Abstract
Repair of DNA damage is critical to genomic integrity and frequently disrupted in cancers. Ubiquitin-specific protease 1 (USP1), a nucleus-localized deubiquitinase, lies at the interface of multiple DNA repair pathways and is a promising drug target for certain cancers. Although multiple inhibitors of this enzyme, including one in phase 1 clinical trials, have been established, their binding mode is unknown. Here, we use cryo–electron microscopy to study an assembled enzyme-substrate-inhibitor complex of USP1 and the well-established inhibitor, ML323. Achieving 2.5-Å resolution, with and without ML323, we find an unusual binding mode in which the inhibitor disrupts part of the hydrophobic core of USP1. The consequent conformational changes in the secondary structure lead to subtle rearrangements in the active site that underlie the mechanism of inhibition. These structures provide a platform for structure-based drug design targeting USP1.
USP1, a cancer target, is inhibited by ML323 replacing part of the protein fold, allosterically disrupting the active site.
INTRODUCTION
Human DNA is under constant assault from external and endogenous agents that induce damage, including ultraviolet radiation, smoking, replication stress, cellular metabolites, and the radiation and chemotherapeutics widely used in targeted cancer treatments. In response, multiple organized DNA repair pathways have evolved, with different types of DNA damage eliciting different responses. Ubiquitin-specific protease 1 (USP1) is a nucleus-localized deubiquitinase and a well-established component of DNA repair, acting in the Fanconi anemia (FA) pathway (1) and translesion synthesis (TLS) (2) to catalyze the removal of specific monoubiquitin signals (Fig. 1A). In the FA pathway, USP1 deubiquitinates the heterodimer of FA groups I and D2 proteins (FANCI-FANCD2) to release the complex from DNA (1, 3), while in TLS it acts on proliferating cell nuclear antigen (PCNA) to regulate polymerase recruitment (2). USP1 has also been shown to deubiquitinate several oncogenic proteins to prevent their degradation (4, 5).
Fig. 1. Cryo-EM structures of inhibitor-bound and unbound USP1.
(A) Schematic representation of USP1-UAF1 and its substrates. (B) ML323 inhibits deubiquitination of FANCI-FANCD2Ub by USP1-UAF1 in vitro. Reactions of USP1-UAF1 at 0.1 μM and FANCI-FANCD2Ub at 1 μM in the presence or absence of 10 μM ML323 [0.25% dimethyl sulfoxide (DMSO)] assessed by SDS–polyacrylamide gel electrophoresis and Coomassie staining. Image is representative of two technical replicates. See also fig. S1. (C) Cryo-EM map of reconstituted FANCI (purple), FANCD2 (green) conjugated with ubiquitin (yellow), USP1 (pink), UAF1 (orange), and ML323. (D) Focused map of USP1-ubiquitin without ML323. (E) Focused map of USP1-ubiquitin with ML323 (blue). A refined model of ML323 and surrounding map contoured at 6.8 root mean square deviation (RMSD) is highlighted.
USP1 is a promising target for the treatment of several types of cancer. USP1 expression is up-regulated in certain breast cancers (6, 7), ovarian cancers (5), colorectal cancers (8), and bone cancers (4), often correlated with poor prognosis (5, 7, 8). Reduction of USP1 activity via small molecules or at the level of gene expression has been shown to decrease growth of cancer cells (4), particularly when in combination with other DNA-targeting treatments (5, 6, 8–10). This has fueled interest in development of USP1 inhibitors (9–11), with the first phase 1 clinical trials commencing in 2021 (12). However, the mechanism for inhibition remains unknown because of a lack of structural data. In particular, there are no available enzyme-inhibitor structures. ML323, a well-established inhibitor, is thought to interact with USP1 allosterically (10) and, at the cellular level, has been suggested to cause replication stress via trapping of USP1 on DNA (13). Identification of the proposed allosteric binding site has remained elusive, hampering efforts to develop the next generation of inhibitors.
Recently, we determined structures of USP1 using crystallography and cryo–electron microscopy (cryo-EM) at resolutions sufficient to resolve protein side chains (3.2 to 3.7 Å) (14). Although such resolution provides understanding of protein-protein interfaces, it is not high enough to model features required in structure-guided drug design, which typically needs 2.5 Å or better to unambiguously identify small-molecule ligands and their interactions (15). Despite extensive efforts, we have found it challenging to produce crystals of USP1 diffracting to high resolution, likely due to inherent flexibility. The small size of USP1 (90 kDa) also makes it a difficult target for cryo-EM. However, when assembled with both its activating partner UAF1 (USP1 associated factor 1) (16), and its largest substrate, the FANCI-FANCD2 DNA clamp (17–20), the complex reaches 0.5 MDa. We recently reported the structure of this complex, mutated at the active site cysteine to limit catalysis (Cys90Ser) (14). Although still at limited global resolution, we were able to resolve the active site, including the isopeptide bond of the ubiquitinated substrate, so we wondered whether this scaffold would be amenable to identifying the binding site and mechanism of USP1 inhibition by ML323.
RESULTS AND DISCUSSION
Previous studies have focused on diubiquitin and ubiquitinated PCNA as substrates in inhibitor screens; therefore, we first asked whether ML323 inhibits FANCI-FANCD2 deubiquitination in vitro. We find that, in our reconstituted assay, the FANCI-FANCD2 heterodimer, with monoubiquitin on FANCD2, is readily deubiquitinated by USP1-UAF1, but this is notably reduced in the presence of ML323 (Fig. 1B and fig. S1). We then assembled the enzyme-substrate-inhibitor complex from the six individual components (USP1C90S-UAF1-FANCI-FANCD2Ub-dsDNA-ML323). Single-particle analysis of this complex with cryo-EM yields a consensus reconstruction to 2.7 Å (Fig. 1C, fig. S2, and table S1). Focused reconstruction of USP1-ubiquitin and classification into two states yields reconstructions of USP1 with and without ML323, each to 2.5 Å (Fig. 1, D and E, and fig. S3). At this resolution, the inhibitor can be unambiguously identified and modeled as well as several water molecules within the structures (Fig. 1E and fig. S4).
