Abstract
Induction of the torCAD operon, encoding the trimethylamine N-oxide (TMAO) respiratory system, is tightly controlled by the TorS-TorR phosphorelay system in response to TMAO availability. TorS is an unorthodox sensor that contains three phosphorylation sites and transphosphorylates TorR via a four-step phosphorelay, His443→Asp723→His850→Asp(TorR). In this study, we provide genetic evidence that TorS can dephosphorylate phospho-TorR when TMAO is removed. Dephosphorylation probably occurs by a reverse phosphorelay, Asp(TorR)→His850→Asp723, since His850 and Asp723 are both essential in this process. By using reverse transcriptase PCR, we also show that TMAO removal results in shutoff of tor operon transcription in less than 2 min. Based on our results and on analogy to other phosphorelay signal transduction systems, we propose that reverse phosphotransfer could be a rapid and efficient mechanism to inactivate response regulators.
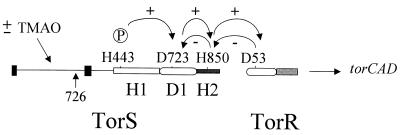
Two-component signal transduction systems are widespread in bacteria, archaebacteria, plants, and lower eucaryotes, and they enable cells to adapt to changing environments (3, 10, 26, 28). The simplest two-component systems contain a sensor protein and a cognate response regulator (11, 23). The sensor protein is usually a transmembrane protein whose N-terminal domain, oriented toward the exterior, detects specific environmental stimuli and whose C-terminal domain, located in the cytoplasm, is a histidine autokinase domain called the transmitter. Once stimulated, the sensor autophosphorylates and transphosphorylates its cognate response regulator on a conserved aspartate of the receiver domain. The response regulator, thus activated, generally regulates expression of target genes by binding to specific DNA sequences. Accordingly, in Escherichia coli, the TorS sensor detects the presence of trimethylamine N-oxide (TMAO) in the medium and transphosphorylates its cognate response regulator, TorR, which in turn activates the torCAD operon, encoding the main TMAO anaerobic respiratory system (16, 20, 31). Interestingly, TorS belongs to a subfamily of sensor proteins, including ArcB of E. coli and BvgS of Bordetella pertussis, called the unorthodox or tripartite sensors because members of this family contain three phosphorylation sites (12, 15, 35). TorS thus possesses two histidine phosphorylation sites (His443 and His850) and an aspartate phosphorylation site (Asp723), which are located on a primary transmitter (H1), a C-terminal secondary transmitter (H2), and a receiver domain (D1), respectively (Fig. 1). We have previously shown that activation of TorR involves the following four-step phosphorelay: His443→Asp723→His850→Asp(TorR) (15). As several unorthodox sensors, including ArcB, BvgS, and EvgS, have been shown to follow the same phosphorylation pathway (9, 17, 25, 34, 35), the four-step phosphorelay is most probably the usual phosphotransfer mechanism shared by two-component systems comprising unorthodox sensors.
FIG. 1.
Schematic representation of the TorS-TorR phosphorelay system. TorS contains a N-terminal TMAO detector region, a primary transmitter domain (H1) with a histidine phosphorylation site (H443), a receiver domain (D1) with an aspartate phosphorylation site (D723), and a secondary transmitter domain (H2) with a histidine phosphorylation site (H850) (15). 726, location of the TMAO-constitutive mutation of TorS726; dark bars, putative transmembrane segments (16). The TorR response regulator contains a phosphoaccepting site (D53) in its receiver domain and a C-terminal DNA-binding domain. In the presence of TMAO, TorR is activated by phosphorylation via a four-step phosphorelay, as shown by the arrows overlined by a plus sign, leading to torCAD induction. The circled P corresponds to the phosphoryl group. The arrows underlined by minus signs indicate the reverse phosphorelay model for the dephosphorylation of TorR-P when TMAO is removed.
