Abstract
We used a photoactivatable, lipophilic reagent, 3′-(trifluoromethyl)-3-(m-[125I]iodophenyl)diazirine, to label proteins in the outer membrane of elementary bodies of Chlamydia trachomatis LGV serovar L2 and mass spectrometry to identify the labeled proteins. The identified proteins were polymorphic outer membrane proteins E, G, and H, which were made late in the developmental cycle, the major outer membrane protein, and a mixture of 46-kDa proteins consisting of the open reading frame 623 protein and possibly a modified form of the major outer membrane protein.
The cell envelope of Chlamydia species resembles that of other gram-negative bacteria, consisting of an outer membrane (OM), an inner membrane (IM), and a periplasmic space. Two unusual features of the chlamydial envelope are a deficient and perhaps novel peptidoglycan structure (reviewed in references 6, 10, and 21) and the presence of a disulfide-bond-cross-linked major outer membrane protein (MOMP) in the OM and cross-linked cysteine-rich proteins (CRPs) in the periplasm (reviewed in reference 12).
The MOMP is the only chlamydial OM protein that has been well characterized. It is a porin (2, 30), is surface exposed (7, 28), and may play a role in the attachment of Chlamydia trachomatis to host cells (27, 28). Genes that potentially encode a family of proteins, referred to as polymorphic outer membrane proteins (Pomps), have been identified in the genomes of Chlamydia species: nine Pomp genes (pmpA to pmpH) are present in the genomes of C. trachomatis serovars D and L2, 21 genes (pmp1 to pmp21) are in C. pneumoniae, and at least six genes (Pomp90 and Pomp98 families) are encoded by the genomes of ovine strains of C. psittaci (15, 20, 26). However, only two Pomps have been shown to be produced in C. trachomatis and C. pneumoniae, and only four Pomps have been identified in C. psittaci (11, 16, 20, 22, 29). The functions of the Pomps are not known, nor is it known when C. trachomatis Pomps are made during the developmental cycle. Several other OM proteins can be predicted from chlamydial genomic sequences (15, 26); however, none of these proteins have been experimentally identified in OMs.
The OM of chlamydiae is poorly characterized, in part because of the difficulty of growing large quantities of chlamydiae but mainly because chlamydial OMs cannot be separated from IMs by density gradient centrifugation. Criteria that have been used to identify chlamydial OM proteins include surface exposure, as detected by susceptibility to trypsin or reaction with antibodies following treatment of infectious elementary bodies (EBs) with these reagents, and insolubility in the weak anionic detergent sodium lauryl sarcosinate (Sarkosyl). Although these methodologies have been useful, they can yield deceptive results. For example, damage to EBs or contamination of EBs with osmotically fragile reticulate bodies during harvesting and purification can expose proteins that are not on the surface of EBs, and the failure to observe positive reactions will result if a surface-exposed protein lacks a trypsin-sensitive site or an immunodominant epitope. Insolubility in Sarkosyl is also subject to misinterpretation. The technique was originally developed by Filip et al. (9) to remove cytoplasmic membrane proteins from well-characterized integral OM proteins of Escherichia coli and was adapted to chlamydial studies by Caldwell et al. (4) and Hatch et al. (14). However, the reason for the differential solubility of IM and OM proteins in Sarkosyl is not known, and it is likely that some non-OM proteins fractionate in the Sarkosyl-insoluble fraction and that some OM proteins are released from OMs by Sarkosyl. For these reasons we chose to identify C. trachomatis LGV serovar L2 OM proteins on the basis of their reaction with 3′-(trifluoromethyl)-3-(m-[125I]iodophenyl)diazirine ([125I]TID), a photoactivatable lipophilic reagent developed by Brunner and Semenza (3) to label amino acid side chains and other ligands in the lipophilic environment of cytoplasmic membranes of eukaryotic cells. Because TID is lipophilic, it penetrates the OM of gram-negative bacteria, where it becomes trapped and incapable of passing through the hydrophilic periplasm and beyond (8).