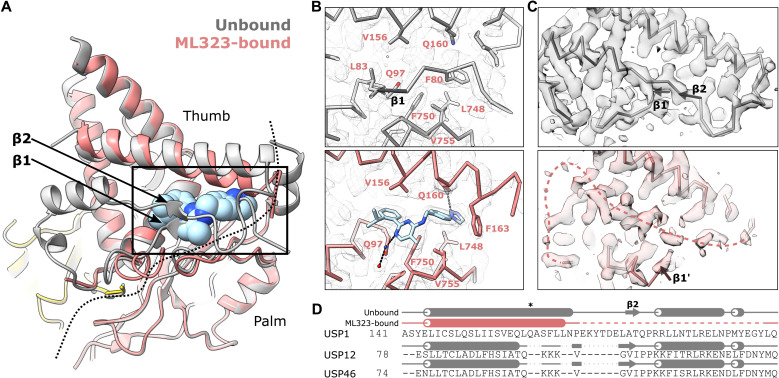
Rather than binding to a pocket on the surface of USP1 or in the active site, the inhibitor binds within the hydrophobic core of USP1, replacing elements of the USP fold (Fig. 2, A and B). This cryptic site lies between the palm and thumb subdomains of the USP fold, ~9 Å from the active site cysteine (Cys90 mutated to Ser; Cβ to the nearest nonhydrogen atom of ML323). Superposition of the ML323 binding site onto the unbound structure reveals >80% of the ML323 atoms clash with atoms of the unbound structure, as measured by solvent-excluded surface area calculations (fig. S5). This extensive overlap highlights the requirement for drastic rearrangement of the protein structure to accommodate the inhibitor. ML323 displaces two segments of the polypeptide containing two short β strands of the thumb (β1 and β2; Fig. 2A). β1 resides at the N terminus of the USP fold and is completely displaced together with adjacent residues (76 to 88; Fig. 2B). β2 becomes less ordered together with adjacent residues (168 to 195), and we were unable to unambiguously model this region (Fig. 2, C and D). In crystal structures of USP1 [Protein Data Bank (PDB) IDs: 7AY0 and 7AY2] (14), these segments are partially disordered in the absence of crystal contacts, suggesting inherent flexibility even in the absence of ML323. Hence, this region may undergo conformational exchange between a compact state, observed in the unbound cryo-EM structure, and an open state in which the ML323 binding site is exposed. In effect, the inhibitor replaces a buried segment of the protein structure and increases disorder in the surrounding region.
Fig. 2. Inhibitor binding disrupts the hydrophobic packing of USP1.
(A) Superposition of the ML323-bound (colored) and unbound (gray) structures. ML323 is shown as spheres. A dashed line represents the interface between the palm and thumb of USP1. The displaced β strands of the thumb (β1: residues 82 to 83, β2: residues 175 to 176) are indicated. (B) Comparison of the ML323 binding site between the bound (colored) and unbound (gray) states. Q97 and Q160 form potential hydrogen bonds with ML323, shown as dashed lines. Cα trace and side chains involved in ML323 binding are shown. Cryo-EM maps are shown as mesh contoured at 6.8 RMSD. (C) Comparison of the region surrounding β2 that becomes less ordered upon ML323 binding (residues 168 to 195). Cα trace is shown and cryo-EM maps as transparent solid contoured at 6.8 RMSD. In the ML323-bound state, this region, which we were unable to model unambiguously, is represented as a pink dashed line. (D) Structure-based sequence alignment of USP1 with close homologs. Q160 is marked by an asterisk. Helices are represented as tubes, β strands as arrows, loops as solid lines, and unmodeled residues as a dashed line.
ML323 contacts numerous hydrophobic residues of both the palm and thumb that were previously part of the hydrophobic core (Fig. 2B). In addition, the side chain of Gln97 is displaced by ML323 to form a hydrogen bond with a water molecule and may form a strained hydrogen bond with the pyrimidine of ML323. The isopropyl group of ML323 occupies the same position of the equivalent group of Leu83 in the unbound structure, while the triazole contributes a hydrogen bond with Gln160, explaining why these modifications improved inhibition compared to the initial inhibitor on which ML323 was based (Fig. 2B and fig. S6) (10, 11). The helix on which Gln160 is located bends to form additional van der Waals contacts with ML323. The corresponding helix in the closely related USP12 (21, 22) and USP46 (23), which both share UAF1 as a cofactor, is shorter (Fig. 2D). This helix and the adjoining loop are also smaller in other USPs compared to USP1 (fig. S7). Hence, this region of USP1 may mediate the selectivity of ML323 for USP1 over other USPs (10).
