Abstract
We report on the successful delivery of the Cre recombinase enzyme in the neural cells of mice in vivo by simple coinjection with peptides derived from HIV-TAT. Cre delivery activates the expression of a reporter gene in both neurons and astrocytes of the cortex without tissue damage and with a transduction efficiency that parallels or exceeds that of a commonly used adeno-associated virus. Our data indicate that the delivery peptides mediate efficient endosomal leakage and cytosolic escape in cells that have endocytosed Cre. The peptides, therefore, act in trans and do not require conjugation to the payload, greatly simplifying sample preparation. Moreover, the delivery peptides are exclusively composed of natural amino acids and are consequently readily degradable and processed by cells. We envision that this approach will be beneficial to applications that require the transient introduction of proteins into cells in vivo.
Peptides that promote rapid endosomal escape achieve intracellular protein delivery in mouse brain tissue.
INTRODUCTION
Understanding the physiology of the central nervous system, particularly the brain, is one of the current and most pressing challenges in medicine. This is exemplified by The Brain Initiative, a large-scale effort that aims at accelerating discoveries in neuroscience (https://braininitiative.nih.gov/). Probing how cells interact in both time and space, in health or disease, and in the context of neural networks is complex. In turn, it requires new research tools that allow the intracellular delivery of probes that can report on cells. Intracranial stereotactic injections of viruses, adeno-associated viruses (AAVs) in particular, are often used in studies that aim at manipulating and understanding cells of the central nervous system (CNS) in vivo (1–4). Genes transferred by these viruses may code for fluorescent reporters or genome editing tools, enabling the monitoring and modulation of cell processes. Despite their widespread use, AAVs have several limitations, including a relatively low packing capacity, a slow onset of transgene expression, low transduction efficiency in some pseudotypes, potential integration in the host genome, and induction of neuroinflammatory responses (5–8). Moreover, the sustained expression of transferred genes can be beneficial for certain applications but problematic for others. This is the case in gene-editing experiments, where the persistent presence of gene-editing machinery in cells can lead to off-target genome modifications (9–11). Conversely, the expression of some proteins of interest, such as transcription factors, often needs to be transient and temporally controlled (12–16). In principle, the direct delivery of proteins could circumvent some of these issues. In particular, proteins can initiate intracellular activities without transcription/translation delays. Proteins also have limited half-lives inside cells, thereby avoiding long-term effects (17, 18). A major limitation to using proteins as intracellular delivery payloads is the current lack of technologies that can introduce these macromolecules into the cytosol of cells with both high efficiency and low toxicity (19–21). Still, many delivery agents are capable of promoting the endocytic uptake of proteins into cells, yet protein payloads often remain trapped inside endosomes (22–25). These protein payloads are therefore unable to reach cytosolic or nuclear targets, and hence, they are unable to induce desired biological activities. To address this problem, we have developed peptides that selectively permeabilize the membrane of late endosomes. These peptides include dfTAT, a dimer of the cell-penetrating peptide (CPP) TAT (26). When externally administered to cells, dfTAT and protein payloads both accumulate into endosomal compartments. They then traffic together to late endosomes where dfTAT mediates membrane permeabilization and allows the escape of the protein payload into the cytosol (27). Notably, dfTAT is used without being conjugated or bound to a payload. Cytosolic delivery succeeds as long as both molecules are endocytosed by cells and reach the same endosome. This approach is a clear shift from strategies where carrier systems encapsulate a payload and carry it into cells (e.g., liposomes and nanoparticles). The uncoupling of the delivery agent and the payload, in turn, provides major benefits that include simple sample preparation, control over the amount of payload delivered, and delivery of chemically and functionally unaltered protein payloads (26). While useful in the context of in vitro cell cultures, it is unclear whether such a strategy can work in vivo, as delivery agent and payload may diffuse away from one another in a tissue and reach different cells separately. In addition, a major difference between in vitro tissue culture and in vivo applications is the time in which cells are exposed to delivery agents and payloads. In tissue culture, the concentration of delivery agent and cargo is relatively constant, whereas in vivo, these concentrations are likely changing because of inherent pharmacokinetic properties. It can be expected that this difference would have a major impact on delivery and toxicity. Considering these critical differences between in vitro and in vivo approaches, our goal in this study was to test this CPP platform in the context of a stereotactic intracranial injection and establish the feasibility of an in vivo protein delivery approach as effective as AAV transduction. We envision that this technology will have immediate impact by allowing the use of protein-based tools and probes for the manipulation and study of tissue physiology in vivo. Future work will determine the potential impact of this method as a therapeutic application, as it presents several advantages over viral and state-of-the art payload delivery.
RESULTS
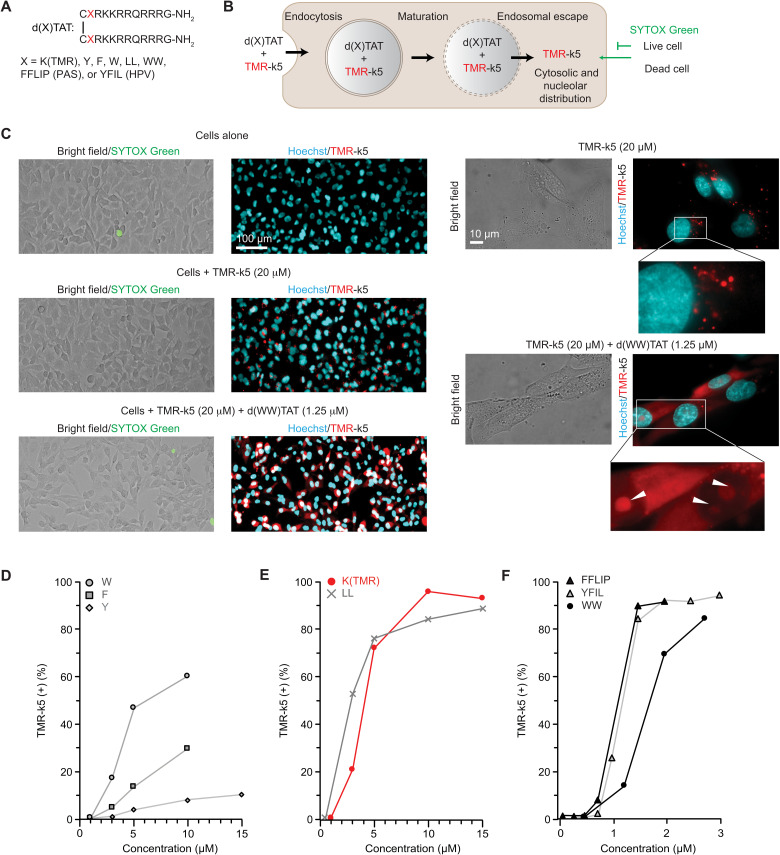
One of the benefits of protein delivery is their rapid intracellular action followed by their intracellular clearance and the absence of prolonged deleterious effects. In this context, it seems appropriate to use a delivery tool that would also degrade quickly once inside cells and leave little to no residue. In this regard, dfTAT is a peptide that degrades quickly during and after cell penetration (fig. S1). Originally, dfTAT contained two copies of carboxytetramethylrhodamine (TMR), a fluorophore that allowed microscopy visualization of the cell penetration process and mechanistic studies. However, the presence of TMR unnecessarily uses a fluorescence channel for microscopy applications in general. Moreover, fluorescent puncta, presumably from fragments degraded during endosomal transit and after cytosolic access, persist in and out of cells after delivery (fig. S1). We were concerned that this accumulation of fluorescent material may result in cellular artifacts in the context of in vivo injections. To avoid this potential problem, we aimed to develop dfTAT analogs that would not contain material foreign to cells and instead decompose into natural amino acids. However, initial testing established that simply removing TMR from dfTAT yields a relatively ineffective delivery agent (fig. S2). Given that hydrophobic moieties often modulate the activity of membrane active peptides (28–31), we hypothesized that the relative hydrophobicity of TMR may contribute to the endosomal membrane disruption activity of dfTAT. We synthesized a small library of analogs with the general structure of d(X)TAT where “X” is hydrophobic residues (Fig. 1A). “X” also includes short hydrophobic peptide sequences—the peptide accelerating sequence Pas (amino acid sequence: FFLIP) and a peptide derived from the capsid protein L2 of human papillomavirus type 33 (amino acid sequence: YFIL)—previously identified to enhance the cell penetration of CPPs (29, 30). To assess the delivery activity of these peptides (which are not directly visible by fluorescence microscopy like dfTAT), TMR-k5 was used as a model payload coincubated with d(X)TAT (to prevent proteolytic degradation of the peptide in cells, k5 is synthesized with d-lysine residues, as denoted by lowercase lettering). TMR-k5 is readily endocytosed by cells but lacks endosomal escape activity on its own (fig. S3). However, when delivered into the cytosol of cells, TMR-k5 distributes throughout the cytoplasm, reaches the nucleus, and stains nucleoli (Fig. 1B and fig. S4) (27, 32, 33). With this distinctive feature confirming intracellular access, the number of cells displaying fluorescent nucleoli in the presence of d(X)TAT reagents was counted (fig. S5). In turn, the percentage of cells with TMR-k5–stained nuclei was used as a proxy to measure the relative delivery efficiency of each d(X)TAT peptide (the detection of TMR-k5 itself being dependent on the detection threshold of the microscope used and the TMR-k5 concentration used during incubation). A SYTOX exclusion assay was used to assess peptide toxicity and to exclude dead cells from delivery quantification. Several peptides [d(X)TAT, X = Y, F, and W] remained less active overall than dfTAT (Fig. 1C). In contrast, d(LL)TAT, which contains two consecutive leucine residues, recapitulated the delivery activity of dfTAT (Fig. 1D). Last, several peptides (X = WW, FFLIP, and YFIL) were overall more active than dfTAT, achieving nuclear/nucleolar delivery of the TMR-k5 payload in a higher percentage of cells at extracellular concentrations of d(X)TAT 5- to 10-fold lower than those required for dfTAT (Fig. 1E). As previously shown with dfTAT (26), the d(X)TAT peptides lose activity when in their monomeric form (fig. S6). This shows that, from a structure-activity point of view, both position “X” and dimerization modulate the delivery activity of the peptides. Notably, d(X)TAT peptides showed little cytotoxicity at concentrations where cytosolic delivery is achieved in a high percentage of cells (fig. S7). The peptides d(LL)TAT and d(WW)TAT were chosen for subsequent studies as representative members of d(X)TAT peptides with moderate to high delivery activities.