Identification of OM proteins in C. trachomatis L2.
EBs were harvested at 48 h after infection of L929 cells, treated with 1% Nonidet P-40 (Sigma Chemical Co., St. Louis, Mo.) to eliminate osmotically fragile reticulate bodies, and purified by centrifugation for 30 min at 80,000 × g on a three-step gradient of 29, 34, and 40% Hypaque-76 (Nycomed Inc., Princeton, N.J.). Purified EBs from 2 × 108 cells were reacted with 25 μCi of [125I]TID, as previously described (8), and incubated for 30 min at 37°C in the presence or absence of 12 μg of trypsin (type III from bovine pancreas; Sigma Chemical Co.) in 200 μl of phosphate-buffered saline (pH 7.4), followed by the addition of 24 μg of trypsin inhibitor (type II-O from chicken egg white; Sigma). After the EBs were washed once in trypsin inhibitor, one half of the EB preparation was solubilized by heating to 90°C in Laemmli buffer (18) containing 5% β-mercaptoethanol and 10 mM dithiothreitol, and the other half was extracted with 500 μl of 0.5% Sarkosyl (Sigma) in phosphate-buffered saline for 30 min at 37°C. The Sarkosyl-insoluble fraction was collected by centrifugation (14,500 × g for 20 min) and washed twice in 500 μl of 0.5% Sarkosyl before it was solubilized in Laemmli buffer with reducing agents. Proteins in whole EBs and the Sarkosyl-insoluble fraction of EBs were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 7.5 to15% gradient gels, and incorporation of 125I into proteins was detected by phosphorimaging of the dried gels.
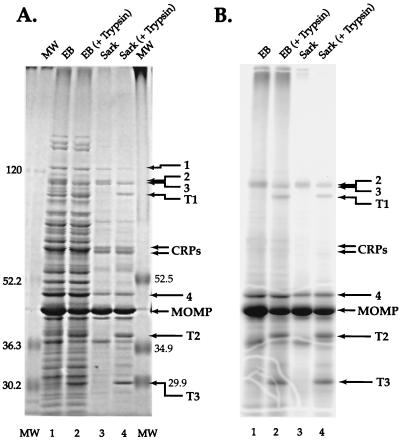
The protein profiles of whole EBs and the Sarkosyl-insoluble fraction of EBs are shown in Fig. 1. A number of proteins were seen in the Coomassie brilliant blue-stained Sarkosyl-insoluble fraction (Fig. 1A, lane 3), including the 40-kDa MOMP and its likely degradation products (28), the 60-kDa CRP doublet, and several unknown proteins. The unknown proteins included one of about 120 kDa (protein 1), a poorly resolved doublet of about 100 kDa (proteins 2 and 3), and a protein of approximately 46 kDa (protein 4). Only the MOMP, likely degradation products of the MOMP, and proteins 2 to 4 were labeled extensively with TID; the periplasmic CRPs, notably, were not labeled (Fig. 1B, lane 3). The TID-labeled, Sarkosyl-insoluble profile was similar to that of the TID-labeled whole EB protein profile (Fig. 1B, lanes 1 and 3). Treatment of EBs with trypsin prior to the preparation of the Sarkosyl-insoluble fraction reduced the intensity of the protein doublet 2-3, with the appearance of new peptides, designated tryptic fragments T1 to T3.
FIG. 1.
Identification of proteins labeled with [125I]TID by SDS-PAGE. (A) Coomassie-stained 7.5 to 15% polyacrylamide gel, with prestained protein standard sizes shown in kilodaltons at each side; (B) phosphorimage of the gel. Lanes: 1, whole EBs; 2, whole EBs following treatment with trypsin; 3, the Sarkosyl-insoluble fraction; 4, the Sarkosyl-insoluble fraction of EBs treated with trypsin. Arrows point to proteins discussed in the text.