The cryo-EM structures also explain the mechanism of inhibition by ML323. Although the active site cysteine is not appreciably perturbed in the ML323-bound structure, there is a subtle change in the loop containing the catalytic aspartate, Asp751, on the palm subdomain (Fig. 3A). This disruption repositions Asp751 further from the catalytic histidine, His593, breaking a hydrogen bond between them. An adjacent aspartate, Asp752, moves closer to His593 forming a new hydrogen bond with the imidazole ring. On the basis of this hydrogen bonding geometry, the imidazole ring of His593 flips and forms an additional hydrogen bond with a water molecule when ML323 is bound (Fig. 3B). In the unbound state, this histidine is poised to deprotonate what would be the catalytic cysteine, which would then allow nucleophilic attack on the isopeptide (Fig. 3B). However, in the flipped conformation, when ML323 is bound, the histidine is unable to deprotonate the cysteine. This flipping of the imidazole ring likely underlies the reduction in the catalytic rate by ML323 (Fig. 1B) (10). ML323 can therefore be classified as a type IV deubiquitinase inhibitor (24)—binding outside of the ubiquitin-binding site and allosterically inhibiting catalysis.
Fig. 3. Inhibitor binding disrupts the catalytic site.
(A) The loop on which the catalytic aspartate, D751, is pushed out by ML323. (B) The catalytic histidine, H593, and cysteine, C90S, remain in approximately the same position; however, D752 replaces the hydrogen bond with H593, causing it to flip. Hydrogen bonds are shown as dashed lines. Nucleophilic attack is shown by a solid arrow.
ML323-induced changes remain localized around the ML323 binding site and USP1 active site. We generated consensus reconstructions of USP1-UAF1-FANCI-FANCD2Ub-dsDNA with and without ML323 based on the classified particles (fig. S8, A to D, and table S1). On FANCD2, adjacent to Lys561, which is conjugated with ubiquitin, Arg560 forms a hydrogen bond with Asp751 of USP1 when ML323 is absent (14). When ML323 is bound, the side chain of Arg560 is somewhat disordered, likely arising from the movement of the Asp751 loop and breaking of the hydrogen bond between Arg560 and Asp751 (fig. S8E). Further, the displaced β1 region moves between FANCD2 and USP1 (fig. S8E). We speculate that these changes may be suboptimal for USP1 binding to substrate and therefore explain the reduced binding of ML323 to substrate-bound USP1 that has previously been observed (10). The remainder of the FANCD2-USP1 interface is effectively unchanged between ML323-bound and unbound structures, as well as the USP1-UAF1 interface (fig. S8F). These interfaces may be able to partially compensate for disruption at the ML323 binding site. ML323 at 10 μM is sufficient to reduce the activity of USP1-UAF1 on FANCI-FANCD2Ub by approximately 50% in gel-based assays (fig. S1). Hence, the ability of ML323 to inhibit USP1 deubiquitination of FANCI-FANCD2Ub appears reduced compared to ubiquitinated PCNA [median inhibitory concentration (IC50) = 820 nM] and further still compared to diubiquitin (IC50 = 174 nM) (10). We speculate that the extensive interfaces that form when USP1-UAF1 recognizes FANCI-FANCD2Ub that are not perturbed by ML323 may contribute to this reduced inhibition. Overall, the structures provide an allosteric mechanism that rationalizes the mixed mode of inhibition.
USP1 contains multiple insertions in the USP core fold (Fig. 4A). While these are predicted to be structurally disordered and are not tightly associated with the USP fold (14), they play important roles in regulating USP1 function (2, 13, 25, 26). The N-terminal extension (NTE) (residues 1 to 75) of USP1 has previously been shown to assist substrate discrimination (25). Deletion of the NTE markedly reduces USP1-mediated deubiquitination of FANCD2 but has little effect on deubiquitination of FANCI or PCNA (25). However, the mechanism of this discrimination is unknown. We integrated AlphaFold predictions together with the cryo-EM data to elucidate the structural basis of FANCD2 interaction with the USP1 NTE. We used AlphaFold-Multimer (27, 28) to generate models of USP1NTE with FANCD2 (Fig. 4 and fig. S9). A short segment of USP1NTE, residues 21 to 25, exhibited good confidence scores [pLDDT (predicted local-distance difference test) > 70, Fig. 4B] and relatively low predicted aligned error relative to FANCD2 (Fig. 4C). This region maps well to previous biochemical experiments in which mutation of these residues to alanine disrupted FANCD2 deubiquitination (25). Focused reconstruction and LocSpiral postprocessing (29) of the USP1-UAF1-FANCD2Ub-FANCI-dsDNA-ML323 cryo-EM data around this region showed additional density in the same position as the NTE of the AlphaFold model (Fig. 4D and fig. S10). In the AlphaFold model, Leu23 of the NTE inserts into a hydrophobic pocket of FANCD2, with Arg22 forming a salt bridge with Asp667 on FANCD2. The importance of positive charge at residue 22 in the NTE has previously been established via mutagenesis analysis (25). Mutation of Arg22 to glutamate in which the charge is reversed drastically reduced the rate of FANCD2 deubiquitination, mutation to alanine in which the charge is removed had a moderate effect, and mutation to lysine in which the charge is conserved had a negligible effect (25). This is consistent with the salt bridge interaction observed in the AlphaFold models. The hydrophobic pocket of FANCD2 in which Leu23 of USP1NTE binds is absent in the equivalent region of the FANCD2 paralog, FANCI (fig. S11). Hence, it is unlikely that the NTE can interact with FANCI in the same way it binds to FANCD2. This explains how the NTE of USP1 acts as an allosteric adapter to target FANCD2.
Fig. 4. AlphaFold confidently predicts an interaction between USP1NTE and FANCD2.
(A) Schematic representation of the USP1 primary structure. (B) USP1NTE pLDDT confidence scores of five AlphaFold-Multimer replicate models. (C) AlphaFold-Multimer predicted aligned error for the USP1NTE-FANCD2 complex, highlighting the region of USP1 confidently predicted to interact with FANCD2. Gray lines separate the two sequences. (D) Rigid-body fit of the AlphaFold-Multimer (27) models of USP1NTE (pink) with FANCD2 (green) into the cryo-EM map generated by focused refinement on FANCD2 with LocSpiral (29) postprocessing. The confidently predicted region of USP1NTE agrees well with the cryo-EM map (contoured at 4 RMSD).