One obvious advantage of a multistep relay system over a classical two-component system may be that it provides additional regulatory checkpoints. For example, phosphatases acting on the intermediate receiver or on the secondary transmitter might reduce or prevent phosphorylation of the response regulator. In Bacillus subtilis, at least three phosphatases (Rap family) can dephosphorylate the intermediate receiver Spo0F-P of the phosphorelay system involved in initiation of sporulation (14, 24). These phosphatases are inhibited by specific peptide pheromones and thus probably link sporulation and quorum sensing. In E. coli, no such acyl phosphate phosphatase has been found yet, but a phosphohistidine phosphatase, SixA, acting on the secondary transmitter of ArcB has recently been described (22). SixA might modulate the activity of the ArcB-ArcA system according to the availability of exogenous electron acceptors in anaerobiosis (19). When overproduced, SixA does not seem to down regulate the Tor system (our unpublished results). Moreover, we were unable to discover genes encoding TorS phosphatases either by a multicopy approach or after random mutagenesis (1, 2).
The Lin group has reported that the two additional phosphorylation sites of ArcB are involved in the transphosphorylation of ArcA not only but also in its dephosphorylation (8). Indeed, it has been clearly shown that, at least in vitro, the phosphoryl group of ArcA can return to the secondary transmitter and then to the receiver domain of ArcB, from which it is finally released. Similarly, the secondary transmitter domain of BvgS plays the role of a shuttle that can give or receive phosphoryl groups from the receiver domains of either BvgS or BvgA (34). In this study, we show that, in vivo, the receiver (D1) and secondary transmitter (H2) domains of TorS are also involved in the down regulation of the tor operon after TMAO removal. We thus propose that TorS can dephosphorylate TorR-P.
Transcription of the torCAD operon is rapidly stopped when TMAO is removed from the medium.
To monitor the effect of TMAO exhaustion on torCAD expression, we used strain LCB620, which contains a torA-lacZ chromosomal fusion (20). Cells were first grown anaerobically in rich medium supplemented with 10 mM TMAO at 37°C. When the culture reached an optical density at 600 nm of about 0.7, the bacteria were pelleted and washed two times in cold medium. The bacteria then were resuspended in prewarmed rich medium supplemented or not with 10 mM TMAO, and expression of the fusion was determined after various incubation times. In the presence of TMAO, the β-galactosidase activity measured on whole cells (21) increased slightly until the culture reached stationary phase, whereas in the absence of TMAO, the activity declined as growth continued, indicating that expression of the tor operon decreased when TMAO was removed from the growth medium (data not shown).
However, it was difficult to carry out a kinetic analysis of the effect of TMAO removal based on β-galactosidase activity, because this enzyme is stable and persists even if the tor operon is no longer expressed. Consequently, we carried out reverse transcriptase (RT) PCR experiments with converging oligonucleotides that hybridize to torC and lacZ gene sequences. Cells were treated as indicated above, and total RNA was extracted (High Pure RNA isolation kit; Roche Diagnostic) just before the washing step (Fig. 2, time zero) and 2, 10, or 20 min after cell resuspension in the prewarmed medium (Fig. 2). As expected, when cells were grown in the presence of TMAO, a strong amplification product corresponding to RT-PCR of tor operon transcripts was observed at all incubation times (Fig. 2, lanes 3, 7, 11, and 15). In contrast, no band appeared on the gel when cells were incubated for 10 or 20 min without TMAO, and a faint band was observed for cells incubated 2 min without TMAO (Fig. 2). These results clearly show that transcription of the tor operon is rapidly arrested when TMAO is removed. As the band intensity measured by densitometry from cells incubated for 2 min without TMAO is at least 10-fold lower than that of the positive control (Fig. 2, compare lanes 5 and 7), we propose that transcription of the tor operon is shut off in less than 2 min after TMAO removal and that the faint band comes from the small amount of RNA that has escaped degradation (the half-life of mRNA is generally very short [4]).