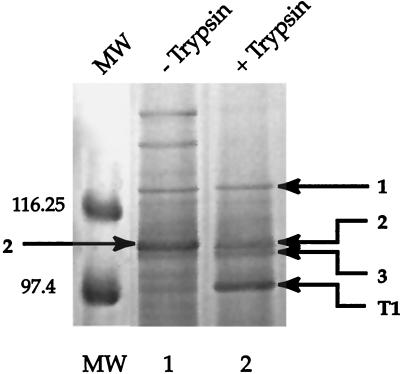
In order to better resolve the high-molecular-weight proteins, the Sarkosyl-insoluble fraction of EBs was fractionated on a 5.0 to 7.5% gel and proteins were visualized with the reversible, negative zinc-imidazole stain described by Castellanos-Serra et al. (5). Under this condition of analysis, the intensity of protein 2 was reduced when the EBs were treated with trypsin, suggesting that the band may consist of a mixture of two or more proteins (Fig. 2). Gel slices containing proteins 1 to 3 and trypsin fragment T1 were excised from the gel shown in Fig. 2 and prepared for analysis by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry after in-gel digestion with trypsin, as described by Shevchenko et al. (25). Protein 4, the MOMP, and trypsin fragments T2 and T3 were similarly analyzed following electrophoresis of the Sarkosyl-insoluble fraction on 7.5 to 15% gels. Because the C. trachomatis L2 genome has not been completely sequenced and annotated, we identified proteins with the C. trachomatis D database located on the Protein Prospector website (http://prospector.ucsf.edu). Our findings are summarized in Table 1.
FIG. 2.
Zinc-imidazole-stained 5 to 7.5% polyacrylamide gel of high-molecular-weight proteins present in the Sarkosyl-insoluble fraction of EBs. Lanes: 1, Sarkosyl-insoluble fraction of EBs; 2, Sarkosyl-insoluble fraction of EBs treated with trypsin. Sizes of prestained protein standards in kilodaltons are shown on the left. Arrows point to bands (numbered the same as in Fig. 1) that were analyzed by mass spectrometry.
TABLE 1.
Proteins identified in the Sarkosyl-insoluble fraction by mass spectometrya
| Band | Identification |
|---|---|
| 1 | ORF 664 |
| 2 (−trypsin) | Mixture of Pomps E, G, and H |
| 2 (+trypsin) | Mixture of Pomps E and H |
| 3 (−trypsin) | Mixture of Pomp G and H |
| 3 (+trypsin) | PompH |
| T1 | PompG |
| 4 | Mixture of MOMP and ORF 623 |
| T2 | MOMP |
| T3 | MOMP |
Zinc-imidazole negatively stained bands were excised from gels, and samples were prepared for MALDI-TOF mass spectrometry by in-gel digestion with trypsin as described by Shevchenko et al. (25).
When EBs were not first treated with trypsin, protein bands 2 and 3 contained a mixture of Pomps E, G, and H. After treatment of EBs with trypsin, these bands were resolved to a mixture Pomps E and H, with PompE being the predominant protein in band 2 and PompH being the predominant protein in band 3; fragment T1 was identified as a degradation product of PompG. We concluded that the trypsin-sensitive PompG and the trypsin-insensitive PompE comigrate as band 2, with contamination with Pomp H (protein 3) due to smearing during electrophoresis. Pomps G and H were previously identified by Mygind et al. (22) in the Sarkosyl-insoluble fraction of C. trachomatis L2.
The best identification of band 1 was C. trachomatis D open reading frame (ORF) 664 (26). The predicted peptide of ORF 664 possesses a potential FHA (forkhead-associated) domain, thought to be important in protein-protein interactions, and is weakly homologous to several proteins found in eukaryotes and prokaryotes; however, its function in chlamydiae is not clear. PSORT analysis (http://psort.nibb.ac.jp) predicted an uncleavable signal sequence and an IM location for the protein encoded by ORF 664, which is consistent with the lack of labeling of protein 1 by [125I]TID. The predicted molecular weight of the mature protein encoded by ORF 664 is 89,649, considerably less than the relative molecular weight (120,000) determined by SDS-PAGE. The predicted pI of the mature protein is 4.32, and the acidic nature of the protein, including several stretches of high-density negative charge, may be responsible for its anomalous migration rate. Interestingly, ORF 664 is weakly paralogous to PompC (E value = 5e-06); however MALDI-TOF analysis failed to identify PompC of either C. trachomatis D or C. trachomatis L2 (only a partial sequence is available) in band 1.