We have revealed an unexpected mode of binding to USP1 by a small-molecule inhibitor, disrupting a substantial portion of the hydrophobic packing. Although there are other examples of an inhibitor displacing protein structure [e.g., (30)], the extent to which the USP1 structure is altered is remarkable. The displacement is likely facilitated by intrinsic plasticity in this region exposing the ML323 binding site. It is tempting to speculate that there may be a native metabolite that regulates USP1 activity at this site. At the resolution achieved, we define key interactions that rationalize previous medicinal chemistry optimizations and reveal conformational changes induced in the catalytic site that underpin inhibition. The ML323-bound structure offers the potential for design of new inhibitors that recapitulate and improve the protein-ligand interactions but have superior drug-like properties. Given the role of the NTE in regulation and the allosteric mode of ML323 inhibition, there are exciting opportunities for the next generation of inhibitors of this enzyme and in the development of future cancer treatments.
MATERIALS AND METHODS
Protein expression and purification
Proteins, all human homologs, were prepared as described previously (14). Protein purification buffers and columns used are provided in table S2. Briefly, His6-TEV-USP1G670A,G671A, His6-TEV-USP1G670A,G671A,C90S, His6-3C-UAF1, His6-3C-FANCD2, His6-FANCI, and His6-TEV-V5-FANCI were expressed separately in Sf21 insect cells. Cells were lysed by sonication, clarified, and purified by Ni–nitrilotriacetic acid (NTA) affinity and then anion exchange chromatography. At this stage, protein aliquots were occasionally flash-frozen in liquid nitrogen and stored −80°C. For His6-TEV (Tobacco Etch Virus)-USP1G670A,G671A and His6-TEV-USP1G670A,G671A,C90S TEV protease treatment was performed overnight at 1:10 protease to target protein with gentle agitation before subtractive Ni-NTA affinity chromatography. Flow through was concentrated to ~10 mg/ml and separated by gel filtration. Purified protein was concentrated to 5 to 15 mg/ml, flash-frozen, and stored at −80°C in 10- to 20-μl single-use aliquots. All steps were performed on ice or at 4°C and completed within 24 to 36 hours of lysis. FANCD2 was ubiquitinated and purified using an engineered Ube2T (Ubiquitin-conjugating enzyme E2 T) and SpyCatcher-SpyTag setup described in detail elsewhere (31, 32). For FANCD2, the His6-3C tag was removed by 3C protease treatment during preparation of the monoubiquitinated version.
Protein concentrations were determined using the predicted extinction coefficients at 280 nm (33) and absorbance via a NanoDrop. The ratio of 260 nm/280 nm was ≤0.65 for all protein batches used in subsequent experiments.
Cryo-EM sample preparation
The USP1C90S-UAF1-FANCI-FANCD2Ub complex was prepared by mixing the four individually purified subunits 5:5:1:1. The complex was exchanged into EM buffer [20 mM tris (pH 8.0), 150 mM NaCl, and 2 mM dithiothreitol (DTT)] using a Bio-Spin P-30 column (Bio-Rad). The concentration of complex was estimated from absorbance at 280 nm (assuming no loss of any of the protein components), and 1.2 equivalents of double-stranded DNA (dsDNA) (61 base pairs; TGATCAGAGGTCATTTGAATTCATGGCTTCGAGCTTCATGTAGAGTCGACGGTGCTGGGAT; IDT) per FANCI-FANCD2Ub was added. ML323 was then added at 2 equivalents of USP1-UAF1. Immediately before preparing grids, the sample was equilibrated to room temperature for 5 min. UltrAuFoil R1.2/1.3 300 mesh grids were glow-discharged twice at 45 mA for 60 s. A 3.5-μl aliquot of 9.3 μM USP1-UAF1, 1.8 μM FANCI-FANCD2Ub, 2.2 μM dsDNA, and 18.3 μM ML323 was applied. The grids were blotted for 3.0 s and vitrified in liquid ethane using a Vitrobot (Thermo Fisher Scientific) operating at ~95% humidity at 15°C.
Cryo-EM sample data collection and processing
Initial grid screening was performed on a JEM-F200 (JEOL) equipped with a DE-20 detector (Direct Electron) at the Scottish Centre for Macromolecular Imaging (SCMI). For data collection, a Titan Krios (Thermo Fisher Scientific) located at the Electron Bio-imaging Centre (eBIC) (Diamond Light Source) equipped with a K3 detector (Gatan) was used. A total of 10,998 movies were collected using beam-image shift. All movies were collected in superresolution mode with 2× binning and a calibrated pixel size of 1.06 Å using EPU (Thermo Fisher Scientific). Movies were collected with a total dose of ~40 e−/Å2 over 40 frames at a rate of 15.46 e− per pixel per s.