Fig. 1. Structure-activity relationships of d(X)TAT establish impact of X on delivery activity.
(A) General structure of the d(X)TAT constructs where X consists of single amino acids or peptide sequences with various hydrophobicity. (B) Scheme highlighting the assay used to quantify the cell delivery activity of d(X)TAT peptides. TMR-k5 is used as a proxy to measure the relative delivery activity of d(X)TAT peptides. An endosomal escape mechanism is shown on the basis of previous results obtained with dfTAT. (C) Bright-field and fluorescence images showing cell morphology and TMR-k5 staining in MDA-MB-231 cells treated with d(WW)TAT. Cells are incubated with d(X)TAT and the probe TMR-k5 for 1 hour. Delivery of TMR-k5 into the cytosol of cells is detected by the diffusion of the probe into the cytosol and its distinct nucleolar staining. The viability of cells is quantified with a SYTOX exclusion assay immediately following incubation. Representative results are provided with ×100 fluorescence microscopy images of TMR-k5 incubated with and without d(WW)TAT. The fluorescence of TMR-k5 is pseudocolored red, while the nuclear stain Hoechst is pseudocolored cyan. A zoom-in inset shows nucleolar staining by TMR-k5, as highlighted by white arrows. Quantification of cell penetration of d(X)TAT peptides (D) for X = W, F, or Y; (E) for X = K(TMR) or LL; and (F) for X = WW, FFLIP (PAS peptide), or YFIL (HPV peptide). TMR-k5 (+) (%) corresponds to the percentage of cells detected as positive for nuclear/nucleolar TMR-k5 staining (percent of total). The data represent the average of biological N = 3 triplicates, with approximately 2700 cells being imaged and counted for each experiment. SDs are omitted for clarity, but all conditions had ≤12% SD, with the exception of d(W)TAT, which had ≤25%.
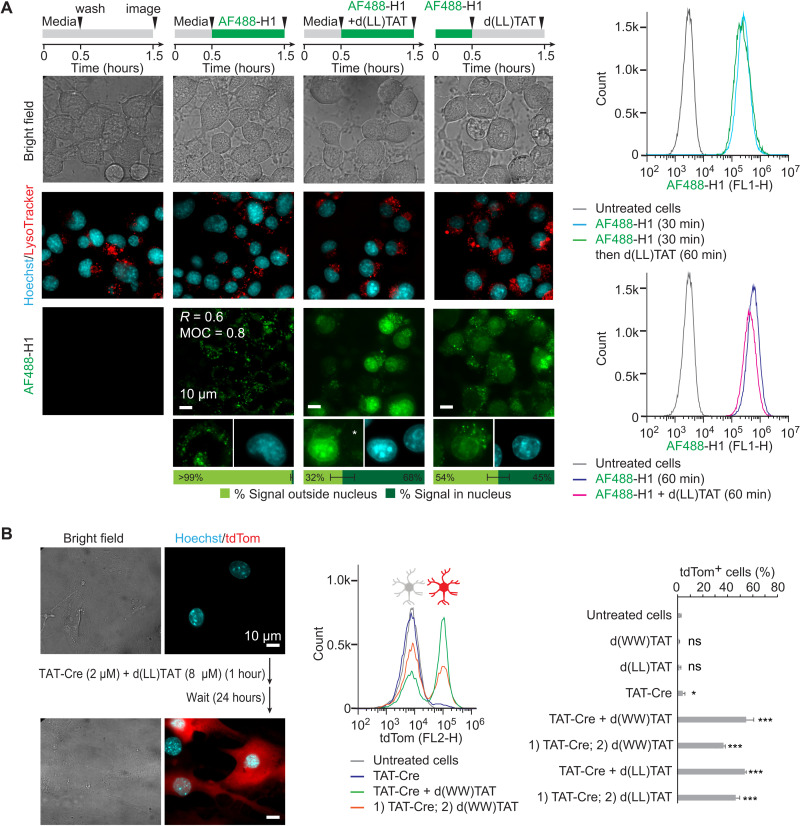
We first tested d(LL)TAT and d(WW)TAT in the context of protein payload delivery in vitro. We have previously reported that dfTAT achieves the endosomal release and cytosolic delivery of various proteins without requiring conjugation to or complexation with the payload itself (26). To test whether the new analogs d(LL)TAT and d(WW)TAT retain the same features, a fluorescently labeled histone [Alexa Fluor 488 (AF488)–H1] was selected as a model payload. Successful delivery of AF488-H1 was assessed by visualizing the nuclear accumulation of the protein by fluorescence microscopy (Fig. 2A). Incubation of Neuro2a cells with AF488-H1 alone leads to a punctate distribution consistent with endosomal entrapment, as demonstrated by colocalization with late endosome/lysosome markers (Fig. 2A and S8). In contrast, coincubation with d(LL)TAT leads to the nuclear accumulation of AF488-H1 with approximately two-thirds of the AF488-H1 fluorescence signal being nuclear while one-third remains endosomal. Incubation with AF488-H1 followed by incubation with d(LL)TAT also yields nuclear staining by AF488-H1. However, the portion of signal remaining trapped in endosomes is higher compared to the coincubation condition [similar results were obtained with d(WW)TAT; figs. S8 and S9]. Notably, the amount of AF488-H1 associated with cells is not increased by the presence of d(LL)TAT (as monitored by flow cytometry; Fig. 2A). Moreover, the nuclear fluorescent signal was confirmed to originate from intact AF488-H1, and AF488-H1 was estimated to accumulate at a low micromolar concentration in nuclei at the conditions tested (figs. S10 to S12). Overall, these data indicate that payload initially trapped in endosomes after preincubation can subsequently be released from endosomes into the cell cytoplasm and nucleus by the action of d(X)TAT delivery peptides. Cytosolic delivery is less efficient in the preincubation format, as shown by lower nuclear staining by AF488-H1. This is expected, as delivery peptide and payload are less likely to be present in the same endocytic organelles at the same time when endocytosed sequentially. These data support the notion that endosomal escape and cytosolic delivery remain possible even when endocytosis of the peptide and payload is not simultaneous. These data also demonstrate that d(X)TAT does not require complexation or interactions between peptide and payload for efficient uptake into cells. In addition, live cells displaying successful AF488-H1 nuclear delivery after 1 hour of incubation (as shown in Fig. 2A) conserve their morphology and do not die at later time points (as monitored 24 hours after performing the 1-hour delivery protocol; fig. S13). This indicates that the d(X)TAT peptides deliver AF488-H1 in a manner that does not kill cells immediately or with a delay.
Fig. 2. d(X)TAT constructs successfully deliver the histone H1 and the gene-editing enzyme TAT-Cre recombinase.