Protein band 4 (relative molecular weight of about 46,000) was found to be a mixture of the MOMP and ORF 623 (26) by MALDI-TOF analysis, with the tryptic peptides with the highest counts being assigned to the MOMP. N-terminal amino acid analysis confirmed the presence of the MOMP and ORF 623 in protein band 4, with the MOMP being present in a ratio of about 2 to 1. ORF 623 encodes a predicted chlamydia-specific protein of unknown function; it possesses a predicted signal sequence, which was confirmed by the N-terminal sequence analysis, and the characteristics of an OM protein as determined by PSORT analysis. The predicted molecular weight of the mature form is 45,669. The presence of the MOMP in protein band 4 may be the result of smearing of the abundant 40-kDa MOMP during electrophoresis or may be due to the presence of a posttranslationally modified form of the MOMP in the band. Kuo and colleagues (17) have presented evidence that the 40-kDa MOMP of C. trachomatis L2 is N-linked glycosylated with a high-mannose oligosaccharide. It is possible the carbohydrate identified by Kuo et al. (17) was the result of contamination of unmodified 40-kDa MOMP with modified 46-kDa MOMP. However, in preliminary studies, we failed to detect alteration in the migration rate of any protein in the Sarkosyl-insoluble fraction when EBs were treated with calf intestinal alkaline phosphatase (Boehringer-Mannheim; 5 U in 200 μl of phosphatase buffer for 1 h at 37°C), N-glycosylase F (Boehringer-Mannheim; 2 U in 200 μl of phosphate-buffered saline for 1 h at room temperature, followed by the addition of Triton X-100 to 0.25% and incubation at room temperature for 30 min and then overnight at 4°C), and endoglycosylase H (Boehringer-Mannheim; 2 mU, same incubation conditions as for N-glycosylase F) (data not shown).
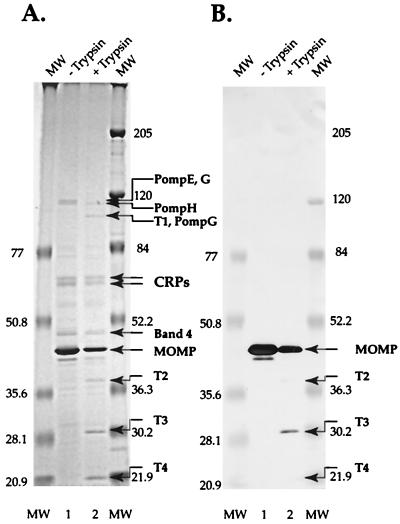
In a further attempt to identify the MOMP in band 4, an immunoblot was treated with monoclonal antibody L2I-45, which recognizes the serovar-specific epitope in the MOMP of C. trachomatis L2 (1). Only the abundant 40-kDa MOMP band and degradation products of the MOMP were recognized by the antibody (Fig. 3).
FIG. 3.
SDS-PAGE (A) and immunoblot (B) analysis of the Sarkosyl-insoluble fraction of EBs incubated in the presence (lane 2) and absence (lane 1) of trypsin. The blot was treated with monoclonal antibody L2I-45, which reacts with the serovar-specific epitope in the MOMP of C. trachomatis L2 (1). Trypsin fragment T4 was not analyzed by mass spectrometry; however, the intensity of the band suggests that it was derived from the MOMP.
Stage-specific synthesis of Pomps.
