Abstract
To evaluate the role of uridylyl-transferase, the Sinorhizobium meliloti glnD gene was isolated by heterologous complementation in Azotobacter vinelandii. The glnD gene is cotranscribed with a gene homologous to Salmonella mviN. glnD1::Ω or mviN1::Ω mutants could not be isolated by a powerful sucrose counterselection procedure unless a complementing cosmid was provided, indicating that glnD and mviN are members of an indispensable operon in S. meliloti.
Sinorhizobium meliloti forms a symbiosis with Medicago sativa (alfalfa) in which nitrogen is fixed by the bacteria and released to the plant in exchange for photosynthates. After infection of plants, bacterial cells differentiate into bacteroids contained within plant membrane-enclosed organelles, symbiosomes, located within root nodules. Establishment of a successful symbiosis involves a shift in bacterial metabolism from assimilation of ammonia to export of nitrogenous compounds to the host plant. Therefore, ascertaining the features regulating this particular metabolic switch may provide valuable insight into symbiotic nitrogen fixation. As in many other bacteria, S. meliloti assimilates ammonia through the glutamine synthetase (GS)/glutamate synthase cycle. Unusually, members of the Rhizobiaceae carry three genes encoding isoforms of GS at separate loci (8). The major enzyme, GSI, is similar to GS of the enteric bacteria and is susceptible to posttranslational adenylylation, which reduces the rate of ammonia assimilation in vivo (1). As in enteric bacteria, a specialized protein called PII regulates the level of GSI adenylylation (2). In Escherichia coli, a model organism for studies on nitrogen regulation, the interactions of PII with its targets depend on the uridylylation state of PII, which responds to intracellular concentrations of the key metabolites glutamine and α-ketoglutarate (for current reviews, see references 10 and 12). Depletion of glutamine is sensed by the GlnD protein (9), which carries uridylyl-transferase/uridylyl-removing (UTase) activities, resulting in uridylylation of PII (10).
The role of PII in S. meliloti was previously examined by construction of two alleles of the corresponding glnB gene: ΔglnB10, a nonpolar null mutation, and glnBP5, a second allele that encodes a protein altered at the site of uridylylation. With respect to symbiosis, PII was required to efficiently transfer fixed nitrogen to the plant but not for nitrogenase expression (2). Predictions were that glnD mutants would exhibit phenotypes similar to glnBP5 mutants; however, this is complicated by the occurrence of multiple PII-like proteins in S. meliloti (D. Kahn and P. Rudnick, unpublished data) as in many other organisms (reviewed in reference 12). Therefore, the focus of this work was to directly address the role of the nitrogen-sensing UTase in the regulation of PII and its homologues in S. meliloti. We report here the cloning, sequencing, and mutagenesis of the S. meliloti glnD gene as well as evidence that this region of the chromosome is essential.
Cloning and sequence analysis of the glnD region of the chromosome.
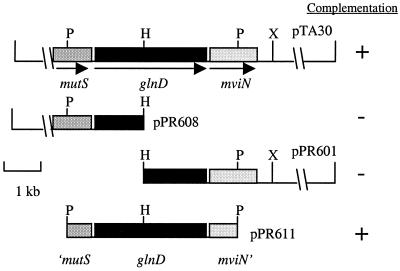
To isolate a library clone encoding a functional UTase, a pLAFR3 genomic library was mobilized into two Nif− glnD strains of Azotobacter vinelandii, MV17 (16) and MV71 (5) (Table 1). Several cosmids were isolated that complemented MV17 and MV71 by restoring their ability to fix nitrogen on N-free Burk's agar medium (11). One complementing cosmid, pTA30, contained a single central HindIII site within the insert. One half of the insert of pTA30 was cloned as a HindIII fragment into pBluescriptII KS+ to form plasmid pPR604. This 13-kb fragment was then cloned into the broad-host-range vector pJRD215 to form pPR608 and mobilized into Azotobacter vinelandii strain MV71 by triparental mating. Neither this plasmid nor pPR601, containing the remaining portion, complemented this strain for growth on N2 (Fig. 1). Indeed, sequencing of the HindIII