FIG. 2.
Kinetic analysis of tor operon transcription followed by 1% agarose gel electrophoresis. To evaluate tor operon expression of strain LCB620 (torA-lacZ), we carried out RT-PCR with two converging oligonucleotides, one corresponding to the torC coding sequence (positions 640 to 667) and the other complementary to the beginning of lacZ (positions 64 to 41). Reverse transcription was performed with Superscript II RT (Gibco BRL) from 100 ng of total RNA prepared with the High Pure RNA isolation kit (Roche Diagnostic). PCR amplification (30 cycles) was carried out from 1/10 of the product of reverse transcription. Lane 1, 1-kb DNA ladder (Gibco BRL); lane 3, RT-PCR experiment with total RNA prepared from cells grown in the presence of TMAO just before the washing step; lanes 5, 9, and 13, RT-PCR experiments with total RNA prepared from cells grown (respectively) 2, 10, and 20 min after TMAO removal; lanes 7, 11, and 15, same experiments as for lanes 5, 9, and 13, respectively, but with TMAO added again after the washing step; lanes 2, 4, 6, 8, 10, 12, and 14, same experiments as for lanes 3, 5, 7, 9, 11, 13, and 15, respectively, but without RT; lane 16, control PCR with genomic DNA.
Since TMAO induction of the tor operon is mediated by phosphorylated TorR (15), rapid dephosphorylation of TorR-P is probably necessary to stop tor operon expression when TMAO is removed. The time for spontaneous dephosphorylation of response regulators ranges from seconds to hours depending on the response regulator (13, 33). To define the half-life of TorR-P, we incubated purified TorR with a constitutively active form of TorS (TorS726) in the presence of [γ-33 P]ATP as previously described (15). Labeled TorR-P was then purified using a heparin column, and the half-life of the phosphoprotein was assessed after incubation in phosphorylation buffer (5 mM MgCl2) at room temperature and quantification of the remaining labeled TorR-P by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Surprisingly, the half-life of TorR-P was found to exceed 20 min (data not shown). The fact that the phospho-aspartyl bond of TorR-P is stable in vitro implies that another protein probably dephosphorylates TorR-P after TMAO removal.
Roles of sensor TorS and of its phosphorylation sites in the dephosphorylation of TorR-P in vivo.
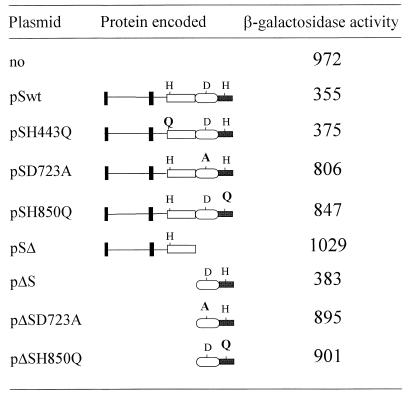
As many sensor proteins also play the role of phosphatase for their cognate response regulators in the absence of stimuli (5, 8, 30, 36), we hypothesized that TorS could dephosphorylate TorR-P when TMAO is missing. To study the role of TorS in vivo, we used strain LCB726 pcnB, which carries a constitutive allele of TorS (TorS726) and a torA-lacZ fusion (16). In this strain, TorS726 phosphorylates TorR even in the absence of TMAO and, as a result, β-galactosidase activity is always very high. Production of a phospho-TorR phosphatase should lower the amount of TorR-P and thus decrease expression of the fusion. As shown in Fig. 3, production of the wild-type TorS protein from plasmid pSwt led to a 2.7-fold decrease of the β-galactosidase activity of the torA-lacZ fusion. This result indicates that in the absence of TMAO, TorS can down regulate the tor operon, most probably by dephosphorylating TorR-P.
FIG. 3.