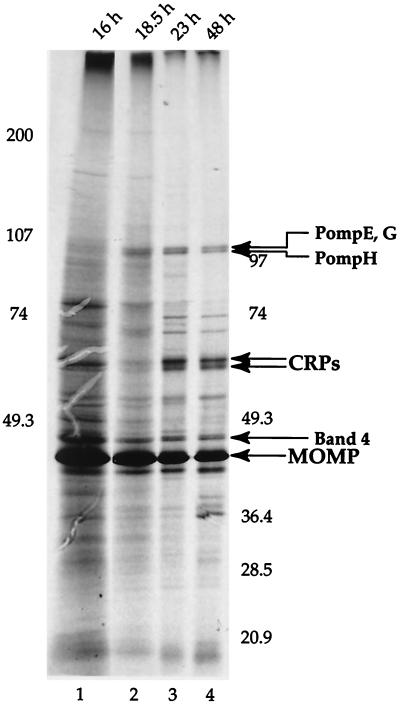
We recently demonstrated that three Pomps of the ovine C. psittaci Pomp90 family and three other potential Pomps are synthesized cotemporally with the late-stage-specific CRPs in C. psittaci 6BC (29). We examined stage-specific synthesis of C. trachomatis L2 Pomps by adding 275 μCi of [35S]Cys-[35S]Met (Protein Labeling Mix; NEN, Boston, Mass.) to 150-cm2 monolayers of infected cells (2 × 107) at 14 h postinfection (p.i.) and preparing a Sarkosyl-insoluble fraction directly from the infected cells at 16, 18.5, and 23 h p.i. and from the harvested medium supernatant fluid and cells remaining attached to the flasks at 48 h p.i. Cycloheximide (0.5 μg/ml) was present during the entire infection to inhibit incorporation of label into host cells. Incorporation of label into chlamydial proteins in the Sarkosyl-insoluble fraction was assessed by autoradiography of a dried polyacrylamide gel, with the amount of material loaded onto the gel adjusted so that the amount of MOMP (present in relatively constant amounts at all times p.i.) for each sample was the same (Fig. 4). The results indicated that the Pomps are made at 18.5 h and later times, whereas synthesis of the late-stage-specific CRPs (13, 23) is not obvious until 23 h. The 16 h p.i. sample was difficult to interpret; the large amount of material loaded resulted in a high background and included proteins which comigrated with the CRPs that were not present at late times p.i. Nonetheless, it appears that the Pomps, at least relative to the MOMP, are not made in significant amounts between 14 and 16 h p.i.
FIG. 4.
Autoradiograph of a 7.5 to 15% polyacrylamide gel showing incorporation of [35S]Cys-[35S]Met into the Sarkosyl-insoluble fraction from 14 to 48 h p.i. The Pomps appear to be made in greater amounts late in the cycle, relative to the MOMP, but at detectable levels sooner than the late-stage-specific 60-kDa CRPs.
Conclusions.
[125I]TID labeling of C. trachomatis L2 EBs identified three Pomps (E, G, and H), the MOMP, and another protein or proteins that migrate during SDS-PAGE with a relative molecular mass of about 46 kDa. It is likely that additional proteins are present in the OM but in insufficient amounts to be detected by the amount of TID label that we used. This is almost certainly the case for PompF, which is encoded immediately upstream of PompE on the same operon, with the UAA stop codon of PompF located only 2 bp upstream of the AUG start codon of PompE (26). Also, in a preliminary, study Lindquist and Stephens (19) detected transcripts of the genes encoding Pomps B, D, I, and G by reverse transcription-PCR in C. trachomatis L2. The Pomps that we detected are encoded by genes that are clustered in the same region of the genome: the operon encoding Pomps F and E (pmpFE) and three divergently transcribed genes encoding Pomps G, H, and I. It is interesting that the divergently transcribed Pomp genes are cotemporally expressed.