(A) Neuro2a cells were exposed to AF488-H1 (0.75 μM) and d(LL)TAT (8 μM) in coincubation or preincubation formats indicated. Representative bright-field and fluorescence images are provided, with AF488-H1 being pseudocolored green and Hoechst being pseudocolored cyan (bleed-through is not present as indicated by the white asterisk). For incubation of AF488-H1 alone, Pearson’s colocalization coefficient (R) and Manders’ overlap coefficient (MOC) were determined for colocalization with LysoTracker red. The average AF488-H1 signal intensity inside and outside nuclei for imaged cells is provided as quantified using total integrated intensity within manually drawn regions of interest in ImageJ. These data were obtained from biological N = 2 duplicates, with 100 cells being imaged per experiment. Flow cytometry was performed to show the relative fluorescence intensity of AF488-H1 in each condition. (B) Delivery of Cre recombinase into primary mouse cortical astrocytes. Cells only express the red fluorescent protein tdTom upon excision of a LoxP-Stop signal by Cre recombinase. Representative bright-field and fluorescence images (tdTom pseudocolored red and Hoechst pseudocolored cyan) and flow cytometry analysis of astrocytes exposed to TAT-Cre, with and without d(X)TAT peptides, are provided. Cells are analyzed 24 hours following Cre delivery to allow for tdTom expression. Delivery was performed using either the co- or preincubation format described in (A). The data represented are the averages and corresponding SDs of biological N = 2 duplicates. *P ≤ 0.05 and ***P ≤ 0.01. ns, not significant.
To further validate these results, we assessed the delivery of the gene-modifying enzyme Cre recombinase (Fig. 2B). The ability for the protein transduction domain (PTD) TAT to enhance endocytic uptake was exploited by fusion with the Cre recombinase enzyme (23). TAT-Cre delivery was tested in primary cortical astrocytes obtained from transgenic ROSA26-LSL-tdTomato (tdTom) mice (34). Intracellular delivery of Cre in these cells yields recombination of a reporter gene and subsequent expression of fluorescent tdTom. A 1-hour incubation of TAT-Cre alone with primary astrocytes yielded activation of tdTom expression in less than 5% of cells. This activation can be increased to 8 and 35% using TAT-Cre alone, but it requires incubation times of 16 and 24 hours, respectively (fig. S14). In contrast, addition of both d(WW)TAT and d(LL)TAT during the 1-hour incubation increases the percentage of tdTom-positive cells to 50% or greater. While approximately 15% of astrocytes were positive for SYTOX green staining immediately after delivery, the surviving cells remained viable 24 hours after the delivery protocol (fig. S15). Notably, TAT-Cre delivery was also improved by the addition of the delivery peptides after incubation with payload, suggesting that TAT-Cre and d(X)TAT do not need to interact to achieve cell entry. Consistent with this idea, TAT-Cre and dfTAT do not appear to bind one another in vitro, as shown in fig. S16. Moreover, Western blot analysis of TAT-Cre present in whole cells indicates that the protein is present in cells at similar levels with or without d(X)TAT (fig. S17). However, analysis of nuclear extracts shows that the portion of TAT-Cre that reaches the nucleus is substantially higher (and estimated to be in the low micromolar range) in the presence of d(X)TAT (fig. S17). As with AF488-H1, these results are consistent with TAT-Cre being endocytosed by cells independently of d(X)TAT. Hence, d(X)TAT does not change the level of endocytic uptake of TAT-Cre but simply causes the redistribution of the protein from endosomes to nucleus.
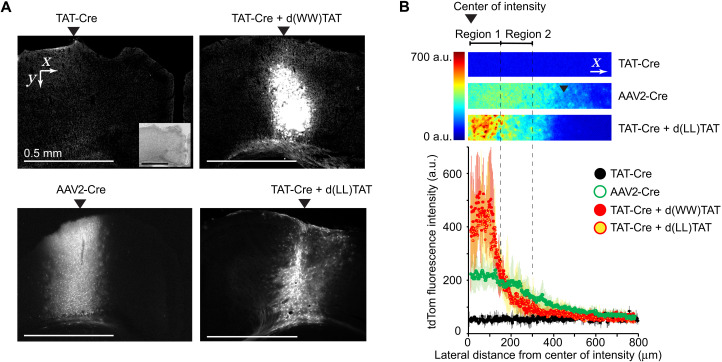
Having established successful Cre recombinase delivery in cell cultures, we next tested the performance of d(X)TAT/TAT-Cre cocktails in vivo using transgenic ROSA26-LSL-tdTom mice. Small volumes (0.4 μl) of d(X)TAT/TAT-Cre [200 μM d(WW)TAT or 400 μM d(LL)TAT, with 40 μM TAT-Cre] were injected into the sensorimotor portion of the cerebral cortex of anesthetized animals (Fig. 3). Stereotactic injections of an AAV engineered to deliver the Cre gene (AAV2-Cre and AAV pseudotype 2) were used for performance comparison (0.4 μl of AAV2-Cre, 0.5 × 1012 transduction units per milliliter as determined via quantitative polymerase chain reaction) (35). Two weeks after injections, animals were euthanized, and 25-μm-thick sections of brain tissues were analyzed by immunohistochemistry and fluorescence microscopy. As expected, injection of AAV2-Cre activated tdTom expression (Fig. 3A). Injection of TAT-Cre alone did not yield an observable red fluorescence, indicating that this PTD conjugate is ineffective on its own in vivo (Fig. 3A). In comparison, injection of d(LL)TAT/ TAT-Cre or d(WW)TAT/ TAT-Cre cocktail yielded tdTom expression at a region spanning several hundred micrometers (Fig. 3, A and B). The d(X)TAT peptides therefore enable the delivery of TAT-Cre into the cytosol and nucleus of cells in vivo at levels similar to those observed with viral transduction of Cre.
Fig. 3. Stereotactic injections of TAT-Cre/d(X)TAT cocktails into the brain of live mice achieve Cre-dependent recombination in vivo.
(A) Representative fluorescence images at ×2.5 magnification of brain slices of mice injected with TAT-Cre, with and without d(WW)TAT or d(LL)TAT. Injection of the AAV2-Cre is provided for comparison with state-of-the-art viral delivery technology. Inset: Bright-field image of the imaged brain slice. (B) Quantification of the tdTom fluorescence intensity in ×2.5 images. The results represent the average and corresponding SD of N = 3 independent injections (cropped images provided as examples, pseudocolored by fluorescence intensity). Measurements are made from the center of the area with the highest intensity in the direction perpendicular to the injection. a.u., arbitrary units.
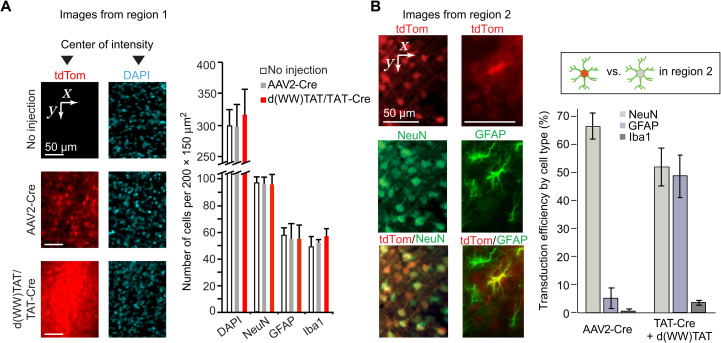
Two distinct regions of tdTom fluorescence were observed, region 1 and region 2, moving laterally away from the injection site, respectively (Fig. 3B). In region 1, tdTom fluorescence intensity is higher for d(X)TAT/TAT-Cre than for AAV2-Cre (Fig. 3B). It is the opposite in region 2, suggesting a more contained diffusion in the d(X)TAT/TAT-Cre groups. To further characterize the differences between d(X)TAT/TAT-Cre and AAV2-Cre, immunostaining of neuronal nuclear protein (NeuN), glial fibrillary acidic protein (GFAP), ionized calcium-binding adapter molecule 1 (Iba1), CD68, and 4′,6-diamidino-2-phenylindole (DAPI) was performed to identify neurons, astrocytes, microglia, infiltrated macrophages, and nuclei, respectively. Given that d(X)TAT displayed some toxicity at high concentrations in vitro, a concern was that the high fluorescence intensity of region 1 arises from tissue damage (e.g., tissue remodeling and increase in cell density). However, the density of cells, as detected by DAPI staining, was not different between tissue injected with d(X)TAT/TAT-Cre, tissue injected with AAV2-Cre, and noninjected tissue (Fig. 4A). Moreover, the densities of neurons, astrocytes, and microglia were also consistent between these conditions (Fig. 4A and fig. S18). As expected, the mechanical disruption caused by the needle does damage the tissue (36, 37), and the regions at close proximity to the needle tracts display some inflammatory response in all the groups. However, no CD68 staining was observed in regions of tdTom expression away from the needle tracts (fig. S19). In addition, no significant differences were observed in between the peptides or AAV-injected animals in the number of neurons and astrocytes expressing proapoptotic marker cleaved caspase 3 (fig. S20) or in the expression of proinflammatory markers [interleukin-6 (IL-6), tumor necrosis factor–α (TNF-α), and IL-1β; fig. S21]. Together, this demonstrates that injected brains did not show signs of major tissue damage, cell death, or active immune responses (besides the needle tract) and indicates that the d(X)TAT/TAT-Cre cocktails were relatively innocuous similar to what was observed after AAV injections.