Subsequent processing was performed in cryoSPARC v2.13.2 and v3.3 (figs. S2, S3, and S10) (34). Patch motion correction, patch CTF (contrast transfer function) estimation, and manual curation was performed resulting in 9344 dose-weighted, motion-corrected micrographs. A maximum alignment resolution of 3 Å was used during patch motion correction. Particle picking, ab initio model generation, and cleaning to remove junk picks were performed similarly to previously described (14). Blob picking was performed on a subset of micrographs with minimum and maximum particle diameters of 150 and 250 Å, respectively, using an elliptical blob, followed by two-dimensional (2D) classification and ab initio reconstruction. Templates from 2D classification were then used to pick from all micrographs, and 6.7 million images were extracted with a box size of 320 × 320 pixels. Cleaning was performed iteratively and in batches by heterogeneous refinement with one good starting model and two or three “junk” starting models distinct from the protein complex of interest, all low-pass–filtered to 20 Å. Further rounds of heterogeneous refinement were performed using the same starting model low-pass–filtered at 12, 15, 20, and 30 Å. Between one and eight final full passes through the dataset were used in heterogeneous refinements. Particles corresponding to the highest resolution class were then reextracted yielding 1.3 million particles. Another round of heterogeneous refinement yielded 1.15 million particles reaching 2.82 Å after nonuniform refinement (35). Local motion correction was performed, again with a maximum alignment resolution of 3 Å improving the resolution to 2.76 Å. Global CTF refinement of beam tilt and trefoil (36) against the pooled images was performed improving the resolution to 2.72 Å. All Fourier shell correlation (FSC) calculations were performed using masks without autotightening as the autotightened mask tended to exclude some side chains and water molecules. Local resolutions were calculated using an adaptive window factor of 20.
Local refinement was performed with a mask covering USP1 and ubiquitin using Gaussian priors of 3° over rotation and 2 Å over shifts with marginalization and nonuniform refinement (fig. S3). The 3D variability analysis of this locally aligned region (two modes and 2.8-Å filter resolution) and clustering was used to yield two states—one with ML323 bound and one unbound. 3D classification with two classes was performed using the two cluster models filtered to 8 Å as inputs. The resulting particles were passed through local refinement again with the mask covering USP1 and ubiquitin. The two particle subsets were also used to generate consensus reconstructions with and without ML323 via nonuniform refinement.
For the FANCD2 helical and C-terminal domain, local refinement was performed with a mask covering this region and using Gaussian priors of 3° over rotation and 2 Å over shifts with marginalization and nonuniform refinement (fig. S10). LocSpiral (29), via the COSMIC2 web platform (https://cosmic-cryoem.org/) (37) was used to postprocess the half maps using resolutions between 2.47 and 30 Å, a bandwidth of 8, threshold for significant comparison 0.95, and a threshold for 3D mask of 0.14.
Model building and refinement
ML323-bound and unbound structures were built into the locally refined maps using USP1 and ubiquitin from the previous structure of USP1-UAF1-FANCI-FANCD2Ub-dsDNA (PDB ID: 7AY1) (14). Manual model editing was performed using Coot (38). ML323 restraints were calculated using the Grade web server (http://grade.globalphasing.org/). Automated refinement against the globally sharpened maps, with the B-factor estimated from the Guinier plot, were performed using Phenix real-space refinement (39). A refinement resolution of 2.8 Å (FSC = 0.5) was used. Bond and angle restraints for the USP1 Zinc finger, as well as secondary structure restraints, were incorporated during automated refinement. To confirm the side-chain conformation of His593 modeled in the ML323-bound state, the conformation was flipped back to the unbound conformation and a 0.5-Å random shift introduced across all atoms via phenix.pdbtools. This model was passed through automated real-space refinement, with NQH-flips enabled, which returned His593 to the inhibited conformation shown in Fig. 3B.
For the consensus reconstructions of the unbound complex, USP1-ubiquitin from the focused reconstruction, together with FANCI, FANCD2, and UAF1 from PDB ID 7AY1 (14), were used as initial models. Manual model editing was performed using Coot (38), with AlphaFold models of FANCI (Q9NVI1), FANCD2 (Q9BXW9), and UAF1 (Q8TAF3) used to guide editing. Automated refinement against the globally sharpened map was performed using Phenix real-space refinement (39). A refinement resolution of 3.1 Å was used. Bond and angle restraints for the USP1 Zinc finger and isopeptide bond, as well as Ramachandran restraints, were incorporated during automated refinement. For the consensus reconstruction of the ML323-bound complex, USP1-ubiquitin-ML323 from the focused reconstruction, together with FANCI, FANCD2, and UAF1 from the consensus model of the unbound complex, were used as initial models. Manual and automated refinement was performed as per the consensus model of the unbound complex. The dsDNA from PDB ID 6VAE (17) was rigid body–fitted into the unsharpened maps, and a final round of atomic displacement parameter refinement was performed. The nucleotides were then stubbed before deposition to the PDB. Cryo-EM data and model statistics are reported in table S1. The FSCs between the models and maps were computed using Phenix (fig. S3) (40).
Structures and maps were analyzed, and figures were produced using ChimeraX (41). Volume operations and calculations were performed in Blender (https://blender.org/) on solvent-excluded surfaces that were computed in ChimeraX and exported as Wavefront files (.obj). A Boolean modifier was used to generate the difference between the ML323-alone volume (V1) and the unbound USP1 volume (V2), which was taken to be the nonoverlapping volume of ML323 (V3). The volume overlap was then calculated as (V1 − V3)/V1. Structure-informed multiple sequence alignments were performed using PROMALS3D (42).
AlphaFold models were generated using the full-length human FANCD2 and the NTE of human USP1 (residues 1 to 75) sequences. AlphaFold v2.1.0 (27, 28) with multimer model preset was used to generate five relaxed models with all other settings as default. The AlphaFold models were superimposed onto the FANCD2 chain from the cryo-EM built model using matchmaker in ChimeraX, and the position was further refined by rigid-body fitting of FANCD2 residues 650 to 800 into the LocSpiral map.