junction revealed that this restriction site cleaves the glnD gene. The sequence of a 5.2-kb PstI fragment containing the HindIII site was determined, showing that the S. meliloti glnD gene is located between mutS, encoding DNA mismatch repair enzymes in many organisms, and a gene homologous to mviN from Salmonella enterica serovar Typhimurium (hereafter called mviN) (4). Sequence similarities of MutS, GlnD, and MviN are, respectively, 40, 32, and 32% amino acid identity with the corresponding proteins from E. coli. An ATGA sequence overlap between the glnD TGA stop codon and the mviN ATG start codon suggests that glnD and mviN are translationally coupled and part of the same operon.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype | Phenotype or characteristic | Reference |
|---|---|---|---|
| A. vinelandii | |||
| MV17 | nfrX16::Tn5a | Nif sup? (fast growing) | 16 |
| MV71 | glnD1::Ω glnY1 | Nif GSc | 5 |
| S. meliloti | |||
| GMI708 | Strain 2011 derivative | Rifr | 3 |
| GMI3105 | glnA1 | GSI unadenylylatable | 1 |
| Plasmids | |||
| pBluescriptII KS+ | Ampr; cloning vector | Stratagene | |
| pGEM-T Easy | Ampr; cloning vector self-ligated at EcoRV | Promega | |
| pHP45Ω | Ampr Spr; carries Ω interposon | 14 | |
| pJRD215 | IncQ broad-host-range vector; Kanr Smr | 7 | |
| pJQ200KS | Gmr; sacB mobilizable cloning vector | 15 | |
| pLAFR3 | IncP cosmid; Tetr | 13 | |
| pPR601 | Derivative of pTA30 with HindIII fragment deleted | This study | |
| pPR602 | pBluescript KS+ with 5.2-kb PstI fragment | This study | |
| pPR604 | pBluescript KS+ with 13-kb HindIII fragment | This study | |
| pPR608 | pJRD215 with 13-kb HindIII fragment | This study | |
| pPR611 | pJRD215 with 5.2-kb ClaI/SacI fragment; glnD+ | This study | |
| pPR612 | pGEM-T Easy carrying 5.2-kb PstI fragment | This study | |
| pPR613 | pPR612 with Ω inserted at HindIII; glnD1::Ω | This study | |
| pPR614 | pJQ200KS carrying glnD1::Ω | This study | |
| pPR615 | pBluescriptII KS+ with 3.6-kb HindIII/SacI fragment | This study | |
| pPR617 | pPR615 with Ω inserted at PstI blunt end; mviN1::Ω | This study | |
| pPR618 | pJQ200KS with mviN1::Ω | This study | |
| pTA30 | Library clone with mutS glnD mviN | This study |
nfrX was the previous designation for A. vinelandii glnD (6).
FIG. 1.
Restriction map of the cloned glnD region of the chromosome. P, PstI; H, HindIII; X, XhoI. The column on the right indicates whether the corresponding plasmid complements A. vinelandii glnD.
Complementation of an A. vinelandii glnD mutant with S. meliloti glnD.
The 5.2-kb PstI fragment of pPR602, containing partial open reading frames for mutS and mviN and the complete glnD gene, was subcloned into pJRD215 to give plasmid pPR611 (Fig. 1). This plasmid was conjugated into A. vinelandii glnD strain MV71. Transconjugants harboring pPR611 had their ability to grow on N-free medium restored, indicating that S. meliloti glnD was the only gene required by this strain to restore growth on this medium and that glnD encodes a functional UTase.
Mutagenesis of glnD.
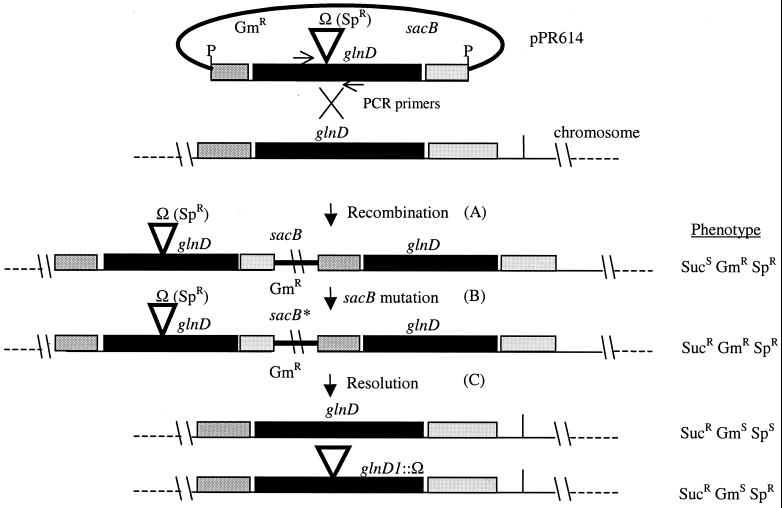
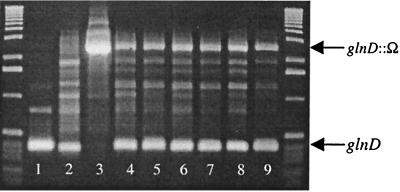
To examine its role, the glnD gene was mutagenized by insertion of an Ω element from pHP45Ω conferring resistance to spectinomycin into the central HindIII site of pPR612, creating a polar mutation in glnD (pPR613). The PstI fragment from pPR613, containing the mutation, was cloned into pJQ200KS, a vector carrying gentamicin resistance and conferring sensitivity to sucrose (sacB), to form plasmid pPR614 (15). pPR614 cannot be replicated in S. meliloti and is a suicide vector. This plasmid was mobilized into wild-type S. meliloti GMI708 by conjugation followed by selection on spectinomycin, which will select for strains in which homologous recombination at the target locus has occurred. A single Spr colony was propagated overnight without antibiotics to facilitate excision of vector sequences and replacement of the wild-type allele with glnD1::Ω by homologous recombination (Fig. 2). The overnight culture was plated on medium containing spectinomycin and sucrose. Surprisingly, 88% of the Sucr Spr isolates were also Gmr, indicating that the vector had not excised. To examine the structure of the chromosome targeted by the mutagenesis, primers were designed to amplify a 400-bp product from wild-type glnD and a 2.4-kb product from glnD1::Ω (Fig. 3, lanes 1 to 3). Long-range PCR analysis of the Sucr Spr Gmr isolates (lanes 6 to 8) generated products identical to Sucs Spr Gmr isolates, which have an integrated copy of pPR614 (lanes 4 and 5). This indicates a strong selective pressure driving Sucr by mutation in sacB rather than by recombination that would excise glnD. The remaining 12% Sucr Spr Gms clones also retained both mutant and wild-type copies of glnD (Fig. 3, lane 9), similar to the above-described isolates. In an experiment reported by Quandt and Hynes in which a construct carrying a similar amount of homologous DNA flanking the cassette was used, only 0 to 2% Gmr colonies were isolated following selection on sucrose and spectinomycin compared to 88% for this experiment (15). Our inability to isolate allelic replacement null mutants of glnD prompted us to investigate further the possibility that glnD, or downstream genes occurring in the same transcriptional unit, may be essential.
FIG. 2.
Schematic representation of events following conjugation of plasmid pPR614 into S. meliloti. (A) Following homologous recombination, the corresponding strain would contain one copy of each of glnD, glnD1::Ω, sacB, and the gentamicin resistance gene, aacC1, giving the indicated phenotype. (B) Loss of sacB (sacB* indicates a mutated form which does not confer sensitivity to sucrose) by mutation would allow both copies of glnD to reside on the chromosome along with aacC1. (C) Resolution of the single recombination resulting in excision of the vector might have one of the two possible outcomes illustrated. The second, resulting in the loss of wild-type glnD, was never observed without another source of the target gene. The locations of the PCR primers used to generate the products seen in Fig. 3 are shown for reference.
FIG. 3.
Long-range PCR amplification of a central fragment of glnD or glnD1::Ω from various sources during mutagenesis. A single isolate was grown overnight without antibiotics and plated on Luria-Bertani medium plus 5% sucrose and spectinomycin (100 μg/ml). Isolates were then screened for resistance to gentamicin. PCR products were amplified from single colonies unless otherwise indicated. Lane 1, pPR602 (plasmid DNA); lane 2, GMI708; lane 3, pPR614 (plasmid DNA); lanes 4 and 5, Sucs Spr Gmr isolates; lanes 6 to 8, Sucr Spr Gmr isolates; lane 9, rare Sucr Spr Gms isolate which unexpectedly retained wild-type glnD. Unlabeled lanes contain the 1-kb DNA ladder (Life Technologies).
In the next experiment, a single clone was selected on gentamicin after conjugation of pPR614. This isolate was then grown overnight without antibiotics and plated on sucrose as the sole selective pressure. Figure 2 illustrates possible events stemming from integration of pPR614 and outcomes. In this experiment, we predicted that resolution of the single crossover would occur at similar frequencies on either side of the cassette if no pressure was applied for maintenance of the wild-type target gene. For example, considering an equal amount of flanking DNA on either side of the cassette, sucrose-resistant isolates should arise at a frequency of 50% Sucr Sps, 50% Sucr Spr, and 0% Sucr Gmr. Very few, if any, should be Gmr, having occurred by presumptive mutations in sacB. However, in contrast to this prediction, between 95 and 99% of the Sucr isolates were Sps, indicating preferential loss of glnD1::Ω and restoration of the wild-type copy by recombination. Of the approximate 5% Spr isolates, all were Gmr, indicating retention of vector sequences (Table 2). These results demonstrate strong selective pressure to maintain the wild-type locus.