Fig. 4. In vivo d(X)TAT protein delivery targets both neurons and astrocytes.
(A) Analysis of cell density in the area of highest tdTom fluorescence intensity, defined as region 1 in Fig. 3B. DAPI is used to stain all cells, while GFAP and Iba1 are used as markers of astrocytes and microglia cells, respectively. Brain tissue injected with d(WW)TAT/TAT-Cre is compared to AAV2-Cre injections and noninjected tissue. The data presented are the average cell densities in 200 μm–by–200 μm areas from injection slices obtained from two different animals. No statistical significance was seen between regions for each type of marker. (B) Identification and quantification of the cells edited by Cre in region 2 (as defined in Fig. 3B). Brain slices immunostained for markers of neurons (NeuN), astrocytes (GFAP), and microglia (Iba1) were imaged for colocalization with tdTom by fluorescence microscopy. Representative images are provided for TAT-Cre/d(WW)TAT injections, with tdTom pseudocolored red and NeuN and GFAP stains pseudocolored green. Colocalization of cells positive for tdTom fluorescence and cell-specific markers was used to determine the percentage of neurons and astrocytes successfully gene edited within region 2. Conversely, the transduction efficiency, using tdTom expression as a proxy for Cre delivery, was assessed by measuring the percentage of tdTom cells among all cells stained for cell-specific markers. The data represent the averages obtained from N = 3 animals per group, with each injection being imaged at a minimum of five distinct locations showing tdTom fluorescence. Coordinates are overlaid in each panel to indicate the direction of the images in relation to the center of intensities.
Consistent with the virus pseudotype used and its reported tropism, the control AAV2-Cre transduced approximately 70% of the neurons present in regions 1 and 2 but less than 5% of astrocytes and microglia (Fig. 4B). The red fluorescence observed in region 1 for d(X)TAT/TAT-Cre is densely distributed, making the identification of individual tdTom cells difficult. In particular, all cells overlap with some red signal, with nonfluorescent cells being either absent or masked by their fluorescent neighbors. Consequently, we were unable to reliably quantify the number of tdTom-positive cells and the type of cells successfully transduced in region 1. In contrast, the tdTom fluorescence of region 2 was more dispersed, permitting the identification of distinct cells and the quantification of colocalization with NeuN, GFAP, and Iba1. d(WW)TAT transduced TAT-Cre in approximately 50% of the neurons and astrocytes present in region 2 (Fig. 4B). The percentage of tdTom-positive microglia cells was below 5%. The ability of d(X)TAT to transduce multiple cell types likely contributes to the high fluorescence signal observed in region 1. It is also likely that this high signal is the result of transduction efficiencies that surpass those detected in region 2 and surpass those detected for AAV2-Cre.
DISCUSSION
We present a new nonviral method for efficient protein delivery into cells of the CNS. This method involves an endosomolytic peptide simply mixed with a protein payload. We use the payload Cre recombinase as a tool to spatially turn on gene expression in a genetically modified organism. We demonstrate that this method is as efficient at activating tdTom expression as an AAV gene delivery protocol. This approach offers some potential benefits: (i) It allows the rapid onset of intracellular biological activity (e.g., Cre enzyme is delivered in its active form and induces recombination rapidly after entering a cell, while AAV2-Cre requires a time delay for Cre recombinase to be expressed from the cre gene). (ii) Proteins are delivered directly and transiently into cells and, in the context of transcription factors or gene-editing tools, can potentially avoid unwanted off-target and knock-on effects otherwise caused by prolonged expression (9–11, 16). (iii) Delivery peptides and protein payloads are straightforward to produce by standard solid-phase peptide synthesis and recombinant technologies, which considerably reduce production time and costs associated with viral strategies. (iv) The delivery approach does not involve potential risks associated with DNA integration with the host genome. Please note that several widely used natural or engineered AAVs (e.g., AAV9, AAV5, AAV-DJ, etc.) have benefits that the current approach does not have: desirable biodistribution in large areas of tissue, controlled tropism, and the ability to cross the blood-brain barrier (this eliminates the need for invasive and time-consuming stereotactic brain injections procedures) (38–40). The protocols described here are therefore not meant to supplant the AAV technology but rather to complement it. For instance, delivery tools such as d(X)TAT could provide new opportunities in terms of the types of cargos that can be delivered to cells of the CNS. It would be of particular interest to deliver payloads that are not packaged by AAVs (e.g., DNA > ~5 kb, mRNA or long noncoding RNAs, large transcription regulator, etc.) (41). In this regard, various proteins, peptides, oligonucleotides, cell-impermeable small molecules, and large nanoparticles (~100 nm in diameter) have been successfully delivered into cells in vitro using dfTAT coincubation protocols (26, 33, 42, 43). It will be interesting to test in the future whether these in vitro results can be replicated in vivo. Notably, previous studies have also achieved gene editing in the CNS by delivery of Cas9 ribonucleoproteins or Cre recombinase in vivo (44, 45). However, these approaches require the modification of proteins with fusion tags that can potentially interfere with protein function and the use of lipid-based transfection agents, which may persist inside cells for long periods of time and elicit a variety of cellular responses (46, 47). In contrast, the CPPs used here can be quickly degraded following delivery (dfTAT has already been established to have little impact on cell physiology), and the protein cargo does not require modifications (26). We therefore expect to assess whether these features yield benefits for in vivo gene-editing applications in future studies.
The reagent dfTAT was originally designed to mediate the cellular delivery of macromolecules into cell cultures by a simple coincubation protocol. The delivery format of coincubation between this endosomolytic agent and macromolecular payload seems appropriate in this context, as cells within a dish are continually exposed to the incubation cocktail. Delivery agent and payload can therefore be endocytosed together without having to interact physically with one another, as dfTAT renders late endosomes leaky, allowing the payload to be released into the cytosol of cells. As a matter of fact, binding interactions between peptide and extracellular components tend to result in a decrease in delivery activity (fig. S16D) (26, 43). Therefore, the ideal cocktail for this approach is likely one where there is no or little interaction between delivery peptide and payload. This format offers several benefits including the elimination of conjugation protocols between delivery agent and payload, the function of the delivered payloads not being perturbed by labeling with the delivery agent, and the concentrations of payload that enter cells being independent of delivery agent and therefore tunable (26, 43). However, stereotactic injections are different, as delivery agent and payload are, in principle, free to diffuse away from the site of injection and, importantly, away from each other. Therefore, cells that internalize enough delivery agent but little payload, or vice versa, can fail to show a positive cytosolic delivery outcome (see the Supplementary Materials for additional discussion accompanying fig. S20). Nonetheless, our data show that, under the conditions used, the cocktail delivery format is capable of achieving successful protein delivery in a region of tissue that spans a distance comparable to that achieved with an all-in-one AAV delivery vector. The in vitro data presented here support the notion that d(X)TAT reagents function similarly to dfTAT, i.e., following a two-step process of endocytosis followed by endosomal escape. In this regard, glial cells have an endocytic pathway with unique features (e.g., high pH and high levels of proteases), and this may explain the low transduction observed in microglia in vivo (48). However, we do not rule out the possibility that other mechanisms of cell entry may be involved in vivo, especially in areas of high local peptide concentration. For instance, it is possible that the d(X)TAT reagents could promote cytosolic delivery of payloads by transiently disrupting the plasma membrane of cells (a process reported for CPPs) (49–51). d(X)TAT and payload could also potentially form particles in vivo that are not detected in vitro before injection, giving rise to the possibility that d(X)TAT and TAT-Cre could travel together to reach the same cells.
We currently do not know how much peptide or payload is present in the interstitial space of injected tissue in vivo and how this changes over time. As seen in fig. S14, TAT-Cre alone must be exposed to cells for an extended period of time to induce tdTom expression in vitro. The mice treated with TAT-Cre alone did not show any tdTom fluorescence. Together, these results suggest that, in vivo, delivery of TAT-Cre alone does not work because the enzyme is not in contact with cells long enough to enter cells. In the case of injection with d(X)TAT peptides, it is possible that the exposure of TAT-Cre to cells is lengthened. Instead, we propose that d(X)TAT delivers TAT-Cre in the narrow time frame during which cells are exposed to the enzyme in vivo. This is because the peptides deliver protein payloads in less than an hour in vitro. Overall, this may prove to be useful for biologics that are poorly stable in vivo and that would require rapid delivery. Note that d(WW)TAT displayed some toxicity against astrocytes in vitro after 1 hour of incubation (<15% cell death). In contrast, we did not detect deleterious effects on injected tissue. It is therefore possible that the contact time between peptide and cells is shorter in vivo than in vitro and that a short in vivo exposure time also mitigates potentially harmful effects. In addition to exposure time, it is likely that several factors modulate the efficiency of delivery as peptide and payload diffuse away from the injection site (a simple model is provided in fig. S20). Peptide stability is also likely a major factor in delivery efficiency in vivo. Ideally, the peptide used should be fully degradable to prevent unwanted downstream effects yet stable enough to successfully deliver the payload in a timely manner. While intracellular degradability was a feature that we sought for in this study, it is possible that formulations that reduce extracellular degradation could enhance the ability of the peptide/payload cocktail to diffuse further away from the site of injection and expand the region of successful delivery. The results presented here reveal how the residues present at position X in the d(X)TAT constructs modulate the efficiency of endosomal escape of these peptides. An area of possible improvement for future d(X)TAT-like delivery agents is to tune this position to further decrease the concentration threshold at which cytosolic delivery is achieved. On the basis of the simple model presented in fig. S22, this would likely increase the size of the region of successful delivery for the same given concentration of peptide. Last, it is interesting to speculate that the cocktail format presented could be modified to achieve cell-specific delivery. In particular, we envision that payloads targeted for endocytosis by specific cell surface receptors could be coincubated with d(X)TAT. Delivery could then be achieved into neurons or astrocytes specifically. Multiple payloads may also be able to enter different cell types simultaneously during a single injection. We will pursue testing such possibilities in future studies.