Effect of plasmid-borne TorS derivatives on expression of the torA-lacZ chromosomal fusion of strain LCB726 pcnB. Cells were grown anaerobically on minimal medium supplemented with glucose (0.2%). No TMAO was added to the growth medium since LCB726 pcnB carries a TMAO-constitutive chromosomal mutation (torS726) (16). The construction of the pS plasmid series has been described previously (15), whereas that of the pΔS plasmid series is described in the text. The representation of each TorS allele encoded by the plasmids is shown. To avoid artifactual effects due to overproduction of the plasmid-borne TorS alleles, a pcnB mutation was transduced into strain LCB726 to lower the plasmid copy number (18). Moreover, as the plasmidic torS alleles are under the control of the leaky tac promoter, only 0.005 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was used for the pS-plasmid series and no IPTG was added for the pΔS plasmid series. β-Galactosidase activities were measured on whole cells by the method of Miller (21); values represent the averages of at least three determinations with a variation of no more than 15% from the mean.
As TorS contains three phosphorylation sites, we investigated the possible role of each site in the dephosphorylation of TorR-P. From Fig. 3 it is clear that TorSH443Q encoded by pSH443Q still affected expression of the fusion, whereas the presence of either TorSD723A (pSD723A) or TorSH850Q (pSH850Q) only slightly decreased the fusion activity. Furthermore, production from pSΔ of a C-terminally truncated form of TorS, devoid of the D1 and H2 domains but containing an intact H1 domain, did not modify torA-lacZ expression. We concluded from these data that the primary phosphorylation site (H443) of TorS is not involved in the dephosphorylation of TorR-P whereas the other two are involved. Alternatively, mutations in the TorS phosphorylation sites D723 and H850 might result in structural changes in the TorS protein. However, this situation is reminiscent of that reported for the ArcB-ArcA system (8). Indeed, ArcA-P dephosphorylation occurs via a reverse phosphotransfer mechanism involving the phosphorylation sites of the secondary transmitter and the receiver domains of ArcB. Similar to the case for TorS, the histidine phosphorylation site of the primary transmitter is not involved in this process.
The TorS C-terminal D1-H2 region alone can dephosphorylate TorR-P in vivo.
To confirm that the TorS receiver (D1) and secondary transmitter (H2) domains are responsible for the dephosphorylation of TorR-P, we constructed the pΔS plasmid series by cloning different alleles of the torS 3′-end sequence (positions 2001 to 2712), encoding the D1-H2 region, downstream from the tac promoter of plasmid pJF119EH (7). The fragments were PCR amplified from chromosomal, pSD723A, or pSH850Q DNA with converging primers. The downstream primer (BamSH2) contains a BamHI site, whereas the upstream primer (EcoSD1) contains an EcoRI site, a methionine codon instead of the codon for Arg666, and a strong ribosome-binding site (AAGGAG) upstream from the start codon. The cloned regions of the resulting plasmids were sequenced to verify the absence of unexpected mutations. As shown in Fig. 3, production of the C-terminal region (D1-H2) of TorS from pΔS decreased the activity of the torA-lacZ fusion. Strikingly, the β-galactosidase activities are similar in the presence of either the entire TorS protein or the TorS C-terminal region. This suggests that the D1-H2 region and intact TorS dephosphorylate TorR-P with equal efficiencies. When either phosphorylation site of the TorS C-terminal region was mutated, expression of the fusion was almost unaffected. This confirms the key role of D723 and H850 in the dephosphorylation of TorR-P.