TABLE 2.
Gene replacement analysis of the glnD-mviN region of the S. meliloti chromosome
| Strain | Plasmid(s) | Primary selectiona | Secondary selectionb | Fractionc Sucr Spr | Fractiond of Sucr Spr that are Gmr |
|---|---|---|---|---|---|
| GMI708 | pPR614 (glnD1::Ω) | Gm | Suc | 5/100 | 5/5 |
| GMI3105 | pPR614 | Gm | Suc | 6/100 | 6/6 |
| GMI708 | pPR618 (mviN1::Ω) | Gm | Suc | 7/100 | 7/7 |
| Mutagenesis of strains harboring complementing plasmids | |||||
| GMI708 | pPR614, pPR611 (glnD+) | Gm, Km | Suc, Km | 58/100 | 58/58 |
| GMI708 | pPR614, pTA30 (glnD+ mviN+) | Gm, Tet | Suc, Tet | 33/100 | 0/33 |
| GMI3105 | pPR614, pPR611 | Gm, Km | Suc, Km | 22/100 | 22/22 |
| GMI3105 | pPR614, pTA30 | Gm, Tet | Suc, Tet | 31/69 | 1/31 |
| GMI708 | pPR618, pPR611 | Gm, Km | Suc, Km | 94/100 | 33/94 |
| GMI708 | pPR618, pTA30 | Gm, Tet | Suc, Tet | 24/99 | 0/24 |
| GMI3105 | pPR618, pPR611 | Gm, Km | Suc, Km | 65/100 | 65/65 |
| GMI3105 | pPR618, pTA30 | Gm, Tet | Suc, Tet | 10/76 | 0/10 |
Indicates original selection of transconjugants.
Selection after growth of transconjugants overnight in the absence of antibiotics.
Represents fraction of Sucr isolates that have retained Spr from glnD1::Ω or mviN1::Ω.
Fraction of Spr isolates that have retained Gmr encoded on the vector.
In other organisms, GlnD regulates the activity of GS through PII, which also seems to be the case for S. meliloti, since cells that encode a uridylylation-deficient PII are unable to deadenylylate GSI (2). Interestingly, A. vinelandii glnD null mutations are lethal in the absence of a suppressor mutation (5, 6) because GS is constitutively inactivated by adenylylation; GS is an essential enzyme in this organism probably because the organism lacks other means to assimilate ammonia, as well as a transport mechanism for glutamine (17). We therefore tested the possibility that S. meliloti glnD mutants could not be isolated because of a requirement for glutamine, either by supplementing all media with 0.4% glutamine, a concentration sufficient to allow growth of GS mutants (data not shown) or by using the glnA1 strain GMI3105, in which GSI is insensitive to adenylylation (Table 2) (1). In both cases, Sucr Spr Gms colonies could not be isolated which lacked the wild-type copy of glnD, further supporting an essential role for the glnD locus.
mviN mutagenesis.
In parallel, an mviN mutagenesis construct, pPR617, was made by insertion of a SmaI-digested Ω element into the single blunt-ended PstI site of mviN, carried on plasmid pPR615. The entire 4.6-kb XhoI fragment containing mviN1::Ω was subcloned into pJQ200KS to form plasmid pPR618. This plasmid was used exactly as pPR614 was (Fig. 2) except for inactivation of mviN. Surprisingly, there was a strong bias towards maintenance of wild-type mviN as well (Table 2).
glnD and mviN mutants can be readily isolated with a cosmid carrying multiple genes in trans.
To support our claim that the glnD-mviN operon may encode essential gene products in S. meliloti, complementing plasmids were introduced into a strain carrying either pPR614 or pPR618 integrated on the chromosome. For these experiments, plasmid pPR611 and cosmid pTA30 were used as a source of the wild-type gene(s). The mutagenesis was carried out as described above with the exception that all plates and cultures were supplemented with the appropriate antibiotic for maintenance of the complementing plasmid. Mutagenesis of glnD carrying pPR611 gave no isolates that were Sucr Spr Gms, indicating that the mutation in glnD has polar effects on essential linked genes and that glnD and mviN belong to the same operon (Table 2). In contrast, glnD or mviN could be easily replaced by gene replacement if pTA30 was in the background. For the glnD mutagenesis, approximately 33% of the Sucr isolates were Spr and Gms and 0% of the Spr isolates were Gmr. This is as predicted when there is little or no selective pressure to maintain the wild-type target gene. Mutagenesis of mviN in the presence of pTA30 gave similar results: 45% of the Sucr isolates were Spr Gms and 1% were Spr Gmr. We conclude that glnD and mviN are members of an essential operon in S. meliloti.