Deubiquitination assays
Deubiquitination reactions were performed by preparing a 2× substrate mix and a 2× enzyme mix and mixing these 1:1 to initiate the reaction. Both mixes were set up on ice and then incubated at room temperature for at least 20 min before reaction initiation and during the reaction. The 2× substrate mix was prepared by diluting stocks (≥30 μM) of FANCD2Ub, His6-V5-TEV-FANCI, and dsDNA (61 base pairs) with Deubiquitination buffer [20 mM tris (pH 8.0), 75 mM NaCl, 5% glycerol, and 1 mM DTT]. The resulting 2× mix was composed 2 μM FANCD2Ub, 2 μM FANCI, and 8 μM dsDNA. The 2× enzyme mixes were prepared by diluting concentrated stocks (≥30 μM) of USP1, His6-3C-UAF1, and ML323 or dimethyl sulfoxide (DMSO) control with DUB buffer. The resulting 2× mixes were composed of 200 nM USP1, 200 nM UAF1, 0.5% DMSO, and 20 μM ML323, where included. Aliquots of 4 μl of reaction were terminated at the indicated time points by addition of 20 μl of 1.2× NuPAGE LDS buffer (Thermo Fisher Scientific) supplemented with DTT (final concentration 100 mM). SDS–polyacrylamide gel electrophoresis was performed using Novex 4 to 12% tris-glycine plus gels (Thermo Fisher Scientific) and subsequent staining of the gels with Instant-Blue Coomassie stain (Expedeon).
For Western blotting, 1:1000 dilution rabbit anti-FANCD2 (Abcam, ab108928) and 1:8000 dilution mouse anti-V5 (Abcam, ab27671) were used for detection of FANCD2 and FANCI, respectively. Secondary antibodies labeled with IRDye 680RD or 800CW [donkey anti-rabbit immunoglobulin G (IgG) or anti-mouse IgG, LI-COR, 926-68073, 926-32212) were used at 1:10,000 dilution. Bands were subsequently visualized on the Odyssey CLX Infrared Imaging System (LI-COR). Each band was quantified using Image Studio with horizontal background subtraction (LI-COR). Percentage ubiquitination was calculated from the ratio of the ubiquitinated band to the sum of the nonubiquitinated and ubiquitinated bands. Replicate assays were performed from different thawed protein aliquots, each from the same protein preparation. Plots were prepared using matplotlib (43).
Acknowledgments
We thank past and current members of the Walden laboratory for experimental suggestions, comments on the manuscript, and support. We acknowledge Diamond Light Source for access and support of the cryo-EM facilities at the UK’s national eBIC (under proposal BI24557), funded by the Wellcome Trust, MRC, and BBRSC. We thank P. da Fonseca and E. Morris for assistance with collecting the data at the eBIC. We acknowledge the SCMI and M. Clarke and J. Streetley for assistance with cryo-EM experiments and access to instrumentation, funded by the MRC (MC_PC_17135) and SFC (H17007). We thank D. Bhella for access to computing resources to run AlphaFold. We thank R. Toth for the expression plasmids. We thank M. Meenan, P. McLaughlin, and I. Sim for maintenance of the GPU server running cryoSPARC.
Funding: This work was supported by the European Research Council (ERC-2015-CoG-681582) ICLUb consolidator grant to H.W. and the Medical Research Council (MC_UU_120164/12) to H.W.
Author contributions: M.L.R. and H.W. conceived this work. M.L.R., C.A., and V.K.C. purified the proteins. M.L.R. performed all the experiments and analyzed all data. M.L.R. wrote the manuscript and prepared figures with contributions from H.W. and C.A. H.W. secured funding and supervised the project.
Competing interests: H.W. is a member of the scientific advisory board of Ubiquigent. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The atomic coordinates have been deposited to the PDB under accession codes 8A9J, 8A9K, 7ZH3, and 7ZH4. The cryo-EM maps have been deposited to the EMDB under accession codes EMD-15284, EMD-14722, EMD-14720, EMD-14721, and EMD-14719.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Tables S1 and S2
References
Other Supplementary Material for this manuscript includes the following:
Data S1 to S5
REFERENCES AND NOTES
- 1.Nijman S. M. B., Huang T. T., Dirac A. M. G., Brummelkamp T. R., Kerkhoven R. M., D’Andrea A. D., Bernards R., The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell 17, 331–339 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Huang T. T., Nijman S. M. B., Mirchandani K. D., Galardy P. J., Cohn M. A., Haas W., Gygi S. P., Ploegh H. L., Bernards R., D’Andrea A. D., Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 8, 339–347 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Sims A. E., Spiteri E., Sims R. J., Arita A. G., Lach F. P., Landers T., Wurm M., Freund M., Neveling K., Hanenberg H., Auerbach A. D., Huang T. T., FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 14, 564–567 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Williams S. A., Maecker H. L., French D. M., Liu J., Gregg A., Silverstein L. B., Cao T. C., Carano R. A. D., Dixit V. M., USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell 146, 918–930 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Sonego M., Pellarin I., Costa A., Vinciguerra G. L. R., Coan M., Kraut A., D’Andrea S., Dall’Acqua A., Castillo-Tong D. C., Califano D., Losito S., Spizzo R., Couté Y., Vecchione A., Belletti B., Schiappacassi M., Baldassarre G., USP1 links platinum resistance to cancer cell dissemination by regulating snail stability. Sci. Adv. 5, eaav3235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim K. S., Li H., Roberts E. A., Gaudiano E. F., Clairmont C., Sambel L. A., Ponnienselvan K., Liu J. C., Yang C., Kozono D., Parmar K., Yusufzai T., Zheng N., D’Andrea A. D., USP1 is required for replication fork protection in BRCA1-deficient tumors. Mol. Cell 72, 925–941.