The fact that the D1-H2 region alone can dephosphorylate TorR-P in vivo gave us the opportunity to study the dephosphorylation process in a more relevant genetic context, i.e., a torS wild-type strain. When plasmid pΔS was introduced into strain LCB640 (torS+ torA-lacZ pcnB) (32), the β-galactosidase activity decreased fivefold in the presence of TMAO, whereas plasmids pΔSD723A or pΔSH850Q did not affect the activity of the fusion (Fig. 4). The negative effect of the D1-H2 region on the expression of the tor operon is thus more apparent in a torS wild-type context than in a torS constitutive strain. This difference probably originates from the fact that the phosphorylation capacity of TorS726 is higher than that of the wild-type TorS protein (compare the β-galactosidase activities in the absence of any plasmid in Fig. 3 and 4). In contrast, the dephosphorylation capacity of the D1-H2 domains probably remains constant in both strains. In conclusion, the D1-H2 region of TorS can efficiently dephosphorylate TorR-P in vivo, and this activity requires both phosphorylation sites of the D1-H2 domains.
FIG. 4.
Effect of plasmid-borne TorS truncated derivatives on expression of the torA-lacZ chromosomal fusion of strain LCB640 (torS+ pcnB). The experimental conditions were identical to those described for Fig. 3 except that 10 mM TMAO was added to the medium when indicated.
Dephosphorylation of response regulators by reverse phosphotransfer in phosphorelay systems: a general mechanism?
The data presented here together with our previous findings (15) strongly suggest that the TorS sensor catalyzes both the phosphorylation and dephosphorylation of its cognate response regulator, TorR. Under inducing conditions, TorS transphosphorylates TorR via a four-step phosphorelay in which His443, Asp723, and His850 of TorS are sequentially phosphorylated and phospho-His850 is the phosphodonor site for TorR. When TMAO is removed, TorS behaves as a phosphatase for TorR-P, but this activity requires only His850 and Asp723. As in vitro studies have clearly shown that certain unorthodox sensors can dephosphorylate their cognate response regulators by reverse phosphotransfer (8), we propose a similar pathway from TorR D53 to TorS H850 and then TorS D723 (Fig. 1).
In several phosphorelay signal transduction systems, the H1-D1 domains and the H2 domain are found on distinct proteins (3, 27, 29). Interestingly, in Vibrio harveyi, the receiver (D1) domain of hybrid (H1-D1) sensor LuxN is again involved in the dephosphorylation of response regulator LuxO (6). This additional example reinforces the idea that reverse phosphotransfer from the phospho-response regulator to the receiver of the hybrid or unorthodox sensor partner is a general mechanism of phosphorelay systems. A striking feature in at least several of these systems is that the half-lives of the phospho-response regulators are very long, whereas those of the sensor phospho-receivers are quite short (8, 13, 34). These findings could explain the reason for the existence of reverse phosphorelays. Indeed, as the response regulator cannot dephosphorylate easily by itself, the reverse phosphorelay allows the transfer of the phosphoryl group to a receiver domain from which the phosphate can readily be released. Finally, our kinetic analysis revealed that, at least for the TorS-TorR system, the proposed reverse phosphorelay is both efficient and rapid, since expression of the target genes was shut off in less than 2 min after stimulus removal. Therefore, in our model, based on our results and on analogy to other phosphorelay signal transduction systems, reverse phosphorelay is a general and powerful mechanism to dephosphorylate stable phospho-response regulators.
Acknowledgments
We thank D. Dubnau, C. Iobbi-Nivol, and L. Olivera for reviewing the manuscript and for helpful suggestions and B. Py for fruitful discussions.
This work was supported by grants from the Centre National de la Recherche Scientifique, the Université de la Méditerranée, and the MENRT (Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires). M.A. was supported by grants from the MENRT and from the Fondation pour la Recherche Médicale (FRM).