MATERIALS AND METHODS
Peptide design, synthesis, purification, and quantitation
All peptides were synthesized on a rink amide 4-methylbenzhydrylamine (MBHA) resin (Novabiochem, San Diego, CA) by solid-phase peptide synthesis using previously described protocols (52). Briefly, the TAT sequence on which all peptides were built on was assembled using standard 9-fluorenyl methoxycarbonyl (Fmoc) protocols containing Fmoc-Lys-(Boc)-OH, Fmoc-Gly-OH, Fmoc-Arg(Pbf)-OH, and Fmoc-Gln-(Trt)-OH (Novabiochem). To this TAT backbone, Fmoc-Tyr(tBu)-OH, Fmoc-Phe-OH, Fmoc-Trp(Boc)-OH, Fmoc-Leu-OH, Fmoc-Ile-OH, Fmoc-Lys(Ac)-OH, and Fmoc-Pro-OH were added where applicable for specific peptides. For fluorophore coupling, Fmoc-Lys(Mtt)-OH was added for selectable deprotection using 1.5% trifluoroacetic acid (TFA) and subsequent coupling [5(6)-carboxytetramethylrhodamine (TMR; Novabiochem)] to the ɛ-amine using standard coupling protocols. Boc-Cys(Trt)-OH was added to the N terminus of all peptides for downstream dimerization following TFA cleavage from the resin. Cleaved monomers were dissolved in aerated phosphate-buffered saline (PBS) at pH 7.6 (~1 mg/ml), and the reaction was agitated on a nutator overnight. The pentalysine constructs were synthesized in the same manner using Fmoc-d-Lys(Boc)-OH with either 7-diethylaminocoumarin-3-carboxylic acid (DEAC) or TMR conjugated directly to the N terminus.
The peptides dfTAT [NH2-CK(ε-NH-TMR)RKKRRQRRRG-NH2], d(F)TAT (NH2-CFRKKRRQRRRG-NH2), d(Y)TAT (NH2-CYRKKRRQRRRG-NH2), d(W)TAT (NH2-CWRKKRRQRRRG-NH2), d(WW)TAT (NH2-CWWRKKRRQRRRG-NH2), d(LL)TAT (NH2-CLLRKKRRQRRRG-NH2), d(PAS)TAT (NH2-CFFLIPRKKRRQRRRG-NH2), d(HPV)TAT (NH2-CYFILRKKRRQRRRG-NH2), dK(Ac)TAT [NH2-CK(Ac)RKKRRQRRRG-NH2], DEAC-k5 (DEAC-kkkkk-NH2), and TMR-k5 (TMR-kkkkk-NH2) were analyzed by reverse-phase high-performance liquid chromatography (HPLC) on an Agilent HP 1200 series instrument and an analytical Phenomenex Luna Omega Polar C18 column (5 μm, 4.6 mm by 250 mm). The flow rate was 2 ml/min, and detection was at 214, 430, and 550 nm. Peptide purification was performed on a Thermo Fisher Scientific UltiMate 3000 HPLC on a Phenomenex Luna Omega Polar C18 column (5 μm, 21.2 mm by 250 mm). The flow rate was 20 ml/min, and detection was at 214, 430, and 550 nm. All runs used linear gradients of 0.1% aqueous TFA (solvent A) and 90% acetonitrile, 9.9% water, and 0.1% TFA (solvent B). The correct identity of all peptides was confirmed by electrospray ionization on a Thermo Fisher Scientific Exactive Orbitrap. For concentration determination, all peptides were reduced to their monomeric form using tris(2-carboxyethyl)phosphine. Concentrations were then obtained using quantitative amino acid analysis (Protein Chemistry Lab, Texas A&M University; figs. S21 to S29).
Expression and purification of TAT-Cre recombinase
The gene for TAT-Cre from pTriEx-HTNC (Addgene) was cloned into the vector pTXB1 and transformed into Escherichia coli BL21 (DE3) cells (Agilent Technologies) using a standard heat shock method. LB media containing ampicillin (100 μg/ml) were inoculated with colonies containing the plasmid and allowed to grow shaking at 37°C overnight. Cultures were then used to inoculate 1 liter of LB containing ampicillin and allowed to grow until OD600 (optical density at 600 nm) = 0.6. Cultures were then induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (Fisher Scientific) and grown, shaking, at 37°C for 3 hours. E. coli cells were then pelleted by centrifugation using a J2-21 (Beckman) centrifuge and a JA-10 rotor at 4000 rpm for 30 min at 4°C. Cell pellets were then resuspended in buffer containing 20 mM NaH2PO4, 500 mM NaCl, and 20 mM imidazole, and then, cells were lysed by sonication for a total sonication time of 8 min. Cell debris was removed from the whole-cell lysate by centrifugation using a JA-20 rotor at 17,000 rpm for 45 min at 4°C. TAT-Cre was then purified using metal affinity chromatography (HisTrap HP; GE Healthcare).
Adeno-associated virus
AAV2-Cre was produced at the Salk Institute Viral Vector Core as described previously (35, 53).
Cell line culturing and maintenance
MDA-MB-231 (breast cancer–derived epithelial), HeLa (cervical cancer–derived epithelial), and Neuro2a (mouse neuroblast) cells were obtained from the American Type Culture Collection. Ai14 astrocytes were isolated from the brain cortex of Ai14flox/flox mice (from JAX, #007908), according to reported protocols (34). All cells were cultured in Dulbecco’s modified Eagle medium (DMEM; Thermo Fisher Scientific, catalog no. SH3002201) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) and penicillin-streptomycin (Thermo Fisher Scientific) in a humidified incubator at 5% CO2 and 37°C. All cell lines tested negative for mycoplasma via MycoAlert (Lonza). For experiments, cells were seeded in a plastic 48-well dish and a Lab-Tek II glass eight-well plate for high-magnification imaging (Fisher).
Fluorescence microscopy
For quantitative analysis, cells were imaged using an EVOS M7000 microscope (Thermo Fisher Scientific) using ×10, ×20, and ×40 objectives. The microscope is equipped with a heated chamber maintained at 37°C. Images were acquired using bright-field imaging and five fluorescence filter sets: cyan fluorescent protein [CFP; excitation (Ex) = 445 ± 22 nm and emission (Em) = 510 ± 21 nm], red fluorescent protein (RFP; Ex = 531 ± 20 nm and Em = 593 ± 20 nm), green fluorescent protein (Ex = 470 ± 11 nm and Em = 525 ± 25 nm), DAPI (Ex = 357 ± 22 nm and Em = 447 ± 30 nm), and Cyanine 5 (Ex = 628 ± 20 nm and Em = 685 ± 20 nm). High-magnification (×100) imaging was performed on a model IX81 Olympus microscope equipped with a Retiga R6 charge-coupled device camera (Teledyne QImaging) and a heated stage maintained at 37°C. Images were acquired using bright-field imaging and four fluorescence filter sets: CFP (Ex = 436 ± 10 nm and Em = 480 ± 20 nm), RFP (Ex = 560 ± 20 nm and Em = 630 ± 35 nm), fluorescein isothiocyanate (Ex = 488 ± 10 nm and Em = 520 ± 20 nm), and DAPI (Ex = 350 ± 50 nm and Em = 460 ± 25 nm). For z stacks and rendering of three-dimensional volumes at ×100, images were taken at 0.2-μm intervals on the z axis with the disk-spinning unit (IX2-DSU) engaged. All images captured for an experiment at a particular magnification were collected using identical neutral density, gain, and exposure times. These values were set to yield the highest intensity achievable without pixel saturation. Images were analyzed and processed using either SlideBook 6.0 (3i), Celleste 4.0 software (Thermo Fisher Scientific), or ImageJ (National Institutes of Health). The reproducibility of all the experiments was assessed by performing experiments with independent batches of cell cultures on three different days (i.e., biological triplicates).