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma A., Tang M., Zhang L., Wang B., Yang Z., Liu Y., Xu G., Wu L., Jing T., Xu X., Yang S., Liu Y., USP1 inhibition destabilizes KPNA2 and suppresses breast cancer metastasis. Oncogene 38, 2405–2419 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Xu X., Li S., Cui X., Han K., Wang J., Hou X., Cui L., He S., Xiao J., Yang Y., Inhibition of ubiquitin specific protease 1 sensitizes colorectal cancer cells to DNA-damaging chemotherapeutics. Front. Oncol. 9, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J., Dexheimer T. S., Ai Y., Liang Q., Villamil M. A., Inglese J., Maloney D. J., Jadhav A., Simeonov A., Zhuang Z., Selective and cell-active inhibitors of the USP1/UAF1 deubiquitinase complex reverse cisplatin resistance in non–small cell lung cancer cells. Chem. Biol. 18, 1390–1400 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang Q., Dexheimer T. S., Zhang P., Rosenthal A. S., Villamil M. A., You C., Zhang Q., Chen J., Ott C. A., Sun H., Luci D. K., Yuan B., Simeonov A., Jadhav A., Xiao H., Wang Y., Maloney D. J., Zhuang Z., A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat. Chem. Biol. 10, 298–304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dexheimer T. S., Rosenthal A. S., Luci D. K., Liang Q., Villamil M. A., Chen J., Sun H., Kerns E. H., Simeonov A., Jadhav A., Zhuang Z., Maloney D. J., Synthesis and structure–activity relationship studies of N -benzyl-2-phenylpyrimidin-4-amine derivatives as potent USP1/UAF1 deubiquitinase inhibitors with anticancer activity against nonsmall cell lung cancer. J. Med. Chem. 57, 8099–8110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.“A Phase 1 Study of KSQ-4279 Alone and in Combination in Patients With Advanced Solid Tumors.” 2021;https://clinicaltrials.gov/ct2/show/NCT05240898.
- 13.Coleman K. E., Yin Y., Lui S. K. L., Keegan S., Fenyo D., Smith D. J., Rothenberg E., Huang T. T., USP1-trapping lesions as a source of DNA replication stress and genomic instability. Nat. Commun. 13, 1740 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rennie M. L., Arkinson C., Chaugule V. K., Toth R., Walden H., Structural basis of FANCD2 deubiquitination by USP1-UAF1. Nat. Struct. Mol. Biol. 28, 356–364 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Renaud J. P., Chari A., Ciferri C., Liu W. T., Rémigy H. W., Stark H., Wiesmann C., Cryo-EM in drug discovery: Achievements, limitations and prospects. Nat. Rev. Drug Discov. 17, 471–492 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Cohn M. A., Kowal P., Yang K., Haas W., Huang T. T., Gygi S. P., D’Andrea A. D., A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell 28, 786–797 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Wang R., Wang S., Dhar A., Peralta C., Pavletich N. P., DNA clamp function of the monoubiquitinated Fanconi anaemia ID complex. Nature 580, 278–282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alcón P., Shakeel S., Chen Z. A., Rappsilber J., Patel K. J., Passmore L. A., FANCD2–FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair. Nat. Struct. Mol. Biol. 27, 240–248 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan W., van Twest S., Leis A., Bythell-Douglas R., Murphy V. J., Sharp M., Parker M. W., Crismani W. M., Deans A. J., Monoubiquitination by the human Fanconi anemia core complex clamps FANCI:FANCD2 on DNA in filamentous arrays. eLife 9, e54128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rennie M. L., Lemonidis K., Arkinson C., Chaugule V. K., Clarke M., Streetley J., Spagnolo L., Walden H., Differential functions of FANCI and FANCD2 ubiquitination stabilize ID2 complex on DNA. EMBO Rep. 21, e50133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Lim K. S., Kim H., Hinds T. R., Jo U., Mao H., Weller C. E., Sun J., Chatterjee C., D’Andrea A. D., Zheng N., Allosteric activation of ubiquitin-specific proteases by β-propeller proteins UAF1 and WDR20. Mol. Cell 63, 249–260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dharadhar S., Clerici M., van Dijk W. J., Fish A., Sixma T. K., A conserved two-step binding for the UAF1 regulator to the USP12 deubiquitinating enzyme. J. Struct. Biol. 196, 437–447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin J., Schoeffler A. J., Wickliffe K., Newton K., Starovasnik M. A., Dueber E. C., Harris S. F., Structural insights into WD-Repeat 48 activation of ubiquitin-specific protease 46. Structure 23, 2043–2054 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Lange S. M., Armstrong L. A., Kulathu Y., Deubiquitinases: From mechanisms to their inhibition by small molecules. Mol. Cell 82, 15–29 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Arkinson C., Chaugule V. K., Toth R., Walden H., Specificity for deubiquitination of monoubiquitinated FANCD2 is driven by the N-terminus of USP1. Life Sci. Alliance 1, e201800162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dharadhar S., Van Dijk W. J., Scheffers S., Fish A., Sixma T. K., Insert L1 is a central hub for allosteric regulation of USP1 activity. EMBO Rep. 22, e51749 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R. Evans, M. O’Neill, A. Pritzel, N. Antropova, A. W. Senior, T. Green, A. Žídek, R. Bates, S. Blackwell, J. Yim, O. Ronneberger, S. Bodenstein, M. Zielinski, A. Bridgland, A. Potapenko, A. Cowie, K. Tunyasuvunakool, R. Jain, E. Clancy, P. Kohli, J. Jumper, D. Hassabis, Protein complex prediction with AlphaFold-Multimer. bioRxiv 2021.10.04.463034 [Preprint]. 10 March 2022. 10.1101/2021.10.04.463034. [DOI]