Mass spectrometric and N-terminal amino acid analysis indicated that the MOMP was a major constituent of the TID-labeled, 46-kDa protein band, suggesting that the protein may be a posttranscriptionally modified form of the MOMP, perhaps identical to the modified form described by Kuo et al. (17). However, the presence of the MOMP was not confirmed by immunoblot analysis and we found no evidence of glycosylated or phosphorylated protein in either the 46- or the 40-kDa protein. Alternatively, the TID-labeled protein may be encoded by ORF 623, a predicted OM protein of unknown function. Definitive identification of the proteins in the 46-kDa band will require additional investigation. Whatever the identity of the TID-labeled 46-kDa protein is, there is evidence that it may be antigenic in that Newhall et al. (24) identified a band of similar size that reacted on immunoblots of multiple serovars of C. trachomatis with the sera of infected patients. Although the proteins in the 46-kDa band did not possess a surface-exposed trypsin-sensitive site, their presence in the OM suggests they may be useful as diagnostic tools or in the development of a subunit vaccine.
Acknowledgments
We thank Harlan Caldwell for the provision of monoclonal antibody L2I-45.
This work was supported by Public Health Service grant AI-19570 from the National Institute of Allergy and Infectious Diseases. Mass spectrometric studies were carried out in the Stout Neuroscience Laboratory at the University of Tennessee, which is supported by Public Health Service grant RR 105222 and National Science Foundation grant DBI 9604633.
REFERENCES
- 1.Baehr W B, Zhang Y-X, Joseph T, Hua S, Nano F E, Everett K D E, Caldwell H D. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc Natl Acad Sci USA. 1988;85:4000–4004. doi: 10.1073/pnas.85.11.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bavoil P, Ohlin A, Schachter J. Role of disulfide bonding in outer membrane structure and permeability in Chlamydia trachomatis. Infect Immun. 1984;44:479–485. doi: 10.1128/iai.44.2.479-485.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunner J, Semenza G. Selective labeling of the hydrophobic core of membranes with 3-(trifluoromethyl)-3-(m-[125I]iododiazirine, a carbene-generating reagent. Biochemistry. 1981;20:7174–7182. doi: 10.1021/bi00528a019. [DOI] [PubMed] [Google Scholar]
- 4.Caldwell H D, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castellanos-Serra L, Proenza W, Huerta V, Moritz R L, Simpson R J. Proteome analysis of polyacrylamide gel-separated proteins visualized by reversible negative staining using imidazole-zinc salts. Electrophoresis. 1999;20:732–737. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<732::AID-ELPS732>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 6.Chopra I, Storey C, Falla T J, Pearce J H. Antibiotics, peptidoglycan synthesis and genomics: the chlamydial anomaly revisited. Microbiology. 1998;144:2673–2678. doi: 10.1099/00221287-144-10-2673. [DOI] [PubMed] [Google Scholar]
- 7.Collett B A, Newhall W J, Jersild V, R A, Jr, Jones R B. Detection of surface-exposed epitopes on Chlamydia trachomatis by immune electron microscopy. J Gen Microbiol. 1989;135:85–94. doi: 10.1099/00221287-135-1-85. [DOI] [PubMed] [Google Scholar]
- 8.Everett K D E, Hatch T P. Architecture of the cell envelope of Chlamydia psittaci 6BC. J Bacteriol. 1995;177:877–882. doi: 10.1128/jb.177.4.877-882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filip C, Fletcher G, Wulff J L, Earhart C F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghuysen J M, Goffin C. Lack of cell wall peptidoglycan versus penicillin sensitivity: new insights into the chlamydial anomaly. Antimicrob Agents Chemother. 1999;43:2339–2344. doi: 10.1128/aac.43.10.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giannikopoulou P, Bini K, Simitsek O D, Pallini V, Vretou E. Two-dimensional electrophoretic analysis of the protein family at 90 kDa of abortifacient Chlamydia psittaci. Electrophoresis. 1997;18:2104–2108. doi: 10.1002/elps.1150181137. [DOI] [PubMed] [Google Scholar]