Nucleotide sequence accession number.
The sequence of the 5.2-kb PstI fragment containing the HindIII site was deposited in GenBank under accession no. AF227730.
Acknowledgments
We thank the European Union BIOTECH program for support (FIXNET program). P. Rudnick was supported in part by a graduate Chateaubriand fellowship from the French Embassy to the United States.
REFERENCES
- 1.Arcondéguy T, Huez I, Fourment J, Kahn D. Symbiotic nitrogen fixation does not require adenylylation of glutamine synthetase I in Rhizobium meliloti. FEMS Microbiol Lett. 1996;145:33–40. doi: 10.1111/j.1574-6968.1996.tb08553.x. [DOI] [PubMed] [Google Scholar]
- 2.Arcondéguy T, Huez I, Tillard P, Gangneux C, de Billy F, Gojon A, Truchet G, Kahn D. The Rhizobium meliloti PII protein, which controls bacterial nitrogen metabolism, affects alfalfa nodule development. Genes Dev. 1997;11:1194–1206. doi: 10.1101/gad.11.9.1194. [DOI] [PubMed] [Google Scholar]
- 3.Batut J, Terzaghi B, Gherardi M, Huguet M, Terzaghi E, Garnerone A M, Boistard P, Huguet T. Localization of a symbiotic fix region on Rhizobium meliloti pSym megaplasmid more than 200 kilobases from the nod-nif region. Mol Gen Genet. 1985;199:232–239. [Google Scholar]
- 4.Carsiotis M, Stocker B A, Weinstein D L, O'Brien A D. A Salmonella typhimurium virulence gene linked to flg. Infect Immun. 1989;57:3276–3280. doi: 10.1128/iai.57.11.3276-3280.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colnaghi, R., P. Rudnick, L. He, A. Green, D. Yan, E. Larson, and C. Kennedy. Characterization of glnD null mutations in Azotobacter vinelandii: lethality is suppressible by prevention of glutamine synthetase adenylylation. Microbiology, in press. [DOI] [PubMed]
- 6.Contreras A, Drummond M, Bali A, Blanco G, Garcia E, Bush G, Kennedy C, Merrick M. The product of the nitrogen fixation regulatory gene nfrX of Azotobacter vinelandii is functionally and structurally homologous to the uridylyltransferase encoded by glnD in enteric bacteria. J Bacteriol. 1991;173:7741–7749. doi: 10.1128/jb.173.24.7741-7749.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davison J, Heusterspreute M, Chevalier N, Ha-Thi V, Brunel F. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene. 1987;51:275–280. doi: 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- 8.de Bruijn F J, Rossbach S, Schneider M, Ratet P, Messmer S, Szeto W W, Ausubel F M, Schell J. Rhizobium meliloti 1021 has three differentially regulated loci involved in glutamine biosynthesis, none of which is essential for symbiotic nitrogen fixation. J Bacteriol. 1989;171:1673–1682. doi: 10.1128/jb.171.3.1673-1682.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang P, Peliska J A, Ninfa A J. Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry. 1998;37:12782–12794. doi: 10.1021/bi980667m. [DOI] [PubMed] [Google Scholar]
- 10.Merrick M J, Edwards R A. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newton J W, Wilson P W, Burris R H. Direct demonstration of ammonia as an intermediate in nitrogen fixation by Azotobacter. J Biol Chem. 1953;204:445–451. [PubMed] [Google Scholar]
- 12.Ninfa A J, Atkinson M R. PII signal transduction proteins. Trends Microbiol. 2000;8:172–179. doi: 10.1016/s0966-842x(00)01709-1. [DOI] [PubMed] [Google Scholar]
- 13.Peet R C, Lindgren P B, Willis D K, Panopoulos N J. Identification and cloning of genes involved in phaseolotoxin production by Pseudomonas syringae pv. “phaseolicola.”. J Bacteriol. 1986;166:1096–1105. doi: 10.1128/jb.166.3.1096-1105.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 15.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 16.Santero E, Toukdarian A, Humphrey R, Kennedy C. Identification and characterization of two nitrogen fixation regulatory regions, nifA and nfrX, in Azotobacter vinelandii and Azotobacter chroococcum. Mol Microbiol. 1988;2:303–314. doi: 10.1111/j.1365-2958.1988.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 17.Toukdarian A, Saunders G, Selman-Sosa G, Santero E, Woodley P, Kennedy C. Molecular analysis of the Azotobacter vinelandii glnA gene encoding glutamine synthetase. J Bacteriol. 1990;172:6529–6539. doi: 10.1128/jb.172.11.6529-6539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]