobacteria—a source book. New York, N.Y: Marcel Dekker, Inc; 1984. pp. 547–573. [Google Scholar]
- 35.Sherman D R, Mdluli K, Hickey M J, Barry C E, 3rd, Stover C K. AhpC, oxidative stress and drug resistance in Mycobacterium tuberculosis. Biofactors. 1999;10:211–217. doi: 10.1002/biof.5520100219. [DOI] [PubMed] [Google Scholar]
- 36.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 37.Stover C K, Warrener P, Vandevanter D R, Sherman D R, Arain T M, Langhorne M H, Anderson S W, Towell J A, Yuan Y, McMurray D N, Kreiswirth B N, Barry C E, 3rd, Baker W R. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature. 2000;405:962–966. doi: 10.1038/35016103. [DOI] [PubMed] [Google Scholar]
- 38.Tabira Y, Ohara N, Kitaura H, Matsumoto S, Naito M, Yamada T. The 16-kDa α-crystallin-like protein of Mycobacterium bovis BCG is produced under conditions of oxygen deficiency and is associated with ribosomes. Res Microbiol. 1998;149:255–264. doi: 10.1016/s0923-2508(98)80301-x. [DOI] [PubMed] [Google Scholar]
- 39.Wayne L G, Salkin D. The bacteriology of resected tuberculous pulmonary lesions. I. The effect of interval between reversal of infectiousness and subsequent surgery. Am Rev Respir Dis. 1956;74:376–387. doi: 10.1164/artpd.1956.74.3.376. [DOI] [PubMed] [Google Scholar]
- 40.Wayne L G. The bacteriology of resected tuberculous pulmonary lesions. II. Observations on bacilli which are stainable but which cannot be cultured. Am Rev Respir Dis. 1960;82:370–377. doi: 10.1164/arrd.1960.82.3.370. [DOI] [PubMed] [Google Scholar]
- 41.Wayne L G. Dynamics of submerged growth of Mycobacterium tuberculosis under aerobic and microaerophilic conditions. Am Rev Respir Dis. 1976;114:807–811. doi: 10.1164/arrd.1976.114.4.807. [DOI] [PubMed] [Google Scholar]
- 42.Wayne L G. Synchronized replication of Mycobacterium tuberculosis. Infect Immun. 1977;17:528–530. doi: 10.1128/iai.17.3.528-530.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wayne L G, Sramek H A. Antigenic differences between extracts of actively replicating and synchronized resting cells of Mycobacterium tuberculosis. Infect Immun. 1979;24:363–370. doi: 10.1128/iai.24.2.363-370.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wayne L G, Lin K Y. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect Immun. 1982;37:1042–1049. doi: 10.1128/iai.37.3.1042-1049.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wayne L G. Dormancy of Mycobacterium tuberculosis and latency of disease. Eur J Clin Microbiol Infect Dis. 1994;13:908–914. doi: 10.1007/BF02111491. [DOI] [PubMed] [Google Scholar]
- 46.Wayne L G, Sramek H A. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:2054–2058. doi: 10.1128/aac.38.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wayne L G, Hayes L G. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wayne L G, Hayes L G. Nitrate reduction as a marker for hypoxic shift down of Mycobacterium tuberculosis. Tubercle Lung Dis. 1998;79:127–132. doi: 10.1054/tuld.1998.0015. [DOI] [PubMed] [Google Scholar]
- 49.Weber I, Fritz C, Ruttkowski S, Kreft A, Bange F C. Anaerobic nitrate reductase (narGHJI) activity of Mycobacterium bovis BCG in vitro and its contribution to virulence in immunodeficient mice. Mol Microbiol. 2000;35:1017–1025. doi: 10.1046/j.1365-2958.2000.01794.x. [DOI] [PubMed] [Google Scholar]
- 50.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 51.Yuan Y, Crane D D, Barry C E., 3rd Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial α-crystallin homolog. J Bacteriol. 1996;178:4484–4492. doi: 10.1128/jb.178.15.4484-4492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan Y, Crane D D, Simpson R M, Zhu Y, Hickey M J, Sherman D R, Barry C E., 3rd The 16-kDa α-crystallin (Acr) protein of Mycobacterium tuberculosis is required for growth in macrophages. Proc Natl Acad Sci USA. 1998;95:9578–9583. doi: 10.1073/pnas.95.16.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]