- 12.Hatch T P. Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? J Bacteriol. 1996;178:1–5. doi: 10.1128/jb.178.1.1-5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatch T P, Miceli M, Sublett J E. Synthesis of disulfide-bonded outer membrane proteins during the developmental cycle of Chlamydia psittaci and Chlamydia trachomatis. J Bacteriol. 1986;165:379–385. doi: 10.1128/jb.165.2.379-385.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatch T P, Vance D W, Al-Hossainy E. Identification of a major envelope protein in Chlamydia spp. J Bacteriol. 1981;146:426–429. doi: 10.1128/jb.146.1.426-429.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman R W, Olinger L, Grimwood J, Davis R W, Stephens R S. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat Genet. 1999;21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- 16.Knudsen K, Madsen A S, Mygind P, Christiansen G, Birkelund S. Identification of two novel genes encoding 97- to 99-kilodalton outer membrane proteins of Chlamydia pneumoniae. Infect Immun. 1999;67:375–383. doi: 10.1128/iai.67.1.375-383.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo C-C, Takahashi N, Swanson A F, Ozeki Y, Hakomori S-I. An N-linked high-mannose type oligosaccharide, expressed at the major outer membrane protein of Chlamydia trachomatis mediates attachment and infectivity of the microorganism to HeLa cells. J Clin Investig. 1996;98:2813–2818. doi: 10.1172/JCI119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Lindquist E A, Stephens R S. Transcriptional activity of a sequence variable protein family in Chlamydia trachomatis. In: Stephens R S, Byrne G I, Christiansen G, Clarke I N, Grayston J T, Rank R G, Ridgway G L, Saikku P, Schachter J, Stamm W E, editors. Chlamydial Infections: Proceedings of the Ninth International Symposium on Human Chlamydial Infections. San Francisco, Calif: International Chlamydia Symposium; 1998. pp. 259–262. [Google Scholar]
- 20.Longbottom D, Russell M, Dunbar S M, Jones G E, Herring A J. Molecular cloning and characterization of the genes coding for the highly immunogenic cluster of 90-kilodalton envelope proteins from the Chlamydia psittaci subtype that causes abortion in sheep. Infect Immun. 1998;66:1317–1324. doi: 10.1128/iai.66.4.1317-1324.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moulder J W. Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan? Infect Agents Dis. 1993;2:87–99. [PubMed] [Google Scholar]
- 22.Mygind P H, Christiansen G, Roepstorff P, Birkelund S. Membrane proteins PmpG and PmpH are major constituents of Chlamydia trachomatis L2 outer membrane complex. FEMS Microbiol Lett. 2000;186:163–169. doi: 10.1111/j.1574-6968.2000.tb09098.x. [DOI] [PubMed] [Google Scholar]
- 23.Newhall W J, V. Biosynthesis and disulfide cross-linking of outer membrane components during the growth cycle of Chlamydia trachomatis. Infect Immun. 1987;55:162–168. doi: 10.1128/iai.55.1.162-168.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newhall W J, Batteiger V, B, Jones R P. Analysis of the human serological response to proteins of Chlamydia trachomatis. Infect Immun. 1982;38:1181–1189. doi: 10.1128/iai.38.3.1181-1189.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 26.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, Koonin E V, Davis R W. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 27.Su H, Caldwell H D. In vitro neutralization of Chlamydia trachomatis by monovalent Fab antibody specific to MOMP. Infect Immun. 1991;59:2843–2845. doi: 10.1128/iai.59.8.2843-2845.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su H, Zhang Y-X, Barrera O, Watkins N G, Caldwell H D. Differential effects of trypsin on infectivity of Chlamydia trachomatis: loss of infectivity requires cleavage of major outer membrane protein variable domains II and IV. Infect Immun. 1988;56:2094–2100. doi: 10.1128/iai.56.8.2094-2100.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanzer R J, Longbottom D, Hatch T P. Identification of polymorphic outer membrane proteins of Chlamydia psittaci 6BC. Infect Immun. 2001;69:2428–2434. doi: 10.1128/IAI.69.4.2428-2434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wyllie S, Ashley R H, Longbottom D, Herring A J. The major outer membrane protein of Chlamydia psittaci functions as a porin-like ion channel. Infect Immun. 1998;66:5202–5207. doi: 10.1128/iai.66.11.5202-5207.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]