Determination of delivery efficiencies in cell culture
MDA-MB-231 cells were washed three times with Leibovitz’s nrL15 media (nr, without cysteine; nrL15, Thermo Fisher Scientific). Cells were then incubated with peptide and 20 μM of the pentalysine probe at 37°C for 1 hour. This was followed by washing the cells two times with nrL15 supplemented with heparin (1 mg/ml; Sigma-Aldrich) to remove extracellular peptides and one time with nrL15 to remove heparin. Cells were treated with the cell-impermeable nuclear stain SYTOX Blue (Thermo Fisher Scientific), SYTOX Green (Thermo Fisher Scientific), or DRAQ7 (Invitrogen) to identify cells that had a compromised plasma membrane (i.e., dead cells). In parallel, the cell-permeable dye Hoechst 33342 (Thermo Fisher Scientific) was used for nuclear staining. Cells were imaged as described above. Briefly, cells that displayed nucleolar staining by the pentalysine peptide, while excluding SYTOX or DRAQ7 stain, were considered positive for delivery if above a threshold determined by TMR-k5 alone. Cells stained by the SYTOX or DRAQ dyes were considered dead. Penetration efficiency was calculated by dividing the number of cells that show pentalysine nuclear staining by the total number of cells present (Hoechst-stained cells, ×20 images). Likewise, cells stained with SYTOX or DRAQ7 dyes were counted to determine the percentage of dead cells. Penetration was not quantified for conditions where d(X)TAT peptides cause more than 20% cell death, as fluorescent cellular debris renders analysis unreliable. In the TMR-k5 delivery assay, the number of molecules that enter cells and reach the nucleus/nucleolus is not directly quantified. Instead, what is quantified is the number of cells in which TMR-k5 is detected to be present above a detection threshold. This threshold is dependent on the microscope, imaging conditions, and the concentration of TMR-k5 used during incubation (fig. S5).
Histone AF488-H1 delivery
Neuro2a were washed three times with nrL15 media (Thermo Fisher Scientific). Following washing, the wells were either (i) preincubated for 30 min with AF488-labeled histone H1 at 0.75 μM followed by a 1-hour peptide incubation with 2.5 μM d(WW)TAT or 8 μM d(LL)TAT or (ii) coincubated for 1 hour with histone and peptide at the same concentrations. After the incubation with histone and/or peptide, the cells were washed two times with nrL15 supplemented with heparin (1 mg/ml; Sigma-Aldrich) and one final nrL15 wash to remove heparin. For fluorescence imaging, a live/dead stain DRAQ7 (BD Accuri) was used to stain dead cells after the final wash. Cells were then imaged as described above.
Cre recombinase splicing assay in cell culture
Ai14 astrocytes were washed three times with L15. TAT-Cre (2 μM) recombinase was either preincubated or coincubated with either 2.5 μM d(WW)TAT or 8 μM d(LL)TAT for 1 hour. Cells were washed with heparin (1 mg/ml) and nrL15 and imaged for tdTom fluorescence. Following delivery of TAT-Cre, cells were returned to DMEM/10% fetal bovine serum and incubated overnight in normal growth conditions for later imaging. For wells used for imaging, the following day (24 hours), cells were washed with nrL15 and imaged for the presence of tdTom fluorescence. After imaging, cells were trypsinized in 75 μl of 0.5% trypsin for 5 min, resuspended in 300 μl of nrL15, and analyzed using a BD Accuri C6 flow cytometer. Cytometric readings were gated for cell events based on an untreated sample and tdTom fluorescence. Flow cytometry data were processed using FlowJo.
Stereotactic injections in mice
Mice used for stereotactic injections were 8- to 10-week-old male and female Ai14flox/flox (from JAX, #007908). Mice were anesthetized with isoflurane anesthetic gas (3% induction and 1 to 2% maintenance face mask). During the surgery, mice were kept on a warming pad at 37°C. The hair overlying the skull was removed with an electric shaver. The surgical site was prepared with Betadine followed by medical-grade, prepackaged isopropanol wipes. The mice were placed on a stereotaxic device (Harvard Apparatus), and ocular lubricant was applied to both eyes to prevent drying. The scalp was incised (midline incision), and small burr holes were drilled in the skull overlying the appropriate areas. Small volumes (0.4 μl) of appropriate delivery mix were injected into the sensorimotor portion of the cerebral cortex. The injections were made via a Hamilton micro syringe modified with glass capillaries. The right sensorimotor cortex was injected 0.7 mm below the cortical surface at one coordinate relative to bregma: 0.1 mm anterior (AP)/1.2 mm lateral (ML). A total of 0.4 μl of AAV-Cre or payload/peptide cocktails was injected per injection site over a duration of 5 min [peptide concentrations were chosen relative to activity seen in cells: 200 μM for d(WW)TAT, 400 μM for d(LL)TAT, and 40 μM for TAT-Cre]. After injection, the scalp was closed with silk sutures (Monocryl 5.0 absorbable) and Dermabond. For perioperative analgesia, mice received 0.5 ml of 0.9% sodium chloride solution, 0.1 ml of buprenorphine (0.1 mg/kg body weight dissolved in 0.9% NaCl solution), and 0.1 ml of penicillin. All procedures and instruments used for the surgery were sterilized before the surgery to prevent secondary infection. After surgery, mice were allowed to recover in a warmed cage and then were moved to a normal cage provided with food and water.
Immunohistochemistry
Fourteen days after injection, mice were euthanized. Mice were given a lethal dose of Fatal-Plus and perfused transcardially with 4% paraformaldehyde, consistent with the recommendations of the American Veterinary Medical Association (AVMA) Guidelines for the Euthanasia of Animals. Semiserial coronal sections of the brains (0.5 mm rostrocaudal, centered around the injection site) were cut with a cryostat (50 μm thick). Every other section per animal (one section every 50 μm) was stained with Iba1, GFAP, and NeuN using different combinations of primary and secondary antibodies for costaining (four different channels) to maximize the use of the tissue. After washing with PBS, sections were treated with a blocking buffer containing 5% normal horse serum (Sigma-Aldrich) and 0.4% Triton X-100 (Sigma-Aldrich) for 1 hour. Incubation with primary antibodies was conducted overnight at 4°C with dilutions as follows: rat anti-GFAP (1:500; Thermo Fisher Scientific, 130300), rabbit anti-NeuN (1:500; Cell Signaling Technology, 24307S), rat anti-CD68 (1:500; Bio-Rad, MCA1957), and rabbit anti-Iba1 (1:500; FUJIFILM Wako Chemicals, 019-19741). After washing with PBS, sections were incubated with goat anti-rat AF488 (1:500; Thermo Fisher Scientific, A11006) and goat anti-rabbit AF647 (1:500; Thermo Fisher Scientific, A32732) secondary antibodies for 1.5 hours at room temperature, washed again then stained with DAPI (1:5000; Thermo Fisher Scientific, D3571) for 10 min, washed, and lastly mounted in Fluoromount-G (Southern Biotechnology, 0100-01). Negative controls for unspecific binding of the secondary antibody were conducted in parallel sections following the same procedure described above, except for the incubation in primary antibodies. Sections were imaged on a Zeiss Axio Observer fluorescent microscope and processed using Zeiss ZEN Blue software.
Quantifications were performed using ZEN Blue or ImageJ. The total number of NeuN/tdTom coexpressing cells, the total number of GFAP/tdTom coexpressing cells, and the total number of Iba1/tdTom coexpressing cells were quantified in sections displaying tdTom fluorescence. Relative region intensities were quantified using ImageJ. Images from different injections were compared with identical contrast settings in ImageJ. The “Jet” LUT was applied to more easily distinguish the regions of highest and lowest intensity. The “Plot Profile” tool was then used to quantify the intensity over the regions of interest, measuring outward from the epicenter of fluorescence.
Three brain sections from each injected animal were costained for cleaved caspase-3 (1:500; Cell Signaling Technology, 9661), NeuN, GFAP, and DAPI, as described above. These were imaged at ×20 using a Zeiss Axio Observer fluorescent microscope, and quantifications were performed using ZEN Blue and ImageJ. The number of caspase-3–expressing cells, tdTom-expressing cells, tdTom/caspase-3 dual-positive cells, NeuN/caspase-3 or GFAP/caspase-3 dual-positive cells, and triple-positive cells expressing NeuN/tdTom/caspase-3 or GFAP/tdTom/caspase-3 was counted over the entire injection site (two to three fields of view averaged) over three sections per stain per animal. The percentage of triple-positive cells over total tdTom-expressing cells was calculated for both NeuN and GFAP population and compared between peptide/cargo and AAV-Cre injection sites. Data were analyzed using analysis of variance (ANOVA) and t tests in GraphPad Prism v9.