REFERENCES
- 1.Ansaldi M. Régulation multifactorielle du système respiratoire triméthylamine N-oxyde (TMAO) réductase chez Escherichia coli. Ph.D. thesis. Marseille, France: Faculté des Sciences de Luminy, Université Aix-Marseille II; 2000. [Google Scholar]
- 2.Ansaldi M, Bordi C, Lepelletier M, Méjean V. TorC apocytochrome negatively autoregulates the trimethylamine N-oxide (TMAO) reductase operon in Escherichia coli. Mol Microbiol. 1999;33:284–295. doi: 10.1046/j.1365-2958.1999.01468.x. [DOI] [PubMed] [Google Scholar]
- 3.Appleby J L, Parkinson J S, Bourret R B. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- 4.Arraiano C M, Yancey S D, Kushner S R. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J Bacteriol. 1988;170:4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castelli M E, Garcia V E, Soncini F C. The phosphatase activity is the target for Mg2+ regulation of the sensor protein PhoQ in Salmonella. J Biol Chem. 2000;275:22948–22954. doi: 10.1074/jbc.M909335199. [DOI] [PubMed] [Google Scholar]
- 6.Freeman J A, Lilley B N, Bassler B L. A genetic analysis of the functions of LuxN: a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol Microbiol. 2000;35:139–149. doi: 10.1046/j.1365-2958.2000.01684.x. [DOI] [PubMed] [Google Scholar]
- 7.Furste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host- range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 8.Georgellis D, Kwon O, De Wulf P, Lin E C C. Signal decay through a reverse phosphorelay in the Arc two-component signal transduction system. J Biol Chem. 1998;273:32864–32869. doi: 10.1074/jbc.273.49.32864. [DOI] [PubMed] [Google Scholar]
- 9.Georgellis D, Lynch A S, Lin E C C. In vitro phosphorylation study of the Arc two-component signal transduction system of Escherichia coli. J Bacteriol. 1997;179:5429–5435. doi: 10.1128/jb.179.17.5429-5435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoch J A. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 11.Hoch J A, Silhavy T J. Two-component signal transduction. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 12.Ishige K, Nagasawa S, Tokishita S, Mizuno T. A novel device of bacterial signal transducers. EMBO J. 1994;13:5195–5202. doi: 10.1002/j.1460-2075.1994.tb06850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janiak-Spens F, Sparling J M, Gurfinkel M, West A H. Differential stabilities of phosphorylated response regulator domains reflect functional roles of the yeast osmoregulatory SLN1 and SSK1 proteins. J Bacteriol. 1999;181:411–417. doi: 10.1128/jb.181.2.411-417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang M, Grau R, Perego M. Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis. J Bacteriol. 2000;182:303–310. doi: 10.1128/jb.182.2.303-310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jourlin C, Ansaldi M, Méjean V. Transphosphorylation of the TorR response regulator requires the three phosphorylation sites of the TorS unorthodox sensor in Escherichia coli. J Mol Biol. 1997;267:770–777. doi: 10.1006/jmbi.1997.0919. [DOI] [PubMed] [Google Scholar]
- 16.Jourlin C, Bengrine A, Chippaux M, Méjean V. An unorthodox sensor protein (TorS) mediates the induction of the tor structural genes in response to trimethylamine N-oxide in Escherichia coli. Mol Microbiol. 1996;20:1297–1306. doi: 10.1111/j.1365-2958.1996.tb02648.x. [DOI] [PubMed] [Google Scholar]
- 17.Kwon O, Georgellis D, Lin E C C. Phosphorelay as the sole physiological route of signal transmission by the Arc two-component system of Escherichia coli. J Bacteriol. 2000;182:3858–3862. doi: 10.1128/jb.182.13.3858-3862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopilato J, Bortner S, Beckwith J. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 19.Matsubara M, Mizuno T. The SixA phospho-histidine phosphatase modulates the ArcB phosphorelay signal transduction in Escherichia coli. FEBS Lett. 2000;470:118–124. doi: 10.1016/s0014-5793(00)01303-x. [DOI] [PubMed] [Google Scholar]