- 28.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaur S., Gomez-Blanco J., Khalifa A. A. Z., Adinarayanan S., Sanchez-Garcia R., Wrapp D., McLellan J. S., Bui K. H., Vargas J., Local computational methods to improve the interpretability and analysis of cryo-EM maps. Nat. Commun. 12, 1240 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcin E. D., Arvai A. S., Rosenfeld R. J., Kroeger M. D., Crane B. R., Andersson G., Andrews G., Hamley P. J., Mallinder P. R., Nicholls D. J., St-Gallay S. A., Tinker A. C., Gensmantel N. P., Mete A., Cheshire D. R., Connolly S., Stuehr D. J., Åberg A., Wallace A. V., Tainer J. A., Getzoff E. D., Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase. Nat. Chem. Biol. 4, 700–707 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaugule V. K., Arkinson C., Toth R., Walden H., Enzymatic preparation of monoubiquitinated FANCD2 and FANCI proteins. Methods Enzymol. 618, 73–104 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Chaugule V. K., Arkinson C., Rennie M. L., Kamarainen O., Toth R., Walden H., Allosteric mechanism for site-specific ubiquitination of FANCD2. Nat. Chem. Biol. 16, 291–301 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pace N. C., Vajdos F., Fee L., Grimsley G., Gray T., How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4, 2411–2423 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., CryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Punjani A., Zhang H., Fleet D. J., Non-uniform refinement: Adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Zivanov J., Nakane T., Scheres S. H. W., Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.M. A. Cianfrocco, M. Wong-Barnum, C. Youn, R. Wagner, A. Leschziner, COSMIC2: A Science Gateway for Cryo-Electron Microscopy Structure Determination, in Practice & Experience in Advanced Research Computing (2017), New Orleans, LA, 9 to 13 July 2017.
- 38.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. Sect. D Struct. Biol. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Afonine P. V., Poon B. K., Read R. J., Sobolev O. V., Terwilliger T. C., Urzhumtsev A., Adams P. D., Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. Sect. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Afonine P. V., Klaholz B. P., Moriarty N. W., Poon B. K., Sobolev O. V., Terwilliger T. C., Adams P. D., Urzhumtsev A., New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. Sect. D Struct. Biol. 74, 814–840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goddard T. D., Huang C. C., Meng E. C., Pettersen E. F., Couch G. S., Morris J. H., Ferrin T. E., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pei J., Kim B. H., Grishin N. V., PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunter J. D., Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007). [Google Scholar]
- 44.Ernst A., Avvakumov G., Tong J., Fan Y., Zhao Y., Alberts P., Persaud A., Walker J. R., Neculai A., Neculai D., Vorobyov A., Garg P., Beatty L., Chan P., Juang Y., Landry M., Yeh C., Zeqiraj E., Karamboulas K., Allali-hassani A., Vedadi M., Tyers M., Moffat J., Sicheri F., Pelletier L., Durocher D., Raught B., Rotin D., Yang J., Moran M. F., Dhe-Paganon S., Sidhu S. S., A strategy for modulation of enzymes in the ubiquitin system. Science 339, 590–595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clerici M., Luna-Vargas M. P. A., Faesen A. C., Sixma T. K., The DUSP-Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange. Nat. Commun. 5, 5399 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teyra J., Singer A. U., Schmitges F. W., Jaynes P., Lui S. K. L., Polyak M. J., Fodil N., Krieger J. R., Tong J., Schwerdtfeger C., Brasher B. B., Ceccarelli D. F. J., Moffat J., Sicheri F., Moran M. F., Gros P., Eichhorn P. J. A., Lenter M., Boehmelt G., Sidhu S. S., Structural and functional characterization of ubiquitin variant inhibitors of USP15. Structure 27, 590–605.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Molland K., Zhou Q., Mesecar A. D., A 2.2 Å resolution structure of the USP7 catalytic domain in a new space group elaborates upon structural rearrangements resulting from ubiquitin binding. Acta Crystallogr. Sect. F Struct. Biol. Commun. 70, 283–287 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pascoe N., Seetharaman A., Teyra J., Manczyk N., Satori M. A., Tjandra D., Makhnevych T., Schwerdtfeger C., Brasher B. B., Moffat J., Costanzo M., Boone C., Sicheri F., Sidhu S. S., Yeast two-hybrid analysis for ubiquitin variant inhibitors of human deubiquitinases. J. Mol. Biol. 431, 1160–1171 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Leznicki P., Natarajan J., Bader G., Spevak W., Schlattl A., Rehman S. A. A., Pathak D., Weidlich S., Zoephel A., Bordone M. C., Barbosa-Morais N. L., Boehmelt G., Kulathu Y., Expansion of DUB functionality generated by alternative isoforms—USP35, a case study. J. Cell Sci. 131, jcs212753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paudel P., Zhang Q., Leung C., Greenberg H. C., Guo Y., Chern Y. H., Dong A., Li Y., Vedadi M., Zhuang Z., Tong Y., Crystal structure and activity-based labeling reveal the mechanisms for linkage-specific substrate recognition by deubiquitinase USP9X. Proc. Natl. Acad. Sci. U.S.A. 116, 7288–7297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gersch M., Gladkova C., Schubert A. F., Michel M. A., Maslen S., Komander D., Mechanism and regulation of the Lys6-selective deubiquitinase USP30. Nat. Struct. Mol. Biol. 24, 920–930 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu M., Li P., Song L., Jeffrey P. D., Chenova T. A., Wilkinson K. D., Cohen R. E., Shi Y., Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 24, 3747–3756 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Tables S1 and S2
References
Data S1 to S5