Cell density analysis in brain slices
Intact brain slices that displayed tdTom expression from peptide and AAV injections were selected for cell density analysis. The total number of cells was determined by Hoechst staining and quantified by counting the number of fluorescent nuclei present in fluorescence images using the ZEN Blue edition software. Similarly, the number of microglia stained with Iba1, astrocytes stained with GFAP, and neurons stained with NeuN were counted in the same area. At least five regions of 40,000 μm2 (200 μm by 200 μm) were selected from three different slices across two animals within each injection type and compared to the number of these cells in untouched tissue.
Proinflammatory cytokine ELISA for acute inflammatory response
Six additional 10-week-old male Ai14flox/flox mice were injected with either d(WW)TAT:TAT-Cre (n = 3) or AAV-Cre (n = 3) bilaterally in the cortex, as described above. Both hemispheres of the sensorimotor cortex were injected 0.7 mm deep at one coordinate relative to bregma: 0.1 mm anterior (AP) and ±1.3 mm lateral (ML). A total of 0.5 μl of AAV-Cre or payload/peptide cocktails was injected per injection site. At 3 days after injection, mice were euthanized via a lethal dose of Fatal-Plus and perfused transcardially with 4% heparinized PBS. A 3 mm–by–3 mm section of the cortex centered on the injection site, and extending the depth of the entire cortex, was microdissected out from each hemisphere. Both hemispheres from each mouse were combined into one sample. Samples were homogenized in an extraction buffer [100 mM tris (pH 7.4), 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, and Halt Protease and Phosphatase Inhibitor] using brief manual tissue dissociation (Drimmel Tissue Tearor Homogenizer) followed by 1 hour on ice on a shaker. After homogenization, the samples were centrifuged, and the supernatant was collected. Protein concentration was measured using NanoDrop, and aliquots were prepared at 10 μg/ml for analysis by enzyme-linked immunosorbent assay (ELISA).
Three separate solid-phase sandwich ELISA plates were performed concurrently on all six animals in triplicate for each sample: TNF-α (R&D Systems, #DY410), IL-1β (R&D Systems, #DY401), and IL-6 (R&D Systems, #DY406). These assays were performed to the manufacturer’s recommended protocol. Briefly, the plates were incubated with the capture antibody overnight at room temperature. After three washes to remove the excess capture antibody, the plates were blocked for 2 hours in Reagent Diluent (1% bovine serum albumin in PBS). After three further washes, 100 μl of the samples (at a protein concentration of 10 μg/ml) and the seven protein standards (prepared as recommended by the manufacturer) in duplicate were added to the appropriate wells. Samples and standards were incubated for 2 hours at room temperature and then washed three times. A total of 100 μl of detection antibody was then incubated for 2 hours at room temperature. After three washes, 100 μl of streptavidin–horseradish peroxidase (1:40) was added to the wells and incubated for 20 min in the dark. Three more washes were performed, followed by 100 μl of substrate solution in each well. After 20 min of incubation in the dark, 50 μl of stop solution was added to each well and the absorbance was read at 450 nm (with 540-nm correction) using a Tecan Infinite M200 plate reader with Magellan software. GraphPad Prism v9 software was used to produce linear standard curves. Absorbance data were converted to protein concentration using the linear equation and analyzed using t tests (Prism).
Statistical analysis
Statistical analysis was performed in Excel using the Analysis ToolPak. All significance was determined using a two-tailed t test and a null hypothesis of zero, assuming equal variances. SDs were calculated in Excel using the “stdev.s” function. The label “ns” corresponds to P > 0.05, * corresponds to P ≤ 0.05, ** corresponds to P ≤ 0.01, and *** corresponds to P ≤ 0.001 via two-tailed t test. The t test analysis is performed between two populations, i.e., untreated control and sample tested.
Acknowledgments
The study was conducted and approved by the Institutional Animal Care Committee at Texas A&M University (IACUC 2021-0025 to C.G.G.). All the experiments reported here were reviewed and approved by and were consistent with the ARRIVE guidelines for animal care and use. All methods were carried out in accordance with the guidelines and regulations from the Institutional Biosafety Committee at Texas A&M. We thank K. Rajewsky and F. Edenhofer for the plasmid encoding TAT-Cre (Addgene, plasmid #13763). We would also like to thank G. Wright of the Texas A&M University Flow Cytometry Facility for help with nuclei isolation.
Funding: This work was supported by award R01GM110137 (to J.-P.P.) from the U.S. National Institute of General Medical Sciences, grant RP190655 from the Cancer Prevention and Research Institute of Texas (to J.-P.P.), the Craig H. Neilsen Foundation grant 650332 (to C.G.G.), and the TIRR foundation (to C.G.G.).
Author contributions: All authors assisted in experimental design. J.K.A. and A.R.P. generated and processed data involving in vitro cell culture, while T.C.S. and C.G.G. were responsible for handling in vivo experiments. J.K.A., J.-P.P., and C.G.G. wrote the manuscript.
Competing interests: J.-P.P. is an inventor on a patent related to this work filed by The Texas A&M University System (no. U.S. 9,662,404 B2, filed 10 September 2014 and published 30 May 2017). J.-P.P. and C.G.G. are cofounders of TANA Therapeutics and declare a competing interest with equity in this company. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The d(X)TAT peptides can be provided by J.-P.P. pending scientific review and a completed material transfer agreement. Requests for the d(X)TAT peptides should be submitted to pellois@tamu.edu.
Supplementary Materials
This PDF file includes:
Figs. S1 to S31
Discussion of nuclear extraction and quantification of delivered protein
Discussion of d(X)TAT binding and diffusion
Supplementary Materials and Methods
REFERENCES AND NOTES
- 1.Daya S., Berns K. I., Gene therapy using adeno-associated virus vectors. Clin. Microbiol. Rev. 21, 583–593 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu K., Lu Y., Lee J. K., Samara R., Willenberg R., Sears-Kraxberger I., Tedeschi A., Park K. K., Jin D., Cai B., Xu B., Connolly L., Steward O., Zheng B., He Z., PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat. Neurosci. 13, 1075–1081 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haery L., Deverman B. E., Matho K. S., Cetin A., Woodard K., Cepko C., Guerin K. I., Rego M. A., Ersing I., Bachle S. M., Kamens J., Fan M., Adeno-associated virus technologies and methods for targeted neuronal manipulation. Front. Neuroanat. 13, 93 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meves J. M., Geoffroy C. G., Kim N. D., Kim J. J., Zheng B., Oligodendrocytic but not neuronal Nogo restricts corticospinal axon sprouting after CNS injury. Exp. Neurol. 309, 32–43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zittersteijn H. A., Goncalves M., Hoeben R. C., A primer to gene therapy: Progress, prospects, and problems. J. Inherit. Metab. Dis. 44, 54–71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McClements M. E., MacLaren R. E., Adeno-associated virus (AAV) dual vector strategies for gene therapy encoding large transgenes. Yale J. Biol. Med. 90, 611–623 (2017). [PMC free article] [PubMed] [Google Scholar]
- 7.Colella P., Ronzitti G., Mingozzi F., Emerging issues in AAV-mediated in vivo gene therapy. Mol. Ther. Methods Clin. Dev. 8, 87–104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez B. A., Shutterly A., Chan Y. K., Byrne B. J., Corti M., Management of neuroinflammatory responses to AAV-mediated gene therapies for neurodegenerative diseases. Brain Sci. 10, 119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naeem M., Majeed S., Hoque M. Z., Ahmad I., Latest developed strategies to minimize the off-target effects in CRISPR-Cas-mediated genome editing. Cell 9, 1608 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papathanou M., Dumas S., Pettersson H., Olson L., Wallen-Mackenzie A., Off-target effects in transgenic mice: Characterization of dopamine transporter (DAT)-Cre transgenic mouse lines exposes multiple non-dopaminergic neuronal clusters available for selective targeting within limbic neurocircuitry. eNeuro 6, ENEURO.0198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song A. J., Palmiter R. D., Detecting and avoiding problems when using the Cre-lox system. Trends Genet. 34, 333–340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amsellem S., Pflumio F., Bardinet D., Izac B., Charneau P., Romeo P. H., Dubart-Kupperschmitt A., Fichelson S., Ex vivo expansion of human hematopoietic stem cells by direct delivery of the HOXB4 homeoprotein. Nat. Med. 9, 1423–1427 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Krosl J., Austin P., Beslu N., Kroon E., Humphries R. K., Sauvageau G., In vitro expansion of hematopoietic stem cells by recombinant TAT-HOXB4 protein. Nat. Med. 9, 1428–1432 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Bani-Yaghoub M., Tremblay R. G., Lei J. X., Zhang D., Zurakowski B., Sandhu J. K., Smith B., Ribecco-Lutkiewicz M., Kennedy J., Walker P. R., Sikorska M., Role of Sox2 in the development of the mouse neocortex. Dev. Biol. 295, 52–66 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Braun D. A., Fribourg M., Sealfon S. C., Cytokine response is determined by duration of receptor and signal transducers and activators of transcription 3 (STAT3) activation. J. Biol. Chem. 288, 2986–2993 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan C., Lu B., Chen H., Bishop C. E., Reprogramming human fibroblasts using HIV-1 TAT recombinant proteins OCT4, SOX2, KLF4 and c-MYC. Mol. Biol. Rep. 37, 2117–2124 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fornasiero E. F., Mandad S., Wildhagen H., Alevra M., Rammner B., Keihani S., Opazo F., Urban I., Ischebeck T., Sakib M. S., Fard M. K., Kirli K., Centeno T. P., Vidal R. O., Rahman R. U., Benito E., Fischer A., Dennerlein S., Rehling P., Feussner I., Bonn S., Simons M., Urlaub H., Rizzoli S. O., Precisely measured protein lifetimes in the mouse brain reveal differences across tissues and subcellular fractions. Nat. Commun. 