- 20.Méjean V, Iobbi-Nivol C, Lepelletier M, Giordano G, Chippaux M, Pascal M C. TMAO anaerobic respiration in Escherichia coli: involvement of the tor operon. Mol Microbiol. 1994;11:1169–1179. doi: 10.1111/j.1365-2958.1994.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 21.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 22.Ogino T, Matsubara M, Kato N, Nakamura Y, Mizuno T. An Escherichia coli protein that exhibits phosphohistidine phosphatase activity towards the HPt domain of the ArcB sensor involved in the multistep His-Asp phosphorelay. Mol Microbiol. 1998;27:573–585. doi: 10.1046/j.1365-2958.1998.00703.x. [DOI] [PubMed] [Google Scholar]
- 23.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 24.Perego M, Glaser P, Hoch J A. Aspartyl-phosphate phosphatases deactivate the response regulator components of the sporulation signal transduction system in Bacillus subtilis. Mol Microbiol. 1996;19:1151–1157. doi: 10.1111/j.1365-2958.1996.tb02460.x. [DOI] [PubMed] [Google Scholar]
- 25.Perraud A L, Kimmel B, Weiss V, Gross R. Specificity of the BvgAS and EvgAS phosphorelay is mediated by the C-terminal HPt domains of the sensor proteins. Mol Microbiol. 1998;27:875–887. doi: 10.1046/j.1365-2958.1998.00716.x. [DOI] [PubMed] [Google Scholar]
- 26.Perraud A L, Weiss V, Gross R. Signalling pathways in two-component phosphorelay systems. Trends Microbiol. 1999;7:115–120. doi: 10.1016/s0966-842x(99)01458-4. [DOI] [PubMed] [Google Scholar]
- 27.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 28.Robinson V L, Buckler D R, Stock A M. A tale of two components: a novel kinase and a regulatory switch. Nat Struct Biol. 2000;7:626–633. doi: 10.1038/77915. [DOI] [PubMed] [Google Scholar]
- 29.Rodrigue A, Quentin Y, Lazdunski A, Méjean V, Foglino M. Two component systems in Pseudomonas aeruginosa: why so many? Trends Microbiology. 2000;8:498–504. doi: 10.1016/s0966-842x(00)01833-3. [DOI] [PubMed] [Google Scholar]
- 30.Shi L, Liu W, Hulett F M. Decay of activated Bacillus subtilis Pho response regulator, PhoP∼P, involves the PhoR∼P intermediate. Biochemistry. 1999;38:10119–10125. doi: 10.1021/bi990658t. [DOI] [PubMed] [Google Scholar]
- 31.Simon G, Jourlin C, Ansaldi M, Pascal M C, Chippaux M, Méjean V. Binding of the TorR regulator to cis-acting direct repeats activates tor operon expression. Mol Microbiol. 1995;17:971–980. doi: 10.1111/j.1365-2958.1995.mmi_17050971.x. [DOI] [PubMed] [Google Scholar]
- 32.Simon G, Méjean V, Jourlin C, Chippaux M, Pascal M C. The torR gene of Escherichia coli encodes a response regulator protein involved in the expression of the trimethylamine N-oxide reductase genes. J Bacteriol. 1994;176:5601–5606. doi: 10.1128/jb.176.18.5601-5606.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanism of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D. C.: ASM Press; 1995. pp. 25–51. [Google Scholar]
- 34.Uhl M A, Miller J F. Central role of the BvgS receiver as a phosphorylated intermediate in a complex two-component phosphorelay. J Biol Chem. 1996;271:33176–33180. doi: 10.1074/jbc.271.52.33176. [DOI] [PubMed] [Google Scholar]
- 35.Uhl M A, Miller J F. Integration of multiple domains in a two-component sensor protein: the Bordetella pertussis BvgAS phosphorelay. EMBO J. 1996;15:1028–1036. [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Y, Qin L, Yoshida T, Inouye M. Phosphatase activity of histidine kinase EnvZ without kinase catalytic domain. Proc Natl Acad Sci USA. 2000;97:7808–7813. doi: 10.1073/pnas.97.14.7808. [DOI] [PMC free article] [PubMed] [Google Scholar]