9, 4230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorrbaum A. R., Kochen L., Langer J. D., Schuman E. M., Local and global influences on protein turnover in neurons and glia. eLife 7, e34202 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y. W., Luther D. C., Kretzmann J. A., Burden A., Jeon T., Zhai S., Rotello V. M., Protein delivery into the cell cytosol using non-viral nanocarriers. Theranostics 9, 3280–3292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolhassani A., Jafarzade B. S., Mardani G., In vitro and in vivo delivery of therapeutic proteins using cell penetrating peptides. Peptides 87, 50–63 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Stewart M. P., Sharei A., Ding X., Sahay G., Langer R., Jensen K. F., In vitro and ex vivo strategies for intracellular delivery. Nature 538, 183–192 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Smith S. A., Selby L. I., Johnston A. P. R., Such G. K., The endosomal escape of nanoparticles: Toward more efficient cellular delivery. Bioconjug. Chem. 30, 263–272 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Wadia J. S., Stan R. V., Dowdy S. F., Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat. Med. 10, 310–315 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Raagel H., Saalik P., Pooga M., Peptide-mediated protein delivery-which pathways are penetrable? Biochim. Biophys. Acta 1798, 2240–2248 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Abes S., Williams D., Prevot P., Thierry A., Gait M. J., Lebleu B., Endosome trapping limits the efficiency of splicing correction by PNA-oligolysine conjugates. J. Control. Release 110, 595–604 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Erazo-Oliveras A., Najjar K., Dayani L., Wang T. Y., Johnson G. A., Pellois J. P., Protein delivery into live cells by incubation with an endosomolytic agent. Nat. Methods 11, 861–867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erazo-Oliveras A., Najjar K., Truong D., Wang T. Y., Brock D. J., Prater A. R., Pellois J. P., The late endosome and its lipid BMP act as gateways for efficient cytosolic access of the delivery agent dfTAT and its macromolecular cargos. Cell Chem. Biol. 23, 598–607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lönn P., Kacsinta A. D., Cui X.-S., Hamil A. S., Kaulich M., Gogoi K., Dowdy S. F., Enhancing endosomal escape for intracellular delivery of macromolecular biologic therapeutics. Sci. Rep. 6, 32301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamper N., Day P. M., Nowak T., Selinka H. C., Florin L., Bolscher J., Hilbig L., Schiller J. T., Sapp M., A membrane-destabilizing peptide in capsid protein L2 is required for egress of papillomavirus genomes from endosomes. J. Virol. 80, 759–768 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takayama K., Nakase I., Michiue H., Takeuchi T., Tomizawa K., Matsui H., Futaki S., Enhanced intracellular delivery using arginine-rich peptides by the addition of penetration accelerating sequences (Pas). J. Control. Release 138, 128–133 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Jones A. T., Sayers E. J., Cell entry of cell penetrating peptides: Tales of tails wagging dogs. J. Control. Release 161, 582–591 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Brock D. J., Kustigian L., Jiang M., Graham K., Wang T. Y., Erazo-Oliveras A., Najjar K., Zhang J., Rye H., Pellois J. P., Efficient cell delivery mediated by lipid-specific endosomal escape of supercharged branched peptides. Traffic 19, 421–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondow-McConaghy H. M., Muthukrishnan N., Erazo-Oliveras A., Najjar K., Juliano R. L., Pellois J. P., Impact of the endosomal escape activity of cell-penetrating peptides on the endocytic pathway. ACS Chem. Biol. 15, 2355–2363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R., Lein E. S., Zeng H., A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geoffroy C. G., Hilton B. J., Tetzlaff W., Zheng B., Evidence for an age-dependent decline in axon regeneration in the adult mammalian central nervous system. Cell Rep. 15, 238–246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casanova F., Carney P. R., Sarntinoranont M., Effect of needle insertion speed on tissue injury, stress, and backflow distribution for convection-enhanced delivery in the rat brain. PLOS ONE 9, e94919 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki T., Sakata H., Kato C., Connor J. A., Morita M., Astrocyte activation and wound healing in intact-skull mouse after focal brain injury. Eur. J. Neurosci. 36, 3653–3664 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuster D. J., Dykstra J. A., Riedl M. S., Kitto K. F., Belur L. R., McIvor R. S., Elde R. P., Fairbanks C. A., Vulchanova L., Biodistribution of adeno-associated virus serotype 9 (AAV9) vector after intrathecal and intravenous delivery in mouse. Front Neuroanat 8, 42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castle M. J., Turunen H. T., Vandenberghe L. H., Wolfe J. H., Controlling AAV tropism in the nervous system with natural and engineered capsids. Methods Mol. Biol. 1382, 133–149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grimm D., Lee J. S., Wang L., Desai T., Akache B., Storm T. A., Kay M. A., In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 82, 5887–5911 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Z., Yang H., Colosi P., Effect of genome size on AAV vector packaging. Mol. Ther. 18, 80–86 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lian X., Erazo-Oliveras A., Pellois J. P., Zhou H. C., High efficiency and long-term intracellular activity of an enzymatic nanofactory based on metal-organic frameworks. Nat. Commun. 8, 2075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen J., Najjar K., Erazo-Oliveras A., Kondow-McConaghy H. M., Brock D. J., Graham K., Hager E. C., Marschall A. L. J., Dubel S., Juliano R. L., Pellois J. P., Cytosolic delivery of macromolecules in live human cells using the combined endosomal escape activities of a small molecule and cell penetrating peptides. ACS Chem. Biol. 14, 2641–2651 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang M., Zuris J. A., Meng F., Rees H., Sun S., Deng P., Han Y., Gao X., Pouli D., Wu Q., Georgakoudi I., Liu D. R., Xu Q., Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 113, 2868–2873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staahl B. T., Benekareddy M., Coulon-Bainier C., Banfal A. A., Floor S. N., Sabo J. K., Urnes C., Munares G. A., Ghosh A., Doudna J. A., Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol. 35, 431–434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lv H., Zhang S., Wang B., Cui S., Yan J., Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release 114, 100–109 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Kedmi R., Ben-Arie N., Peer D., The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials 31, 6867–6875 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Majumdar A., Cruz D., Asamoah N., Buxbaum A., Sohar I., Lobel P., Maxfield F. R., Activation of microglia acidifies lysosomes and leads to degradation of Alzheimer amyloid fibrils. Mol. Biol. Cell 18, 1490–1496 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Pisa M., Chassaing G., Swiecicki J. M., Translocation mechanism(s) of cell-penetrating peptides: Biophysical studies using artificial membrane bilayers. Biochemistry 54, 194–207 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Pae J., Saalik P., Liivamagi L., Lubenets D., Arukuusk P., Langel U., Pooga M., Translocation of cell-penetrating peptides across the plasma membrane is controlled by cholesterol and microenvironment created by membranous proteins. J. Control. Release 192, 103–113 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Najjar K., Erazo-Oliveras A., Mosior J. W., Whitlock M. J., Rostane I., Cinclair J. M., Pellois J. P., Unlocking endosomal entrapment with supercharged arginine-rich peptides. Bioconjug. Chem. 28, 2932–2941 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Najjar K., Erazo-Oliveras A., Pellois J. P., Delivery of proteins, peptides or cell-impermeable small molecules into live cells by incubation with the endosomolytic reagent dfTAT. J. Vis. Exp. , 53175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geoffroy C. G., Lorenzana A. O., Kwan J. P., Lin K., Ghassemi O., Ma A., Xu N., Creger D., Liu K., He Z., Zheng B., Effects of PTEN and nogo codeletion on corticospinal axon sprouting and regeneration in mice. J. Neurosci. 35, 6413–6428 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S31
Discussion of nuclear extraction and quantification of delivered protein
Discussion of d(X)TAT binding and diffusion
Supplementary Materials and Methods